This study identifies and characterizes marvelD3, a novel tight junction protein that contains a conserved MARVEL domain. Analyses using phylogenetic, expression profiling, microscopic, and functional approaches show that marvelD3, occludin, and tricellulin are related and have distinct but overlapping functions at the tight junction.
Abstract
In vitro studies have demonstrated that occludin and tricellulin are important for tight junction barrier function, but in vivo data suggest that loss of these proteins can be overcome. The presence of a heretofore unknown, yet related, protein could explain these observations. Here, we report marvelD3, a novel tight junction protein that, like occludin and tricellulin, contains a conserved four-transmembrane MARVEL (MAL and related proteins for vesicle trafficking and membrane link) domain. Phylogenetic tree reconstruction; analysis of RNA and protein tissue distribution; immunofluorescent and electron microscopic examination of subcellular localization; characterization of intracellular trafficking, protein interactions, dynamic behavior, and siRNA knockdown effects; and description of remodeling after in vivo immune activation show that marvelD3, occludin, and tricellulin have distinct but overlapping functions at the tight junction. Although marvelD3 is able to partially compensate for occludin or tricellulin loss, it cannot fully restore function. We conclude that marvelD3, occludin, and tricellulin define the tight junction–associated MARVEL protein family. The data further suggest that these proteins are best considered as a group with both redundant and unique contributions to epithelial function and tight junction regulation.
INTRODUCTION
The tight junction is a network of transmembrane and peripheral proteins that form a semipermeable barrier to paracellular flux (Farquhar and Palade, 1963; Claude and Goodenough, 1973). The functions of some of these proteins have been defined in recent years, including roles for the peripheral membrane proteins ZO-1 and -2 in recruiting transmembrane claudin proteins to the tight junction and the critical responsibilities of the latter in defining ion selectivity (Itoh et al., 1999; Umeda et al., 2006). However, the specific contributions of most tight junction–associated proteins remain enigmatic. Foremost among these is occludin, the first transmembrane protein to be associated with the tight junction (Furuse et al., 1993).
Uncertainty regarding occludin function has been magnified by contrasting results of in vitro and in vivo studies. The former have shown that occludin can mediate intercellular adhesive interactions, that modulation of occludin expression impacts tight junction barrier function, and that occludin-derived peptides can disrupt the tight junction (Furuse et al., 1993; Balda et al., 1996; McCarthy et al., 1996; Wong and Gumbiner, 1997; Yu et al., 2005). Moreover, both in vitro and in vivo studies have associated occludin endocytosis with pathophysiological and pharmacological tight junction barrier loss (Massoumi and Sjolander, 2001; Clayburgh et al., 2005; Shen and Turner, 2005; Utech et al., 2005; Schwarz et al., 2007). These data are difficult to reconcile with the observations that occludin knockout mice are viable, fail to display defective epidermal, respiratory, renal, or intestinal barrier function, and do not develop spontaneous skin, airway, kidney, or gut disease (Saitou et al., 2000). As a result, many have concluded that occludin is unnecessary for normal epithelial tight junction function. However, occludin knockout mice do have significant pathologies, including small size, testicular atrophy, male infertility, salivary gland dysfunction, atrophic gastritis, thinning of compact bone, and brain calcifications. Thus, two potential explanations must be considered. First, occludin may contribute to epithelial activities other than development of basal barrier function, and second, that other tight junction proteins may partially compensate for occludin loss.
The occludin-related protein tricellulin has recently been reported (Ikenouchi et al., 2005). Both occludin and tricellulin contain the tetra-spanning MARVEL (MAL and related proteins for vesicle trafficking and membrane link) domain that is present in proteins involved in membrane apposition and concentrated in cholesterol-rich microdomains (Sanchez-Pulido et al., 2002). Like occludin (McCarthy et al., 1996; Yu et al., 2005), tricellulin knockdown interferes with tight junction assembly and enhances solute diffusion across cultured epithelial monolayers (Ikenouchi et al., 2005), while tricellulin overexpression enhances barrier function (Krug et al., 2009). Human tricellulin mutations are associated with nonsyndromic hearing loss, but with no other pathologies (Riazuddin et al., 2006; Chishti et al., 2008). Tricellulin is, therefore, not essential for function of epidermal, respiratory, renal, or intestinal tight junctions. Given that both occludin and tricellulin are MARVEL domain–containing proteins, we sought additional members of this family that participate in tight junction function.
To comprehensively define the members of the MARVEL domain–containing protein family present at the tight junction, we performed bioinformatic analyses. These identified the previously uncharacterized protein marvelD3 as a potential tight junction component. Analyses of evolutionary conservation, tissue distribution, subcellular localization, trafficking, protein interactions, dynamic behavior, siRNA knockdown, and in vivo responses to immune activation were used to characterize marvelD3 as well as tricellulin and occludin. The data define marvelD3, occludin, and tricellulin as the only members of the tight junction–associated MARVEL protein (TAMP) family and show that although marvelD3 may partially compensate for occludin or tricellulin loss, it is insufficient to fully restore function in the absence of other TAMPs. We conclude that these proteins are best considered as a group with parallel, but nonredundant, functions.
MATERIALS AND METHODS
Bioinformatic Analyses
As MARVEL domain–containing protein sequences are highly divergent, alignment was performed using the E-INS-i algorithm in the MAFFT package (Katoh and Toh, 2008), one of the most accurate algorithms for alignment of sequences with conserved residues scattered across an extended region. Transmembrane topology for the alignment was establishing using PolyPhobius (Kall et al., 2005) and the TMHMM package (Sonnhammer et al., 1998), which generated similar predictions. However, as the former algorithm is known to produce more reliable results for transmembrane topology, especially for eukaryotic proteins (Jones, 2007), PolyPhobius predictions are shown.
ProtTest was used to fit models to the alignment (Abascal et al., 2005), and Blosum62 with site heterogeneity and empirical residue frequency was found to be the optimal phylogenetic model for tree reconstruction. Toward this goal, PhyML (Guindon and Gascuel, 2003) with 500 bootstrap replicates and MrBayes (Huelsenbeck et al., 2001) with a single Markov chain for 1 million generations were each conducted. Both algorithms yielded highly convergent trees, but the latter typically yields a more accurate result and is presented in Figure 1.
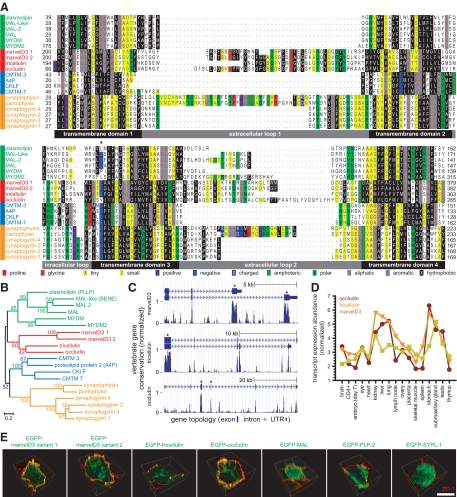
Figure 1.
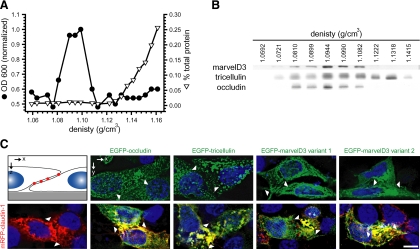
MarvelD3, tricellulin, and occludin are the only members of the tight junction–associated MARVEL protein (TAMP) subfamily. (A) MARVEL domain alignment was ordered after model fitting and tree reconstruction and shaded within each of the four MARVEL protein lineages according to the Blosum62 score. Amino acids are shaded according to physiochemical properties as indicated in the figure. The characteristic acidic residue in the third transmembrane domain (Sanchez-Pulido et al., 2002) is indicated by an asterisk (*). (B) The alignment was used for tree reconstruction, and disparities were calculated according to the fraction of mutated residues. Posterior values are shown as 100-fold original predictions, implying that a bootstrap value of 60 is highly reliable. (C) Gene conservation analysis demonstrates that exons encoding MARVEL domains (∗) show the highest degree of evolutionary conservation within the marvelD3, tricellulin, and occludin genes. Data are presented as a base-by-base conservation measurement, with 0 indicating a neutral region and 1 indicating perfect conservation, from 28 vertebrate species including marsupial, monotreme, and placental mammals, as well as reptile, amphibian, bird, and fish clades. (D) Analysis of transcript expression in the Affymetrix GeneChip Mouse Genome 430 2.0 Array (Santa Clara, CA) reveals a highly correlated (p < 0.001), though nonredundant, pattern of expression for marvelD3, tricellulin, and occludin within epithelial organs. (E) EGFP-MARVEL domain–containing fusion proteins (green) were transfected into Caco-2 human intestinal epithelial cells. Polarized monolayers were grown on Transwell supports, and ZO-1 (red) was detected by immunofluorescent staining. Three-dimensional reconstruction shows that EGFP-marvelD3 splice variants 1 and 2, EGFP-tricellulin, and EGFP-occludin localize to tight junctions and intracellular vesicles. Both EGFP-marvelD3 splice variants are enriched along bicellular tight junctions, similar to occludin, whereas tricellulin is specifically concentrated at tricellular regions. MARVEL proteins from other lineages were found to decorate the apical plasma membrane and intracellular transport vesicles, but not the tight junction. Images are representative of ≥3 experiments, all with similar results. Bar, 10 μm.
Gene conservation data were extracted using the PhastCons track from the UCSC Genome Browser (Kuhn et al., 2009). This hidden Markov model-based analysis, which considers both individual alignment columns as well as flanking sequences, approximates the likelihood that a nucleotide belongs to a conserved region based on a multispecies alignment (Siepel et al., 2005).
Using the updated annotation file for the Affymetrix Mouse Genome 430 2.0 Array, Mouse4302_Mm_ENSG (Dai et al., 2005), we reanalyzed the tissue expression profile database GEO series GSE1986 (Barrett et al., 2007) using GCRMA and Bioconductor (Gentleman et al., 2004; Wu and Irizarry, 2004) and extracted the expression information for marvelD3, tricellulin, and occludin.
Cell Culture
Caco-2BBe cells were grown on collagen-coated polycarbonate Transwells (Corning Life Sciences, Corning, NY), as previously described (Turner et al., 1997) and studied 14 d after confluence. Transepithelial resistance (TER) was measured using an epithelial volt-ohm meter (EVOM; World Precision Instruments, Sarasota, FL). Fluid resistance was subtracted from all readings. L929 cells were grown on glass coverslips, maintained in DMEM, and studied 1 d after confluence.
Plasmids
Fluorescent fusion constructs were generated by joining the cDNA sequences of genes into pEGFP-C1 (BD Biosciences Clontech, San Jose, CA) and pcDNA3 (Invitrogen, Carlsbad, CA) containing mRFP1 sequence (Campbell et al., 2002), a kind gift from R. Tsien (University of California at San Diego). MarvelD3 splice variant 1, marvelD3 splice variant 2, tricellulin, occludin, claudin-1, and ZO-1 coding sequences were cloned from Caco-2BBe cDNA by PCR amplification. Human MAL (I.M.A.G.E. clone 2822079), proteolipid protein 2 (I.M.A.G.E. clone 40033888), and pantophysin (I.M.A.G.E. clone 3883697) cDNAs were amplified and cloned from vectors purchased from American Type Culture Collection (Manassas, VA). Glutathione S-transferase (GST) and VSVG (vesicular stomatitis virus glycoprotein) fusion constructs were generated by joining cDNA sequences into pGEX-4T3 (GE Healthcare Bio-sciences, Piscataway, NJ), and removing the GST coding sequence by site-directed mutagenesis (Stratagene, La Jolla, CA) for VSVG constructs. Plasmid integrity and directionality were confirmed by restriction digestion and sequencing of expression constructs and adjacent regions. Constructs were transfected into Caco-2BBe cells with Lipofectamine 2000 (Invitrogen), and transgene expression was enhanced by culture with 7.5 mM sodium butyrate in antibiotic-free media for 20 h. Monolayers were cultured in sodium butyrate–free media for 3 h before experimental use.
Immunofluorescence Microscopy
Caco-2BBe monolayers grown in Transwells and 5-μm sections of snap-frozen mouse tissue were fixed with 1% paraformaldehyde and stained as described previously (Shen et al., 2006; Su et al., 2009). Immunofluorescence was performed using occludin, tricellulin, and ZO-1 antibodies (Invitrogen), as well as anti-marvelD3 polyclonal antisera (described below). Primary incubations were followed by Hoechst 33342 and Alexa dye–conjugated secondary antibodies or phalloidin (Invitrogen). After mounting in ProLong Gold (Invitrogen), samples were examined using a DMLB epifluorescence microscope (Leica Microsystems, Bannockburn, IL) equipped with an 88000 filter set (Chroma Technology, Rockingham, VT), 63× PLAN APO 1.32 NA oil immersion objective, Retiga EXi camera (QImaging, Surrey, BC, Canada), and Metamorph 7 acquisition software (Molecular Devices, Sunnyvale, CA). Z-stacks were deconvolved with Autodeblur X (Media Cybernetics, Bethesda, MD) for 10 iterations.
Polyclonal Anti-marvelD3 Antisera Development
Anti-marvelD3 antisera were developed in rabbits immunized with two marvelD3-directed synthetic peptides (residues 13-26, RPRERDPGRRPHPD; and residues 174-187, EVEYYQSEAEGLLE) in combination (YenZym, South San Francisco, CA). These peptides, which share 89% homology between mouse and human sequences, are common to both splice variants, and were selected based on predicted immunogenicity. Immune responses were assessed by ELISA. Polyclonal antisera were affinity-purified.
Immunoblotting
Lysates of mouse jejunal enterocytes and Caco-2BBe cells were separated by SDS-PAGE and transferred to PVDF membranes. Sample protein concentration was determined by dye-binding protein assay (Bio-Rad, Hercules, CA), and equal quantities of protein were loaded in each lane. Immunoblots were performed using the previously described antibodies, as well as GAPDH (Invitrogen), VSVG, and β-actin (Sigma-Aldrich, St. Louis, MO), followed by either HRP- (Cell Signaling Technology, Boston, MA) or infrared dye-conjugated (Li-Cor, Lincoln, NE) secondary antibodies. Protein was detected either by chemiluminescence or using an Odyssey System (Li-Cor). Densitometry of immunoblot data were performed using ImageJ software (Abramoff et al., 2004).
Animals
Seven- to 10-wk-old C57BL/6 mice were used for all studies, which were conducted in an AAALAC-accredited facility under protocols approved by The University of Chicago IACUC.
Epithelial Isolation
Mouse jejunum was opened lengthwise, and epithelial cells were isolated at 4°C as described previously (Clayburgh et al., 2005). Cells were resuspended and lysed in Laemmli sample buffer or buffer RLT (Qiagen, Valencia, CA) for protein or RNA analysis, respectively.
Immunoelectron Microscopy
Seven-week-old C57BL/6 mice were perfused with 1% paraformaldehyde in 0.1 M phosphate buffer, pH 7.4, for 10 min, and jejuna were fixed in 1% paraformaldehyde for 20 min at 4°C. Immunoelectron microscopy using ultrathin cryosections was performed as described previously (Tokuyasu, 1980). Ultrathin sections were blocked with 10% normal goat serum in PBS for 30 min at room temperature, incubated with anti-marvelD3 polyclonal antisera at 1:100 for 17 h at 4°C, rinsed, and then incubated with 10-nm gold conjugated goat anti-rabbit IgG (BB International, Llanishen, Cardiff, United Kingdom) for 1 h at room temperature. Samples were examined with an H-7500 transmission electron microscope (Hitachi, Tokyo, Japan) at an acceleration voltage of 100 kV.
Immunoprecipitation
All steps were performed at 4°C. Confluent Caco-2BBe monolayers were washed thrice in PBS and incubated in RIPA buffer (Santa Cruz Biotechnology, Santa Cruz, CA) for 10 min. Cells were scraped from filters and passed five times through a 21-gauge needle. Lysates were precleared with rabbit IgG for 30 min and centrifuged for 5 min at 2500 rpm. The supernatant was immunoprecipitated using the previously described antibodies with protein A/G PLUS-Agarose beads (Santa Cruz Biotechnology).
GST Pulldown
BL21(DE3)pLysS competent bacteria transformed with GST and VSVG fusion constructs were induced and processed according to the GST Protein Interaction Pull-Down Kit (Pierce, Rockford, IL). Immobilized bait and soluble prey protein lysates were combined, incubated at 4°C while gently rocking overnight, washed three times in 10 mM NaCl, and eluted using 10 mM reduced glutathione.
Subcellular Fractionation
All steps were performed at 4°C. Confluent Caco-2BBe monolayers were rinsed two times in HBSS. Cells were isolated and lysed by scraping with a rubber policeman into HBSS containing 1% Triton X-100 in TBS, and passed 10 times through a 25-gauge needle. Lysates were separated on a continuous sucrose gradient by ultracentrifugation, and fractions were analyzed for sucrose concentration by refractometry as described previously (Shen and Turner, 2005).
Quantitative Real-Time RT-PCR
Caco-2BBe and mouse jejunal epithelial samples were lysed in buffer RLT. RNA was purified from each source using the RNeasy Mini Kit with DNase treatment (Qiagen). The Protoscript First-Strand cDNA Synthesis Kit (New England Biolabs, Ipswitch, MA) was used for reverse transcriptase reaction with poly-T primers. Primers for quantitative real-time RT-PCR (qRT-PCR) were designed to amplify 75-250 base pair regions spanning intron-exon boundaries within genes of interest and validated for amplification efficiency between 0.9 and 1.1 (human marvelD3 splice variant 1 sense GAACCCCCTTCGGAGAGATA, antisense CGGCAAGGACAAAGTAGGAG; human marvelD3 splice variant 2 sense TTACCAGTCAGAGGCGGAAG, antisense CCCCCTGTGGAACTGTAAGA; human tricellulin sense TCAGACAGATGATGAGCGAGA, antisense ATGTTCCTGTCGGCTTTCC; human occludin sense TCCAATGGCAAAGTGAATGA, antisense AGTCCTCCTCCAGCTCATCA; human claudin-1 sense CGATGAGGTGCAGAAGATGA, antisense CCAGTGAAGAGAGCCTGACC; mouse marvelD3 splice variant 1 sense GGGCTTCGGAAAGATACGTG, antisense CACCGTCAAAGCCACTATAAG; mouse marvelD3 splice variant 2 sense CTCCTGGATTGCCACAAATG, antisense GTGCCCTCAAAAGGTGAGTA; mouse tricellulin sense CTCGGAGACATCGGGAGTTC, antisense CCTGATCCCTCTGTCGATCACT; mouse occludin sense GCGGAAGAGGTTGACAGTCC, antisense ACTCCCCACCTGTCGTGTAG; mouse and human GAPDH sense CTTCACCACCATGGAGAAGGC, antisense GGCATGGACTGTGGTCATGAG). Transcripts were quantified using SYBR Green PCR (Bio-Rad, Hercules, CA).
TAMP Knockdown
Caco-2BBe cells were transfected with small interfering RNA (siRNA) using Lipofectamine 2000 (Invitrogen) as previously described (Hu et al., 2006). siRNAs against marvelD3 (GAGAGGAGGUGGAAUAUUA and GCGCAGGUGAACACGGAGU), tricellulin (AGAACCAGCUAUAGCGCCA), and occludin (GAAGAAAGAUGGACAGGUA) were purchased from Dharmacon (Lafayette, CO). Scrambled siRNAs with similar GC content were used as controls.
Fluorescence Recovery after Photobleaching
Transfected Caco-2BBe cells were grown as confluent monolayers on Transwells and imaged in nominally bicarbonate-free HBSS supplemented with 15 mM HEPES, pH 7.4, and 4.5 g/l glucose, on a 37°C temperature-controlled stage. Fluorescence bleaching and imaging were performed using a DM4000 epifluorescence microscope (Leica Microsystems) with a MicroPoint system (Photonic Instruments, St. Charles, IL), CoolSnap HQ camera (Roper Scientific, Trenton, NJ), and 63× U-V-I 1.2 NA water immersion objective. Images were collected every 10 s until steady-state fluorescence was reached.
Raw data were aligned and mean fluorescence of background, whole-cell, and bleached bicellular or tricellular tight junction regions were quantified over time using Metamorph 7 software. For all fluorescence recovery after photobleaching (FRAP) experiments, background fluorescence was subtracted from regions of interest and from total-cell fluorescence at each time point. Data correction, mobile fraction, and halftime of recovery were determined as described previously (Yguerabide et al., 1982; Shen et al., 2008) using Sigma Plot for curve fitting (SPSS, Chicago, IL).
Statistical Analysis
All data are presented as mean ± SE. Student's unpaired t test was used to compare means, with statistical significance taken as p < 0.05.
RESULTS
Occludin and Tricellulin Are Members of a MARVEL Subfamily That Includes MarvelD3
The tight junction proteins occludin and tricellulin each contain MARVEL domains (Sanchez-Pulido et al., 2002). To identify additional tight junction–associated MARVEL domain–containing proteins, a minimally redundant collection of human MARVEL domain protein sequences was obtained from the EMBL-EBI InterPro database. Alignment of these sequences (Figure 1A) and tree reconstruction (Figure 1B) suggested the existence of four subfamilies within the greater MARVEL family. One subfamily was composed of the tight junction proteins occludin and tricellulin and two splice variants of marvelD3, a previously uncharacterized protein. MarvelD3 splice variants 1 and 2 have identical amino-terminal cytoplasmic tails, but due to translation of either exon 4 or 3, respectively, differ in the sequence of their MARVEL domains and short cytoplasmic carboxy-termini. Although alternative splicing also generates variants of tricellulin and occludin (Muresan et al., 2000; Gu et al., 2008), these differ only in cytoplasmic amino- and carboxy-terminal regions and contain identical MARVEL domains.
Gene conservation analysis of marvelD3, tricellulin, and occludin across vertebrate species demonstrated a higher degree of evolutionary conservation within exons encoding MARVEL domains than in those encoding cytoplasmic tails or within noncoding genomic sequences, suggesting that the MARVEL domain itself significantly influences the function of these proteins (Figure 1C). Moreover, analysis of murine tissue transcript profiles revealed a highly correlated (p < 0.001), although nonredundant, pattern of marvelD3, tricellulin, and occludin expression, consistent with shared functions among members of this MARVEL subfamily (Figure 1D). The analysis also indicated enrichment of these proteins in epithelial organs, including lung, stomach, kidney, and liver. In sum, these results suggest that marvelD3, tricellulin, and occludin represent a functionally unique subfamily within the broader family of MARVEL domain–containing proteins.
MarvelD3, Tricellulin, and Occludin are the Only Members of the Tight Junction–associated MARVEL Protein Subfamily
Previous analyses have identified MARVEL family members within cellular structures that include intracellular transport vesicles, the apical plasma membrane, and the endoplasmic reticulum (Zacchetti et al., 1995; Haass et al., 1996; Breitwieser et al., 1997). Although occludin and tricellulin are the only MARVEL domain–containing proteins reported to concentrate at the tight junction (Furuse et al., 1993; Ikenouchi et al., 2005), our evolutionary analyses suggest that marvelD3 may also be associated with tight junctions. To test this hypothesis, marvelD3, tricellulin, occludin, and representative proteins from each of the three additional MARVEL subfamilies (Figure 1B) were expressed as enhanced green fluorescent protein (EGFP)-fusion constructs in Caco-2 human intestinal epithelial cells. EGFP-tricellulin and EGFP-occludin were present at junctions, where they colocalized with ZO-1 (Figure 1E), and were concentrated at tricellular (where three cells meet) and bicellular (where two cells meet) junction regions, respectively (Furuse et al., 1993; Ikenouchi et al., 2005). Consistent with the evolutionary relationship between marvelD3, tricellulin, and occludin suggested by MARVEL domain alignment and tree reconstruction, EGFP fusion proteins of both marvelD3 splice variants also colocalized with ZO-1 at junctions and were primarily localized to bicellular regions (Figure 1E). In contrast, EGFP-MAL concentrated at apical membranes, whereas proteolipid protein 2 and pantophysin predominantly localized to cytoplasmic vesicles (Figure 1E), consistent with previous studies of each protein (Zacchetti et al., 1995; Haass et al., 1996; Breitwieser et al., 1997). All three members of a single subfamily of MARVEL domain–containing proteins are, therefore, concentrated at the tight junction, whereas representative members of other subfamilies are not. These data indicate that marvelD3, tricellulin, and occludin constitute the complete tight junction–associated MARVEL protein (TAMP) family. Moreover, they support the hypothesis that marvelD3 is a functional component of the tight junction.
TAMPs Are Differentially Expressed at the Tight Junction across a Wide Variety of Epithelial Tissues
Occludin and tricellulin are expressed in a variety of epithelial tissues (Ikenouchi et al., 2005), and in silico analysis of transcript profiles (Figure 1D) suggests that marvelD3 expression may be similar. Semiquantitative RT-PCR was used to assess marvelD3, occludin, and tricellulin mRNA content in Caco-2 monolayers, where all TAMP transcripts were found to be expressed, but at different levels (Figure 2A). In particular, marvelD3 variant 2 message was less abundant than marvelD3 variant 1, tricellulin, and occludin transcripts. TAMP transcripts were also found to be differentially expressed throughout mouse intestine, liver, and kidney (Figure 3A). MarvelD3 variant 1 message was more abundant within mouse liver, marvelD3 splice variant 2 transcript predominated in mouse kidney, and expression of the two forms was comparable in small intestine and colon.
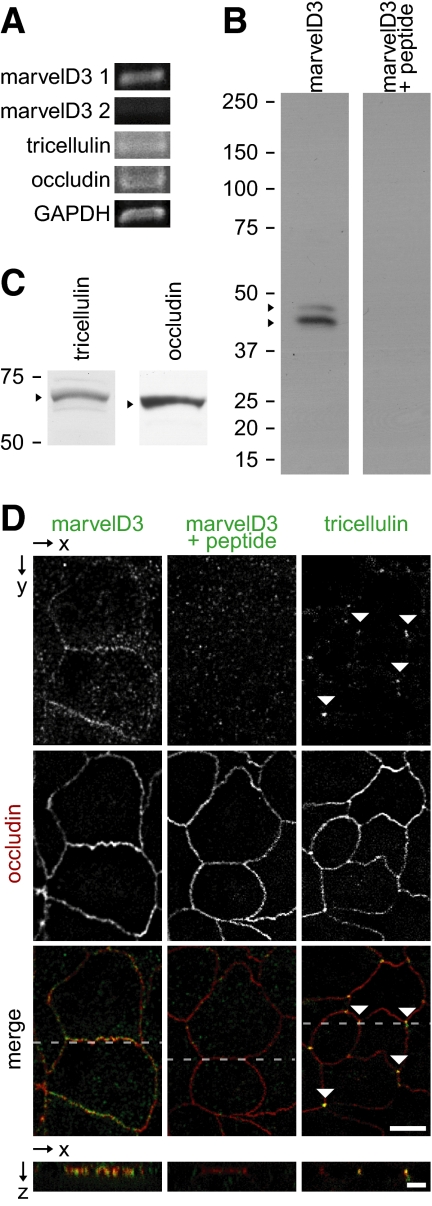
Figure 2.
TAMPs are differentially expressed at intercellular junctions of Caco-2 human intestinal epithelial cell monolayers. (A) Analysis of TAMP transcript expression by RT-PCR reveals differential expression of marvelD3 splice variants, tricellulin, and occludin in Caco-2 cells. (B) MarvelD3 proteins (arrowheads) were detected in Caco-2 cells as determined by SDS-PAGE immunoblot analysis. Preincubation of polyclonal antisera with peptides used as immunogens eliminated the marvelD3 bands. (C) Tricellulin and occludin proteins (arrowheads) were also detected in Caco-2 cells by SDS-PAGE immunoblot. (D) Immunofluorescence of polarized Caco-2 monolayers demonstrates junctional and vesicular localization of all TAMP family members. MarvelD3 (green) staining was abrogated following preincubation of antisera with competing peptides. Tricellulin (green) specifically localized to tricellular junctions (arrowheads). Occludin (red) is shown for orientation in each image. Bar,10 μm. Imaging along the z-axis at the indicated position (dashed line) reveals colocalization of marvelD3 and tricellulin with occludin at apical intercellular junctions. Bar, 5 μm.
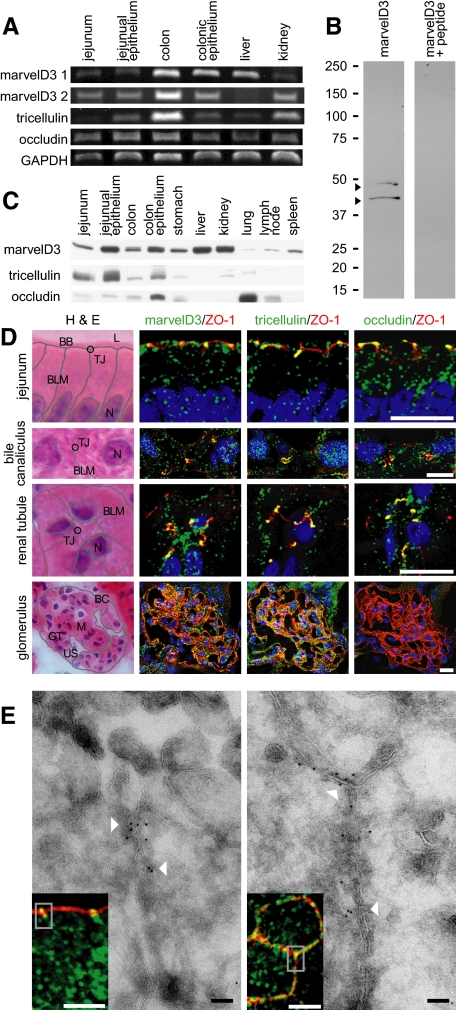
Figure 3.
TAMPs are differentially expressed across epithelial tissues. (A) Analysis of TAMP transcript expression by semiquantitative RT-PCR reveals differential expression of marvelD3 splice variants, tricellulin, and occludin message in epithelial-rich mouse organs. (B) MarvelD3 splice variants (arrowheads) were each detected in mouse jejunal epithelium samples as determined by SDS-PAGE immunoblot analysis. Preincubation of polyclonal antisera with peptides used as immunogens eliminated the marvelD3 bands. (C) Analysis of TAMP expression across an array of mouse organs showed that marvelD3, tricellulin, and occludin proteins are differentially expressed in epithelium-rich tissues. (D) Immunofluorescence shows differential localization of marvelD3, tricellulin, and occludin (green) in vivo. All TAMP family members are concentrated at the apical intercellular junction of jejunal epithelia, hepatocytes, and renal tubular epithelium. In contrast, glomerular epithelial cells are enriched in marvelD3 and tricellulin, but largely lack occludin immunofluorescence. Nuclei (blue), ZO-1 (red), and H&E images (BB, brush border; TJ, tight junction; L, lumen; BLM, basolateral membrane; N, nucleus; BC, Bowman's capsule; M, mesenchyme; GT, glomerular tuft; US, urinary space) are shown for orientation. Bars, 10 μm. (E) Transmission immunoelectron microscopy of mouse jejunum demonstrates specific localization of marvelD3 to the tight junction in sagittal and en face orientations (arrowheads). Immunofluorescence insets with marvelD3 (green) and actin (red) are shown for orientation. Gray boxes approximate electron microscopic fields. White bars, 10 μm; black bars, 100 nm. Data are representative of ≥2 experiments, all with similar results.
Polyclonal antisera against peptides derived from cytoplasmic regions common to both marvelD3 splice variants were developed. SDS-PAGE immunoblot of Caco-2 cell lysates using these antisera demonstrated two bands consistent with the predicted molecular mass of human marvelD3 splice variants 1 and 2 of 46 and 45 kDa, respectively (Figure 2B). The antisera were specific, because preincubation with the peptides used as immunogens eliminated these bands (Figure 2B). Similarly, SDS-PAGE immunoblot analysis of murine jejunal epithelium using marvelD3 polyclonal antisera produced two bands consistent with the predicted molecular mass of mouse marvelD3 splice variants 1 and 2 at 42 and 44 kDa, respectively (Figure 3B). Detection of both bands was eliminated by preincubation of antisera with peptide immunogens (Figure 3B). Along with immunoblots using commercially available antisera to tricellulin and occludin (Figure 2C), these data show that all three TAMPs are expressed in Caco-2 cell monolayers as well as within various murine tissues including jejunal and colonic epithelial cells, stomach, small intestine, colon, liver, kidney, and lungs (Figure 3C). In contrast, expression of marvelD3, tricellulin, and occludin was limited or absent in organs lacking significant epithelial tissue, such as lymph nodes and spleen.
Immunofluorescent microscopy showed that endogenous marvelD3 was concentrated at the apical junctional complex of polarized Caco-2 cell monolayers and was also present within intracellular vesicles (Figure 2D). Like occludin, marvelD3 at the apical junctional complex was predominantly at bicellular regions, whereas tricellulin was concentrated at tricellular regions. The subcellular distributions of marvelD3, tricellulin, and occludin within murine jejunal enterocytes were similar to those in Caco-2 monolayers (Figure 3D), and all three TAMPs were associated with hepatocyte junctional complexes (located at the interface between the apical canalicular and basolateral sinusoidal plasma membrane domains; Figure 3D). MarvelD3, tricellulin, and occludin were also all expressed in renal tubular epithelium, where they concentrated at the apical junctional complex. In contrast, marvelD3 and tricellulin were expressed in glomerular epithelium, but occludin was not (Figure 3D).
Although the immunofluorescence analyses above suggest that marvelD3 localizes to the tight junction, it is possible that these fluorescent signals emanate from other apical junctional complex structures such as the adherens junction. To resolve this, cryo-ultrathin sections of mouse jejunum were analyzed by transmission immunoelectron microscopy. The results (Figure 3E) definitively show that marvelD3 is specifically localized to the tight junction. Overall, these data demonstrate that the TAMP subfamily of MARVEL proteins are associated with the tight junction in a broad range of epithelia, but that, as a reflection of their distinct expression patterns, marvelD3, tricellulin, and occludin may serve unique functions.
TAMPs Demonstrate Unique Interactions with Each Other and ZO-1
The overlapping morphological location of TAMP family members suggests that marvelD3, tricellulin, and occludin may physically associate with one another. To determine whether TAMPs engage in heterotypic interactions, marvelD3, tricellulin, and occludin were immunoprecipitated from Caco-2 lysates. Tricellulin and marvelD3 coimmunoprecipitated with one another, suggesting that interactions between these proteins are common (Figure 4A). In contrast, occludin immunoprecipitates contained marvelD3, but marvelD3 immunoprecipitates did not contain occludin. These data indicate that occludin and marvelD3 interact, but suggest that the fraction of the total marvelD3 pool bound to occludin may be greater than the fraction of the total occludin pool bound to marvelD3. Tricellulin and occludin did not coimmunoprecipitate, suggesting that these TAMP family members do not physically interact or that associations are either unstable or uncommon. These data lend themselves to a model of TAMP interaction at the tight junction, whereby marvelD3 associates with both occludin and tricellulin, whereas the latter TAMPs do not interact directly.
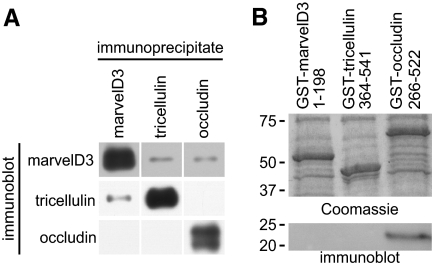
Figure 4.
TAMPs demonstrate unique interactions with each other and ZO-1. (A) Coimmunoprecipitation of TAMPs from Caco-2 lysates reveals interactions among marvelD3, tricellulin, and occludin. The data are consistent with a model of TAMP interaction, where marvelD3 physically associates with both occludin and tricellulin, but the latter two TAMPs do not interact. (B) TAMP cytoplasmic tails were expressed as GST fusion constructs, and equal quantities of each were loaded onto GST-agarose, as confirmed by the Coomassie blue–stained gel. Pulldown experiments using VSVG-ZO-1-GuK as prey demonstrate interaction with the carboxy-terminal tail of occludin (immunoblot). In contrast, neither cytoplasmic tails of tricellulin nor marvelD3 were able to pull down VSVG-ZO-1-GuK. Data are representative of ≥2 experiments, each in duplicate, with similar results.
The cytoplasmic carboxy-terminal tail of occludin is known to interact with the GuK domain of the plaque protein ZO-1 (Furuse et al., 1994). Moreover, the carboxy-terminal tail of tricellulin has been shown to interact with the amino-terminal half of ZO-1 (Riazuddin et al., 2006) which, in addition to the GuK domain, contains three PDZ domains. To investigate the direct ZO-1 binding of TAMP family members, GST-fusion constructs of occludin (266-522) and tricellulin (364-551) carboxy-terminal, and marvelD3 amino-terminal (1-198) tails were expressed as recombinant proteins and were bound to GST-agarose (Figure 4B). These were used to assess binding of a recombinant fusion protein consisting of the VSVG epitope tag (Kreis, 1986) and the ZO-1 GuK domain (VSVG-GuK). Consistent with previous studies (Furuse et al., 1994), VSVG-GuK interacted with the carboxy-terminal domain of occludin (Figure 4B). However, the carboxy terminus of tricellulin was not able to bind VSVG-GuK, suggesting that tricellulin may interact with ZO-1 at another site. Alternatively, minor differences between the tricellulin tail constructs used here and in a previous study (Riazuddin et al., 2006) may explain the disparate results. In contrast to occludin, the GST fusion construct of the marvelD3 cytoplasmic tail failed to associate with VSVG-GuK (Figure 4B). These data highlight important distinctions between the interactions formed by ZO-1, marvelD3, tricellulin, and occludin, and again support a nonredundant model for TAMP function at the tight junction.
TAMPs Partition into Detergent-insoluble Membrane Microdomains and Claudin-based Tight Junction-like Strands
Tight junction proteins are concentrated in cholesterol and sphingolipid rich, detergent-insoluble membrane microdomains (Nusrat et al., 2000). Subcellular fractionation showed that, like occludin, marvelD3 and tricellulin are enriched in these low-density membranes (Figure 5, A and B). Tricellulin distributed across a greater density range than other TAMP family members, consistent with its distribution along lateral membranes at tricellular junction sites (Ikenouchi et al., 2005).
Figure 5.
TAMPs partition into detergent-insoluble membrane microdomains and claudin-based tight junction-like strands. (A) Caco-2 monolayers were solubilized at 4°C in Triton X-100, and lysates were separated on sucrose density gradients. OD600, a measurement of the light scattering property of intact membranes, and protein recovery, are shown as a function of density. (B) Immunoblot analysis demonstrated that TAMPs are enriched in the light-scattering fraction of detergent-resistant membranes. (C) EGFP-TAMP constructs (green) were transfected into L929 human fibroblast cells alone or with mRFP1-claudin-1 (red). Monolayers were grown on glass coverslips, and nuclei (blue) were detected by staining with Hoechst 33342. EGFP-marvelD3 splice variants 1 and 2, EGFP-tricellulin, and EGFP-occludin localize to intracellular vesicles when expressed independently, but are recruited to tight junction-like strands (yellow) along areas of membrane overlap (arrowheads) in cells expressing claudin-1. Images are representative of ≥3 experiments, all with similar results. Bar, 10 μm.
Like occludin, members of the claudin tight junction protein family are also concentrated into detergent-insoluble membrane microdomains (Shen et al., 2006). When expressed in fibroblasts, claudin proteins are able to form tight junction-like strands (Furuse et al., 1998; Sasaki et al., 2003). In contrast, occludin and tricellulin are recruited to claudin-based strands, but are unable to form these structures in the absence of claudin family members (Furuse et al., 1998; Ikenouchi et al., 2008). To determine whether marvelD3 can form tight junction-like strands, EGFP-marvelD3 was expressed in L929 fibroblasts cells in the presence and absence of mRFP1-claudin-1. As previously shown, claudin-1 was able to form strands (Furuse et al., 1998). However, neither marvelD3 splice variant formed intercellular strands when expressed individually. Likewise, occludin and tricellulin also failed to form intercellular strands. However, all three TAMPs, including both marvelD3 splice variants, were incorporated into mRFP1–claudin-1-based, tight junction-like strands (Figure 5C). These data suggest that, much like other members of the TAMP family, marvelD3 is a component of tight junction strands but is unable to form stands independently.
Epithelial Barrier Development Coincides with TAMP Expression and Trafficking to the Tight Junction
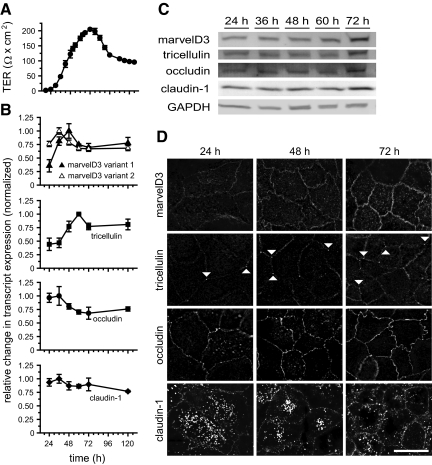
To assess TAMP expression and trafficking during tight junction assembly, mRNA content, and protein expression and localization were monitored along with TER (Figure 6A). qRT-PCR analysis (Figure 6B) showed that marvelD3 splice variant 1 and tricellulin transcription increased significantly ahead of TER development (2.9 ± 0.50- and 2.3 ± 0.03-fold increases, respectively; p < 0.05 for each). In contrast, marvelD3 splice variant 2 mRNA, which was poorly expressed in mature Caco-2 monolayers (Figure 2A), was not significantly up-regulated during barrier development (1.2 ± 0.20-fold increase; p > 0.2). Occludin and claudin-1 transcript levels also remained relatively constant during tight junction assembly (1.1 ± 0.03- and 1.0 ± 0.06-fold increases, respectively; p > 0.4 for each). Tricellulin and marvelD3 transcription was accompanied by increased protein expression, which coincided with development of peak TER 72 h after plating (Figure 6C). Consistent with qRT-PCR data, claudin-1 protein content was stable throughout monolayer maturation. In contrast, occludin protein expression increased during tight junction assembly despite constant mRNA content. These data suggest that marvelD3 and tricellulin protein expression is primarily regulated by transcriptional mechanisms during tight junction assembly, whereas expression of occludin is controlled by modifications to translation or turnover rate.
Figure 6.
Epithelial barrier development coincides with TAMP expression and trafficking to the tight junction. (A) TER was monitored during barrier development in Caco-2 monolayers. Cells were collected for qRT-PCR, immunofluorescence, and SDS-PAGE immunoblot analyses at the indicated times. (B) qRT-PCR data show that tricellulin and marvelD3 splice variant 1 mRNA synthesis is induced during TER development. (C) SDS-PAGE immunoblot shows that TAMP protein expression increases during barrier development. (D) Immunofluorescence of Caco-2 monolayers demonstrates that marvelD3, tricellulin, occludin, and claudin-1 traffic to the tight junction during TER development. Matched exposures are shown for each protein. Arrowheads indicate tricellular regions. Bar, 20 μm. Data are representative of ≥3 experiments, each in duplicate, all with similar results.
TAMPs exist in multiple subcellular locations, including tight junction, lateral membrane, and vesicular pools. Consistent with previous studies of tricellulin and occludin (Ikenouchi et al., 2008), these proteins, as well as marvelD3, are trafficked to the tight junction in concert with TER development (Figure 6D). MarvelD3 and tricellulin accumulated at the tight junction over time, and this correlated with increased protein expression. As has been noted previously (Ikenouchi et al., 2008), tricellulin was present at bicellular as well as tricellular tight junction regions of immature monolayers (compare with Figure 2D). In contrast, occludin and claudin-1 trafficking to the tight junction during barrier development coincided with reduced intracellular pools (Figure 6D). Together with the qRT-PCR and immunoblot data, these results show that occludin and claudin-1 are redistributed from cytoplasmic stores to the tight junction during barrier development. In contrast, marvelD3 and tricellulin are delivered to the tight junction immediately after synthesis. Moreover, the correlation of increased TER with progressive enrichment of each TAMP family member at the tight junction is consistent with the hypothesis that marvelD3, like occludin, tricellulin, and claudin-1, participates in the development and maintenance of epithelial barrier function.
TAMP Knockdown Delays Tight Junction Assembly
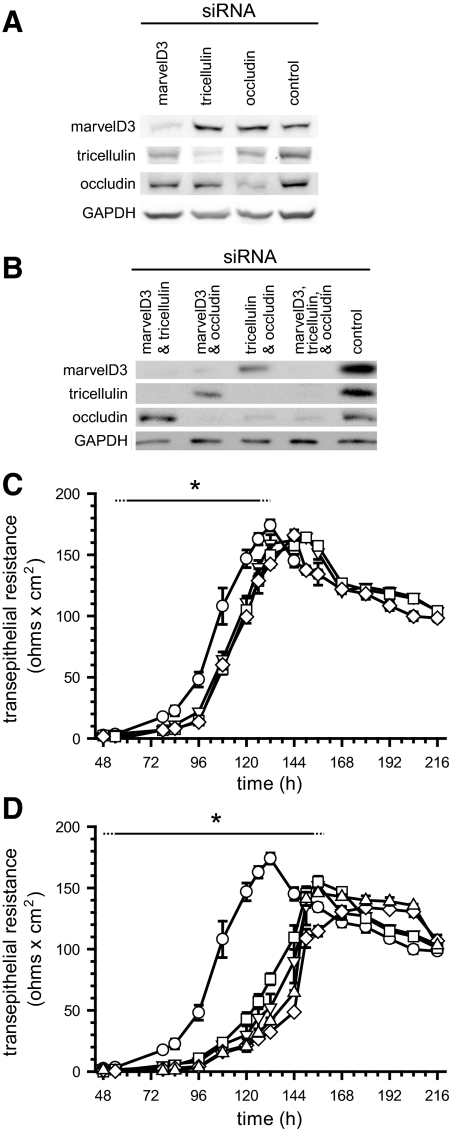
To directly assess the role of marvelD3 in tight junction assembly and barrier development, Caco-2 cells were transfected with TAMP-targeting or control siRNAs, and TER was measured at regular intervals. siRNA transfection reduced TAMP protein expression by more than 60% at 60 h after plating (Figure 7, A and B). Suppression of individual TAMPs produced modest delays in TER development, averaging 18 ± 4% (p < 0.01), relative to monolayers treated with control siRNA (Figure 7C). Similar findings have been reported for tricellulin and occludin knockdown (Ikenouchi et al., 2005; Yu et al., 2005), but this is the first demonstration of a functional role for marvelD3. Interestingly, the TER delay incurred by knockdown of any one TAMP was indistinguishable from that caused by knockdown of any other. TAMP knockdown did not induce compensatory upregulation of remaining family members. This suggests that although similar, the roles of TAMPs at the tight junction are unique. Consistent with nonredundant function, simultaneous knockdown of multiple TAMPs caused greater delays in barrier development than knockdown of any single TAMP (Figure 7D; 53 ± 12% delay; p < 0.01 relative to monolayers transfected with control siRNA; p < 0.05 relative to monolayers transfected with siRNA against a single TAMP). However, the delays for combined knockdown of any two (or all three) TAMPs were similar. Taken as a whole, these data suggest that TAMPs make both overlapping and unique contributions to tight junction assembly and barrier function.
Figure 7.
TAMP knockdown delays tight junction assembly. Caco-2 cells were treated with either control or TAMP-targeted siRNAs and plated on Transwell supports. (A and B) SDS-PAGE immunoblot analysis revealed that TAMP-targeted siRNAs specifically knocked down expression of only the targeted TAMP and that treatment was not associated with compensatory upregulation of other family members. (C) Knockdown (KD) of single TAMPs (marvelD3 KD, ▿; tricellulin KD, □; occludin KD, ◇) significantly (*p < 0.01) delayed barrier development relative to control monolayers (○). (D) Simultaneous knockdown of multiple TAMPs (control, ○; marvelD3 and tricellulin KD, ▿; marvelD3 and occludin KD, □; tricellulin and occludin KD, ◇; marvelD3, tricellulin and occludin KD, ▵) produced greater delays than loss of any one TAMP (*p < 0.05). Solid line: significant TER delay for all knockdown conditions; dashed line: significant TER delay for at least one knockdown condition. Data are representative of ≥5 experiments, each in quadruplicate, all with similar results.
TAMPs Have Unique Dynamic Behaviors at Bicellular and Tricellular Tight Junction Regions
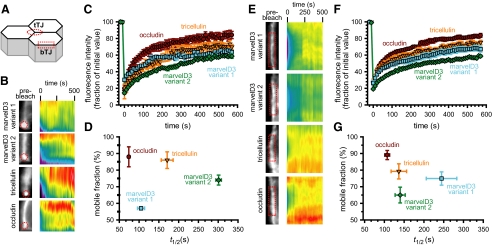
The inability of TAMPs to compensate for loss of other subfamily members during barrier development, as well as their differential tissue expression, discrete patterns of synthesis during tight junction assembly, distinct subcellular distributions, and selective interactions all suggest that members of this MARVEL subfamily may have unique biophysical behaviors. Our previous analyses established that the tight junction is subject to continuous dynamic remodeling at steady state and that occludin is highly mobile within the tight junction and lateral membrane (Shen et al., 2008). However, the dynamic behaviors of neither marvelD3 nor tricellulin have been studied. To investigate the mobility of TAMP family members, Caco-2 epithelial cells expressing EGFP-fusion proteins were grown as polarized monolayers and protein dynamics were assessed by FRAP at bicellular and tricellular tight junction regions (Figure 8A). TAMP mobility profiles and times of half-maximal recovery at bicellular and tricellular tight junction regions spanned a broad spectrum (Table 1). Dynamic behavior of marvelD3 splice variants was a function of position within the tight junction; marvelD3 variant 1 was more stable at tricellular regions (Figure 8, B–D) whereas variant 2 was more stable at bicellular regions (Figure 8, E–G). In contrast, tricellulin and occludin mobilities were similar at bicellular and tricellular regions. The distinct dynamic behaviors of marvelD3 splice variants at bicellular and tricellular regions suggest distinct roles at each site. Moreover, the unique behaviors of each TAMP lend further support to the hypothesis that these proteins have distinct functions at the tight junction.
Figure 8.
TAMPs have unique dynamic behaviors at bicellular and tricellular tight junction regions. (A) Caco-2 cells were transfected with EGFP-TAMP fusion proteins, and protein dynamics at bicellular (bTJ) and tricellular (tTJ) tight junction regions were assessed by FRAP. TAMPs exhibit distinct and unique dynamic behaviors at tricellular (B–D) and bicellular (E–G) areas of the tight junction. Occludin and tricellulin are highly mobile at both regions. In contrast, mobility of both marvelD3 splice variants varied between bicellular and tricellular tight junction regions (see Table 1). Data are representative of ≥3 experiments, each in triplicate, all with similar results.
Table 1.
The dynamic behavior of TAMP proteins
| TAMP | Tight junctionregion | Mobilefraction (%) | t1/2 (s) |
|---|---|---|---|
| MarvelD3 splice variant 1 | Bicellular | 75 ± 4*† | 245 ± 39*† |
| Tricellular | 57 ± 1*‡ | 105 ± 9*‡ | |
| MarvelD3 splice variant 2 | Bicellular | 61 ± 3*† | 149 ± 13*† |
| Tricellular | 74 ± 3*‡ | 301 ± 7*‡ | |
| Tricellulin | Bicellular | 79 ± 5 | 137 ± 20 |
| Tricellular | 86 ± 5 | 171 ± 15 | |
| Occludin | Bicellular | 89 ± 3 | 107 ± 6* |
| Tricellular | 88 ± 6 | 74 ± 3* |
* Data are significantly different (p < 0.05) between bicellular and tricellular regions.
† Data are significantly different (p < 0.05) between marvelD3 splice variants at bicellular regions.
‡ Data are significantly different (p < 0.05) between marvelD3 splice variants at tricellular regions.
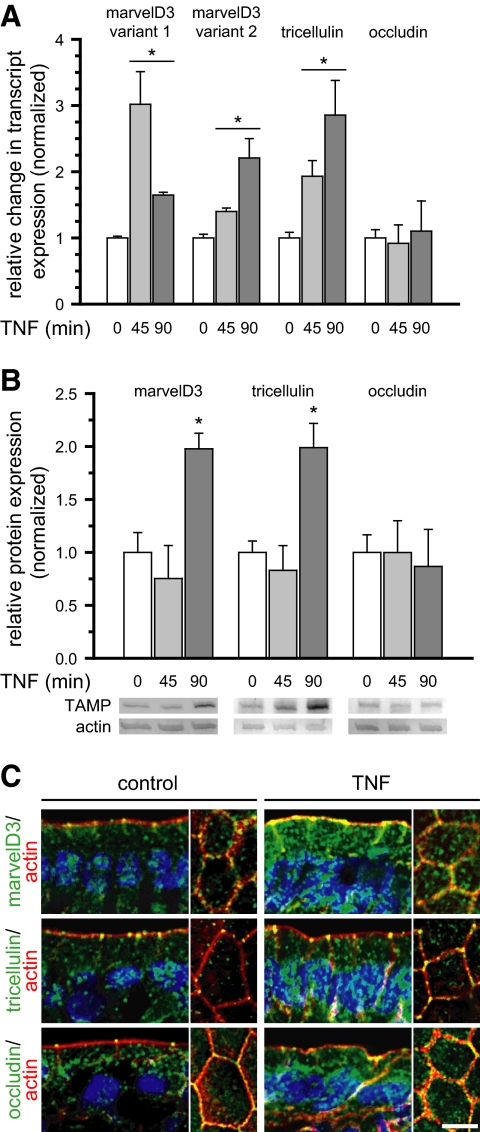
TAMPs Are Differentially Expressed and Redistributed in Response to In Vivo Tumor Necrosis Factor Treatment
We have previously shown that in vivo tumor necrosis factor (TNF)-induced diarrhea requires myosin light-chain kinase activation, tight junction reorganization, and increased paracellular permeability that is accompanied by occludin internalization (Clayburgh et al., 2005, 2006). To assess the in vivo regulation of marvelD3 and tricellulin, murine jejunal epithelial cells and intact jejunum were isolated at intervals after intraperitoneal TNF injection. MarvelD3 and tricellulin transcripts increased rapidly within enterocytes after TNF administration (p < 0.01; Figure 9A). In contrast, occludin mRNA remained constant (p > 0.40). Consistent with this, expression of both marvelD3 and tricellulin proteins increased in response to in vivo TNF treatment (p < 0.05), whereas occludin protein content did not change (p > 0.37; Figure 9B). These data suggest that increases in marvelD3 and tricellulin transcription within 45 min of TNF exposure lead to the increased translation of these TAMP family members detected after 90 min. Immunofluorescence of mouse jejunum demonstrated differential redistribution of these related tight junction members after TNF treatment. Although occludin was redistributed into cytoplasmic vesicles (Figure 9C), increased marvelD3 protein was detected at the tight junction, apical and lateral membranes, and within cytoplasmic vesicles. Similarly, tricellulin was enriched at both tricellular and bicellular tight junctions, as well as within the cytoplasm, after TNF treatment (Figure 9C). The similarity between in vivo marvelD3 and tricellulin synthesis and trafficking and that which occurred during in vitro barrier development suggests that these responses may reflect an effort to stabilize the tight junction and compensate for occludin removal. However, the fact that barrier loss still occurs (Clayburgh et al., 2005), as well as the distinct responses of each TAMP, suggest that marvelD3, tricellulin, and occludin serve unique functions in the epithelial response to proinflammatory stimuli.
Figure 9.
TAMPs are differentially expressed and redistributed in response to in vivo TNF treatment. Mice were sacrificed at the indicated times after intraperitoneal injection of 5 μg of TNF. (A) Epithelial cells were isolated, and RNA was harvested for qRT-PCR analysis. MarvelD3 and tricellulin message increased 45 min after TNF treatment and remained elevated after 90 min (*p < 0.01), whereas occludin message content was not changed (p > 0.40). (B) SDS-PAGE immunoblot and densitometric analyses of jejunal epithelial lysates demonstrate increased marvelD3 and tricellulin protein after 90 min of TNF exposure (*p < 0.05). (C) Jejunum was frozen 120 min after TNF injection and immunostained for TAMPs (green), F-actin (red), and nuclei (blue). MarvelD3 and tricellulin protein expression increased and was enriched at the tight junction, lateral membrane, and cytoplasm. Bar, 5 μm. Data are representative of ≥3 experiments, all with similar results.
DISCUSSION
As the first identified transmembrane component at the tight junction, occludin was initially thought to be essential to structure and function (Furuse et al., 1993). However, studies since that time have demonstrated that occludin is not required for tight junction barrier function (Saitou et al., 1998, 2000; Yu et al., 2005). Moreover, although occludin knockdown does reduce barrier function in vitro, it does not alter ion selectivity (Yu et al., 2005). In contrast, claudin proteins, which assemble into tight junction-like strands, are critical determinants of barrier function and paracellular ion selectivity (Simon et al., 1999; Sonoda et al., 1999; Furuse et al., 2001, 2002; Van Itallie et al., 2001; Hamazaki et al., 2002; Colegio et al., 2003; Umeda et al., 2006; Van Itallie et al., 2008).
Despite the central role of the claudin protein family, in vivo analyses suggest that TAMP family members do play critical roles in some organs. For example, occludin knockout mice suffer from growth retardation and a variety of other defects (Saitou et al., 2000). In addition, although tricellulin knockout mice have not been described, mutations in human patients with isolated nonsyndromic hearing loss suggest a similar view of tricellulin function (Riazuddin et al., 2006; Chishti et al., 2008). Furthermore, substantial in vitro data suggest that both occludin and tricellulin are important to tight junction function (Balda et al., 1996; Chen et al., 1997; Wong and Gumbiner, 1997; Ikenouchi et al., 2005; Yu et al., 2005). Based on these observations and the presence of MARVEL domains in both occludin and tricellulin (Sanchez-Pulido et al., 2002), we hypothesized that additional TAMPs might compensate for occludin or tricellulin loss. We began with bioinformatic analyses of human MARVEL domain sequences and then developed novel tools to study these proteins. The data demonstrate that a previously uncharacterized protein, marvelD3, is a component of the tight junction and that marvelD3, occludin, and tricellulin constitute the complete TAMP subfamily of MARVEL proteins. However, the functional analyses and in vivo regulation of these proteins suggest that marvelD3, occludin, and tricellulin are not able to complement one another functionally. This is consistent with the significant structural differences outside of the MARVEL domains of each TAMP family member. Perhaps most notable is the presence of ELL domains in the cytoplasmic carboxy-terminal tails of occludin and tricellulin, but not within the short carboxy-terminus or longer amino-terminal cytoplasmic tails of either marvelD3 splice variant.
Previous unpublished and published analyses have speculated both in favor (http://www.pdg.cnb.uam.es/MARVEL/tree.gif) and against (Riazuddin et al., 2006) the existence of an evolutionary relationship between marvelD3 and tricellulin and occludin. However, these studies were based on analyses that excluded the third transmembrane-spanning sequence of the MARVEL domain itself and used evolutionary trees that lacked posterior values at branch points. The utility of previous analyses is also limited because the approaches used were not optimized for proteins with highly divergent sequences, as seen in the MARVEL family. After narrowing the search space to include only human MARVEL domain–containing proteins, we constructed a phylogeny of complete MARVEL domains using an algorithm suitable for alignment of sequences with large, unalignable regions. The results suggest the existence of four subfamilies, one of which is composed of marvelD3, tricellulin, and occludin. Although our study has focused on the tight junction–associated MARVEL protein subfamily, it will be of interest for future studies to determine if functional themes characterize the other three MARVEL subfamilies defined here.
Our studies using antisera and EGFP-tagged proteins in murine tissues and a human intestinal epithelial cell line show that TAMP proteins concentrate at the tight junction and within cytoplasmic vesicles. Together with the relationships indicated by the evolutionary analysis, these data suggest that marvelD3, tricellulin, and occludin may fulfill similar roles in epithelial cells. This hypothesis is supported by several findings beyond our initial observation. First, TAMPs are predominantly expressed in epithelium-rich organs. Second, members of this protein subfamily interact physically (Figure 10), are concentrated in cholesterol-rich membrane microdomains of epithelial cells, and are incorporated into claudin-based tight junction-like strands between fibroblasts. Finally, the delays in development of barrier function after knockdown of any one TAMP are similar. Moreover, although more pronounced, the delays caused by knockdown of different combinations of TAMPs are comparable. It should be noted that the effect of tricellulin and occludin siRNA knockdown in the present study is different from previously published reports, which showed decreases in both baseline and peak TER (Ikenouchi et al., 2005; Yu et al., 2005; Krug et al., 2009). There are several possible explanations for the differences between our study using transient knockdown in human intestinal epithelial (Caco-2) cells and the previous studies that used stable knockdowns generated in mouse mammary (Eph4) and Madin-Darby canine kidney cells. First, cell tissue of origin may be critical. Such a hypothesis is consistent with the absence of gut pathology or intestinal epithelial barrier defects in human subjects with tricellulin mutations or in occludin knockout mice (Saitou et al., 2000; Schulzke et al., 2005). Alternatively, although our transient knockdown efficiency was more than 60% and significant knockdown was maintained for at least 6 d, we cannot exclude the possibility that residual TAMP expression or waning of the knockdown effect contributed to the barrier development that occurred in our studies. Nevertheless, the delay in barrier development demonstrates that, like the other TAMPs, marvelD3 is an important, but not essential, component of the tight junction.
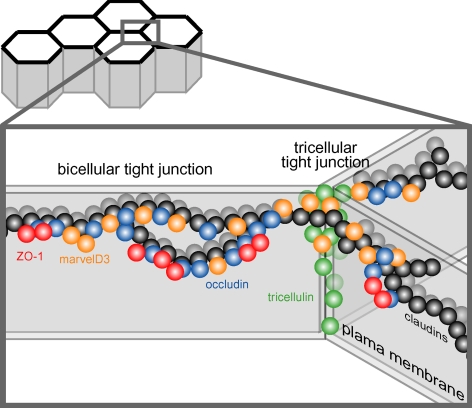
Figure 10.
Model of TAMP locations and interactions at the tight junction. The figure shows the organization of proteins within a small area of a confluent monolayer (see image at the top). The bicellular tight junction is the interface between two cells, whereas the vertex where three cells meet is termed the tricellular tight junction. The tight junction strands within both bicellular and tricellular regions are composed of claudins (black spheres). All three TAMP proteins, marvelD3 (orange spheres), tricellulin (green spheres), and occludin (blue spheres) incorporate into claudin-based tight junction strands. Occludin and tricellulin are primarily found at bicellular and tricellular regions, respectively, whereas marvelD3 is present at both sites. This may explain why marvelD3 binds to both tricellulin and occludin, but tricellulin and occludin do not interact with one another. Tricellulin is unique in that it is present at the tight junction and along the lateral membrane. Only occludin interacts with the ZO-1 (red spheres) via the GuK domain. Thus, although there are similarities, each TAMP can also be defined by unique characteristics.
The data presented here also suggest that marvelD3, tricellulin, and occludin make distinct contributions to tight junction structure and function. First, there are subtle differences between tissue distribution and subcellular localization. Moreover, deficits in tight junction assembly after knockdown of individual TAMPs cannot be compensated for by other family members. This observation might be expected for tricellulin, which has a unique affinity for tricellular tight junctions and the lateral membranes beneath these regions and, perhaps as a reflection of this localization, distributes into detergent insoluble membrane fractions of higher density than do marvelD3 and occludin. However, the data also indicate differences between the roles of marvelD3 and occludin. For example, coimmunoprecipitation studies show that marvelD3 interacts with both tricellulin and occludin, but that the latter two TAMPs do not associate. This suggests that marvelD3 is present at both bicellular and tricellular tight junction regions and may play a broader organizational role than other TAMP family members. Such a role would also be consistent with the increased stability of marvelD3 at the tight junction, relative to tricellulin and occludin; the distinct behaviors of marvelD3 splice variants at bicellular versus tricellular tight junction regions; and, most strikingly, the unique responses of each TAMP to in vivo TNF exposure. As a whole, these data suggest that TAMP family members are functionally nonredundant and may be involved in distinct aspects of tight junction assembly, maintenance, or regulation.
In conclusion, these data demonstrate that marvelD3, tricellulin, and occludin define the TAMP family of tight junction proteins and that these proteins have overlapping, but nonredundant, expression, subcellular localization, and function. The results suggest that, as is true for claudin proteins, the role of TAMPs in paracellular barrier function is best considered in the context of a complete protein family. Finally, the identification of both the TAMP family and marvelD3 provide a new framework with which to understand the contributions of occludin and tricellulin to tight junction biology as well as tissue-specific pathologies that follow mutation or loss of individual TAMPs.
ACKNOWLEDGMENTS
We thank Lindsay Finger for support and encouragement; Anthony Chang, W. Vallen Graham, and Sam C. Nalle for generously sharing advice; and Sachiko Tsukita for facilitating the immunoelectron microscopic studies. This work was supported by the National Institutes of Health Grants R01DK61931, R01DK68271, P01DK67887, T32HL007237, and T32GM07281 and the University of Chicago Cancer Center Grant P30CA14599.
Abbreviations used:
- MARVEL
MAL and related proteins for vesicle trafficking and membrane link
- TAMP
tight junction–associated MARVEL protein
- TER
transepithelial resistance.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-08-0734) on February 17, 2010.
REFERENCES
- Abascal F., Zardoya R., Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Abramoff M. D., Magelhaes P. J., Ram S. J. Image processing with Image. J. Biophoton. Int. 2004;11:36–42. [Google Scholar]
- Balda M. S., Whitney J. A., Flores C., Gonzalez S., Cereijido M., Matter K. Functional dissociation of paracellular permeability and transepithelial electrical resistance and disruption of the apical-basolateral intramembrane diffusion barrier by expression of a mutant tight junction membrane protein. J. Cell Biol. 1996;134:1031–1049. doi: 10.1083/jcb.134.4.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T., Troup D. B., Wilhite S. E., Ledoux P., Rudnev D., Evangelista C., Kim I. F., Soboleva A., Tomashevsky M., Edgar R. NCBI GEO: mining tens of millions of expression profiles—database and tools update. Nucleic Acids Res. 2007;35:D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitwieser G. E., McLenithan J. C., Cortese J. F., Shields J. M., Oliva M. M., Majewski J. L., Machamer C. E., Yang V. W. Colonic epithelium-enriched protein A4 is a proteolipid that exhibits ion channel characteristics. Am. J. Physiol. 1997;272:C957–C965. doi: 10.1152/ajpcell.1997.272.3.C957. [DOI] [PubMed] [Google Scholar]
- Campbell R. E., Tour O., Palmer A. E., Steinbach P. A., Baird G. S., Zacharias D. A., Tsien R. Y. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Merzdorf C., Paul D. L., Goodenough D. A. COOH terminus of occludin is required for tight junction barrier function in early Xenopus embryos. J. Cell Biol. 1997;138:891–899. doi: 10.1083/jcb.138.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chishti M. S., Bhatti A., Tamim S., Lee K., McDonald M. L., Leal S. M., Ahmad W. Splice-site mutations in the TRIC gene underlie autosomal recessive nonsyndromic hearing impairment in Pakistani families. J. Hum. Genet. 2008;53:101–105. doi: 10.1007/s10038-007-0209-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claude P., Goodenough D. A. Fracture faces of zonulae occludentes from “tight” and “leaky” epithelia. J. Cell Biol. 1973;58:390–400. doi: 10.1083/jcb.58.2.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayburgh D. R., Barrett T. A., Tang Y., Meddings J. B., Van Eldik L. J., Watterson D. M., Clarke L. L., Mrsny R. J., Turner J. R. Epithelial myosin light chain kinase-dependent barrier dysfunction mediates T cell activation-induced diarrhea in vivo. J. Clin. Invest. 2005;115:2702–2715. doi: 10.1172/JCI24970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayburgh D. R., Musch M. W., Leitges M., Fu Y. X., Turner J. R. Coordinated epithelial NHE3 inhibition and barrier dysfunction are required for TNF-mediated diarrhea in vivo. J. Clin. Invest. 2006;116:2682–2694. doi: 10.1172/JCI29218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colegio O. R., Van Itallie C., Rahner C., Anderson J. M. Claudin extracellular domains determine paracellular charge selectivity and resistance but not tight junction fibril architecture. Am. J. Physiol. Cell Physiol. 2003;284:C1346–C1354. doi: 10.1152/ajpcell.00547.2002. [DOI] [PubMed] [Google Scholar]
- Dai M., et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farquhar M., Palade G. Junctional complexes in various epithelia. J. Cell Biol. 1963;17:375–412. doi: 10.1083/jcb.17.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M., Furuse K., Sasaki H., Tsukita S. Conversion of zonulae occludentes from tight to leaky strand type by introducing claudin-2 into Madin-Darby canine kidney I cells. J. Cell Biol. 2001;153:263–272. doi: 10.1083/jcb.153.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M., Hata M., Furuse K., Yoshida Y., Haratake A., Sugitani Y., Noda T., Kubo A., Tsukita S. Claudin-based tight junctions are crucial for the mammalian epidermal barrier: a lesson from claudin-1-deficient mice. J. Cell Biol. 2002;156:1099–1111. doi: 10.1083/jcb.200110122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M., Hirase T., Itoh M., Nagafuchi A., Yonemura S., Tsukita S. Occludin: a novel integral membrane protein localizing at tight junctions. J. Cell Biol. 1993;123:1777–1788. doi: 10.1083/jcb.123.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M., Itoh M., Hirase T., Nagafuchi A., Yonemura S., Tsukita S. Direct association of occludin with ZO-1 and its possible involvement in the localization of occludin at tight junctions. J. Cell Biol. 1994;127:1617–1626. doi: 10.1083/jcb.127.6.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M., Sasaki H., Fujimoto K., Tsukita S. A single gene product, claudin-1 or -2, reconstitutes tight junction strands and recruits occludin in fibroblasts. J. Cell Biol. 1998;143:391–401. doi: 10.1083/jcb.143.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman R. C., et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J. M., Lim S. O., Park Y. M., Jung G. A novel splice variant of occludin deleted in exon 9 and its role in cell apoptosis and invasion. FEBS J. 2008;275:3145–3156. doi: 10.1111/j.1742-4658.2008.06467.x. [DOI] [PubMed] [Google Scholar]
- Guindon S., Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Haass N. K., Kartenbeck M. A., Leube R. E. Pantophysin is a ubiquitously expressed synaptophysin homologue and defines constitutive transport vesicles. J. Cell Biol. 1996;134:731–746. doi: 10.1083/jcb.134.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamazaki Y., Itoh M., Sasaki H., Furuse M., Tsukita S. Multi-PDZ domain protein 1 (MUPP1) is concentrated at tight junctions through its possible interaction with claudin-1 and junctional adhesion molecule. J. Biol. Chem. 2002;277:455–461. doi: 10.1074/jbc.M109005200. [DOI] [PubMed] [Google Scholar]
- Hu Z., Wang Y., Graham W. V., Su L., Musch M. W., Turner J. R. MAPKAPK-2 is a critical signaling intermediate in NHE3 activation following Na+-glucose cotransport. J. Biol. Chem. 2006;281:24247–24253. doi: 10.1074/jbc.M602898200. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck J. P., Ronquist F., Nielsen R., Bollback J. P. Bayesian inference of phylogeny and its impact on evolutionary biology. Science. 2001;294:2310–2314. doi: 10.1126/science.1065889. [DOI] [PubMed] [Google Scholar]
- Ikenouchi J., Furuse M., Furuse K., Sasaki H., Tsukita S. Tricellulin constitutes a novel barrier at tricellular contacts of epithelial cells. J. Cell Biol. 2005;171:939–945. doi: 10.1083/jcb.200510043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikenouchi J., Sasaki H., Tsukita S., Furuse M. Loss of occludin affects tricellular localization of tricellulin. Mol. Biol. Cell. 2008;19:4687–4693. doi: 10.1091/mbc.E08-05-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M., Furuse M., Morita K., Kubota K., Saitou M., Tsukita S. Direct binding of three tight junction-associated MAGUKs, ZO-1, ZO-2, and ZO-3, with the COOH termini of claudins. J. Cell Biol. 1999;147:1351–1363. doi: 10.1083/jcb.147.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D. T. Improving the accuracy of transmembrane protein topology prediction using evolutionary information. Bioinformatics. 2007;23:538–544. doi: 10.1093/bioinformatics/btl677. [DOI] [PubMed] [Google Scholar]
- Kall L., Krogh A., Sonnhammer E. L. An HMM posterior decoder for sequence feature prediction that includes homology information. Bioinformatics. 2005;21(Suppl 1):i251–i257. doi: 10.1093/bioinformatics/bti1014. [DOI] [PubMed] [Google Scholar]
- Katoh K., Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- Kreis T. E. Microinjected antibodies against the cytoplasmic domain of vesicular stomatitis virus glycoprotein block its transport to the cell surface. EMBO J. 1986;5:931–941. doi: 10.1002/j.1460-2075.1986.tb04306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug S. M., Amasheh S., Richter J. F., Milatz S., Gunzel D., Westphal J. K., Huber O., Schulzke J. D., Fromm M. Tricellulin forms a barrier to macromolecules in tricellular tight junctions without affecting ion permeability. Mol. Biol. Cell. 2009;20:3713–3724. doi: 10.1091/mbc.E09-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn R. M., et al. The UCSC Genome Browser Database: update 2009. Nucleic Acids Res. 2009;37:D755–D761. doi: 10.1093/nar/gkn875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massoumi R., Sjolander A. Leukotriene D(4) affects localisation of vinculin in intestinal epithelial cells via distinct tyrosine kinase and protein kinase C controlled events. J. Cell Sci. 2001;114:1925–1934. doi: 10.1242/jcs.114.10.1925. [DOI] [PubMed] [Google Scholar]
- McCarthy K. M., Skare I. B., Stankewich M. C., Furuse M., Tsukita S., Rogers R. A., Lynch R. D., Schneeberger E. E. Occludin is a functional component of the tight junction. J. Cell Sci. 1996;109:2287–2298. doi: 10.1242/jcs.109.9.2287. [DOI] [PubMed] [Google Scholar]
- Muresan Z., Paul D. L., Goodenough D. A. Occludin 1B, a variant of the tight junction protein occludin. Mol. Biol. Cell. 2000;11:627–634. doi: 10.1091/mbc.11.2.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusrat A., Parkos C. A., Verkade P., Foley C. S., Liang T. W., Innis-Whitehouse W., Eastburn K. K., Madara J. L. Tight junctions are membrane microdomains. J. Cell Sci. 2000;113:1771–1781. doi: 10.1242/jcs.113.10.1771. [DOI] [PubMed] [Google Scholar]
- Riazuddin S., et al. Tricellulin is a tight-junction protein necessary for hearing. Am. J. Hum. Genet. 2006;79:1040–1051. doi: 10.1086/510022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou M., Fujimoto K., Doi Y., Itoh M., Fujimoto T., Furuse M., Takano H., Noda T., Tsukita S. Occludin-deficient embryonic stem cells can differentiate into polarized epithelial cells bearing tight junctions. J. Cell Biol. 1998;141:397–408. doi: 10.1083/jcb.141.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou M., Furuse M., Sasaki H., Schulzke J. D., Fromm M., Takano H., Noda T., Tsukita S. Complex phenotype of mice lacking occludin, a component of tight junction strands. Mol. Biol. Cell. 2000;11:4131–4142. doi: 10.1091/mbc.11.12.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Pulido L., Martin-Belmonte F., Valencia A., Alonso M. A. MARVEL: a conserved domain involved in membrane apposition events. Trends Biochem. Sci. 2002;27:599–601. doi: 10.1016/s0968-0004(02)02229-6. [DOI] [PubMed] [Google Scholar]
- Sasaki H., Matsui C., Furuse K., Mimori-Kiyosue Y., Furuse M., Tsukita S. Dynamic behavior of paired claudin strands within apposing plasma membranes. Proc. Natl. Acad. Sci. USA. 2003;100:3971–3976. doi: 10.1073/pnas.0630649100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulzke J. D., Gitter A. H., Mankertz J., Spiegel S., Seidler U., Amasheh S., Saitou M., Tsukita S., Fromm M. Epithelial transport and barrier function in occludin-deficient mice. Biochim. Biophys. Acta. 2005;1669:34–42. doi: 10.1016/j.bbamem.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Schwarz B. T., Wang F., Shen L., Clayburgh D. R., Su L., Wang Y., Fu Y. X., Turner J. R. LIGHT signals directly to intestinal epithelia to cause barrier dysfunction via cytoskeletal and endocytic mechanisms. Gastroenterology. 2007;132:2383–2394. doi: 10.1053/j.gastro.2007.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L., Black E. D., Witkowski E. D., Lencer W. I., Guerriero V., Schneeberger E. E., Turner J. R. Myosin light chain phosphorylation regulates barrier function by remodeling tight junction structure. J. Cell Sci. 2006;119:2095–2106. doi: 10.1242/jcs.02915. [DOI] [PubMed] [Google Scholar]
- Shen L., Turner J. R. Actin depolymerization disrupts tight junctions via caveolae-mediated endocytosis. Mol. Biol. Cell. 2005;16:3919–3936. doi: 10.1091/mbc.E04-12-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L., Weber C. R., Turner J. R. The tight junction protein complex undergoes rapid and continuous molecular remodeling at steady state. J. Cell Biol. 2008;181:683–695. doi: 10.1083/jcb.200711165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon D. B., et al. Paracellin-1, a renal tight junction protein required for paracellular Mg2+ resorption. Science. 1999;285:103–106. doi: 10.1126/science.285.5424.103. [DOI] [PubMed] [Google Scholar]
- Sonnhammer E. L., von Heijne G., Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- Sonoda N., Furuse M., Sasaki H., Yonemura S., Katahira J., Horiguchi Y., Tsukita S. Clostridium perfringens enterotoxin fragment removes specific claudins from tight junction strands. Evidence for direct involvement of claudins in tight junction barrier. J. Cell Biol. 1999;147:195–204. doi: 10.1083/jcb.147.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L., Shen L., Clayburgh D. R., Nalle S. C., Sullivan E. A., Meddings J. B., Abraham C., Turner J. R. Targeted epithelial tight junction dysfunction causes immune activation and contributes to development of experimental colitis. Gastroenterology. 2009;136:551–563. doi: 10.1053/j.gastro.2008.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuyasu K. T. Immunochemistry on ultrathin frozen sections. Histochem. J. 1980;12:381–403. doi: 10.1007/BF01011956. [DOI] [PubMed] [Google Scholar]
- Turner J. R., Rill B. K., Carlson S. L., Carnes D., Kerner R., Mrsny R. J., Madara J. L. Physiological regulation of epithelial tight junctions is associated with myosin light-chain phosphorylation. Am. J. Physiol. 1997;273:C1378–C1385. doi: 10.1152/ajpcell.1997.273.4.C1378. [DOI] [PubMed] [Google Scholar]
- Umeda K., Ikenouchi J., Katahira-Tayama S., Furuse K., Sasaki H., Nakayama M., Matsui T., Tsukita S., Furuse M. ZO-1 and ZO-2 independently determine where claudins are polymerized in tight-junction strand formation. Cell. 2006;126:741–754. doi: 10.1016/j.cell.2006.06.043. [DOI] [PubMed] [Google Scholar]
- Utech M., Ivanov A. I., Samarin S. N., Bruewer M., Turner J. R., Mrsny R. J., Parkos C. A., Nusrat A. Mechanism of IFN-gamma-induced endocytosis of tight junction proteins: myosin II-dependent vacuolarization of the apical plasma membrane. Mol. Biol. Cell. 2005;16:5040–5052. doi: 10.1091/mbc.E05-03-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Itallie C., Rahner C., Anderson J. M. Regulated expression of claudin-4 decreases paracellular conductance through a selective decrease in sodium permeability. J. Clin. Invest. 2001;107:1319–1327. doi: 10.1172/JCI12464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Itallie C. M., Holmes J., Bridges A., Gookin J. L., Coccaro M. R., Proctor W., Colegio O. R., Anderson J. M. The density of small tight junction pores varies among cell types and is increased by expression of claudin-2. J. Cell Sci. 2008;121:298–305. doi: 10.1242/jcs.021485. [DOI] [PubMed] [Google Scholar]
- Wong V., Gumbiner B. M. A synthetic peptide corresponding to the extracellular domain of occludin perturbs the tight junction permeability barrier. J. Cell Biol. 1997;136:399–409. doi: 10.1083/jcb.136.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Irizarry R. A. Preprocessing of oligonucleotide array data. Nat. Biotechnol. 2004;22:656–658. doi: 10.1038/nbt0604-656b. [author reply, 658] [DOI] [PubMed] [Google Scholar]
- Yguerabide J., Schmidt J. A., Yguerabide E. E. Lateral mobility in membranes as detected by fluorescence recovery after photobleaching. Biophys. J. 1982;40:69–75. doi: 10.1016/S0006-3495(82)84459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A. S., McCarthy K. M., Francis S. A., McCormack J. M., Lai J., Rogers R. A., Lynch R. D., Schneeberger E. E. Knockdown of occludin expression leads to diverse phenotypic alterations in epithelial cells. Am. J. Physiol. Cell Physiol. 2005;288:C1231–C1241. doi: 10.1152/ajpcell.00581.2004. [DOI] [PubMed] [Google Scholar]
- Zacchetti D., Peranen J., Murata M., Fiedler K., Simons K. VIP17/MAL, a proteolipid in apical transport vesicles. FEBS Lett. 1995;377:465–469. doi: 10.1016/0014-5793(95)01396-2. [DOI] [PubMed] [Google Scholar]