Abstract
Freshwater planarians are able to regenerate any missing part of their body and have extensive tissue turnover because of the action of dividing cells called neoblasts. Neoblasts provide an excellent system for in vivo study of adult stem cell biology. We identified the Smed-CHD4 gene, which is predicted to encode a chromatin-remodeling protein similar to CHD4/Mi-2 proteins, as required for planarian regeneration and tissue homeostasis. Following inhibition of Smed-CHD4 with RNA interference (RNAi), neoblast numbers were initially normal, despite an inability of the animals to regenerate. However, the proliferative response of neoblasts to amputation or growth stimulation in Smed-CHD4(RNAi) animals was diminished. Smed-CHD4(RNAi) animals displayed a dramatic reduction in the numbers of certain neoblast progeny cells. Smed-CHD4 was required for the formation of these neoblast progeny cells. Together, these results indicate that Smed-CHD4 is required for neoblasts to produce progeny cells committed to differentiation in order to control tissue turnover and regeneration and suggest a crucial role for CHD4 proteins in stem cell differentiation.
Keywords: CHD4, Differentiation, Planaria, Regeneration, Stem cells
INTRODUCTION
Stem cells have diverse and important roles in animal development and are required for the maintenance of cell numbers during tissue homeostasis in adult organisms. The hallmark properties of stem cells are long-term self-renewal and differentiation potential (Wagers and Weissman, 2004; Weissman et al., 2001). Regulation of these two processes under normal and abnormal conditions is crucial, although still poorly defined.
Planarians are one of the simplest organisms known to have robust regeneration capacities and extensive tissue turnover as adults (Reddien and Sánchez Alvarado, 2004); these properties require active dividing cells known as neoblasts. Neoblasts have been identified by their stem-cell-like morphology and their proliferative capacity (Dubois, 1949; Reddien and Sánchez Alvarado, 2004). Although neoblasts were first described to have stem-cell-like properties more than 100 years ago, the mechanisms regulating their proliferation and differentiation still remain largely unknown. Neoblasts are constantly dividing to maintain tissue turnover in the animals. In addition, injury (e.g. amputation) stimulates neoblast proliferation. Neoblasts are required for the generation of a blastema, an unpigmented epithelial/mesenchymal bud at the site of injury, within which missing body parts are formed (Dubois, 1949; Reddien and Sánchez Alvarado, 2004).
Many newly developed resources and methodologies have made the study of stem cells in planarians possible (Newmark and Sánchez Alvarado, 2000; Newmark et al., 2003; Reddien et al., 2005a; Sánchez Alvarado et al., 2002). For example, whole-mount in situ hybridizations and high-throughput RNAi screens have resulted in the identification of genes required for normal neoblast function (Guo et al., 2006; Reddien et al., 2005a; Reddien et al., 2005b). Because neoblasts are the major category of proliferating cells in the animal, they can be labeled with the thymidine analog bromodeoxyuridine (BrdU) and can also be isolated using a DNA-labeling dye by flow cytometry (Newmark and Sánchez Alvarado, 2000; Hayashi et al., 2006; Reddien et al., 2005b). More recently, a panel of neoblast progeny markers have been identified (Eisenhoffer et al., 2008). The abundance of neoblasts, the prominent role they have in regeneration and the newly available experimental tools for the study of neoblasts, make these cells a powerful new setting for in vivo stem cell experimentation.
In several stem cell systems, transcription factors regulate differentiation towards a particular lineage (Bonifer et al., 2008; Fuchs and Horsley, 2008; Le Grand and Rudnicki, 2007; Orkin and Zon, 2008). The accessibility of target genes to these transcription factors can be dictated by chromatin structure; therefore, regulation of chromatin structure can contribute to changes in gene activity (Bibikova et al., 2008; Georgopoulos, 2002; Kingston and Narlikar, 1999; Kouzarides, 2007; Li et al., 2007; Lunyak and Rosenfeld, 2008; Narlikar et al., 2002). Changes in chromatin structure can be accomplished by both post-translational modifications of histone tails and by ATP-dependent chromatin remodeling proteins (Becker and Horz, 2002; Kouzarides, 2007).
The Mi-2 protein is a central component of the nucleosome remodeling deacetylase (NuRD) complex (Tong et al., 1998; Wade et al., 1998; Zhang et al., 1998). There are two Mi-2 proteins: Mi-2α, also known as the chromodomain helicase DNA binding protein 3 (CHD3) and Mi-2β, also known as CHD4 (Seelig et al., 1996). The latter is the predominant Mi-2 protein associated with the mammalian NuRD complex (Zhang et al., 1998). Chromatin remodeling by the NuRD complex usually results in transcriptional repression due to the histone deacetylase activity residing in the complex (Ng and Bird, 2000; Tong et al., 1998). However, Mi-2β can also associate with histone acetyltransferases and methyl transferases, resulting in these cases in transcriptional activation (Denslow and Wade, 2007; Shimono et al., 2003; Williams et al., 2004). The NuRD complex can be recruited to a specific promoter directly by one of its subunits or indirectly by interaction with a sequence-specific DNA-binding protein (Ahringer, 2000; Denslow and Wade, 2007; Kim et al., 1999). The CHD4 protein belongs to an evolutionarily conserved family and contains two plant homeodomain (PHD) zinc-finger motifs, two chromodomains and an ATPase/helicase domain of the SWI2/SNF family. The NuRD complex, and in particular Mi-2, is required for normal development and cell fate control in several organisms such as Caenorhabditis elegans, Drosophila melanogaster, Arabidopsis thaliana and mammals (Kehle et al., 1998; Ogas et al., 1997; Ogas et al., 1999; Unhavaithaya et al., 2002; von Zelewsky et al., 2000; Yoshida et al., 2008). The general role(s) for Mi-2 in adult stem cell biology has remained unresolved.
Here, we show that Smed-CHD4 is essential for regeneration in the planarian Schmidtea mediterranea. Many of the defects observed in intact Smed-CHD4(RNAi) animals are hallmarks of neoblast dysfunction. Our experiments indicate that Smed-CHD4 is required for the ability of neoblasts to produce progeny cells committed to differentiation, suggesting that chromatin regulation by CHD4 is an important aspect of adult stem cell lineage specification.
MATERIALS AND METHODS
CHD4 and RNAi experiments
RT-PCR was used to amplify the CHD4 gene. Complete gene sequence was determined using 5′ and 3′ RACE PCR (Ambion). CHD4 and the control gene unc-22 from C. elegans were cloned into the pPR244 RNAi expression vector using Gateway recombination reactions as described (Reddien et al., 2005a). RNAi experiments were performed by feeding the animals with a mixture of liver and bacteria expressing CHD4 dsRNA (Reddien et al., 2005a). Ten milliliters of bacteria culture was pelleted and resuspended in 100 μl of liver or in 30 μl of liver. Three feeding protocols were used: animals were fed on day 0, day 4 and day 7, or on day 0 and day 4, or on day 0 only. For regeneration studies, animals were cut at day 8 (for the three-feedings protocol) or at day 5 (for the two-feedings protocol). For homeostasis studies, one extra feeding was added to the three-feedings protocol at day 14.
In situ hybridizations
In situ hybridizations on whole animals, cell macerates or isolated cells were performed as described (Reddien et al., 2005b). CHD4 whole-mount in situ hybridizations and fluorescence in situ hybridizations (FISH) were performed as described (Pearson et al., 2009). To quantify data from in situ hybridizations on cells, the percentage of DAPI-positive cells with signal was determined. To determine the number of Smed-AGAT-1-expressing cells present following irradiation, the number of Smed-AGAT-1+ cells within 0.3 mm by 0.3 mm tissue from the dorsal side and anterior to the pharynx was determined and normalized by animal length.
Antibody stainings
Animals were treated with 10% N-acetyl-cysteine in PBS for 3-5 minutes at room temperature (RT) and fixed in Carnoy’s solution. Animals were then labeled as previously described (Newmark and Sánchez Alvarado, 2000). Numbers of mitoses were divided by animal surface area. For SMEDWI-1 antibody stainings, animals were fixed in a 4% formaldehyde, PBST solution following FISH with the Smed-AGAT-1 riboprobe, blocked and labeled with 1:2000 α-SMEDWI-1.The SMEDWI-1 polyclonal antibody was raised in rabbits using the peptide previously described (Guo et al., 2006). Total numbers of Smed-AGAT-1+ cells, SMEDWI-1; Smed-AGAT-1 double-positive cells, or SMEDWI-1-positive cells were determined in optical sections.
BrdU labeling
Animals were fed with RNAi food at days 0 and 4, and injected at day 6 following initial RNAi feeding with a solution of 5 mg/ml of BrdU (Fluka) in planarian water. Animals were then killed 4 days later in 5% N-acetyl-cysteine in PBS for 5 minutes at RT and fixed as described (Pearson et al., 2009). FISH using the Smed-AGAT-1 riboprobe were performed as described (Pearson et al., 2009). Following FISH, animals were fixed, treated with 2N HCl for 45 minutes at RT and labeled with a 1:100 rat anti-BrdU antibody (Oxford Biotech) as previously described (Newmark and Sánchez Alvarado, 2000). Apotome images were obtained from BrdU-labeled and FISH animals using an Axiocam digital camera, a Zeiss AxioImager with use of an Apotome, and Axiovision software. Total numbers of Smed-AGAT-1-expressing cells containing a BrdU-labeled nucleus were determined in optical sections.
FACS
Planarians were cut into small pieces and incubated in calcium-free, magnesium-free medium plus BSA (CMFB) containing 1 mg/ml of collagenase for 45 minutes at RT as described (Reddien et al., 2005b). Cells were centrifuged at 1250 rpm for 5 minutes and resuspended in CMFB containing Hoechst 33342 (10 μg/ml) for 45 minutes at RT. Calcein (0.5 μg/ml) was incubated with the cells for 15 minutes at RT. Finally, 5 μg/ml of propidium iodide was added to the cell suspension prior to flow cytometry analysis or sorting. Analyses and sorts were performed using a MoFlo3 FACS sorter.
CHD4 qPCR
Total RNA was isolated from wild-type or lethally irradiated animals and from isolated X1 and X2 cells. cDNA was prepared and quantitative PCR (qPCR) was performed using SYBR Green (Applied Biosystems). Data were normalized to the expression of UDP (clone H.55.12e, accession number AY068123). UDP was selected as an internal control as a constitutively expressed gene. Sets of specific CHD4-primers were used to evaluate gene expression (5′-GTCAGCAAAATGTTTACACTGAGG-3′ and 5′-ACGTCTTGCTTCATGTCTGATAGT-3′). Samples without reverse transcriptase served as the negative control template.
RESULTS
CHD4 is required for planarian regeneration
We identified the S. mediterranea Smed-CHD4 gene (chromodomain helicase DNA-binding protein 4) in a screen for genes expressed in neoblasts after wounding (data not shown). SMED-CHD4 is highly similar to proteins within the conserved CHD family of chromatin regulatory proteins; by phylogenetic analysis it clustered to the CHD3/4/5 members of the family, and using BLAST we found that it is most similar to the mammalian CHD4 (see Fig. S1 and Fig. S2 in the supplementary material). CHD4/Mi-2β is a central component of the nucleosome remodeling histone deacetylase complex NuRD (Tong et al., 1998; Wade et al., 1998; Zhang et al., 1998). The Smed-CHD4 gene will be hereafter referred to in the text in short as CHD4.
CHD4(RNAi) worms were unable to form a blastema following amputation, demonstrating that CHD4 is required for planarian regeneration (Fig. 1A). The inability to form a blastema was noticeable as early as 5 days following initial double-stranded (ds) RNA delivery (see Fig. S3 in the supplementary material). Compared with other genes required for regeneration in planarians (Reddien et al., 2005a), the onset of regeneration failure in CHD4(RNAi) animals is very fast. The rapid appearance and strength of the CHD4(RNAi) phenotype is consistent with the possibility that CHD4 has a primary function in a crucial aspect of neoblast function.
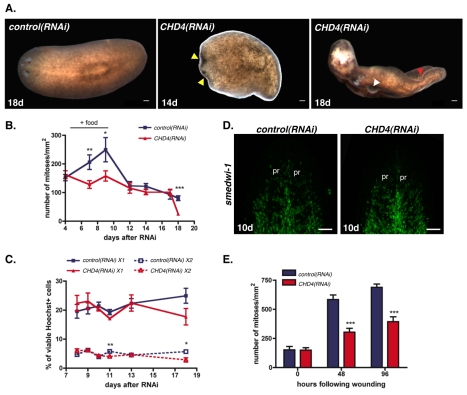
Fig. 1.
CHD4 is required for planarian regeneration and is expressed in neoblasts. (A) As shown in comparison with a control animal, a CHD4(RNAi) worm did not regenerate 7 days following amputation (41/41 were similar). Worms were fed three times and amputated at day 8 after initial RNAi. White dotted line represents the blastema boundary. Anterior is to the left. (B) Expression of CHD4 in wild-type and irradiated (6000 rads 5 days prior) animals, dorsal and ventral views (7/7 were similar). Expression of an irradiation-insensitive gene, PC2 (expressed in the nervous system) served as a control. Anterior is to the left. (C) Expression of CHD4 in neoblast subsets, X1 (1472 cells counted) and X2 (655 cells counted), and radiation-insensitive cells, Xins (1117 cells counted). Scale bars: 0.1 mm.
CHD4 was widely expressed in the planarian parenchyma (a region containing neoblasts), brain and ventral nerve cords, as determined with in situ hybridizations (Fig. 1B). CHD4 mRNA was reduced in CHD4(RNAi) worms following dsRNA feeding (see Fig. S4A in the supplementary material). A lethal dose of γ-irradiation (6000 rads) depletes planarians of neoblasts, eliminating the ability to regenerate and to replace aged cells during homeostasis (Bardeen and Baetjer, 1904; Dubois, 1949; Reddien et al., 2005b). Five days after lethal irradiation, expression of CHD4 in the nervous system persisted but the expression in the parenchyma was reduced, suggesting that CHD4 was expressed in neoblasts or in their immediate descendants (Fig. 1B). Expression of non-neoblast genes remained unchanged following irradiation (Fig. 1B; see Fig. S4B in the supplementary material).
Neoblasts are actively dividing cells that are required for the production of all cells of the adult organism (Reddien and Sánchez Alvarado, 2004). Studies to uncover their lineage potential are emerging (Eisenhoffer et al., 2008). Flow cytometry has been used to isolate cells with neoblast properties (Hayashi et al., 2006; Reddien et al., 2005b). Using vital dyes, two populations of irradiation-sensitive cells can be purified. X1 cells are actively dividing cells (in S and G2/M stages of the cell cycle) with a higher DNA content (>2n); X2 cells contain postmitotic neoblast descendants (Eisenhoffer et al., 2008) and might contain neoblasts in the G1 stage of the cell cycle as well. A population of irradiation-insensitive cells, Xins, has been used as a control population in flow cytometry experiments (Hayashi et al., 2006; Reddien et al., 2005b). To determine whether CHD4 was expressed in neoblasts, we performed in situ hybridizations with X1, X2 and Xins cells. CHD4 was indeed expressed in neoblasts as well as in presumptive differentiated cells: 53% of X1 cells, 78% of X2 cells and 69% of Xins cells were labeled with the CHD4 riboprobe (Fig. 1C). We further confirmed that CHD4 is expressed in neoblasts using quantitative PCR (qPCR; see Fig. S4C in the supplementary material). Moreover, CHD4 was highly expressed at wound sites 3 days following amputation and this expression was not detected in irradiated worms that lack neoblasts (see Fig. S4D in the supplementary material). It is not known whether CHD4 expression was upregulated following wounding or whether the high abundance of CHD4 expression near wounds was due to migration of cells that already expressed this gene. Because irradiation eliminates neoblasts, this result suggests that neoblasts and/or their progeny near wounds express CHD4 during regeneration.
CHD4 is required for tissue turnover
Neoblasts constantly proliferate in adult planarians in order to replace aged tissues (Reddien and Sánchez Alvarado, 2004). To investigate the role of CHD4 during normal tissue turnover, we performed RNAi experiments in intact (i.e. non-amputated) animals. Head regression is a characteristic sign of neoblast dysfunction that occurs between 10 and 12 days following a lethal dose of irradiation that eliminates all neoblasts. Head regression is also observed in animals in which genes required for neoblast function have been inhibited with RNAi (Guo et al., 2006; Reddien et al., 2005a; Reddien et al., 2005b). Intact CHD4(RNAi) worms started to display head regression 14 days after initial dsRNA feeding (Fig. 2A, middle panel). Head regression in some intact CHD4(RNAi) animals started in the center of the head instead of being initiated at the tip of the head, as is typically the case for irradiated animals. Another well-documented defect of irradiated animals is curling around the ventral surface of the animal, probably stemming from the failure of neoblast progeny to replace normally aging differentiated tissue (Reddien and Sánchez Alvarado, 2004). Curling occurs by approximately 14 days following irradiation. Following head regression, intact CHD4(RNAi) animals also displayed curling around their ventral surface (Fig. 2A, right panel). In addition to these classic hallmarks of neoblast dysfunction, intact CHD4(RNAi) worms showed tissue protrusions after 16-18 days following the first dsRNA feeding (Fig. 2A, right panel). These protrusions will be discussed below. Shortly after curling, the CHD4(RNAi) animals died by lysis. Together, these observations demonstrate that CHD4 is required for normal tissue homeostasis.
Fig. 2.
CHD4 is required for homeostasis and normal stimulation of neoblast proliferation. (A) Control and CHD4(RNAi) animals during normal tissue turnover 14 and 18 days after initial RNAi. Yellow arrowhead, head regression; white arrowhead, lesions; red arrowhead, curling (10/10 were similar). Anterior is to the left. (B) Numbers of mitoses labeled with an anti-phospho histone 3 (αH3P) antibody were divided by animal surface area. Animals were fed three times and fixed at time points after initial RNAi. ‘+ food’ indicates the proliferation response following feedings. Data are means ± s.e.m. (n=2 experiments, >8 worms per time point). *, P<0.05; **, P<0.001, ***, P<0.0001, Student’s t-test. (C) Animals were fed three times with dsRNA and cell macerates were generated at multiple time points after initial RNAi. Cells were labeled with Hoechst 33342, calcein, and propidium iodide; X1 and X2 cell percentages (in the total population of Hoechst-labeled cells) were determined by flow cytometry. Data are means ± s.e.m. (n=3 experiments). *, P<0.05; **, P<0.001, Student’s t-test. (D) Fluorescence in situ hybridizations of animals fixed 10 days after multiple dsRNA feedings using the smedwi-1 riboprobe (>12 worms per condition were similar). pr, photoreceptors. Anterior is up. (E) Numbers of mitoses labeled with αH3P were divided by animal surface area. Animals were fed three times, amputated 8 days after initial feeding and fixed at different times following wounding. Data are means ± s.e.m.; n=2 experiments, >8 animals per time point. ***, P<0.0001, Student’s t-test. Scale bars: 0.1 mm.
CHD4 is required for an increase in neoblast proliferation in response to feeding and wounding
One simple hypothesis to explain the similarity of the CHD4(RNAi) phenotype to that of irradiated animals lacking neoblasts would be that CHD4(RNAi) causes a decrease in the presence of neoblasts or in the capacity of neoblasts to proliferate. To test this hypothesis, we quantified the number of mitotic neoblasts (Newmark and Sánchez Alvarado, 2000) in intact animals using an RNAi feeding approach (see Materials and methods). Food stimulates neoblast proliferation, and proliferation returns to the previous steady-state levels a few days later (Baguñà, 1976). The elevation of mitotic numbers immediately following feeding failed to robustly occur in CHD4(RNAi) animals (days 7 and 9 after RNAi; Fig. 2B). However, steady-state numbers of mitotic cells were comparable with those of control animals, including at time points after RNAi at which animals completely failed to regenerate (between days 12 and 16 following the first RNAi feeding), suggesting that neoblasts were maintained in CHD4(RNAi) animals (Fig. 2B). Much later, when severe head regression and curling was occurring (approximately between 16-18 days following the first dsRNA feeding), a significant decrease in mitotic cells was observed (Fig. 2B). A drop in neoblast population numbers late in the progression of an RNAi phenotype has also been observed following RNAi of a different gene (smedwi-2) required for homeostasis but not for neoblast maintenance (Reddien et al., 2005b). Therefore, this late mitotic decrease might be a consequence of, rather than cause of, failed homeostasis.
Because the number of steady-state mitotic neoblasts was normal at time points at which regeneration failed in CHD4(RNAi) animals, we sought to determine whether the total number of neoblasts was unaffected. Indeed, approximately normal numbers of X1 and X2 cells following RNAi were present at all time points examined, except near the time at which animals were dying (Fig. 2C). A transient decrease in the number of X2 cells at day 11 following the first dsRNA feeding was observed. Because X2 cells appear to include non-dividing neoblasts as well as neoblast progeny (Eisenhoffer et al., 2008), a defect in neoblast proliferation following feeding could explain this slight decrease in X2 cells. Moreover, the gross expression of markers of X1 cells, smedwi-1 and Smed-histone H2B (Smed-H2B), appeared normal in CHD4(RNAi) animals 10 days after the initial dsRNA feeding (Fig. 2D; see Fig. S5 in the supplementary material). Together, these observations indicate that the regeneration defect of CHD4(RNAi) animals is not a consequence of a loss of neoblasts or a loss of their capacity to proliferate during homeostasis prior to injury.
In addition to the sustained neoblast proliferation in intact planarians that is required for replacement of aging cells during normal turnover, neoblasts undergo an increase in proliferation after wounding (Saló and Baguñà, 1984). We found that although neoblasts in CHD4(RNAi) worms were able to respond to wounding by elevating mitotic numbers at 48 hours and 96 hours following amputation, the response was diminished compared with control RNAi animals (Fig. 2E). Because neoblast numbers in CHD4(RNAi) animals were normal in steady-state, a partially defective proliferative response to injury is probably not sufficient to explain the complete absence of a blastema in CHD4(RNAi) worms. For example, prior RNAi screening indicates that animals with reduced, but not eliminated, mitoses can be capable of making small blastemas (Reddien et al., 2005a). Thus, given the severe blastema failure of CHD4(RNAi) animals, it is probable that an additional and robust defect exists to explain the regeneration phenotype. We therefore examined the ability of neoblasts in CHD4(RNAi) animals to produce progeny cells.
CHD4(RNAi) animals die more rapidly than do normal animals following lethal irradiation
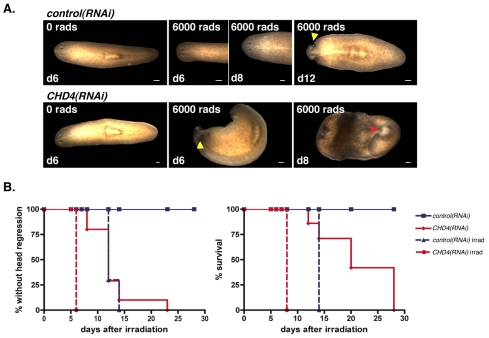
Lethally irradiated and intact wild-type worms develop the first signs of defects due to neoblast loss approximately 10-12 days after irradiation (Reddien et al., 2005b). This phenotype involves darkening of the tip of the head, followed by head regression. A few days later, the worms curl around their ventral surface and finally they lyse. Although actively dividing neoblasts completely disappear within 1 day after irradiation, the initial 10- to 12-day window during which no phenotype is apparent might in part rely on the replacement of normally dying aged differentiated cells by existing non-dividing progenitors (Reddien and Sánchez Alvarado, 2004). Therefore, the time of survival after irradiation could be affected by alterations in numbers or function of neoblast progeny cells that can act as progenitors of differentiated cells. We lethally irradiated control and CHD4(RNAi) worms 8 days after the initial dsRNA feeding. As expected, irradiated control(RNAi) worms showed head regression by 12 days following irradiation (Fig. 3A,B, left graph), followed by curling and lysis (Fig. 3A,B, right graph). By contrast, irradiated CHD4(RNAi) animals displayed head regression 6 days following irradiation, and curled and lysed 8 days after irradiation (Fig. 3). The rate of appearance of defects in irradiated CHD4(RNAi) animals was faster than that observed in starved or fed irradiated wild-type or control(RNAi) worms (Reddien et al., 2005b) (Fig. 3). Most of the non-irradiated CHD4(RNAi) animals showed head regression by 12 days after the irradiation day (Fig. 3B, left graph) and they curled and lysed between days 20 and 28 after the irradiation day (Fig. 3B, right graph). Similar results were obtained with CHD4(RNAi) animals irradiated at 10 days and 12 days after the initial dsRNA feeding, times at which normal steady-state neoblast numbers were reached following feeding (data not shown). This indicates that the faster appearance of the irradiation phenotype in CHD4(RNAi) animals is not a consequence of the failure to boost proliferation immediately after feeding. The rapid appearance of head regression following irradiation of CHD4(RNAi) animals suggests a possible decreased number of non-dividing progeny cells that are neoblast descendants or impaired neoblast differentiation. We therefore directly examined neoblast progeny cells in CHD4(RNAi) animals (see below).
Fig. 3.
CHD4(RNAi) intact animals die faster following irradiation. (A) Control and CHD4(RNAi) animals were lethally irradiated with 6000 rads 8 days following initial RNAi. Irradiated control(RNAi) animals showed head regression by 12 days following irradiation (7/10 animals) and curling by 14 days (10/10), whereas irradiated CHD4(RNAi) animals showed head regression by 6 days following irradiation (10/10) and curling by 8 days (10/10). Yellow arrowhead, head regression; red arrowhead, curling. Anterior is to the left. (B) Percentages of control or CHD4(RNAi) animals without head regression (left) and still alive (right) following irradiation are shown. Scale bars: 0.1 mm.
CHD4(RNAi) animals have decreased numbers of Smed-AGAT-1-expressing cells
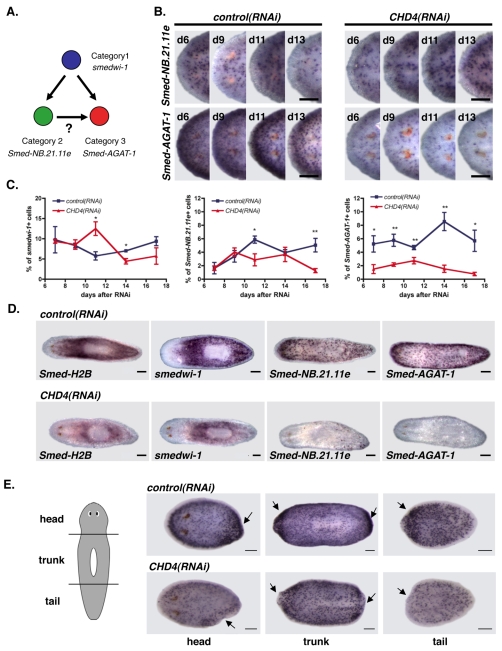
Previous studies identified three irradiation-sensitive, lineage-related cell subsets (Eisenhoffer et al., 2008) (Fig. 4A). During neoblast differentiation, Category 1 gene-expressing cells differentiate into cells expressing Category 2 and 3 genes (Eisenhoffer et al., 2008) (Fig. 4A). Category 1 gene-expressing cells are self-renewing neoblasts (i.e. X1 cells) (Eisenhoffer et al., 2008), which are found in the parenchyma but excluded from the region anterior to the photoreceptors and the pharynx (Newmark and Sánchez Alvarado, 2000). Category 1 gene-expressing cells disappear 1 day after irradiation. Category 1 genes include the known stem cell genes smedwi-1 and Smed-bruli-1, as well as cell cycle regulatory genes (Eisenhoffer et al., 2008; Guo et al., 2006; Reddien et al., 2005b). Genes from Categories 2 and 3 are expressed in X2 and Xins cells but not X1 cells, and are found anterior to the photoreceptors and close to the animal margins (regions where no mitotic cells are found). Similar to the case for Category 1 genes, the cells expressing Category 2 genes (e.g. the two novel genes Smed-NB.21.11e and Smed-NB.32.1g) rapidly decrease in number 1 day following irradiation (Eisenhoffer et al., 2008). Cells expressing Category 3 genes are lost within 7 days following irradiation. Category 3 genes include those that encode L-arginine:glycine amidinotransferase (Smed-AGAT)-1, -2 and -3, a ras-related protein family member (Smed-ras-related), a mitochondrial carrier protein 1 (Smed-MCP-1) and a novel protein (Smed-NB.52.12f), among others. Based on BrdU labeling experiments, Category 2 and 3 gene-expressing cells are rapidly produced descendants of neoblasts (Eisenhoffer et al., 2008). Although the precise nature of these cells is still unclear, they might either represent a population of non-mitotic progenitors that will generate terminal differentiated cells or a very short-lived population of differentiated cells. Regardless, these markers provide a useful tool to study neoblast progeny cells and, therefore, neoblast differentiation.
Fig. 4.
CHD4(RNAi) animals have reduced numbers of Smed-AGAT-1-expressing cells. (A) Lineage-related cells and the markers expressed in each cell type. Other models are possible, see Eisenhoffer et al. (Eisenhoffer et al., 2008). (B) Head regions of whole-mount in situ hybridizations using a Category 2 probe Smed-NB.21.11e (upper row) and a Category 3 probe Smed-AGAT-1 (lower row) of control and CHD4(RNAi) animals at several time points following initial RNAi (>4 worms/time point displayed similar results). Anterior is to the left. (C) Cell in situ hybridizations using a Category 1 probe (smedwi-1; >823 cells per time point), a Category 2 probe (Smed-NB.21.11e; >954 cells per time point) and a Category 3 probe (Smed-AGAT-1; >1157 cells per time point) of control and CHD4(RNAi) worm macerates at multiple times after initial RNAi. Percentages of smedwi-1+, Smed-NB.21.11e+, and Smed-AGAT-1+ cells within the total DAPI-positive cell population are shown (means ± s.e.m., n=3 experiments). *, P<0.05; **, P<0.001, Student’s t-test. (D) Whole-mount in situ hybridizations with Category 1 probes (Smed-H2B and smedwi-1), a Category 2 probe (Smed-NB.21.11e) and a Category 3 probe (Smed-AGAT-1) of control and CHD4(RNAi) animals 20 days following initial RNAi. At this late time point, CHD4(RNAi) animals displayed head regression, had lesions and began to curl (>7 worms displayed similar results). Anterior is to the left. (E) Whole-mount in situ hybridizations with the Smed-AGAT-1 riboprobe of control and CHD4(RNAi) head, trunk and tail pieces fixed 3 days following amputation. Animals were amputated 7 days following initial RNAi (>11 regenerating pieces displayed similar results). Arrows indicate the site of amputation. Anterior is to the left. Scale bars: 0.1 mm.
We utilized expression of these genes as markers for distinct cell types to determine if CHD4(RNAi) animals have a defect in the production or maintenance of neoblast progeny cells. We examined the expression of the Category 2 gene Smed-NB.21.11e and the Category 3 gene Smed-AGAT-1 by in situ hybridizations. CHD4(RNAi) worms had approximately normal numbers of Smed-NB.21.11e-expressing cells (Fig. 4B,C; see Fig. S6 in the supplementary material). However, there was a marked decrease in the number of Category 3 Smed-AGAT-1-expressing cells in CHD4(RNAi) worms, as early as 6 days after initial dsRNA feeding (Fig. 4B,C; see Fig. S6 in the supplementary material).
We quantified numbers of Category 1, 2 and 3 cell types by performing in situ hybridizations on total cells from macerated planarians and counting the fraction of total cells that expressed smedwi-1 (Category 1), Smed-NB.21.11e (Category 2) or Smed-AGAT-1 (Category 3). CHD4(RNAi) animals had significantly reduced numbers of Smed-AGAT-1-expressing cells at every time point analyzed and largely normal numbers of Smed-NB.21.11e-expressing cells (Fig. 4C). There were lower numbers of Smed-NB.21.11e-expressing cells at 11 days following RNAi (Fig. 4C). The small defects in the numbers of Category 2-expressing cells observed were possibly due to defects in responses to feeding used in the RNAi experiments. smedwi-1-expressing cells were present in CHD4(RNAi) animals in approximately normal numbers initially, elevated in number at day 11 after the first dsRNA feeding, and low in number late following RNAi (Fig. 4C). The increased number of smedwi-1-expressing cells in CHD4(RNAi) animals at day 11, considered together with the associated decrease in Category 3-expressing cells, is consistent with a possible accumulation of smedwi-1-expressing cells because of failure to differentiate. Overall, the earliest, most prominent and most constant abnormality observed in CHD4(RNAi) animals was the pronounced low number of Smed-AGAT-1-expresssing cells.
Twenty days after the initial dsRNA feeding, when the CHD4(RNAi) animals displayed head regression and were dying, the animals were mostly depleted of Smed-NB.21.11e-expressing cells and entirely lacked Smed-AGAT-1-expressing cells (Fig. 4D). At this late time point, similar to the observed decrease in mitotic cells (Fig. 2B), smedwi-1-expressing cells and Smed-H2B-expressing cells were greatly reduced as well (Fig. 4D). Because this defect occurred late following RNAi, it probably occurred as a consequence of an earlier primary defect in the number of neoblast progeny cells. At this 20-day time point, CHD4(RNAi) animals had protrusions; however, we saw no accumulation of dividing cells, Smed-NB.21.11e or Smed-AGAT-1-expressing cells into foci that could correspond to protrusions (Fig. 4D).
Smed-AGAT-1-expressing cells normally accumulate at the wound site and in blastemas after amputation, providing evidence for an important role for this population in regeneration (Eisenhoffer et al., 2008). Perhaps as a consequence of the reduced number of Smed-AGAT-1-expressing cells, CHD4(RNAi) animals failed to display accumulation of Smed-AGAT-1-expressing cells at wound sites to the extent observed in the control (Fig. 4E). Our data suggest that a failure to produce sufficient numbers of neoblast progeny cells committed to differentiation, such as Smed-AGAT-1-expressing cells, results in insufficient neoblast progeny for blastema formation in CHD4(RNAi) animals.
CHD4 is required for the formation of Smed-AGAT-1-expressing cells
The decreased numbers of Smed-AGAT-1-expressing cells in CHD4(RNAi) animals suggests that CHD4 is required either for the normal generation or maintenance of these cells. First, in order to determine if the CHD4(RNAi) phenotype is explained by a defect in the maintenance of these cells, we determined the rate at which Smed-AGAT-1-expressing cells disappeared following irradiation. Irradiation kills neoblasts and therefore blocks the generation of new Smed-AGAT-1-expressing cells. Animals were fed only once with CHD4 dsRNA or control food and irradiated the same day (Fig. 5A, left graph), or fed 3 times over 7 days and irradiated 7 days following the initial feeding (Fig. 5A, right graph). The numbers of Smed-AGAT-1-expressing cells were determined at different time points following irradiation. In both experiments, Smed-AGAT-1-expressing cells disappeared at similar rates following irradiation in both control and CHD4(RNAi) animals, suggesting that the CHD4(RNAi) phenotype is not explained by a defect in the maintenance of these cells. Therefore, these data point to a defect in the generation of Smed-AGAT-1-expressing cells.
Fig. 5.
CHD4 is required for the formation of Smed-AGAT-1-expressing cells. (A) Smed-AGAT-1+ cell numbers normalized by animal length from whole-mount in situ hybridizations are shown at time points following irradiation. Animals (>4 worms per group) were fed once and lethally irradiated the same day (left) or fed three times and lethally irradiated at day 7 (right). Data are means ± s.e.m. (B) Percentage of double-labeled SMEDWI-1; Smed-AGAT-1-expressing cells in the total number of Smed-AGAT-1+ cells in control and CHD4(RNAi) animals (upper graph), and the percentage of double-labeled SMEDWI-1; Smed-AGAT-1-expressing cells normalized by the area of the head region (lower graph). Data are means ± s.e.m. (>5 worms per group). *, P<0.05; **, P<0.001; ***, P<0.0001, Student’s t-test. Animals were fed twice and fixed at time points following the initial RNAi. Fluorescence in situ hybridizations using Smed-AGAT-1 riboprobe (green) and SMEDWI-1 antibody (red) are shown. Arrowheads indicate cells that co-express Smed-AGAT-1 and SMEDWI-1. (C) SMEDWI-1-expressing cell numbers in control and CHD4(RNAi) animals. pr, photoreceptors. Circled area, region counted. Data are means ± s.e.m. (>8 worms per group). **, P<0.001, Student’s t-test. A second experiment had similar results. Animals were fed twice and fixed at time points following initial RNAi. Anterior is up. Scale bars: 0.1 mm.
To determine if CHD4 is required for the generation of Smed-AGAT-1-expressing cells directly, two different experiments were performed. First, the formation of Smed-AGAT-1-expressing cells in CHD4(RNAi) or control animals following one pulse with the thymidine analog BrdU was observed (see Fig. S7 in the supplementary material). BrdU is incorporated into neoblasts within a few hours following injection (Newmark and Sánchez Alvarado, 2000), and 4 days later, double-labeled BrdU-Smed-AGAT-1-expressing cells are found (Eisenhoffer et al., 2008). Four days after the BrdU pulse, most of the Smed-AGAT-1-expressing cells were labeled with BrdU (see Fig. S7 in the supplementary material). We found that the number of newly formed Smed-AGAT-1-expressing cells was significantly lower in CHD4(RNAi) animals compared with control animals 4 days after the BrdU pulse and 10 days following the initial RNAi feeding (see Fig. S7 in the supplementary material).
Second, we determined the number of SMEDWI-1; Smed-AGAT-1 double-positive cells in CHD4(RNAi) and control animals. The SMEDWI-1 protein has been demonstrated to be present not only in neoblasts, which also express smedwi-1 mRNA, but also in non-mitotic neoblast progeny that no longer express smedwi-1 mRNA (Guo et al., 2006). This is presumably due to perdurance of the SMEDWI-1 protein into immediate neoblast descendants. Moreover, three days following lethal irradiation, all SMEDWI-1-expressing cells disappeared from the animal, indicating that only neoblasts or their immediate progeny cells express this protein (Guo et al., 2006). We found that CHD4(RNAi) animals have a decreased number of SMEDWI-1; Smed-AGAT-1 double-positive cells at 7 days following initial RNAi and an even greater defect at 10 days following the initial RNAi feeding (Fig. 5B). In summary, these experiments demonstrate that the CHD4(RNAi) phenotype is explained by a defect in the generation of Smed-AGAT-1-expressing cells rather than by a defect in their perdurance.
If neoblasts are defective in production of Smed-AGAT-1-positive cells, where is the process of differentiation affected? We found a transient increase in the number of SMEDWI-1 protein-expressing cells in animal head regions in CHD4(RNAi) animals at day 7 following RNAi (Fig. 5C). These SMEDWI-1 protein-positive cells in head tips represent immediate non-mitotic neoblast progeny. Therefore, coincident with a decrease in Smed-AGAT-1-positive cells, there existed an increase in number of immediate neoblast progeny. This observation indicates that immediate neoblast progeny might accumulate because of a failure to differentiate.
CHD4(RNAi) animals have a decreased number of neoblast progeny cells
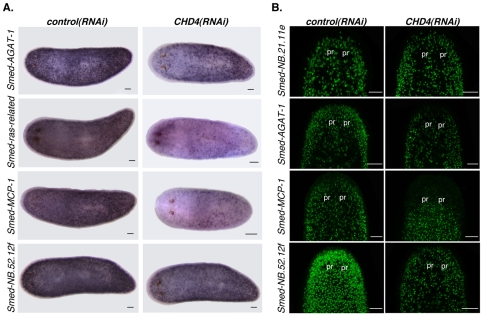
To more definitively determine the role of CHD4 in neoblast differentiation, and to exclude any potential role of CHD4 in specifically regulating Smed-AGAT-1 expression, we examined the expression of three additional Category 3 markers in CHD4(RNAi) animals (Eisenhoffer et al., 2008). CHD4(RNAi) animals also had a robust decrease in the number of cells expressing the Smed-ras-related, Smed-MCP-1 and Smed-NB.52.12f genes (Fig. 6A,B). However, the existing cells that expressed these genes displayed no obvious defect in expression levels for these genes. Previous data have shown that not all Smed-AGAT-1-expressing cells co-express the Smed-ras-related gene (Eisenhoffer et al., 2008). In addition, almost no Smed-AGAT-1-expressing cells co-express the Smed-MCP-1 gene (Eisenhoffer et al., 2008), indicating that there is a heterogeneous population of neoblast progeny cells expressing Category 3 genes. Our results demonstrate that CHD4 is required for the generation of a heterogeneous population of neoblast progeny cells committed to differentiation.
Fig. 6.
CHD4(RNAi) animals have decreased numbers of several Category 3-expressing cells. (A) Whole-mount in situ hybridizations using Category 3 probes 10 days following initial RNAi (>4 worms per riboprobe were similar). Anterior is to the left. (B) Fluorescence in situ hybridizations of animals fixed 10 days after multiple dsRNA feedings using the Smed-NB.21.11e, Smed-AGAT-1, Smed-MCP-1 and Smed-NB.52.12f riboprobes (>12 worms per condition were similar). The head region is shown. pr, photoreceptors. Anterior is up. Scale bars: 0.1 mm.
DISCUSSION
Planarians are an emerging in vivo model system for the study of mechanisms that regulate stem cell proliferation, maintenance and differentiation because of their remarkable regenerative capacities and extensive tissue turnover. These two processes are driven by the numerous neoblasts that reside throughout the animal (Reddien and Sánchez Alvarado, 2004). The use of adult stem cells to replace aged or damaged cells and tissues is essential for most metazoans but is still poorly understood at the molecular level.
To date, very few planarian genes have been described to have a function crucial for neoblast biology. Two examples are the Smed-bruli-1 and smedwi-2 genes (Guo et al., 2006; Reddien et al., 2005b). Smed-bruli-1 is not needed for neoblast proliferation in response to wounding and differentiation. Instead, Smed-bruli-1 has been shown to play a role in neoblast self-renewal. Smed-bruli(RNAi) animals are gradually depleted of neoblasts and are therefore unable to regenerate and survive during homeostasis (Guo et al., 2006). By contrast, smedwi-2 is not required for neoblast maintenance but instead is needed for the production of cells capable of replacing aged differentiated cells. In smedwi-2(RNAi) animals, aged differentiated cells are not replaced during homeostasis and regeneration, resulting in the incapacity of these animals to survive and regenerate missing tissues (Reddien et al., 2005b). Few genes have yet been described to have a crucial role in the production of differentiation-committed neoblast progeny cells (Pearson and Sánchez Alvarado, 2010).
The usage of RNAi and lineage markers helps to establish S. mediterranea as a powerful in vivo model system for the study of stem cell proliferation and lineage differentiation during tissue homeostasis and regeneration. Here, we show the existence of a gene that is required for the transition of neoblasts from self-renewing stem cells into more committed non-dividing cells during regeneration and homeostasis.
We found that a CHD4-like gene is required for planarian regeneration and neoblast differentiation. Numerous abnormalities were observed in CHD4(RNAi) animals. These included a robust block in regeneration, failure in the maintenance of differentiated tissues (which normally requires the neoblasts) and ultimately lesions/protrusions in the body of the RNAi animals before animal death. The explanation for the protrusions is unknown. Our data indicate that these protrusions are not explained by an accumulation of proliferating cells or any known neoblast progeny cells. Therefore, this defect might reflect a requirement for CHD4 in differentiated tissue or the aberrant action of unidentified neoblast progeny cells.
CHD4(RNAi) animals cannot regenerate as early as 5 days following the initial dsRNA feeding (see Fig. S8 in the supplementary material). This failure to regenerate occurs rapidly following RNAi when compared with worms where neoblast function has been disrupted by RNAi of other genes (Reddien et al., 2005b). The inability of CHD4(RNAi) animals to regenerate and to replace normally aging differentiated tissue cannot be explained by a loss of neoblasts because these animals initially have normal or slightly higher numbers of actively dividing neoblasts (smedwi-1-expressing cells). By contrast, our data indicate that the failure to regenerate and to maintain normal tissue turnover is due to a marked reduction in numbers of neoblast progeny cells (such as cells expressing Category 3 genes). Furthermore, our results indicate that the reduced number of neoblast progeny cells found in CHD4(RNAi) animals was due to a defect in the generation of these cells as opposed to a defect in their maintenance. The neoblasts of CHD4(RNAi) animals produced immediate progeny cells (not expressing Category 3 genes) that transiently accumulated, presumably as a consequence of the differentiation failure. Because no other markers besides the Categories 2 and 3 markers used in this study have been identified to be expressed in neoblast progeny cells, and no accumulation of germ cells has been observed in CHD4(RNAi) animals (not shown), it remains unresolved if neoblasts in CHD4(RNAi) animals differentiate into a particular lineage different from the ones tested here. At least some neoblast progeny cells have been shown to localize to the blastema (Eisenhoffer et al., 2008); however, perhaps because of the limited number of these cells in CHD4(RNAi) animals, neoblast progeny cells did not localize to the wound sites in appropriate numbers. These observations, together with the finding that CHD4 is expressed near the wound site in neoblasts or neoblast progeny cells 3 days following amputation, suggest a crucial role for CHD4 early in the regeneration process.
Changes in chromatin structure have been implicated in the regulation and maintenance of many cell fate decisions. Chromatin modifiers can be essential to keep stem cells in a pluripotent stage or to drive them into a differentiation pathway. For example, in the case of mouse and human embryonic stem (ES) cells, components of the polycomb complex can silence developmental regulatory genes that are preferentially expressed during differentiation (Boyer et al., 2006; Lee et al., 2006). These results suggest that polycomb proteins maintain the pluripotent state of ES cells. Ezh, a component of the PRC2 polycomb complex, is also active in epidermal progenitors (Ezhkova et al., 2009). However, in general, how the regulation of differentiation in ES cells corresponds to what occurs in adult stem cell types in vivo remains unresolved. In the case of the NuRD complex component CHD4, our data indicate a role in promoting, rather than repressing, stem cell differentiation. The NuRD complex has been shown to regulate cell fate decisions and development in multiple contexts. In D. melanogaster for example, Mi-2 can function with polycomb genes to maintain repression of homeotic genes during embryonic patterning (Kehle et al., 1998). In C. elegans, the CHD4 homolog let-418, is required for vulval cell fate determination and for maintenance of germline-soma distinctions (Unhavaithaya et al., 2002; von Zelewsky et al., 2000). Specifically, in let-418-defective animals, somatic cells express germ cell markers (e.g. the P granule component PGL-1 and Vasa homologs), indicating that a CHD4 protein in C. elegans is required to repress germline cell identity in differentiated cells. Interestingly, a CHD4 homolog in the plant A. thaliana, pickle, is required for repression of embryonic-like characteristics after germination (Ogas et al., 1997; Ogas et al., 1999). In pickle mutants, the fatty acid composition of roots was shown to resemble that of seeds, and roots formed callus growths and embryo-like physical structures. Therefore, in both C. elegans and Arabidopsis, these data suggest a function in repression of germ cell/embryonic-like features in differentiated tissue. It will be interesting to determine in the future how these roles compare with the action of CHD4 in stem cells in other organisms. For instance, it is possible that CHD4 represses stem cell/embryonic-like genes in differentiated tissues in these organisms and in planarian neoblasts to promote neoblast differentiation.
In the mouse, knockout of Mi-2β (CHD4) has complex impacts on the functioning of hematopoietic stem cells (HSCs) (Yoshida et al., 2008). Mi-2β-deficient HSCs displayed increased proliferation and were able to begin differentiation into erythroid, but not myeloid and lymphoid, lineages. Specifically, conditional knockout Mi-2β animals showed depletion of granulocytes (myeloid) and newly formed B cells (lymphocytes). Furthermore, although pro-erythroblasts were formed, the differentiation of these pro-erythroblasts into basophilic and other cells was aberrant in these mice. Therefore, in the case of this stem cell type, Mi-2β appears to be required for lineage decisions that could reflect a role in differentiation. Expression analyses indicated that Mi-2β did not play a global role in gene expression regulation but affected specific transcripts. The gene expression defects in these Mi-2β-deficient HSCs also indicated a potential additional role for Mi-2β in self-renewal. A different component of the NuRD complex, Mbd3, was found to be required for ES cells to differentiate in vitro (Kaji et al., 2006); Mbd3 was also required for proliferation of epiblast cells in culture and derivation of ES cells from inner-cell-mass cells (Kaji et al., 2007). The data from adult planarian neoblasts described here, taken in the context of the results describing the function of CHD4 and other NuRD components in other organisms, suggest that a major and broadly utilized role for this complex is in promoting stem cell differentiation.
Supplementary Material
Acknowledgments
We thank Laurie Boyer, Ezequiel Alvarez-Saavedra, Daniel Wagner and all members of the Reddien lab for manuscript comments, support and discussions. We thank Danielle Wenemoser for collaboration with microarray experiments and the SMEDWI-1 antibody. This work was supported by NIH R01GM080639. We acknowledge support by the Keck, Smith, Searle and Rita Allen Foundations. M.L.S. was supported by a Jane Coffin Childs Memorial Fund Fellowship. P.W.R. is an Early Career Scientist of the Howard Hughes Medical Institute and holds a Thomas D. and Virginia W. Cabot Career Development Professorship. Deposited in PMC for release after 6 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.042051/-/DC1
References
- Ahringer J. (2000). NuRD and SIN3 histone deacetylase complexes in development. Trends Genet. 16, 351-356 [DOI] [PubMed] [Google Scholar]
- Baguñà J. (1976). Mitosis in the intact and regenerating planarian Dugesia mediterranea n.sp. I. Mitotic studies during growth, feeding and starvation. J. Exp. Zool. 195, 53-64 [Google Scholar]
- Bardeen C. R., Baetjer F. H. (1904). The inhibitive action of the Roentgen rays on regeneration in planarians. J. Exp. Zool. 1, 191-195 [Google Scholar]
- Becker P. B., Horz W. (2002). ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 71, 247-273 [DOI] [PubMed] [Google Scholar]
- Bibikova M., Laurent L. C., Ren B., Loring J. F., Fan J. B. (2008). Unraveling epigenetic regulation in embryonic stem cells. Cell Stem Cell 2, 123-134 [DOI] [PubMed] [Google Scholar]
- Bonifer C., Hoogenkamp M., Krysinska H., Tagoh H. (2008). How transcription factors program chromatin-lessons from studies of the regulation of myeloid-specific genes. Semin. Immunol. 20, 257-263 [DOI] [PubMed] [Google Scholar]
- Boyer L. A., Plath K., Zeitlinger J., Branbrink T., Medeiros L. A., Lee T. I., Levine S. S., Wernig M., Tajonar A., Ray M. K., et al. (2006). Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441, 349-353 [DOI] [PubMed] [Google Scholar]
- Denslow S. A., Wade P. A. (2007). The human Mi-2/NuRD complex and gene regulation. Oncogene 26, 5433-5438 [DOI] [PubMed] [Google Scholar]
- Dubois F. (1949). Contribution á l ’ètude de la migration des cellules de règènèration chez les Planaires dulcicoles. Bull. Biol. Fr. Belg. 83, 213-283 [Google Scholar]
- Eisenhoffer G. T., Kang H., Sánchez Alvarado A. (2008). Molecular analysis of stem cells and their descendants during cell turnover and regeneration in the planarian Schmidtea mediterranea. Cell Stem Cell 3, 327-339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E., Pasolli H. A., Parker J. S., Stokes N., Su I. H., Hannon G., Tarakhovsky A., Fuchs E. (2009). Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell 136, 1122-1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E., Horsley V. (2008). More than one way to skin. Genes Dev. 22, 976-985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopoulos K. (2002). Haematopoietic cell-fate decisions, chromatin regulation and ikaros. Nat. Rev. Immunol. 2, 162-174 [DOI] [PubMed] [Google Scholar]
- Guo T., Peters A. H., Newmark P. A. (2006). A Bruno-like gene is required for stem cell maintenance in planarians. Dev. Cell 11, 159-169 [DOI] [PubMed] [Google Scholar]
- Hayashi T., Asami M., Higuchi S., Shibata N., Agata K. (2006). Isolation of planarian X-ray-sensitive stem cells by fluorescence-activated cell sorting. Dev. Growth Differ. 48, 371-380 [DOI] [PubMed] [Google Scholar]
- Kaji K., Caballero I. M., MacLeod R., Nichols J., Wilson V. A., Hendrich B. (2006). The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat. Cell Biol. 8, 285-292 [DOI] [PubMed] [Google Scholar]
- Kaji K., Nichols J., Hendrich B. (2007). Mbd3, a component of the NuRD co-repressor complex, is required for development of pluripotent cells. Development 134, 1123-1132 [DOI] [PubMed] [Google Scholar]
- Kehle J., Beuchle D., Treuheit S., Christen B., Kennison J. A., Bienz M., Muller J. (1998). dMi-2, a hunchback-interacting protein that functions in polycomb repression. Science 282, 1897-1900 [DOI] [PubMed] [Google Scholar]
- Kim J., Sif S., Jones B., Jackson A., Koipally J., Heller E., Winandy S., Viel A., Sawyer A., Ikeda T., et al. (1999). Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity 10, 345-355 [DOI] [PubMed] [Google Scholar]
- Kingston R. E., Narlikar G. J. (1999). ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 13, 2339-2352 [DOI] [PubMed] [Google Scholar]
- Kouzarides T. (2007). Chromatin modifications and their function. Cell 128, 693-705 [DOI] [PubMed] [Google Scholar]
- Lee T. I., Jenner R. G., Boyer L. A., Guenther M. G., Levine S. S., Kumar R. M., Chevalier B., Johnstone S. E., Cole M. F., Isono K., et al. (2006). Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125, 301-313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Grand F., Rudnicki M. A. (2007). Skeletal muscle satellite cells and adult myogenesis. Curr. Opin. Cell Biol. 19, 628-633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Carey M., Workman J. L. (2007). The role of chromatin during transcription. Cell 128, 707-719 [DOI] [PubMed] [Google Scholar]
- Lunyak V. V., Rosenfeld M. G. (2008). Epigenetic regulation of stem cell fate. Hum. Mol. Genet. 17, R28-R36 [DOI] [PubMed] [Google Scholar]
- Narlikar G. J., Fan H. Y., Kingston R. E. (2002). Cooperation between complexes that regulate chromatin structure and transcription. Cell 108, 475-487 [DOI] [PubMed] [Google Scholar]
- Newmark P. A., Sánchez Alvarado A. (2000). Bromodeoxyuridine specifically labels the regenerative stem cells of planarians. Dev. Biol. 220, 142-153 [DOI] [PubMed] [Google Scholar]
- Newmark P. A., Reddien P. W., Cebria F., Sánchez Alvarado A. (2003). Ingestion of bacterially expressed double-stranded RNA inhibits gene expression in planarians. Proc. Natl. Acad. Sci. USA 100, 11861-11865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng H. H., Bird A. (2000). Histone deacetylases: silencers for hire. Trends Biochem. Sci. 25, 121-126 [DOI] [PubMed] [Google Scholar]
- Ogas J., Cheng J. C., Sung Z. R., Somerville C. (1997). Cellular differentiation regulated by Gibberellin in the Arabidopsis thaliana pickle mutant. Science 277, 91-94 [DOI] [PubMed] [Google Scholar]
- Ogas J., Kaufmann S., Henderson J., Somerville C. (1999). PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc. Natl. Acad. Sci. USA 96, 13839-13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin S. H., Zon L. I. (2008). Hematopoiesis: an evolving paradigm for stem cell biology. Cell 132, 631-644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson B. J., Sánchez Alvarado A. (2010). A planarian p53 homolog regulates proliferation and self-renewal in adult stem cell lineages. Development 137, 213-221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson B. J., Eisenhoffer G. T., Gurley K. A., Rink J. C., Miller D. E., Sánchez Alvarado A. (2009). Formaldehyde-based whole-mount in situ hybridization method for planarians. Dev. Dyn. 238, 443-450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien P. W., Sánchez Alvarado A. (2004). Fundamentals of planarian regeneration. Annu. Rev. Cell Dev. Biol. 20, 725-757 [DOI] [PubMed] [Google Scholar]
- Reddien P. W., Bermange A. L., Murfitt K. J., Jennings J. R., Sánchez Alvarado A. (2005a). Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Dev. Cell 8, 635-649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien P. W., Oviedo N. J., Jennings J. R., Jenkin J. C., Sánchez Alvarado A. (2005b). SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells for regeneration and homeostasis. Science 310, 1327-1330 [DOI] [PubMed] [Google Scholar]
- Saló E., Baguñà J. (1984). Regeneration and pattern formation in planarians. I. The pattern of mitosis in anterior and posterior regeneration in Dugesia (G) tigrina, and a new proposal for blastema formation. J. Embryol. Exp. Morphol. 83, 63-80 [PubMed] [Google Scholar]
- Sánchez Alvarado A., Newmark P. A., Robb S. M., Juste R. (2002). The Schmidtea mediterranea database as a molecular resource for studying platyhelminthes, stem cells and regeneration. Development 129, 5659-5665 [DOI] [PubMed] [Google Scholar]
- Seelig H. P., Renz M., Targoff I. N., Ge Q., Frank M. B. (1996). Two forms of the major antigenic protein of the dermatomyositis-specific Mi-2 autoantigen. Arthritis Rheum. 39, 1769-1771 [DOI] [PubMed] [Google Scholar]
- Shimono Y., Murakami H., Kawai K., Wade P. A., Shimokata K., Takahashi M. (2003). Mi-2 beta associates with BRG1 and RET finger protein at the distinct regions with transcriptional activating and repressing abilities. J. Biol. Chem. 278, 51638-51645 [DOI] [PubMed] [Google Scholar]
- Tong J. K., Hassig C. A., Schnitzler G. R., Kingston R. E., Schreiber S. L. (1998). Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature 395, 917-921 [DOI] [PubMed] [Google Scholar]
- Unhavaithaya Y., Shin T. H., Miliaras N., Lee J., Oyama T., Mello C. C. (2002). MEP-1 and a homolog of the NURD complex component Mi-2 act together to maintain germline-soma distinctions in C. elegans. Cell 111, 991-1002 [DOI] [PubMed] [Google Scholar]
- von Zelewsky T., Palladino F., Brunschwig K., Tobler H., Hajnal A., Muller F. (2000). The C. elegans Mi-2 chromatin-remodelling proteins function in vulval cell fate determination. Development 127, 5277-5284 [DOI] [PubMed] [Google Scholar]
- Wade P. A., Jones P. L., Vermaak D., Wolffe A. P. (1998). A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr. Biol. 8, 843-846 [DOI] [PubMed] [Google Scholar]
- Wagers A. J., Weissman I. L. (2004). Plasticity of adult stem cells. Cell 116, 639-648 [DOI] [PubMed] [Google Scholar]
- Weissman I. L., Anderson D. J., Gage F. (2001). Stem and progenitor cells: origins, phenotypes, lineage commitments, and transdifferentiations. Annu. Rev. Cell Dev. Biol. 17, 387-403 [DOI] [PubMed] [Google Scholar]
- Williams C. J., Naito T., Arco P. G., Seavitt J. R., Cashman S. M., De Souza B., Qi X., Keables P., Von Andrian U. H., Georgopoulos K. (2004). The chromatin remodeler Mi-2beta is required for CD4 expression and T cell development. Immunity 20, 719-733 [DOI] [PubMed] [Google Scholar]
- Yoshida T., Hazan I., Zhang J., Ng S. Y., Naito T., Snippert H. J., Heller E. J., Qi X., Lawton L. N., Williams C. J., et al. (2008). The role of the chromatin remodeler Mi-2beta in hematopoietic stem cell self-renewal and multilineage differentiation. Genes Dev. 22, 1174-1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., LeRoy G., Seelig H. P., Lane W. S., Reinberg D. (1998). The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 95, 279-289 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.