Abstract
Recent humanitarian efforts have led to the widespread release of antiretroviral drugs for the treatment of the more than 33 million HIV afflicted people living in resource-scarce settings. Here, the enumeration of CD4+ T lymphocytes is required to establish the level at which the immune system has been compromised. The gold standard method used in developed countries, based on flow cytometry, though widely accepted and accurate, is precluded from widespread use in resource-scarce settings due to its high expense, high technical requirements, difficulty in operation-maintenance and the lack of portability for these sophisticated laboratory-confined systems. As part of continuing efforts to develop practical diagnostic instrumentation, the integration of semiconductor nanocrystals (quantum dots, QDs) into a portable microfluidic-based lymphocyte capture and detection device is completed. This integrated system is capable of isolating and counting selected lymphocyte sub-populations (CD3+CD4+) from whole blood samples. By combining the unique optical properties of the QDs with the sample handling capabilities and cost effectiveness of novel microfluidic systems, a practical, portable lymphocyte measurement modality that correlates nicely with flow cytometry (R2 = 0.97) has been developed. This QD-based system reduces the optical requirements significantly relative to molecular fluorophores and the mini-CD4 counting device is projected to be suitable for use in both point-of-need and resource-scarce settings.
Introduction
Of the more than 33 million people infected with HIV globally, more than 85% live in developing countries with significant resource limitations. While portable lateral immunoassay kits are available and can be used to diagnose patients with HIV, a missing link remains with respect to the treatment and management of HIV patients in these developing countries.1,2
As HIV infection progresses into active AIDS, clinicians must frequently monitor the patient’s CD4+ T lymphocytes to assess the degree of immunological deterioration and determine the effectiveness of antiretroviral therapy.3,4 According to the Centers for Disease Control and Prevention guidelines, absolute T cell counts of 200 cells/μL or 14% CD3+CD4+ serve as the reference values at which HIV is considered to become active AIDS.5,6 To determine the status of immune function, a multi-color signaling approach to identify and differentiate between the different cluster of differentiation (CD) markers of interest, is typically obtained using flow cytometers (FC), or on double platforms involving FC and hematology analyzers.
A number of more compact FC instruments have been developed, such as Guava EasyCD4 and the PointCare instrument, however, these alternative flow methods have not been widely adopted.7,8 Nonflow alternatives have also been developed based on impedance, microfluidics, or magnetic beads, yet these methods suffer from lower throughput and more labor-intensive manipulations.9-13 These alternatives are also less accurate than FC and have not received endorsement from the World Health Organization.9,12 The development of increasingly smaller bio-analysis devices is attractive for several factors, including enhanced portability, decreased sample size, and reduced turnaround times.14-17 Nano-bio technologies, advanced microfluidics, and lab-on-a-chip sensor systems offer interesting and exciting new prospects for these humanitarian diagnostic challenges.18 However, despite the remarkable advances in the development of miniaturized sensing, and analytical components for use in a variety of biomedical and clinical applications, the ability to assemble and interface individual components in order to achieve a high level of integration in complete working systems continues to pose daunting challenges for the scientific community as a whole. For example, the optical requirements present in many miniaturized systems frequently challenge their development.
Fluorescence, as a common transduction analytical signal, presents unique opportunities, as well as demands, in a miniaturized assay platform. A robust excitation source and high performance filters, used in conjunction with ideal reagent components, to generate a strong target-specific signal and a reduced non-specific background, is crucial for the miniaturized system’s ability to detect the intended target with sensitivity and accuracy. Current efforts to overcome the optical challenges present in point-of-need devices include advanced excitation sources such as lasers and powerful light emitting diodes (LEDs), miniaturized charge coupled devices (CCDs), as well as next generation fluorophores.
For the analyte label area, one of the most promising fluorescent probes is the semiconductor nanocrystal quantum dot (QD).19 These inorganic compounds are synthesized from group II–VII elements through a number of synthetic routes with CdSe particles being the most common.20,21 One or more passivation layers are generally coated around the fluorescent core serving to increase hydrophilicity, reduce agglomeration and increase fluorescence intensity, resulting in particles with diameters between 5–20 nm depending on the size of the original core semiconductor.22-26 Attributes, such as long-term photostability, resistance to quenching, and narrow emission profiles, have made QDs a popular alternative to traditional organic fluorophores, which often suffer from photobleaching, broad bandwidth emission, and specific excitation ranges.27,28
In contrast, QDs possess a broad excitation spectrum spanning the UV and near visible range, and their emission wavelength is dependent solely on the size of the inorganic core, with larger particles fluorescing closer to the IR, and the smaller, at shorter wavelengths. The resulting extremely long Stokes shift combined with narrow emission profiles and the ability to simultaneously excite a number of different size nanoparticles with characteristic emission wavelengths, make QDs ideal fluorophores for multiplexed experiments such as CD3+CD4+ dual labeling.
Coupling QDs to biological recognition moieties, such as antibodies, remains a challenge and the area continues to evolve with a number of different techniques now being explored in the literature.29,30 These efforts have contributed to some bio-assay successes whereby QDs have demonstrated their use in such applications as Förster resonance energy transfer (FRET),31 cellular imaging,32-34 immunolabeling,35,36 and protein assays.37,38 Although not all areas have harnessed the full capability of QDs, these particles have emerged as strong candidates for signal generation in devices dedicated to a number of important diagnostic applications.39 Currently, conjugation strategies involve diverse reagents and methodologies, including electrostatic interactions,40 avidin/biotin affinity,41 and a variety of covalent techniques.30,42 Current promising conjugation schemes include 1-ethyl-3-[3-dimethylamino propyl]carbodiimide/N-hydroxysulfosuccinimide (EDC/NHS) coupling of carboxyl coated QDs to free antibody amines,43 creating free sulfhydryl on the biomolecule with succinimidyl 4-(N-maleimidomethyl) cyclohexanecarboxylate (SMCC) facilitating QD linkage,44 and hydrazine modified QDs with periodate oxidized antibodies.45 Common to all covalent conjugation schemes is a modification of both the antibody structure and of the QD surface, with an inherent risk to alter the recognition specificity of the biomolecule and fluorescence characteristics of the nanoparticle. No clear universal bioconjugation methodology has evolved to date. Rather, the most appropriate methods have shown to be dependent upon the specific end application and have been defined on a case-by-case basis.
For over a decade, our laboratory has sustained work utilizing microelectromechanical systems (MEMS)-based components for studies of the design, fabrication, and testing of microfluidic, nano-bio-chip (NBC) structures with integrated fluid handling and optical detection capabilities.46 The NBC ensembles consist of a closed, miniaturized system (chip) that allows for specific biological analytes (bio) to be immobilized from complex matrices and quantitated via a colorimetric or fluorescent signal arising from nanoparticles that are entrapped in a nano-net generated in agarose microspheres (nano). Immobilization of the target entity is performed either via an antigen- or antibody-specific bead array or an analyte size-specific membrane. Sample types amenable to the NBC include, but are not limited to, serum, plasma, saliva, and whole blood, as well as environmental samples. These structures have been shown to be suitable for the identification and quantitation of C-reactive protein,47 other cardiac risk markers,48 pH, and physiologically relevant cations.49 More recently, we have developed miniaturized microfluidic systems that service cellular analysis applications.50 These membrane-based micro-systems have been shown to be suitable for the determination of lymphocytes for HIV immune function testing and total white blood cell counts for cardiac risk assessment.51,52
In this manuscript, efforts are devoted to creating a bridge between QD-based detection methods and the integrated membrane-based NBC systems. The efforts strive to relieve the optical requirements and reduce the overall engineering demands for both the integrated NBC sample processing modality along with the associated fluorescence-based detection system. Specifically, the photostability of QDs and the ability to excite and image two different colors concurrently with a single epifluorescent arrangement are exploited to reduce the optical pathway and thus decrease the overall analyzer size. These efforts strive to bring these evolving diagnostic tools one step closer to widespread use in resource-scarce settings.
Materials and methods
Reagents and samples
Primary mouse antibody specific to CD3 (OEM, Toms River, NJ) and rat antibody to CD4 (Genetex, San Antonio, TX) were purchased and centrifuged briefly prior to use. Goat anti-mouse secondary antibody conjugated to QD with peak fluorescence intensity at 655 nm (QD 655), goat anti-rat QD 565, and goat anti-mouse QD 565 (Invitrogen, Carlsbad, CA), all H + L and highly cross-absorbed, were obtained. For comparison studies, goat anti-mouse Alexa Fluor 647, goat anti-mouse Alexa Fluor 488, and goat anti-mouse FITC (Invitrogen) were also procured. Fixative and FC reagents were also acquired (Beckman Coulter, Fullterton, CA). InSpeck calibration beads with a green-channel fluorescence fingerprint similar to that of FITC, and reagents for a SMCC and EDC/NHS facilitated conjugation were purchased from Invitrogen. For the EDC conjugation, EviTag Hops Yellow QDs were purchased from Evident Technologies (Troy, NY). These conjugations were performed as described previously.29,33
Whole blood samples were collected from anonymous donors at the University of Texas at Austin in accordance with Institutional Review Board procedures by standard venipuncture technique using dilute EDTA as an anticoagulant. Samples were evaluated within 24 hours of collection.
Nano-bio-chip construction
The research grade NBC ensemble was constructed from a polymethyl methacrylate (PMMA) base machined in-house. Stainless steel support frits were photofabricated and treated with a non-reflective coating. Vinyl adhesive layers were precision-cut according to evolving computer-aided designs and served to focus fluid flow through the nucleopore track-etched membrane (Whatman/GE Healthcare, Piscataway, NJ). Stainless steel conduits were purchased from Small Parts, Inc. (Miramar, FL). Each unit was typically assembled in under five minutes. The research prototype was affixed beneath a customized optical station (see ESI‡).
The integrated, disposable NBC was constructed from a plastic base and cover and various levels of precision cut laminate adhesive (part number 9500PC, 3M, St. Paul, MN). A Luer lock adapter served as a sample introduction port. Home-made polydimethylsiloxane (PDMS)-based blister packs with an internal volume of ~500 μL containing liquid reagents and buffers, were affixed onto the card and their output directed into the fluid channels created by the adhesive layers. Fluidic networks were composed of channels of various widths from 200 to 2000 μm. The membrane cell capture mechanism in the integrated NBC remained the same. Waste reservoirs were created by forming chambers between the top and bottom layers.
Cell immunolabeling
Aliquots of 50 μL from a thoroughly homogeneous whole blood reservoir were briefly treated with 5 μL Cyto-Chex fixative (Streck, Omaha, NE) before dilution to 1 mL with 1% bovine serum albumin in phosphate buffered saline (PBS/BSA) purchased from Sigma Aldrich (St. Louis, MO) and Thermo Scientific (Waltham, MA), respectively. The prepared sample was injected into the NBC ensemble with a flow rate sufficient to separate the lymphocytes from the remaining blood components. The sample was then subjected to a 500 μL treatment of 10 μg/mL primary antibody at a flow rate of ~17 μL/min for 10 minutes. Unbound antibody was subsequently removed with PBS rinse at which point 500 μL of 10 nM secondary antibody was introduced, incubated, and rinsed similarly to the conditions described above. Alternatively, this labeling could be performed in microcentrifuge tubes prior to delivery to the NBC; total assay time was under 30 minutes. For SEM images, the membrane was removed from the NBC and fixed with 4% glutaraldehyde followed by ultrapure water washes. This preparation was additionally stabilized with OsO4 and hexamethyldisilazane.
As a reference method, flow cytometry data was collected on a Cytomics FC 500 (Beckman Coulter, Fullerton, CA) using a protocol provided by the manufacturer. Aliquots from the same blood reservoir were prepared with Immuno-Prep lysing reagents (Beckman Coulter) and labeled with CD3-Alexa Fluor 488 and CD4-Alexa Fluor 647 (Invitrogen). Immuno-Trol and Cyto-Comp standards and Flow-Count calibration microspheres (Beckman Coulter) as well as mouse IgG negative control Alexa Fluor 647 and 488 (Serotec, Raleigh, NC) were used for negative control experiments and instrument calibration.
Results
Nano-bio-chip devices
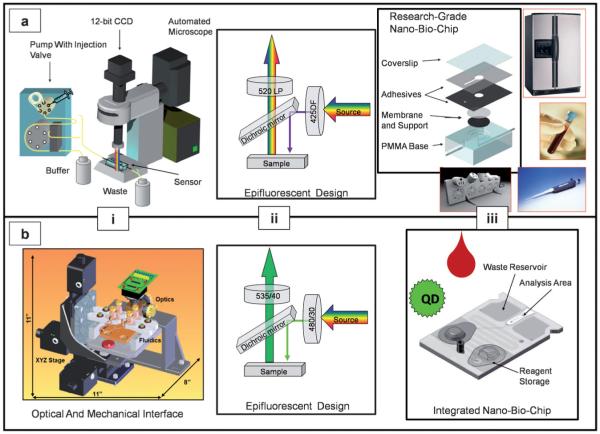
Through our previous studies, we defined the conceptual basis for cell counting in a microfluidic packaged membrane-based system using the benchtop microscope seen in Fig. 1(a). Like most current bioMEMs sensors, this system utilizes external sample processing steps, such as pipette-based liquid transfer. Fluid motion inside the device is controlled with external pumps and manual volume metering of venipuncture blood specimens is employed. Refrigerated liquid-phase antibody reagents, in tandem with molecular fluorophores, are utilized to complete the cell counting assays. These experimental manipulations, while arguably more convenient than flow cytometry, still require a lab-based infrastructure and trained operators, both of which are problematic for use in resource-scarce settings.
Fig. 1.
Schematics are provided for the subject matter related to analyzer and NBC condensation. (a) The bench top model is used to optimize individual components of the overall miniaturized device and employs an automated microscope (i), in tandem with support components including Hg lamp, CCD camera, peristaltic pump and other fluid handling elements. The adapted microscope used herein contains an epifluorescent arrangement customized for QD image capture (ii). This long-pass filter is selective for light above 520 nm which, in tandem with the simultaneous excitation of two different QD color regimes, results in a reduction of optical hardware by one half. (iii) The cell capture device is constructed from a custom-machined PMMA base fitted with stainless steel inlet and outlet ports onto which a support frit is placed. The base structure is fitted with a polycarbonate track etched membrane for lymphocyte capture and stabilization. This membrane is sealed to the underlying support via two layers of double-sided adhesive acrylic laminate. (iii) Insets: requirements of the bench-top microscope analysis system include refrigeration for reagents, fluid metering secondary hardware, and a reliable power supply. (b) After optimization, individual components are integrated into the prototype self-contained analysis platform that combines battery power supply, optics, and image analysis software into a portable device (i). This device employs singly selective fluorescent filter cubes specific to each color channel (ii). Sample metering, on-board reagent supply, and waste disposal are all contained within this highly integrated NBC system (iii).
Since the initial report, we have devoted significant efforts to develop a simplified, compact, yet highly integrated measurement tool set that is shown in Fig. 1(b). The new integrated sample processing and detection modalities shown in Fig. 1(b)i contains both mechanical and optical components such as battery supply, compact LED illumination package, and minimicroscope. Complementary to this analyzer is a disposable labcard (Fig. 1(b)iii) containing reagents, fluid-holding blister packs, waste reservoir, and plastic mother board with integrated microfluidic circuitry that is compatible with high fidelity fluorescence measurements. This selection of components eliminates the requirements for external pumps, high voltage power supplies, pipetting steps, waste handling, and antibody storage requirements. Sample collection is performed via a needle stick to finger with transfer of blood to labcard by capillary tube and antibody reagents and labels are stored within the cartridge as solid layers. This last specification serves to eliminate the cold chain, i.e. refrigeration, which creates a significant burden in resource-scarce settings.
In Fig. 1(b)ii the optical arrangement of excitation, emission, and beam filtering optics inherent to an epifluorescent design is shown. Optical wavelength requirements are determined by the molecular structure, and are specific to each fluorophore. In a multi-color analysis, separate filter cubes would be necessary for each color regime. In contrast, the QD-based system utilizes a reduced optical pathway (Fig. 1(a)ii) in which a single UV excitation source and a long-pass emission filter replaces the stringent optical hardware requirements of multiple organic fluorophores. More details on the construction of the integrated NBC will be reported elsewhere. In this report, the focus is the integration of nanoparticle QDs into a NBC sensor system for the determination of CD4+ T lymphocyte counts and percentages using unprocessed whole blood samples.
Complementary to the condensed optics, the NBC miniaturized cell capture device serves to sequester specific cells of interest from the whole blood matrix. The NBC used herein was constructed internally and is based on an evolution from designs described previously, and illustrated in Fig. 1(a)iii.50,51 The rectangular base houses the complete assembly including the support frit, which serves to maintain a leveled surface and provide for membrane integrity, while allowing for waste flow through. A polycarbonate track-etched membrane with pore size determined by experiment is positioned upon the support. For this lymphocyte-specific application, 3 μm diameter holes within the membrane serve a dual role of both retaining structures larger than that of the pores while allowing the sample matrix to pass through. Thus, the more rigid and structured lymphocytes become immobilized here while the remaining blood components including the more supple erythrocytes and small platelets, as well as plasma components, are allowed to pass; effectively isolating the cells of interest while allowing the remaining blood components and unbound antibody to be relegated to waste without lysis or additional preparatory steps (Fig. 2). Capture efficiency for lymphocytes is greater than 95%. This was determined by injecting the sample eluate, or flow through, into a second NBC. After support frit and membrane are inserted, a precision-cut layer of double-sided vinyl adhesive is affixed to the top of the device effectively immobilizing the separation mechanism. The adhesive vinyl layer seals the membrane securely around the surface of the underlying support, and also fastens a cover slip to top the system. The membrane-based NBC is serviced by inlet and outlet channels fitted with stainless steel tubing facilitating fluid flow including sample introduction and waste removal.
Fig. 2.
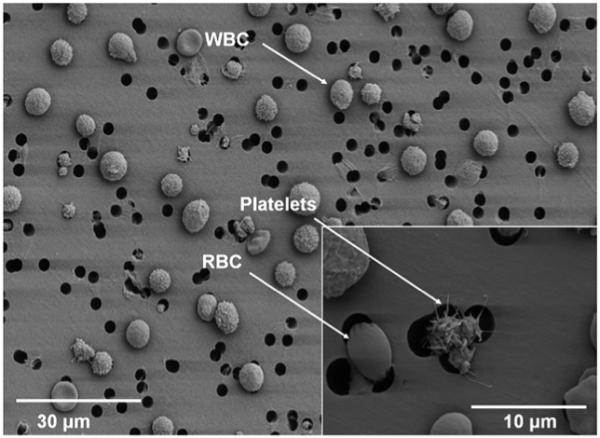
An image recorded by SEM is shown of the blood components immobilized on the capture membrane within the NBC. The more rigid leukocytes are retained by the porous membrane while the more flexible erythrocytes, as well as plasma are relegated to waste. Inset shows platelet aggregate and erythrocyte passing through overlapping pores. Note rarity of pore overlap in lower magnification image.
QD evaluation
For our initial attempts to explore QD-labeled lymphocytes, we completed survey studies using reactions in microcentrifuge tubes whereby direct conjugation schemes were employed including EDC and SMCC-facilitated coupling of antibody to nanoparticle surface. These studies served as a reliable and simple method to decouple the reagent transport issues as they occur within the microfluidic elements, from the intrinsic issues related to the coupling chemistry. Problems with conjugation procedures were noted with respect to nonspecific binding as well as with the formation of precipitates.
The simple conjugation test procedure involved an incubation of 50 μL whole blood with the QD-antibody adduct, subsequent dilution with buffer, and injection of the sample into the membrane-based NBC. After a wash to ensure separation of blood components, images were captured with both a 1 and 3 s exposure. This assay procedure was repeated with 50 μL of buffer replacing blood to serve as a blank control. After these steps the resulting photomicrographs were then analyzed for cell-like objects and precipitates using digital image processing. Parameters of size (70–500 pixels), circularity (0.6–1.0; defined as 4π(area/perimeter2), and intensity (2σ above mean of image) were used to establish cell containing areas of interest (AOIs). A more detailing description of the image analysis protocols used may be found in the ESI.‡
The 2σ discrimination above the background of the image was found to be sufficient to identify 95% of fluorescent objects on the membrane. The pixel size range was determined empirically and evolved from experiments with 6 μm fluorescent polystyrene beads serving as a model for the lymphocytes. This range was sufficiently broad to identify particles immobilized in slightly different focal planes as well as those partially overlapping.
The fluorescent objects including cells and precipitates were further characterized by comparing the signal intensity present in the AOI to the signal of membrane surrounding the area, i.e. the background signal. Variation in this background membrane signal was defined as noise. Signal to background (SBR) ratios could then be determined by subtracting background from signal, with the resulting value divided by background. Signal to noise (SNR) ratios were calculated by taking the background corrected signal over noise. In this application, SBR served as the more relevant figure-of-merit as it was the basis for identification of the AOI as a cell; particles with an SBR well above a value of 1 were considered to be an authentic event (i.e. an actual lymphocyte). Electron microscopy and the use of multiple surface markers were used to confirm the identity of authentic cells as described previously.51
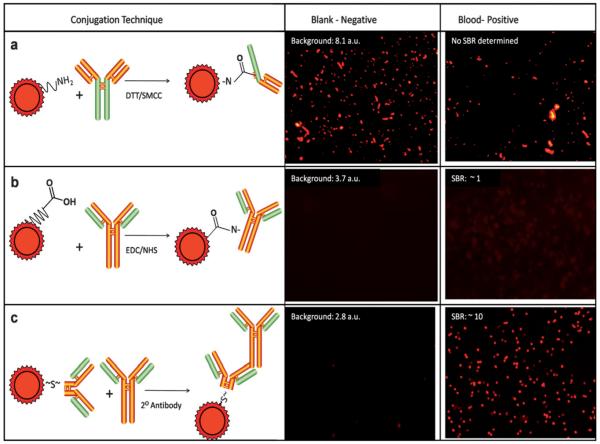
With these standard image analysis methods in hand, the results of different QD–antibody conjugation methods were compared qualitatively and quantitatively to establish information relative to labeling performance for each approach. With the SMCC conjugation method, in addition to particles matching the expected lymphocyte qualification descriptors, additional fluorescent artitifacts with irregular patterns, very large size, and extremely high intensity were seen as shown in Fig. 3(a). Especially important here were the blank experiments in which only QD–antibody conjugate, in the absence of blood, was introduced into the membrane-based NBC. Here, total mean membrane fluorescence without blood was actually slightly higher than in the presence of sample at 8.08 vs. 5.11 a.u., respectively. This aggregation may be due to either passivation layer defect in the QD, denaturation of the biomolecule, or both. The same problematic background issues persisted despite attempts to solve the matter with filtration, sonication, centrifugation, and further purification procedures. Although it is possible that some of the particles in the sample-positive experiment were authentic lymphocytes, discriminating between artifacts and labeled cells using only morphological characteristics was considered to be prone to errors and not reliable.
Fig. 3.
Three QD–antibody conjugation strategies are shown. (a) First, DTT treatment cleaves disulfide bonds within the intact IgG species allowing free sulfhydryl linkage to QD surface after SMCC activation of amino-PEG functionalized nanoparticles. (b) Second, an alternative approach utilizing an NHS-enhanced EDC linkage is illustrated. (c) Third, a secondary antibody based approach using commercially available F(ab’)2 fragments specific to primary antibody, with QD thiol-linked is shown and yielded a strong, specific signal without particle aggregation.
The EDC technique (Fig. 3b), though less problematic in terms of precipitation (negative control mean membrane fluorescence 3.66 a.u.) yielded only vague and dim labeling. This conjugate behaved well for labeling stabilized, FC blood-type products (i.e. commercial blood standards), but obtained results were found to be unacceptable with traditional fresh blood specimens derived from standard venipuncture collection. As linkage to biomolecule is through any free amine, a variety of antibody orientations of mixed avidity are available with this technique, which could potentially reduce specificity.29 Mean SBR ratios were near unity, and although some lymphocyte identification was possible, further optimization was not performed. This poor performance of established conjugation methods may be attributable to the specific structure of the antibodies employed and reinforces the necessity to separately consider each application in terms of conjugation scheme. Although other methods of QD conjugation exist including biotin–avidin interactions and use of a Histag, they were not employed for the purposes of this study.29
Effective in eliminating the obtrusive precipitate while concurrently increasing the overall signal, were commercially available QDs, thiol-linked to secondary antibodies (Fig. 3c). Here, unlabeled primary antibodies specific to CD markers were applied first, followed by a 2 minute on-membrane wash; secondary QD-labeled antibody specific to primary was then applied. Clone specificity of the secondary antibodies to their respective primaries was exploited to allow simultaneous staining of two different lymphocyte populations. To find experimental conditions optimized for highest cell intensity with low membrane staining and low non-specific signal, a series of titrations were performed using varying concentrations of secondary antibody versus a constant primary and vice versa (data not shown); 10 μg/mL primary antibody and 10 nM secondary QD conjugate were used for the remainder of the experiments. These conditions were repeated in the absence of any blood components and a nearly complete lack of precipitate was observed. Negative and positive control mean membrane fluorescence was 2.80 and 5.28 a.u, respectively for QD 655 and 15.01 and 19.56 a.u for QD 565. The SBR ratios were well above 4 for QD 565 and 10 for QD 655.
Optimization and validation
To validate the secondary antibody-QDs, we first labeled blood samples with CD4 mouse primary antibody labeled with Alexa Fluor 488 and then QD 655 secondary antibody specific to the mouse IgG. Red and green color channel overlap was >99%, verifying this secondary antibody QD visualization methodology. When secondary antibodies were incubated with blood samples devoid of primary antibody, and subsequently injected into the NBC, fluorescence signal was reduced by ~90% relative to the positive controls, as the unbound fluorescent antibody is passed on to waste. This background signal is attributable to membrane staining and stray, conspicuous aggregate—both easily identified and removed via image processing.
Monocytes, like CD4+ T lymphocytes, possess the CD4 surface marker and, as such, are a critical consideration in the membrane-based NBC analysis. The presence of monocytes can lead to artificially high CD4 counts. Fig. 4 details both the spatial and intensity gating strategies employed to diminish any artificial monocyte contribution to desired cell count. First, the CD4+ cells are divided into two categories, dim and bright, with only the bright cells subjected to further processing (Fig. 4a). Second, the presence of the CD3 marker is required to confirm the cell’s authenticity as a T lymphocyte (Fig. 4b) Only cells positive for both markers are then counted (Fig. 4c). Monocytes and lymphocytes are clearly separated by the membrane-based NBC into two distinct populations with a binomial distribution as seen in the histogram in Fig. 4d. The distribution gives baseline separation between the two populations of CD4+ cells. Flow cytometry gives a similar distribution, but also contains a low level population of non-specifically stained cells (Fig. 4e). Monocytes in the membrane-based NBC were found to have a fluorescent intensity approximately 4 times less than lymphocytes. This is consistent with their decreased expression of the CD4 epitope relative to the T cells.53
Fig. 4.
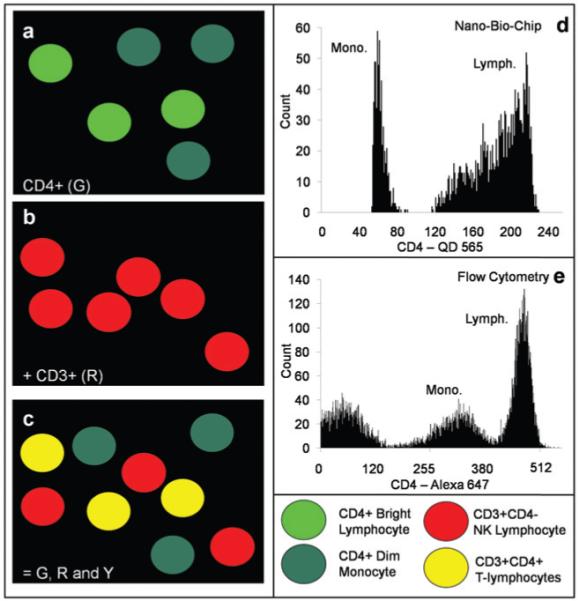
Graphical depiction of cells and the spatial and intensity gating procedures used for selected lymphocyte and monocyte populations. (a) The CD4+ cells fall into two distinct populations: CD4+ bright, lymphocytes, and CD4+ dim, monocytes, as shown in the schematic. The intensity of this CD4-specific signal is an initial descriptor to remove monocytes from analysis. (b) The CD3+ cells include T and NK lymphocytes. (c) Overlap of the two images results in red, green, and yellow cells. Cells doubly positive for red and green, and thus yellow, are the CD4+ T lymphocytes of interest. Cells remaining green are the interfering monocytes. (d) A green color channel 8-bit intensity histogram is shown for the CD4+ cells as analyzed by the membrane-based NBC. The cells are labeled with QD 565. (e) A similar 10-bit histogram is shown for the populations as determined by FC. (FC histogram truncated from 1024 to ~550 to maximize plot area).
As clone specificity between the primary and secondary antibodies is of utmost importance in this dual color staining approach, any non-specific binding within the system was also investigated. Whole blood samples were first incubated with CD4 primary antibody raised in rat; washed, blocked, and incubated with QD 655 labeled secondary antibody specific to mouse. Non-specific signal was 9.1% of specific signal (90.9% reduction of signal) in such a trans-species experiment. The CD3 mouse primary/QD 565 goat anti-rat secondary experiment was performed analogously to that described above with similar results. Thus, for the remainder of the experiments, CD4+ lymphocytes appear green (Fig. 5a) and CD3+ cells red (Fig. 5b). Upon digital overlay of these two images, the dually stained cells of interest emerge yellow (Fig. 5c). Dim green cells in the image are monocytes and cells remaining solely red represent CD8+ killer T lymphocytes and NK cells. Images were also obtained with a long-pass emission filter similar to those acquired with individual filters (Fig. 5d). Image collection and processing time as well as the optical hardware spatial requirements are reduced with the single emission experimental design and SNR and SBR values validated the long-pass filter scheme (further analysis below).
Fig. 5.
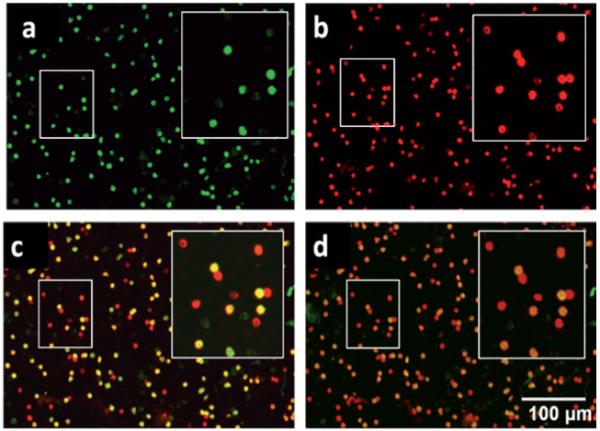
Representative photomicrographs of whole blood labeled with CD specific antibody and QD fluorescently tagged secondary antibody taken with a 10× objective and 3 seconds of exposure time. The QD 565 labels CD4+ cells including monocytes and T lymphocytes in the green channel (a) while QD 655 stains CD3+ lymphocytes red (b), as observed through separate filter cubes specific to each fluorophore. A digital overlap of the red and green images (c) shows monocytes (CD3−CD4+, green), T lymphocytes (CD3+CD4+, yellow) resulting from signal both in red and green channels, and remaining NK and CD8+CD3+ T-killer lymphocytes (CD3+CD4−, red). An alternative approach (d) utilizes a long-pass emission filter cube allowing for a single capture event to produce a similar image to that generated by separate photomicrographs. Insets illustrate highlighted areas with 2× digital zoom.
Absolute determinations
Having validated that the fluorescent particles seen on the membrane were indeed authentic lymphocytes of identifiable subsets, next the ability of the membrane-based NBC system to count QD-labeled cells was explored. Critical to this success was a NBC design in which a near uniform distribution of sample is achieved across the entire membrane area. To evaluate whether this was indeed the case, photomicrographs in 5 different fields of view (FOVs) across the membrane were captured, with one in the center of the membrane and 4 additional locations arranged orthogonally from each other and surrounding the center image. This consistent method of defining FOVs across the membrane involves analysis areas both in the center and near the edges of the membrane and creates a broad sampling of surface area. We have investigated the use of different membrane sizes, pore sizes, and flow rates and found that with the parameters used herein, low CVs were routinely obtained, consistent with our previous work.52 Cell counting and profiling studies were done to show that a small fraction of the membrane could be used to accurately represent the total count within the entire volume. Standard deviation and mean when 5 FOVs were used result in a %CV value that was typically 5–10% (see below). These measurements serve to establish that the cell distribution is optimized across the membrane structure. No further decrease in variance was seen upon the inclusion of more than 5 FOVs.
Also critical was the consistency in the background autofluorescence of the membrane, as this served to define the SBR ratio. Autofluorescence is reduced in the red QD 655 channel, relative to the green QD 565 channel. Fortunately, this was negated by the high QD 565 signal which still allowed high SBR measurements. An alternative method used image capture before introduction of sample and ensuing background subtraction.
To demonstrate the ability of the membrane-based NBC ensemble to count across a wide linear dynamic range of cells, increasing volumes of whole blood ranging from 5 to 100 μL were stained for CD4 cells and injected into the mini cell capture system. The resulting cell count values (see ESI‡ for analysis protocol) were plotted versus volume and yielded a linear response over this sample volume range (R2 = 0.99, Fig. 6a). The linearity of this dose response curve provides strong evidence for the analytical capabilities of the approach and suggests that the fluorescent nanoparticles and automated digital counting methodologies here defined may be used to measure lymphocyte populations under these conditions. As might be expected, at either very high or very low blood volumes, the %CV of the measurements increases. This behavior is due to overlap and clumping at the high end, and lack dissemination across the entire membrane at low volumes. For the remainder of the experiments 50 μL of sample was used.
Fig. 6.
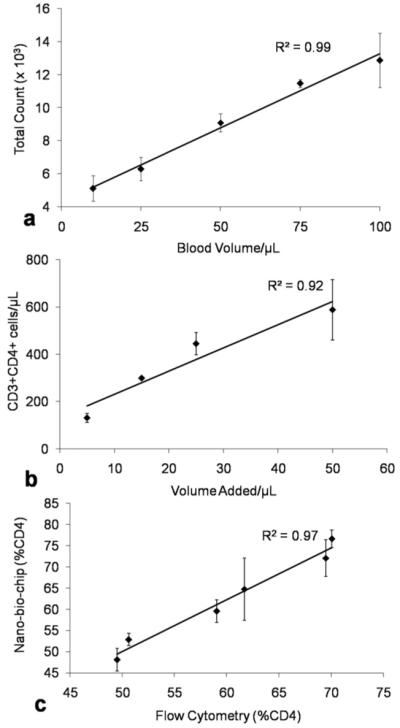
Whole blood of varying volume was subjected to the staining process and membrane-based NBC analysis. (a) Undiluted blood was stained for CD4+ lymphocytes and counts per field of view translated into total number of cells distributed across the entire membrane. (b) To simulate immuno-compromised samples, blood volumes were subjected to PBS/BSA dilution to a total volume of 50 μL and stained for CD3+CD4+ lymphocytes with total membrane area and dilution correction factors utilized to estimate cells per μL in the original 50 μL sample. (c) Small pilot study comparison of the membrane-based NBC method to flow cytometry for %CD4 determinations correlates at R2 = 0.97.
To simulate the ability to perform cell counts over the range of values typically encountered in both healthy and immunocompromised individuals, a dilution study was performed in which blood volumes of 5–50 μL were diluted with 1% PBS/BSA to a total volume to 50 μL. Cells were stained for both CD3 and CD4 and doubly positive cells (yellow) from 5 FOVs were counted. This was again extrapolated via entire analysis area to give a total cell count which was divided by the 50 μL sample volume resulting in a measurement of counts per μL. Good linearity was observed across the 350 to 500 cells/μL region where increased monitoring is recommended.5 Lymphocyte counts to below 200 cells/μL were measured with acceptable accuracy indicating that the diagnostic criterion for the separation barrier between HIV and AIDS, in addition to measurement of healthy cell count values, can be achieved with this approach (R2 = 0.92, Fig. 6b).
Flow cytometry correlation
To compare the membrane-based NBC to the gold standard of cellular determinations, flow cytometry, parallel experiments were executed to measure values of %CD4 (i.e. CD3+CD4+ cells per total CD3+ cells). Similar measurements have been completed previously using a combination of red encoded CD3, CD19 and CD56 cell surface markers to secure the entire population of lymphocytes. For the purpose of these experiments, whole blood samples were collected from 6 different healthy donors by venipuncture, divided into 2 aliquots, and analyzed the same day as collection on both the FC and membrane-based NBC systems. Through the image analysis protocol described above, the number of cells displaying both the CD3 and CD4 receptors can be determined and compared to CD3 counts. In the membrane-based NBC, this corresponds to the total number of dually-positive yellow cells divided by total red cell counts. In FC, lymphocytes were first identified from monocytes and granulocytes via their unique side and forward scatter signature. These cells were further characterized by the number of CD4 and CD3 surface receptors present, as determined by red and green fluorescence intensity, respectively. Clear separation between dually positive and CD3+CD4− cells was seen and cell counts per μL derived via calibrator bead standards. The values of %CD4 could then be determined by number of cells CD3+CD4+ divided by total CD3+. Linear regression analysis gave an R2 = 0.97 correlation of the NBC to FC (Fig. 6c).
To assess the precision of both approaches, one sample was repeatedly assayed 6 different times with the membrane-based NBC. Here, the NBC system yielded a mean value of 53% CD4 with a trial to trial variance of 5%; this %CD4 value is statistically the same as the measurement value determined by flow cytometry, which yielded a value of 51% CD4. Typical trial to trial variance values for flow cytometry are in the range of 5 to 8%.5 The FC procedure takes > 60 minutes from startup to results acquisition as its’ elaborate optical and fluid handling design require daily maintenance. The NBC system produced results in < 30 minutes.
Comparison to molecular fluorophores
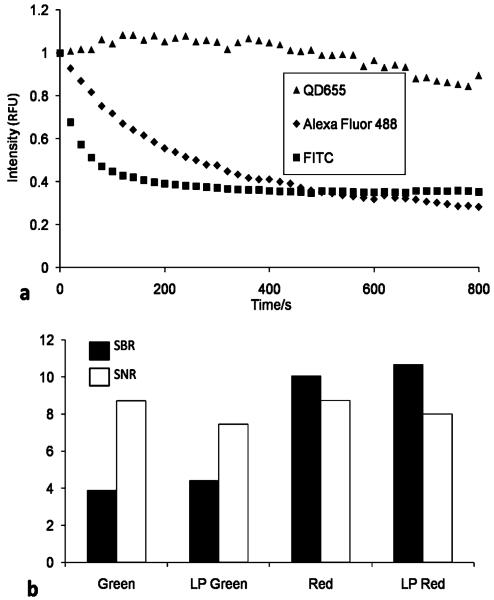
Previously, organic fluorophores have been used to label CD4 and CD3 cells for membrane-based NBC HIV immune function testing. To explore the performance of the QDs relative to their organic counterparts, a series of image analysis and photostability studies were completed with the two labels. Here 50 μL whole blood was labeled with mouse CD3 primary antibody and then fluorescently tagged with equi-molar quantities of goat anti-mouse secondary antibody conjugated to QD 655, Alexa Fluor 488, or FITC. A sequence of images was acquired with photomicrographs taken every 20 s for 15 minutes. The AOIs, specific to each experiment, were created from the initial and most intense image, and then applied to subsequent photomicrographs recording the mean intensity of each AOI collection. Intensity values for each fluorophore were plotted relative to their unique maximum signal versus time for the 3 fluorophores (Fig. 7a). The attributes of the fluorescent nanoparticles become clear as the molecular dyes exhibit exponential signal intensity decay within minutes. Signal intensity advantage at the end of the 15 minute experiment is a factor of 6 greater for the QD probes. Excitation power values were comparable for both the molecular dyes and the quantum particles in these measurements. The Alexa Fluor dyes used in these comparisons are provided with a fluorophore to antibody ratio of ~ 7 : 1. Conversely, due to the large size of the quantum particles relative to the molecular dyes, this ratio drops to ~ 0.3 : 1. This stability is particularly advantageous to the membrane-based NBC system as multiple FOVs are captured, during which time any loss of fluorescence would directly translate into a reduced number of lymphocytes imaged. Furthermore, additional reagents targeting other intra, or extra, cellular moieties of interest could be added after this initial analysis gleaning supplementary information from the sample while retaining the stable QD signal.
Fig. 7.
(a) The CD3+ cells were processed with equimolar concentrations of both primary and secondary antibody–fluorophore conjugate, which was normalized to fluorophore. This was injected into the NBC, and images captured every 20 s for 15 min. Intensities from each fluorophore were plotted relative to their individual maximum, initial intensity. Conventional fluorophores exhibit exponential decay versus QD photostability that is critical for capturing a number of FOVs in the NBC system. This stability may also be exploited to add additional reagents for further cell analysis. (b) Signal to noise and signal to background ratios for individual filter cubes versus a single long pass filter for staining CD3+ cells. Individual cell intensities were evaluated relative to immediate environment in terms of membrane auto-fluorescence and variation in background signal. For this application, SBR proved critical in identifying authentic events.
Reduced optical pathway evaluation
To assess the performance characteristics between individual filter cubes for red and green QDs versus a single long pass filter, images were captured under both conditions. Photomicrographs were split into their RGB components, AOIs determined as described above, and average pixel intensity in the gated regions recorded. The SNR and SBR values were calculated, the results of which may be seen in Fig. 7b. Increase in the SBR value by 14% is observed with the long-pass filter for QD 565, 6% for QD 655. The SBR values for the green channel QDs are 60% lower than red channel QDs. This observation is attributable not to decreased intensity, but rather to increased autofluorescence of the membrane. Capturing an image before sample introduction with resulting background subtraction negates this effect. The membrane-based NBC system was capable of adequately identifying cells even at SBR values approaching unity. The similarity between the individual filter cubes and a condensed optical pathway demonstrates the ability to develop QD-based NBC systems utilizing only a single light conduit for reduced overall analyzer footprint.
Discussion
Despite the availability of antiretroviral therapy (ART) as treatment for declining immunity, use of ART as a tool to decrease morbidity among the more than 33 million people infected with HIV has not become widespread in resource-scarce settings. Lack of CD4 HIV immune function testing has been described as the “missing link” in the management of HIV+ patients in resource-scarce settings.1 Problems with CD4 measurements here can be attributed to the above highlighted infrastructure requirements including a lack of skilled operators, high cost, constant power supplies, the need for refrigerated reagents, and lengthy sample preparation. Identifying those individuals most at need for ART as determined by the immunological profile remains the ultimate goal of our sustained efforts in this area.
In this paper we describe the conceptual basis for the creation of a highly integrated sample handling and cell counting modality that is compatible with use in resource-scarce settings. Such a simplified infrastructure and ease of user interface are critical requirements for health monitoring devices that will be deployed in these environments. Likewise, with the appropriate experimental design and optimized microfluidic structures, in tandem with robust optical components, it is possible to make high quality measurements in a miniaturized test modality while concurrently reducing overall cost per assay. Assay costs, combined with infrastructure considerations, and overall performance, serve as some of the key considerations in the definition of the conceptual basis of new test modalities that may be used in resource-scarce settings. It should be clear that the use of mini-systems here described has the potential to lead to a significant reduction of reagent amounts (by 10 to 100 fold) to a degree that they are no longer major considerations for the overall test cost. More importantly, the development of a fully integrated approach that provides increased access for these critical measurements requires all aspects of sample collection/handling, reagent processing, and cell separation/counting to be completed in a convenient manner. By incorporating all aspects of analysis into a single analyzer, the necessity of the laboratory is eliminated. The NBC cards are completely disposable reducing risk of biological contamination and allows needle stick-sized samples to be analyzed in under an hour, with CD4 percentages obtained to a degree similar to traditional flow cytometry without the need for erythrocyte lysis, manual counting, or internal bead standardization. This design no longer produces a “chip-in-a-lab”, but rather a true “lab-on-a-chip”.
Further, we demonstrate here the ability of the membrane-based NBC system to determine CD4 absolute counts up to 600/μL and below 200/μL whereby the disease diagnostic decision threshold between HIV and AIDS is established. Through these activities, an important step has been taken in the direction of defining methods for inclusion of inorganic quantum particles as selective and stable cellular labels in the context of HIV immune function testing. The efforts demonstrate the feasibility of incorporating next generation, nanometer-sized fluorophores via a secondary antibody linkage into a microfluidic device to monitor the progression of HIV and other immunologically relevant diseases.
A definitive explanation for the failure of established direct conjugation protocols in this application may be impossible; however, it may be attributable to the specific types of antibody used in the application or the generation of QD probes obtained from the vendor. As the EDC technique links non-specifically through free amines, a variety of antibody orientations is possible. This method yields high antibody to QD ratio which could potentially induce steric hindrance of active sites. Further, this EDC can potentially induce cross-linking of non-targeted amines. The SMCC method employs DTT, a strong reducing agent. While DTT may cleave the disulfide linkages along the IgG backbone, it may also break other S–S bonds, damaging tertiary structure and thus antigen specificity. This damage may go so far as to denature the protein and, perhaps, explains the aggregates seen. It should be noted, however, that this phenomenon is likely specific to individual antibody structure as we have since used the SMCC method with excellent results in other non-cellular applications. Poor performance with directly bound QDs is not uncommon, and the secondary antibody method thus employed proved to be a specific and effective transduction pathway.29,33 In addition, more than one secondary antibody may be bound to each primary, further increasing the number of fluorophores per lymphocyte and thus signal output.
An accurate comparison of the performance features between biologically active QDs and molecular dyes is admittedly difficult.54 Although information describing the attributes of QDs in their native state abounds in the literature,55-57 the spectroscopic data characteristic of the particles upon synthesis including molar extinction coefficient, quantum yield, FWHM, emission maximum, and average radius can radically change after passivation and further functionalization with biological ligands. Evaluation versus organic fluorophores adds the additional challenge of uncertain antibody to fluorophore ratio and differences in excitation intensity. Nevertheless, we observed photostability in the QDs superior to their molecular counterparts. In the time dependent study in which relative intensities were calculated via each fluorophores initial intensity, Alexa Fluor 488 diminished to an intensity one half maximum in 280 s with Alexa Fluor 647 and FITC achieving this value in less than 80 s (Fig. 7a). Neither QD 565 nor QD 655 displayed significant intensity loss during this time frame.
Analogous to the gates, or data discriminators, placed on data by flow cytometrists, with parameters such as scatter and intensity, the membrane-based NBC and subsequent image processing allows individual components of a complex sample to be identified and quantified. In lieu of sample lysing to isolate the fraction of interest, the membrane-based NBC approach utilizes a size gate, i.e. membrane, to isolate white blood cells from the greater sample matrix. The scatter gate, typically related to cell morphology in FC, is replaced by both the size gate described above as well as a fluorescent intensity discriminator. The intensity gate has two important applications. First is the SBR ratio, used to discriminate between authentic and non-authentic events. Secondly, the intensity gate addresses monocytes, an import consideration in these types of cell assays as the presence of such cells in samples could artificially contribute to the total CD4 count. Thus, low level CD4 expressing monocytes are effectively eliminated in the digital image analysis method here devised so as to leave only green channel positive lymphocytes without the use of additional CD14 labeling. Literature reference values of approximately 50 000 copies of the CD4 antigen on lymphocytes, with 3000–9000 copies on monocytes,53 corresponds nicely to the factor of 4–5 intensity difference between monocytes and lymphocytes observed in the membrane-based NBC. Intensity histograms for all CD4 positive cells show baseline resolution between lymphocytes and monocytes. From these histograms the ratio of monocytes to lymphocytes could be determined for each image and compared to the ratio determined by flow cytometry. The difference between the two methods was <6%. Finally, cell location on the membrane serves as a spatial gate to identify doubly positive CD3 +CD4+ T lymphocytes of interest and a circularity gate in the image analysis protocol here used rejects the vast majority of the debris that may be present on membrane surface.
Although many reports have been published in which QDs are used for cellular imaging,30,31 to the best of our knowledge, this is the first report in which QDs are used for cellular quantitation in an enclosed, disposable device. Other methods using interference or impedance are frequently incompatible with peripheral blood samples as they are overwhelmed by the presence of erythrocytes.10,13 Manual counting methods involving magnetic beads11 often fail to correlate to established methods above R2 = 0.90, perhaps due to residual monocyte contamination. Other alternatives to FC using QDs are not self-contained,34 an important consideration when dealing with HIV-positive samples. Literature reports using QDs quantitatively in blood is limited to protein viral disease markers.39 The membrane-based NBC correlates to FC at levels similar to non-labeling methods, based on cell affinity immobilization,9,12 but also has the built-in capability for further analysis including ratios and percentages along with the capacity to exclude monocyte contamination in a rigorous way. Indeed, the membrane-based NBC can be extended to measurements of CD4/CD8 ratios, rare cell event detection,58,59 panel evaluations similar to complete blood counts60,61 and other advanced FC experiments.62,63 As such, the membrane-based NBC serves as an interesting new approach that can service the requirements for a “personal flow cytometer” for use in both resource-scarce environments as well as in numerous research settings where the cost and complex nature of FC limits its current utility.
Conclusion
In summary, these experiments demonstrate the viability of incorporating quantum dot detection schemes into a microfluidic analysis device for lymphocyte enumeration as a tool for monitoring HIV progression. Through the use of QDs, the optical requirements of the system are reduced significantly allowing for small components in a smaller analyzer for point-of-need analysis. Cell counts are obtained through interpretation of the fluorescent images with automated software and require no manual counting. The correlation of results to flow cytometry in a pilot study (R2 = 0.97) suggests that a more exhaustive study is warranted now so as to include samples from HIV+ individuals. The new method is compatible with %CD4 determinations, absolute CD4 counts, CD4/CD8 ratios and other cellular analyses. The integrated approach here defined spans from the nano regime with the use of QDs to the global health care area thereby addressing important gaps in health care. Advances of this type in nanotechnology, materials chemistry, and microfluidics, promise to yield diagnostic tools with relaxed optical requirements, which can provide essential information rapidly, robustly, and reliably. Such devices have the potential to serve as transformative tools in joining currently available antiretroviral therapies with those individuals in need of such treatment, especially in resource-scarce environments.
Acknowledgements
This work was supported by grants from the National Institutes of Health (1 U01 DE017793-01), the Bill and Melinda Gates Foundation, and the Welch Foundation (F-1193).
Footnotes
Part of a special issue on Point-of-care Microfluidic Diagnostics; Guest Editors—Professor Kricka and Professor Sia.
Electronic supplementary information (ESI) available: See DOI: 10.1039/b817116e
References
- 1.Cohen J. Science. 2004;304:1936–1936. doi: 10.1126/science.304.5679.1936. [DOI] [PubMed] [Google Scholar]
- 2.U.N. Program on HIV/AIDS. World Health Organization . AIDS Epidemic Update. Geneva, Switzerland: 2007. [Google Scholar]
- 3.Obrien WA, Hartigan PM, Daar ES, Simberkoff MS, Hamilton JD. Ann. Intern. Med. 1997;126:939–945. doi: 10.7326/0003-4819-126-12-199706150-00002. [DOI] [PubMed] [Google Scholar]
- 4.Yarchoan R, Tosato G, Little RF. Nat. Clin. Pract. Oncol. 2005;2:406–415. doi: 10.1038/ncponc0253. [DOI] [PubMed] [Google Scholar]
- 5.CDC MMWR. 1997;46 [Google Scholar]
- 6.Bofill M, Janossy G, Lee CA, MacDonald-Burns D, Phillips AN, Sabin C, Timms A, Johnson MA, Kernoff PB. Clin. Exp. Immunol. 1992;88:243–252. doi: 10.1111/j.1365-2249.1992.tb03068.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.W.H.O. UNAIDSCD4+ T-Cell Enumeration Technologies. 2005. a. [Google Scholar]
- 8.Zijenah LS, Kadzirange G, Madzime S, Borok M, Mudiwa C, Tobaiwa O, Mucheche M, Rusakaniko S, Katzenstein DA. J. Trans. Med. 2006;4 doi: 10.1186/1479-5876-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thorslund S, Larsson R, Nikolajeff F, Bergquist J, Sanchez J. Sens. Actuators, B. 2007;B123:847–855. [Google Scholar]
- 10.Mishra NN, Retterer S, Zieziulewicz TJ, Isaacson M, Szarowski D, Mousseau DE, Lawrence DA, Turner JN. Biosens. Bioelectron. 2005;21:696–704. doi: 10.1016/j.bios.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 11.Truett April A, Letizia A, Malyangu E, Sinyangwe F, Morales Brandi N, Crum Nancy F, Crowe Suzanne M. J. Acquir. Immune Defic. Syndr. 2006;41:168–174. doi: 10.1097/01.qai.0000191282.77954.fc. [DOI] [PubMed] [Google Scholar]
- 12.Cheng X, Irimia D, Dixon M, Sekine K, Demirci U, Zamir L, Tompkins RG, Rodriguez W, Toner M. Lab Chip. 2007;7:170–178. doi: 10.1039/b612966h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ozcan A, Demirci U. Lab Chip. 2008;8:98–106. doi: 10.1039/b713695a. [DOI] [PubMed] [Google Scholar]
- 14.Reyes DR, Iossifidis D, Auroux PA, Manz A. Anal. Chem. 2002;74:2623–2636. doi: 10.1021/ac0202435. [DOI] [PubMed] [Google Scholar]
- 15.Vilkner T, Janasek D, Manz A. Anal. Chem. 2004;76:3373–3385. doi: 10.1021/ac040063q. [DOI] [PubMed] [Google Scholar]
- 16.Tudos AJ, Besselink GAJ, Schasfoort RBM. Lab Chip. 2001;1:83–95. doi: 10.1039/b106958f. [DOI] [PubMed] [Google Scholar]
- 17.Whitesides GM. Nat. Biotechnol. 2003;21:1161–1165. doi: 10.1038/nbt872. [DOI] [PubMed] [Google Scholar]
- 18.Yager P, Edwards T, Fu E, Helton K, Nelson K, Tam MR, Weigl BH. Nature. 2006;442:412–418. doi: 10.1038/nature05064. [DOI] [PubMed] [Google Scholar]
- 19.Jovin TM. Nat. Biotechnol. 2003;21:32–33. doi: 10.1038/nbt0103-32. [DOI] [PubMed] [Google Scholar]
- 20.Murray CB, Norris DJ, Bawendi MG. J. Am. Chem. Soc. 1993;115:8706–8715. [Google Scholar]
- 21.Murray CB, Kagan CR, Bawendi MG. Ann. Rev. Mater. Sci. 2000;30:545–610. [Google Scholar]
- 22.Liu W, Choi HS, Zimmer JP, Tanaka E, Frangioni JV, Bawendi M. J. Am. Chem. Soc. 2007;129:14530–14531. doi: 10.1021/ja073790m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hines MA, Guyot-Sionnest P. J. Phys. Chem. 1996;100:468–471. [Google Scholar]
- 24.Skaff H, Sill K, Emrick T. J. Am. Chem. Soc. 2004;126:11322–11325. doi: 10.1021/ja047260r. [DOI] [PubMed] [Google Scholar]
- 25.Potapova I, Mruk R, Prehl S, Zentel R, Basche T, Mews A. J. Am. Chem. Soc. 2003;125:320–321. doi: 10.1021/ja028641c. [DOI] [PubMed] [Google Scholar]
- 26.Hines MA, Guyot-Sionnest P. J. Phys. Chem. 1996;100:468–471. [Google Scholar]
- 27.Chan WCW, Maxwell DJ, Gao X, Bailey RE, Han M, Nie S. Curr. Opin. Biotechnol. 2002;13:40–46. doi: 10.1016/s0958-1669(02)00282-3. [DOI] [PubMed] [Google Scholar]
- 28.Parak WJ, Gerion D, Pellegrino T, Zanchet D, Micheel C, Williams SC, Boudreau R, Le Gros MA, Larabell CA, Alivisatos AP. Nanotechnology. 2003;14:R15–R27. [Google Scholar]
- 29.Xing Y, Chaudry Q, Shen C, Kong KY, Zhau HE, Chung LW, Petros JA, O’Regan RM, Yezhelyev MV, Simons JW, Wang MD, Nie S. Nat. Protoc. 2007;2:1152–1165. doi: 10.1038/nprot.2007.107. [DOI] [PubMed] [Google Scholar]
- 30.Goldman ER, Uyeda HT, Hayhurst A, Mattoussi H. Methods Mol. Biol. 2007;374:207–227. doi: 10.1385/1-59745-369-2:207. [DOI] [PubMed] [Google Scholar]
- 31.Medintz IL, Mattoussi H, Goldman ER, Clapp AR, Mauro JM. PMSE Preprints. 2003;89:205–206. [Google Scholar]
- 32.Liu W, Howarth M, Greytak AB, Zheng Y, Nocera DG, Ting AY, Bawendi MG. J. Am. Chem. Soc. 2008;130:1274–1284. doi: 10.1021/ja076069p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alivisatos AP, Gu W, Larabell C. Annu. Rev. Biomed. Eng. 2005;7:55–76. doi: 10.1146/annurev.bioeng.7.060804.100432. [DOI] [PubMed] [Google Scholar]
- 34.Bocsi J, Mittag A, Varga VS, Molnar B, Tulassay Z, Sack U, Lenz D, Tarnok A. Proc. SPIE. 2006;6096:60960Z/60961–60960Z/60911. [Google Scholar]
- 35.Sukhanova A, Devy J, Venteo L, Kaplan H, Artemyev M, Oleinikov V, Klinov D, Pluot M, Cohen JHM, Nabiev I. Anal. Biochem. 2004;324:60–67. doi: 10.1016/j.ab.2003.09.031. [DOI] [PubMed] [Google Scholar]
- 36.Xiao Y, Gao X, Gannot G, Emmert-Buck MR, Srivastava S, Wagner PD, Amos MD, Barker PE. Int. J. Cancer. 2008;122:2178–2186. doi: 10.1002/ijc.23320. [DOI] [PubMed] [Google Scholar]
- 37.Medintz IL, Goldman ER, Clapp AR, Mattoussi H. PMSE Preprints. 2005;93:5–6. [Google Scholar]
- 38.Zajac A, Song D, Qian W, Zhukov T. Colloids Surf. B. 2007;58:309–314. doi: 10.1016/j.colsurfb.2007.02.019. [DOI] [PubMed] [Google Scholar]
- 39.Klostranec JM, Xiang Q, Farcas GA, Lee JA, Rhee A, Lafferty EI, Perrault SD, Kain KC, Chan WCW. Nano Letters. 2007;7:2812–2818. doi: 10.1021/nl071415m. [DOI] [PubMed] [Google Scholar]
- 40.Clapp AR, Goldman ER, Mattoussi H. Nat. Protoc. 2006;1:1258–1267. doi: 10.1038/nprot.2006.184. [DOI] [PubMed] [Google Scholar]
- 41.Goldman ER, Balighian ED, Mattoussi H, Kuno MK, Mauro JM, Tran PT, Anderson GP. J. Am. Chem. Soc. 2002;124:6378–6382. doi: 10.1021/ja0125570. [DOI] [PubMed] [Google Scholar]
- 42.Medintz IL, Uyeda HT, Goldman ER, Mattoussi H. Nat. Mater. 2005;4:435–446. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- 43.Hua X-F, Liu T-C, Cao Y-C, Liu B, Wang H-Q, Wang J-H, Huang Z-L, Zhao Y-D. Anal. Bioanal. Chem. 2006;386:1665–1671. doi: 10.1007/s00216-006-0807-5. [DOI] [PubMed] [Google Scholar]
- 44.Pathak S, Davidson MC, Silva GA. Nano Letters. 2007;7:1839–1845. doi: 10.1021/nl062706i. [DOI] [PubMed] [Google Scholar]
- 45.Song E-Q, Wang G-P, Xie H-Y, Zhang Z-L, Hu J, Peng J, Wu D-C, Shi Y-B, Pang D-W. Clin. Chem. 2007;53:2177–2185. doi: 10.1373/clinchem.2007.092023. [DOI] [PubMed] [Google Scholar]
- 46.Lavigne JJ, Savoy S, Clevenger MB, Ritchie JE, McDoniel B, Yoo SJ, Anslyn EV, McDevitt JT, Shear JB, Neikirk D. J. Am. Chem. Soc. 1998;120:6429–6430. [Google Scholar]
- 47.Christodoulides N, Mohanty S, Miller CS, Langub MC, Floriano PN, Dharshan P, Ali MF, Bernard B, Romanovicz D, Anslyn E, Fox PC, McDevitt JT. Lab Chip. 2005;5:261–269. doi: 10.1039/b414194f. [DOI] [PubMed] [Google Scholar]
- 48.Christodoulides N, Tran M, Floriano PN, Rodriguez M, Goodey A, Ali M, Neikirk D, McDevitt JT. Anal. Chem. 2002;74:3030–3036. doi: 10.1021/ac011150a. [DOI] [PubMed] [Google Scholar]
- 49.Goodey A, Lavigne JJ, Savoy SM, Rodriguez MD, Curey T, Tsao A, Simmons G, Wright J, Yoo SJ, Sohn Y, Anslyn EV, Shear JB, Neikirk DP, McDevitt JT. J. Am. Chem. Soc. 2001;123:2559–2570. doi: 10.1021/ja003341l. [DOI] [PubMed] [Google Scholar]
- 50.Weigum SE, Floriano PN, Christodoulides N, McDevitt JT. Lab Chip. 2007;7:995–1003. doi: 10.1039/b703918b. [DOI] [PubMed] [Google Scholar]
- 51.Rodriguez WR, Christodoulides N, Floriano PN, Graham S, Mohanty S, Dixon M, Hsiang M, Peter T, Zavahir S, Thior I, Romanovicz D, Bernard B, Goodey AP, Walker BD, McDevitt JT. Plos Medicine. 2005;2:663–672. doi: 10.1371/journal.pmed.0020182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Christodoulides N, Floriano PN, Acosta SA, Ballard KL, Weigum SE, Mohanty S, Dharshan P, Romanovicz D, McDevitt JT. Clin. Chem. 2005;51:2391–2395. doi: 10.1373/clinchem.2005.054882. [DOI] [PubMed] [Google Scholar]
- 53.Davis KA, Abrams B, Iyer SB, Hoffman RA, Bishop JE. Cytometry. 1998;33:197–205. doi: 10.1002/(sici)1097-0320(19981001)33:2<197::aid-cyto14>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 54.Resch-Genger U, Grabolle M, Cavaliere-Jaricot S, Nitschke R, Nann T. Nat. Methods. 2008;5:763–775. doi: 10.1038/nmeth.1248. [DOI] [PubMed] [Google Scholar]
- 55.Gao X, Nie S. Anal. Chem. 2004;76:2406–2410. doi: 10.1021/ac0354600. [DOI] [PubMed] [Google Scholar]
- 56.Empedocles SA, Norris DJ, Bawendi MG. Phys. Rev. Lett. 1996;77:3873–3876. doi: 10.1103/PhysRevLett.77.3873. [DOI] [PubMed] [Google Scholar]
- 57.Cottingham K. Anal. Chem. 2005;77:354A–357A. doi: 10.1021/ac053380l. [DOI] [PubMed] [Google Scholar]
- 58.Kyewski B. Science. 2008;321:776–777. doi: 10.1126/science.1162966. [DOI] [PubMed] [Google Scholar]
- 59.Dainiak MB, Kumar A, Galaev IY, Mattiasson B. Adv. Biochem. Eng./Biotech. 2007;106:1–18. doi: 10.1007/10_2007_069. [DOI] [PubMed] [Google Scholar]
- 60.Tanabe R, Hata S, Shimokohbe A. Microelectron. Eng. 2006;83:1646–1650. [Google Scholar]
- 61.Zheng S, Lin JC-H, Kasdan HL, Tai Y-C. Sens. Actuators B. 2008;B132:558–567. [Google Scholar]
- 62.Chattopadhyay PK, Price DA, Harper TF, Betts MR, Yu J, Gostick E, Perfetto SP, Goepfert P, Koup RA, De Rosa SC, Bruchez MP, Roederer M. Nat. Med. 2006;12:972–977. doi: 10.1038/nm1371. [DOI] [PubMed] [Google Scholar]
- 63.Simonnet C, Groisman A. Anal. Chem. 2006;78:5653–5663. doi: 10.1021/ac060340o. [DOI] [PubMed] [Google Scholar]