Abstract
We have built a novel computational microscopy platform that integrates image acquisition, storage, processing and analysis to study cell populations in situ. This platform allows high-content studies where multiple features are measured and linked at multiple scales. We used this approach to study the cellular composition and architecture of the mouse mammary gland by quantitatively tracking the distribution and type, position, proliferative state, and hormone receptor status of epithelial cells that incorporated bromodeoxyuridine while undergoing DNA synthesis during puberty and retained this label in the adult gland as a function of tissue structure. Immunofluorescence was used to identify label-retaining cells, as well as epithelial cells expressing the proteins progesterone receptor and P63. Only 3.6% of luminal cells were label-retaining cells, the majority of which did not express the progesterone receptor. Multi-scale in situ analysis revealed that luminal label-retaining cells have a distinct nuclear morphology, are enriched 3.4-fold in large ducts, and are distributed asymmetrically across the tissue. We postulated that LRC enriched in the ventral mammary gland represent progenitor cells. Epithelial cells isolated from the ventral versus the dorsal portion of the gland were enriched for the putative stem cell markers CD24 and CD49f as measured by fluorescence activated cell sorting. Thus, quantitative analysis of the cellular composition of the mammary epithelium across spatial scales identified a previously unrecognized architecture in which the ventral-most, large ducts contain a reservoir of undifferentiated, putative stem cells.
Introduction
Microscopy is the tool of choice to study cells in complex tissues, their interrelationships and their responses to complex signals from the microenvironment. Spatially restricted events largely determine the biology of cells, tissues and organs, while heterogeneity and three-dimensionality are two essential features of a functional tissue. Microscopy allows the observation of specific proteins using antibodies and other reagents, their distribution and abundance, and the spatial determinants, whether subcellular, cellular, or multicellular, within a sample. Although images are provocative to the non-expert, some features are difficult to readily extract as critical information. Analytical methods based on immunohistochemistry and conventional microscopy account for heterogeneity, but they frequently neglect spatial organization and statistical analysis. Large-scale morphological patterns, such as spatial patterning across tissues or relationships between specific cell types are thus lost. This type of cataloguing also fails to relate features in novel and meaningful ways that will further our understanding of basic biology. Tools to integrate three-dimensional information from tissue at multiple levels of resolution would be of benefit for understanding how cells behave as a function of changing microenvironments within the same tissue.
Another significant aspect of microscopy of fixed specimens is that changes in shape, response, and organization in a population of cells, whether in vivo or in culture, are distributed statistically and that some observations cannot take place on the same sample over time.1 It is therefore necessary to conduct large population studies and correlate distinct features measured from images with annotation data in order to fully characterize a pattern of response. In this context image analysis aids in the segmentation of sub-cellular components, e.g. nuclei, and further definition of these components with their attributes such as size, texture, or shape.2
The mammary gland is unique in that the epithelium develops postnatally during puberty under the control of ovarian steroid hormones. Mammary development gives rise to an epithelial tissue where cells can be functionally classified according to their type (luminal vs. myoepithelial), cell-cycle status (proliferative vs. quiescent) and hormone-responsiveness (hormone receptor positive or negative). Several studies have suggested that the spatial organization of cells may dictate the proliferative response to ovarian hormones, such that hormone receptor positive cells signal to hormone receptor negative cells to proliferate.3,4 During pregnancy, the mammary epithelium undergoes a remarkable cycle of growth, differentiation and involution that indicates the presence of a reservoir of undifferentiated cells in the adult tissue.5
To address these issues, we have developed multi-scale in situ analysis to integrate information about frequency, location, morphological features, and neighbor relationships of cell classes at different organizational scales (single cell, population of ducts, entire tissue). We used semi-automated image analysis to measure the nuclear properties exhibited in the epithelial cells of the mouse mammary gland and to classify cells by defining distinct features of different subpopulations. We then used the M-function analysis to quantitatively determine the degree of spatial clustering of a cell population and its statistical significance.6 Different morphological structures were compared using the M-function to determine whether particular cell classes cluster preferentially, as has been observed for stem cells in the hair follicle bulge.7 We validated the model suggested by computational analysis using fluorescence activated cell sorting of disaggregated cells based on cell surface markers reported to be associated with undifferentiated, putative stem cells.8 Our quantitative analysis revealed an underlying organization of the mammary epithelium consistent with a differential distribution of proliferative and differentiation potentials across the mouse mammary gland.
Insight, innovation, integration
We have developed novel image analysis tools to characterize cell populations at multiple scales. We have applied these tools to cells that actively divide during mammary development and then remain quiescent for months (label-retaining cells, LRC). Morphometric analysis and Monte Carlo simulations demonstrated that LRC have characteristic nuclear features, form small, non-random, clusters in large mammary ducts, are undifferentiated and remain proliferation competent. Tissue-wide quantification showed that LRC are asymmetrically distributed in the tissue, suggesting patterning of the progenitor cell reservoir. We confirmed this hypothesis using fluorescence activated cell sorting of mammary stem cell markers. The high through-put image analysis of putative progenitor cells in the murine mammary gland revealed a previously unrecognized architecture of the mammary gland.
Results
Multi-scale in situ analysis of mouse mammary gland
We used a computational microscopy platform, R3D2, that integrates image acquisition, processing and analysis, and hierarchical storage.6,9 Mouse mammary glands were serially-sectioned, and the sections stained using fluorochrome-labeled antibodies and a nuclear counter stain. Every section was automatically imaged at low magnification. Areas of interest defined by the investigator were re-imaged at high magnification and linked to the low-magnification images within R3D2 (see Materials and Methods). Linking micrographs from serial sections allows partial reconstruction of the three-dimensional architecture of the sample. Subsequent analysis steps were compartmentalized by classifying high-magnification images according to the morphological structure that they correspond to in the tissue. A feature vector was computed for each cell, including nuclear size, shape factor, bending energy and chromatin texture (see Materials and Methods), as well as positive/negative status for protein markers detected by immunofluorescence. M-Function analysis was used to compute several indexes of local tissue organization for each cell, to determine whether they were part of clustered or regularly spaced populations or whether they were localized in the proximity of other cell populations.6 The statistical significance of these indexes was assessed via Monte Carlo simulations. The global distribution of the cells across the tissue was calculated with respect to different morphological structures and/or tissue axes (e.g. anterior/posterior or dorsal/ventral).
The mammary epithelium is composed of luminal cells surrounded by basal myoepithelial cells. Occasional suprabasal cells are found between the myoepithelial and the luminal layers.10 The epithelium is ensheathed by the periductal stroma, embedded itself in the adipose stroma.11 The mammary anlage present at each nipple proliferates during puberty in response to ovarian hormones to elaborate a branched ductal tree in the mammary fat pad. Label-retaining cells (LRC) represent a population of quiescent, long-lived or functionally distinct mammary cells.12 We hypothesized that linking information about the morphological properties of LRC, their expression of proliferation and differentiation markers, and their local and global distribution within the tissue would allow us to formulate testable hypotheses about the architecture of the mouse mammary gland.
Bromodeoxyuridine (BrdU) was administered continuously for 2 weeks at the onset of puberty, labeling cells that incorporated BrdU into newly synthesized DNA. At the end of this period, 80–90% of mammary epithelial cells were positive for an antibody that recognized BrdU-substituted DNA. LRC were defined as those cells that retained the label during a subsequent 13 week chase.
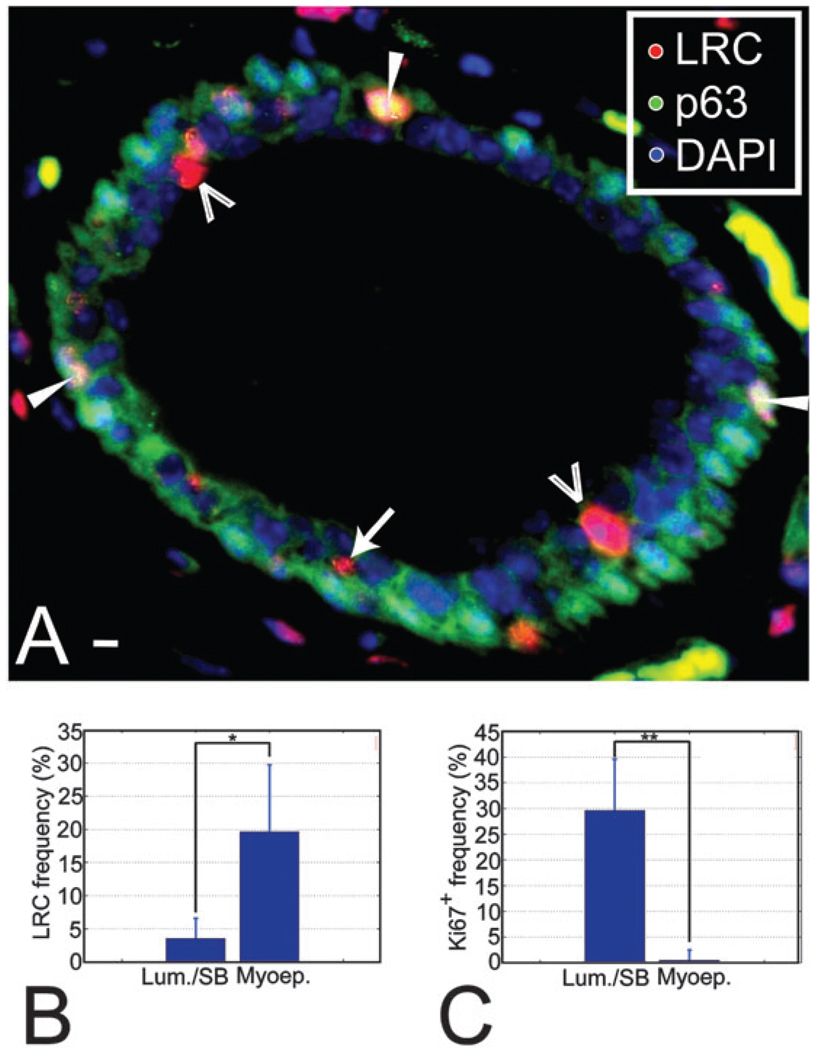
Myoepithelial cells are long-lived, differentiated contractile cells13 that can be identified by location, shape and the expression of the protein P63 in the mammary gland.14 The myoepithelial population contained a high frequency of LRC (19.7 ± 10.1%, Fig. 1A and B). We speculated that terminal differentiation during administration of the label or at the beginning of the chase was responsible for the high LRC frequency among myoepithelial cells. If so, myoepithelial cells should have little potential to proliferate in response to hormonal signals. We quantified the proliferative potential of myoepithelial cells in glands collected at mid-pregnancy stages, when proliferation occurs throughout the mammary tree. At mid-pregnancy 29.5 ± 8.6% of luminal/suprabasal epithelial cells expressed the proliferation marker Ki67+, against only 0.5 ± 0.4% of myoepithelial cells (Fig. 1C), supporting the idea that the myoepithelial LRC are largely terminally differentiated cells.
Fig. 1.
LRC are a heterogeneous population. (A) Section through a mammary duct where LRC and myoepithelial cells have been labeled using fluorescent markers. The image was background-corrected to improve visualization. Open arrowheads indicate the position of luminal LRC, an arrow points at a suprabasal LRC and white arrowheads show the location of myoepithelial LRC. Saturated green areas in the stroma correspond to non-specific staining. They can be identified by their yellow, flat appearance and non-nuclear localization versus the punctate nuclear pattern of staining in true P63+ cells. Scale bar, 10 µm. (B) LRC frequencies in the luminal and the myoepithelial compartment show that LRC are more abundant among myoepithelial cells (*P = 0.027), possibly reflecting early differentiation of these cells. The number of cells counted was 25 317 and 2975 for the luminal and myoepithelial compartments, respectively, in n=4 animals. (C) At mid-pregnancy, proliferating cells are 59-fold more abundant in the luminal layer than in the myoepithelial layer (**P = 0.0065). The number of cells counted was 10 567 and 6902 for luminal and myoepithelial compartments, respectively, in n = 4 animals.
The percentage of LRC among luminal cells was 3.6 ± 3.1% (Fig. 1A and B), consistent with their greater proliferative potential relative to myoepithelial cells (Fig. 1C).15 We used dual immunofluorescence staining to co-localize luminal LRC with the progesterone receptor (PR), a marker predominantly expressed by non-proliferating and differentiated cells.3,16–19 The frequency of PR+ cells among luminal LRC was half of that observed in the luminal epithelium at this age (11.3 ± 5.2% vs. 22.4 ± 3.7%) suggesting that luminal LRC are a relatively undifferentiated population.
Luminal LRC have a distinct nuclear signature
Nuclear morphology can be used to classify cell types and differentiation levels.20–22 Even within a fixed and sectioned tissue, nuclear cross-sections can reveal characteristic differences when measured across large populations. We analyzed several morphological features of luminal LRC and compared them to cells that did not contain any label after the chase (non-LRC), which were assumed to be mostly transit cells or terminally differentiated. Because our analysis excluded explicitly proliferating cells, nuclear signatures were not affected by cell cycle.
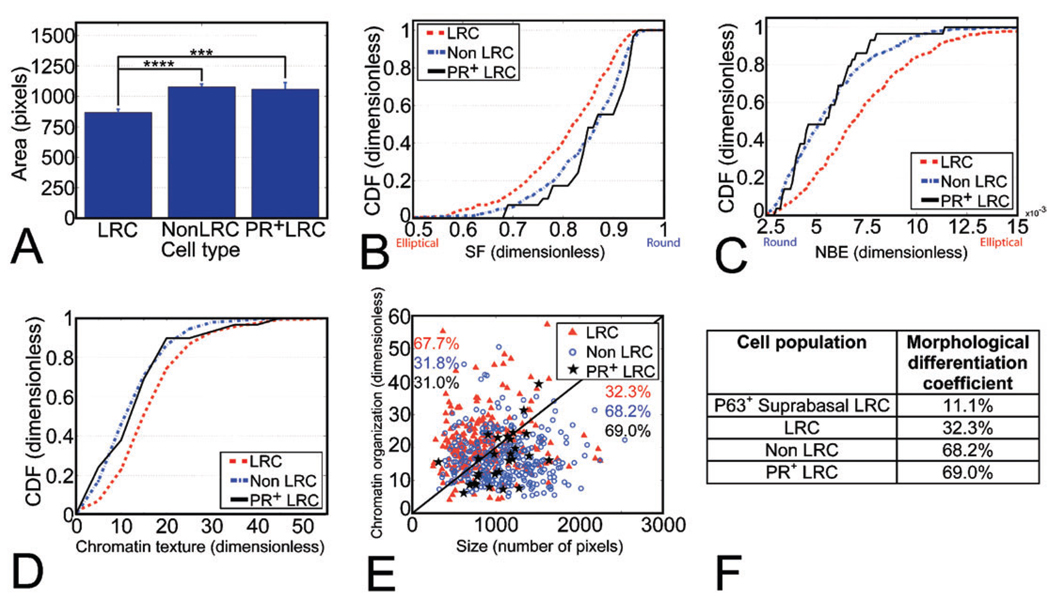
LRC nuclei were significantly smaller than non-LRC nuclei (Fig. 2A). We used a shape factor (SF) to measure nuclear roundness and the normalized bending energy (NBE) to quantify perimeter irregularities (see Materials and Methods). LRC nuclei were significantly more elliptical than non-LRC nuclei (smaller SF, larger NBE, Fig. 2B and C, respectively). Changes in nuclear organization have been associated with differentiation in mammary cells.22 Consistent with this, the chromatin texture distribution for LRC was significantly more heterogeneous than for non-LRC (Fig. 2D). Displaying texture information as a function of nuclear size showed that LRC and non-LRC have distinct nuclear signatures. A linear threshold readily separated two-thirds of the LRC with small nuclei and heterogeneous chromatin texture from two-thirds of non-LRC displaying large nuclei and more homogeneous chromatin texture values (Fig. 2E).
Fig. 2.
Multi-feature image analysis shows that LRC have a distinct nuclear signature. (A) Nuclear size analysis shows that LRC nuclei are smaller than non-LRC (****P =3.34 ± 10−9) and PR+ LRC (***P =0.003) nuclei. The number of nuclei analyzed was n= 226, 321 and 29 for LRC, non-LRC and PR+ LRC, respectively. (B, C) LRC have smaller SF and larger NBE than non-LRC (P=7.11 × 10−6 and P=3.78 × 10−11, respectively) and PR+ LRC nuclei (P = 0.004 and P = 0.003, respectively), as shown by their cumulative distribution functions (CDF). (D) Chromatin texture cumulative distribution functions (CDF) show a different chromatin organization in LRC with respect to non-LRC (P = 3.09 × 10−9) and PR+ LRC (P = 0.025). (E) Nuclear size vs. chromatin texture. The numbers indicate the percentage of cells in each population above and below the line, which can be quantitatively described by eqn (4). The line delineates the bulk of the differentiated and undifferentiated populations. (F) Degree of morphological differentiation for the populations characterized in situ. The number of nuclei analyzed was n = 9, 226, 321 and 29 for P63+ suprabasal LRC, all LRC, non-LRC and PR+ LRC, respectively.
Since not all differentiated cells in the mammary gland are PR+, it is likely that there are differentiated LRC that do not express PR. In order to refine our assessment of the frequency of differentiated cells among LRC, we measured the nuclear features of PR+ LRC. The size, shape and chromatin texture distributions of PR+ LRC nuclei were significantly different from total LRC (Fig. 2A–C) and overlapped those of non-LRC, suggesting that these features represent the signature of differentiated cell populations. Based on nuclear size and chromatin texture criteria, 32.3% of luminal LRC were morphologically differentiated (Fig. 2E and F), compared to 68.2% of non-LRC and 69% of PR+ LRC (Fig. 2F).
Suprabasal cells, identified by their intermediate location between luminal and myoepithelial cells, are very rare (100 out of 4211 cells counted in 3 glands) and have been postulated to be the most undifferentiated mammary cells based on ultra-structural studies.10 Consistent with this, a high frequency of LRC was found in the suprabasal compartment (28 out 100 suprabasal cells) and also showed a low differentiation nuclear signature similar to that of luminal LRC (Fig. 2F). It has been hypothesized that suprabasal cells contain bilineage progenitors that can give rise to the luminal and the myoepithelial populations.10,23 Consistent with this model, 9% of suprabasal LRC expressed the myoepithelial marker P63.
Luminal LRC form clusters in larger ducts and contain proliferation competent, undifferentiated cells
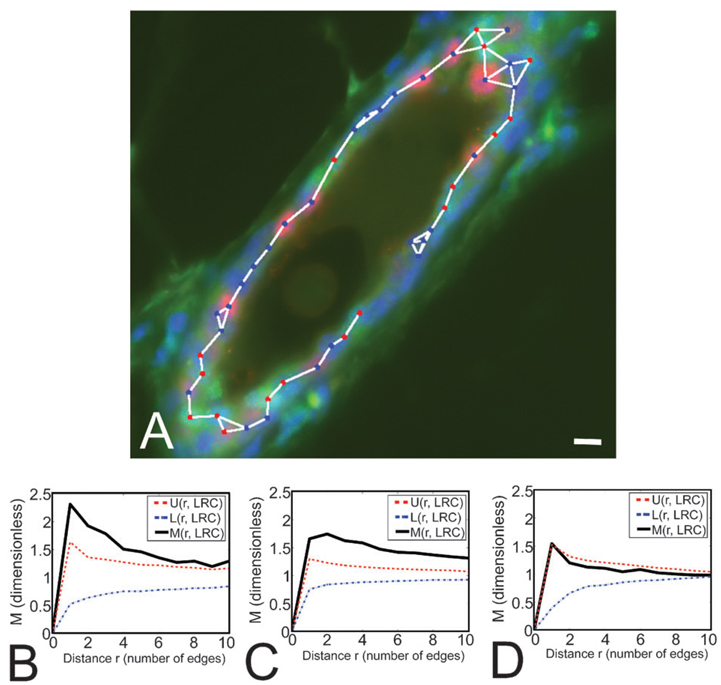
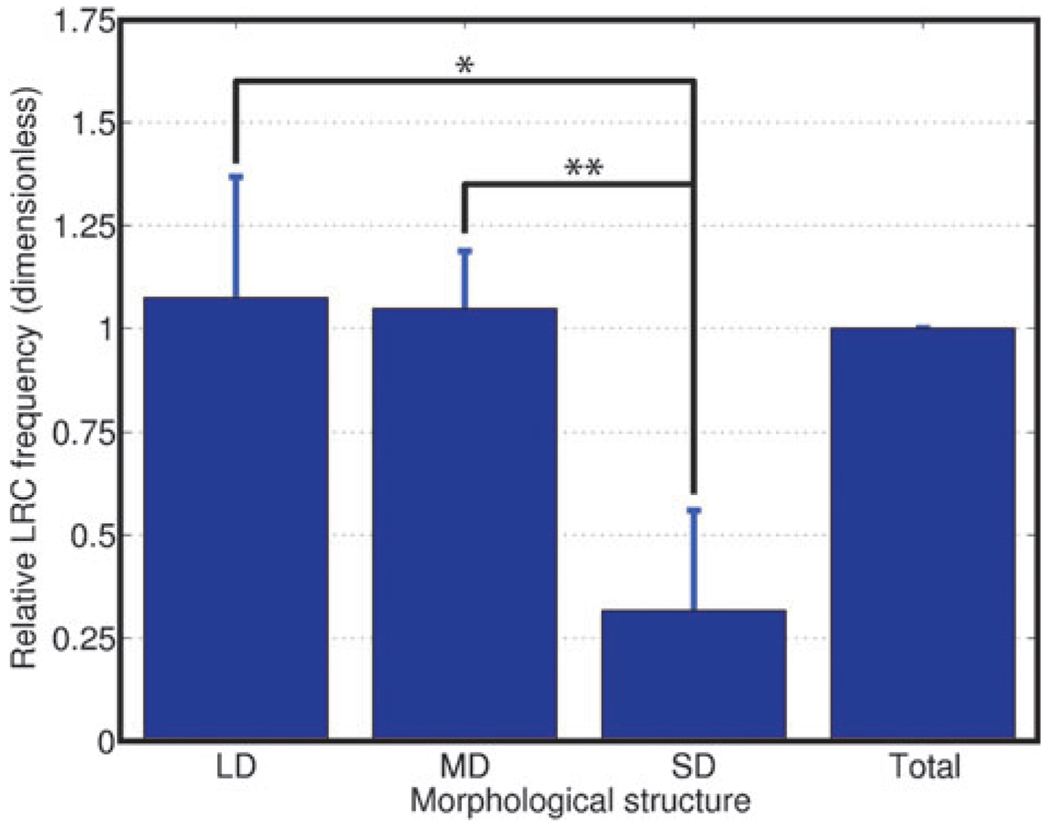
We examined the distribution of luminal LRC in different morphological structures in the mammary gland. The mammary gland consists of primary, secondary and tertiary ducts that roughly correspond to large, medium and small ducts (LD, MD and SD, respectively). We quantified LRC distribution using the M-function, which indicates the fold-increase in clustering with respect to a distribution with homogeneous density6 (see Materials and Methods). LRC formed small clusters in larger ducts (Fig. 3A–C). The average cluster size (number of LRC per cluster) was 2.79 cells in LD, and 2.66 cells in MD. In SD, LRC appeared to be randomly distributed (Fig. 3D), although the robustness of the measurement may be affected by the low frequency of LRC in this type of duct. These data show for the first time a preferential location of a functional subpopulation of mammary cells, which suggests that mammary gland architecture might dictate the organization of different types of cells.
Fig. 3.
Monte Carlo simulations demonstrate that LRC form small clusters in larger ducts. (A) Medium-sized duct where LRC (green nuclei) form small clusters. The duct was also stained for PR (red nuclei) and counterstained with DAPI (blue). The graph connecting neighbor cells used to evaluate the M-function (see Materials and Methods) is overlaid. Scale bar, 10 µm. (B–D) M-Function analyses for LRC in LD, MD and SD (respectively) reveal significant clustering in LD and MD.Mindicates the degree of LRC clustering observed in tissue samples, while U and L show the upper and lower limits for the range of clustering values obtained from simulations of random LRC distributions. M values above U indicate that the degree of clustering of LRC observed in situ cannot be recapitulated in random simulations. Therefore LRC are significantly clustered in LD and MD. In SD, M values remain within the range obtained in simulations (U and L), indicating that the pattern of LRC distribution is likely to be random in smaller ducts. The number of cells analyzed was n = 127, 295 and 18 for LD, MD and SD, respectively.
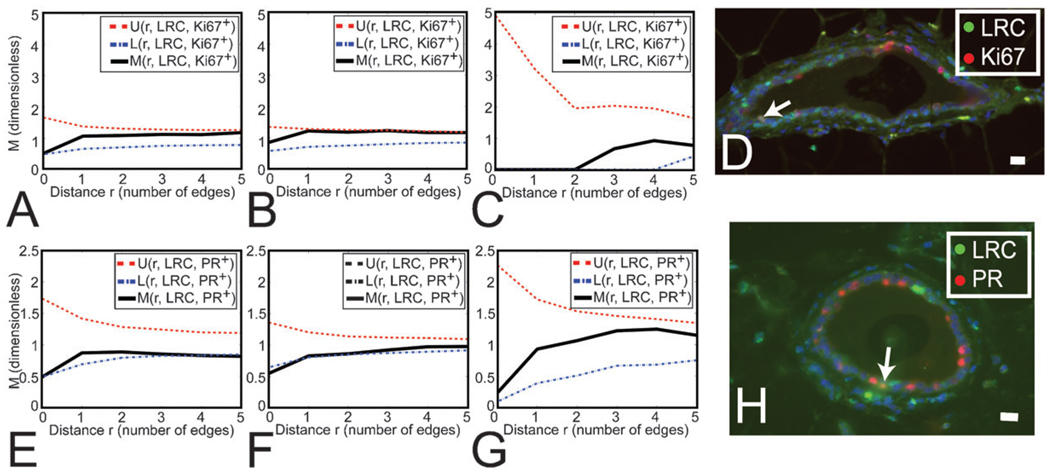
We then investigated a potential association between the location of luminal LRC and actively proliferating cells using M-function analysis to determine whether LRC co-localized with proliferation markers or if LRC were in the close proximity of dividing cells. We did not find a significant neighborhood relationship between luminal LRC and actively dividing Ki67+ cells, or significant co-localization of both markers in single cells (Fig. 4A–C). Although these data suggest that luminal LRC were primarily quiescent, a small percentage of luminal LRC were Ki67+ (3.1 ± 1.2% of all LRC, Fig. 4D), similar to the overall percentage of Ki67+ cells in the luminal epithelium. Thus, LRC are proliferation-competent cells.
Fig. 4.
LRC do not preferentially associate with the proliferation marker Ki67, but they significantly exclude the differentiation marker PR. (A–C, E–G) M Indicates the degree of association between LRC and Ki67+ (A–C) or PR+ (E–G) cells. U and L show the upper and lower limits for the range of association values obtained from simulations of independent distributions of LRC and Ki67+ (A–C) or PR+ (E–G) cells. (A–C) Distribution of Ki67+ cells with respect to LRC in LD, MD and SD (respectively) shows that both populations are independently arranged. The number of cells analyzed was n = 220, 334 and 6 for LD, MD and SD, respectively. (D) Dual staining for LRC and Ki67. Nuclei were counterstained with DAPI (blue). Arrow points at a cell where both markers co-localize. Scale bar, 20 µm. (E–G) Distribution of PR+ cells with respect to LRC in LD, MD and SD (respectively) shows that retention of BrdU correlates with the absence of PR expression in LD and MD (where M has a minimum below L at r=0, and L is the lower limit of clustering density values obtained in the simulations). At all other distances (r > 1) there is no relationship (aggregation or separation) between LRC and PR+ cells. The number of cells analyzed was n=129, 310 and 20 for LD, MD and SD, respectively. (H) Dual staining for LRC and PR. Nuclei were counterstained with DAPI (blue). Arrow points at a cell where both markers co-localize. Scale bar, 20 µm.
Proliferation in the mammary gland is stimulated by the ovarian steroid hormones, which act through cognate hormone receptors that are restricted to a subset of luminal epithelial cells.24,25 Because hormone receptor-positive cells rarely co-localize with proliferation markers but are often direct neighbors of dividing cells,3 it has been proposed that hormone receptor positive cells regulate proliferation of their neighbors through juxtacrine mechanisms.26 We investigated a relationship between the location of LRC and hormone receptor-positive cells (i.e., PR immunoreactive). PR+ frequency in the LRC pool was half of that observed in the luminal epithelium (Fig. 4H). M-Function analysis confirmed that LRC were less likely to express PR in LD and MD (Fig. 4E and F). Neighborhood analysis showed that LRC and PR+ cell populations were independent (Fig. 4E–G). Thus, while proliferating cells are near hormone receptor-positive cells during estrus,3 the locations of LRC and PR+ cells are not associated.
Mammary LRC are differentially distributed across the gland
Mammary LRC contain cells with stem/progenitor potential. 27–29 Zones of adult stem cell (ASC) enrichment have been described at the bottom of the intestinal crypts30,31 the bulge of the hair follicle32 or the prostate tissue around the urethra.33 Since mammary fragments from any portion of the gland generate outgrowths when transplanted to parenchyma- free fat pads,34 we hypothesized that LRC would be homogeneously distributed throughout the ductal tree. However we found that LRC were significantly more frequent in LD (3.4-fold) and MD (3.3-fold) than they were in SD (Fig. 5). These data are consistent with a model where tissue architecture restricts the location of certain cell populations.
Fig. 5.
Tissue-wide analysis shows that LRC are more abundant in larger ducts. Relative LRC frequency in each type of duct normalized to the LRC frequency in the entire gland shows LRC enrichment in LD (* P = 0.015) and MD (** P = 0.006). The number of nuclei counted was 7623, 12 551 and 1305 for LD, MD and SD, respectively in n = 4 animals.
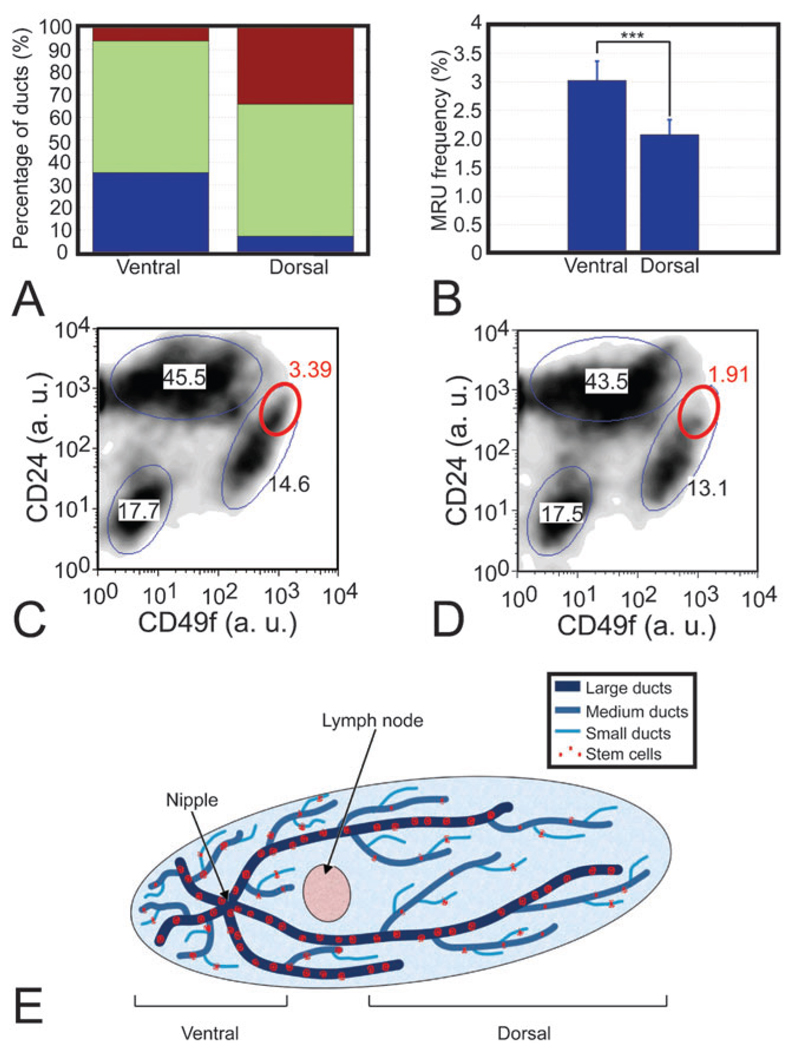
The prevalence of undifferentiated mammary LRC in large and medium ducts suggested that the larger ducts act as a reservoir of proliferative potential. As larger ducts were more abundant in the ventral-most region of the tissue, close to the nipple, and small ducts are more profuse in the dorsal region (Fig. 6A), we postulated that the differential distribution of mammary LRC would be paralleled by a dorsal-ventral difference in the localization of putative mammary progenitor cells. We used fluorescence-activated cell sorting of mammary epithelial cells on the basis of their expression of CD24 and CD49f (i.e. CD24+CD49fhigh), which identifies a population highly enriched for mouse mammary ASC.8 The fraction of CD24+CD49fhigh was 50% greater in ventral tissue with respect to dorsal tissue (3.0 ± 0.3% vs. 2.1 ± 0.3%, Fig. 6B), indicating that the distribution of CD24+CD49fhigh cells was skewed towards the large duct-rich ventral region of the gland, proximal to the nipple. Notably, when we examined different populations previously characterized for their expression of CD24 and CD49f,8 CD24+CD49fhigh was the only population that showed a significant difference between ventral and dorsal tissue (Fig. 6C and D), supporting the idea of a differential distribution of the mammary cells with the ability to proliferate and differentiate into multiple lineages (Fig. 6E).
Fig. 6.
Fluorescence-activated cell sorting reveals an asymmetric distribution of mammary repopulating cells. (A) Ventral tissue was mainly composed of large (blue) and medium (green) ducts, while dorsal tissue was principally formed by medium and small (red) ducts. Ducts were selected and imaged with a homogeneous spatial distribution across n = 4 mammary glands of 18-week-old females. (B) Frequency of CD24+CD49fhigh cells is higher in ventral tissue than in dorsal tissue (***P=0.0048). (C, D) Density plots representing the cell number in each gated population (ellipses) in dorsal and ventral tissue, respectively, show that CD24+CD49fhigh cells (red ellipses) are the only population that shows a significant change. Notice how the saturated area within the CD24+CD49fhigh gate for ventral cells is absent in the plot for dorsal cells. Axes indicate arbitrary intensity units (a. u.). (E) Cartoon representing our model for stem cell distribution along the mouse mammary gland with a reservoir of proliferative potential in larger ducts.
Discussion
Multi-scale in situ sorting of epithelial cells used in conjunction with statistical modeling revealed several new aspects of murine mammary biology. First, we defined the nuclear morphology of an undifferentiated luminal LRC population; then we showed that suprabasal cells are enriched for both luminal and myoepithelial features; and finally, we demonstrated that LRC are clustered in the ventral-most, larger ducts, proximal to the site of embryonic origin of the gland. We tested the implication of these data by using fluorescence activated cell sorting to show ventral enrichment of CD24+CD49fhigh cells, which have significant mammary repopulating potential.8,27 The differential distributions of LRC and CD24+CD49fhigh cells are consistent with an architectural reservoir of proliferative potential in larger mammary ducts.
LRC have been the basis for identifying ASC distribution in situ in many tissues.7,35–39 LRC proliferate during tissue development, are long-lived, and retain a nuclear label for long periods of time (> 13 weeks). Mammary LRC have been shown to co-segregate their DNA strands,28 a property attributed to ASC both in vitro40 and in vivo.36,41 Furthermore, a recent study using cell surface marker-based sorting has shown that LRC are enriched several fold in CD24+CD49fhigh cells, a population shown to contain mammary ASC.27 However, no information was available until now about the morphology of LRC or their distribution in the different morphological structures that form the mammary gland. If understanding heterogeneity is essential for understanding how normal tissues function, it becomes critical in cancer, where cell heterogeneity and clonal selection are at the very core of carcinogenesis and cancer progression. In breast cancer, little is known about why some parts of the mammary gland are more susceptible than others to developing neoplastic lesions or about what drives the evolution of some neoplastic clones to aggressive phenotypes while their neighbors remain unaltered.
Our analysis showed that LRC nuclei were smaller and more elliptical than those of non-LRC. These features are similar to those of muscle ASC and might reflect a distinct nuclear configuration that favors efficient transcription or silencing of certain genes involved in cell division or differentiation. 20,21 Interestingly, recent evidence has shown increased deformability of the nuclei of adult hematopoietic stem cells compared to differentiated cells.42 Our data indicate that this might be the case for mammary LRC as well. We also found that DAPI staining was less rigidly patterned in LRC than in differentiated cells. DAPI intensity is high in heterochromatin and low in euchromatin, and thus, the different DAPI patterns observed in LRC and non-LRC nuclei suggest differences in chromatin organization, which have been observed during morphological differentiation in the mammary gland.22 The small nuclei and heterogeneous chromatin pattern of LRC are consistent with the nuclear features of small light cells, a putative mammary progenitor population whose morphological features have been investigated using electron microscopy.10
Hormone receptors are generally considered markers of quiescent, differentiated cells.16,29 The frequency of PR+ cells in the LRC compartment was half that of the luminal epithelium overall, indicating that LRC were relatively undifferentiated. However, a recent study found a similar frequency of PR+ cells in the entire epithelium and among LRC (32% vs. 28%).11 It is likely that the much longer labeling strategies that we employed (14 vs. 5 days labeling and 13 vs. 6 weeks chase) provided opportunity for turnover of differentiated cells during the additional weeks of chase. We found agreement between the nuclear signatures of PR+ LRC and non-LRC. Using nuclear morphology to approximate the degree of differentiation of several mammary subpopulations in situ, we estimated that two thirds of the LRC were relatively undifferentiated. Our data analysis shows that many LRC represent a long-lived, largely undifferentiated population in the mouse mammary gland.
Spatial analysis between ducts and across the tissue revealed a previously unrecognized organization of mammary LRC. We found that LRC were more frequently located in larger ducts, where they formed small clusters (two to three cells), as found in other epithelial tissues.32,43,44 This suggests that large ducts have an increased proliferation and differentiation potential. Consistent with this model, tissue from central areas of the gland (where larger ducts are more abundant) can be serially transplanted more times than tissue obtained from peripheral areas (mostly SD), which exhausts its regeneration potential more rapidly.34 Taken together these data suggest the presence of LRC niches along the larger ducts of the mouse mammary gland, and establish a candidate region for the location of the mammary ASC niche. Further investigation using bona fide mammary ASC markers will be necessary to confirm the location of this niche and to elucidate the molecular cues that regulate the behavior of the cells included in the niche.
Differential distribution of LRC may reflect an underlying, previously unrecognized, architecture of the gland where the density of quiescent, undifferentiated cells is higher in the larger mammary ducts. However, some recent reports have suggested that LRC may not be enriched for ASC.45,46 Our analysis demonstrates that LRC are heterogeneous with respect to cell type, nuclear signature, and expression of differentiation, proliferation and lineage markers, which could explain the contradictory reports regarding the relationship between LRC and ASC. Fluorescence activated cell sorting analysis of CD24+CD49fhigh cells, which have high mammary repopulating potential,8 demonstrated significant enrichment in cells isolated from ventral versus dorsal tissue; importantly, CD24+CD49fhigh cells were the only population that showed differential distribution when we investigated several populations with diverse levels of expression of CD24 and CD49f.8 These data confirm our prediction of tissue architecture using LRC and validate the use of multiscale in situ analysis to characterize cell populations within their tissue context. The differential distribution of CD24+CD49fhigh cells is consistent with a graded localization of mammary ASC along the ductal tree and suggests that the mammary gland architecture contains a reservoir of proliferative and differentiation potential in the larger ducts. Importantly, recent studies in the human breast also show evidence of a differential distribution of putative stem cells.47 The human breast is formed by ducts, which are analogous to larger mouse mammary ducts, and lobules, which correspond to small ducts in the mouse gland. The findings of Villadsen and colleagues suggest that human mammary stem cells are located in the ducts rather than the lobules, a distribution similar to that described here for mouse LRC and CD24+CD49fhigh cells.
The novel computational tools for quantitative in situ analysis of cell morphology and localization developed in this study lead us to conclude that mammary gland architecture is more complex than previously appreciated. Gene expression profiling of cells sorted on the basis of putative stem cell markers48 might soon reveal proteins for the unequivocal identification of mouse mammary ASC in situ, thus allowing the use of the computational tools described here to contrast bona fide ASC to LRC by comparing nuclear features or localization patterns. In the interim, analysis of immunolocalization and nuclear features in conjunction with statistical modeling provides a new tool to discriminate cell populations in vivo and to define the basic architecture of the mammary gland.
Materials and methods
Mice
All experiments described below were executed with the approval of the animal care committees at Lawrence Berkeley National Laboratory and the University of California, San Francisco. Three-week-old, wild-type FVB female mice (Charles River) were implanted with miniosmotic pumps containing BrdU as described before.29 After 2 weeks, the pumps were removed and the animals housed for chase periods of 9 or 13 weeks. Groups consisted of 4 animals. One group of mice was bred after the 9-week chase and the pregnant females were euthanized ten days after the first day of mating. The fourth, inguinal mammary glands were collected, fixed in 10% buffered formalin overnight and stored in 70% ethanol until embedding.
For mammary cell isolation, the inguinal mammary glands of 8-week-old (adult) FVB female mice (Charles River) were collected and dissected into three fragments: ventral, between the nipple and the lymph node; lateral, containing the lymph node; and dorsal, between the lymph node and the periphery of the gland. The lateral fragments were discarded and the dorsal and ventral parts were collected separately. Mammary epithelial cells were isolated from these fragments as described before.27 Cell viability was determined by Trypan Blue exclusion.
Tissue processing and immunostaining
Tissues were paraffin-embedded and serially sectioned at 4 µm along the longitudinal axis of the gland. Histoclear (National Diagnostics, #HS-200) was used to remove paraffin. Sections were subjected to antigen retrieval by microwaving for 21 min in antigen unmasking solution (Vector, #H-3300), and blocked in 0.5% casein–0.1% Tween in phosphate-buffered saline (PBS) for 1 h. Double immunofluorescence staining for BrdU and an additional marker was performed in sequence. The BrdU primary antibody was incubated for 1 h at 37 °C after incubation with the additional primary (overnight at 4 °C) and its corresponding secondary (1 h at room temperature). The slides were washed for 5 min with 0.1% Tween–PBS between incubations. Finally, the sections were counterstained with DAPI (Sigma, #D9542, 1 : 1000) for 10 min and mounted with Vectashield (Vector, #H-1000).
The primary antibodies used were rat anti-BrdU (1 : 100, Serotec, #MCA2060), rabbit anti-Ki67 (1 : 800, Novocastra, #NCL-Ki67p), rabbit anti-PR (1 : 800, a gift from Dr Shyamala Harris) and mouse anti-P63 (1 : 200, Lab Vision, #MS-1081-P). The secondary antibodies were used at a concentration of 1 : 100, and included goat anti-rat FITC (Pierce, #31629), goat anti-rat Texas red (Molecular Probes, #T-6392), goat anti-rabbit Texas red (Molecular Probes, #T-2767) and goat anti-mouse FITC (Pierce, #31541).
Image acquisition and preprocessing
A Zeiss Axioplan I microscope coupled with a 12 bit, monochrome Orca-ER CCD camera (Hamamatsu) was used to acquire images using a multiple bandpass Pinkel filter set (Chroma Technologies), by consecutively imaging while exciting the sample at 360 nm (DAPI), 490 nm (FITC) and 560 nm (Texas red). The microscope was controlled using journals written in the Metamorph scripting language (Molecular Devices). Routines were programmed to scan the entire area of the slide occupied by the tissue at low magnification (2.5×). To do this, single camera snapshots were automatically tiled together into a large image. Microscope focusing was automatically corrected whenever necessary to ensure high-contrast in the entire area under study. New journals were written to automatically revisit ducts chosen from the low magnification tissue images at high magnification (40×).
An in-house developed computational microscopy system, R3D2,9 was used for image processing and analysis. The low magnification tissue images were aligned and used as references to map the positions of ducts in high magnification images. All images were corrected for illumination inhomogeneities using background images.
Nuclear signature analysis
The nuclear contours of epithelial LRC or non-LRC that were Ki67− and presented clean boundaries were randomly chosen and manually traced on high magnification (40×) images. The nuclear curves were then measured within R3D2 using DIPImage (Technical University of Delft), a Matlab (Mathworks) toolbox for image analysis. The measurements obtained included area, shape factor—a measurement of circularity—, normalized bending energy—an estimate of contour irregularity—, and chromatin texture, obtained as the standard deviation of the counterstaining (DAPI) intensity within each nucleus to provide an index of chromatin organization. These magnitudes are defined as follows:
| (1) |
| (2) |
| (3) |
where A is the area of the nucleus, P its perimeter, and κ(p) the curvature of the nucleus at a certain point, p, on its perimeter. The term IDAPI is a vector of counterstaining intensities per pixel within a given nucleus, and σ represents the standard deviation of a sample. A linear threshold to obtain maximum separation between LRC and non-LRC on the basis of their nuclear size and chromatin texture distributions was empirically established as:
| (4) |
with x, nuclear size, and y, chromatin texture values, respectively. The degree of morphological differentiation for a certain population was defined as the percentage of cells in that population that falls below the threshold. Nuclear properties from different cell populations were compared using the Kolmogorov–Smirnov test to evaluate differences between distributions.
Spatial analysis
M-function analysis was used as described6 to assess clustering or regularity in the pattern of one population, and aggregation or separation between two populations. Briefly, a graph was automatically generated to determine neighborhood relationships between cell nuclei in ducts imaged at high magnification (40×). These graphs provided faithful models of tissue topology as well as grids to measure distances between cells. A clustering-density function, M, was evaluated on top of the graphs. This function is defined for a population, C (e.g. LRC) at distance r (with r an integer number of cells) as:
| (5) |
where niCr is the number of neighbors of cell i within distance r belonging to population C; nir is the total number of neighbors of cell i within distance r; NC is the total number of cells belonging to population C in the image under analysis; and N is the total number of cells in the same image. This function can be easily extended to consider multiple populations. The M-function describes the fold-increase in clustering (or aggregation, for multiple populations) with respect to a homogeneous distribution. Values larger than one indicate clustering (aggregation), while values smaller than one show isolation (separation). The statistical significance of these phenomena was assessed using Monte Carlo simulations of random or independent distributions to determine whether the observed M values where within the range generated in the simulations (upper and lower limits depicted by the U and L functions). For the case of clustering, for example, a number of cells equal to the number of LRC found in situ was randomly distributed in the same graphs used for the in situ measurements. The M function was then re-evaluated for this random distribution. This process was repeated 199 times per area assessed, thus obtaining the range of M values that could be obtained if LRC were randomly distributed. By comparing these values to the value measured in situ we determined the significance of the clustering observed as:
| (6) |
where q is the number of M values obtained from random simulations that are above the value observed in situ. A similar analysis was used to assess the statistical significance of the spatial association of multiple cell populations.
Frequency analysis
The positions of the luminal and suprabasal cell nuclei were manually annotated using the high magnification DAPI images. The frequency of positive nuclei for a given marker was calculated based on thresholds determined from primary antibody negative control slides included in each experiment. To assign frequency values to different morphological structures, ducts were classified as small, medium or large based on their position in the fat pad, tracked across 10 consecutive sections, with larger ducts closer to the nipple and smaller ducts branching off of larger ducts. A quantitative assessment based on the thickness of the periductal stroma at the duct was used to confirm this classification. Small ducts were defined as those with stroma less than 8.5 mm-thick, large ducts as those with more than 15.3 µm, and medium ducts were defined as those with stromal thickness intermediate between both thresholds. These definitions were empirically selected for one of the glands and maintained for all the other ones. Information about the position of the different tree levels relative to the organ periphery was also incorporated into this classification.
Quantification of LRC frequencies was carried out in areas of the tissue that contained BrdU+ cells at the end of the labeling period, i.e., in the ventral region of the gland (between the nipple and the lymph node). Thus we controlled for the fact that mammary ducts may have not elongated until the ends of the fat pad at the time of pump extraction (five weeks of age). If the entire gland were taken into account, this could result in a disadvantage for small ducts, more abundant at the dorsal region of the tissue. LRC frequencies in each structure (LD, MD, SD) were normalized to the total LRC frequency in each gland. The variances of these normalized values across samples were then compared using an F-test. Holm’s t-test for comparison of multiple groups was used to contrast the means of the distributions.
Fluorescence-activated cell sorting
Single cell suspensions were isolated from dorsal and ventral tissue fragments independently and stained as described before.8 The experiments were repeated four times, and 60 000 cells were counted each time. Several populations were defined on the basis of their expression levels of CD24 and CD49f. The variances of the frequencies of these populations were compared between the dorsal and the ventral ends of the mammary gland using the F-test. Student’s t-test was then used to compare the mean frequency of these populations in the dorsal and ventral experimental groups.
Statistical analysis
Errors are given as standard deviations. Statistical significance was established at P < 0.05 in all cases.
Acknowledgements
The authors would like to thank Rosie Chau, Claudia Kuper and Shraddha Ravani for technical help, Dr Daniel Medina for continued discussion and advice, and Drs Mohamed Bentires-Alj and Mark LaBarge for critical reading of this manuscript. We thank Ignacio Arganda-Carreras for generating the image in the graphical abstract using R3D2. This work was supported by funds from the Department of Defense Breast Cancer Research Program (predoctoral fellowship DAMD 17-03-1-0594 to RFG; grants DAMD 17-00-1-0227 and DAMD 17-00-1-0306 COS) and a grant jointly funded by the National Institute of Environmental Health Sciences and the National Cancer Institute (U01 ES012801 to MHBH and ZW).
List of Abbreviations
- ASC
Adult stem cells
- a. u.
Arbitrary units
- BrdU
Bromodeoxyuridine
- LD
Large ducts
- LRC
Label-retaining cells
- MD
Medium ducts
- NBE
Normalized bending energy
- PBS
Phosphate-buffered saline
- PR
Progesterone receptor
- SD
Small ducts
- SF
Shape factor
- SLC
Small light cells
Footnotes
The authors declare no competing interests.
References
- 1.Fowlkes CC, Hendriks CL, Keranen SV, Weber GH, Rubel O, Huang MY, Chatoor S, DePace AH, Simirenko L, Henriquez C, Beaton A, Weiszmann R, Celniker S, Hamann B, Knowles DW, Biggin MD, Eisen MB, Malik J. Cell. 2008;133:364–374. doi: 10.1016/j.cell.2008.01.053. [DOI] [PubMed] [Google Scholar]
- 2.Luengo Hendriks CL, Keranen SV, Fowlkes CC, Simirenko L, Weber GH, DePace AH, Henriquez C, Kaszuba DW, Hamann B, Eisen MB, Malik J, Sudar D, Biggin MD, Knowles DW. Genome Biol. 2006;7:R123. doi: 10.1186/gb-2006-7-12-r123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clarke RB, Howell A, Potten CS, Anderson E. Cancer Res. 1997;57:4987–4991. [PubMed] [Google Scholar]
- 4.Ewan KB, Oketch-Rabah HA, Ravani SA, Shyamala G, Moses HL, Barcellos-Hoff MH. Am. J. Pathol. 2005;167:409–417. doi: 10.1016/s0002-9440(10)62985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Topper YJ, Freeman CS. Physiol. Rev. 1980;60:1049–1105. doi: 10.1152/physrev.1980.60.4.1049. [DOI] [PubMed] [Google Scholar]
- 6.Fernandez-Gonzalez R, Barcellos-Hoff MH, Ortiz-de-Solorzano C. IEEE Trans. Image Process. 2005;14:1300–1313. doi: 10.1109/tip.2005.852466. [DOI] [PubMed] [Google Scholar]
- 7.Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, Fuchs E. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, Li HYI, Eaves CJ. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 9.Fernandez-Gonzalez R, Jones A, Garcia-Rodriguez E, Chen PY, Idica A, Lockett SJ, Barcellos-Hoff MH, Ortiz de Solorzano C. Microsc. Res. Tech. 2002;59:522–530. doi: 10.1002/jemt.10233. [DOI] [PubMed] [Google Scholar]
- 10.Chepko G, Smith GH. Tissue Cell. 1997;29:239–253. doi: 10.1016/s0040-8166(97)80024-9. [DOI] [PubMed] [Google Scholar]
- 11.Booth BW, Smith GH. Breast Cancer Res. 2006;8:R49. doi: 10.1186/bcr1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith GH, Medina D. Breast Cancer Res. 2008;10:6. doi: 10.1186/bcr1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adriance MC, Inman JL, Petersen OW, Bissell MJ. Breast Cancer Res. 2005;7:190–197. doi: 10.1186/bcr1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barbareschi M, Pecciarini L, Cangi MG, Macri E, Rizzo A, Viale G, Doglioni C. Am. J. Surg. Pathol. 2001;25:1054–1060. doi: 10.1097/00000478-200108000-00010. [DOI] [PubMed] [Google Scholar]
- 15.Pechoux C, Gudjonsson T, Ronnov-Jessen L, Bissell MJ, Petersen OW. Dev. Biol. 1999;206:88–99. doi: 10.1006/dbio.1998.9133. [DOI] [PubMed] [Google Scholar]
- 16.Clarke RB, Butterfield P, Sanderson C, Howell A, Potten CS, Anderson E. Br. J. Cancer. 1999;80:78–78. [Google Scholar]
- 17.Brisken C, Park S, Vass T, Lydon JP, O’Malley BW, Weinberg RA. Proc. Natl. Acad. Sci. U. S. A. 1998;95:5076–5081. doi: 10.1073/pnas.95.9.5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Russo J, Ao X, Grill C, Russo IH. Breast Cancer Res. Treat. 1999;53:217–227. doi: 10.1023/a:1006186719322. [DOI] [PubMed] [Google Scholar]
- 19.Kariagina A, Aupperlee MD, Haslam SZ. Endocrinology. 2007;148:2723–2736. doi: 10.1210/en.2006-1493. [DOI] [PubMed] [Google Scholar]
- 20.Schultz E, McCormick KM. Rev. Physiol. Biochem. Pharmacol. 1994;123:213–257. doi: 10.1007/BFb0030904. [DOI] [PubMed] [Google Scholar]
- 21.Powell JA, Molgo J, Adams DS, Colasante C, Williams A, Bohlen M, Jaimovich E. J. Neurosci. 2003;23:8185–8192. doi: 10.1523/JNEUROSCI.23-23-08185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaminker P, Plachot C, Kim SH, Chung P, Crippen D, Petersen OW, Bissell MJ, Campisi J, Lelievre SA. J. Cell Sci. 2005;118:1321–1330. doi: 10.1242/jcs.01709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smalley M, Ashworth A. Nat. Rev. Cancer. 2003;3:832–844. doi: 10.1038/nrc1212. [DOI] [PubMed] [Google Scholar]
- 24.Shyamala G, Barcellos-Hoff MH, Toft D, Yang X. J. Steroid Biochem. Mol. Biol. 1997;63:251–259. doi: 10.1016/s0960-0760(97)00128-3. [DOI] [PubMed] [Google Scholar]
- 25.Shyamala G, Chou YC, Louie SG, Guzman RC, Smith GH, Nandi S. J. Steroid Biochem. Mol. Biol. 2002;80:137–148. doi: 10.1016/s0960-0760(01)00182-0. [DOI] [PubMed] [Google Scholar]
- 26.Clarke RB. Maturitas. 2006;54:327–334. doi: 10.1016/j.maturitas.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 27.Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat ML, Wu L, Lindeman GJ, Visvader JE. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 28.Smith GH. Development. 2005;132:681–687. doi: 10.1242/dev.01609. [DOI] [PubMed] [Google Scholar]
- 29.Welm BE, Tepera SB, Venezia T, Graubert TA, Rosen JM, Goodell MA. Dev. Biol. 2002;245:42–56. doi: 10.1006/dbio.2002.0625. [DOI] [PubMed] [Google Scholar]
- 30.Potten CS. Philos. Trans. R. Soc. London, Ser. B. 1998;353:821–830. doi: 10.1098/rstb.1998.0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barker N, van de Wetering M, Clevers H. Genes Dev. 2008;22:1856–1864. doi: 10.1101/gad.1674008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morris RJ, Liu YP, Marles L, Yang ZX, Trempus C, Li SL, Lin JS, Sawicki JA, Cotsarelis G. Nat. Biotechnol. 2004;22:411–417. doi: 10.1038/nbt950. [DOI] [PubMed] [Google Scholar]
- 33.Xin L, Lawson DA, Witte ON. Proc. Natl. Acad. Sci. U. S. A. 2005;102:6942–6947. doi: 10.1073/pnas.0502320102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Daniel CW, Young LJT. Exp. Cell Res. 1971;65:27. doi: 10.1016/s0014-4827(71)80046-0. [DOI] [PubMed] [Google Scholar]
- 35.Potten CS, Booth C, Tudor GL, Booth D, Brady G, Hurley P, Ashton G, Clarke R, Sakakibara S, Okano H. Differentiation. 2003;71:28–41. doi: 10.1046/j.1432-0436.2003.700603.x. [DOI] [PubMed] [Google Scholar]
- 36.Shinin V, Gayraud-Morel B, Gomes D, Tajbakhsh S. Nat. Cell Biol. 2006;8:677–682. doi: 10.1038/ncb1425. [DOI] [PubMed] [Google Scholar]
- 37.Blanpain C, Horsley V, Fuchs E. Cell. 2007;128:445–458. doi: 10.1016/j.cell.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuwahara R, Kofman AV, Landis CS, Swenson ES, Barendswaard E, Theise ND. Hepatology. 2008;47:1994–2002. doi: 10.1002/hep.22218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szotek PP, Chang HL, Brennand K, Fujino A, Pieretti-Vanmarcke R, Lo Celso C, Dombkowski D, Preffer F, Cohen KS, Teixeira J, Donahoe PK. Proc. Natl. Acad. Sci. U. S. A. 2008;105:12469–12473. doi: 10.1073/pnas.0805012105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Merok JR, Lansita JA, Tunstead JR, Sherley JL. Cancer Res. 2002;62:6791–6795. [PubMed] [Google Scholar]
- 41.Potten CS, Owen G, Booth D. J. Cell Sci. 2002;115:2381–2388. doi: 10.1242/jcs.115.11.2381. [DOI] [PubMed] [Google Scholar]
- 42.Pajerowski JD, Dahl KN, Zhong FL, Sammak PJ, Discher DE. Proc. Natl. Acad. Sci. U. S. A. 2007;104:15619–15624. doi: 10.1073/pnas.0702576104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Potten CS, Loeffler M. Development. 1990;110:1001–1020. doi: 10.1242/dev.110.4.1001. [DOI] [PubMed] [Google Scholar]
- 44.Savill NJ, Sherratt JA. Dev. Biol. 2003;258:141–153. doi: 10.1016/s0012-1606(03)00107-6. [DOI] [PubMed] [Google Scholar]
- 45.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 46.Kiel MJ, He S, Ashkenazi R, Gentry SN, Teta M, Kushner JA, Jackson TL, Morrison SJ. Nature. 2007;449:238–242. doi: 10.1038/nature06115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Villadsen R, Fridriksdottir AJ, Ronnov-Jessen L, Gudjonsson T, Rank F, LaBarge MA, Bissell MJ, Petersen OW. J. Cell Biol. 2007;177:87–101. doi: 10.1083/jcb.200611114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Asselin-Labat ML, Shackleton M, Stingl J, Vaillant F, Forrest NC, Eaves CJ, Visvader JE, Lindeman GJ. J. Natl. Cancer Inst. 2006;98:1011–1014. doi: 10.1093/jnci/djj267. [DOI] [PubMed] [Google Scholar]