Abstract
Plants exhibit different developmental strategies than animals; these are characterized by a tight linkage between environmental conditions and development. As plants have neither specialized sensory organs nor a nervous system, intercellular regulators are essential for their development. Recently, major advances have been made in understanding how intercellular regulation is achieved in plants on a molecular level. Plants use a variety of molecules for intercellular regulation: hormones are used as systemic signals that are interpreted at the individual-cell level; receptor peptide-ligand systems regulate local homeostasis; moving transcriptional regulators act in a switch-like manner over small and large distances. Together, these mechanisms coherently coordinate developmental decisions with resource allocation and growth.
Keywords: Development, Hormones, Intercellular regulation, Plant biology, Protein movement, Signaling
Introduction
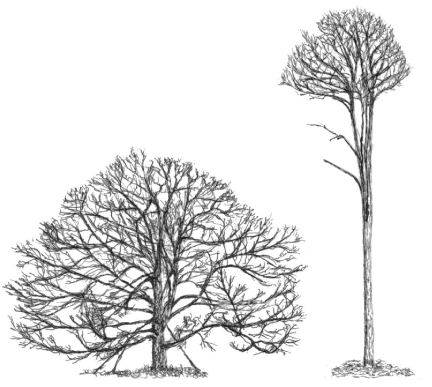
Multicellularity requires elaborate communication between the different cells and tissues of an organism. Most animals have developed highly complex systems of cell-to-cell communication through efficient circulatory and chemo-electrical systems, with various specialized organs and tissues dedicated to collecting, integrating and processing endogenous and environmental information, as well as to releasing instructive signals to effector cells and tissues. Plants lack such a sophisticated infrastructure. Nevertheless, they are far from being uninformed about or unresponsive to internal or external changes. Indeed, owing to the challenges of their sessile lifestyle, powerful interfaces between the regulation of their development and external conditions could evolve. The resulting developmental plasticity is readily apparent by looking at plants of the same species that grow in different ecological niches, for example a forest-growing versus a freestanding white oak tree (Fig. 1). Numerous complex responses to environmental factors, such as light availability, contribute to the development of such divergent phenotypes.
Fig. 1.
Schematic representation of two different specimens of white oak (Quercus alba). The specimen on the left is a free-standing tree, whereas the tall slender tree on the right grew in a forest. Reproduced with permission from Holdrege (Holdrege, 2005).
The implementation of developmental plasticity in plants is attributable to regulators that move between cells and that orchestrate developmental responses to a changing environment. Intercellular communication is a prerequisite for the complex development that multicellular organisms undergo. Individual aspects of intercellular regulation in plants have been reviewed extensively (see De Smet et al., 2009; Giakountis and Coupland, 2008; Mitchum et al., 2008; Wolters and Jurgens, 2009); here, we aim to provide an overarching view of recent advances in understanding plant intercellular regulators and their integration in developmental plasticity. We discuss a variety of ways in which plant cells orchestrate growth and developmental decisions by communicating and regulating biological processes over short and long distances, and cover classical plant hormones, receptor kinases and peptide-ligand systems, as well as moving regulators of transcription. Moving RNA molecules, however, are not discussed; we point the interested reader to a recent review on this topic (see Kehr and Buhtz, 2008). We conclude with a section that describes the fascinating exploitation of these communication systems by a variety of intruding organisms.
Plant hormones serve as global signals
The most powerful players in intercellular regulation are plant hormones. These low-molecular-weight compounds act as systemic signals that can transmit information over large distances. This information is interpreted at the cellular level, and different types of cells respond differently to the same hormones. Cells are confronted with signals from different hormones at the same time and translate this cumulative systemic information into coherent responses. Owing to the broad and diverse functions of hormones, we first briefly introduce different hormones and then discuss two examples to highlight the complex interplay of hormone signaling pathways.
Plant hormones can cause dramatic phenotypic effects. Thus, it is not surprising that many of them were discovered before the dawn of molecular genetics (Sachs and Thimann, 1967; Thimann and Skoog, 1933). Modern transcriptome profiling technologies have provided a global view of their effects at the molecular level and identified hundreds to thousands of genes, the expression levels of which are modified by individual hormones (Goda et al., 2008).
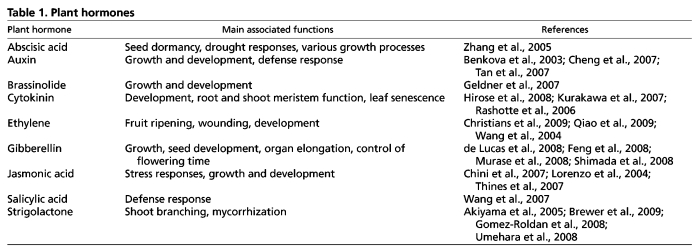
Traditionally, hormones have been implicated primarily in developmental and stress-response processes. The phytohormones auxin, cytokinin, brassinosteroid, gibberellin and strigolactone appeared to be mainly involved in developmental processes, whereas the compounds abscisic acid, jasmonic acid and ethylene were associated with stress responses (Table 1). More recently, however, a large body of evidence has revealed significant interplay between different hormones (Brewer et al., 2009; Grunewald et al., 2009b; Ruzicka et al., 2009; Stepanova et al., 2008), as well as the involvement of ‘stress hormones’ in developmental processes and vice versa. As plants respond to changing environmental conditions, including stress, by altering development, this complex interplay is not surprising.
Table 1.
Plant hormones
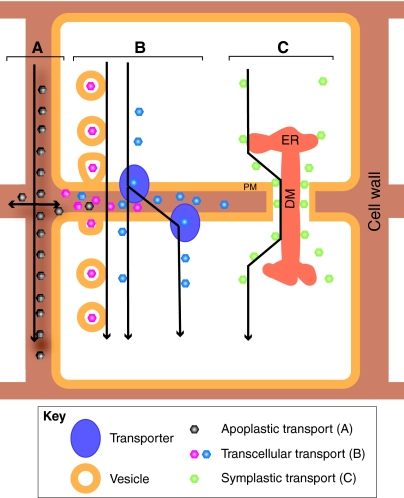
The physiology of higher plants is very different to that of animals and this is reflected in hormone signaling. There is no central nervous system that could orchestrate the release of hormones, nor are there specialized glands in which hormones are synthesized. Instead, these compounds are made in many places and transported throughout the plant by a combination of long-range transport through the vasculature (see Glossary, Box 1) and short-range modes of transport, including apoplastic (see Glossary, Box 1), symplastic (see Glossary, Box 1) and transcellular transport (Fig. 2). Whereas hormones that interact with a cell-surface receptor, such as brassinosteroids (Symons et al., 2008) and cytokinins (Heyl et al., 2007), operate at least partly in the apoplast, for others, like auxin, transcellular transport appears to be more important (Blilou et al., 2005), even though there is some evidence for extracellular auxin signaling involving an apoplastic auxin receptor (Yamagami et al., 2004). Transcellular transport ties hormone action not only to its synthesis and reception, but also provides other levels of regulation, such as the expression of transporters and the compartmentalization of different types of the same hormone.
Box 1. Glossary
Apex. The tip of a structure, such as shoot or root.
Apical dominance. The phenomenon wherein the apex controls the outgrowth of axillary buds.
Apoplast. Extracellular space between plant cells that allows the free diffusion of molecules. This space includes the cell wall.
Axillary bud. Predominantly dormant meristem associated with the base of leaves.
Boolean switch. A switch that has two states.
Fasciation. The enlargement of the plant apex by unregulated proliferative growth.
Florigen. A mobile signal that is produced in the leaves and that leads to the initiation of flowering in the shoot apex.
Grafting. The transplantation of a plant part, such as the root or shoot, to another individual.
Meristem. Stem cell-containing structures located at the growing points in plants. Meristems provide an instructive environment for long-term stem cell maintenance and are thus regarded as stem cell niches.
Photoperiod. The daily duration of light that an organism is exposed to.
Plasmodesma. A plasma membrane-enclosed pore that crosses the cell wall and contains a central element of endoplasmic reticulum.
Symplast. Intracellular space that is separated from the apoplast by the plasma membrane and that consists of the cytoplasm of multiple cells that are connected by plasmodesmata.
Vasculature. Specialized transport tissues that consist of two separate systems in plants. The xylem transports water and nutrients from root to shoot, whereas the phloem transports the products of photosynthesis and other metabolites from source to sink.
Fig. 2.
Modes of intercellular movement. (A) The apoplastic transport of molecules occurs by diffusion in the extracellular space. (B) Transcellular transport involves passage through two plasma membranes and can occur by secretion and subsequent endocytosis, diffusion or transporter activity. (C) Symplastic transport is facilitated by plasmodesmata that connect adjacent cells. They consist of endoplasmic reticulum (ER) that is appressed into the central axial desmotubule (DM) and flanked by the plasma membrane (PM). Transport can occur through the cytoplasmic sleeve between DM and PM.
Even though it is fascinating to look at the action of individual hormones, in terms of signal transmission between different tissues of the plant, their complex interplay might hold the key to understanding their activity. The past few years have uncovered several processes in which the combinatorial action of hormones governs developmental processes. For example, the interplay of auxin and cytokinin is important for setting the border of cell proliferation and cell differentiation in the root. In the root meristem (see Glossary, Box 1), auxin application promotes cell division, whereas the addition of cytokinin promotes cell differentiation and decreases meristem size. At the border between the proliferation and differentiation zones, cytokinin signaling leads to the induction of the SHORT HYPOCOTYL 2 (SHY2) gene, which encodes an inhibitor of the auxin response. The SHY2 protein, however, is degraded in an auxin-mediated fashion, thus demonstrating the convergence of auxin and cytokinin on a single gene for regulating crucial aspects of root development (Dello Ioio et al., 2007; Dello Ioio et al., 2008).
Another process that has been intensively studied for several decades, and for which the complexity of hormonal interplay has been largely resolved, is the control of shoot branching, i.e. the emergence of new shoots from buds in the axils of leaves. Most of the meristems that are located in these buds are dormant under a wide range of environmental conditions. It was recognized early on that auxin plays an important role in the state of axillary buds (see Glossary, Box 1). Upon removal of the auxin-rich primary apex (see Glossary, Box 1), bud outgrowth could be observed. The phenomenon that the apex prevents other buds from growing out is called ‘apical dominance’ (see Glossary, Box 1). The inhibitory effect of the apex could be mimicked by the application of auxin (Thimann and Skoog, 1933). Later, it was shown that auxin is synthesized in the shoot apex and that it is actively transported down the stem (Ljung et al., 2001). The direction of transport is mediated by the basal localization of the PIN auxin efflux carriers in the vasculature (Galweiler et al., 1998). However, it became clear that the effect of auxin on bud dormancy is not direct because radioactively labeled auxin led to outgrowth inhibition before the labeled auxin could be detected in the buds (Hall and Hillman, 1975).
One candidate for mediating the auxin response in the context of bud activation was cytokinin, which had been shown to be an activator of bud outgrowth (Sachs and Thimann, 1967). A tight link between cytokinin and auxin has been characterized on the molecular scale, as auxin generally represses cytokinin signaling by rapidly downregulating cytokinin biosynthesis in the shoot (Nordstrom et al., 2004). Moreover, it was shown that after the removal of the primary apex, transcripts of cytokinin biosynthesis genes and cytokinin concentrations rise very quickly in the nodal stem, and that the application of auxin inhibits this (Tanaka et al., 2006). This led to a model according to which auxin from the primary shoot apical meristem constantly downregulates local cytokinin synthesis in the shoot. The removal of the auxin source would therefore result in the elevated synthesis of cytokinin; cytokinin signaling would then activate the dormant buds.
More recently, a novel signal was identified that contributes to axillary bud activation and that moves from the root to the shoot. This signal has been identified as a strigolactone (Gomez-Roldan et al., 2008; Umehara et al., 2008). Strigolactones are a group of substances that have been previously identified as being secreted by plants and used as signals by parasitic weeds as well as by fungi in symbiotic mycorrhiza interactions (Akiyama et al., 2005). Exogenously applied strigolactone inhibited or significantly reduced bud outgrowth after auxin depletion in several species and stopped previously initiated bud outgrowth more efficiently than auxin transport inhibitors, which supports the notion that the strigolactones act independently of auxin transport (Brewer et al., 2009).
Despite this large body of research, many details remain unclear. For instance, strigolactones move from the root to the shoot. However, the young buds are not connected to the stem through the vasculature, and thus one would not expect strigolactone levels to be very high in the bud. Moreover, this three-hormone system displays a substantial amount of emerging complexity because of internal interactions. Auxin has been shown to regulate cytokinin biosynthesis and there is evidence for auxin positively regulating strigolactone synthesis (Bainbridge et al., 2005; Foo et al., 2005; Zou et al., 2006). It is also very probable that additional hormones or signals are involved in shoot branching.
The case of shoot branching is a marvelous example of the intertwined role of hormones as intercellular regulators. First of all, hormones function as reporters of significant changes in internal or external conditions. In axillary buds, this leads to the activation of a developmental program upon removal of the primary apex. This developmental program is only activated in axillary buds and not in other tissue types. This response thus depends on both the hormonal signals and the potential of the bud. The same signals, however, are also perceived and interpreted in other cells and can lead to many other developmental decisions. Thus, hormones appear to report the systemic status of the plant rather than deliver specific instructions. Plant hormones therefore coordinate many responses, such as resource usage and growth, throughout the plant and serve as chemical coordinators that allow plant developmental decisions to act in concert.
Receptor-like kinases and peptide ligands in local homeostasis
Receptor-like kinase (RLK) peptide systems are of particular importance in plants. In addition to their fundamental role in the plant pathogen response, they play a prominent role in local developmental decisions and in maintaining local homeostasis. This is owing to the intrinsic properties of RLK peptide systems, which typically constrain them to short-range signaling. A recent review has covered the broad role of RLKs in plant development (see De Smet et al., 2009); here, we provide some instructive examples of these functions in selected systems.
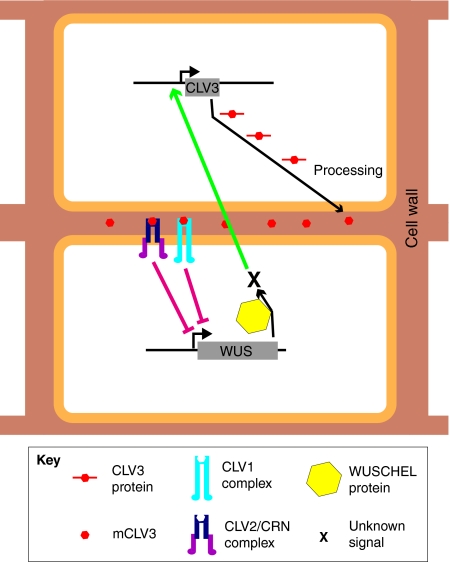
The best-understood RLK model system is the CLAVATA (CLV) system, which is essential for regulating the size of the stem cell population in the shoot apical meristem (SAM). Loss-of-function mutants for each of the three CLV genes in Arabidopsis exhibit shoot fasciation (see Glossary, Box 1), enlarged floral meristems and increased numbers of floral organs (Clark et al., 1993; Clark et al., 1995; Kayes and Clark, 1998). CLV1 and CLV2 encode proteins with a high similarity, but whereas the CLV1 gene encodes a leucine-rich repeat RLK, CLV2 encodes an LRR receptor-like protein (RLP) that lacks a kinase domain (Clark et al., 1997; Jeong et al., 1999). The third gene, CLV3, encodes a small peptide (Fletcher et al., 1999) that is secreted into the extracellular space (Rojo et al., 2002). Even though the molecular details were unclear, it was hypothesized that the CLV3 peptide binds the CLV1/CLV2 receptor complex and that this functional complex leads to signaling that eventually causes the repression of the WUSCHEL (WUS) transcription factor gene. This, in turn, is required to specify stem cells in the SAM. How WUS activity is translated into stem cell maintenance is not certain, because both WUS mRNA and protein are restricted to a domain directly beneath the stem cells. Intriguingly, WUS positively regulates the expression of CLV3 in the stem cell domain, an interaction that creates an autoregulatory loop that fine-tunes stem cell homeostasis (Brand et al., 2000; Schoof et al., 2000) (Fig. 3). The molecular details of the interactions of the CLV proteins were unknown for a long time. Recently, it has been shown that not only must CLV3 be processed into a 13-amino-acid peptide (MCLV3), but it also needs to be post-translationally modified for efficient binding to the CLV1 receptor (Ohyama et al., 2009).
Fig. 3.
The CLAVATA signaling module in the shoot apical meristem. The action of the transcription factor WUSCHEL (WUS) leads to the production of an unknown signal (X) that results in the transcription of CLV3 in neighboring cells. CLV3 protein is processed into the mature CLV3 peptide (mCLV3), which subsequently binds to CLV1 and to CLV2/CRN receptor complexes. This event eventually causes the repression of WUS.
Even though early studies of the local homeostasis of stem cell regulation in the SAM focused primarily on the role of the CLV system, other important components have been discovered. Recently, it has been proposed that CLV2 and a novel RLK, CORYNE (CRN), interact via their transmembrane domains, and that this complex transmits the CLV3 signal independently of CLV1 (Muller et al., 2008). In the periphery of the SAM, the BARELY ANY MERISTEM (BAM) receptors are proposed to sequester CLV3-like ligands and thus to contribute to the complexity of receptor-mediated meristem homeostasis (Deyoung and Clark, 2008). Studies in rice (Oryza sativa) indicate that this basic framework of meristem maintenance is conserved across angiosperms. Based on the similarity of the intron/exon structure and the high homology of a conserved 14-amino-acid sequence (CLE domain) of CLV3, four CLE genes were found in rice and Arabidopsis that seem to have evolved from a common ancestor. Generally, all four genes are implicated in meristem maintenance; however, in rice, the roles of these CLE peptides seem to have diversified, as different CLE peptides appear to regulate different types of meristems (Suzaki et al., 2008).
Although the function of receptor-peptide pairs is understood most clearly in the SAM, there are other tissues in which similar pairs seem to play an important role. In the root meristem, a different RLK peptide pair has been linked to stem cell maintenance: the RLK ARABIDOPSIS CRINKLY4 (ACR4) and the CLE40 peptide control the maintenance of a stem cell population in the root (Stahl et al., 2009). Interestingly, here, the CLE signal originates from the differentiated cells and not from the stem cells, as in the SAM. This difference at the operational level might be due to differences in the structural organization of the root and shoot meristems. Additionally, the receptor ACR4 has also been implicated in lateral root formation, which involves the initiation of a new meristem. In this context, it first promotes formative cell divisions for lateral root development and then constrains the number of these divisions after the onset of organogenesis (De Smet et al., 2008).
This root-specific RLK peptide pair aside, parts of the regulatory circuits that are active in the shoot, such as the CLAVATA signaling module, fulfill similar purposes in the root. CRN as well as CLV2 have been implicated in the maintenance of the root apical meristem, because both crn and clv2 mutant plants are somewhat resistant to the application of synthetic CLE dodeca-peptides (Miwa et al., 2008). The important role of diverse CLEs is underlined by the conservation of the family in various plant species, including very basal plants, like green algae and moss (Kinoshita et al., 2007; Oelkers et al., 2008).
Further evidence for an important role for other CLE peptides in Arabidopsis is provided by the fact that many CLE genes are transcribed in distinct tissues during development (Sharma et al., 2003). This supports the hypothesis that CLE peptides are widely employed as intercellular regulators. However, with the exception of clv3, no loss-of-function phenotype of a single CLE has been reported. Nevertheless, the overexpression and application of numerous CLE peptides result in developmental phenotypes in the root and shoot meristems (Kinoshita et al., 2007; Strabala et al., 2006). These data support a general role for CLE peptide signaling in growth and differentiation processes. However, they also indicate that various CLE peptides are functionally redundant or that there is receptor promiscuity for different CLE ligands.
To better understand the role of RLK peptide signaling on local homeostasis, a good knowledge of the interactions between specific mobile CLEs and stationary RLKs is crucial. Unfortunately, this has proven be a very difficult task, mainly because the specificity of CLE-RLK interactions does not seem to be very high, and also because of the enormous size of the RLK family, which includes more than 600 genes in Arabidopsis (Shiu and Bleecker, 2001). For instance, CLV2 can perceive several CLEs (Fiers et al., 2005), whereas several CLEs, including CLE1 to CLE7, can at least partially rescue the SAM phenotypes of clv3 mutants through a CLV1-dependent pathway (Ni and Clark, 2006). This supports the idea that it is not the individual CLE that is important, but rather that its expression domain and local concentration, as well as its post-translational modifications (as seen in MCLV3), are key to determining the specificity of RLK-CLE interactions.
Yet another layer of complexity comes into play when considering the processing of CLE peptides. The cleavage of CLE peptides to their mature form requires proteases. To date, only one subtilase has been demonstrated to cleave a growth-promoting pro-peptide (Srivastava et al., 2008); however, there are several other proteases implicated in RLK-CLE signaling processes. Very recently, the ectopic overexpression of the subtilase AtSBT5.4 was reported to cause a clv-like phenotype, which is dependent on the protease activity of the subtilase. However, AtSBT5.4 did not cleave the CLV3 peptide in an in vitro assay (Liu et al., 2009). One possibility is that this subtilase processes other signal peptides in the SAM, which might disturb the signaling balance in the tissue, possibly by competing with CLV3 for receptor binding.
RLK peptide systems act over small distances. The peptides seem to diffuse freely in the apoplast, but the diffusion rate for small molecules in the apoplastic space appears to be much lower than in water (Kramer et al., 2007), which would restrict peptide-mediated signaling to a very limited space. If the constant sequestration of the peptide ligands is assumed, a steep concentration gradient at the border of the domain from which the peptide is secreted would be established. Such a gradient would produce a high sensitivity for changes in peptide production. This property makes RLK peptide systems well-suited for regulating local homeostasis, which requires groups of cells to readjust their activity constantly to avoid losing equilibrium.
Moving transcriptional regulators mediate short- and long-range developmental decisions
Several transcriptional regulators have been shown to move between cells and tissues. Whereas hormone and RLK-peptide pairs act indirectly on gene transcription, moving transcriptional regulators directly participate in changing the transcriptional programs of target cells. Interestingly, many of these genes are involved in fundamental developmental decisions. Most transcription factors have been described to have a range of movement of one or at most a few cell layers (Kim et al., 2002; Kim et al., 2003; Lucas et al., 1995; Nakajima et al., 2001; Perbal et al., 1996; Sessions et al., 2000; Wada et al., 2002), but there is one reported case in which a transcriptional regulator moves over longer distances (Corbesier et al., 2007; Jaeger and Wigge, 2007; Mathieu et al., 2007). Here, we first describe the structural basis for protein movement in plants, then give an overview of moving transcriptional regulators and finally discuss two cases in detail, one involving short-distance movement and the other long-distance movement.
A network of cytoplasmic connections called plasmodesmata (see Glossary, Box 1) links plant cells. Various molecules, including transcription factors, can use this system to move between cells. Primary plasmodesmata are assembled during cytokinesis and form a plasma membrane-enclosed pore that crosses the cell wall and that contains a central element of endoplasmatic reticulum. Secondary plasmodesmata have the same structure but can be formed later in development, even if two cells are not clonally related. Unlike gap junctions in animal systems, the size limits for the passage through plasmodesmata vary greatly between different tissues, ranging from 30 to 50 kDa (Crawford and Zambryski, 2001; Kim et al., 2005).
There are two modes of movement between cells via plasmodesmata: selective movement that is determined by specific sequences within the transported protein (Lucas et al., 1995) and non-selective movement without such requirements (Wu et al., 2003). A variety of molecules have been shown to move through plasmodesmata. The first proteins that were recognized to use these connections for movement between cells were viral particles that spread throughout the plant (Samuel, 1934). Later on, it was shown that plant regulatory proteins also use this path. The first of these moving regulatory proteins to be identified was the KNOTTED1 (KN1) protein in maize (Zea mays) (Lucas et al., 1995). It has been reported that KN1 modulates the size exclusion limit of plasmodesmata (Kragler et al., 2000; Lucas et al., 1995). KNOTTED1 in maize and its homologue STM in Arabidopsis are expressed in the SAM and both are downregulated in incipient leaf primordia and developing leaves. STM was shown to play a crucial role in maintaining stem cell homeostasis and cell indeterminacy in the SAM (Lenhard et al., 2002). To date, several other functionally characterized transcriptional regulators have also been described to move between cells (Table 2). Interestingly, these factors also play important roles in cell fate decisions. In the root, the single-repeat R3 MYB transcription factor CAPRICE is involved in the cell fate determination of hair-forming cells (Wada et al., 2002), whereas SHORTROOT (SHR), which belongs to the plant-specific GRAS family of transcription factors, controls aspects of radial patterning as well as the maintenance of stem cells in the quiescent center (QC) of the root meristem (Nakajima et al., 2001). Moving transcriptional regulators also control various important aspects of flower development. First of all, a small globular transcriptional regulator called FLOWERING LOCUS T (FT) triggers flowering (Corbesier et al., 2007; Jaeger and Wigge, 2007; Mathieu et al., 2007). Further downstream of this triggering event, the moving transcription factor LFY functions as a developmental switch that is necessary and sufficient to initiate floral fate in shoot meristems (Blazquez and Weigel, 1999; Weigel and Nilsson, 1995). Two other moving transcription factors, GLOBULOSA and DEFICIENS, control organ identity in the flower (Perbal et al., 1996). In addition to these well-characterized examples, further insight into the extent of transcription factor movement came from a comparison of the expression of 24 transcriptional and translational transcription factor gene GFP-fusion constructs that were expressed in a tissue-specific manner in the Arabidopsis root. Surprisingly, in 17% of the cases investigated, transcription occurred in a cell type that was different from the cell type in which the protein was localized (Lee et al., 2006). The molecular details and the significance of this extensive transcription factor movement are not well understood. However, two moving regulators of transcription that trigger important developmental decisions in plants have been characterized in depth.
Table 2.
Developmentally important moving transcriptional regulators in plants
SHR regulates the asymmetric cell divisions that occur in the stem cell population known as the cortex/endodermal initials (CEI) found in the root meristem. The first hint that SHR is an intercellular regulator was the fact that its mRNA was exclusively expressed in the stele of the root and that it was required for the asymmetric cell division involved in the formation of the adjacent ground tissue (endodermis and cortex), as well as for the specification of endodermis (Helariutta et al., 2000), with shr mutants lacking a cell layer between the epidermis and the stele. Further studies using immunolocalization and SHR-GFP fusion proteins showed that the SHR protein moves from the root stele into the adjacent cell layers and triggers the formative cell divisions that give rise to the endodermis and cortex (Nakajima et al., 2001). This function depends on SCARECROW (SCR), another GRAS transcription factor that is expressed in the endodermis, the CEI cell and the QC, i.e. the tissues that SHR proteins move to. Intriguingly, SHR movement is constrained to one cell layer by SCR-mediated protein sequestration to the endodermal nuclei (Cui et al., 2007). Additionally, SHR and SCR are necessary for the specification and patterning of the QC (Sabatini et al., 2003). In the ground tissue as well as in the QC, two zinc finger proteins, JACKDAW (JKD) and MAGPIE (MGP), regulate the range of SHR action. Interestingly, both JKD and MGP are also regulated by SHR and SCR (Welch et al., 2007). This reciprocal interaction highlights the fine-grained regulation of this complex system.
The movement of SHR depends on its amino acid sequence. The substitution of a single amino acid in the VHIID domain inhibits SHR movement completely (Gallagher et al., 2004). Furthermore, proper sub-cellular localization is necessary for SHR movement. A model for SHR trafficking has been proposed according to which the balance between nuclear localization and nuclear export promotes SHR movement into the ground tissue (Gallagher and Benfey, 2009).
Whereas most cases of regulatory protein movement appear to occur over short distances, as is the case for SHR, there is at least one example of a transcriptional co-activator traveling long distances to trigger an essential event in the life of a plant. Plants need to synchronize their development with environmental conditions. One crucial developmental decision for flowering plants is the switch from the vegetative to the flowering state. To produce offspring at the right time and in a synchronous manner is a complex task that involves the detection of cues that indicate the season. Among others, day length is an important parameter. However, there is a morphological problem between perceiving light signals and making the switch to the flowering stage: the switch is made in the SAM, but this is typically overgrown by emerging leaves and thus is not optimally located to perceive light. As early as the 19th century, it was recognized that the signal for flowering comes from the leaves (Hofmeister et al., 1865). More sophisticated experiments in the first half of the 20th century led to the proposal of the ‘florigen’ hypothesis (see Glossary, Box 1). This model defined florigen as a substance that was generated under inductive photoperiod conditions (see Glossary, Box 1) in the leaf and subsequently traveled to the apex to trigger flowering (Cajlachjan, 1936). More than two thirds of a century and a battery of new techniques were required to identify the molecular identity of the florigen. Grafting experiments (see Glossary, Box 1) showed that the photoperiodically controlled nuclear protein CONSTANS (CO) was upstream of this leaf-derived signal (An et al., 2004; Ayre and Turgeon, 2004). FT was a suitable candidate for the leaf-derived signal because of its known role in promoting flowering and its activation by a dexamethasone-inducible version of CO (Samach et al., 2000). The activation of FT by CO occurs in the leaf vascular tissue and leads to floral induction (An et al., 2004; Takada and Goto, 2003). A strong body of evidence for FT protein movement from the leaves to the shoot apex was generated (Corbesier et al., 2007; Jaeger and Wigge, 2007; Lifschitz et al., 2006; Lin et al., 2007; Mathieu et al., 2007; Tamaki et al., 2007) (Fig. 4), where it triggers the switch from the vegetative state of the meristem to the floral state. The bZIP protein FLOWERING LOCUS D (FD), which is exclusively expressed in the SAM, was found to interact with FT. Moreover, mutations in FD repress the overexpression phenotype of FT and show it to be required for FT activity. Chromatin immunoprecipitation and expression analysis demonstrated that FT, together with FD, activates the transcription of the floral marker gene APETALA1 (AP1), which supports a role for FT as a transcriptional co-activator (Abe et al., 2005; Wigge et al., 2005). Despite its prominent role as a trigger for flowering, recent evidence in tomatoes (Solanum lycopersicum) indicates that the FT homolog does not only act as a switch for flowering. Rather, it has been proposed that it acts, like a hormone, as a general systemic regulator of growth and termination (Shalit et al., 2009).
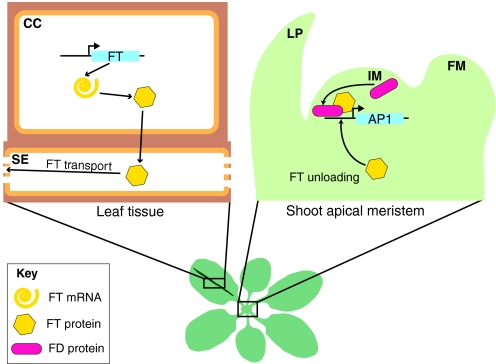
Fig. 4.
FLOWERING LOCUS T (FT) protein movement. In suitable conditions, FT is transcribed in the phloem companion cells (CC) of leaves (left). FT is then transported into the sieve elements (SE). There, long-distance transport of FT occurs toward sink tissues. In the shoot apex (right), FT is unloaded from the phloem and acts together with FLOWERING LOCUS D (FD) to induce APETALA 1 (AP1) expression. FM, floral meristem; IM, inflorescence meristem; LP, leaf primordium.
These examples of moving transcriptional regulators have several characteristics in common. First, at least part of their function seems to be comparable to a Boolean switch (see Glossary, Box 1) in the target cells as opposed to gradual regulators or global signals that are interpreted at the single-cell level. In the case of SHR, a formative cell division is triggered when the protein moves to the target cell type; in the case of FT, the switch from a vegetative to a flowering meristem can be triggered only after FT reaches its target tissue. Second, they generally appear to function together with a local co-factor that is required for their activity and that probably acts as a safeguard against the erroneous activation of target genes. For example, SHR regulates genes cooperatively with SCR, whereas SCR itself sequestrates SHR to the nucleus and prevents further movement. FT interacts with FD, which is only expressed in the target tissue, and this leads to the induction of downstream targets. Taken together, the movement of transcriptional regulators seems to be used to orchestrate the coordinated expression of a set of genes for switching cellular programs at the right time and place.
Invading organisms use intercellular signaling systems to manipulate plant development
Complex systems are prone to manipulative perturbation by imposters that deploy components similar or equal to those present in the system itself. In the case of plants, intercellular regulators are often used to modify the endogenous biological system of the plant to the advantage of pathogenic organisms (Fig. 5). Probably the most common targets for this molecular hijacking are hormonal signaling pathways. Owing to their morphogenetic power, auxin and cytokinin pathways are used by plant pathogens of all major branches of life. The disease caused by the tobacco mosaic virus is induced in part by the misregulation of auxin response genes. The mechanism that underpins this is an interaction of the virus replicase with several members of the AUX/IAA family of nuclear proteins that negatively regulate the auxin response. It was suggested that the virus uses this as a means to reprogram the cellular environment of older cells to one that is more favorable for virus replication and spread (Padmanabhan et al., 2005; Padmanabhan et al., 2008). Bacteria also often interfere with auxin and cytokinin signaling, in part by synthesizing auxins and cytokinins that contribute to tumor gall formation (Lichter et al., 1995; Vandeputte et al., 2005). These galls seem to provide a favorable microenvironment and contain large numbers of cells that produce metabolites used by the bacteria. A well-characterized example of this process is the bacterial infection of Arabidopsis by Rhodococcus fascians, a Gram-positive phytopathogenic actinomycete. The host plant displays some distinct developmental and morphological changes upon infection, including serrated leaves and a stunted appearance of the aerial parts, abnormal flowers, multiple rosettes and inflorescences (de Manes et al., 2004; Vereecke et al., 2000). Rhodococcus infection is dependent on the fas operon, which contains cytokinin synthesis genes (Crespi et al., 1992). Detailed molecular analysis led to the model that Rhodococcus perturbs the hormonal balance of the host plant by secreting cytokinins, which cause the activation of KNOX genes in the plant (Depuydt et al., 2008). In Arabidopsis, KNOX genes are required for meristem function and are thought to keep cells in an undifferentiated state. The activity of these genes at the infection site keeps cells in a juvenile state and maintains the production of metabolites that are beneficial for the pathogen. Interestingly, KNOX gene expression and cytokinin addition result in different phenotypes, which could indicate that additional pathways are required for pathogenesis (Depuydt et al., 2008). Recent findings that a sophisticated mixture of cytokinins is used by Rhodococcus (Pertry et al., 2009) indicate that there is an elaborate synchronization between cytokinin secretion and the plant response. This is probably a key to successful pathogenesis.
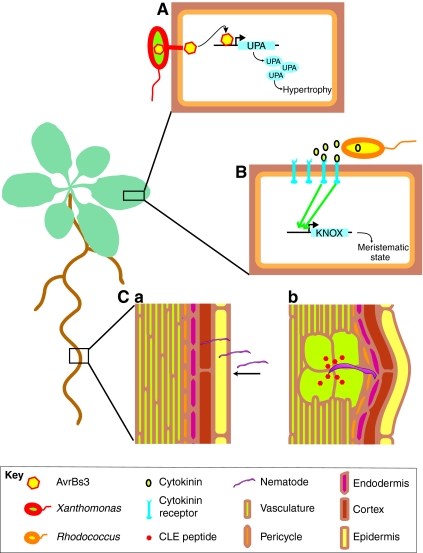
Fig. 5.
Some pathogens can usurp intercellular regulatory pathways. Three strategies through which pathogens alter plant development are shown. (A) Xanthomonas bacteria secrete AvrBs3 protein into plant cells. This directly induces upregulated by AvrBs3 (UPA) genes that eventually lead to hypertrophy. (B) The Rhodococcus bacteria produce cytokinin, which activates class-I KNOTTED-like homeobox genes (KNOX) that convert differentiated tissue into a meristematic state. (C) Root-knot nematode worms invade the root (a) and excrete CLE peptides that lead to the dedifferentiation and the enlargement of cells that develop into feeding cells (large cells in green) for the nematodes (b).
Fungi also have the ability to synthesize plant hormones and to manipulate host development. There is a large body of evidence that biotrophic fungal pathogens, such as powdery mildews, rusts and downy mildews, use cytokinins to generate ‘green islands’ (Walters and McRoberts, 2006; Walters et al., 2008). Cells in the green islands remain healthy but the surrounding tissue senesces and turns yellow. The green islands are leaf parts that are transformed into strong nutrient sinks on which the biotrophs feed. It has been shown that the concentration of nutrients within these green islands is significantly increased compared with non-infected surrounding tissues, which enables the pathogens to remove nutrients continuously (Angra and Mandahar, 1991). Some animals seem to have adopted the same trick: leafminer caterpillars can also induce green islands by cytokinin accumulation, similar to those seen at fungal infection sites (Giron et al., 2007).
Among animals, nematodes constitute a large group of plant pathogens. These roundworms have gained a remarkable ability to manipulate plant root morphology. They induce the formation of abnormally large cells in the root and then feed on these. Not surprisingly, they use auxin pathways to dedifferentiate root cells for this purpose (Grunewald et al., 2009a). Moreover, some nematodes have adopted a fascinating trick. HgSYV46 is a gene unique to the soybean cyst nematode (Wang et al., 2001) and encodes a protein with a conserved 14-amino-acid C-terminal motif that is characteristic for the plant CLE peptide family (Olsen and Skriver, 2003), which, as discussed above, is involved in stem cell maintenance. Upon infection, this protein is highly expressed within the single dorsal esophageal gland cell of the soybean cyst nematode during syncytium formation in plants (Wang et al., 2001). Even though direct evidence on the plant side is still lacking, the location of the HgSYV46 protein along the dorsal gland extension to the base of the nematode stylet (the oral feeding spear) in parasitic life stages suggests that HgSYV46 is secreted into plant tissues. When expressed in Arabidopsis, HgSYV46 can rescue the clv3 mutant phenotype and lead to a CLV3 overexpression phenotype in wild-type plants (Wang et al., 2005). In the soybean cyst nematode, a second CLE gene has been identified (Gao et al., 2003), and CLE peptide-encoding genes have been detected in beet cyst nematodes and potato cyst nematodes (Mitchum et al., 2008).
In another group of nematodes, the 16D10 gene, which encodes a 13-amino-acid secretory peptide, also displays sequence similarities to plant CLE domains. Its overexpression in Arabidopsis stimulates root growth but does not rescue the clv3 mutant phenotype. In line with the root phenotype, it has been shown that 16D10 can interact with SCR-like transcription factors (Huang et al., 2006b). The silencing of 16D10 leads to decreased root-knot nematode infectivity (Huang et al., 2006a).
Not only are hormones and small peptides used to manipulate the developmental programs of the host, but transcription factors are also delivered from pathogens into plant cells. The protein AvrBs3 from the Gram-negative bacterium Xanthomonas campestris pv. vesicatoria belongs to a large family also known as transcription activator-like (TAL) effectors. After delivery into the host cell, it acts as a transcription factor and directly induces plant gene expression (Kay et al., 2007; Romer et al., 2007). In susceptible hosts, this eventually leads to a leaf hypertrophy (i.e. enlargement) phenotype (Marois et al., 2002). This appears to allow higher bacterial transmission rates by decreasing the volume of the intercellular space at the end of the bacterial growth phase, which creates pressure that might expel the bacteria out of the leaves (Wichmann and Bergelson, 2004). Members of the AvrBs3 family and related proteins are composed of a central repeat region that mediates protein dimerization (Gurlebeck et al., 2005) and DNA binding (Kay et al., 2007). Nuclear localization signals enable entry into the eukaryotic nucleus and an acidic activation domain in the C-terminus is essential for the activation of plant gene expression (Gurlebeck et al., 2006). On the host side, transcript profiling has identified sets of AvrBs3-like protein target genes in rice (Sugio et al., 2007; Yang et al., 2006) and bell peppers (Capsicum annuum) (Kay et al., 2007; Marois et al., 2002). These bacterial proteins are not unique in acting as plant transcription factors. There are other candidates in bacteria that might mimic eukaryotic transcription factors as well, such as HsvB and HsvG from Pantoea agglomerans (Nissan et al., 2006).
Unsurprisingly, plants have evolved mechanisms to counter the pathogen-induced manipulation of their development. The treatment of plants with bacterial flaggelin leads to the expression of the microRNA miR393 in Arabidopsis, which negatively modulates auxin signaling and contributes to the resistance to the Gram-negative bacterium Pseudomonas syringae (Navarro et al., 2006). Salicylic acid, a hormone involved in the pathogen response, also modifies the auxin response (Wang et al., 2007). Abnormal cytokinin levels are equally monitored by plants, and unusually high concentrations of cytokinin can induce programmed cell death (Carimi et al., 2003; Mlejnek and Prochazka, 2002) and the expression of the pathogen resistance gene PR1 (Memelink et al., 1987).
Pathogens have found many ways to manipulate developmental aspects of their host plant by using pathways employed by plant intercellular regulators. It seems that rather than modifying one aspect of plant growth, they predominantly manipulate pathways that allow them to influence various aspects of growth and development. Given that these general target pathways are nearly universal in higher plants, it is unlikely that the plant will counterattack by altering the pathways themselves. Instead, plants use surveillance mechanisms that can detect when the target pathways are modulated and then activate pathogen defense mechanisms.
Conclusions
By making extensive use of intercellular regulation, plants are able to undergo complex developmental processes. This enables the enormous phenotypic plasticity seen in plants, as epitomized by the aforementioned white oak trees growing in different environments (Fig. 1). Even though the mechanistic details of how such adaptations are implemented on a molecular scale over many years are not yet understood, they probably involve the combined action of all the mechanisms discussed in this review.
Recently, we have seen tremendous advances in our understanding of how intercellular signaling is controlled. This is in part owing to a variety of new and rapidly advancing technologies, including expression profiling and cell type-specific technologies and measurements (Brady et al., 2007). It seems probable that observing phenomena at the level of individual cell types, as opposed to whole organs or whole plants, will dramatically increase our understanding of many regulatory processes, including intercellular regulation. Different types of cells respond very differently to common signals such as hormones. This difference is probably attributable to their different fates and cell cycle states. Combining signal perturbations with expression profiling following fluorescence-activated cell sorting (FACS) or even single-cell RNA extraction will lead to a better understanding of the cellular aspects of intercellular regulation. From a technological perspective, deep sequencing technologies promise to eliminate current blind spots and will help to profile additional classes of RNAs that are mobile or that are regulated by moving signals.
Other technologies, including those deployed for proteomic and metabolomic studies, applied at the level of individual cell types might help to model and infer the movement of molecules between cells, as well as their regulatory functions. The large-scale interactome project for Arabidopsis that is currently underway (http://interactome.dfci.harvard.edu/A_thaliana/index.php) will, in conjunction with cell-type specific ‘-omic’ datasets, provide the information needed to model intercellular regulation on a systems scale.
Another field that will strongly enhance our comprehension of intercellular regulation is high-throughput laser confocal microscopy combined with automated image processing. Currently, technologies are emerging that enable researchers to quantitatively investigate reporter gene expression and tagged protein localization (Mace et al., 2006) as well as metabolite fluxes (Wiechert et al., 2007) over extended periods of time. With such technologies, novel screens for moving regulators could be conducted. Moreover, the movement and fluxes of various kinds of molecules could be continuously tracked at a cellular resolution.
The integrative analysis of these data will enhance our comprehension of plant development and intercellular regulation. Eventually, this might lead to computer models with predictive power that take short- and long-range regulatory mechanisms into account, which in turn will provide the opportunity to delve more deeply into the systems dynamics involved in plant development.
Acknowledgments
We are grateful to members of the Benfey laboratory for helpful comments. Our work in this area is funded by the NIH and the NSF AT2010 program. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
References
- Abe M., Kobayashi Y., Yamamoto S., Daimon Y., Yamaguchi A., Ikeda Y., Ichinoki H., Notaguchi M., Goto K., Araki T. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309, 1052-1056 [DOI] [PubMed] [Google Scholar]
- Akiyama K., Matsuzaki K., Hayashi H. (2005). Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435, 824-827 [DOI] [PubMed] [Google Scholar]
- An H., Roussot C., Suarez-Lopez P., Corbesier L., Vincent C., Pineiro M., Hepworth S., Mouradov A., Justin S., Turnbull C., et al. (2004). CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131, 3615-3626 [DOI] [PubMed] [Google Scholar]
- Angra R., Mandahar C. L. (1991). Pathogenesis of barley leaves by Helminthosporium teres I: Green island formation and the possible involvement of cytokinins. Mycopathologia 114, 21-27 [Google Scholar]
- Ayre B. G., Turgeon R. (2004). Graft transmission of a floral stimulant derived from CONSTANS. Plant Physiol. 135, 2271-2278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainbridge K., Sorefan K., Ward S., Leyser O. (2005). Hormonally controlled expression of the Arabidopsis MAX4 shoot branching regulatory gene. Plant J. 44, 569-580 [DOI] [PubMed] [Google Scholar]
- Benkova E., Michniewicz M., Sauer M., Teichmann T., Seifertova D., Jurgens G., Friml J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591-602 [DOI] [PubMed] [Google Scholar]
- Blazquez M. A., Weigel D. (1999). Independent regulation of flowering by phytochrome B and gibberellins in Arabidopsis. Plant Physiol. 120, 1025-1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou I., Xu J., Wildwater M., Willemsen V., Paponov I., Friml J., Heidstra R., Aida M., Palme K., Scheres B. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433, 39-44 [DOI] [PubMed] [Google Scholar]
- Brady S. M., Orlando D. A., Lee J. Y., Wang J. Y., Koch J., Dinneny J. R., Mace D., Ohler U., Benfey P. N. (2007). A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318, 801-806 [DOI] [PubMed] [Google Scholar]
- Brand U., Fletcher J. C., Hobe M., Meyerowitz E. M., Simon R. (2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289, 617-619 [DOI] [PubMed] [Google Scholar]
- Brewer P. B., Dun E. A., Ferguson B. J., Rameau C., Beveridge C. A. (2009). Strigolactone acts downstream of auxin to regulate bud outgrowth in pea and Arabidopsis. Plant Physiol. 150, 482-493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajlachjan M. C. (1936). New facts in support of the hormonal theory of plant development. Comptes Rendus De L Academie Des Sciences De L Urss 13, 79-83 [Google Scholar]
- Carimi F., Zottini M., Formentin E., Terzi M., Lo, Schiavo F. (2003). Cytokinins: new apoptotic inducers in plants. Planta 216, 413-421 [DOI] [PubMed] [Google Scholar]
- Cheng Y., Dai X., Zhao Y. (2007). Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19, 2430-2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernandez G., Adie B., Chico J. M., Lorenzo O., Garcia-Casado G., Lopez-Vidriero I., Lozano F. M., Ponce M. R., et al. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666-671 [DOI] [PubMed] [Google Scholar]
- Christians M. J., Gingerich D. J., Hansen M., Binder B. M., Kieber J. J., Vierstra R. D. (2009). The BTB ubiquitin ligases ETO1, EOL1 and EOL2 act collectively to regulate ethylene biosynthesis in Arabidopsis by controlling type-2 ACC synthase levels. Plant J. 57, 332-345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S. E., Running M. P., Meyerowitz E. M. (1993). CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 119, 397-418 [DOI] [PubMed] [Google Scholar]
- Clark S. E., Running M. P., Meyerowitz E. M. (1995). CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 121, 2057-2067 [Google Scholar]
- Clark S. E., Williams R. W., Meyerowitz E. M. (1997). The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89, 575-585 [DOI] [PubMed] [Google Scholar]
- Corbesier L., Vincent C., Jang S., Fornara F., Fan Q., Searle I., Giakountis A., Farrona S., Gissot L., Turnbull C., et al. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316, 1030-1033 [DOI] [PubMed] [Google Scholar]
- Crawford K. M., Zambryski P. C. (2001). Non-targeted and targeted protein movement through plasmodesmata in leaves in different developmental and physiological states. Plant Physiol. 125, 1802-1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespi M., Messens E., Caplan A. B., van Montagu M., Desomer J. (1992). Fasciation induction by the phytopathogen Rhodococcus fascians depends upon a linear plasmid encoding a cytokinin synthase gene. EMBO J. 11, 795-804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H., Levesque M. P., Vernoux T., Jung J. W., Paquette A. J., Gallagher K. L., Wang J. Y., Blilou I., Scheres B., Benfey P. N. (2007). An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 316, 421-425 [DOI] [PubMed] [Google Scholar]
- de Lucas M., Daviere J. M., Rodriguez-Falcon M., Pontin M., Iglesias-Pedraz J. M., Lorrain S., Fankhauser C., Blazquez M. A., Titarenko E., Prat S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451, 480-484 [DOI] [PubMed] [Google Scholar]
- de Manes O. C. L., Beeckman T., Ritsema T., Van Montagu M., Goethals K., Holsters M. (2004). Phenotypic alterations in Arabidopsis thaliana plants caused by Rhodococcus fascians infection. J. Plant Res. 117, 139-145 [DOI] [PubMed] [Google Scholar]
- De Smet I., Vassileva V., De Rybel B., Levesque M. P., Grunewald W., Van Damme D., Van Noorden G., Naudts M., Van Isterdael G., De Clercq R., et al. (2008). Receptor-like kinase ACR4 restricts formative cell divisions in the Arabidopsis root. Science 322, 594-597 [DOI] [PubMed] [Google Scholar]
- De Smet I., Voss U., Jurgens G., Beeckman T. (2009). Receptor-like kinases shape the plant. Nat. Cell Biol. 11, 1166-1173 [DOI] [PubMed] [Google Scholar]
- Dello Ioio R., Linhares F. S., Scacchi E., Casamitjana-Martinez E., Heidstra R., Costantino P., Sabatini S. (2007). Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr. Biol. 17, 678-682 [DOI] [PubMed] [Google Scholar]
- Dello Ioio R., Nakamura K., Moubayidin L., Perilli S., Taniguchi M., Morita M. T., Aoyama T., Costantino P., Sabatini S. (2008). A genetic framework for the control of cell division and differentiation in the root meristem. Science 322, 1380-1384 [DOI] [PubMed] [Google Scholar]
- Depuydt S., Dolezal K., Van Lijsebettens M., Moritz T., Holsters M., Vereecke D. (2008). Modulation of the hormone setting by Rhodococcus fascians results in ectopic KNOX activation in Arabidopsis. Plant Physiol. 146, 1267-1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyoung B. J., Clark S. E. (2008). BAM receptors regulate stem cell specification and organ development through complex interactions with CLAVATA signaling. Genetics 180, 895-904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Martinez C., Gusmaroli G., Wang Y., Zhou J., Wang F., Chen L., Yu L., Iglesias-Pedraz J. M., Kircher S., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451, 475-479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiers M., Golemiec E., Xu J., van der Geest L., Heidstra R., Stiekema W., Liu C. M. (2005). The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway. Plant Cell 17, 2542-2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher J. C., Brand U., Running M. P., Simon R., Meyerowitz E. M. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283, 1911-1914 [DOI] [PubMed] [Google Scholar]
- Foo E., Bullier E., Goussot M., Foucher F., Rameau C., Beveridge C. A. (2005). The branching gene RAMOSUS1 mediates interactions among two novel signals and auxin in pea. Plant Cell 17, 464-474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher K. L., Benfey P. N. (2009). Both the conserved GRAS domain and nuclear localization are required for SHORT-ROOT movement. Plant J. 57, 785-797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher K. L., Paquette A. J., Nakajima K., Benfey P. N. (2004). Mechanisms regulating SHORT-ROOT intercellular movement. Curr. Biol. 14, 1847-1851 [DOI] [PubMed] [Google Scholar]
- Galweiler L., Guan C., Muller A., Wisman E., Mendgen K., Yephremov A., Palme K. (1998). Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282, 2226-2230 [DOI] [PubMed] [Google Scholar]
- Gao B., Allen R., Maier T., Davis E. L., Baum T. J., Hussey R. S. (2003). The parasitome of the phytonematode Heterodera glycines. Mol. Plant Microbe Interact. 16, 720-726 [DOI] [PubMed] [Google Scholar]
- Geldner N., Hyman D. L., Wang X., Schumacher K., Chory J. (2007). Endosomal signaling of plant steroid receptor kinase BRI1. Genes Dev. 21, 1598-1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giakountis A., Coupland G. (2008). Phloem transport of flowering signals. Curr. Opin. Plant Biol. 11, 687-694 [DOI] [PubMed] [Google Scholar]
- Giron D., Kaiser W., Imbault N., Casas J. (2007). Cytokinin-mediated leaf manipulation by a leafminer caterpillar. Biol. Lett. 3, 340-343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H., Sasaki E., Akiyama K., Maruyama-Nakashita A., Nakabayashi K., Li W., Ogawa M., Yamauchi Y., Preston J., Aoki K., et al. (2008). The AtGenExpress hormone and chemical treatment data set: experimental design, data evaluation, model data analysis and data access. Plant J. 55, 526-542 [DOI] [PubMed] [Google Scholar]
- Gomez-Roldan V., Fermas S., Brewer P. B., Puech-Pages V., Dun E. A., Pillot J. P., Letisse F., Matusova R., Danoun S., Portais J. C., et al. (2008). Strigolactone inhibition of shoot branching. Nature 455, 189-194 [DOI] [PubMed] [Google Scholar]
- Grunewald W., Cannoot B., Friml J., Gheysen G. (2009a). Parasitic nematodes modulate PIN-mediated auxin transport to facilitate infection. PLoS Pathog. 5, e1000266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald W., Vanholme B., Pauwels L., Plovie E., Inze D., Gheysen G., Goossens A. (2009b). Expression of the Arabidopsis jasmonate signalling repressor JAZ1/TIFY10A is stimulated by auxin. EMBO Rep. 10, 923-928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurlebeck D., Szurek B., Bonas U. (2005). Dimerization of the bacterial effector protein AvrBs3 in the plant cell cytoplasm prior to nuclear import. Plant J. 42, 175-187 [DOI] [PubMed] [Google Scholar]
- Gurlebeck D., Thieme F., Bonas U. (2006). Type III effector proteins from the plant pathogen Xanthomonas and their role in the interaction with the host plant. J. Plant Physiol. 163, 233-255 [DOI] [PubMed] [Google Scholar]
- Hall S. M., Hillman J. R. (1975). Correlative inhibition of lateral bud growth in Phaseolus vulgaris L. timing of bud growth following decapitation. Planta 123, 137-143 [DOI] [PubMed] [Google Scholar]
- Helariutta Y., Fukaki H., Wysocka-Diller J., Nakajima K., Jung J., Sena G., Hauser M. T., Benfey P. N. (2000). The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101, 555-567 [DOI] [PubMed] [Google Scholar]
- Heyl A., Wulfetange K., Pils B., Nielsen N., Romanov G. A., Schmulling T. (2007). Evolutionary proteomics identifies amino acids essential for ligand-binding of the cytokinin receptor CHASE domain. BMC Evol. Biol. 7, 62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose N., Takei K., Kuroha T., Kamada-Nobusada T., Hayashi H., Sakakibara H. (2008). Regulation of cytokinin biosynthesis, compartmentalization and translocation. J. Exp. Bot. 59, 75-83 [DOI] [PubMed] [Google Scholar]
- Hofmeister W. F. B., Irmisch T., Sachs J., Bary A. D. (1865). Handbuch der physiologischen botanik, in verbindung mit A. de Bary, Th. Irmisch und J. Sachs Leipzig: W. Engelmann; [Google Scholar]
- Holdrege C. (2005). The forming tree. In Context – A Publication of The Nature Institute 14, 18-22 [Google Scholar]
- Huang G., Allen R., Davis E. L., Baum T. J., Hussey R. S. (2006a). Engineering broad root-knot resistance in transgenic plants by RNAi silencing of a conserved and essential root-knot nematode parasitism gene. Proc. Natl. Acad. Sci. USA 103, 14302-14306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G., Dong R., Allen R., Davis E. L., Baum T. J., Hussey R. S. (2006b). A root-knot nematode secretory peptide functions as a ligand for a plant transcription factor. Mol. Plant Microbe Interact. 19, 463-470 [DOI] [PubMed] [Google Scholar]
- Jaeger K. E., Wigge P. A. (2007). FT protein acts as a long-range signal in Arabidopsis. Curr. Biol. 17, 1050-1054 [DOI] [PubMed] [Google Scholar]
- Jeong S., Trotochaud A. E., Clark S. E. (1999). The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 11, 1925-1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay S., Hahn S., Marois E., Hause G., Bonas U. (2007). A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science 318, 648-651 [DOI] [PubMed] [Google Scholar]
- Kayes J. M., Clark S. E. (1998). CLAVATA2, a regulator of meristem and organ development in Arabidopsis. Development 125, 3843-3851 [DOI] [PubMed] [Google Scholar]
- Kehr J., Buhtz A. (2008). Long distance transport and movement of RNA through the phloem. J. Exp. Bot. 59, 85-92 [DOI] [PubMed] [Google Scholar]
- Kim I., Cho E., Crawford K., Hempel F. D., Zambryski P. C. (2005). Cell-to-cell movement of GFP during embryogenesis and early seedling development in Arabidopsis. Proc. Natl. Acad. Sci. USA 102, 2227-2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. Y., Yuan Z., Cilia M., Khalfan-Jagani Z., Jackson D. (2002). Intercellular trafficking of a KNOTTED1 green fluorescent protein fusion in the leaf and shoot meristem of Arabidopsis. Proc. Natl. Acad. Sci. USA 99, 4103-4108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. Y., Yuan Z., Jackson D. (2003). Developmental regulation and significance of KNOX protein trafficking in Arabidopsis. Development 130, 4351-4362 [DOI] [PubMed] [Google Scholar]
- Kinoshita A., Nakamura Y., Sasaki E., Kyozuka J., Fukuda H., Sawa S. (2007). Gain-of-function phenotypes of chemically synthetic CLAVATA3/ESR-related (CLE) peptides in Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol. 48, 1821-1825 [DOI] [PubMed] [Google Scholar]
- Kragler F., Monzer J., Xoconostle-Cazares B., Lucas W. J. (2000). Peptide antagonists of the plasmodesmal macromolecular trafficking pathway. EMBO J. 19, 2856-2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer E. M., Frazer N. L., Baskin T. I. (2007). Measurement of diffusion within the cell wall in living roots of Arabidopsis thaliana. J. Exp. Bot. 58, 3005-3015 [DOI] [PubMed] [Google Scholar]
- Kurakawa T., Ueda N., Maekawa M., Kobayashi K., Kojima M., Nagato Y., Sakakibara H., Kyozuka J. (2007). Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445, 652-655 [DOI] [PubMed] [Google Scholar]
- Lee J. Y., Colinas J., Wang J. Y., Mace D., Ohler U., Benfey P. N. (2006). Transcriptional and posttranscriptional regulation of transcription factor expression in Arabidopsis roots. Proc. Natl. Acad. Sci. USA 103, 6055-6060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhard M., Jurgens G., Laux T. (2002). The WUSCHEL and SHOOTMERISTEMLESS genes fulfil complementary roles in Arabidopsis shoot meristem regulation. Development 129, 3195-3206 [DOI] [PubMed] [Google Scholar]
- Lichter A., Barash I., Valinsky L., Manulis S. (1995). The genes involved in cytokinin biosynthesis in Erwinia herbicola pv. gypsophilae: characterization and role in gall formation. J. Bacteriol. 177, 4457-4465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifschitz E., Eviatar T., Rozman A., Shalit A., Goldshmidt A., Amsellem Z., Alvarez J. P., Eshed Y. (2006). The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc. Natl. Acad. Sci. USA 103, 6398-6403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M. K., Belanger H., Lee Y. J., Varkonyi-Gasic E., Taoka K., Miura E., Xoconostle-Cazares B., Gendler K., Jorgensen R. A., Phinney B., et al. (2007). FLOWERING LOCUS T protein may act as the long-distance florigenic signal in the cucurbits. Plant Cell 19, 1488-1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. X., Srivastava R., Howell S. (2009). Overexpression of an Arabidopsis gene encoding a subtilase (AtSBT5.4) produces a clavata-like phenotype. Planta 230, 687-697 [DOI] [PubMed] [Google Scholar]
- Ljung K., Bhalerao R. P., Sandberg G. (2001). Sites and homeostatic control of auxin biosynthesis in Arabidopsis during vegetative growth. Plant J. 28, 465-474 [DOI] [PubMed] [Google Scholar]
- Lorenzo O., Chico J. M., Sanchez-Serrano J. J., Solano R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16, 1938-1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas W. J., Bouche-Pillon S., Jackson D. P., Nguyen L., Baker L., Ding B., Hake S. (1995). Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science 270, 1980-1983 [DOI] [PubMed] [Google Scholar]
- Mace D. L., Lee J. Y., Twigg R. W., Colinas J., Benfey P. N., Ohler U. (2006). Quantification of transcription factor expression from Arabidopsis images. Bioinformatics 22, e323-e331 [DOI] [PubMed] [Google Scholar]
- Marois E., Van den Ackerveken G., Bonas U. (2002). The xanthomonas type III effector protein AvrBs3 modulates plant gene expression and induces cell hypertrophy in the susceptible host. Mol. Plant Microbe Interact. 15, 637-646 [DOI] [PubMed] [Google Scholar]
- Mathieu J., Warthmann N., Kuttner F., Schmid M. (2007). Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr. Biol. 17, 1055-1060 [DOI] [PubMed] [Google Scholar]
- Memelink J., Hoge J. H., Schilperoort R. A. (1987). Cytokinin stress changes the developmental regulation of several defence-related genes in tobacco. EMBO J. 6, 3579-3583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchum M. G., Wang X., Davis E. L. (2008). Diverse and conserved roles of CLE peptides. Curr. Opin. Plant Biol. 11, 75-81 [DOI] [PubMed] [Google Scholar]
- Miwa H., Betsuyaku S., Iwamoto K., Kinoshita A., Fukuda H., Sawa S. (2008). The receptor-like kinase SOL2 mediates CLE signaling in Arabidopsis. Plant Cell Physiol. 49, 1752-1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlejnek P., Prochazka S. (2002). Activation of caspase-like proteases and induction of apoptosis by isopentenyladenosine in tobacco BY-2 cells. Planta 215, 158-166 [DOI] [PubMed] [Google Scholar]
- Muller R., Bleckmann A., Simon R. (2008). The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell 20, 934-946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase K., Hirano Y., Sun T. P., Hakoshima T. (2008). Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456, 459-463 [DOI] [PubMed] [Google Scholar]
- Nakajima K., Sena G., Nawy T., Benfey P. N. (2001). Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413, 307-311 [DOI] [PubMed] [Google Scholar]
- Navarro L., Dunoyer P., Jay F., Arnold B., Dharmasiri N., Estelle M., Voinnet O., Jones J. D. (2006). A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312, 436-439 [DOI] [PubMed] [Google Scholar]
- Ni J., Clark S. E. (2006). Evidence for functional conservation, sufficiency, and proteolytic processing of the CLAVATA3 CLE domain. Plant Physiol. 140, 726-733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissan G., Manulis-Sasson S., Weinthal D., Mor H., Sessa G., Barash I. (2006). The type III effectors HsvG and HsvB of gall-forming Pantoea agglomerans determine host specificity and function as transcriptional activators. Mol. Microbiol. 61, 1118-1131 [DOI] [PubMed] [Google Scholar]
- Nordstrom A., Tarkowski P., Tarkowska D., Norbaek R., Astot C., Dolezal K., Sandberg G. (2004). Auxin regulation of cytokinin biosynthesis in Arabidopsis thaliana: a factor of potential importance for auxin-cytokinin-regulated development. Proc. Natl. Acad. Sci. USA 101, 8039-8044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelkers K., Goffard N., Weiller G. F., Gresshoff P. M., Mathesius U., Frickey T. (2008). Bioinformatic analysis of the CLE signaling peptide family. BMC Plant Biol. 8, 1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama K., Shinohara H., Ogawa-Ohnishi M., Matsubayashi Y. (2009). A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat. Chem. Biol. 5, 578-580 [DOI] [PubMed] [Google Scholar]
- Olsen A. N., Skriver K. (2003). Ligand mimicry? Plant-parasitic nematode polypeptide with similarity to CLAVATA3. Trends Plant Sci. 8, 55-57 [DOI] [PubMed] [Google Scholar]
- Padmanabhan M. S., Goregaoker S. P., Golem S., Shiferaw H., Culver J. N. (2005). Interaction of the tobacco mosaic virus replicase protein with the Aux/IAA protein PAP1/IAA26 is associated with disease development. J. Virol. 79, 2549-2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan M. S., Kramer S. R., Wang X., Culver J. N. (2008). Tobacco mosaic virus replicase-auxin/indole acetic acid protein interactions: reprogramming the auxin response pathway to enhance virus infection. J. Virol. 82, 2477-2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perbal M. C., Haughn G., Saedler H., Schwarz-Sommer Z. (1996). Non-cell-autonomous function of the Antirrhinum floral homeotic proteins DEFICIENS and GLOBOSA is exerted by their polar cell-to-cell trafficking. Development 122, 3433-3441 [DOI] [PubMed] [Google Scholar]
- Pertry I., Vaclavikova K., Depuydt S., Galuszka P., Spichal L., Temmerman W., Stes E., Schmulling T., Kakimoto T., Van Montagu M. C., et al. (2009). Identification of Rhodococcus fascians cytokinins and their modus operandi to reshape the plant. Proc. Natl. Acad. Sci. USA 106, 929-934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H., Chang K. N., Yazaki J., Ecker J. R. (2009). Interplay between ethylene, ETP1/ETP2 F-box proteins, and degradation of EIN2 triggers ethylene responses in Arabidopsis. Genes Dev. 23, 512-521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte A. M., Mason M. G., Hutchison C. E., Ferreira F. J., Schaller G. E., Kieber J. J. (2006). A subset of Arabidopsis AP2 transcription factors mediates cytokinin responses in concert with a two-component pathway. Proc. Natl. Acad. Sci. USA 103, 11081-11085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo E., Sharma V. K., Kovaleva V., Raikhel N. V., Fletcher J. C. (2002). CLV3 is localized to the extracellular space, where it activates the Arabidopsis CLAVATA stem cell signaling pathway. Plant Cell 14, 969-977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romer P., Hahn S., Jordan T., Strauss T., Bonas U., Lahaye T. (2007). Plant pathogen recognition mediated by promoter activation of the pepper Bs3 resistance gene. Science 318, 645-648 [DOI] [PubMed] [Google Scholar]
- Ruzicka K., Simaskova M., Duclercq J., Petrasek J., Zazimalova E., Simon S., Friml J., Van Montagu M. C., Benkova E. (2009). Cytokinin regulates root meristem activity via modulation of the polar auxin transport. Proc. Natl. Acad. Sci. USA 106, 4284-4289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S., Heidstra R., Wildwater M., Scheres B. (2003). SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev. 17, 354-358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs T., Thimann V. (1967). Role of auxins and cytokinins in release of buds from dominance. Am. J. Bot. 54, 136-144 [Google Scholar]
- Samach A., Onouchi H., Gold S. E., Ditta G. S., Schwarz-Sommer Z., Yanofsky M. F., Coupland G. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288, 1613-1616 [DOI] [PubMed] [Google Scholar]
- Samuel G. (1934). The movement of tobacco mosaic virus within the plant. Ann. App. Biol. 21, 90-111 [Google Scholar]
- Schoof H., Lenhard M., Haecker A., Mayer K. F., Jurgens G., Laux T. (2000). The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100, 635-644 [DOI] [PubMed] [Google Scholar]
- Sessions A., Yanofsky M. F., Weigel D. (2000). Cell-cell signaling and movement by the floral transcription factors LEAFY and APETALA1. Science 289, 779-782 [DOI] [PubMed] [Google Scholar]
- Shalit A., Rozman A., Goldshmidt A., Alvarez J. P., Bowman J. L., Eshed Y., Lifschitz E. (2009). The flowering hormone florigen functions as a general systemic regulator of growth and termination. Proc. Natl. Acad. Sci. USA 106, 8392-8397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V. K., Ramirez J., Fletcher J. C. (2003). The Arabidopsis CLV3-like (CLE) genes are expressed in diverse tissues and encode secreted proteins. Plant Mol. Biol. 51, 415-425 [DOI] [PubMed] [Google Scholar]
- Shimada A., Ueguchi-Tanaka M., Nakatsu T., Nakajima M., Naoe Y., Ohmiya H., Kato H., Matsuoka M. (2008). Structural basis for gibberellin recognition by its receptor GID1. Nature 456, 520-523 [DOI] [PubMed] [Google Scholar]
- Shiu S. H., Bleecker A. B. (2001). Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA 98, 10763-10768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava R., Liu J. X., Howell S. H. (2008). Proteolytic processing of a precursor protein for a growth-promoting peptide by a subtilisin serine protease in Arabidopsis. Plant J. 56, 219-227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl Y., Wink R. H., Ingram G. C., Simon R. (2009). A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr. Biol. 19, 909-914 [DOI] [PubMed] [Google Scholar]
- Stepanova A. N., Robertson-Hoyt J., Yun J., Benavente L. M., Xie D. Y., Dolezal K., Schlereth A., Jurgens G., Alonso J. M. (2008). TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133, 177-191 [DOI] [PubMed] [Google Scholar]
- Strabala T. J., O’Donnell P. J., Smit A. M., Ampomah-Dwamena C., Martin E. J., Netzler N., Nieuwenhuizen N. J., Quinn B. D., Foote H. C., Hudson K. R. (2006). Gain-of-function phenotypes of many CLAVATA3/ESR genes, including four new family members, correlate with tandem variations in the conserved CLAVATA3/ESR domain. Plant Physiol. 140, 1331-1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugio A., Yang B., Zhu T., White F. F. (2007). Two type III effector genes of Xanthomonas oryzae pv. oryzae control the induction of the host genes OsTFIIAgamma1 and OsTFX1 during bacterial blight of rice. Proc. Natl. Acad. Sci. USA 104, 10720-10725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzaki T., Yoshida A., Hirano H. Y. (2008). Functional diversification of CLAVATA3-related CLE proteins in meristem maintenance in rice. Plant Cell 20, 2049-2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons G. M., Ross J. J., Jager C. E., Reid J. B. (2008). Brassinosteroid transport. J. Exp. Bot. 59, 17-24 [DOI] [PubMed] [Google Scholar]
- Takada S., Goto K. (2003). Terminal flower2, an Arabidopsis homolog of heterochromatin protein1, counteracts the activation of flowering locus T by constans in the vascular tissues of leaves to regulate flowering time. Plant Cell 15, 2856-2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki S., Matsuo S., Wong H. L., Yokoi S., Shimamoto K. (2007). Hd3a protein is a mobile flowering signal in rice. Science 316, 1033-1036 [DOI] [PubMed] [Google Scholar]
- Tan X., Calderon-Villalobos L. I., Sharon M., Zheng C., Robinson C. V., Estelle M., Zheng N. (2007). Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446, 640-645 [DOI] [PubMed] [Google Scholar]
- Tanaka M., Takei K., Kojima M., Sakakibara H., Mori H. (2006). Auxin controls local cytokinin biosynthesis in the nodal stem in apical dominance. Plant J. 45, 1028-1036 [DOI] [PubMed] [Google Scholar]
- Thimann K. V., Skoog F. (1933). Studies on the growth hormone of plants: III. The inhibiting action of the growth substance on bud development. Proc. Natl. Acad. Sci. USA 19, 714-716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G., Nomura K., He S. Y., Howe G. A., Browse J. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448, 661-665 [DOI] [PubMed] [Google Scholar]
- Umehara M., Hanada A., Yoshida S., Akiyama K., Arite T., Takeda-Kamiya N., Magome H., Kamiya Y., Shirasu K., Yoneyama K., et al. (2008). Inhibition of shoot branching by new terpenoid plant hormones. Nature 455, 195-200 [DOI] [PubMed] [Google Scholar]
- Vandeputte O., Oden S., Mol A., Vereecke D., Goethals K., El Jaziri M., Prinsen E. (2005). Biosynthesis of auxin by the gram-positive phytopathogen Rhodococcus fascians is controlled by compounds specific to infected plant tissues. Appl. Environ. Microbiol. 71, 1169-1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vereecke D., Burssens S., Simon-Mateo C., Inze D., Van Montagu M., Goethals K., Jaziri M. (2000). The Rhodococcus fascians-plant interaction: morphological traits and biotechnological applications. Planta 210, 241-251 [DOI] [PubMed] [Google Scholar]
- Wada T., Kurata T., Tominaga R., Koshino-Kimura Y., Tachibana T., Goto K., Marks M. D., Shimura Y., Okada K. (2002). Role of a positive regulator of root hair development, CAPRICE, in Arabidopsis root epidermal cell differentiation. Development 129, 5409-5419 [DOI] [PubMed] [Google Scholar]
- Walters D. R., McRoberts N. (2006). Plants and biotrophs: a pivotal role for cytokinins? Trends Plant Sci. 11, 581-586 [DOI] [PubMed] [Google Scholar]
- Walters D. R., McRoberts N., Fitt B. D. (2008). Are green islands red herrings? Significance of green islands in plant interactions with pathogens and pests. Biol. Rev. Camb. Philos. Soc. 83, 79-102 [DOI] [PubMed] [Google Scholar]
- Wang D., Pajerowska-Mukhtar K., Culler A. H., Dong X. (2007). Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr. Biol. 17, 1784-1790 [DOI] [PubMed] [Google Scholar]
- Wang K. L., Yoshida H., Lurin C., Ecker J. R. (2004). Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein. Nature 428, 945-950 [DOI] [PubMed] [Google Scholar]
- Wang X., Allen R., Ding X., Goellner M., Maier T., de Boer J. M., Baum T. J., Hussey R. S., Davis E. L. (2001). Signal peptide-selection of cDNA cloned directly from the esophageal gland cells of the soybean cyst nematode Heterodera glycines. Mol. Plant Microbe Interact. 14, 536-544 [DOI] [PubMed] [Google Scholar]
- Wang X. H., Mitchum M. G., Gao B. L., Li C. Y., Diab H., Baum T. J., Hussey R. S., Davis E. L. (2005). A parasitism gene from a plant-parasitic nematode with function similar to CLAVATA3/ESR (CLE) of Arabidopsis thaliana. Mol. Plant Pathol. 6, 187-191 [DOI] [PubMed] [Google Scholar]
- Weigel D., Nilsson O. (1995). A developmental switch sufficient for flower initiation in diverse plants. Nature 377, 495-500 [DOI] [PubMed] [Google Scholar]
- Welch D., Hassan H., Blilou I., Immink R., Heidstra R., Scheres B. (2007). Arabidopsis JACKDAW and MAGPIE zinc finger proteins delimit asymmetric cell division and stabilize tissue boundaries by restricting SHORT-ROOT action. Genes Dev. 21, 2196-2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichmann G., Bergelson J. (2004). Effector genes of Xanthomonas axonopodis pv. vesicatoria promote transmission and enhance other fitness traits in the field. Genetics 166, 693-706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiechert W., Schweissgut O., Takanaga H., Frommer W. B. (2007). Fluxomics: mass spectrometry versus quantitative imaging. Curr. Opin. Plant Biol. 10, 323-330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge P. A., Kim M. C., Jaeger K. E., Busch W., Schmid M., Lohmann J. U., Weigel D. (2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309, 1056-1059 [DOI] [PubMed] [Google Scholar]
- Wolters H., Jurgens G. (2009). Survival of the flexible: hormonal growth control and adaptation in plant development. Nat. Rev. Genet. 10, 305-317 [DOI] [PubMed] [Google Scholar]
- Wu X., Dinneny J. R., Crawford K. M., Rhee Y., Citovsky V., Zambryski P. C., Weigel D. (2003). Modes of intercellular transcription factor movement in the Arabidopsis apex. Development 130, 3735-3745 [DOI] [PubMed] [Google Scholar]
- Yamagami M., Haga K., Napier R. M., Iino M. (2004). Two distinct signaling pathways participate in auxin-induced swelling of pea epidermal protoplasts. Plant Physiol. 134, 735-747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., Sugio A., White F. F. (2006). Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc. Natl. Acad. Sci. USA 103, 10503-10508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Garreton V., Chua N. H. (2005). The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev. 19, 1532-1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J., Zhang S., Zhang W., Li G., Chen Z., Zhai W., Zhao X., Pan X., Xie Q., Zhu L. (2006). The rice HIGH-TILLERING DWARF1 encoding an ortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds. Plant J. 48, 687-698 [DOI] [PubMed] [Google Scholar]