Abstract
The ligand-gated ion channels that participate in fast synaptic transmission comprise the nicotinic acetylcholine, 5-hydroxytryptamine3 (5-HT3), γ-aminobutyric acidA (GABAA), glycine, ionotropic glutamate and P2X receptor families. A consistent and systematic nomenclature for the individual subunits that comprise these receptors and the receptors that result from their co-assembly is highly desirable. There is also a need to develop criteria that aid in deciding which of the vast number of heteromeric combinations of subunits that can be assembled in heterologous expression systems in vitro, are known, or likely, to exist as functional receptors in vivo. The aim of this short article is to summarize the progress being made by the nomenclature committee of IUPHAR (NC-IUPHAR) in formulating recommendations that attempt to address these issues.
Keywords: Ligand-gated ion channels, Nomenclature
1. Introduction
The heteromeric nature of most ligand-gated ion channels (Fig. 1), with their accessory proteins, and the multiple proteins involved in receptor trafficking and responses to receptor activation pose multiple challenges to the definition of their pharmacology. Furthermore, the receptors must be well characterized for definition of their functional roles in normal brain and in disease states and for new drug discovery. To this end the journal Neuropharmacology and The International Union of Basic and Clinical Pharmacology (IUPHAR) have joined forces in this Special Issue to address the nomenclature, the structures, the pharmacology, the roles, and therapeutic opportunities of ligand-gated ion channels (LGICs) that are activated by neurotransmitters (Fig. 1).
Fig. 1.
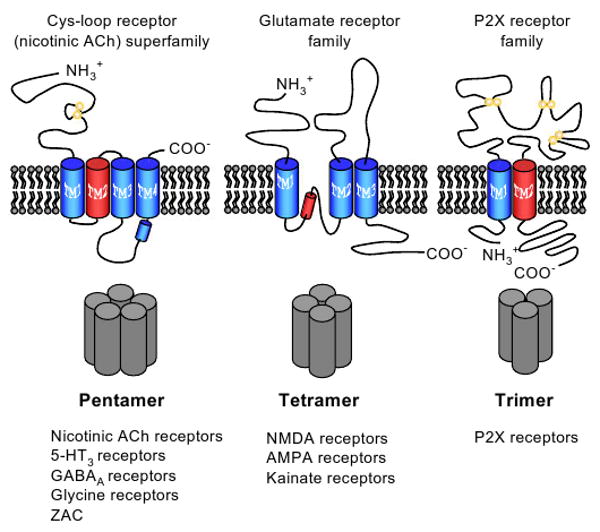
Schematic representation of the three structural categories of ligand-gated ion channel subunit. The pentameric Cys-loop receptor superfamily comprises the nicotinic acetylcholine (ACh) receptors, 5-hydroxytryptamine3 (5-HT3) and a zinc-activated channel that form cation selective ion channels and the γ-aminobutyric acidA and strychnine-sensitive glycine receptors that conduct anions. The tetrameric ionotropic glutamate receptors are subdivided into N-methyl-d-aspartate (NMDA), α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid (AMPA) and kainate receptor subfamilies. The highly schematic topography of each receptor category indicates the locations of the extracellular and intracellular termini, the number of transmembrane spans (large colored cylinders), and cysteine residues participating in disulphide bond formation (yellow circles). Red cylinders indicate α-helical regions participating in ion conduction/selectivity.
2. NC-IUPHAR
The International Union of Pharmacology Committee on Receptor Nomenclature and Drug Classification (NC-IUPHAR) is a body that issues guidelines for receptor and ion channel classification. It addresses the main issues in pharmacology today, classifying the major receptor and ion channel systems in the human genome and depositing the data on a freely available web site (http://www.iuphar-db.org). NC-IUPHAR has >50 subcommittees with expert scientists freely giving up their time in order to facilitate the interface between the discovery of new sequences from the Human Genome Project and the designation of the derived proteins as functional receptors and ion channels.
Furthermore, the multitude of factors between a published genomic sequence and an assigned receptor function in a given tissue (epigenetics, alternative splicing, messenger RNA editing, polymorphisms, the combinatorial nature of subunit association) ensures that there are multiple drug targets. The practical implications of the new pharmacology are immense, particularly for drug discovery where the magnitude of the variables affecting drug response is only now becoming fully appreciated. NC-IUPHAR needs input from motivated scientists interested in receptors, so if you are interested please contact us! NC-IUPHAR works in coordination with the Human Genome Organisation (HUGO) Gene Nomenclature Committee (HGNC).
The goals of NC-IUPHAR include: (i) establishing, as far as possible, an overall consistent classification and nomenclature for the LGICs; and (ii) developing a subunit list (with template information for a database). Table 1 presents such a list of the genes encoding LGIC subunits that are expressed in humans. Thus, certain subunits, such as the nicotinic acetylcholine receptor α8 subunit (Schoepfer et al., 1990) that has not been identified in the mammalian brain, and the glycine receptor α4 subunit (Matzenbach et al., 1994), which is likely to be a pseudogene in man, are not listed. Similarly, the avian GABAA receptor β4 and γ4 subunits, which may have evolved into the mammalian GABAA receptor θ and ε subunits, respectively, are not tabulated (Simon et al., 2004). At this point in time we also do not consider intracellular ion channels such as the inositol trisphosphate (IP3) or ryanodine receptors that are gated by ligands. Other classes of cell surface ion channel that are activated, or modulated, by ligands, such as the cyclic nucleotide regulated ion channels and numerous members of the transient receptor potential family have been the subject of previous NC-IUPHAR recommendations (Clapham et al., 2005; Hofmann et al., 2005).
Table 1.
NC-IUPHAR list of ligand-gated ion channel subunits
| Receptor family | NC-IUPHAR subunit nomenclature | Human gene name | Human chromosomal location |
|---|---|---|---|
| A. Cys-loop superfamily | |||
| 5-HT3 | 5-HT3A | HTR3A | 11q23.1 |
| 5-HT3B | HTR3B | 11q23.1 | |
| 5-HT3C | HTR3C | 3q27.1 | |
| 5-HT3D | HTR3D | 3q27.1 | |
| 5-HT3E | HTR3E | 3q27.1 | |
| Nicotinic ACh | α1 | CHRNA1 | 2q24–q32 |
| α2 | CHRNA2 | 8p21 | |
| α3 | CHRNA3 | 15q24 | |
| α4 | CHRNA4 | 20q13.2–q13.3 | |
| α5 | CHRNA5 | 15q24 | |
| α6 | CHRNA6 | 8p11.21 | |
| α7 | CHRNA7 | 15q14 | |
| α9 | CHRNA9 | 4p14 | |
| α10 | CHRNA10 | 11p15.5 | |
| β1 | CHRNB1 | 17p13.1 | |
| β2 | CHRNB2 | 1q21.3 | |
| β3 | CHRNB3 | 8p11.2 | |
| β4 | CHRNB4 | 15q24 | |
| γ | CHRNG | 2q33–q34 | |
| δ | CHRND | 2q33–q34 | |
| ε | CHRNE | 17p13–p12 | |
| GABAA | α1 | GABRA1 | 5q34–q35 |
| α2 | GABRA2 | 4p12 | |
| α3 | GABRA3 | Xq28 | |
| α4 | GABRA4 | 4p12 | |
| α5 | GABRA5 | 15q11.2–q12 | |
| α6 | GABRA6 | 5q34 | |
| β1 | GABRB1 | 4p12 | |
| β2 | GABRB2 | 5q34 | |
| β3 | GABRB3 | 15q11.2–q12 | |
| γ1 | GABRG1 | 4p12 | |
| γ2 | GABRG2 | 5q31.1–q33.1 | |
| γ3 | GABRG3 | 15q12 | |
| δ | GABRD | 1p36.3 | |
| ε | GABRE | Xq28 | |
| θ | GABRQ | Xq28 | |
| π | GABRP | 5q33–q34 | |
| ρ1 | GABRR1 | 6q13–q16.3 | |
| ρ2 | GABRR2 | 6q13–q16.3 | |
| ρ3 | GABRR3 | 3q11.2 | |
| Glycine | α1 | GLRA1 | 5q32 |
| α2 | GLRA2 | Xp22.1–p21.3 | |
| α3 | GLRA3 | 4q33–q34 | |
| β | GLRB | 4q31.3 | |
| Zinc-activated | ZAC | ZACN | 17q25.3 |
| B. P2X family | |||
| P2X | P2X1 | P2RX1 | 17p13.3 |
| P2X2 | P2RX2 | 12q24.33 | |
| P2X3 | P2RX3 | 11q12 | |
| P2X4 | P2RX4 | 12q24.32 | |
| P2X5 | P2RX5 | 17p13.3 | |
| P2X6 | P2RX6 | 22q11.21 | |
| P2X7 | P2RX7 | 12q24 | |
| C. Ionotropic glutamate family | |||
| AMPA | GluA1 | GRIA1 | 5q31.1 |
| GluA2 | GRIA2 | 4q32–q33 | |
| GluA3 | GRIA3 | Xq25–q26 | |
| GluA4 | GRIA4 | 11q22 | |
| Kainate | GluK1 | GRIK1 | 21q22.11 |
| GluK2 | GRIK2 | 6q16.3–q21 | |
| GluK3 | GRIK3 | 1p34–p33 | |
| GluK4 | GRIK4 | 11q22.3 | |
| GluK5 | GRIK5 | 19q13.2 | |
| NMDA | GluN1 | GRIN1 | 9q34.3 |
| GluN2A | GRIN2A | 16p13.2 | |
| GluN2B | GRIN2B | 12p12 | |
| GluN2C | GRIN2C | 17q25 | |
| GluN2D | GRIN2D | 19q13.1 | |
| GluN3A | GRIN3A | 9q31.1 | |
| GluN3B | GRIN3B | 19p13.3 | |
| ‘Orphan’ (GluD) | GluD1 | GRID1 | 10q22 |
| GluD2 | GRID2 | 4q22 | |
Note, the entries in this table do not attempt to address the multiple subunits that frequently arise from a single gene as a consequence of alternative splicing and editing of RNA transcripts.
In recommending a consistent nomenclature for LGIC subunits, it is appropriate to reflect upon the acceptance, or otherwise, of previous NC-IUPHAR recommendations and current practice in the literature. Lukas et al. (1999) in an interim NC-IUPHAR statement on the nomenclature of nicotinic acetylcholine receptor subunits stated that ‘the 16 nACh receptor subunits identified to date are defined using a Greek letter sometimes followed by an Arabic numeral (neither subscripted nor superscripted)’. A survey of the literature indicates this formalism to be widely employed. By contrast, in an extensive and still valuable review of the classification of GABAA receptors, Barnard et al. (1998) indicated that Greek subunit letters should be followed by a subscripted Arabic numeral, where appropriate. However, a representative search of the literature subsequent to that publication indicates no consistent usage of subscripts even, in some instances, between contributions emanating from the same laboratory. A similar situation is apparent for the strychnine-sensitive glycine receptors, upon which NC-IUPHAR have yet to issue guidance. By contrast, subscripted numbers and letters are almost universally used to denote the 5-HT3 and P2X receptor subunits (e.g. 5-HT3A; P2X3) in accordance with previous NC-IUPHAR guidelines (Hoyer et al., 1994; Khakh et al., 2001).
A revised nomenclature of the ionotropic glutamate receptors subunits triggered NC-IUPHAR to reconsider the naming of LGIC subunits in general, but in particular with regard to the use of subscripts. Each of the LGIC subcommittees were consulted in an attempt to reach an overall consensus. Various reasons were elaborated for the continued use (largely historical), or not (consistency across receptor families, reserving subscript to specify receptor stoichiometry, difficulties in database searches, formatting issues) of subscript notation. After considerable deliberation the NC-IUPHAR Committee sets out the following which is a recommendation for implementation:
The use of subscript may be retained specifically for the receptor names GABAA and 5-HT3. For historical reasons this would be difficult, if not impossible, to change.
Subunits within a receptor should not be denoted by subscripts.
Stoichiometry, where known, should be indicated by placing the subunit in parenthesis and indicating the number of subunits by use of a subscripted number following the close of the parenthesis (where the number of subunits is greater than one). This is already a formal recommendation of the NC-IUPHAR nicotinic acetylcholine receptor subcommittee (Lukas et al., 1999). However, stoichiometry should not be indicated unnecessarily.
Subunits should be listed in alphabetical, or numerical, sequence without punctuation between subunits. An exception arises in the case of subunits types denoted by a numeral (e.g. P2X2; P2X3), where a solidus should be placed between the subunits as previously recommended when describing receptors of unspecified stoichiometry (Khakh et al., 2001).
Examples of the recommended nomenclature are given in Tables 1 and 2.
Table 2.
NC-IUPHAR recommendations on receptor nomenclature
| Receptor with unspecified stoichiometry | Receptor with specified stoichiometry |
|---|---|
| Nicotinic AChα4β2 | Nicotinic ACh(α4)2(β2)3 |
| 5-HT3AB | 5-HT3(A)2(B)3 |
| GABAAα1β2γ2 | GABAA(α1)2(β2)2γ2 |
| Glyα1β | Gly(α1)2(β)3 |
| GluA1A2 | Glu(A1)2(A2)2 |
| P2X2/3 | P2X(2)23 |
Stoichiometry should not be indicated unnecessarily.
3. Ionotropic glutamate receptors (iGluRs)
The ionotropic glutamate receptors posed a special case to its subcommittee,1 due to historical circumstances (see Lodge, submitted for publication). The receptors had been classified by pharmacologists and named after the synthetic agonists AMPA, kainate and NMDA and by the end of the 1980's this terminology was firmly established (Watkins and Jane, 2006). The cloning of the subunits confirmed this pharmacological classification but, of course, added a wealth of complexity by virtue of the identification of the many constituent proteins. Various nomenclatures were introduced by the laboratories that cloned the subunits, so, for example, the same AMPA receptor subunit was called either GluR1 (Boulter et al., 1990), or GluR-A (Keinanen et al., 1990), and the same NMDA receptor subunit NMDAR1 (Moriyoshi et al., 1991), or ζ1 (Meguro et al., 1992). Table 3 presents the currently recommended subunit nomenclature together with a list of former appellations that should be avoided in the future. The kainate receptor subunits had a more consistent, but illogical, nomenclature starting at GluR5. The challenge was two-fold: to obtain a nomenclature that was logical for the ionotropic GluRs and one that was as consistent as possible with the general principles of the nomenclature for the LGIC superfamilies.
Table 3.
NC-IUPHAR recommended and previous nomenclature of ionotropic glutamate receptor subunits
| NC-IUPHAR subunit nomenclature | Previous nomenclatures |
|---|---|
| GluA1 | GLUA1, GluR1, GluRA, GluR-A, GluR-K1, HBGR1 |
| GluA2 | GLUA2, GluR2, GluRB, GluR-B, GluR-K2, HBGR2 |
| GluA3 | GLUA3, GluR3, GluRC, GluR-C, GluR-K3 |
| GluA4 | GLUA4, GluR4, GluRD, GluR-D |
| GluK1 | GLUK5, GluR5, GluR-5, EAA3 |
| GluK2 | GLUK6, GluR6, GluR-6, EAA4 |
| GluK3 | GLUK7, GluR7, GluR-7, EAA5 |
| GluK4 | GLUK1, KA1, KA-1, EAA1 |
| GluK5 | GLUK2, KA2, KA-2, EAA2 |
| GluN1 | GLUN1, NMDA-R1, NR1, GluRξ1 |
| GluN2A | GLUN2A, NMDA-R2A, NR2A, GluRε1 |
| GluN2B | GLUN2B, NMDA-R2B, NR2B, hNR3, GluRε2 |
| GluN2C | GLUN2C, NMDA-R2C, NR2C, GluRε3 |
| GluN2D | GLUN2D, NMDA-R2D, NR2D, GluRε4 |
| GluN3A | GLUN3A, NMDA-R3A, NMDAR-L, chi-1 |
| GluN3B | GLUN3B, NMDA-R3B |
| GluD1 | GluRδ1 |
| GluD2 | GluRδ2 |
Greek symbols in NMDA receptor subunit names were applied to the mouse orthologue only.
The committee took no time to reach the consensus that the AMPARs subunits should be renamed GluA1, GluA2, GluA3 and GluA4. An interim recommendation (Lodge and Dingledine, 2000) had concluded that these subunits be named GLUA1, GLUA2, GLUA3 and GLUA4 (Table 3). The decision to omit “R” conformed to the NC-IUPHAR general recommendation that it is preferable not to label a subunit as a receptor, given that many of these subunits do not form functional receptors when expressed alone. The new nomenclature adopted the same general principle but made two changes. First, it was agreed to adopt Glu, the three letter amino acid code for glutamate, rather than GLU, to identify the neurotransmitter. Secondly it was agreed to drop the use of subscripts (for the reasons set out above). This new nomenclature has two important attributes: first, it harmoniously combines the two commonly used nomenclatures (e.g. GluR1 and GluRA become GluA1). Second the protein name can be instantly derived from the gene name by the conversion to two letters: “RI” becoming “lu”: Thus GRIA1 translates to GluA1, GRIA2 translates to GluA2, etc. (Table 1). There was a discussion whether, indeed, the two names should be identical but the general consensus was that this could be confusing.
The NMDA receptor was similarly non-contentious and adopted the same pattern: NR1 becoming GluN1, NR2A becoming GluN2A and so forth (Table 3). Once again, the protein name mirrors the gene name, with just the two letter code difference (i.e., GRIN1 translates to GluN1, GRIN2A translates to GluN2A).
The problem with the kainate receptors is that the first subunit cloned was named GluR5 (Bettler et al., 1990) and this name has been widely adopted in the field (Table 3). Clearly, however, there are no functional reasons for considering the kainate receptor subunits as a continuum of the AMPAR subunits. Despite some structural and pharmacological similarities there is no evidence that the two receptor families co-assemble. Several solutions were put forward by the subcommittee, each with its own merits. After considerable deliberation, however, it was decided to take the radical step to rename the subunits as follows: GluK1, GluK2, GluK3, GluK4 and GluK5 to replace the names GluR5, GluR6, GluR7, KA-1 and KA-2, respectively (Table 3). This again has the virtue that the protein names mirror the gene names, which are GRIK1, GRIK2, GRIK3, GRIK4, GRIK5, respectively (Table 1). Of course, the committee realizes that there will be a period of adjustment whilst the users equate GluK1 with GluR5, etc., but our brains are highly plastic and so such adjustment should not pose a great difficulty. Indeed, the precedent has already been set by the voltage-gated ion channel community who readily embraced a new more logical nomenclature for K+ and Ca2+ channels (Catterall et al., 2005; Goldstein et al., 2005; Gutman et al., 2005; Kubo et al., 2005; Wei et al., 2005).
There are, of course, challenges ahead. In particular is the need that the community embraces the new nomenclature. Whether the subcommittee has reached the correct recommendation remains to be seen, but what is clear is that a consistent and logical nomenclature that is widely adopted is urgently required. There is also the need to establish a consistent nomenclature for the alternative splice variants and for the edited states of the subunits.
4. Criteria for identifying native receptors
We have suggested criteria for selecting receptor subtype heteromer candidates for inclusion on a native receptor list that is currently under development by the LGIC subcommittees of NC-IUPHAR. These criteria include two categories for recombinant receptors: showing that a given combination of subunits is expressed as a pentamer (Cys-loop family), tetramer (ionotropic glutamate receptors), or trimer (P2X receptors) (see Fig. 1) and that it has unique biophysical and/or pharmacological properties. There are three criteria for native receptors: (i) evidence for co-localization of subunits; (ii) physical evidence for subunit–subunit type interactions; and (iii) demonstration of specific function. The mechanism for selection involves a subcommittee of NC-IUPHAR who evaluate the quality of the evidence that receptor subtype candidates meet the criteria for inclusion on the receptor list. Our tentative selections utilize the new principle of three categories of receptors (Olsen and Sieghart, in press):
identified;
existence with high probability;
tentative.
The same review includes a working receptor list for GABAA receptors, with 26 total entries. We continue the use of a wild card nomenclature (see, for example, Lukas et al., 1999), when a subunit is not clearly identified.
5. Concluding remarks
Now that we know that there are virtually no more LGICs to be discovered in the human genome, we hope that the proposed nomenclature will be used for all mammalian species for a very long time.
Acknowledgments
We thank the Chairs and members of all the LGIC subcommittees of NC-IUPHAR for their work in developing a receptor nomenclature and database.
Footnotes
NC-IUPHAR subcommittee membership: Bernhard Bettler, Graham Collingridge (Chair), Ray Dingledine, Stephen F. Heinemann, Michael Hollmann, Juan Lerma, David Lodge, Mark Mayer, Masayoshi Mishina, Christophe Mulle, Shigetada Nakanishi, Richard Olsen, John A. Peters, Peter Seeburg, Michael Spedding, Jeffrey C. Watkins, Robert J. Wenthold.
References
- Barnard EA, Skolnick P, Olsen RW, Mohler H, Sieghart W, Biggio G, Braestrup C, Bateson AN, Langer SZ. International Union of Pharmacology. XV. Subtypes of gamma-aminobutyric acid A receptors: classification on the basis of subunit structure and receptor function. Pharmacol Rev. 1998 Jun;50(2):291–313. [PubMed] [Google Scholar]
- Bettler B, Boulter J, Hermans-Borgmeyer I, O'Shea-Greenfield A, Deneris ES, Moll C, Borgmeyer U, Hollmann M, Heinemann S. Cloning of a novel glutamate receptor subunit, GluR5: expression in the nervous system during development. Neuron. 1990;5:583–595. doi: 10.1016/0896-6273(90)90213-y. [DOI] [PubMed] [Google Scholar]
- Boulter J, Hollmann M, O'Shea-Greenfield A, Hartley M, Deneris E, Maron C, Heinemann S. Molecular cloning and functional expression of glutamate receptor subunit genes. Science. 1990;249:1033–1037. doi: 10.1126/science.2168579. [DOI] [PubMed] [Google Scholar]
- Catterall WA, Perez-Reyes E, Snutch TP, Striessnig J. International Union of Pharmacology. XLVIII. Nomenclature and structure-function relationships of voltage-gated calcium channels. Pharmacol Rev. 2005;57:411–425. doi: 10.1124/pr.57.4.5. [DOI] [PubMed] [Google Scholar]
- Clapham DE, Julius D, Montell C, Schultz G. International Union of Pharmacology. XLIX. Nomenclature and structure-function relationships of transient receptor potential channels. Pharmacol Rev. 2005;57:427–450. doi: 10.1124/pr.57.4.6. [DOI] [PubMed] [Google Scholar]
- Goldstein SAN, Bayliss DA, Kim D, Lesage F, Plant LD, Rajan S. International Union of Pharmacology. LV. Nomenclature and molecular relationships of two-P potassium channels. Pharmacol Rev. 2005;57:527–540. doi: 10.1124/pr.57.4.12. [DOI] [PubMed] [Google Scholar]
- Gutman GA, Chandy KG, Grissmer S, Lazdunski M, Mckinnon D, Pardo LA, Robertson GA, Rudy B, Sanguinetti MC, Stühmer W, Wang X. International Union of Pharmacology. LIII. Nomenclature and molecular relationships of voltage-gated potassium channels. Pharmacol Rev. 2005;57:473–508. doi: 10.1124/pr.57.4.10. [DOI] [PubMed] [Google Scholar]
- Hofmann F, Biel M, Kaupp UB. International Union of Pharmacology. LI. Nomenclature and structure-function relationships of cyclic nucleotide-regulated channels. Pharmacol Rev. 2005;57:455–462. doi: 10.1124/pr.57.4.8. [DOI] [PubMed] [Google Scholar]
- Hoyer D, Clarke DE, Fozard JR, Hartig PR, Martin GR, Mylecharane EJ, Saxena PR, Humphrey PPA. VII. International Union of Pharmacology classification of receptors for 5-hydroxytryptamine (Serotonin) Pharmacol Rev. 1994;46:157–203. [PubMed] [Google Scholar]
- Keinanen K, Wisden W, Sommer B, Werner P, Herb A, Verdoorn TA, Sakmann B, Seeburg PH. A family of AMPA-selective glutamate receptors. Science. 1990;249:556–560. doi: 10.1126/science.2166337. [DOI] [PubMed] [Google Scholar]
- Khakh BS, Burnstock G, Kennedy C, King BF, North RA, Séguéla P, Voigt M, Humphrey PPA. International Union of Pharmacology. XXIV. Current status of the nomenclature and properties of P2X receptors and their subunits. Pharmacol Rev. 2001;53:107–118. [PubMed] [Google Scholar]
- Kubo Y, Adelman JP, Clapham DE, Jan LY, Karschin A, Kurachi Y, Lazdunski M, Nichols CG, Seino S, Vandenberg CA. International Union of Pharmacology. LIV. Nomenclature and molecular relationships of inwardly rectifying potassium channels. Pharmacol Rev. 2005;57:509–526. doi: 10.1124/pr.57.4.11. [DOI] [PubMed] [Google Scholar]
- Lodge D. The history of the pharmacology and cloning of ionotropic glutamate receptors and the development of idiosyncratic nomenclature. Neuropharmacology. doi: 10.1016/j.neuropharm.2008.08.006. submitted for publication. [DOI] [PubMed] [Google Scholar]
- Lodge D, Dingledine R. The IUPHAR Receptor Compendium. IUPHAR Media; 2000. Ionotropic glutamate receptors; pp. 189–194. [Google Scholar]
- Lukas RJ, Changeux JP, Le Novere N, Albuquerque EX, Balfour DJK, Berg DK, Bertrand D, Chiappinelli VA, Clarke PBS, Collins AC, Dani JA, Grady SR, Kellar KJ, Lindstrom JM, Marks MJ, Quik M, Taylor PW, Wonnacott S. International Union of Pharmacology. XX. Current status of the nomenclature for nicotinic acetylcholine receptors and their subunits. Pharmacol Rev. 1999;51:397–401. [PubMed] [Google Scholar]
- Matzenbach B, Maulet Y, Sefton L, Courtier B, Avner P, Guénet JL, Betz H. Structural analysis of mouse glycine receptor alpha subunit genes. Identification and chromosomal localization of a novel variant. J Biol Chem. 1994;269:2607–2612. [PubMed] [Google Scholar]
- Meguro H, Mori H, Araki K, Kushiya E, Kutsuwada T, Yamazaki M, Kumanishi T, Arakawa M, Sakimura K, Mishina M. Functional characterization of a heteromeric NMDA receptor channel expressed from cloned cDNAs. Nature. 1992;357:70–74. doi: 10.1038/357070a0. [DOI] [PubMed] [Google Scholar]
- Moriyoshi K, Masu M, Ishii T, Shigemoto R, Mizuno N, Nakanishi S. Molecular cloning and characterization of the rat NMDA receptor. Nature. 1991;354:31–37. doi: 10.1038/354031a0. [DOI] [PubMed] [Google Scholar]
- Olsen RW, Sieghart W. International Union of Pharmacology. (***) Subtypes of γ-aminobutyric acidA receptors: classification on the basis of subunit composition, pharmacology, and function. Update. Pharmacol Rev. doi: 10.1124/pr.108.00505. in press. [DOI] [PubMed] [Google Scholar]
- Schoepfer R, Conroy WG, Whiting P, Gore M, Lindstrom J. Brain alpha-bungarotoxin binding protein cDNAs and MAbs reveal subtypes of this branch of the ligand-gated ion channel gene superfamily. Neuron. 1990;5:35–48. doi: 10.1016/0896-6273(90)90031-a. [DOI] [PubMed] [Google Scholar]
- Simon JA, Wakimoto H, Fujita N, Lalande M, Barnard EA. Analysis of the set of GABAA receptor genes in the human genome. J Biol Chem. 2004;279:41422–41435. doi: 10.1074/jbc.M401354200. [DOI] [PubMed] [Google Scholar]
- Watkins JC, Jane DE. The glutamate story. Br J Pharmacol. 2006;147:S100–S108. doi: 10.1038/sj.bjp.0706444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei AD, Gutman GA, Aldrich R, Chandy KG, Grissmer S, Wulff H. International Union of Pharmacology. LII. Nomenclature and molecular relationships of calcium-activated potassium channels. Pharmacol Rev. 2005;57:463–472. doi: 10.1124/pr.57.4.9. [DOI] [PubMed] [Google Scholar]


