Abstract
We identified Pumilio (Pum), a Drosophila translational repressor, in a computational search for metazoan proteins whose activities might be regulated by assembly into ordered aggregates. The search algorithm was based on evolutionary sequence conservation patterns observed for yeast prion proteins, which contain aggregation-prone glutamine/asparagine (Q/N)-rich domains attached to functional domains of normal amino acid composition. We examined aggregation of Pum and its nematode ortholog PUF-9 by expression in yeast. A domain of Pum containing the Q/N-rich sequence, denoted as NQ1, the entire Pum N terminus, and the complete PUF-9 protein localize to macroscopic aggregates (foci) in yeast. NQ1 and PUF-9 can generate the yeast Pin+ trait, which is transmitted by a heritable aggregate. NQ1 also assembles into amyloid fibrils in vitro. In Drosophila, Pum regulates postsynaptic translation at neuromuscular junctions (NMJs). To assess whether NQ1 affects synaptic Pum activity in vivo, we expressed it in muscles. We found that it negatively regulates endogenous Pum, producing gene dosage-dependent pum loss-of-function NMJ phenotypes. NQ1 coexpression also suppresses lethality and NMJ phenotypes caused by overexpression of Pum in muscles. The Q/N block of NQ1 is required for these phenotypic effects. Negative regulation of Pum by NQ1 might be explained by formation of inactive aggregates, but we have been unable to demonstrate that NQ1 aggregates in Drosophila. NQ1 could also regulate Pum by a “dominant-negative” effect, in which it would block Q/N-mediated interactions of Pum with itself or with cofactors required for translational repression.
Introduction
Accumulation of intracellular or extracellular aggregates is a feature of many diseases. At least 40 human diseases are associated with the formation of amyloid fibrils, deposits, or inclusions (Chiti and Dobson, 2006). To understand these diseases, it is necessary to define the normal functions of aggregation-prone regions of proteins and how these proteins are perturbed in disease states.
Sequences found in normal proteins, including sequences enriched in glutamine (Q) and asparagine (N), have a propensity to assemble into ordered aggregates. Protein misfolding also generates aggregates. Some aggregates are toxic, whereas others might be formed as a protective response to a pathogenic event.
The study of fungal prions has provided evidence for the idea that controlled aggregation can have regulatory functions. In Podospora anserina, aggregation of the prion protein [HET-s] controls heterokaryon incompatibility during the normal life cycle. In yeast (Saccharomyces), aggregation of the Q/N-rich domain of the Sup35p prion protein, which encodes a translation termination factor, produces the [PSI+] phenotype (for review, see Wickner et al., 2007).
In this paper, we examine the properties of a Q/N-rich domain of the Drosophila Pumilio (Pum) protein. During early development, Pum controls pattern formation by binding to the 3′ untranslated region (UTR) of hunchback mRNA and repressing its translation in the posterior half of the embryo. It also has a variety of functions in the nervous system. At the larval neuromuscular junction (NMJ), Pum represses postsynaptic eIF-4E and GluRIIA expression and binds directly to the 3′ UTRs of both mRNAs. eIF-4E, the cap-binding protein, is often limiting for translation, so Pum might repress postsynaptic translation of many mRNAs by controlling the levels of eIF-4E. NMJ bouton numbers are increased in larvae lacking postsynaptic Pum, and this is at least partially attributable to an increase in synaptic eIF-4E levels (Menon et al., 2004, 2009).
Pum is a large (1533 aa) protein with a conserved C-terminal PUF RNA-binding domain (RBD) that has 80% sequence identity to human Pum2. The N-terminal 1092 aa of Pum contains the Q/N-rich domain and is required for rescue of pum NMJ phenotypes (Menon et al., 2004) and repression of Na+ channel expression (Muraro et al., 2008).
Materials and Methods
Yeast strains and methods.
Plasmids and strains were obtained from Jonathan Weissman's group at University of California, San Francisco. The aggregation-prone domain (PrD) of New1p is fused to enhanced cyan fluorescent protein (CFP) under control of the CUP1 promoter within the pRS426 (URA3) plasmid. We substituted Pum and PUF-9 sequences for New1p sequences in this plasmid. For focus formation and Pin+ assays, yeast expressing CUP1 promoter-driven fusion proteins were induced at an optical density of 6 and grown at 30°C. For Pin+ assays, they were plated after 48 h of growth onto −Ade plates to select the [PSI+] phenotype.
[psi−] [pin−], [psi−] [PIN+], and [PSI+] [PIN+] yeast strains used in this study bear the ade1–14(UGA) mutation and are also ura3- and leu2 (Osherovich and Weissman, 2001). We used two sets of strains for these experiments. The first set are isogenic [psi−] and [PSI+] derivatives of 74D-694 [MATa, his3, leu2, trp1, ura3, with the suppressible marker ade1–14(UGA)] (Chernoff et al., 1995). We also used strains created by Osherovich and Weissman by introducing 74D markers into the W303 background. These are as follows: YJW 508 ([PSI+] [PIN+] MATa, ade1-14, his3-11,15, leu2-3, trp1-1, ura3-1); YJW 509, a [psi−] [pin−] derivative of this strain; and YJW 564, a [psi−] [PIN+] derivative. Data in Figure 1 were obtained using the W303-derived strains, but the 74D strains gave equivalent results. Sup35NM–yellow fluorescent protein (YFP), used for assays of Pin+, is in the pRS425 (LEU2) plasmid.
Figure 1.
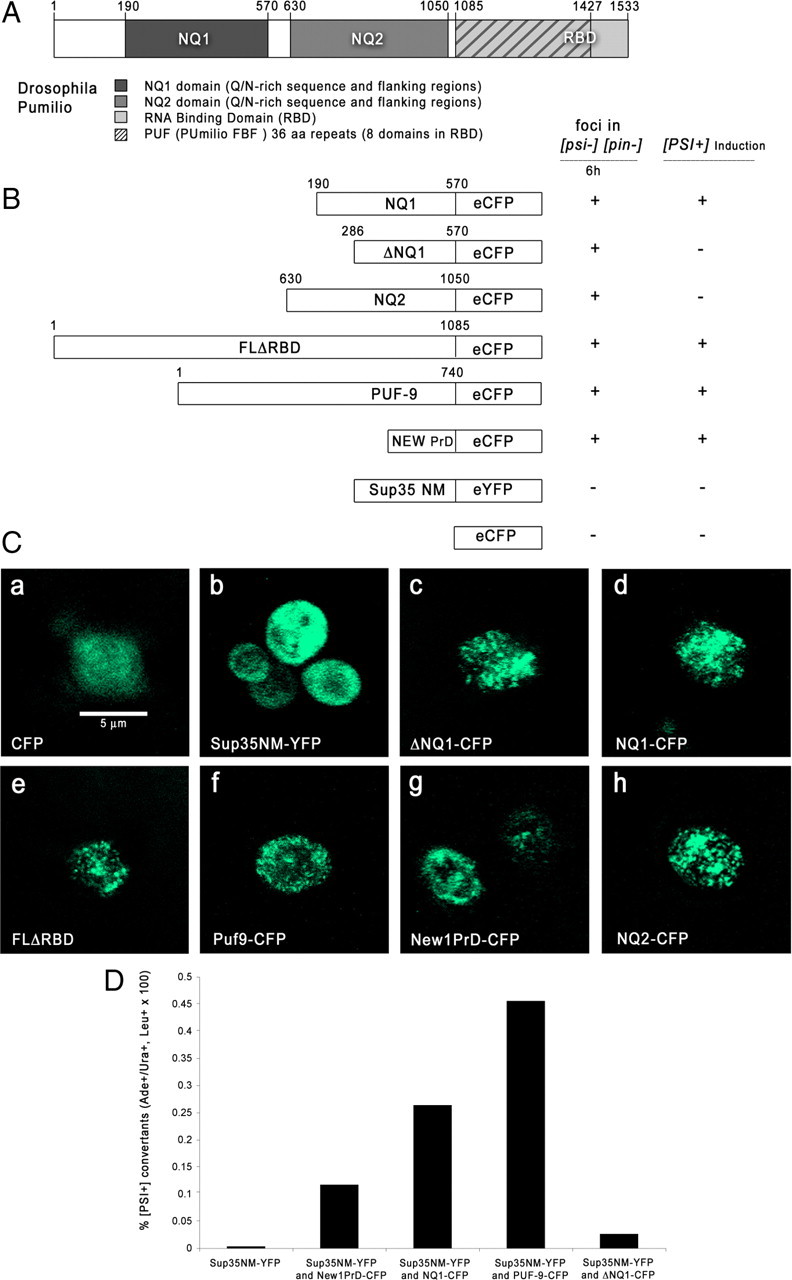
Pum and PUF-9 domains aggregate and generate Pin+ in yeast. A, Diagram of Drosophila Pum protein. Segments used for constructs are as indicated. For locations and sequences of Q/N blocks, see Results and supplemental Figure S1 (available at www.jneurosci.org as supplemental material). B, Diagrams of Pum and PUF-9 constructs and summary of aggregation and Pin+ data. FLΔRBD contains the whole N terminus but not the RBD domain. C, Aggregation of Pum proteins in yeast. Representative live cells expressing the indicated proteins were imaged by confocal microscopy using intrinsic GFP fluorescence after 6 h of copper induction. Scale bar (in a), 5 μm. D, Bar graph of percentage conversion of [psi−] [pin−] yeast to [PSI+] by transient expression of Sup35NM–YFP alone or in combination with the indicated constructs.
We tried to generate a chimera that could produce [PSI+] directly by substituting an NQ1–Sup35–EF chimeric gene for one copy of the Sup35 gene in diploids. A chromosomally integrated gene fusion of the NQ1–Sup35–EF chimera was generated by homologous recombination (gamma integration) of PCR-amplified sequences containing a His tag. These were transformed into diploid yeast strains and grown on −His plates. Resulting colonies were PCR screened for the presence of NQ1. The diploid strains, with both NQ1–Sup35–EF and endogenous Sup35, could be converted to [PSI+] by transformation with NQ1–CFP and Sup35NM–YFP.
To generate haploid lines with only NQ1–Sup35–EF, the diploid strains were sporulated using standard procedures, dissected, and grown on yeast/peptone/dextrose (YPD) plates. However, we could not obtain any viable haploid NQ1–Sup35 lines, indicating that the NQ1–Sup35p–EF chimera is not a functional translation termination factor. It might be completely sequestered into an inactive aggregate (total loss of Sup35p is lethal) or simply have the wrong conformation.
For guanidine hydrochloride (GdHCl) curing of [PSI+], yeast were grown on minimal media plates containing 5 mm GdHCl for 5–7 d at 30°C. Colonies were then streaked out on −Ade plates to evaluate the loss of [PSI+] and on YPD plates to visualize color reversion. Red colonies picked off the YPD plates were retested for failure to grow on −Ade plates.
In vitro conversion experiments.
pJC45–NM and M constructs (Lindquist Group, Massachusetts Institute of Technology, Cambridge, MA) were obtained from Addgene. NQ1 and ΔNQ1 were also cloned into pJC45 (Clos and Brandau, 1994), which produces protein fusions with a 10× His tag at the N terminus of the proteins.
Fusion proteins were induced in the BL21(DE3) strain with 100 mm isopropyl-β-d-thiogalactopyranoside for 4 h, and the cells were stored as a pellet. His tag purification on Ni-NTA agarose (Qiagen) in 8 m urea and 20 mm Tris, pH 8, using standard denaturing protocols was used to purify the protein (Serio et al., 1999). After elution in 400 mm imidazole, the fractions were precipitated in 100% MeOH and then washed and stored frozen in 70% MeOH.
Samples were resuspended at concentrations of 5–10 μm. An appropriate amount of slurry was centrifuged at high speed to make a pellet that would be briefly allowed to dry. A small amount of a solution (1:20 final volume) containing 8 m urea and 20 mm Tris, pH 8, was applied to the pellet to resuspend it. After this, PBS was added (19:20 final volume). The resuspension was centrifuged to remove insoluble material. Soluble fractions were then aliquoted into fresh tubes; for each time point to be assayed, 25 μl of sample was used. For NQ1, it was necessary to siliconize the tube with Sigmacote (Sigma), because the aggregates adhered to the walls of the vessel. The samples were then subjected to slow rotation at various temperatures for the indicated amount of time and measured for SDS solubility (Serio et al., 1999).
Negative staining of fibrils was done essentially as described previously (Glover et al., 1997; Serio et al., 1999). Five microliters of a 5 μm solution of fibrils were applied to a glow discharged, carbon-coated copper grid for 1 min; the liquid was removed and the preparation was stained with 2% uranyl acetate, followed by 1% uranyl acetate, with liquid removal using a paper wick after each staining application. Grids were allowed to air dry and then examined in a Tecnai T12 electron microscope. For NQ1, at this concentration of protein, the stain had a tendency to obscure the fibrils. Thus, to obtain images of fibrils, a 1:5 dilution in PBS (1 μm fibers) was used.
Drosophila genetics and molecular biology.
The UAS–NQ1–CFP and UAS–ΔNQ1–CFP plasmids are pUAST-based vectors. Injections into embryos were performed by Rainbow Genetics. We generated >100 lines for each plasmid and screened these for CFP expression after crossing to 24B–GAL4 and other drivers. We picked a subset of lines that had bright CFP expression and mapped them to chromosomes. NQ1–CFP and ΔNQ1–CFP were expressed at approximately equivalent levels in these lines. pumET9 was recombined with the 24B–GAL4 insert (both are on the third chromosome), and the recombinant line was crossed to UAS–NQ1–CFP and UAS–ΔNQ1–CFP lines with inserts on the second or third chromosomes. For the lethality rescue experiment (Table 1), we used second chromosome lines (from R. Wharton, Duke University, Durham, NC) bearing UAS–Pum with its own 3′ UTR or with a tubulin 3′ UTR, which increases translation efficiency. These were recombined with second chromosome UAS–NQ1–CFP inserts or combined with third chromosome inserts, and the resulting lines were crossed to 24B–GAL4. Data reported in Figures 4 and 5 are for second chromosome inserts of each type, but others had qualitatively similar phenotypes. Data in Table 1 are for one third chromosome insert and one second chromosome insert. The table lists the genotypes examined. UAS–Pum constructs were on the second chromosome and were either recombined with UAS–NQ1 inserts on the second chromosome or combined with inserts on the third chromosome. Both sets gave equivalent results when crossed to 24B–GAL4. The numbers reported are pupae counted from crosses with the same number of females and males.
Table 1.
Rescue of Pum overexpression lethality by coexpression of NQ1
| Genotype | Numbers of pupae counted |
|---|---|
| 24B–GAL4/+ | 324 |
| UAS–Pum/24B–GAL4 | 0 |
| UAS–Pum-tub3′UTR/24B–GAL4 | 0 |
| UAS–GFP/+; UAS–Pum/24B–GAL4 | 0 |
| UAS–GFP/+; UAS–Pum–tub3′UTR/24B–GAL4 | 0 |
| UAS–ΔNQ1–CFP/+; UAS–Pum/24B–GAL4 | 0 |
| UAS–NQ1–CFP, UAS–Pum/24B–GAL4 | 405 |
| UAS–NQ1–CFP, UAS–Pum–tub3′UTR/24B–GAL4 | 262 |
| UAS–NQ1–CFP/+; UAS–Pum/24B–GAL4 | 159 |
| UAS–NQ1–CFP/+; UAS–Pum–tub3′UTR/24B–GAL4 | 169 |
Figure 4.
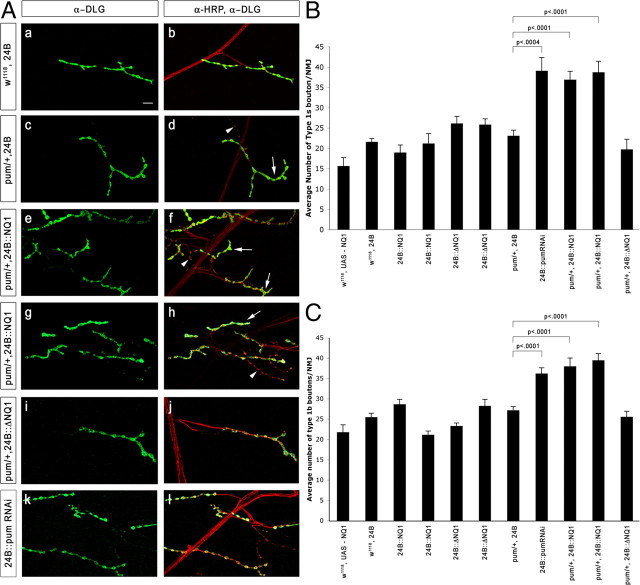
NMJ bouton number increases in pum/+ larvae expressing Pum NQ1 in muscles. A, Segment A2 or A3 muscle 4 NMJs from third-instar larvae of the indicated genotypes were stained with antibodies and imaged by confocal microscopy. Green, Anti-Dlg (Discs-large, a postsynaptic marker); red, anti-HRP (a presynaptic membrane marker). 1b boutons (arrows) have high levels of Dlg, and 1s boutons (arrowheads) have lower levels. Genotypes: a, b, 24B–GAL4 × w1118; c, d, pumET9, 24B–GAL4/+ larva from pumET9, 24B–GAL4/TM6B × w1118 cross; e–h, UAS–NQ1/pumET9, 24B–GAL4 (two different UAS–NQ1 lines are shown); i, j, UAS–ΔNQ1/pumET9, 24B–GAL4; k, l, 24B–GAL4 × UAS–pum RNAi. Scale bar (in a), 10 μm. B, Bar graph of 1s bouton counts. C, Bar graph of 1b bouton counts. Full genotypes and numbers of A2 and A3 hemisegments scored are in Materials and Methods.
Figure 5.
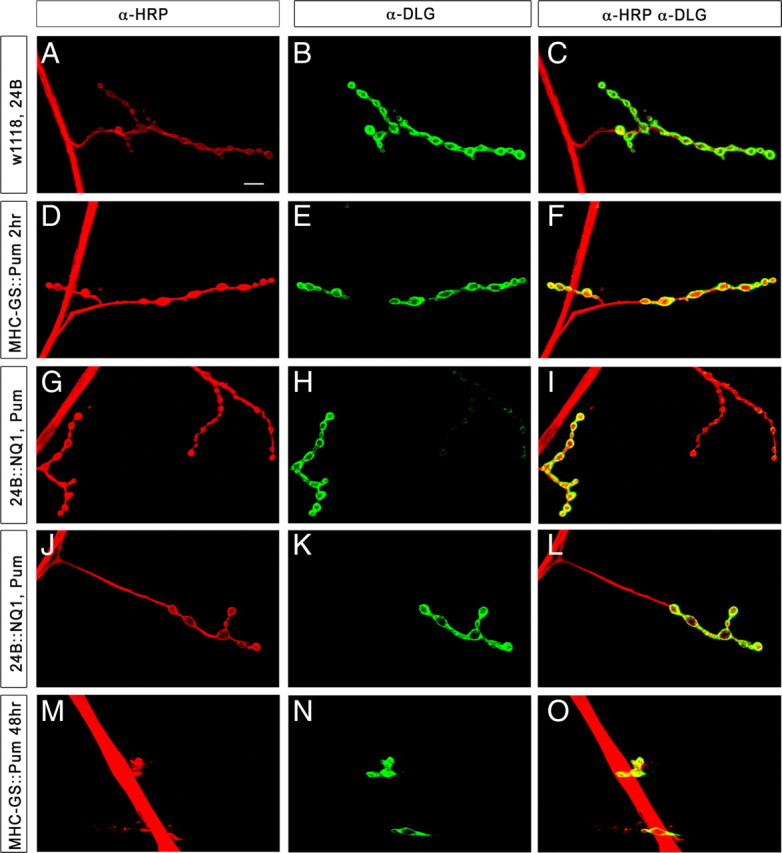
Rescue of the Pum postsynaptic overexpression NMJ phenotype by NQ1 coexpression. Segment A2 or A3, muscle 4 NMJs from third-instar larvae of the indicated genotypes were stained with antibodies and imaged by confocal microscopy. Green, Anti-Dlg (Discs large, a postsynaptic marker); red, anti-HRP (a presynaptic membrane marker). Scale bar (in A), 10 μm. Genotypes: A–C, 24B–GAL4 × w1118; D–F, MHC–GS–GAL4 × UAS–Pum, placed for 2 h on RU486 food; G–I, UAS–NQ1, UAS–Pum(line #1) × 24B–GAL4; J–L, UAS–NQ1, UAS–Pum(line #2) × 24B–GAL4; M–O, MHC–GS–GAL4 × UAS–Pum, incubated for 48 h on RU486 food.
Antibodies, immunocytochemistry, and confocal microscopy.
The following antibodies were used: rat anti-Pum RBD (from R. Wharton), mouse anti-Dlg (Discs large) and anti-GluRIIA (from the Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA), and tetramethylrhodamine isothiocyanate-conjugated HRP (Jackson Laboratories). Secondary antibodies used were AlexaFluor 488 anti-mouse and AlexaFluor 568 anti-rat (Invitrogen). Third-instar larvae were dissected and fixed as described previously (Menon et al., 2004).
A Zeiss LSM 510 inverted microscope was used for obtaining images of foci in yeast. Appropriate lasers were used to excite CFP and YFP fluorescence. A 100× Zeiss oil-immersion objective was used, and projected z-stacks with intervals of 0.25 μm were obtained for final images. Random fields were scanned; the images represent the majority of yeast cells observed.
The same inverted microscope was used for the creation of images of the NMJ. Third-instar larval muscle 4 NMJs from A2 and A3 segments were analyzed by confocal microscopy of antibody-stained preparations. Appropriate lasers were used to excite fluorescently tagged antibodies at either 488 or 568 nm. A 63× Zeiss oil-immersion objective was used, and projected z-stacks with intervals of 0.5 μm were obtained for final images.
Quantitation of bouton number increases in pum/+ larvae expressing NQ1 in muscles.
In Figure 4, genotypes are indicated below the x-axis; full genotypes and numbers of A2 and A3 hemisegments scored are as follows: UAS–NQ1/+ (n = 9); 24B–GAL4/+ (n = 34); UAS–NQ1(line #1)/24B–GAL4 (n = 13); UAS–NQ1(line #2)/24B–GAL4 (n = 9); UAS–ΔNQ1(line #1)/24B–GAL4 (n = 24); UAS–ΔNQ1(line #2)/24B–GAL4 (n = 16); pumET9, 24B–GAL4/+ (n = 34); UAS–PumFLRNAi/24B (n = 18); UAS–NQ1(line #1)/pumET9, 24B–GAL4 (n = 39); UAS-NQ1(line #2)/pumET9, 24B–GAL4 (n = 15); UAS–ΔNQ1(line #1)/pumET9, 24B–GAL4 (n = 17). Error bars are shown, and p values (Student's t test) for differences between pum/+ genotypes expressing versus not expressing postsynaptic NQ1 are indicated by brackets. The two bars from the UAS–NQ1/pum, 24B–GAL4 lines differ significantly from all the others (p < 0.0001), as do the bars for the UAS–RNAi, 24B–GAL4 lines for type 1s (small) boutons (p < 0.0004) and for type 1b (big) boutons (p < 0.0001); no other differences in bar height are significant. Similar results were obtained using another pum allele, pumMsc (data not shown).
Rescue of Pum muscle overexpression phenotype by co-overexpression of NQ1.
Upstream activating sequence (UAS) full-length Pum larvae bearing the strong muscle driver 24B–GAL4 died before third instar at all temperatures. To observe the Pum muscle overexpression phenotype in third-instar larvae, therefore, we used a genetic background in which an attenuated steroid-inducible GAL4 (GS–GAL4) was expressed in muscle via a myosin heavy chain (MHC) promoter (Osterwalder et al., 2001). Muscle overexpression of Pum was transiently induced by growing larvae for either 2 h (control) or 48 h on rich food containing 25 mg/ml of the steroid RU486 (11β-[p-(dimethylamino) phenyl]-17β-hydroxy-17-(1-propynyl)estra-4,9-dien-3-one) at room temperature. Dissection and staining were performed as described above.
Quantitation of NMJ bouton number increases in pum/+ larvae expressing Rox8 in muscles.
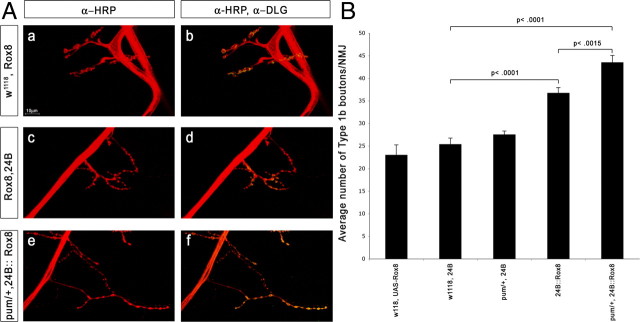
In Figure 6B, genotypes are indicated below the x-axis; full genotypes and numbers of A2 and A3 hemisegments scored are as follows: UAS–Rox8/+ (n = 9); 24B–GAL4/+ (n = 34); pumET9, 24B–GAL4/+ (n = 34); UAS–Rox8/24B–GAL4 (n = 28); UAS–Rox8/pumET9, 24B–GAL4 (n = 32). Error bars are shown, and p values (Student's t test) for differences between genotypes expressing postsynaptic Rox8 are indicated by brackets. The bar with postsynaptic Rox8 expression, UAS–Rox8/24B–GAL4, differs significantly from the bar with postsynaptic Rox8 expression with a copy of pum removed, UAS–Rox8/pumET9 (p < 0.0015). Both of these bars differ highly significantly from all the others (p < 0.0001); no other differences in bar height are significant.
Figure 6.
NMJ bouton number increases in pum/+ larvae expressing Rox8 in muscles. A, Segment A2 or A3 muscle 4 NMJs from third-instar larvae of the indicated genotypes were stained with antibodies and imaged by confocal microscopy. Green, Anti-Dlg (Discs large, a postsynaptic marker); red, anti-HRP (a presynaptic membrane marker). Scale bar (in A), 10 μm. Genotypes: a, b, Rox8 × w1118; c, d, UAS–Rox8 × 24B–GAL4; e, f, UAS–Rox8/pumET9, 24B–GAL4. B, Bar graph of 1b bouton counts. Full genotypes and numbers of hemisegments scored are in Materials and Methods. Rox8 overexpression does not produce major changes in 1s bouton counts (supplemental Fig. S4, available at www.jneurosci.org as supplemental material).
Results
A computational search for aggregation-prone regulatory proteins
A census of metazoan proteomes identified 143 proteins in Caenorhabditis elegans and 472 proteins in Drosophila that have Q/N blocks, defined as regions with 30 Q or N residues within an 80 aa window (Michelitsch and Weissman, 2000). To parse this large collection of proteins for those few that might use aggregation as a normal regulatory mechanism, we used the structures and sequence conservation patterns of yeast prions, which have a bipartite structure in which the Q/N-rich, aggregation-prone domain (PrD) is linked to a functional domain, as a guide.
We focused on regulatory proteins containing annotated nucleic acid-binding domains. Q/N blocks are rapidly evolving elements that are seldom conserved within an orthologous pair of proteins from distantly related species. We reasoned that, if the Q/N-rich amino acid composition of a sequence element is important for the function of a regulatory protein, its Drosophila and C. elegans orthologs would retain both the nucleic acid binding domain (identified by sequence conservation) and a Q/N block (identified by conservation of amino acid composition but not sequence). This search represents a stringent selection, because arthropods and nematodes are very distantly related, and it identified only two protein pairs: Pum (fly):PUF-9 (worm) and Kismet (fly):T04D1.4 (worm). Kismet is a chromatin-binding protein.
The Pum Q/N block identified in our search (amino acids 191-286; 42 Q/N in 95 aa) is upstream of a sequence (amino acids 378-538) that is conserved (33% identity) between arthropods and mammals. Amino acids 652-751 are also Q/N rich (36 Q/N). All vertebrate Pum proteins have Q/N-rich sequences. The N terminus (340 aa) of PUF-9 also contains a Q/N block (37 Q/N in 97 aa) (supplemental Fig. S1, available at www.jneurosci.org as supplemental material).
Pum Q/N sequences aggregate in yeast and form amyloid fibrils in vitro
To assess whether the Q/N domains identified in our search are actually aggregation prone, we examined macroscopic fluorescent protein aggregates (foci) in yeast cells. We attached DNA encoding a region containing the Pum Q/N block and conserved element (NQ1, amino acids 190-571) to the CFP gene and inserted it into a yeast vector with a copper-inducible promoter. A parallel construct, ΔNQ1–CFP, was made by deleting the Q/N block. Similar CFP fusions were made with a Pum region containing the amino acids 652-751 Q/N block (NQ2), the entire Pum N terminus, and the entire PUF-9 protein (Fig. 1A). As controls, we obtained yeast prion constructs in the same vector, encoding New1p–CFP (PrD, amino acids 1-153) and Sup35NM–YFP (PrD, amino acids 1-253) (Osherovich and Weissman, 2001).
Each construct was introduced into [psi−] [pin−] cells, which lack aggregated Rnq1p. Rnq1p aggregation generates “[PSI+] inducibility” ([PIN+]), a prion trait that is required to drive aggregation of the Sup35p PrD and allow conversion to [PSI+] (Derkatch et al., 1997). Generation of foci in [pin−] cells has been used as an initial test of the ability of a protein to form heritable aggregates (Derkatch et al., 2001; Osherovich and Weissman, 2001). When we examined cells shortly after (6 h) the onset of copper induction, so as to avoid expressing the proteins at excessive levels, we observed that discrete foci of CFP appeared in cells expressing all Pum and PUF-9 constructs, indicating that the proteins had coalesced into aggregates (Fig. 1C). Surprisingly, ΔNQ1–CFP also formed foci, indicating that the Q/N block we deleted is not the only sequence in NQ1 that can drive aggregation in yeast. In agreement with published data, New1p–CFP also formed foci, whereas Sup35NM–YFP remained diffuse (Osherovich and Weissman, 2001). Sup35NM–YFP formed foci when coexpressed with New1p, NQ1, or PUF-9 (data not shown).
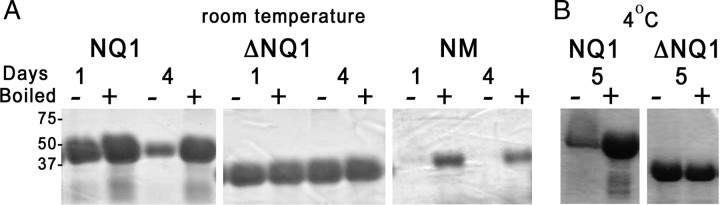
Fungal prion proteins form amyloid fibrils under neutral-pH conditions. To test this property for Pum, we solubilized NQ1, ΔNQ1, and Sup35pNM from Escherichia coli inclusion bodies and slowly rotated the samples, removing aliquots at intervals to assess conversion to amyloid using an SDS gel assay (Fig. 2). We examined the samples at various times during the conversion process by electron microscopy. Sup35NM and NQ1 samples converted for 4 d contained networks of fibrils (Fig. 3), whereas preconversion samples did not (supplemental Fig. S2, available at www.jneurosci.org as supplemental material). ΔNQ1 converted more slowly than NQ1 in the gel assay (Fig. 2) but also generated fibrils. Thus, as in the yeast assay, ΔNQ1 retains the ability to aggregate.
Figure 2.
Conversion of Pum NQ1 to amyloid. A, Polymerization of the indicated proteins into SDS-insoluble aggregates at room temperature, assessed by gel electrophoresis. Protein solutions in PBS were slowly rotated for the indicated times. M is a negative control that does not convert; it is Sup35p PrD amino acids 124-253, C-terminal to the Q/N region. + samples were boiled in 2% SDS for 10 min to dissociate aggregates, whereas − samples were incubated in SDS but not boiled. The differences in band intensity between − and + samples reflects assembly into insoluble aggregates that do not enter the gel unless they are boiled. This is a standard assay for amyloid aggregation. Numerals indicate molecular weight marker positions. B, Polymerization at 4°C. Note that Sup35NM (labeled as NM in the figure) converts within 1 d, whereas 4 d are required to achieve conversion of most of the NQ1. NQ1 does not convert at all within this timeframe but will convert if allowed to rotate for a longer time period.
Figure 3.
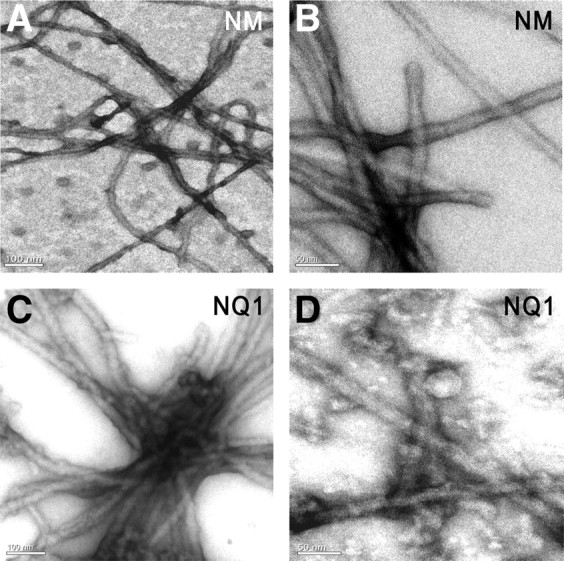
A Pum domain forms amyloid fibrils. A, B, Sup35NM (labeled as NM on the panel) fibrils visualized by transmission electron microscopy after 4 d of polymerization. C, D, NQ1 fibrils after 4 d. Scale bars: A, C, 100 nm; B, D, 50 nm.
Pum Q/N sequences can generate the Pin+ trait in yeast
We first attempted to test whether NQ1 could confer [PSI+] by replacing the PrD of chromosomal Sup35p with NQ1 but found that NQ1–Sup35 haploid yeast were not viable. However, diploids bearing both NQ1–Sup35 and Sup35 could convert to [PSI+] (for details, see Materials and Methods).
We then examined whether NQ1 or PUF-9 can generate Pin+, which is normally carried by aggregated Rnq1p but can also be conferred by other prion proteins such as New1p (Osherovich and Weissman, 2001). To do this, we transiently expressed CFP fusion proteins in ade1–stop, [psi−] [pin−] cells together with Sup35NM–YFP and selected transformants for [PSI+]. [PSI+] cells grow in the absence of adenine because aggregation of Sup35p causes read-through of the stop codon. In a [psi−] [pin−] strain, expression of Sup35NM–YFP cannot cause aggregation of Sup35p unless Rnq1p or a domain that can replace it is coexpressed (Derkatch et al., 2001; Osherovich and Weissman, 2001).
In this assay, we found that NQ1 and PUF-9 transformed yeast to adenine prototrophy with efficiencies equal to or greater than that of New1p (Fig. 1D). New1p is more effective than Rnq1p itself in conferring Pin+ (Osherovich and Weissman, 2001), so our data suggest that NQ1 and PUF-9 can generate this phenotype as efficiently as an endogenous yeast prion. ΔNQ1 produced Ade+ transformants at levels only slightly greater than the background frequency produced by Sup35NM–YFP alone, showing that the Q/N block is required for generation of Pin+. This is interesting because ΔNQ1 is able to generate foci in yeast and generates amyloid fibrils, implying that these assays are less stringent than the Pin+ assay. This is supported by the observations that non-Q/N-rich green fluorescent protein (GFP) fusion proteins can aggregate in yeast if expressed at sufficiently high levels (data not shown) and that many proteins can form amyloid fibrils under the correct conditions. Transformation to Ade+ by NQ1 was reversed by plating cells on low concentrations of guanidine hydrochloride, which inhibits the Hsp104 chaperone and eliminates [PSI+] (data not shown).
The Q/N domain negatively regulates postsynaptic Pumilio function
To examine the properties of NQ1 in its native system, we generated many transgenic Drosophila lines containing constructs with NQ1–CFP and ΔNQ1–CFP in a vector with a “UAS” promoter, which is activated by the yeast transcription factor GAL4, and selected lines for analysis that produced bright CFP expression when crossed to the “driver” line 24B–GAL4, which expresses GAL4 in all muscles. Macroscopic fluorescent foci were not seen in embryos or larvae expressing NQ1–CFP in muscles or in neurons (supplemental Fig. S3, available at www.jneurosci.org as supplemental material).
To analyze the phenotypic consequences of postsynaptic NQ1 expression, we examined the structures of third-instar larval NMJs. Type 1b (big) and 1s (small) bouton branches within an NMJ are both glutamatergic but are from different neurons. Type 1b boutons have higher levels of the postsynaptic marker Dlg, allowing them to be distinguished from type 1s. We initially counted 1b and 1s boutons at NMJs from F1 larvae expressing NQ1 in postsynaptic muscle cells (24B–GAL4 × UAS–NQ1–CFP, henceforth abbreviated as 24B::NQ1) but observed no differences from controls (Fig. 4B).
To sensitize the assay and focus on potential genetic interactions between NQ1 and pum, we then recombined the strong (frame shift) mutation pumET9 with the 24B–GAL4 insert. pumET9/+ animals have normal numbers of NMJ boutons. When several independent UAS–NQ1–CFP lines were crossed to pumET9, 24B–GAL4/TM6B, however, we observed a striking and highly significant (p < 0.0001) NMJ phenotype in heterozygous F1 larvae (pumET9/+, 24B::NQ1) (data shown for two lines in Fig. 4). The number of 1s boutons at the muscle 4 NMJ was increased by up to twofold in these larvae relative to pumET9/+, 24B::ΔNQ1 and to all control animals (pumET9, 24B–GAL4/+; pumET9/+; driver alone; and UAS–NQ1–CFP alone) (Fig. 4Af,Ah,B). The number of 1b boutons was increased by up to 1.5-fold (Fig. 4Ae,Ag,C).
The magnitude of the effect of postsynaptic NQ1 expression in a pum/+ background on 1s bouton number matches the increase in 1s bouton number that our group found previously in pum transheterozygous loss-of-function (LOF) mutants (pumET9/pumET7). This increase was reversed by restoring Pum to muscles in these mutants, showing that it is a postsynaptic phenotype. Type 1b bouton numbers are not increased in pumET9/pumET7 larvae because of a superimposed synaptic growth defect caused by loss of Pum from neurons (Menon et al., 2004). To determine whether the increase in 1b bouton number seen in pumET9/+, 24B::NQ1 larvae corresponds to a postsynaptic pum LOF phenotype, we expressed a UAS–pum double-stranded RNA [RNA interference (RNAi)] in muscles using 24B–GAL4. 24B::pum RNAi larvae had increases in both 1b and 1s bouton numbers (Fig. 4Ak,Al,B,C), indicating that a reduction of Pum function only on the postsynaptic side does increase 1b bouton number.
These results show that NQ1 displays a dosage-dependent genetic interaction with pum. This interaction is dependent on the Q/N block, because postsynaptic expression of ΔNQ1 in the same genetic backgrounds did not produce phenotypes (Fig. 4Aj,B,C).
Rescue of Pumilio gain-of-function phenotypes by NQ1
When full-length Pum is expressed in muscles from a strong, early GAL4 driver like 24B–GAL4, no larvae appear, indicating that overexpression is lethal during embryogenesis or at hatching. To examine whether Pum toxicity could be rescued by NQ1 expression, we made lines containing both UAS–Pum (with its own 3′ UTR or with a tubulin 3′ UTR) and UAS–NQ1–CFP. When UAS–Pum lines were crossed to 24B–GAL4 at 29°C, no 24B::Pum larvae emerged. However, when combined UAS–Pum, UAS–NQ1 lines were crossed under the same conditions, hundreds of 24B::Pum+NQ1 larvae appeared and survived through the pupal stage. The Q/N block is required for this rescue activity, because 24B::Pum+ΔNQ1 larvae did not survive (Table 1). The UAS–ΔNQ1 data also show that rescue by UAS–NQ1 is unlikely to be attributable to titration of GAL4 by the presence of two UAS constructs. We confirmed this by showing that a 24B–GAL4, UAS–GFP line produced complete larval lethality when crossed to both UAS–Pum lines. These results indicate that coexpression of NQ1 produces a strong suppression of the toxic effects caused by muscle overexpression of Pum.
Because 24B–GAL4-driven Pum causes complete larval lethality, we defined the postsynaptic Pum overexpression NMJ phenotype using an inducible (and weaker) muscle driver, MHC–GeneSwitch–GAL 4 (MHC–GS–GAL4), to transiently overexpress Pum in the muscles. This was accomplished through growing larvae for 48 h in the presence of RU486, which activates the GS–GAL4 protein (Osterwalder et al., 2001). These Pum overexpression larvae exhibited dramatic NMJ phenotypes, having stunted presynaptic terminals with only a few boutons (Fig. 5M–O). In contrast, in F1 larvae from crosses of UAS–Pum, UAS–NQ1 to 24B–GAL4, which is a stronger driver than MHC–GS–GAL4 and also expresses continuously from the embryonic stage, we observed normal NMJ bouton morphology (Fig. 5G–L). These data show that both lethality and NMJ morphology are rescued by co-overexpressing NQ1 with full-length Pum in the muscle, providing additional evidence for a genetic interaction between Pum and NQ1.
Genetic interactions between pumilio and Rox8
Mouse Pum2 is localized to stress granules (SGs) in the dendrites of cultured hippocampal neurons, and the Q/N domain of Pum2 forms aggregates in transfected mammalian cells (Vessey et al., 2006). Induction of SGs requires a Q/N-rich domain of the core TIA-1 protein, and the yeast Sup35p PrD can substitute for this TIA-1 domain (Gilks et al., 2004). Pum is also found in granules in cultured Drosophila neurons (Barbee et al., 2006). These data suggest that Q/N sequences might regulate Pum function by controlling its assembly into granular structures. To examine this issue, we overexpressed Rox8, the Drosophila TIA-1 ortholog, in the muscles of Drosophila larvae. Rox8 overexpression increases 1b bouton numbers (Fig. 6Ad,B).
Removing a copy of wild-type pum in a Rox8 overexpression background (pumET9/+, 24B::Rox8) results in an additional increase in 1b bouton number, revealing a genetic interaction between Pum and Rox-8 (Fig. 6Af,B). The numbers of 1b boutons in pumET9/+, 24B::Rox8 and 24B::pum RNAi NMJs are similar, suggesting that postsynaptic Pum function may be reduced when Rox8 is overexpressed in a heterozygous pum background. Antibodies against Rox-8 do not stain tissue, and we did not see coalescence of muscle Pum staining into obvious granules when Rox-8 was overexpressed (data not shown). Although the mechanisms involved in the effects of Rox8 on Pum are unknown, our results do suggest that Pum function can be perturbed by altering expression of the core protein of SGs.
Discussion
We identified an aggregation-prone, Q/N-rich segment of the Drosophila Pum protein, which is a translational repressor, as a potential regulatory domain using a computational search based on evolutionary conservation. We showed that this protein segment, NQ1, downregulates postsynaptic Pum function using three different assays (Figs. 4, 5; Table 1). NQ1 and the nematode Pum ortholog PUF-9 aggregate when expressed in yeast (Fig. 1), and NQ1 forms amyloid fibrils in vitro (Figs. 2, 3). NQ1 and PUF-9 can also generate the yeast Pin+ trait (Fig. 1D). Although Pin+ is normally conferred by aggregation of the Rnq1p prion (Derkatch et al., 2001), these results do not show that NQ1 or PUF-9 act as prions in yeast but only that they can produce a phenotype that is transmitted by a heritable aggregate.
What is the relationship between the aggregation behavior of NQ1 in yeast and its ability to negatively regulate postsynaptic Pum function in Drosophila? In a prion-like regulatory model, NQ1 would sequester Pum into inactive aggregates that are not readily visualized by light microscopy. Studies of the aggregation of the Aplysia RNA-binding protein CPEB (cytoplasmic polyadenylation element binding protein) in yeast were used as the basis for a model in which CPEB aggregation would control its activity at synapses (Si et al., 2003). However, there is no experimental evidence that NQ1, Pum, CPEB, or other translational regulators actually form aggregates in neurons or muscles. We analyzed NQ1-expressing larval body wall lysates using the SDD-AGE gel electrophoresis assay (Bagriantsev et al., 2006) and detected only monomeric NQ1. We saw no slowly migrating smears, which are taken as indicators of aggregation in yeast lysates (data not shown).
It is possible that the similarities between Pum and yeast prions could be informative with regard to the nature of the protein–protein interactions in which Pum participates rather than as direct indicators that Pum has prion-like activity. Our results are also consistent with a model in which NQ1 acts as a “dominant negative,” perturbing Q/N-mediated interactions of Pum with itself or with essential cofactors. The N terminus of Pum containing the Q/N-rich regions is required for rescue of pum NMJ phenotypes (Menon et al., 2004) and repression of Na+ channel expression (Muraro et al., 2008), perhaps through protein partners that interact with the Pum Q/N region. The Q/N-rich N terminus of human Pum2 is required for Pum2 to bind to itself in a yeast two-hybrid assay (Jaruzelska et al., 2003). There is also evidence that Pum activity requires dimer formation, because the Pum RBD forms dimers in vitro when bound to NRE sequences (Gupta et al., 2009).
Mammalian Pum2 is localized to dendritic stress granules in cultured hippocampal neurons (Vessey et al., 2006). Drosophila Pum can also be found in granules in cultured neurons, although these were classified as P-bodies rather than SGs (Barbee et al., 2006). P-bodies are cytoplasmic complexes that are sites for translational repression, deadenylation, and degradation of mRNAs. All core P-body proteins have Q/N-rich sequences. The Q/N-rich domain of the P-body protein Lsm4p can be replaced by the PrD of Rnq1p (Decker et al., 2007), and Lsm4p can substitute for the Rnq1p prion (Derkatch et al., 2001). The Ccr4p deadenylase requires a Q/N-rich domain for incorporation into P-bodies, and this domain forms foci (Reijns et al., 2008). Interestingly, the Ccr4p/Pop2p complex binds to PUF proteins, and this may be a mechanism by which PUF translational repressors recruit deadenylases to mRNAs (Goldstrohm et al., 2007).
SGs and P-bodies are found in dendrites and may be relevant to regulation of synaptic translation. Granules that contain Pum proteins also include a variety of synaptic translational regulators, including the Pum target eIF-4E, PolyA-binding protein, Staufen, and Fragile X protein (Barbee et al., 2006; Vessey et al., 2006).
Finally, given the fact that many known neurodegenerative diseases are linked to protein aggregation, it is interesting to speculate that some aggregation-related neurodegenerative conditions could involve perturbation of Q/N-rich proteins found at synapses, such as Pumilio, or of Q/N-mediated dendritic granule assembly pathways.
Footnotes
This work was supported by a McKnight Foundation Brain Disorders Award and by National Institutes of Health Grants RO1 NS28182, R01 NS62821, and RO1 NS43416 (K.Z.). We thank the following: Erich Schwarz (WormBase at the California Institute of Technology, Pasadena, CA) for conducting the computational search of the fly and worm proteomes for us; Dale Cameron, Lev Osherovich, and Jonathan Weissman for help with gamma integration; James Shorter, Johannes Graumann, Ashley Wright, Nina Sherwood, Alice Schmid, and Nicki Fox for helpful discussions; the Weissman (University of California, San Francisco, San Francisco, CA), Lindquist (Massachusetts Institute of Technology, Cambridge, MA), Deshaies (California Institute of Technology), and Wharton (Duke University, Durham, NC) groups for materials; and Elena Armand, Violana Nesterova, Lisa Nesterova, and Carlos Diaz-Balzac for technical assistance. We thank Bill Tivol for assistance with EM, which was performed at the Broad Center EM facility. Confocal microscopy was performed at the Caltech Biological Imaging Facility. We thank Sean Brennan for assistance with graphics and good music.
References
- Bagriantsev SN, Kushnirov VV, Liebman SW. Analysis of amyloid aggregates using agarose gel electrophoresis. Methods Enzymol. 2006;412:33–48. doi: 10.1016/S0076-6879(06)12003-0. [DOI] [PubMed] [Google Scholar]
- Barbee SA, Estes PS, Cziko AM, Hillebrand J, Luedeman RA, Coller JM, Johnson N, Howlett IC, Geng C, Ueda R, Brand AH, Newbury SF, Wilhelm JE, Levine RB, Nakamura A, Parker R, Ramaswami M. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron. 2006;52:997–1009. doi: 10.1016/j.neuron.2006.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- Clos J, Brandau S. pJC20 and pJC40: two high-copy-number vectors for T7 RNA polymerase-dependent expression of recombinant genes in Escherichia coli. Protein Expr Purif. 1994;5:133–137. doi: 10.1006/prep.1994.1020. [DOI] [PubMed] [Google Scholar]
- Decker CJ, Teixeira D, Parker R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J Cell Biol. 2007;179:437–449. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch IL, Bradley ME, Zhou P, Chernoff YO, Liebman SW. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics. 1997;147:507–519. doi: 10.1093/genetics/147.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: the story of [PIN(+)] Cell. 2001;106:171–182. doi: 10.1016/s0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover JR, Kowal AS, Schirmer EC, Patino MM, Liu JJ, Lindquist S. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell. 1997;89:811–819. doi: 10.1016/s0092-8674(00)80264-0. [DOI] [PubMed] [Google Scholar]
- Goldstrohm AC, Seay DJ, Hook BA, Wickens M. PUF protein-mediated deadenylation is catalyzed by Ccr4p. J Biol Chem. 2007;282:109–114. doi: 10.1074/jbc.M609413200. [DOI] [PubMed] [Google Scholar]
- Gupta YK, Lee TH, Edwards TA, Escalante CR, Kadyrova LY, Wharton RP, Aggarwal AK. Co-occupancy of two Pumilio molecules on a single hunchback NRE. RNA. 2009;15:1029–1035. doi: 10.1261/rna.1327609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaruzelska J, Kotecki M, Kusz K, Spik A, Firpo M, Reijo Pera RA. Conservation of a Pumilio-Nanos complex from Drosophila germ plasm to human germ cells. Dev Genes Evol. 2003;213:120–126. doi: 10.1007/s00427-003-0303-2. [DOI] [PubMed] [Google Scholar]
- Menon KP, Sanyal S, Habara Y, Sanchez R, Wharton RP, Ramaswami M, Zinn K. The translational repressor Pumilio regulates presynaptic morphology and controls postsynaptic accumulation of translation factor eIF-4E. Neuron. 2004;44:663–676. doi: 10.1016/j.neuron.2004.10.028. [DOI] [PubMed] [Google Scholar]
- Menon KP, Andrews S, Murthy M, Gavis ER, Zinn K. The translational repressors Nanos and Pumilio have divergent effects on presynaptic terminal growth and postsynaptic glutamate receptor subunit composition. J Neurosci. 2009;29:5558–5572. doi: 10.1523/JNEUROSCI.0520-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelitsch MD, Weissman JS. A census of glutamine/asparagine-rich regions: implications for their conserved function and the prediction of novel prions. Proc Natl Acad Sci U S A. 2000;97:11910–11915. doi: 10.1073/pnas.97.22.11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraro NI, Weston AJ, Gerber AP, Luschnig S, Moffat KG, Baines RA. Pumilio binds para mRNA and requires Nanos and Brat to regulate sodium current in Drosophila motoneurons. J Neurosci. 2008;28:2099–2109. doi: 10.1523/JNEUROSCI.5092-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osherovich LZ, Weissman JS. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI(+)] prion. Cell. 2001;106:183–194. doi: 10.1016/s0092-8674(01)00440-8. [DOI] [PubMed] [Google Scholar]
- Osterwalder T, Yoon KS, White BH, Keshishian H. A conditional tissue-specific transgene expression system using inducible GAL4. Proc Natl Acad Sci U S A. 2001;98:12596–12601. doi: 10.1073/pnas.221303298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijns MA, Alexander RD, Spiller MP, Beggs JD. A role for Q/N-rich aggregation-prone regions in P-body localization. J Cell Sci. 2008;121:2463–2472. doi: 10.1242/jcs.024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serio TR, Cashikar AG, Moslehi JJ, Kowal AS, Lindquist SL. Yeast prion [psi+] and its determinant, Sup35p. Methods Enzymol. 1999;309:649–673. doi: 10.1016/s0076-6879(99)09043-6. [DOI] [PubMed] [Google Scholar]
- Si K, Lindquist S, Kandel ER. A neuronal isoform of the Aplysia CPEB has prion-like properties. Cell. 2003;115:879–891. doi: 10.1016/s0092-8674(03)01020-1. [DOI] [PubMed] [Google Scholar]
- Vessey JP, Vaccani A, Xie Y, Dahm R, Karra D, Kiebler MA, Macchi P. Dendritic localization of the translational repressor Pumilio 2 and its contribution to dendritic stress granules. J Neurosci. 2006;26:6496–6508. doi: 10.1523/JNEUROSCI.0649-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner RB, Edskes HK, Shewmaker F, Nakayashiki T. Prions of fungi: inherited structures and biological roles. Nat Rev Microbiol. 2007;5:611–618. doi: 10.1038/nrmicro1708. [DOI] [PMC free article] [PubMed] [Google Scholar]