Introduction
Cardiovascular disease is the predominant cause of human morbidity and mortality in developed countries. As such, extraordinary effort has been devoted to determining the molecular and pathophysiological characteristics of the diseased heart and vasculature with the goal of developing innovative diagnostic and therapeutic strategies to combat cardiovascular disease. The collective work of multiple research groups has uncovered a complex transcriptional and post-transcriptional regulatory circuit, the integrity of which is essential for maintenance of cardiac homeostasis. Mutations in or aberrant expression of various transcriptional and post-transcriptional regulators have now been correlated with human cardiac disease and pharmacological modulation of the activity of these target genes is a major focus of ongoing research. Recently, a novel class of small non-coding RNAs termed microRNAs (miRNAs) was identified as important transcriptional and post-transcriptional inhibitors of gene expression, thought to “fine tune” the translational output of target mRNAs 1, 2. MiRNAs are implicated in the pathogenesis of various cardiovascular diseases and have become an intriguing target for therapeutic intervention. This review focuses on the basic biology and mechanism of action of miRNAs specifically pertaining to cardiovascular disorders, and addresses the potential for miRNAs to be targeted therapeutically in the treatment of cardiovascular disease.
miRNA processing and function
MiRNAs originate from longer precursor RNAs, called primary miRNAs (pri-miRNA), which are regulated by conventional transcription factors and transcribed by RNA polymerase II. pri-miRNAs are hundreds to thousands of nucleotides long and are processed in the nucleus into a ~70-100 nucleotide hairpin-shaped precursor miRNA (pre-miRNA) by the RNase III enzyme Drosha and the double-stranded RNA binding protein DGCR8. The pre-miRNA is then transported into the cytoplasm by the nuclear export factor exportin 5 and further processed into an ~19-25 nucleotide double stranded RNA by the RNaseIII enzyme Dicer. This duplex miRNA is then incorporated into the RNA-induced silencing complex (RISC). One strand remains in the RISC and becomes the “mature” miRNA, while the other strand is often rapidly degraded and is called the “star” strand (miRNA*). Upon being loaded into RISC, the mature miRNA associates with target mRNAs and acts as a negative regulator of gene expression by promoting mRNA degradation or inhibiting translation3. Translational inhibition seems to be the predominant mechanism in mammals, however target genes that are strongly downregulated on the protein level often show a reduced mRNA level4, suggesting mRNA destablization is a major contributor to gene silencing.
A mature miRNA typically regulates gene expression via an association with the 3’UTR of an mRNA with complimentary sequence, although emerging evidence suggests miRNAs may also target 5’UTRs or exons, and may potentially even undergo base pairing with regulatory DNA sequences to regulate transcription. Upon miRNA binding to a 3’ UTR, the degree of transcriptional degradation and/or translational repression is affected by multiple mechanisms, including the overall complimentarity between the miRNA and target mRNA, the secondary structure of the adjacent sequences, the distance of the miRNA binding site to the coding sequence of the mRNA, and the number of target sites within the 3’UTR5. Complimentarity between nucleotides 2 through 8 of the miRNA, termed the “seed” region, appears to be essential for 3’UTR identification. Therefore, miRNAs with high sequence homology and identical seed region are commonly grouped into miRNA families that are likely to target similar sets of mRNAs6.
Up to 1000 miRNAs are predicted to exist in the human genome, each of which could potentially target hundreds of mRNAs. Most 3’UTRs contain potential binding sites for a large number of individual miRNAs, allowing for redundancy or cooperative interactions between various seemingly unrelated miRNAs. Furthermore, the targets of many miRNAs can modulate the expression of additional miRNAs or groups of miRNAs, generating positive or negative feedback loops. Finally, miRNA maturation seems to be post-transcriptionally regulated in a sequence specific manner7, potentially explaining why genetically clustered and co-transcribed miRNAs are often expressed at different levels.
Multiple miRNA target prediction tools are now available (summarized in Supplemental Table). Generally, in silico target prediction algorithms use a standard scheme to identify and rank potential targets8. Briefly, potential targets are ranked based on the complimentarity between miRNA and 3’UTR and the degree of conservation of the miRNA and the 3’ UTR target sequence across species. A particular miRNA target is considered to be more meaningful if the sequence is evolutionarily conserved.
Identification and validation of miRNA targets remains a major hurdle in the study of miRNA function since many putative targets display little or no detectable regulation when tested in vitro. This is likely due, at least partially, to the relatively modest effect of any single miRNA on the translational output of the target mRNA. Therefore, many predicted miRNA binding sites are probably not true targets and experimental validation is essential for confirming target genes.
A surprising number of published miRNA targets do not conform to the traditional rules of target prediction outlined above. For example, in several instances, cross-species conservation is not observed in the target sequence, even between rodents and humans. Of course, for a miRNA target to be therapeutically viable, the miRNA target sequence must be conserved in humans, and not simply present in the model organism studied. Therefore, this review primarily focuses on published miRNAs that target 3’UTR binding sites that are conserved in humans.
A key insight into the mechanism of miRNA action has been that a large number of miRNAs apparently target multiple functionally related mRNAs. The coordinated regulation of multiple steps in a complex physiological process by one miRNA or a group of similarly expressed miRNAs is an important characteristic of miRNA biology that lends itself to therapeutic applications. In contrast to therapeutically modulating a single target with a conventional drug, miRNA biology can, in principle, modulate multiple levels of a pathological process by targeting a single nucleic acid molecule.
Expression of miRNAs implicated in CV disease
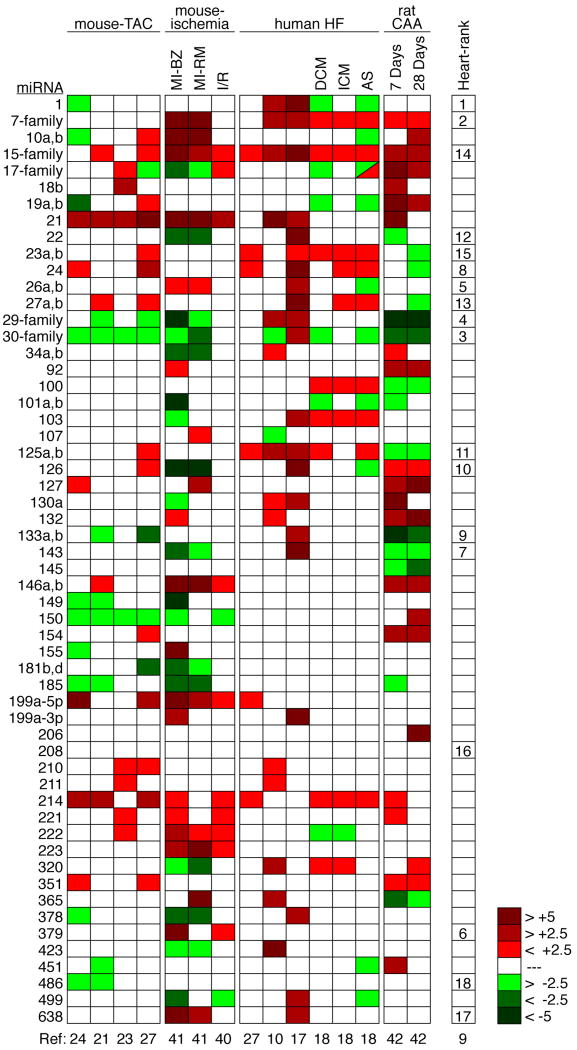
As presented in Figure 1, miRNA expression profiling has identified a subset of miRNAs expressed in the normal heart and modulated during cardiovascular disease. The 18 most strongly expressed miRNAs and miRNA-families account for more than 90% of all miRNA expressed in the adult mouse heart9 (Figure 1), although the specific degree of expression likely varies between species, and depends on the precise location of cardiac tissue harvested. In this respect, it is interesting that several of the most abundant miRNAs in the heart belong to families, with the let-7 family accounting for about 14%, the miR-30 family for about 5% and the miR-29 family for about 4% of all miRNAs expressed in the murine heart9 (Figure 1). Surprisingly, nearly all of the 18 most enriched miRNAs within the heart display altered expression during cardiac disease, indicating an extremely dynamic regulation of miRNAs in the adult heart and pointing towards the importance of microRNAs as modifiers of gene expression programs in cardiovascular disease.
Figure 1.
Regulated miRNAs in cardiovascular disease. Disease state is listed in top row and the particular study for each column is stated below. Heat map illustrates level of up-or down-regulation based on color scheme in legend. All miRNAs that were similarly regulated in at least two independent studies are listed to the left of the heat map. The last column lists the most abundant cardiac miRNAs in order of expression level in mice, which might be slightly different from the expression in humans. miRNAs with the same seed region (nucleotides 2-8) were combined into families due to putatively similar functions and due to technical difficulties in distinguishing very similar miRNAs with micro-arrays, northern-blotting or real-time PCR. The family was labeled as regulated if at least one member was changed. Let-7 family, let-7b,c,d,e,f,g,h,i,j; miR-15 family, miR-15a,b/16/195/424/497, miR-29 family, miR-29a,b,c; miR-30 family, miR-30a,b,c,d,e; miR-17 family, miR-17-5p/20a,b/93/106a,b. TAC, thoracic aortic constriction; MI, myocardial infarction; BZ, border zone; RM, remote myocardium; HF, heart failure; DCM, dilated cardiomyopathy; ICM, ischemic cardiomyopathy; AS, aortic stenosis; CAA, carotid artery angioplasty.
Re-expression of a fetal cardiac gene program is a hallmark of various models of cardiovascular disease and human heart failure. Upregulated genes include the fetal isoform of the MHC gene (βMHC) and the atrial and brain natriuretic factor genes, among others. Recently, it has been suggested there also exists a fetal miRNA program that is re-activated upon cardiac stress10, and may contribute to the re-activation of the fetal gene mRNA program during cardiac disease.
Although some of the miRNAs that are regulated during cardiovascular disease are not highly expressed outside of the heart, a large number of such miRNAs are broadly expressed throughout the body. For example, miR-21, which typically displays dramatic alterations in expression following various cardiac or vascular stresses, is detectable in nearly every tissue analyzed. Inhibition of miRNAs that are specifically expressed, or highly enriched, in the cardiovascular system may circumvent side effects due to miRNA activity in additional organs.
MiRNAs experimentally implicated in various CV disease settings and regeneration Myocardial remodeling
Myocardial remodeling is typically characterized by cardiomyocyte (CMC) hypertrophy, CMC apoptosis, interstitial fibrosis, and aberrant cardiac conduction, which ultimately impair the electrico-mechanical performance of the myocardium. The following sections review the potential roles of miRNAs in each of these processes (see Figure 2) and suggest potential therapeutic implications.
Figure 2.
Published target mRNAs and functional role of disease related microRNAs in cardiomyocytes and fibroblasts. Targets that could not be validated in later publications are labeled with a (?). The arrow adjacent to miRNA illustrates whether the miRNA is up or downregulated in cardiac disease. Green and purple shaded regions indicate stimulation or repression of a process in cardiac disease, respectively. AP-1, activator protein 1; CDC42, cell division cycle 42; CTGF, connective tissue growth factor; Cx43, connexin 43; Hand2, heart and neural crest derivatives expressed 2; HCN2, hyperpolarization activated cyclic nucleotide-gated potassium channel 2; HIF1α, Hypoxia inducible factor; Hop, homeodomain-only protein; Hsp, heat shock protein; Irx5, Iroquois homeobox protein 5; Kcnd2, potassium voltage-gated channel subfamily D member 2; Kcnj2, potassium inwardly-rectifying channel, subfamily J, member 2; MEF2, myocyte enhancing factor-2; MuRF 1, muscle RING-finger protein 1; PDCD4, programmed cell death 4; PHD2, prolyl hydroxylase 2; PP2A, B56a regulatory subunit of protein phosphatase 2a; PTEN, phosphatase and tissue homolog; RhoA, Ras homolog gene family, member A; Sirt1, Sirtuin; SRF, serum response factor; Thrap1, thyroid hormone receptor associated protein1
Cardiomyocyte Hypertrophy
Pathological hypertrophy is a maladaptive process that ultimately leads to reduced cardiac output and is an independent risk factor in heart failure11. Pathological cardiac hypertrophy occurs primarily upon pressure overload due to arterial hypertension or stenosis of the aortic valve, as well as inherited mutations in sarcomeric and cytoskeletal proteins. Myocardial infarction also frequently leads to hypertrophic growth of the remote myocardium as a means of compensating for lost contractile function.
The general importance of miRNAs for the homeostasis of CMCs was demonstrated by CMC-specific deletions of Dicer12 and Dgcr89, two essential components of the machinery that is required to generate miRNAs. Embryonic deletion of Dicer in CMCs resulted in embryonic or early postnatal death - dependent on the time point of Dicer deletion13, 14. Early postnatal deletion of Dicer induced fatal arrhythmias, while depletion of Dicer in adult mice lead to the development of severe heart failure15. Perinatal deletion of Dgcr8 also resulted in rapid development of severe heart failure and premature death9. It is interesting to speculate that the downregulation of Dicer in patients with heart failure might contribute to the disease pathogenesis and progression13.
Several individual miRNAs are transcriptionally regulated during cardiac hypertrophy and heart failure (Figure 1). Some of these have been experimentally verified to play important roles in cardiac development and disease, and are reviewed below.
miR-1
miR-1 is encoded by two genes (miR-1-1 and miR-1-2), each of which is co-expressed bicistronically with one copy of the two miR-133a genes. miR-1 expression is restricted to heart and skeletal muscle and regulated by the transcription factors SRF and MEF2/MyoD, respectively16. During cardiogenesis, miR-1 is believed to control the balance between differentiation and proliferation of cardiac precursor cells by targeting Hand2, a transcription factor that promotes expansion of ventricular CMCs16. Overexpression of miR-1 in embryonic CMCs under the control of the β-myosin heavy chain (βMHC) promoter resulted in thin-walled ventricles and developmental arrest at embryonic day (E)13.516. Homozygous deletion of just one miR-1 gene (miR-1-2) in mice resulted in late embryonic / postnatal mortality of ~50% of the offspring mainly due to ventricular septal defects14. Some of the survivors died postnatally due to heart failure, but the majority succumbed to sudden cardiac death due to arrhythmias with no obvious alterations in cardiac morphology. This severe phenotype seen by deletion of only one miR-1 gene is surprising given that most miRNA knockout mice show only mild phenotypes under basal conditions. One possible explanation might be that miR-1 is by far the strongest expressed miRNA in the murine heart9, 14 (Figure 1).
Besides the crucial role of miR-1 during cardiac development, it is currently unclear whether miR-1 contributes to adverse remodeling in human heart failure. Some studies suggest upregulation of miR-1 expression in heart failure10, 17, while others report downregulation18. Ikeda et al. (2009) also found miR-1 downregulated in hearts of calcineurin transgenic mice, a well-established murine heart failure model19. Adenoviral overexpression of miR-1 in the heart attenuated CMC hypertrophy most likely via downregulation of calmodulin and MEF2a19.
miR-21
miR-21 is the most upregulated miRNA in cardiac disease, although under basal conditions is only weakly expressed (Figure 1). miR-21 is also highly upregulated in several cancers and believed to function as an oncogene by inhibiting apoptosis20. The role of miR-21 in cardiac disease is controversial at present. Some studies find an induction of CMC hypertrophy by miR-21 in vitro21 and indirectly in vivo via fibroblasts22. In contrast, other studies report an antihypertrophic effect of miR-21 in isolated cardiomyocytes23, find a promotion of cellular outgrowths of cardiomyocytes24, a reduction of infarct size by miR-2125 or an inhibition of H2O2-induced apoptosis of isolated cardiomyocytes26. The reasons for the discrepancy between these studies are unclear, however miR-21 is predominantly expressed in cardiac fibroblasts, not CMCs22. It is possible that much of the miR-21 enrichment seen during cardiac disease might be primarily due to an increase in cardiac fibroblast cell number, implying a rather limited physiological relevance of miR-21, at least in CMCs. This is supported by the fact that CMC-specific overexpression of miR-21 in vivo does not evoke an obvious cardiac phenotype22. miR-21 is broadly expressed in multiple tissues including the vasculature, which hampers the discrimination of primary and secondary cardiac effects in models that use ubiquitous knockdown of miR-21, such as intravenous injection of antagomiRs.
miR-23
There are two miR-23 genes that differ by only one nucleotide in the mature miRNA sequence. Each miR-23 gene is closely clustered with a miR-24 and a miR-27 gene, suggesting that they are transcribed as a common transcript. Accordingly, several groups found that miR-23, miR-24 and miR-27 are all upregulated in heart failure and murine cardiac hypertrophy (Figure 1). van Rooij et al. (2006) showed that adenoviral overexpression of miR-23a induces hypertrophy of isolated cardiomyocytes27. Lin et al. (2009) showed recently that miR-23a expression in CMCs is regulated by nuclear factor of activated T cells (NFATc3) and that miR-23a promotes CMC hypertrophy by downregulation of the muscle specific ring finger protein 1 (Murf-1), an anti-hypertrophic protein28. Knockdown of mir-23a by injection of a specific antagomiR attenuated isoproterenol-induced cardiac hypertrophy28.
miR-133
The miR-133 family contains three miRNA genes: miR-133a-1, miR-133a-2, and miR-133b, which are each transcribed as bicistronic transcripts together with miR-1-2, miR-1-1, and miR-206, respectively. The expression of the two miR-1/133a-clusters is regulated by MEF2 and SRF and restricted to cardiac and skeletal muscle29. The miR-206/133b cluster is expressed only in skeletal muscle.
Partial knockdown of miR-133 in mice with specific antagomiRs was shown to induce cardiac hypertrophy, suggesting that pharmacological elevation of miR-133 expression might prevent cardiac hypertrophy during cardiac disease30. In contrast, Liu et al. (2008) reported that mice with a genetic deletion of either miR-133a gene were phenotypically normal and showed a normal hypertrophic response to pressure overload of the left ventricle despite an ~50% decrease of miR-133 expression in the heart31. Deletion of both miR-133a genes resulted in late embryonic or neonatal lethality due to ventricular-septal defects, accompanied by abnormalities in CMC proliferation, apoptosis and aberrant expression of smooth muscle genes in the heart31. About a quarter of the double knockout mice survived to adulthood, developed extensive myocardial fibrosis without evidence of CMC hypertrophy and ultimately died from heart failure and sudden death. Cardiac-restricted overexpression of miR-133a under the control of the βMHC promoter resulted in embryonic lethality due to inhibition of CMC proliferation31.
Many of the phenotypic abnormalities observed in miR-133a knockout mice such as ectopic expression of smooth muscle genes and aberrant CMC proliferation could be ascribed at least partially to inappropriate expression of the miR-133 target genes SRF and cyclin D231. It is not clear why genetic deletion of miR-133 and knockdown by antagomiRs provoke different effects, however a partial and transient knockdown with antagomiRs might have different consequences than a complete genetic deletion in which the gene is eliminated throughout the whole life. Perhaps long-term genetic deletion allows for compensatory mechanisms that do not occur in response to transient miRNA knockdown. Further studies are required to determine whether therapeutic modulation of miR-133 expression might represent an interesting target for the treatment of cardiac disease.
miR-208a
The cardiac-specific miR-208a is encoded by an intron of the alpha myosin heavy chain (αMHC) gene. Thyroid hormone (T3) stimulates the expression of αMHC and miR-208a after birth, while repressing the expression of the embryonically predominant βMHC isoform. Cardiac disease is associated with a re-activation of the fetal gene program with increased expression of βMHC and decreased expression of αMHC, although a corresponding decrease in miR-208a levels is not observed in short-term studies since miR-208a is very stable. The results of van Rooij et al. (2007) demonstrated that miR-208a regulates the induction of βMHC expression upon cardiac stress in adult mice32. Callis et al., (2009) further demonstrate that this results in the concomitant upregulation of miR-208b, which is encoded by an intron of the βMHC gene33. Genetic deletion of miR-208a in mice resulted in blunted hypertrophy and reduced cardiac fibrosis in response to TAC, while cardiomyocyte-specific overexpression of miR-208a induced cardiomyocyte hypertrophy32, 33. The regulation of cardiomyocyte growth by miR-208a might be at least partially due to the repression of thyroid hormone receptor associated protein 1 (THRAP1) and myostatin, two negative regulators of muscle growth and hypertrophy32, 33. Although initially protective against acute cardiac stress-induced remodeling, long-term deletion of miR-208a led to a decrease in cardiac contractility32, possibly resulting from perturbations in the cardiac conduction system resulting in atrial fibrillation of miR-208a knockout mice33. The latter seems to be caused by misregulation of GATA4, homeodomain only protein (Hop), and connexin 40 33. It would be interesting to determine whether partial knockdown of miR-208 might prevent hypertrophy while circumventing the side effects of complete miR-208a knockout, thereby enhancing the potential of miR-208 as a therapeutic target.
Cardiomyocyte apoptosis and regeneration
Since the adult heart has only limited regenerative capacities, an excessive loss of CMCs following myocardial ischemia or infarction can significantly decrease cardiac performance. Some miRNAs seem to play important roles in the regulation of CMC apoptosis in vivo, and will be discussed below. MiRNAs are certainly crucial regulators of cell fate determination and differentiation of stem cells34, however it is currently unclear whether they play a role in the regeneration of the adult heart.
miR-195
miR-195 belongs to a family including miR-15a, miR-15b, miR-16-1, miR-16-2, miR-424 and miR497 (the miR-15 family). The miR-15 family was consistently found to be strongly upregulated in cardiac ischemia and heart failure (Figure 1). CMC-specific overexpression of miR-195 resulted in cardiac hypertrophy and rapid progression to fatal dilated cardiomyopathy27. The exact function of the miR-15 family in the heart is not clear, however studies in other cell types suggest that the miR-15 family might induce apoptosis by downregulation of the anti-apoptotic factor Bcl-220. In this respect, antagomiR-induced knockdown of the miR-15 family might be a means to prevent ischemia-induced CMC apoptosis.
miR-199a
The miR-199 family contains 3 miRs: miR-199a-1, miR-199a-2 and miR-199b that are all encoded by the antisense strand of an intron of a dynamin gene (Dnm2, Dnm3 and Dnm1, respectively). Further, mir-199a-2 is co-transcribed with miR-214. The transcriptional regulation of miR-199 in the heart is unknown, but Rane et al. reported recently that miR-199a was rapidly downregulated in CMCs upon hypoxic conditions, most likely via a post-transcriptional mechanism since the expression level of the miR-199a precursor was unaffected35. Downregulation of miR-199a de-repressed the expression of hypoxia-inducible factor (Hif)-1α, the most important transcription factor for induction of gene expression upon hypoxia35. miR-199a downregulation also resulted in the de-repression of Sirtuin 1, which was responsible for downregulation of prolyl hydroxylase (PHD) 2, the enzyme that hydroxylates Hif-1α to induce its degradation35. The results of this study demonstrated that knockdown of miR-199a during hypoxia induced apoptosis, while knockdown of miR-199a before hypoxia surprisingly mirrored preconditioning and protected CMCs against hypoxic damage35.
miR-320
In contrast to the upregulation of the miR-15 family, miR-320 expression was decreased upon ischemia/reperfusion injury36. Transgenic mice with cardiac-specific overexpression of miR-320 exhibited increased apoptosis and infarct size following ischemia/reperfusion injury. Conversely, administration of miR-320 antagomiRs reduced infarct size, probably at least partially via de-repression of the cardioprotective heat-shock protein 20 (Hsp20)36. It is early to speculate, but targeting miR-320 with antagomiRs following myocardial infarction could be a new treatment option to decrease CMC loss.
Cardiac conduction
Membrane excitability is a special characteristic of CMCs and is regulated via ion channels. Specifically, Na+, Ca2+, and K+-channels and gap junction proteins such as connexin 43 are important regulators of CMC polarization and depolarization during contraction and relaxation, respectively. Several miRNAs, including miR-1 and miR-133, are predicted to target ion channels and might therefore play important roles in cardiac conduction and the onset of arrhythmias during cardiac disease.
Zhao et al., (2007) reported that adult miR-1-2 knockout mice showed several ECG alterations such as reduced heart rate, shortened PR-interval and widened QRS complexes and died due to cardiac arrhythmia14. The ECG alterations were presumably at least partially due to elevated expression of the miR-1 target Irx5, a transcription factor that regulates the expression of Kcnd2, a potassium channel important for normal cardiac repolarization14. Further, Yang et al. (2007) showed that miR-1 is upregulated in coronary artery disease37. Overexpression of miR-1 exacerbated and antagomiR-induced knockdown relieved arrhythmogenesis upon myocardial infarction, most likely via regulation of connexin 43 and the K+-channel Kir2.1 subunit37. Additionally, miR-1 regulates Ca2+-signaling via targeting of the B56a regulatory subunit of the protein phosphatase PP2A38. Downregulation of B56a by miR-1 resulted in hyperphosphorylation of L-type calcium channels and the ryanodine receptor (RyR2) via disrupting localization of PP2A activity to these channels38. These studies suggest that miRNA-1 and potentially other miRNAs might be promising targets to treat cardiac arrhythmia.
Fibrosis
Fibroblast activation and proliferation during cardiac disease leads to inappropriate secretion of extracellular matrix proteins and concomitant interstitial fibrosis. Fibrosis results in impaired cardiac contractility and alters the electromechanical characteristics of the myocardium, often leading to arrhythmias, an important cause of mortality in heart disease. The expression of several miRNAs is altered following myocardial infarction (MI) or other fibrotic pathologies, some of which have been shown to play both direct and indirect roles in the regulation of cardiac fibrosis.
As mentioned above, adult miR-133a double knockout mice develop severe fibrosis and heart failure31. In line with those data, Duisters et al. (2009) showed that miR-133 could target connective tissue growth factor (CTGF)39. Downregulation of miR-133 during cardiac disease might therefore result in increased expression and secretion of CTGF from CMCs, which consecutively stimulates extracellular matrix synthesis in fibroblasts.
The fibroblast-enriched miR-21 is upregulated in failing and hypertrophic myocardium, possibly as a consequence of fibroblast proliferation. Thum et al. (2008) demonstrated that miR-21 increases fibroblast survival and fibrosis possibly via inhibition of sprouty homologue 1 and consecutive ERK-MAP kinase activation22. Knockdown of miR-21 with antagomiRs attenuated interstitial fibrosis and cardiac remodeling after aortic banding22. Mir-21 is also upregulated in the border zone following MI, a fibroblast-rich region adjacent to the infarct, and was shown to promote MMP-2 expression via repression of PTEN40.
van Rooij et al. (2008) found all members of the miR-29-family downregulated after myocardial infarction, particularly in the border zone41. MiR-29 is predicted to target myriad genes that are involved in fibrosis such as collagens, fibrillins, and elastin, and is a prime example of the ability to modulate a large portion of a particular pathology by pharmacologically targeting one miRNA. Knockdown of miR-29 by intravenous injection of an antagomiR resulted in the upregulation of collagens in the heart, suggesting that miR-29 indeed acts as an inhibitor of cardiac fibrosis41. In the future, it will be interesting to determine the effect of miR-29 replacement strategies on the outcome of fibrotic pathologies such as MI.
miRNAs in the Vascular System and Vascular Disease
Vessel injury is characterized by profound phenotypic changes in molecular and physiological identity; vascular smooth muscle cells (VSMCs) in particular undergo a program of de-differentiation and become more proliferative and migratory after injury. These changes contribute to neointimal thickening during proliferative vascular diseases such as atherosclerosis, hypertension, and restenosis. Expression profiling has identified a signature pattern of miRNA expression in VSMCs and endothelial cells of the major vessels that are specifically altered in vessel injury (Figure 1).
Restenosis
miR-21
Several groups have demonstrated roles for various miRNAs in SMC phenotypic modulation and the response of the vasculature to injury. Antisense mediated knockdown of miR-21, which is normally increased following vessel injury (Figure 1), blunted the formation of a neointimal lesion in response to balloon angioplasty of the carotid artery42. The results of Ji et al. (2007) demonstrated that miR-21 promoted SMC proliferation following vessel injury via inhibition of PTEN and the subsequent activation of the PI3K/Akt signaling pathway, which is partially blocked upon knockdown of miR-2142 (Figure 3).
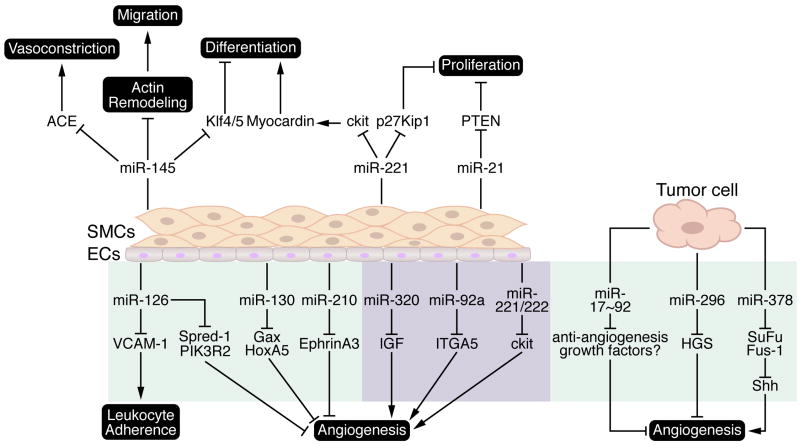
Figure 3.
Published target mRNAs and functional role of disease related miRNAs in vascular remodeling and angiogenesis. Spindle shaped yellow cells represent vascular smooth muscle cells (SMCs) and cuboidal grey cells represent endothelial cells (ECs). Purple and green shaded regions indicate microRNAs with “pro-angiogenic” and “anti-angiogenic” characteristics, respectively. ACE, angiotensin converting enzyme; SuFu, suppressor of fused; Shh, sonic hedgehog; HGS, Hepatocyte growth factor-regulated tyrosine kinase substrate; IGF, insulin-like growth factor; ITGA5, integrin-α5; PIK3R2, phosphoinositol-3 kinase regulatory subunit 2 (p85β); PTEN, phosphatase and tensin homolog; Spred-1, sprouty-related EVH domain-containing protein-1; VCAM-1, vascular cell adhesion molecule.
miR-145
Recently, the SMC enriched miR-143/-145 gene cluster, which is downregulated in the carotid artery after mechanical injury, has been implicated in the regulation of SMC contractility and the stress response to vessel injury. Mice harboring deletions of miR-143, -145, or both reveal a role for these miRNAs in the acquisition of a contractile SMC phenotype; vessels of KO mice are thin and distended and null VSMCs appear to have acquired a “synthetic” phenotype based on reduced actin stress fiber formation and increased rough ER production43, 44. Furthermore, miR-143/-145 null mice display decreased vascular tone and a reduction in systolic blood pressure. However, mutant mice were viable and appeared to have functional SMCs, albeit shifted slightly towards the synthetic phenotype. This is in contrast to in vitro analyses that demonstrated a critical role for miR-145 in the differentiation of cultured SMCs and ES cells43, 45, likely via repression of Klf4 and 5, transcriptional repressors that have previously been shown to inhibit a differentiated state.
Adenoviral-mediated overexpression of miR-145 at the onset of carotid artery injury blunted the phenotypic modulation of VSMCs and reduced the formation of a neointima in response to angioplasty46. Surprisingly, Xin et al. (2009) described a diminution of neointima formation in miR-145 null mice following ligation of the carotid artery43. This disparity might suggest that a fully differentiated state at baseline may be essential for appropriate phenotypic modulation of the VSMC state. It is also possible that the reduced vascular tone in miR-145 mutant mice impinges upon the responsiveness of the vessel wall to vascular injury and the phenotype reflects an inability to “sense” the injury as opposed to an inability to “respond” to injury. Finally, miR-143/-145 targets a disproportionate number of genes involved in actin cytoskeletal rearrangements and SMC migration (Figure 3) and it is possible that deficiencies in stress induced migration account for the protection from neointima formation in mutant mice43. Additional studies should be undertaken to determine the viability of pharmacological modulation of miR-145 as a therapy for restenosis.
miR-221
miR-221, although not VSMC-specific, is induced in VSMCs upon platelet-derived growth factor (PDGF) stimulation47. Activation of the PDGF signaling pathway results in the switch of VSMCs from a fully differentiated, contractile state to a less differentiated, synthetic state typified by increases in proliferation and cell migration, contributing to the formation of a neointimal lesion following arterial injury. Indeed, miR-221 expression is moderately elevated after carotid artery angioplasty (Figure 1)42. In vitro data suggests miR-221 contributes to SMC phenotypic switching by affecting multiple cellular responses; miR-221 targets cKit for repression, resulting in a decrease in SMC differentiation, and also inhibits p27Kip1, thereby increasing SMC proliferation47. Therefore, knockdown of miR-221 following vessel injury may block neointima formation. Further examination in animal models should be forthcoming to ascertain its therapeutic potential.
Angiogenesis
Neoangiogenesis plays an essential role in the process of cardiac repair following ischemic injuries such as myocardial infarction (MI) by promoting vascularization of the infarcted tissue through growth of collateral vessels that bypass the infarcted artery. Various growth factors such as vascular endothelial growth factor (VEGF) and fibroblast growth factor (FGF) are required for proper generation of blood vessels after MI.
Recently a role for miRNAs has been suggested in the regulation of angiogenesis. Genetic deletion of Dicer in mice resulted in abnormal blood vessel formation and embryonic lethality between embryonic day (E)12.5 and E14.548. si-RNA mediated knockdown of Dicer in endothelial cells (ECs) resulted in a failure of capillary sprouting and tube formation49, 50. Individual miRNAs have since been shown to play positive and negative regulatory roles in angiogenesis as presented in Figure 3, and discussed below.
miR-221 and -222
Studies using human umbilical vein endothelial cells (HUVECs) demonstrated miR-221 and -222 regulate angiogenesis in response to Stem Cell Factor (SCF), presumably by directly repressing the levels of cKit and attenuating cell survival, migration, and endothelial tube formation51. Although it was not clear from this study whether the expression of miR-221 /-222 is altered during the physiological process of capillary formation in response to SCF, it is clear from overexpression studies that modulation of miR-221 /-222 affects endothelial tube formation and represents a potential avenue for therapeutic modulation of angiogenesis.
miR-210
miR-210 expression is elevated during hypoxic conditions and has been shown to possess pro-angiogenic properties in vitro52, 53. Overexpression of miR-210 in ECs resulted in accelerated tube formation under normoxic conditions and enhanced VEGF-dependent migration. Conversely, knockdown of miR-210 blocked capillary formation in response to hypoxia. This study demonstrated EphrinA3 as an important target of miR-210, inhibition of which is a major contributor to miR-210 mediated cell survival, migration and tube formation in response to hypoxia (Figure 3). A recent study suggests that cytoprotection afforded by cardiac ischemia/repurfusion may be partially mediated by the induction of miR-210, which promoted mesenchymal stem cell survival by blocking caspase-8 associated protein-254. The results of these studies suggest miR-210 should be further examined for a potential cardioprotective role following ischemic injury.
miR-126
miR-126 has been implicated in the maintenance of vascular integrity and promotion of vessel growth as a pro-angiogenic factor both in vitro and in vivo55, 56. Genetic deletion of miR-126 resulted in profound vascular defects, phenotypes that were previously ascribed to the host gene Egfl7, due to inadvertent deletion of miR-126 in the Egfl7 null mouse57. Although a significant fraction of miR-126 null mice died embryonically due to vessel leakage, those mice that escaped neonatal lethality were particularly susceptible to vascular rupture following MI, due to a deficit in neovascularization of the infarcted tissue. The pro-angiogenic effect of miR-126 was attributed, at least in part, to the repression of Spred-1, an intracellular inhibitor of VEGF and FGF-mediated angiogenesis and phosphotidylinositol-3-kinase regulatory subunit PIK3R2 (p85β) (Figure 3). Interestingly, miR-126 is also expressed in hematopoietic progenitor cells, a circulating stem cell population that contributes to post-MI cardiac regeneration. It is tempting to speculate that miR-126 may also influence cardiac repair by affecting the homing ability of circulating hematopoietic progenitor cells. In addition to a direct role in angiogenesis, miR-126 may play a role in the process of leukocyte infiltration and vascular inflammation. Vascular cell adhesion molecule (VCAM-1) expression is modulated by miR-126, thus affecting TNFα stimulated leukocyte adherence to endothelial cells and vessel inflammation58. Taken together, these studies suggest that pharmacological elevation of miR-126 levels may be a viable therapeutic strategy to enhance neoangiogenesis and cardiac repair in ischemic myocardium.
miR-17~ 92 cluster
Members of the miR-17-92 cluster were demonstrated to either positively or negatively influence angiogenesis, depending on the cellular context. The EC enriched miR-92a, which is upregulated following ischemia, is a negative regulator of vessel growth59. In vivo knockdown of miR-92a using antisense oligonucleotides resulted in improved recovery from ischemic damage due to accelerated vessel growth. The mechanism for the anti-angiogenic role of miR-92a was attributed to repression of the pro-angiogenic factor integrin α5 (ITGA5) (Figure 3). Conversely, other members of the miR-17-92 cluster (particularly miR-18 and miR-19) suppressed anti-angiogenic factors in tumor cells and promoted angiogenesis and the vascularization of tumors in vivo60, 61. These studies demonstrated the importance of miRNAs in the modulation of angiogenesis in a pathological setting and also highlighted the potential for pharmacological intervention by inhibition of a miRNA using an antisense approach.
miRNAs as therapeutic targets in cardiovascular disease
miRNAs are rapidly becoming an intriguing pharmacological target in the treatment of cardiovascular disease. The development of antisense oligonucleotide-mediated knockdown (anti-miR) and miRNA-mimic mediated overexpression techniques might soon allow for the regulation of any given miRNA in cardiovascular disease.
Antisense-mediated miRNA knockdown
Anti-miRs are antisense oligonucleotides with the reverse complimentary sequence of the target miRNA. They can be conjugated to a cholesterol moiety to increase cellular uptake, and often contain a modified sugar backbone that increases stability. Upon cellular uptake, anti-miRs bind to the mature miRNA, thereby inhibiting its activity 62. Since miRNAs typically act as repressors of gene expression, inhibition of a miRNA should result in the de-repression of mRNAs that are directly targeted by the miRNA. Thus, the primary effect of a miRNA inhibitor is activation of gene expression. Krutzfeldt et al. (2005) reported the first mammalian anti-miR knockdown using cholesterol-conjugated, 2’-O-methyl modified anti-miRs, termed antagomiRs, which resulted in de-repression of putative target mRNAs62. In a later study, they showed that systemic antagomiR delivery via intravenous injection efficiently reduced the level of a given miRNA in multiple tissues over an extended period of time63. Several animal model studies have now demonstrated the value of antagomiRs to downregulate the expression of specific miRNAs in the heart and to thereby influence myocardial remodeling22, 28, 30, 36, 41, 59.
Besides antagomiRs, other anti-miRs with different modifications are now being developed, which display distinct pharmacokinetics and possibly mechanisms of action. For example, anti-miRs with 2’-O-methoxyethyl phosphorothioate (MOE) substitutions have proven useful for inhibition of miR-122 in the liver64. A very promising new approach might be the use of locked nucleic acid (LNA) chemistry, which has already been demonstrated in the downregulation of miRNAs in non-human primates65 and are currently being tested in the first human clinical trial of miRNA inhibition.
miRNA mimics
miRNA mimics are synthetic RNA duplexes in which one strand is identical to the mature miRNA sequence (guide strand) and is designed to “mimic” the function of the endogenous miRNA. The other strand (passenger strand) is often only partially complimentary to the guide strand and typically linked to cholesterol to enhance cellular uptake. The double-stranded structure is required for efficient recognition and loading of the guide strand into the RISC66. The use of miRNA-mimics would be particularly useful to enhance the expression of miRNAs that are downregulated in cardiovascular disease, however their in vivo efficacy has not yet been experimentally validated in a pathological setting. It is important to note that the use of miRNA-mimics also results in their uptake in tissues that do not normally express the miRNA and may result in unanticipated side effects, which could be overcome by tissue-specific targeting strategies.
Intravenously injected antagomirs and miRNA-mimics have been the primary method of systemic delivery to date; however they are preferentially targeted to the liver, kidney and spleen. Therefore, a major challenge will be the development of strategies for enrichment of anti-miRs or miR-mimics in the cardiovascular system. One possible approach would be a conjugation strategy with the nucleic acid linked to targeting molecules such as peptides, antibodies, or other bioactive molecules, which may promote homing of the anti-miR/miRNA-mimic to the heart. Alternatively, the anti-miR / miRNA-mimic could be encapsulated into a lipid-based formulation that enhances cardiac uptake. Finally, the specific application of the anti-miR / miR-mimic to the heart e.g. into the coronary vessels during cardiac catheterization upon myocardial infarction might improve cardiac uptake.
There are several difficulties to overcome in order to promote miRNAs as a viable therapeutic target. As stated above, results obtained from pharmacological knockdown or overexpression by administration of anti-miRs and miR-mimics often differ from those observed using genetic mouse models. Several possibilities might explain these findings: 1) miRNAs often have hundreds or even thousands of predicted mRNA targets, but at physiological expression levels a miRNA most likely targets only a small fraction of these. Forced overexpression of a miRNA, however, might result in the regulation of physiologically irrelevant targets. 2) Similarly, anti-miRs may potentially have considerable off-target effects that in some cases might be more responsible for the observed effects than the actual knockdown of the miRNA. 3) Since systemic delivery of antagomiRs and miRNA-mimics affect miRNA expression globally, effects observed in the heart could also be secondary to effects in extra-cardiac tissues e.g. due to altered blood pressure or a change in the level of circulating hormones. 4) Finally, a short-term and partial knockdown with antagomiRs might have different consequences than a complete genetic deletion in which the gene is eliminated from embryogenesis to adulthood; genetic deletion may result in embryonic phenotypes that may complicate the analysis of adult disease models. Further, compensatory mechanisms could be activated in genetic knockout mice that account for mild phenotypes as compared to transient knockdown. Nevertheless, the most valuable model to study the function of a miRNA is genetic deletion, which should specifically de-repress only those mRNAs that are physiologically repressed by the miRNA. In summary, it is apparent that miRNA biology must be examined using a combination of genetic models and pharmacological manipulation.
Concluding Remarks
The demonstration that miRNAs play a crucial role in cardiovascular disease and can be easily regulated in vitro and in vivo by anti-miRs and miR-mimics has tremendously accelerated miRNA research and has nourished hopes that the drugs used and verified in animal models could be equally used in humans. Currently, therapeutic applications of miRNA biology focus only on affecting translational regulation. Besides the use of miRNAs as therapeutic targets, they might serve a valuable diagnostic and prognostic function for various cardiovascular pathologies, since cardiac damage results in the detectable levels of cardiac miRNAs in the blood67. In conclusion, miRNAs represent a relatively young field of basic biological and translational research into new and innovative therapeutic applications. However, the rapid advancement towards viable miRNA-driven therapeutic options increases the odds that the next “small RNA step” will be a “giant leap” for the treatment of cardiovascular disease.
Supplementary Material
Acknowledgments
Funding Sources This work was supported by grants from the National Institutes of Health, the Donald W Reynolds Clinical Cardiovascular Research Center, the Sandler Foundation for Asthma Research, and the Robert A Welch Foundation to ENO.
Footnotes
Conflict of Interest Disclosures Eric M. Small: None; Robert J.A. Frost: None; Eric N. Olson: Co-founder of Miragen Therapeutics.
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 3.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 4.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 6.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 7.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yousef M, Showe L, Showe M. A study of microRNAs in silico and in vivo: bioinformatics approaches to microRNA discovery and target identification. FEBS J. 2009;276:2150–2156. doi: 10.1111/j.1742-4658.2009.06933.x. [DOI] [PubMed] [Google Scholar]
- 9.Rao PK, Toyama Y, Chiang HR, Gupta S, Bauer M, Medvid R, Reinhardt F, Liao R, Krieger M, Jaenisch R, Lodish HF, Blelloch R. Loss of Cardiac microRNA-Mediated Regulation Leads to Dilated Cardiomyopathy and Heart Failure. Circ Res. 2009;105:585–594. doi: 10.1161/CIRCRESAHA.109.200451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thum T, Galuppo P, Wolf C, Fiedler J, Kneitz S, van Laake LW, Doevendans PA, Mummery CL, Borlak J, Haverich A, Gross C, Engelhardt S, Ertl G, Bauersachs J. MicroRNAs in the human heart: a clue to fetal gene reprogramming in heart failure. Circulation. 2007;116:258–267. doi: 10.1161/CIRCULATIONAHA.107.687947. [DOI] [PubMed] [Google Scholar]
- 11.Hill JA, Olson EN. Cardiac plasticity. N Engl J Med. 2008;358:1370–1380. doi: 10.1056/NEJMra072139. [DOI] [PubMed] [Google Scholar]
- 12.Thum T. Cardiac dissonance without conductors: how dicer depletion provokes chaos in the heart. Circulation. 2008;118:1524–1527. doi: 10.1161/CIRCULATIONAHA.108.807230. [DOI] [PubMed] [Google Scholar]
- 13.Chen JF, Murchison EP, Tang R, Callis TE, Tatsuguchi M, Deng Z, Rojas M, Hammond SM, Schneider MD, Selzman CH, Meissner G, Patterson C, Hannon GJ, Wang DZ. Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc Natl Acad Sci U S A. 2008;105:2111–2116. doi: 10.1073/pnas.0710228105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 15.da Costa Martins PA, Bourajjaj M, Gladka M, Kortland M, van Oort RJ, Pinto YM, Molkentin JD, De Windt LJ. Conditional dicer gene deletion in the postnatal myocardium provokes spontaneous cardiac remodeling. Circulation. 2008;118(15):1567–1576. doi: 10.1161/CIRCULATIONAHA.108.769984. [DOI] [PubMed] [Google Scholar]
- 16.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 17.Matkovich SJ, Van Booven DJ, Youker KA, Torre-Amione G, Diwan A, Eschenbacher WH, Dorn LE, Watson MA, Margulies KB, Dorn GW., 2nd Reciprocal regulation of myocardial microRNAs and messenger RNA in human cardiomyopathy and reversal of the microRNA signature by biomechanical support. Circulation. 2009;119:1263–1271. doi: 10.1161/CIRCULATIONAHA.108.813576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ikeda S, Kong SW, Lu J, Bisping E, Zhang H, Allen PD, Golub TR, Pieske B, Pu WT. Altered microRNA expression in human heart disease. Physiol Genomics. 2007;31:367–373. doi: 10.1152/physiolgenomics.00144.2007. [DOI] [PubMed] [Google Scholar]
- 19.Ikeda S, He A, Kong SW, Lu J, Bejar R, Bodyak N, Lee KH, Ma Q, Kang PM, Golub TR, Pu WT. MicroRNA-1 negatively regulates expression of the hypertrophy-associated calmodulin and Mef2a genes. Mol Cell Biol. 2009;29:2193–2204. doi: 10.1128/MCB.01222-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Lee CG. MicroRNA and cancer--focus on apoptosis. J Cell Mol Med. 2009;13:12–23. doi: 10.1111/j.1582-4934.2008.00510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng Y, Ji R, Yue J, Yang J, Liu X, Chen H, Dean DB, Zhang C. MicroRNAs are aberrantly expressed in hypertrophic heart: do they play a role in cardiac hypertrophy? Am J Pathol. 2007;170:1831–1840. doi: 10.2353/ajpath.2007.061170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, Castoldi M, Soutschek J, Koteliansky V, Rosenwald A, Basson MA, Licht JD, Pena JT, Rouhanifard SH, Muckenthaler MU, Tuschl T, Martin GR, Bauersachs J, Engelhardt S. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–984. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 23.Tatsuguchi M, Seok HY, Callis TE, Thomson JM, Chen JF, Newman M, Rojas M, Hammond SM, Wang DZ. Expression of microRNAs is dynamically regulated during cardiomyocyte hypertrophy. J Mol Cell Cardiol. 2007;42:1137–1141. doi: 10.1016/j.yjmcc.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sayed D, Rane S, Lypowy J, He M, Chen IY, Vashistha H, Yan L, Malhotra A, Vatner D, Abdellatif M. MicroRNA-21 targets Sprouty2 and promotes cellular outgrowths. Mol Biol Cell. 2008;19:3272–3282. doi: 10.1091/mbc.E08-02-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yin C, Wang X, Kukreja RC. Endogenous microRNAs induced by heat-shock reduce myocardial infarction following ischemia-reperfusion in mice. FEBS Lett. 2008;582:4137–4142. doi: 10.1016/j.febslet.2008.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng Y, Liu X, Zhang S, Lin Y, Yang J, Zhang C. MicroRNA-21 protects against the H(2)O(2)-induced injury on cardiac myocytes via its target gene PDCD4. J Mol Cell Cardiol. 2009;47:5–14. doi: 10.1016/j.yjmcc.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin Z, Murtaza I, Wang K, Jiao J, Gao J, Li PF. miR-23a functions downstream of NFATc3 to regulate cardiac hypertrophy. Proc Natl Acad Sci U S A. 2009;106:12103–12108. doi: 10.1073/pnas.0811371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu N, Williams AH, Kim Y, McAnally J, Bezprozvannaya S, Sutherland LB, Richardson JA, Bassel-Duby R, Olson EN. An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proc Natl Acad Sci U S A. 2007;104:20844–20849. doi: 10.1073/pnas.0710558105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Care A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, Bang ML, Segnalini P, Gu Y, Dalton ND, Elia L, Latronico MV, Hoydal M, Autore C, Russo MA, Dorn GW, 2nd, Ellingsen O, Ruiz-Lozano P, Peterson KL, Croce CM, Peschle C, Condorelli G. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 31.Liu N, Bezprozvannaya S, Williams AH, Qi X, Richardson JA, Bassel-Duby R, Olson EN. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22:3242–3254. doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 33.Callis TE, Pandya K, Seok HY, Tang RH, Tatsuguchi M, Huang ZP, Chen JF, Deng Z, Gunn B, Shumate J, Willis MS, Selzman CH, Wang DZ. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J Clin Invest. 2009;119:2772–2786. doi: 10.1172/JCI36154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–658. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 35.Rane S, He M, Sayed D, Vashistha H, Malhotra A, Sadoshima J, Vatner DE, Vatner SF, Abdellatif M. Downregulation of miR-199a derepresses hypoxia-inducible factor-1alpha and Sirtuin 1 and recapitulates hypoxia preconditioning in cardiac myocytes. Circ Res. 2009;104:879–886. doi: 10.1161/CIRCRESAHA.108.193102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ren XP, Wu J, Wang X, Sartor MA, Qian J, Jones K, Nicolaou P, Pritchard TJ, Fan GC. MicroRNA-320 is involved in the regulation of cardiac ischemia/reperfusion injury by targeting heat-shock protein 20. Circulation. 2009;119:2357–2366. doi: 10.1161/CIRCULATIONAHA.108.814145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang B, Lin H, Xiao J, Lu Y, Luo X, Li B, Zhang Y, Xu C, Bai Y, Wang H, Chen G, Wang Z. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat Med. 2007;13:486–491. doi: 10.1038/nm1569. [DOI] [PubMed] [Google Scholar]
- 38.Terentyev D, Belevych AE, Terentyeva R, Martin MM, Malana GE, Kuhn DE, Abdellatif M, Feldman DS, Elton TS, Gyorke S. miR-1 overexpression enhances Ca(2+) release and promotes cardiac arrhythmogenesis by targeting PP2A regulatory subunit B56alpha and causing CaMKII-dependent hyperphosphorylation of RyR2. Circ Res. 2009;104:514–521. doi: 10.1161/CIRCRESAHA.108.181651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duisters RF, Tijsen AJ, Schroen B, Leenders JJ, Lentink V, van der Made I, Herias V, van Leeuwen RE, Schellings MW, Barenbrug P, Maessen JG, Heymans S, Pinto YM, Creemers EE. miR-133 and miR-30 regulate connective tissue growth factor: implications for a role of microRNAs in myocardial matrix remodeling. Circ Res. 2009;104:170–178. doi: 10.1161/CIRCRESAHA.108.182535. [DOI] [PubMed] [Google Scholar]
- 40.Roy S, Khanna S, Hussain SR, Biswas S, Azad A, Rink C, Gnyawali S, Shilo S, Nuovo GJ, Sen CK. MicroRNA expression in response to murine myocardial infarction: miR-21 regulates fibroblast metalloprotease-2 via phosphatase and tensin homologue. Cardiovasc Res. 2009;82:21–29. doi: 10.1093/cvr/cvp015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, Hill JA, Olson EN. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ji R, Cheng Y, Yue J, Yang J, Liu X, Chen H, Dean DB, Zhang C. MicroRNA expression signature and antisense-mediated depletion reveal an essential role of MicroRNA in vascular neointimal lesion formation. Circ Res. 2007;100:1579–1588. doi: 10.1161/CIRCRESAHA.106.141986. [DOI] [PubMed] [Google Scholar]
- 43.Xin M, Small EM, Sutherland LB, Qi X, McAnally J, Plato CF, Richardson JA, Bassel-Duby R, Olson EN. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 2009;23:2166–2178. doi: 10.1101/gad.1842409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boettger T, Beetz N, Kostin S, Schneider J, Kruger M, Hein L, Braun T. Acquisition of the contractile phenotype by murine arterial smooth muscle cells depends on the Mir143/145 gene cluster. J Clin Invest. 2009;119:2634–2647. doi: 10.1172/JCI38864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN, Srivastava D. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng Y, Liu X, Yang J, Lin Y, Xu DZ, Lu Q, Deitch EA, Huo Y, Delphin ES, Zhang C. MicroRNA-145, a novel smooth muscle cell phenotypic marker and modulator, controls vascular neointimal lesion formation. Circ Res. 2009;105:158–166. doi: 10.1161/CIRCRESAHA.109.197517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davis BN, Hilyard AC, Nguyen PH, Lagna G, Hata A. Induction of microRNA-221 by platelet-derived growth factor signaling is critical for modulation of vascular smooth muscle phenotype. J Biol Chem. 2009;284:3728–3738. doi: 10.1074/jbc.M808788200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang WJ, Yang DD, Na S, Sandusky GE, Zhang Q, Zhao G. Dicer is required for embryonic angiogenesis during mouse development. J Biol Chem. 2005;280:9330–9335. doi: 10.1074/jbc.M413394200. [DOI] [PubMed] [Google Scholar]
- 49.Suarez Y, Fernandez-Hernando C, Pober JS, Sessa WC. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ Res. 2007;100:1164–1173. doi: 10.1161/01.RES.0000265065.26744.17. [DOI] [PubMed] [Google Scholar]
- 50.Kuehbacher A, Urbich C, Zeiher AM, Dimmeler S. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ Res. 2007;101:59–68. doi: 10.1161/CIRCRESAHA.107.153916. [DOI] [PubMed] [Google Scholar]
- 51.Poliseno L, Tuccoli A, Mariani L, Evangelista M, Citti L, Woods K, Mercatanti A, Hammond S, Rainaldi G. MicroRNAs modulate the angiogenic properties of HUVECs. Blood. 2006;108:3068–3071. doi: 10.1182/blood-2006-01-012369. [DOI] [PubMed] [Google Scholar]
- 52.Fasanaro P, D’Alessandra Y, Di Stefano V, Melchionna R, Romani S, Pompilio G, Capogrossi MC, Martelli F. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J Biol Chem. 2008;283:15878–15883. doi: 10.1074/jbc.M800731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pulkkinen K, Malm T, Turunen M, Koistinaho J, Yla-Herttuala S. Hypoxia induces microRNA miR-210 in vitro and in vivo ephrin-A3 and neuronal pentraxin 1 are potentially regulated by miR-210. FEBS Lett. 2008;582:2397–2401. doi: 10.1016/j.febslet.2008.05.048. [DOI] [PubMed] [Google Scholar]
- 54.Kim HW, Haider HK, Jiang S, Ashraf M. Ischemic preconditioning augments survival of stem cells via MIR-210 expression by targeting caspase-8 associated protein 2. J Biol Chem. 2009;284:33161–33168. doi: 10.1074/jbc.M109.020925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DY, Srivastava D. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15:272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, Hill JA, Richardson JA, Bassel-Duby R, Olson EN. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15:261–271. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuhnert F, Mancuso MR, Hampton J, Stankunas K, Asano T, Chen CZ, Kuo CJ. Attribution of vascular phenotypes of the murine Egfl7 locus to the microRNA miR-126. Development. 2008;135:3989–3993. doi: 10.1242/dev.029736. [DOI] [PubMed] [Google Scholar]
- 58.Harris TA, Yamakuchi M, Ferlito M, Mendell JT, Lowenstein CJ. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc Natl Acad Sci U S A. 2008;105:1516–1521. doi: 10.1073/pnas.0707493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bonauer A, Carmona G, Iwasaki M, Mione M, Koyanagi M, Fischer A, Burchfield J, Fox H, Doebele C, Ohtani K, Chavakis E, Potente M, Tjwa M, Urbich C, Zeiher AM, Dimmeler S. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science. 2009;324:1710–1713. doi: 10.1126/science.1174381. [DOI] [PubMed] [Google Scholar]
- 60.Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, Wentzel E, Furth EE, Lee WM, Enders GH, Mendell JT, Thomas-Tikhonenko A. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38:1060–1065. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suarez Y, Fernandez-Hernando C, Yu J, Gerber SA, Harrison KD, Pober JS, Iruela-Arispe ML, Merkenschlager M, Sessa WC. Dicer-dependent endothelial microRNAs are necessary for postnatal angiogenesis. Proc Natl Acad Sci U S A. 2008;105:14082–14087. doi: 10.1073/pnas.0804597105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 63.Krutzfeldt J, Kuwajima S, Braich R, Rajeev KG, Pena J, Tuschl T, Manoharan M, Stoffel M. Specificity, duplex degradation and subcellular localization of antagomirs. Nucleic Acids Res. 2007;35:2885–2892. doi: 10.1093/nar/gkm024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, Subramaniam A, Propp S, Lollo BA, Freier S, Bennett CF, Bhanot S, Monia BP. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 65.Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, Gullans S, Kearney P, Sarnow P, Straarup EM, Kauppinen S. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 66.Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- 67.Ji X, Takahashi R, Hiura Y, Hirokawa G, Fukushima Y, Iwai N. Plasma miR-208 as a Biomarker of Myocardial Injury. Clin Chem. 2009;55:1944–1949. doi: 10.1373/clinchem.2009.125310. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.