We have established in human breast cancer cells that indole-3-carbinol (I3C), a promising anti-cancer phytochemical from Brassica vegetables, ablates ERα expression by stimulating the Rbx-1 E3 ligase mediated degradation of ERα protein and disruption of a cross-regulatory positive feedback loop involving ERα and the GATA3 transcription factor.
Abstract
Estrogen receptor (ER)α is a critical target of therapeutic strategies to control the proliferation of hormone-dependent breast cancers. Preferred clinical options have significant adverse side effects that can lead to treatment resistance due to the persistence of active estrogen receptors. We have established the cellular mechanism by which indole-3-carbinol (I3C), a promising anticancer phytochemical from Brassica vegetables, ablates ERα expression, and we have uncovered a critical role for the GATA3 transcription factor in this indole-regulated cascade. I3C-dependent activation of the aryl hydrocarbon receptor (AhR) initiates Rbx-1 E3 ligase-mediated ubiquitination and proteasomal degradation of ERα protein. I3C inhibits endogenous binding of ERα with the 3′-enhancer region of GATA3 and disrupts endogenous GATA3 interactions with the ERα promoter, leading to a loss of GATA3 and ERα expression. Ectopic expression of GATA3 has no effect on I3C-induced ERα protein degradation but does prevent I3C inhibition of ERα promoter activity, demonstrating the importance of GATA3 in this I3C-triggered cascade. Our preclinical results implicate I3C as a novel anticancer agent in human cancers that coexpress ERα, GATA3, and AhR, a combination found in a large percentage of breast cancers but not in other critical ERα target tissues essential to patient health.
INTRODUCTION
One of the challenges in developing new therapeutic strategies for human breast cancer is the existence of several distinct classes of mammary tumors that differ in their phenotypes and proliferative responses to hormonal cues. It is critical to distinguish these properties to match the appropriate treatment to the corresponding tumor type (Brenton et al., 2005). Estrogens are a class of steroid hormones that play a critical role in the development of the normal breast and in the genesis of hormone-dependent and -independent breast cancer (Fuqua et al., 1991; Pasqualini and Chetrite, 2005). The existing options for the clinical management of hormone-dependent breast cancers are the use of selective estrogen receptor modulators (SERMs), such as tamoxifen, that can block ligand-dependent receptor activation (Dutertre and Smith, 2000) or inhibitors of aromatase activity that prevent estrogen synthesis, such as exemestane and letrozole (Miller, 1999). Both classes of therapeutic agents are prescribed to treat hormone-dependent early stage breast cancer; however, a limitation of their effectiveness is the development of tumor resistance within ∼5 y of initiating either therapy due to the persistence of functional estrogen receptors (Clarke et al., 2003). These treatments do not alter cellular levels of estrogen receptor protein and estrogen receptor activity is maintained by ligand-independent activation due to selective receptor phosphorylation, which can lead to more aggressive forms of hormone-independent cancers (Badia et al., 2007).
A biological complexity in the development of more effective therapeutic strategies that target estrogen receptor expression in human cancers is the existence of two major estrogen receptor subtypes, estrogen receptor (ER) α and ERβ (Enmark and Gustafsson, 1999). Each receptor subtype is encoded by different genes that trigger distinct cellular and physiological responses (Chang et al., 2006). After ligand activation, ERα and ERβ regulate the transcription of unique but overlapping sets of target genes (Harris et al., 2002), although both receptors are capable of activating synthetic reporter plasmids driven by estrogen response elements (EREs). Normal and cancerous breast epithelium express both ERα and ERβ. A high ERα: ERβ ratio correlates positively with enhanced cellular proliferation (Ali and Coombes, 2000; Lee et al., 2008); whereas predominance of functional ERβ over ERα is associated with decreased proliferation (Campbell-Thompson et al., 2001; Roger et al., 2001; Shaaban et al., 2003). Consistent with the proliferative effect of estrogens being mediated primarily by ERα, ablation of ERα in mouse mammary glands leads to a severely underdeveloped mammary epithelium. Although ERβ expression is preserved in ERα null epithelium, the impaired mammary epithelial cell growth cannot be rescued by administration of pharmacological levels of estrogens (Bocchinfuso and Korach, 1997), suggesting that ERα mediates the proliferative effects of estrogens. This is further strengthened by clinical observations where high-risk precancerous breast lesions were shown to possess elevated ERα levels and declining ERβ expression (Roger et al., 2001; Shaaban et al., 2003), which implicates the direct involvement of ERα with the estrogen-mediated increase in tumorigenicity in humans (Ali and Coombes, 2000).
The majority of ERα-positive breast cancers respond well to antihormonal therapy (Colozza et al., 2008), although only fulvestrant (ICI-182780), a selective estrogen receptor down-regulator (SERD) used in clinical trials, has been shown to reduce the level of total ERα protein to block both ligand-dependent and -independent receptor activation (Osborne et al., 2004). Fulvestrant, similar to the currently used SERMs and aromatase inhibitors, has significant systemic side effects (Mercier et al., 2003), and an emerging priority is the development of new classes of SERDs that effectively target ERα expression selectively in human breast cancers with minimal to no side effects. Epidemiological and physiological studies have suggested that phytochemicals from vegetables and fruits represent intriguing natural sources for new classes of potential anticancer molecules with minimal adverse side effects that function as SERDs. One such phytochemical is indole-3-carbinol (I3C), a natural compound derived by hydrolysis from glucobrassicin produced in Brassica cruciferous vegetables, such as cabbage, broccoli, and Brussels sprouts.
I3C exhibits potent anticarcinogenic properties in a wide range of human cancers such as lung, liver, colon, cervical, endometrial, prostate, and breast cancer (Aggarwal and Ichikawa, 2005; Kim and Milner, 2005; Safe et al., 2008; Weng et al., 2008). Exposure of human cancer cells to I3C triggers complementary sets of transcriptional, cell signaling, enzymatic, and metabolic cascades that directly lead to cell cycle arrest and apoptosis (Cover et al., 1999; Rahman et al., 2004; Garcia et al., 2005). Within the context of this antiproliferative environment, there is compelling evidence in estrogen-sensitive human breast cancer cell lines, such as MCF-7 and T47D, that I3C treatment disrupts estrogen-responsive gene expression (Cover et al., 1998; Auborn et al., 2003; Wang et al., 2006) and inhibits estrogen-dependent proliferation. I3C does not bind to either ERα or ERβ (Cover et al., 1999); the endocrine disrupting effects of this indole are due to its strong down-regulation of ERα protein and transcript expression, as well as the activation of ERβ (Sundar et al., 2006). Consistent with these effects on estrogen-responsive proliferation, I3C cooperates with tamoxifen to more effectively ablate phosphorylation of retinoblastoma protein accompanied by a G1 cell cycle arrest of MCF-7 cells (Cover et al., 1999). In addition, I3C disrupts the estrogenic responses of ERα-selective agonists such as propyl pyrazole triol, and in transient transfections of human breast cancer cells, I3C strongly attenuated ERα gene (ESR1) promoter activity (Sundar et al., 2006). An understanding of the precise pathway by which I3C disrupts ERα expression will potentially lead to the design of new clinical applications for I3C and improve patient outcomes through targeted therapies of human breast cancers. However, identification of the I3C triggered cascade and the indole regulated transcription factors that direct the loss of ERα expression and responsiveness in human breast cancer cells has remained elusive.
One of the critical regulators of ESR1 gene promoter activity that is involved in mammary gland development is the GATA3 transcription factor (Asselin-Labat et al., 2007). GATA3 expression is positively correlated with ERα expression in 97% of breast cancer biopsies (Hoch et al., 1999), suggesting a functional association between GATA3 status and the ER+ breast cancer phenotype. The physiological significance of GATA3 in ERα expression is further strengthened by studies showing that ERα and GATA3 knockout mice exhibit marked similarities in that both mouse mammary glands display severe developmental deficiencies in epithelial morphogenesis (Asselin-Labat et al., 2007). One explanation for these physiological observations is that GATA3 and ERα are involved in a cross-stimulatory positive feedback loop that helps to drive proliferation of estrogen-sensitive breast cancer cells (Eeckhoute et al., 2007). Activated ERα binds to a 3′-enhancer in the GATA3 gene to activate its transcription, and the GATA3 transcription factor stimulates ESR1 transcription through multiple binding sites in the ESR1 gene promoter (Eeckhoute et al., 2007). We now demonstrate in estrogen responsive human breast cancer cells that I3C triggers the ubiquitin 26S proteasome-mediated degradation of ERα protein in a cascade that requires the aryl hydrocarbon receptor (AhR) and the E3 ubiquitin ligase ring box-1 (Rbx1). This I3C-triggered loss of ERα protein then directly disrupts the GATA3/ERα cross-regulatory loop, which results in the down-regulation of ESR1 promoter activity leading to the ablation of ERα expression and loss of ERα-responsive proliferation.
MATERIALS AND METHODS
Reagents
ERα, GATA3, AhR, and Rbx1 antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). I3C, MG132, and dimethyl sulfoxide (DMSO) were obtained from Sigma-Aldrich (St. Louis, MO). All other chemicals were of the highest quality available.
Cell Culture
MCF-7 human breast cancer cells were obtained from American Type Culture Collection (Manassas, VA). Cells were grown in DMEM from Lonza Walkersville (Walkersville, MD), supplemented with 10% fetal bovine serum from Mediatech (Herndon, VA), 10 μg/ml insulin, 50 U/ml penicillin, 50 U/ml streptomycin, and 2 mM l-glutamine from Sigma-Aldrich. Cells were grown to subconfluence in a humidified chamber at 37°C containing 5% CO2. A 200 mM stock solution of I3C was dissolved in DMSO. I3C was then diluted 1:1000 in media before culture plate application.
Western Blotting
After the indicated treatments, Western blots were performed as indicated previously (Sundar et al., 2006). Mouse anti-ERα (sc-8005) and goat anti-Rbx1 (sc-5201) were diluted 1:200 in Tris-buffered saline/Tween 20 (TBST). Rabbit anti GATA3 (sc-9009) was diluted 1:500 in TBST. Hsp90 (610419; BD Biosciences, Franklin Lakes, NJ) and actin (AAN01; Cytoskeleton, Denver, CO) were used as loading controls, and antibodies for these were diluted 1:2000 and 1:1000 respectively, in TBST. Immunoreactive proteins were detected after incubation with horseradish peroxidase-conjugated secondary antibodies diluted 3 × 10−4 in 1% nonfat dry milk in TBST. Blots were then treated with enhanced chemiluminescence reagents (Eastman Kodak, Rochester NY) visualization on film.
Immunoprecipitation
After the indicated treatments, immunoprecipitations were performed as described previously (Failor et al., 2007). Precleared samples were then incubated with 50 μg of mouse anti-ERα or 50 μg of rabbit anti-AhR overnight at 4°C. Immunoprecipitated protein was eluted from beads by addition of gel loading buffer (50 mM Tris-Hcl, pH 6.8, 2% SDS, 10% glycerol, 1% β-mercaptoethanol, 12.5 mM EDTA, 0.02 mM bromophenol blue) and heating the sample at 100°C for 5 min. Samples were analyzed by Western blot as described previously (Sundar et al., 2006). Densitometry analysis was performed by measuring band intensity by using Photoshop version 8 histogram analysis (Adobe Systems, Mountain View, CA) and normalized to loading followed subsequently by normalization to the control treatments. The error represented deviation of values obtained from multiple experiments (three or more) after normalization.
Small Interfering RNA (siRNA), Short Hairpin RNA (shRNA), and Overexpression Plasmid Transfection
Cells were grown and indicated treatments performed on 10-cm tissue culture plates from Nalge Nunc International (Rochester, NY). Once cells reached 50% confluence transfection with siRNA constructs was performed using siRNA transfection reagent (sc-29528; Santa Cruz Biotechnology) using control siRNA (1022076; QIAGEN, Valencia, CA) or Rbx1-specific siRNA (sc-44072; Cruz Biotechnology) after transfection reagent manufacturer protocol. Short-hairpin RNA plasmids directed against AhR (TI378261) or against green fluorescent protein (GFP) (TR30003) were obtained from Origene Technologies (Rockville, MD). Human cytomegalovirus (CMV)-GATA3 overexpression plasmid and CMV-GATA-KRR dominant-negative plasmid were a kind gift from Dr. Astar Winoto (Department of Molecular and Cell Biology, University of California–Berkeley, Berkeley, CA). Human CMV-ERα was a kind gift from Dr. Benita Katzenellenbogen (University of Illinois, Urbana-Champagne, IL). Transfection of expression vectors and shRNA plasmids were performed using Polyfect transfection reagent from QIAGEN per the manufacturers' recommended protocol.
Immunofluorescence
Cells were grown and indicated treatments performed on two-well chamber slides from Nalge Nunc International. The cells were fixed with 3.75% formaldehyde in phosphate-buffered saline (PBS) for 15 min at room temperature. After three additional washes with PBS, the plasma membrane was permeabilized with 0.1% Triton-X-100, 10 mM Tris-HCl, pH 7.5, 120 mM sodium chloride, 25 mM potassium chloride, 2 mM EGTA, and 2 mM EDTA) for 10 min at room temperature. Slides were incubated with 3% bovine serum albumin (Sigma-Aldrich) before incubation with primary antibodies. Rabbit anti-AhR antibody (sc-8088; Santa Cruz Biotechnology) was used at a 1:400 dilution. Secondary Alexa488 anti-rabbit and Texas Red-conjugated phalloidin were used at 1:400 dilutions each. Stained cells were mounted with Vectashield mounting medium containing 4,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Burlingame, CA). Stained and mounted cells were then processed with an Axioplan epifluorescence microscope (Carl Zeiss, Thornwood, NY).
Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
Total RNA from MCF-7 cells treated with indicated compounds was isolated with Tri Reagent according to manufacturer's protocol (Sigma-Aldrich). Total RNA (4 μg) was used to synthesize cDNA using Moloney murine leukemia virus-reverse transcriptase (Promega, (Madison, WI) with random hexamers as primers. The cDNA reaction product (200 ng) was amplified with primers of the following sequences: ERα forward, 5′-AGCACCCAGTGAAGCTACT-3′ and ERα reverse, 5′-TGAGGCACACAAACTCCT-3′; GATA3 forward, 5′-CTCATTAAGCCCAAGCGAAG-3′ and GATA3 reverse, 5′-TTTTTCGGTTTCTGGTCTGG-3′; and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) forward, 5′-TGAAGGTCGGAGTCAACGGATTTG-3′ and GAPDH reverse, 5′-CATGTGGGCCATGAGGTCCACCAC-3′. PCR products were analyzed on 1.2% agarose gel along with 1-kb Plus DNA ladder from Invitrogen (Carlsbad, CA), and the products were visualized with GelRed from Biotium (Hayward, CA). Densitometry analysis was performed by measuring band intensity using Photoshop version 8 histogram analysis and normalized to loading followed subsequently by normalization to the control treatments. The error represented deviation of values obtained from multiple experiments (3 or more) after normalization.
ERα Promoter Fragment and Mutation Generation
pERα-3794-pgl2 and pERα-860-pgl2 plasmids were kind gifts from Dr. Lisa McPherson (Stanford University, Stanford, CA). pERα-2294, pERα-1892, and pERα-985 were amplified from purified genomic DNA isolated from LNCaP prostate cancer cells by using the restriction enzymes MluI and BglII from New England Biolabs (Ipswich, MA). Restriction sites were engineered into the following PCR primers: −2294, 5′-CTCGAGTCGGCCCTTGACTTCTACA-3′ and +46, 5′-GGCGCCTTGCTGCTGTCCAGGTACA-3′; −1892: 5′-TGCCATTCCACGCACAAACACATC-3′ and +109, 5′-TAAGTACTGGTCTCCCGA-3; and −985, 5′-ATGTGTGTGTGTATGTGCGTGT-3′ and +285, 5′-AAAGAGCACAGCCCGAGGTTAGA-3′. For PCR amplification, a 50-μl PCR reaction (1× VENT polymerase buffer Mg2+ free, 0.2 mM dNTPs, 1 U of VENT polymerase (New England Biolabs), 1.5 mM MgCl2, and 0.2 μM each primer) was amplified for 38 cycles (30 s at 94°C, 30 s at 58°C, and 2.5 min at 72°C) with a 94°C 1-min hot start. Sequence was confirmed by automated DNA sequencing (University of California–Berkeley Sequencing Facility). PCR products from all constructs were purified using QIAEXII gel extraction kit (QIAGEN), digested with MluI and BglII, and subcloned into PGL2-basic (Promega).
Mutations in the predicted transcription factor binding sites were introduced using the pERα-2294-pgl2 construct as a template. Primer sequences with the indicated mutations were as follows: GATA3-1 forward, 5′-GGCATTTGATCCACATGGCGCCCAGAAGGCTTTTATTG-3′ and GATA3-1 reverse, 5′-CAATAAAAGCCTTCTGGGCGCCATGTGGATCAAATGCC-3′; GATA3-2-forward, 5′-GTCCTATTTTGTAAACTTGGCGCCCATACACTTTTGACTGG-3′ and GATA3-2 reverse, 5′-CCAGTCAAAAGTGTATGGGCGCCAAGTTTACAAAATAGGAC-3′; and Ap1 forward, 5′-CTATTTTAGCCGTAAGACTATTGATCACAGCAAGCCTGTTTTTCCTC-3′ and Ap1 reverse, 5′-GAGGAAAAACAGGCTTGCTGTGATCAATAGTCTTACGGCTAAAATAG-3′. pERα-2294-pgl2 was used as a template for the mutagenesis reactions. Template DNA (5 ng) was mixed with 125 ng of each primer, and mutagenesis was performed using the Stratagene Lighting QuikChange mutagenesis kit per the manufacturer's instructions.
Luciferase Assays
MCF-7 cells grown to 70% confluence in six-well Nunc plates were transfected with 2 μg/well of the indicated plasmid construct. Transfections were performed in serum-free medium using Superfect (QIAGEN) transfection reagent as per manufacturer's instructions. Cells were treated 24 h posttransfection with DMSO or 200 μM I3C for 24 h. Cells were then lysed and relative luciferase activity was evaluated using the Luciferase assay kit (Promega). Relative luciferase activities were normalized to protein input and pERα-3794-pgl2 construct activity with SE. Reproducibility of these results was verified by three independent experiments performed with triplicate samples of each treatment.
Chromatin Immunoprecipitation (ChIP) Assays
MCF-7 cells were grown to subconfluence and treated for 48 h with 200 μM I3C or DMSO vehicle control. ChIP was performed as described previously (Sundar et al., 2008). Primers for ChIP experiments were as follows: pERα-GATA3-2207 forward, 5′-AATGCCTCTGTTCAGAGACTGGG-3′ and reverse, 5′-GCTTGCTGTGAATCATAGTCTTACGGC-3′; pERα-GATA3-3812 forward, 5′-TTTAATCTGGGTGGCTGGAG-3′ and reverse, 5′-CTCAACTTCCCCGTGTCTGT-3′; and GATA3 3′Enhancer-ERα forward, 5′-GATGTTAGGGAGTGACCAAAGAGG-3′ and reverse, 5′-GTGGACAAGTTTGATCCTTTAAGCTCTG-3′. Products were visualized on a 1.5% agarose gel buffered with Tris borate-EDTA.
RESULTS
I3C Induces the Ubiquitination and Proteasome-mediated Degradation of ERα Protein before Down-Regulation of ERα Transcripts
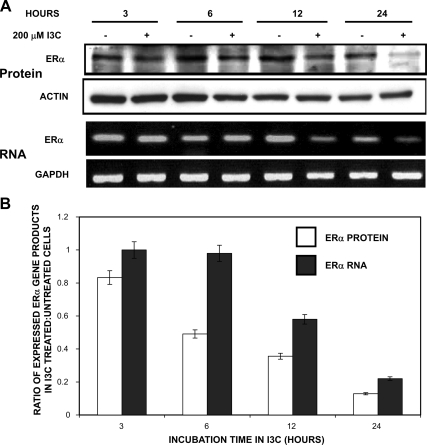
The effects of I3C on the kinetics of ERα transcript and protein down-regulation were examined in a 24-h time course of MCF-7 human breast cancer cells treated with or without 200 μM I3C. This concentration of I3C causes the maximal inhibition of estrogen-dependent proliferation without any apoptotic effects (Sundar et al., 2006). Total ERα protein levels at each time point were determined by Western blot analysis of electrophoretically fractionated cell extracts, and the corresponding levels of ERα transcripts were analyzed by RT-PCR analysis of isolated total RNA. As shown in Figure 1A and quantified in Figure 1B, I3C treatment caused a rapid down-regulation of ERα protein before any effect on ERα-transcript levels. A significant reduction in ERα protein levels was observed within 6 h of indole treatment, and ablation of detectable ERα protein occurred by 24 h. In contrast, the earliest time point in which a significant decrease in ERα transcripts occurred was 12 h (Figure 1, A and B). No changes were observed for gel loading control genes actin (protein) or GAPDH (mRNA) throughout the time course. Thus, I3C exerts a rapid and direct effect on ERα protein levels, which is followed kinetically by the attenuation of ERα transcription. The ablation of ERα protein expression is independent of the overall growth inhibitory effect of I3C, because 3,3′-diindoylylmethane, the self-condensation dimer of I3C (Staub et al., 2002), has no effect on ERα protein levels (Supplemental Figure 1) but does induce a potent G1 cell cycle arrest in MCF7 breast cancer cells (Hong et al., 2002). Also, treatment with tryptophol, an inactive indole that contains one additional carbon group at the C-3 position constituent group compared with I3C, has no effect on ERα protein degradation (Supplemental Figure 1).
Figure 1.
Effects of I3C on kinetics of ERα expression. (A) MCF-7 human breast cancer cells were treated with or without 200 μM I3C through a 24-h time course. At the indicated time points, the levels of ERα protein were monitored by Western blots and the levels of ERα transcripts determined by RT-PCR. Actin was used as protein loading control, and GAPDH was used as a RNA loading control. PCR products were visualized on a 1% agarose gel stained with ethidium bromide. (B) Levels of ERα protein and transcripts were quantified by densitometry of the Western blots and RT-PCR gels shown in A. This result was repeated three times, and a representative blot is shown. The bar graphs represent the ratio of ERα gene expression observed in I3C-treated versus untreated controls.
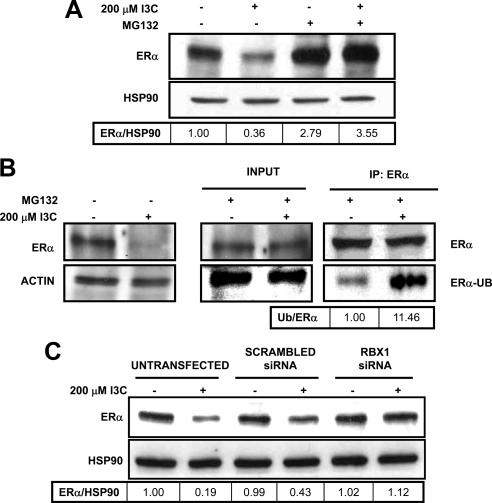
To determine whether the I3C down-regulation of ERα protein was due to induced ubiquitination and 26S proteasome-mediated degradation, MCF-7 cells were treated with or without 200 μM I3C for 6 h in the presence or absence of MG132, an inhibitor of proteasome peptidase enzymatic activity. As shown in Figure 2A, immunoblot analysis revealed that MG132 treatment completely rescued ERα protein from the I3C-induced degradation. Similar to this effect in MCF7 cells, treatment with MG132 blocked I3C-dependent degradation in the hormone-sensitive T47D cell line (Supplemental Figure 3A). Attachment of ubiquitin to ERα protein was examined by Western blot analysis of immunoprecipitated ERα protein in cells treated with or without 200 μM I3C for 6 h in the presence of MG132. As shown in Figure 2B, I3C treatment resulted in a significant increase in ubiquitinated ERα protein levels. Thus, I3C-induced degradation of ERα protein is triggered by ubiquitination that targets ERα for destruction by the 26S proteasome.
Figure 2.
I3C induces the ubiquitination and 26S proteasome degradation of ERα protein and requirement of the Rbx1 E3 ubiquitin ligase this degradative process. (A) MCF-7 cells were treated with the indicated combinations of 200 μM I3C and 5 mM MG132 (a 26S proteasome inhibitor) for 6 h, and the level of ERα protein monitored by Western blots. HSP90 was used as gel loading control. A representative blot of three independent experiments is shown. (B) In MCF-7 cells were treated with the indicated combinations of 200 μM I3C and 5 mM MG132. Total cell extracts were immunoprecipitated with mouse-anti-ERα antibodies and electrophoretically fractionated samples blotted with either rabbit-anti-ERα or rabbit-anti-ubiquitin antibodies (ERα-UB). Result was repeated four times, representative blot shown. (C) MCF-7 cells were transfected with control scrambled siRNA or Rbx1-specific siRNA or remained untransfected for 24 h. Cells were then treated with or without 200 μM I3C for 6 h, and the level of ERα protein determined by Western blot analysis. Result was repeated twice. Densitometry numbers are the ratio of ERα to loading control, normalized to the DMSO ratio.
Several E3 ubiquitin ligases are involved in ERα protein degradation in different cell systems (Osborne et al., 2004; Eakin et al., 2007). The potential role of Rbx1, a RING-domain E3 ligase, in I3C induced-destruction of ERα protein was examined because this E3 ligase has been implicated in xenobiotic-mediated degradation of ERα protein (Ohtake et al., 2007). Rbx1 expression was selectively ablated in MCF-7 cells by transfection of Rbx1-specific siRNA. Control cells were either transfected with scrambled siRNA or remained untransfected. As shown in Figure 2C, I3C treatment failed to induce ERα protein degradation in cells expressing Rbx1 siRNA. In contrast, in control cells expressing scrambled siRNA or in those that remained untransfected this indole efficaciously triggered the degradation of ERα protein. These results demonstrate that Rbx1 is required for the I3C induced degradation of ERα protein in MCF-7 human breast cancer cells. Furthermore, this result differentiates the degradative mechanism of I3C from that of fulvestrant, which is a SERD that binds directly to ERα and causes the receptor to localize to the nuclear periphery and recruit the NEDD8 E3 ligase for receptor degradation (Fan et al., 2003).
Activated AhR Is Required for I3C-induced ERα Protein Degradation
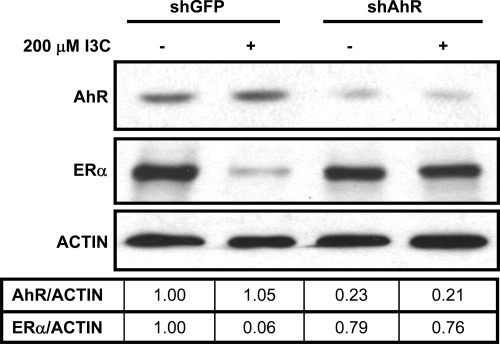
Previous studies have determined that treatment of estrogen-responsive human breast or prostate human cancer cells with the 3-MC or 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) high-affinity xenobiotic ligands for the AhR can cause ERα degradation (Wormke et al., 2003) through recruitment of Rbx-1 (Ohtake et al., 2009). Although I3C has relatively low affinity for AhR (Jellinck et al., 1993), we examined whether the I3C degradation of ERα protein is dependent on AhR in MCF-7 human breast cancer cells by disrupting its expression with AhR-specific shRNA. Expression of shRNA to green fluorescence protein (shGFP) provided a control for the transfection procedure. Transfected MCF-7 cells were treated with or without 200 μM I3C for 6 h, and ERα protein was examined by Western blots. As shown in Figure 3, shRNA targeted to AhR effectively blocked ERα protein degradation by I3C, demonstrating the AhR dependence on this process.
Figure 3.
I3C-mediated degradation of ERα protein requires AhR. MCF-7 cells were transfected with shAhR or shGFP (control plasmid) and then treated with or without or 200 μM I3C for 6 h. Total cell extracts were electrophoretically fractionated and the levels of ERα, AhR, and actin (loading control) analyzed by Western blots. This result was repeated in three independent experiments. Densitometry numbers are the ratio of ERα to loading control, normalized to the DMSO ratio.
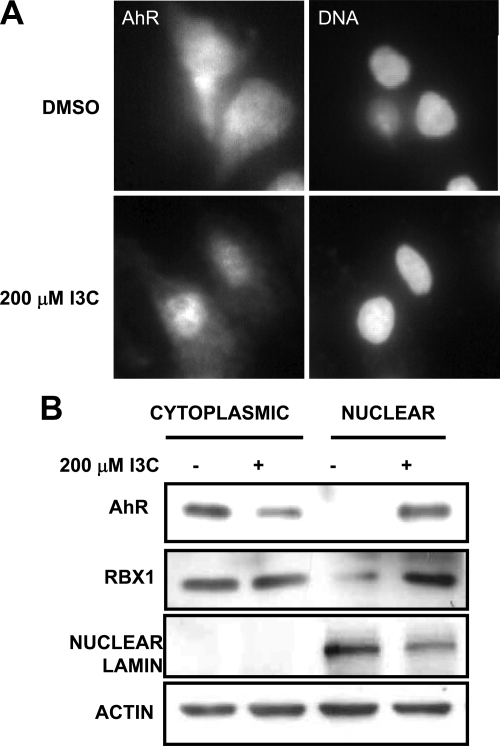
The ligand-dependent activation of AhR leads to its nuclear translocation, which provides access to its gene targets (Richter et al., 2001). To determine whether I3C activates AhR, the nuclear translocation of AhR was examined by indirect immunofluorescence in 6 h I3C treated and untreated MCF-7 cells. As shown in Figure 4A, I3C treatment significantly increased nuclear fluorescence of AhR, whereas in untreated cells AhR was distributed more uniformly throughout the cell. These results suggest that I3C activates AhR, a process that is required for its nuclear translocation, which in turn triggers the Rbx1-mediated ubiquitination and proteasomal degradation of ERα protein.
Figure 4.
I3C induces the nuclear localization of AhR. (A) MCF-7 cells were treated with or without 200 μM I3C for 6 h, and the subcellular localization of AhR determined by indirect immunofluorescence microscopy. DAPI staining was used to visualize DNA stained nuclei (right). (B) MCF-7 cells treated with 200 μM I3C or with the DMSO (vehicle control) for 24 h, and cell extracts fractionated into cytoplasmic and nuclear fractions. Each set of subcellular fractions were electrophoretically fractionated and analyzed by Western blot for the levels of AhR, Rbx1, nuclear lamin (fractionation control), and actin (loading control) in each cellular compartment. The result was repeated in three independent experiments.
The I3C-induced nuclear localization of AhR was confirmed by biochemical fractionation of the MCF-7 cells into nuclear and cytoplasmic fractions. As shown in Figure 4B, AhR was highly enriched in the cytoplasmic fraction of untreated cells, whereas in 24 h I3C-treated cells, the majority of AhR cofractionated with the nuclei. Furthermore, Rbx1, the E3 ubiquitin ligase that is required for the indole induced ERα degradation, also cofractionates with the nuclear compartment in I3C-treated cells but not in untreated cells. The efficiency of the nuclear fractionation in both I3C-treated and untreated cells were verified using lamin as a nuclear marker (Figure 4B).
I3C-induced Degradation of ERα Protein Causes the Down-Regulation of GATA3 Transcription Factor Gene Expression
The temporal delay between I3C-induced ablation of ERα protein and loss of ERα transcripts suggests that either the indole-induced down-regulation of ERα protein and transcripts result from independent I3C-regulated pathways or that the indole-mediated ERα protein degradation initiates a cellular cascade that directly accounts for the loss of ERα transcripts. One candidate transcription factor that may link the I3C down-regulation of ERα protein and transcripts is GATA3, an ERα target gene (Tremblay and Viger, 2001) that has been shown to affect ERα promoter activity in a cross-regulatory feedback loop (Eeckhoute et al., 2007). To initially test whether I3C regulates GATA3 expression, MCF-7 cells were treated with or without 200 μM I3C over a 72-h time course, and the levels of GATA3 and ERα gene products monitored by Western blots and by RT-PCR. As shown in Figure 5, I3C strongly down-regulates GATA3 protein and transcript levels in MCF7 cells as well as in T47D cells (Supplemental Figure 3B). This indole down-regulates GATA3 transcript levels before the protein levels are affected. Importantly, at the 24-h time point when ERα protein is essentially ablated, GATA3 gene products decrease in indole treated cells. The I3C-mediated down-regulation of GATA3 protein and transcripts occurs with similar dose-response profiles as ERα, with an observed half-maximal down-regulation at ∼150 μM I3C (Supplemental Figure 2). These results demonstrate that in MCF-7 human breast cancer cells, GATA3 expression is strongly attenuated by I3C and that the kinetics of this response is consistent with GATA3 expression being regulated by ERα protein.
Figure 5.
Effects of I3C on the expression of GATA3 protein and GATA3 transcripts. MCF-7 cells were treated with or without 200 μM I3C, and at the indicated times the level of GATA3 and ERα protein was monitored by Western blot analysis (top), and GATA3 and ERα transcript expression was determined by RT-PCR (bottom). The PCR products were visualized on a 1% agarose gel stained with ethidium bromide. HSP90 provided a loading control for the western blots and GAPDH provided a gel loading control for the RT-PCR. The results were repeated three times, and representative blots and gels are shown. Densitometry numbers are the ratio of ERα or GATA3 to loading control, normalized to the DMSO ratio.
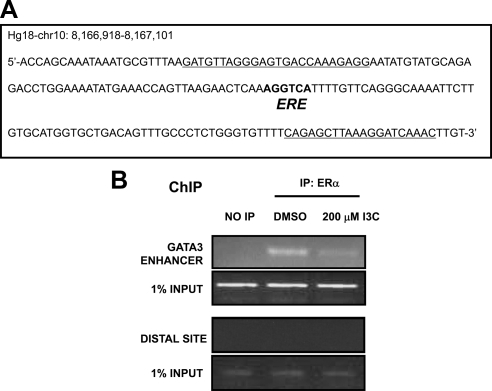
The 3′ enhancer of the GATA3 gene contains many estrogen-regulated elements that are responsible for increased ERα binding to GATA3 cis-regulatory regions (Eeckhoute et al., 2007; Figure 6A). However, the consequences on GATA3 gene transcription of ERα binding to cis-regulatory regions are unknown. Chromatin immunoprecipitation was used to determine whether the I3C down-regulation of ERα protein in MCF-7 cells affects the endogenous ERα interactions with the 3′ enhancer of the GATA3 gene. As shown in Figure 6B, in 24-h I3C-treated cells, binding of ERα to a representative GATA3 3′ enhancer element was significantly decreased in I3C-treated cells compared with untreated cells. Thus, in MCF-7 human breast cancer cells, I3C down-regulates endogenous ERα interactions with the 3′ enhancer region of the GATA3 gene, which accounts for the indole-mediated loss of GATA3 transcripts.
Figure 6.
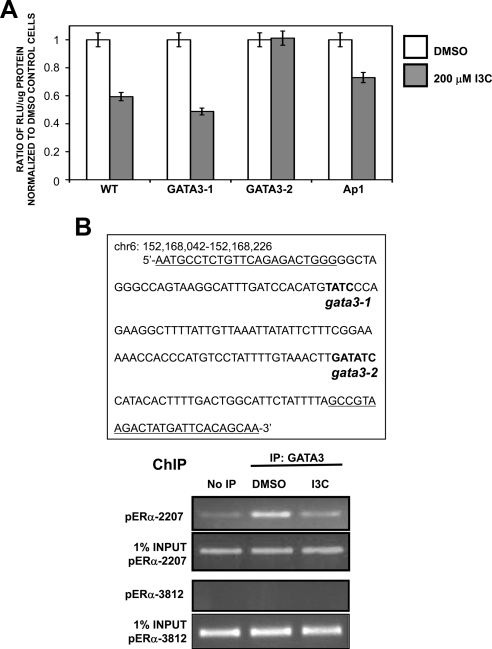
I3C disrupts ERα protein interaction with GATA3 regulatory regions. (A) Genomic sequences of the GATA3 gene enhancer contain a consensus half-ERE site. Primers used to amplify ERE site for chromatin immunoprecipitation are underlined. Sequence and chromosomal location were obtained from the UCSC Genome Browser. (B) ChIP was used to characterize endogenous ERα interactions with the ERE region of the GATA3 enhancer region. Chromatin was isolated from MCF-7 cells treated with or without 200 μM I3C for 24 h. ERα was immunoprecipitated from total cell extracts using Sepharose G bound to anti-ERα antibody, and DNA released from ERα was amplified using the indicated oligonucleotide primers. Control primers directed at downstream site (distance, 1256 base pairs) showed no amplification in immunoprecipitated (IP) samples. Input samples represent total genomic DNA from each treatment (loading control). This result was repeated twice.
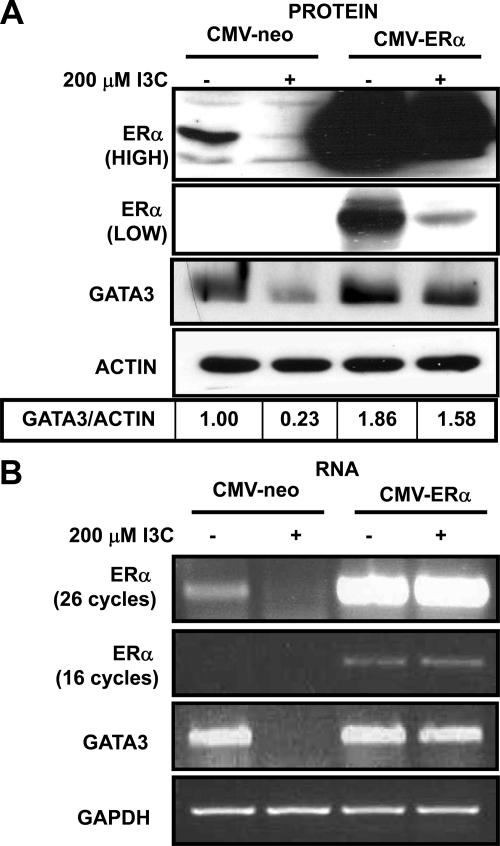
A key prediction of the direct role of ERα in mediating the I3C down-regulation of GATA3 gene expression is that overexpression of exogenous ERα should prevent I3C inhibition of GATA3 protein and transcripts. MCF-7 cells were stably transfected with either a CMV-ERα expression vector or with a CMV-neo empty vector control and examined for ERα gene expression in the presence or absence of I3C. As shown in Figure 7A, the overexpressed exogenous ERα protein is down-regulated in I3C-treated cells as predicted by the indole induced proteasomal degradation of ERα protein. However, enough exogenous ERα protein remains that the overall ERα levels in I3C-treated cells are comparable with endogenous levels in control empty vector-transfected cells treated with the vehicle control. In the presence of exogenous ERα protein, I3C-induced down-regulation of endogenous GATA3 transcripts and protein is effectively blocked (Figure 7B). Ectopic expression of ERα reversed the I3C down-regulation in ERα and GATA3 transcripts, suggesting that the I3C-mediated degradation of ERα protein is a pivotal event that precedes and causes the loss of ERα and GATA3 gene expression.
Figure 7.
I3C down-regulation of GATA3 gene expression requires the I3C mediated loss of ERα protein. (A) MCF-7 cells were transfected with CMV-ERα or the CMV-Neo vector control and treated with or without 200 μM I3C for 48 h. Total Cell lysates were electrophoretically fractionated and analyzed by Western blots for the levels of GATA3, ERα, and actin (loading control) protein. (HIGH) and (LOW) designations refer to film exposure times of the blot. This result was repeated in three independent experiments. Densitometry numbers are the ratio of GATA3 to loading control, normalized to the DMSO ratio. (B) Total RNA was collected from MCF-7 cells treated with or without 200 μM I3C for 48 h, and RT-PCR was used to detect GATA3 and ERα transcripts. GAPDH was used as total RNA loading control. PCR products were visualized on a 1% agarose gel stained with ethidium bromide. This experiment was repeated twice.
Down-Regulation of GATA3 Expression Is Required for I3C Inhibition of ERα Transcription
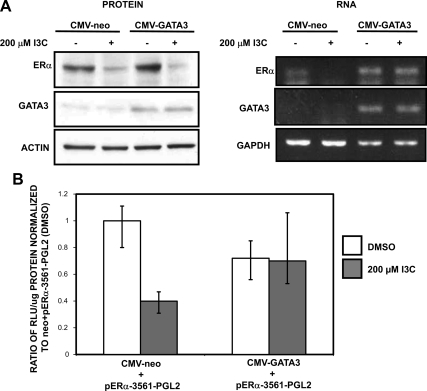
Our results suggest that the I3C-induced ERα protein degradation triggers the loss of GATA3 expression, which then causes the down-regulation of ERα transcription. Consistent with this possibility, transfection of a dominant-negative GATA-KRR, which blocks GATA3 transcription activity, disrupts ERα gene expression (Supplemental Figure 3C). A key test of this mechanism is that the ectopic expression of the GATA3 transcription factor from a constitutive expression vector should override the I3C down-regulation of ERα transcripts, but have no effect on the I3C-induced degradation of ERα protein. MCF-7 cells were transfected with either a CMV-GATA3 expression vector or with the CMV-neo empty vector and analyzed for ERα protein and transcripts in the presence or absence of I3C. As shown in Figure 8A, expression of exogenous GATA3 prevented the I3C down-regulation of endogenous ERα transcripts but failed to alter the I3C-mediated loss of ERα protein levels. In empty vector-transfected cells, I3C down-regulated both ERα protein and transcript levels.
Figure 8.
The I3C inhibition of ERα transcripts levels requires the down-regulation of GATA3 gene expression. (A) MCF-7 cells were transfected with CMV-GATA3 or CMV-neo vector control and treated with or without 200 μM I3C for 48 h. Total Cell lysates were electrophoretically fractionated and analyzed by Western blots for the levels of GATA3, ERα, and actin (loading control) protein (left). Total RNA was collected from MCF-7 cells treated with or without 200 μM I3C for 24 h and RTPCR was used to detect GATA3 and ERα transcripts (right). GAPDH was used as total RNA loading control. PCR products were visualized on a 1% agarose gel stained with ethidium bromide. This result was repeated four times, and representative blots and gels are shown. (B) MCF-7 cells were cotransfected with the I3C-responsive −3561 base pairs fragment of the ERα promoter linked to a luciferase reporter plasmid along with either CMV-GATA3 or CMV-neo (vector control). At 24 h posttransfection, cells were treated with or without 200 μM I3C for 24 h, and the relative luciferase activity was evaluated in lysed cells using the Luciferase assay kit (Promega). The reporter plasmid levels are normalized to the −3561 ERα promoter fragment treated with the DMSO vehicle control. Two additional controls (data not shown) included CMV-luciferase to validate transfection efficiency (positive control) and pgl2 to measure background fluorescence (negative control). Bar graphs indicate relative luciferase activity normalized to the protein input. Error bars were derived from the results of three independent experiments.
We demonstrated previously that I3C inhibits ERα promoter activity (Sundar et al., 2006), which suggests that an indole-responsive transcription factor directly mediates this response. To assess the effects of ectopic GATA3 expression on the I3C inhibition of ERα promoter activity, MCF-7 cells were cotransfected with an ERα promoter-luciferase reporter plasmid containing a 3561 base pairs fragment of the ERα promoter (pERα-3561-pgl2) along with either the CMV-GATA3 expression vector or the CMV-neo empty vector control. As shown in Figure 8B, exogenous GATA3 was able to override the I3C-mediated down-regulation of ERα promoter activity, demonstrating the central role of the GATA3 transcription factor in attenuation of ERα transcription by I3C. Consistent with the effects observed in MCF7 cells, in estrogen-responsive T47D human breast cancer cells ectopic expression of GATA3 was able to override the I3C-mediated down-regulation of ERα promoter activity (Supplemental Figure 3D). Together, our results demonstrate that indole down-regulation of GATA3 is a crucial intermediate step in the I3C-regulated cascade that links I3C-induced ERα protein degradation to the loss of ERα transcripts and promoter activity.
Role of GATA3 in I3C-mediated Inhibition of ERα Promoter Activity
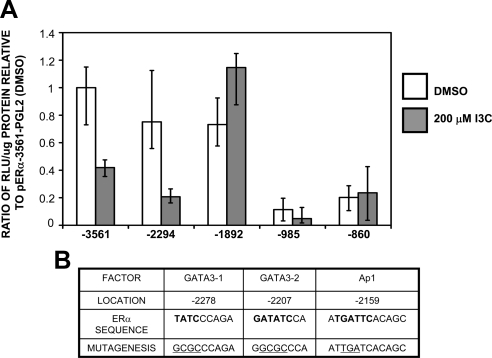
Because the GATA3 transcription factor has many potential target genes, it was important to determine whether I3C treatment disrupts a direct interaction between GATA3 and the ERα promoter, or whether GATA3 mediates its effects through another intermediate transcriptional regulator. To define the I3C-responsive region in the ERα promoter, a series of 5′ deletion mutants were generated and subcloned into luciferase reporter plasmids, and ERα promoter activity was assessed in transfected MCF-7 cells treated with or without 200 μM I3C for 24 h. As shown in Figure 9A, the −2294 promoter fragment was fully indole-responsive, whereas activities of the reporter plasmids linked to promoter fragments lacking the −2294 to −1892 base pairs region were not affected by I3C. This finding localizes the I3C responsive region between −2294 and −1892 base pairs upstream of the ERα transcription start site.
Figure 9.
Identification of the I3C responsive region of the ERα promoter and predicted transcription factor binding sites located within. (A) MCF-7 cells were transfected with the indicated ERα promoter 5′ deletion constructs linked to a luciferase reporter gene, and 24 h posttransfection cells were treated for 24 h with either the DMSO vehicle control or with 200 μM I3C. Relative luciferase activity was evaluated in lysed cells using the Luciferase assay kit (Promega) and normalized to the reporter plasmid activity of the −3561 ERα promoter fragment in cells treated with DMSO. Two controls (data not shown) included CMV-luciferase to validate transfection efficiency (positive control) and pgl2 to measure background fluorescence (negative control). Bar graphs indicate relative luciferase activity normalized to the protein input and error bars were derived from the results of three independent experiments. (B) Transcription factor binding site analysis of the I3C-responsive region was performed using TFSearch program, followed by manual curation of potential sites. Positions displayed are relative to the ERα promoter-A transcription start site. Bold bases indicate consensus sequences of the indicated transcription factor sites within the ERα promoter. Underlined sequence indicates the positions of site-directed mutagenesis and the mutations that were introduced into the ERα promoter.
As diagrammed in Figure 9B, sequence analysis revealed that the I3C-responsive region of the ERα promoter contains predicted DNA binding sites for several families of transcription factors, including two putative binding sites for GATA3 at −2278 (GATA3-1) and at −2207 (GATA3-2). There are also consensus DNA binding sites for NF-κB, Sp1, Ets, and Ap1 transcription factors, which have all been identified previously as regulated by indoles in several human cancer cell lines (Cram et al., 2001; Takada et al., 2005). To determine which elements are responsible for I3C down-regulation of ERα promoter activity in human breast cancer cells, select DNA sites within the −2294 ERα promoter fragment were mutagenized and the entire promoter fragments were inserted into luciferase reporter plasmids. These constructs were then transfected into MCF-7 cells, and effects of I3C on wild-type (WT) and mutated (GATA3-1, GATA3-2, and Ap1) promoter activities were analyzed. As shown in Figure 10A, mutation of the GATA3-2 site at −2207, but not mutation of any other site, prevented I3C down-regulation of ERα promoter activity.
Figure 10.
Identification of GATA3 as the transcription factor responsible for ERα promoter down-regulation by I3C. (A) MCF-7 cells were transfected with luciferase reporter plasmids driven by either the wild-type ERα promoter fragment starting at −2294 upstream of the RNA site start, or with ERα promoter fragments with the designated mutations in the consensus GATA3-1, GATA3-2, or Ap1 transcription factor binding sites. Twenty-four hour-transfected cells were treated for an additional 24 h with either DMSO or 200 μM I3C. Relative luciferase activity was evaluated in lysed cells using the Luciferase assay kit (Promega). Bar graphs indicate relative luciferase activity normalized to the protein input. Values are normalized to DMSO of each transfection. Result was repeated twice. (B) Left, ERα genomic sequence containing both predicted GATA3 binding sites (bold) within the I3C-responsive region of ERα promoter. Primers used to amplify GATA3 sites for chromatin immunoprecipitation are underlined. Chromatin was isolated from MCF-7 cells treated with or without 200 μM I3C for 24 h. GATA3 was immunoprecipitated from total cell extracts using Sepharose G bound to anti-ERα antibody. DNA released from ERα was amplified using indicated primers. Control primers directed at upstream site (−3812 base pairs) showed no amplification in IP samples. Input samples represent total genomic DNA from each treatment (loading control). This result was repeated twice.
ChIP was used to determine whether I3C could affect endogenous GATA3 recruitment to the native ERα promoter. Cells were treated with or without 200 μM I3C for 24 h, DNA derived from chromatin immunoprecipitated with anti-human GATA3 antibody was PCR amplified with primers specific to the GATA3 DNA binding sites contained within the ERα promoter. As shown in Figure 10B, GATA3 is bound to ERα promoter in untreated MCF-7 cells at the predicted consensus sequence, and I3C treatment strongly down-regulated GATA3 binding to this promoter region. Together, our results show that GATA3 directly interacts with the ERα promoter and mediates I3C control of ERα promoter activity.
DISCUSSION
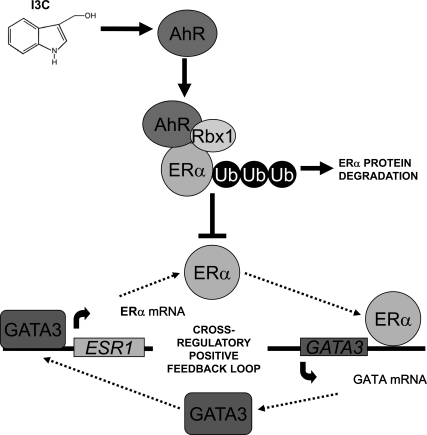
We have established the cellular mechanism by which I3C, a promising anticancer phytochemical from Brassica vegetables, ablates ERα expression and estrogen-dependent proliferation of estrogen responsive human breast cancer cells, and we have uncovered a critical role for the GATA3 transcription factor in this indole-regulated cellular cascade. As summarized in Figure 11, our results demonstrate that I3C triggers the ubiquitin 26S proteasome mediated degradation of ERα protein in a process that requires the AhR and the E3 ubiquitin ligase Rbx1. The I3C-mediated degradation of ERα protein directly down-regulates expression of GATA3, which then disrupts the GATA3/ERα cross-regulatory loop because of the loss of GATA3-stimulated ESR1 promoter activity. The cellular consequences of the I3C triggered transcriptional cascade are the inhibition of ERα expression and consequent loss of ERα responsive proliferation.
Figure 11.
Proposed model for the I3C disruption of a cross-regulatory positive feedback loop involving expression of GATA3 and ERα by stimulating the degradation of ERα protein. In the absence of I3C, ERα and the GATA3 transcription factor maintain a cross-regulatory positive feedback loop that results in a high level of ERα expression. ERα stimulates GATA3 transcription by interacting with an enhancer region in the GATA3 gene, whereas GATA3 stimulates ERα promoter activity by interacting its corresponding binding sites in the ERα promoter. I3C disrupts this feedback loop by inducing the ubiquitination and proteasome-mediated degradation of ERα protein. The I3C-induced degradative pathway requires the Rbx1 E3 ubiquitin ligase and the I3C activation and nuclear localization of AhR. We propose that I3C-activated AhR tethers Rbx1 to its ERα protein substrate for ubiquitination and subsequent destruction.
Ablation of either AhR or Rbx1 expression prevented the I3C induced degradation of ERα protein, demonstrating the necessity of both cellular components in this process. We propose that the I3C activation and nuclear import of the AhR helps to tether the Rbx1 E3 ubiquitin ligase to ERα for its ubiquitination and targeting for destruction. I3C has only a weak affinity for AhR (Chen et al., 1996; Ociepa-Zawal et al., 2007), and in combination with the small fraction of I3C that actually enters a cell (Staub et al., 2002), explains the relatively high concentration (200 μM) of I3C that is needed to observe the maximal degradation of ERα protein. AhR-induced degradation of ERα has been observed previously (Wormke et al., 2003; Ohtake et al., 2007), but with high-affinity ligands such as TCDD that is associated with reproductive defects in a variety of animal species (Antkiewicz et al., 2005). However, I3C is a natural dietary component, and unlike anthropogenic high-affinity AhR ligands, is reported to be beneficial to human health in epidemiological studies (Higdon et al., 2007).
A critical intermediate step within the I3C-triggered transcriptional cascade is the down-regulated expression of the GATA3 transcription factor, which disrupts the maintenance of a positive cross-regulatory between GATA3 and ERα (Eeckhoute et al., 2007). Expression of GATA3 is stimulated by ERα (Figure 11), and the time required for the maximal down-regulation of GATA3 gene products in I3C-treated cells accounts for the kinetic differences between the rapid I3C induced degradation of ERα protein and the longer duration observed for the loss of ERα transcripts. Ecoptic expression of GATA3 reversed the I3C-mediated decrease in ERα promoter activity but had no effect on the indole-induced degradation of ERα protein, suggesting that loss of ERα protein precedes the loss of GATA3 expression. Several recent studies have linked the cellular actions of ERα and GATA3 in mammary epithelial cell proliferation in both human breast cancer cells and in rodent model systems (Asselin-Labat et al., 2008; Kouros-Mehr et al., 2008b). GATA3 controls expression of many genes involved in the differentiation and proliferation of mammary luminal epithelial cells and is a strong predictor of tumor differentiation and estrogen receptor status of breast cancer (Kouros-Mehr et al., 2008a). Consistent with this concept, GATA3 and ERα are coexpressed in human breast cancer, and GATA3 is associated with maintaining estrogen-responsive proliferation (Fang et al., 2009). Furthermore, expression of ERα has been shown to be critical in the development of the normal breast epithelium, and GATA3 −/− mice display mammary glands that are strikingly similar to ERα −/− mammary glands (Kouros-Mehr et al., 2006; Asselin-Labat, 2007). During T cell differentiation, GATA3 levels are increased due to the loss of a transcriptional repressor that interacts with silencer elements in the GATA3 promoter (Gregoire and Romeo, 1999); therefore, the potential clinical use of I3C to enhance ERα protein degradation would not be predicted to block GATA3 expression in immune cells. Thus, we propose that I3C will be most effective in disrupting the estrogen-dependent growth of human cancer cells, such as breast cancer, that coexpress ERα, GATA3, and AhR.
The effects of I3C on ERα protein degradation and disruption of the GATA3-ERα–positive cross-regulatory loop were observed in two distinct human breast cancer cell lines, MCF7 and T47D, which are highly estrogen responsive. In T47D cells, even though ERα is expressed at somewhat lower levels than MCF7 cells, I3C rapidly induced ERα protein degradation and down-regulation of GATA3 gene expression. Furthermore, overexpression of GATA3 prevented the I3C down-regulation of ERα gene expression, whereas ectopic expression of a dominant-negative GATA3 mimicked the inhibitory effect of I3C on ERα expression. We have also observed that in certain other types of estrogen responsive cancer cells, such as human endometrial carcinoma cells, I3C fails to induce a cell cycle arrest and to disrupt ERα protein stability. Although we do not understand the mechanism of resistance to I3C these cell lines, an important clinical issue will be to determine the specificity of the tissues that respond to I3C based on the coexpression levels and activities of ERα, GATA3, and AhR.
ERα is an important target of therapeutic strategies to control the proliferation of hormone dependent breast cancers, although the preferred clinical options have significant adverse side effects and can lead to resistance to the treatments due to the persistence of active estrogen receptors. Aromatase inhibitors can reduce estrogen production at the cancer site, however, extended treatment can lead to bone loss as well as arthralgia due to estrogen requirements in the heart (Safi et al., 2005; Ewer and Gluck, 2009). The adverse side effects of SERMs, such as tamoxifen, are associated with the ligand-independent activation of ERα. Because I3C functions as a potent SERD of ERα expression, this indole can diminish both ligand-dependent and -independent ERα activation in breast cancer cells and consequently disrupt expression of the ERα target genes involved in cell proliferation.
One of the SERDs that is currently in clinical use, fluvestrant, causes the degradation of ERα in breast cancer cells (Osborne et al., 2004); however, this degradative process also occurs in many other tissues and can lead to pulmnar emboli, deep vein thrombosis, osteopenia, osteoporosis, and arthralgia. We propose that I3C will be most effective in disrupting the estrogen-dependent growth of human cancer cells, such as breast cancer, that coexpress ERα, GATA3, and AhR and therefore will have a significantly reduced systemic side effects in tissues such as heart and bone that express and are developmentally regulated by alternate GATA family members (Charron et al., 1999; Garimella et al., 2007; Afouda et al., 2008). Furthermore, in addition to the I3C-dependent ablation of ERα gene expression, this indole also activates ERβ (Sundar et al., 2006), which would allow many beneficial estrogen responses to be maintained in I3C treated cells, including antiproliferative signaling by this ER subtype. The novel properties of I3C, and its strong SERD function, implicate this indole has a highly tissue specific therapeutic potential for estrogen responsive cancers with the promise to improve the overall clinical outcome of patients.
Supplementary Material
ACKNOWLEDGMENTS
We thank A. J. Maxwell for generation of the −2294 ERα promoter construct and feedback. We greatly appreciate the comments and suggest by the entire Firestone laboratory during the course of our work. This study was supported by National Institutes of Health U.S. Public Service grant CA-102360 awarded from the National Cancer Institute. S.N.S. was supported by a postdoctoral fellowship from the Susan G. Komen Breast Cancer Fund, and C.N.M. was supported by a dissertation fellowship from the California Breast Cancer Research Program (13GB-1801).
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-08-0689) on February 3, 2010.
REFERENCES
- Afouda B. A., Martin J., Liu F., Ciau-Uitz A., Patient R., Hoppler S. GATA transcription factors integrate Wnt signalling during heart development. Development. 2008;135:3185–3190. doi: 10.1242/dev.026443. [DOI] [PubMed] [Google Scholar]
- Aggarwal B. B., Ichikawa H. Molecular targets and anticancer potential of indole-3-carbinol and its derivatives. Cell Cycle. 2005;4:1201–1215. doi: 10.4161/cc.4.9.1993. [DOI] [PubMed] [Google Scholar]
- Ali S., Coombes R. C. Estrogen receptor alpha in human breast cancer: occurrence and significance. J Mammary Gland Biol. Neoplasia. 2000;5:271–281. doi: 10.1023/a:1009594727358. [DOI] [PubMed] [Google Scholar]
- Antkiewicz D. S., Burns C. G., Carney S. A., Peterson R. E., Heideman W. Heart malformation is an early response to TCDD in embryonic zebrafish. Toxicol. Sci. 2005;84:368–377. doi: 10.1093/toxsci/kfi073. [DOI] [PubMed] [Google Scholar]
- Asselin-Labat M. L. Mammary stem and progenitor cells: critical role of the transcription factor Gata-3. Med. Sci. 2007;23:1077–1080. doi: 10.1051/medsci/200723121077. [DOI] [PubMed] [Google Scholar]
- Asselin-Labat M. L., et al. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat. Cell Biol. 2007;9:201–209. doi: 10.1038/ncb1530. [DOI] [PubMed] [Google Scholar]
- Asselin-Labat M. L., Vaillant F., Shackleton M., Bouras T., Lindeman G. J., Visvader J. E. Delineating the epithelial hierarchy in the mouse mammary gland. Cold Spring Harb. Symp. Quant. Biol. 2008;73:469–478. doi: 10.1101/sqb.2008.73.020. [DOI] [PubMed] [Google Scholar]
- Auborn K. J., Fan S., Rosen E. M., Goodwin L., Chandraskaren A., Williams D. E., Chen D., Carter T. H. Indole-3-carbinol is a negative regulator of estrogen. J. Nutr. 2003;133:2470S–2475S. doi: 10.1093/jn/133.7.2470s. [DOI] [PubMed] [Google Scholar]
- Badia E., Oliva J., Balaguer P., Cavailles V. Tamoxifen resistance and epigenetic modifications in breast cancer cell lines. Curr. Med. Chem. 2007;14:3035–3045. doi: 10.2174/092986707782794023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocchinfuso W. P., Korach K. S. Mammary gland development and tumorigenesis in estrogen receptor knockout mice. J Mammary Gland Biol. Neoplasia. 1997;2:323–334. doi: 10.1023/a:1026339111278. [DOI] [PubMed] [Google Scholar]
- Brenton J. D., Carey L. A., Ahmed A. A., Caldas C. Molecular classification and molecular forecasting of breast cancer: ready for clinical application? J. Clin. Oncol. 2005;23:7350–7360. doi: 10.1200/JCO.2005.03.3845. [DOI] [PubMed] [Google Scholar]
- Campbell-Thompson M., Lynch I. J., Bhardwaj B. Expression of estrogen receptor (ER) subtypes and ERbeta isoforms in colon cancer. Cancer Res. 2001;61:632–640. [PubMed] [Google Scholar]
- Chang E. C., Frasor J., Komm B., Katzenellenbogen B. S. Impact of estrogen receptor beta on gene networks regulated by estrogen receptor alpha in breast cancer cells. Endocrinology. 2006;147:4831–4842. doi: 10.1210/en.2006-0563. [DOI] [PubMed] [Google Scholar]
- Charron F., Paradis P., Bronchain O., Nemer G., Nemer M. Cooperative interaction between GATA-4 and GATA-6 regulates myocardial gene expression. Mol. Cell. Biol. 1999;19:4355–4365. doi: 10.1128/mcb.19.6.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I., Safe S., Bjeldanes L. Indole-3-carbinol and diindolylmethane as aryl hydrocarbon (Ah) receptor agonists and antagonists in T47D human breast cancer cells. Biochem. Pharmacol. 1996;51:1069–1076. doi: 10.1016/0006-2952(96)00060-3. [DOI] [PubMed] [Google Scholar]
- Clarke R., et al. Antiestrogen resistance in breast cancer and the role of estrogen receptor signaling. Oncogene. 2003;22:7316–7339. doi: 10.1038/sj.onc.1206937. [DOI] [PubMed] [Google Scholar]
- Colozza M., Califano R., Minenza E., Dinh P., Azambuja E. Aromatase inhibitors: a new reality for the adjuvant endocrine treatment of early-stage breast cancer in postmenopausal women. Mini Rev. Med. Chem. 2008;8:564–574. doi: 10.2174/138955708784534472. [DOI] [PubMed] [Google Scholar]
- Cover C. M., Hsieh S. J., Cram E. J., Hong C., Riby J. E., Bjeldanes L. F., Firestone G. L. Indole-3-carbinol and tamoxifen cooperate to arrest the cell cycle of MCF-7 human breast cancer cells. Cancer Res. 1999;59:1244–1251. [PubMed] [Google Scholar]
- Cover C. M., Hsieh S. J., Tran S. H., Hallden G., Kim G. S., Bjeldanes L. F., Firestone G. L. Indole-3-carbinol inhibits the expression of cyclin-dependent kinase-6 and induces a G1 cell cycle arrest of human breast cancer cells independent of estrogen receptor signaling. J. Biol. Chem. 1998;273:3838–3847. doi: 10.1074/jbc.273.7.3838. [DOI] [PubMed] [Google Scholar]
- Cram E. J., Liu B. D., Bjeldanes L. F., Firestone G. L. Indole-3-carbinol inhibits CDK6 expression in human MCF-7 breast cancer cells by disrupting Sp1 transcription factor interactions with a composite element in the CDK6 gene promoter. J. Biol. Chem. 2001;276:22332–22340. doi: 10.1074/jbc.M010539200. [DOI] [PubMed] [Google Scholar]
- Dutertre M., Smith C. L. Molecular mechanisms of selective estrogen receptor modulator (SERM) action. J. Pharmacol. Exp. Ther. 2000;295:431–437. [PubMed] [Google Scholar]
- Eakin C. M., Maccoss M. J., Finney G. L., Klevit R. E. Estrogen receptor alpha is a putative substrate for the BRCA1 ubiquitin ligase. Proc. Natl. Acad. Sci. USA. 2007;104:5794–5799. doi: 10.1073/pnas.0610887104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eeckhoute J., Keeton E. K., Lupien M., Krum S. A., Carroll J. S., Brown M. Positive cross-regulatory loop ties GATA-3 to estrogen receptor alpha expression in breast cancer. Cancer Res. 2007;67:6477–6483. doi: 10.1158/0008-5472.CAN-07-0746. [DOI] [PubMed] [Google Scholar]
- Enmark E., Gustafsson J. A. Oestrogen receptors—an overview. J. Intern. Med. 1999;246:133–138. doi: 10.1046/j.1365-2796.1999.00545.x. [DOI] [PubMed] [Google Scholar]
- Ewer M. S., Gluck S. A woman's heart: the impact of adjuvant endocrine therapy on cardiovascular health. Cancer. 2009;115:1813–1826. doi: 10.1002/cncr.24219. [DOI] [PubMed] [Google Scholar]
- Failor K. L., Desyatnikov Y., Finger L. A., Firestone G. L. Glucocorticoid-induced degradation of GSK3 protein is triggered by Sgk and Akt signaling and controls beta-catenin dynamics and tight junction formation in mammary epithelial tumor cells. Mol. Endocrinol. 2007;21:2403–2415. doi: 10.1210/me.2007-0143. [DOI] [PubMed] [Google Scholar]
- Fan M., Bigsby R. M., Nephew K. P. The NEDD8 pathway is required for proteasome-mediated degradation of human estrogen receptor (ER)-alpha and essential for the antiproliferative activity of ICI 182,780 in ERalpha-positive breast cancer cells. Mol. Endocrinol. 2003;17:356–365. doi: 10.1210/me.2002-0323. [DOI] [PubMed] [Google Scholar]
- Fang S. H., Chen Y., Weigel R. J. GATA-3 as a marker of hormone response in breast cancer. J. Surg. Res. 2009;157:290–295. doi: 10.1016/j.jss.2008.07.015. [DOI] [PubMed] [Google Scholar]
- Fuqua S. A., Fitzgerald S. D., Chamness G. C., Tandon A. K., McDonnell D. P., Nawaz Z., O'Malley B. W., McGuire W. L. Variant human breast tumor estrogen receptor with constitutive transcriptional activity. Cancer Res. 1991;51:105–109. [PubMed] [Google Scholar]
- Garcia H. H., Brar G. A., Nguyen D. H., Bjeldanes L. F., Firestone G. L. Indole-3-carbinol (I3C) inhibits cyclin-dependent kinase-2 function in human breast cancer cells by regulating the size distribution, associated cyclin E forms, and subcellular localization of the CDK2 protein complex. J. Biol. Chem. 2005;280:8756–8764. doi: 10.1074/jbc.M407957200. [DOI] [PubMed] [Google Scholar]
- Garimella R., Kacena M. A., Tague S. E., Wang J., Horowitz M. C., Anderson H. C. Expression of bone morphogenetic proteins and their receptors in the bone marrow megakaryocytes of GATA-1(low) mice: a possible role in osteosclerosis. J. Histochem. Cytochem. 2007;55:745–752. doi: 10.1369/jhc.6A7164.2007. [DOI] [PubMed] [Google Scholar]
- Gregoire J. M., Romeo P. H. T-cell expression of the human GATA-3 gene is regulated by a non-lineage-specific silencer. J. Biol. Chem. 1999;274:6567–6578. doi: 10.1074/jbc.274.10.6567. [DOI] [PubMed] [Google Scholar]
- Harris H. A., Katzenellenbogen J. A., Katzenellenbogen B. S. Characterization of the biological roles of the estrogen receptors, ERalpha and ERbeta, in estrogen target tissues in vivo through the use of an ERalpha-selective ligand. Endocrinology. 2002;143:4172–4177. doi: 10.1210/en.2002-220403. [DOI] [PubMed] [Google Scholar]
- Higdon J. V., Delage B., Williams D. E., Dashwood R. H. Cruciferous vegetables and human cancer risk: epidemiologic evidence and mechanistic basis. Pharmacol. Res. 2007;55:224–236. doi: 10.1016/j.phrs.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch R. V., Thompson D. A., Baker R. J., Weigel R. J. GATA-3 is expressed in association with estrogen receptor in breast cancer. Int. J. Cancer. 1999;84:122–128. doi: 10.1002/(sici)1097-0215(19990420)84:2<122::aid-ijc5>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Hong C., Kim H. A., Firestone G. L., Bjeldanes L. F. 3,3′-Diindolylmethane (DIM) induces a G(1) cell cycle arrest in human breast cancer cells that is accompanied by Sp1-mediated activation of p21(WAF1/CIP1) expression. Carcinogenesis. 2002;23:1297–1305. doi: 10.1093/carcin/23.8.1297. [DOI] [PubMed] [Google Scholar]
- Jellinck P. H., Forkert P. G., Riddick D. S., Okey A. B., Michnovicz J. J., Bradlow H. L. Ah receptor binding properties of indole carbinols and induction of hepatic estradiol hydroxylation. Biochem. Pharmacol. 1993;45:1129–1136. doi: 10.1016/0006-2952(93)90258-x. [DOI] [PubMed] [Google Scholar]
- Kim Y. S., Milner J. A. Targets for indole-3-carbinol in cancer prevention. J. Nutr. Biochem. 2005;16:65–73. doi: 10.1016/j.jnutbio.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Kouros-Mehr H., Bechis S. K., Slorach E. M., Littlepage L. E., Egeblad M., Ewald A. J., Pai S. Y., Ho I. C., Werb Z. GATA-3 links tumor differentiation and dissemination in a luminal breast cancer model. Cancer Cell. 2008a;13:141–152. doi: 10.1016/j.ccr.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouros-Mehr H., Kim J. W., Bechis S. K., Werb Z. GATA-3 and the regulation of the mammary luminal cell fate. Curr. Opin. Cell Biol. 2008b;20:164–170. doi: 10.1016/j.ceb.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouros-Mehr H., Slorach E. M., Sternlicht M. D., Werb Z. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell. 2006;127:1041–1055. doi: 10.1016/j.cell.2006.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W. L., Cheng M. H., Chao H. T., Wang P. H. The role of selective estrogen receptor modulators on breast cancer: from tamoxifen to raloxifene. Taiwan J. Obstet. Gynecol. 2008;47:24–31. doi: 10.1016/S1028-4559(08)60051-0. [DOI] [PubMed] [Google Scholar]
- Mercier I., Mader S., Calderone A. Tamoxifen and ICI 182,780 negatively influenced cardiac cell growth via an estrogen receptor-independent mechanism. Cardiovasc. Res. 2003;59:883–892. doi: 10.1016/s0008-6363(03)00517-0. [DOI] [PubMed] [Google Scholar]
- Miller W. R. Biology of aromatase inhibitors: pharmacology/endocrinology within the breast. Endocr. Relat. Cancer. 1999;6:187–195. doi: 10.1677/erc.0.0060187. [DOI] [PubMed] [Google Scholar]
- Ociepa-Zawal M., Rubis B., Lacinski M., Trzeciak W. H. The effect of indole-3-carbinol on the expression of CYP1A1, CYP1B1 and AhR genes and proliferation of MCF-7 cells. Acta Biochim. Pol. 2007;54:113–117. [PubMed] [Google Scholar]
- Ohtake F., et al. Dioxin receptor is a ligand-dependent E3 ubiquitin ligase. Nature. 2007;446:562–566. doi: 10.1038/nature05683. [DOI] [PubMed] [Google Scholar]
- Ohtake F., Fujii-Kuriyama Y., Kato S. AhR acts as an E3 ubiquitin ligase to modulate steroid receptor functions. Biochem. Pharmacol. 2009;77:474–484. doi: 10.1016/j.bcp.2008.08.034. [DOI] [PubMed] [Google Scholar]
- Osborne C. K., Wakeling A., Nicholson R. I. Fulvestrant: an oestrogen receptor antagonist with a novel mechanism of action. Br. J. Cancer. 2004;1(90 suppl):S2–S6. doi: 10.1038/sj.bjc.6601629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualini J. R., Chetrite G. S. Recent insight on the control of enzymes involved in estrogen formation and transformation in human breast cancer. J. Steroid Biochem. Mol. Biol. 2005;93:221–236. doi: 10.1016/j.jsbmb.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Rahman K. M., Li Y., Sarkar F. H. Inactivation of akt and NF-kappaB play important roles during indole-3-carbinol-induced apoptosis in breast cancer cells. Nutr. Cancer. 2004;48:84–94. doi: 10.1207/s15327914nc4801_12. [DOI] [PubMed] [Google Scholar]
- Richter C. A., Tillitt D. E., Hannink M. Regulation of subcellular localization of the aryl hydrocarbon receptor (AhR) Arch. Biochem. Biophys. 2001;389:207–217. doi: 10.1006/abbi.2001.2339. [DOI] [PubMed] [Google Scholar]
- Roger P., Sahla M. E., Makela S., Gustafsson J. A., Baldet P., Rochefort H. Decreased expression of estrogen receptor beta protein in proliferative preinvasive mammary tumors. Cancer Res. 2001;61:2537–2541. [PubMed] [Google Scholar]
- Safe S., Papineni S., Chintharlapalli S. Cancer chemotherapy with indole-3-carbinol, bis(3′-indolyl)methane and synthetic analogs. Cancer Lett. 2008;269:326–338. doi: 10.1016/j.canlet.2008.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safi R., Kovacic A., Gaillard S., Murata Y., Simpson E. R., McDonnell D. P., Clyne C. D. Coactivation of liver receptor homologue-1 by peroxisome proliferator-activated receptor gamma coactivator-1alpha on aromatase promoter II and its inhibition by activated retinoid X receptor suggest a novel target for breast-specific antiestrogen therapy. Cancer Res. 2005;65:11762–11770. doi: 10.1158/0008-5472.CAN-05-2792. [DOI] [PubMed] [Google Scholar]
- Shaaban A. M., O'Neill P. A., Davies M. P., Sibson R., West C. R., Smith P. H., Foster C. S. Declining estrogen receptor-beta expression defines malignant progression of human breast neoplasia. Am. J. Surg. Pathol. 2003;27:1502–1512. doi: 10.1097/00000478-200312000-00002. [DOI] [PubMed] [Google Scholar]
- Staub R. E., Feng C., Onisko B., Bailey G. S., Firestone G. L., Bjeldanes L. F. Fate of indole-3-carbinol in cultured human breast tumor cells. Chem. Res. Toxicol. 2002;15:101–109. doi: 10.1021/tx010056m. [DOI] [PubMed] [Google Scholar]
- Sundar S. N., Kerekatte V., Equinozio C. N., Doan V. B., Bjeldanes L. F., Firestone G. L. Indole-3-carbinol selectively uncouples expression and activity of estrogen receptor subtypes in human breast cancer cells. Mol. Endocrinol. 2006;20:3070–3082. doi: 10.1210/me.2005-0263. [DOI] [PubMed] [Google Scholar]
- Sundar S. N., Marconett C. N., Doan V. B., Willoughby J. A., Sr., Firestone G. L. Artemisinin selectively decreases functional levels of estrogen receptor-alpha and ablates estrogen induced proliferation in human breast cancer cells. Carcinogenesis. 2008;29:2252–2258. doi: 10.1093/carcin/bgn214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada Y., Andreeff M., Aggarwal B. B. Indole-3-carbinol suppresses NF-kappaB and IkappaBalpha kinase activation, causing inhibition of expression of NF-kappaB-regulated antiapoptotic and metastatic gene products and enhancement of apoptosis in myeloid and leukemia cells. Blood. 2005;106:641–649. doi: 10.1182/blood-2004-12-4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay J. J., Viger R. S. GATA factors differentially activate multiple gonadal promoters through conserved GATA regulatory elements. Endocrinology. 2001;142:977–986. doi: 10.1210/endo.142.3.7995. [DOI] [PubMed] [Google Scholar]
- Wang T. T., Milner M. J., Milner J. A., Kim Y. S. Estrogen receptor alpha as a target for indole-3-carbinol. J. Nutr. Biochem. 2006;17:659–664. doi: 10.1016/j.jnutbio.2005.10.012. [DOI] [PubMed] [Google Scholar]
- Weng J. R., Tsai C. H., Kulp S. K., Chen C. S. Indole-3-carbinol as a chemopreventive and anti-cancer agent. Cancer Lett. 2008;262:153–163. doi: 10.1016/j.canlet.2008.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wormke M., Stoner M., Saville B., Walker K., Abdelrahim M., Burghardt R., Safe S. The aryl hydrocarbon receptor mediates degradation of estrogen receptor alpha through activation of proteasomes. Mol. Cell. Biol. 2003;23:1843–1855. doi: 10.1128/MCB.23.6.1843-1855.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.