Gfer links mitochondrial dynamics with pluripotency in mouse embryonic stem cells through its modulation of the mitochondrial fission GTPase Drp1.
Abstract
The relationship of mitochondrial dynamics and function to pluripotency are rather poorly understood aspects of stem cell biology. Here we show that growth factor erv1-like (Gfer) is involved in preserving mouse embryonic stem cell (ESC) mitochondrial morphology and function. Knockdown (KD) of Gfer in ESCs leads to decreased pluripotency marker expression, embryoid body (EB) formation, cell survival, and loss of mitochondrial function. Mitochondria in Gfer-KD ESCs undergo excessive fragmentation and mitophagy, whereas those in ESCs overexpressing Gfer appear elongated. Levels of the mitochondrial fission GTPase dynamin-related protein 1 (Drp1) are highly elevated in Gfer-KD ESCs and decreased in Gfer-overexpressing cells. Treatment with a specific inhibitor of Drp1 rescues mitochondrial function and apoptosis, whereas expression of Drp1-dominant negative resulted in the restoration of pluripotency marker expression in Gfer-KD ESCs. Altogether, our data reveal a novel prosurvival role for Gfer in maintaining mitochondrial fission–fusion dynamics in pluripotent ESCs.
INTRODUCTION
Pluripotent embryonic stem cells (ESCs) are remarkable in their ability to undergo unlimited proliferation and self-renewal. Consequently, the capacity to proliferate indefinitely in culture in an undifferentiated state confer ESCs with considerable therapeutic potential, especially in the treatment of spinal cord injury and of degenerative diseases such as type 1 diabetes and Parkinson's and Alzheimer's diseases. To preserve their pluripotency, ESCs in prolonged culture conditions must be protected from genomic, epigenetic, oxidative, or mitochondrial damage (Zeng and Rao, 2007). There are significant gaps in our knowledge regarding genes that execute the fundamental task of maintaining ESC stability and consequentially preserving their pluripotency in culture. A comprehensive understanding of the mechanisms by which ESCs maintain their pluripotency is therefore crucial to realize their full potential in therapeutic regenerative medicine.
Growth factor erv1-like (Gfer), or augmenter of liver regeneration, is the mammalian homologue of the evolutionarily conserved yeast erv1 protein, an FAD-dependent sulfhydryl oxidase predominantly located in the intermembrane space (IMS) of mitochondria (Lisowsky et al., 2001). In addition to mitochondrial IMS, Gfer is also present in the cytoplasm and nucleus, and a nonmitochondrial function is associated with its role in spermatogenesis (Klissenbauer et al., 2002). Although its precise function is unknown, Gfer plays roles in cytosolic Fe-S cluster assembly and mitochondrial biogenesis (Polimeno et al., 2000; Lange et al., 2001; Gatzidou et al., 2006). In yeast, deletion of erv1 is lethal with the mutants displaying aberrant mitochondrial morphology and defective biogenesis of cytoplasmic Fe-S clusters (Becher et al., 1999; Lange et al., 2001; Lisowsky et al., 2001). Moreover, Gfer is an important component of a disulfide redox relay system that mediates the import of proteins to the IMS of the mitochondria (Mesecke et al., 2005).
Gfer was identified as one of 216 (Ramalho-Santos et al., 2002) or 283 (Ivanova et al., 2002) common genes that are enriched in ESCs, neuronal and hematopoietic stem cells (SCs). Gfer maps within the t-haplotype region (Silver, 1993) of the SC gene-enriched mouse chromosome 17 (a region syntenic with the proximal end of the short arm of human chromosome 16; Polimeno et al., 1999), suggesting an evolutionary clustering of genes important in SC biology. Gfer has roles in the regeneration of mammalian liver, pancreas and Drosophila leg imaginal discs, and its expression correlates with early spermatogenesis (Klissenbauer et al., 2002; Klebes et al., 2005; Gatzidou et al., 2006).
As no role has been established for Gfer in any stem cell, we used gene depletion and overexpression approaches to evaluate its role in the most primitive of SCs, the pluripotent ESC. Here we show that down-regulation of Gfer in mouse ESCs results in significantly reduced pluripotency marker gene expression, EB formation, and cell survival. Depletion of Gfer also resulted in loss of ESC mitochondrial membrane potential (ΔΨm), fragmentation of mitochondria, and autophagy of the damaged mitochondria (mitophagy). Conversely, mitochondria in ESCs overexpressing Gfer appeared significantly elongated, with well-defined cristae. Interestingly, levels of the mitochondrial fission GTPase Drp1 were highly elevated in Gfer-KD cells and decreased in Gfer-overexpressing cells, indicating that the enhanced Drp1 levels in Gfer-KD ESCs may be responsible for increased mitochondrial fragmentation, loss of ΔΨm, and apoptosis. Consistent with this idea, treatment with mdivi-1, a specific small molecule inhibitor of Drp1 (Cassidy-Stone et al., 2008) or ectopic expression of a dominant negative K38A mutant of Drp1 (Drp1DN) that inhibits GTP binding (Smirnova et al., 2001) rescued mitochondrial dysfunction, apoptosis, and pluripotency in Gfer-KD ESCs. A selective role for Gfer in SCs was corroborated by our observation that depletion of Gfer from differentiated cells such as primary mouse embryonic fibroblasts (MEFs) did not affect mitochondrial morphology, function, or cell survival. Thus, during homeostasis, Gfer modulates the levels of Drp1 to preserve mouse ESC mitochondrial morphology and function and maintain pluripotency marker expression in these primitive cells.
MATERIALS AND METHODS
Cell Culture and Reagents
Mitomycin C and rotenone were purchased from Sigma (St. Louis, MO). Primary C57BL/6 (B6) MEFs were from ATCC (Manassas, VA) and NIH 3T3 cells were a kind gift from Dr. Geoffrey Clark (James Graham Brown Cancer Center). Primary MEFs were cultured on gelatin (0.1% gelatin in water; Millipore, Billerica, MA)–coated dishes, in knockout DMEM (Invitrogen, Carlsbad, CA) containing 15% ES-screened fetal bovine serum (FBS; HyClone, Logan, UT), 0.1 mM 2-mercaptoethanol (βME), 0.1 mM nonessential amino acids (NEAA; all from Millipore), 2 mM l-glutamine (HyClone), and 50 U/ml penicillin/streptomycin (P/S; Invitrogen). For the production of ESC feeder cells, MEFs were treated with 10 μg/ml mitomycin C, in media for 3 h at 37°C. NIH 3T3 cells were grown in DMEM with 10% FCS (Invitrogen), 2 mM l-glutamine, and 50 U/ml P/S. For culture of ESCs, B6 ESCs were plated on feeder cells on gelatin-coated plates, in the presence of 15% ES-FBS, 0.1 mM βME, 2 mM l-glutamine, 0.1 mM NEAA, 50 U/ml P/S, and 1000 U/ml leukemia inhibitory factor (LIF)-ESGROW (Millipore; ESC media). For passaging, ESCs were harvested by adding 0.25% trypsin-EDTA or TrypLE Express (Invitrogen). Residual feeder cells were panned by plating the entire cell suspension on regular tissue culture–coated dishes (not gelatin-coated) for 20 min at 37°C. The floating undifferentiated ESCs were harvested and replated on fresh gelatin-coated dishes, in the presence of feeder cells and ESC media, at required densities (0.8 × 106/ml for normal passage). For rotenone treatment, rotenone was dissolved in dimethyl sulfoxide (DMSO; Sigma) to make 1 mM stock. ESCs were treated with DMSO or rotenone at 10 μM final concentration for 24 h. The cells were then harvested after repeated washing with 1× phosphate-buffered saline (PBS). For culture of EBs, approximately 250,000 ESCs were seeded per well of fibroblast-free Ultra-Low-Attachment six-well dishes (Corning) containing ESC medium without LIF.
Virus Infection of ESCs
FG12-Lenti-green fluorescent protein (GFP)–short hairpin RNA (shRNA) viruses were produced as described (Qin et al., 2003) using 293T cells. Gfer shRNA sequences are as follows: Gfer1: 5′ GGAACAGCTTCCTTAGCGTT 3′ and Gfer2: 5′ CCAGGTGCCTCGTACCCTTCA 3′. Nanog shRNA was previously described (Wang et al., 2007). ESCs were infected with lentiviruses at a multiplicity of infection (MOI) of 5:1 with 4 μg/ml Polybrene (Sigma). GFP+ cells were sorted out 72 h after infection, using a FACSAria (BD Biosciences, San Jose, CA). Full-length mouse Gfer cDNA was purchased from Open Biosystems (Huntsville, AL) and cloned into the murine stem cell virus (MSCV)-internal ribosome entry site (IRES)-GFP system, and viruses were produced as described (Kitsos et al., 2005). Freshly passaged ESCs were infected with MSCV-IRES-GFP (control) or MSCV-IRES-GFP-Gfer (MSCV-Gfer) viruses at a MOI of 5:1 with 4 μg/ml Polybrene (Sigma). GFP+ cells were sorted out 72 h after infection as mentioned before. For ectopic expression of Drp1DN, the Flag-tagged DRP1 K38A pcDNA3.1 expression plasmid was used (a generous gift from Dr. Sally Kornbluth, Department of Pharmacology and Cancer Biology, Duke University Medical Center, Durham, NC). Briefly, GFP+ lentivirus infected ESCs were sorted out, transfected with Drp1DN expression plasmid using the mouse ES Cell Nucleofector Kit (Lonza, Walkersville, MD) using the A23 program and immediately were plated on feeder cells that had been preplated on gelatin-coated eight-well Lab-Tek Chamber Slides (Nalgene Nunc, Rochester, NY).
ESC Proliferation Assay
Ten thousand ES cells were plated per well of a gelatin-coated 24-well dish in the presence of feeder cells and ESC media. The cell numbers and viability (using trypan blue) were monitored for several passages using a Vi-CELL (Beckman Coulter, Fullerton, CA). For the GFP-Competition Assay, approximately 7500 Lenti-GFP-shRNA virus–infected GFP+ ESCs were mixed with 2500 GFP-WT ESCs cells (3:1 ratio) and plated in the presence of feeder cells and ESC media, in gelatin-coated wells. Self-renewal was measured for five passages by analyzing for GFP expression.
Immunoassays
Alkaline Phosphatase.
ES cells were seeded at 5000 cells/well on gelatin-coated 16-well Lab-Tek Chamber Slides (Nalgene Nunc). Alkaline phosphatase activity was measured using a kit (Chemicon International, Temecula, CA). For immunocytochemistry of ES cells, the following primary antibodies were used: Nanog (rabbit polyclonal; Abcam, Cambridge, MA), Oct4 (mouse monoclonal; BD Transduction, Franklin Lakes, NJ), SSEA3 (mouse monoclonal; Millipore), and Gfer (11293–1-AP, rabbit polyclonal; PTG Labs, Chicago, IL). Cy-3 secondary antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA) were used, and cells were mounted using VectaShield Mounting Media with DAPI (Vector Laboratories, Burlingame, CA). Images were captured using the Axio Observer Z1 microscope with ApoTome assembly using a 63× oil-immersion objective (Carl Zeiss, Thornwood, NY). The signal intensities were calculated using the AxioVision 2.0 software. Primary antibodies used for immunoblotting were β-actin (clone AC-15, Sigma), Bax (p-19; Santa Cruz Biotechnology, Santa Cruz, CA), caspase-3 (AB1899, Millipore), Gfer (PTG Labs), Beclin-1 (clone 20, BD Transduction); rabbit polyclonal Atg12 (Cell Signaling Technology, Beverly, MA), Lc3B (Cell Signaling), Drp1 (clone 8/DLP-1, BD Transduction, or H-300 rabbit polyclonal, Santa Cruz), OPA-1 (rabbit polyclonal, Abcam), Mfn-1 (chicken polyclonal, Novus Biologicals, Littleton, CO), and Mfn-2 (M6319, Sigma).
Digital Imaging of Mitochondria.
The mammalian expression vector pDsRed2-Mito (Clontech Laboratories, Mountain View, CA) was introduced into ESCs using the mouse ES cell nucleofector kit, and the cells were immediately plated on feeder cells that were plated on gelatin-coated eight-well Lab-Tek chamber slides. Digital optical sectioning was performed 24 h after nucleofection using Axio Observer Z1 inverted fluorescence microscope with ApoTome assembly using a 100× oil-immersion objective (Plan-Neofluar; Carl Zeiss).
Mdivi-1 Treatment.
Mdivi-1 (Enzo Life Sciences, Farmingdale, NY) was dissolved in DMSO at a concentration of 25 mg/ml. Reconstituted mdivi-1 was added directly to the cell culture medium at a final concentration of 25 μM, and cells were cultured for 24 h.
Flow Cytometry Assays
Apoptosis Assay.
Staining for annexin V/7-AAD was performed using the annexin V-PE apoptosis detection kit I (BD Biosciences). Samples were analyzed using BD FACSAria (BD Biosciences). Analysis of tetramethylrhodamine ethyl ester (TMRE) fluorescence was conducted as follows: cells were loaded with 50 nM TMRE (Molecular Probes, Invitrogen, Eugene, OR) in growth media, for 20 min under normal growth conditions. Cells were then harvested and analyzed for PE fluorescence. The cytochrome c staining assay was performed as described previously (Waterhouse and Trapani, 2003). Briefly, harvested cells were permeabilized with digitonin (Sigma; 50 μg/ml digitonin in PBS with 100 mM KCl) for 5 min on ice, followed by fixation in 4% paraformaldehyde. Cells were incubated overnight at 4°C with 1:200 anti-cytochrome c mAb (BD PharMingen) in blocking buffer (3% BSA, 0.05% Saponin (Sigma) in PBS) followed by incubation with a PE-labeled secondary antibody. The PE-Active caspase-3 Apoptosis kit (BD PharMingen) was used to measure active caspase-3 levels.
Electron Microscopy.
Cell pellets containing 300,000 cells were washed briefly with a 0.1 M sodium cacodylate buffer and then fixed in 3% glutaraldehyde in 0.1 M sodium cacodylate overnight at 4°C. Cells were then postfixed in 1% osmium tetroxide in 0.1 M sodium cacodylate for 1 h. Cells were then briefly rinsed with 0.1 M sodium cacodylate and then dehydrated using a series of graded alcohols. Cell pellets were then embedded in LX-112 epoxy resin (Ladd Research Industries, Burlington, VT), and ∼8-μm sections were cut on an LKB microtome (LKB Instruments, Gaithersburg, MD). Sections were then stained using uradyl acetate and lead citrate before viewing on a CM12 electron microscope equipped with a digital camera (Phillips Electronic Instruments, Mahwah, NJ).
Real-Time RT-PCR Analysis.
Total RNA was prepared using RNAqueous kits (Ambion, Austin, TX). cDNA was prepared using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). Quantitative (q) real-time RT-PCR was performed on the IQ5 I-Cycler (Bio-Rad Laboratories, Hercules CA) using IQ SYBR Green supermix (Bio-Rad). Nanog and Oct4 primers were previously listed (Pasini et al., 2007). Other primer sequences listed in Supplementary Table 1.
Statistical Analysis
Student's t test was used to assess statistical significance, and p < 0.05 was deemed significant.
RESULTS
Knockdown of Gfer in Murine ESCs Results in Diminished Expression of Pluripotency Markers and Impaired Embryoid Body Formation
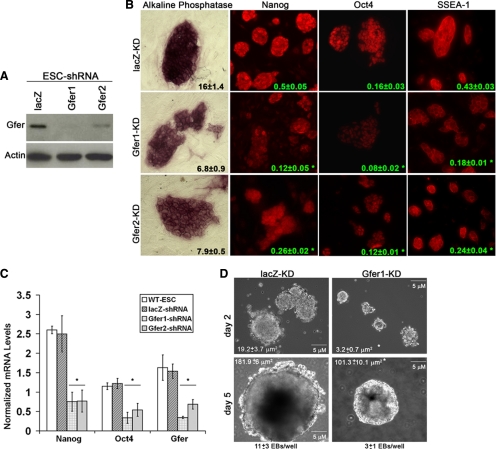
To evaluate the biological function of Gfer in ESCs, we down-regulated its expression in wild-type (WT) ESCs using the FG12-lentivirus-GFP based shRNA system, where the shRNA expression is driven by a human U6 RNA polymerase-III promoter and GFP expression by a modified ubiquitin-C promoter (Qin et al., 2003). We used two different shRNA constructs: Gfer1 shRNA targeting exon 2 and Gfer2 shRNA constructs targeting the 3′ untranslated region (UTR) of Gfer mRNA. We achieved ∼90 and 60% KD of the Gfer protein and a twofold decrease in Gfer mRNA, respectively, using Gfer1 and Gfer2 shRNAs (Figure 1, A and C). We first evaluated colony morphology and alkaline phosphatase activity in ESCs cultured in conditions favoring self-renewal, that is, in the presence of feeder cells and LIF. The control ESC colonies (lacZ-KD, nonspecific shRNA; Qin et al., 2003) were rounded and compact, with well-defined boundaries, and were stained intensely for alkaline phosphatase activity (Figure 1B). In contrast, the decrease of Gfer using either Gfer1 or Gfer2 shRNA resulted in more flattened colonies with reduced (twofold) alkaline phosphatase activity in cells within each colony (Figure 1B). Next, we evaluated by immunocytochemistry the expression of three markers of murine ESC pluripotency, the transcription factors Nanog and Oct-4 and the cell surface protein stage specific embryonic antigen 1 (SSEA1). As shown in Figure 1B, the expression of each of the three proteins was decreased by several fold (p < 0.05) in ESCs with decreased Gfer expression compared with control ESCs. Additionally, Nanog mRNA was reduced threefold and Oct-4 mRNA twofold in Gfer1/2-KD ESCs compared with lacZ-KD ESCs (Figure 1C). These results indicate that Gfer plays an important role in maintaining ESCs in an undifferentiated state.
Figure 1.
A role for Gfer in the maintenance of ESC pluripotency. (A) Representative immunoblots depicting protein levels of Gfer and Actin in Lac-Z, Gfer1, and Gfer2 KD ESCs (n = 3). (B) Digital optical section ApoTome images (magnification, 200×) of day 6 lacZ, Gfer1, and Gfer2 KD ESCs depicting alkaline phosphatase activity and levels of Nanog, Oct-4, and SSEA1 (all in red). Numbers in black and green represent average signal intensities ± SD quantified from at least 30 colonies (n = 3), *p = 0.001. (C) Average mRNA levels of Nanog, Oct-4, and Gfer in day 6 WT, WT-lacZ-KD, Gfer1-KD, and Gfer2-KD ESCs. The mRNA levels measured by qRT-PCR were normalized to actin mRNA (n = 3), *p = 0.01. (D) Embryoid bodies formed by WT-lacZ-KD and Gfer1-KD ESCs cultured in the absence of feeder cells and LIF in the media for 2 or 5 d. Magnification, ×100; scale bar, 5 μm. Average surface area in μm2 ± SD and average numbers of day 5 EBs per well ± SD from each genotype are shown (n = 3); *p = 0.005.
Abnormal morphology and reduced expression of pluripotency markers in Gfer-KD ESCs could be due to premature differentiation. However, KD of Gfer in ESCs resulted in the formation of significantly fewer and smaller EBs (Figure 1D). Two days after culture in the absence of feeder cells and LIF, Gfer1-KD ESCs formed EBs that were sixfold smaller in surface area than lacZ-KD EBs (Figure 1D). Even by day 5 in culture, Gfer-KD EBs were significantly smaller in surface area (100 ± 10 μm2) compared with day 5 lacZ EBs (182 ± 6 μm2). Additionally, Gfer KD ESCs yielded threefold less EBs than lacZ-KD ESCs (Figure 1D). We also analyzed the expression levels of markers of differentiation into the three germ layers, viz., neuroectoderm, endoderm and mesoderm, in day 6 Gfer1/2-KD and LacZ-KD EBs. Diminished Gfer in the EBs resulted in significantly reduced marker gene expression characteristic of all three germ layers (Supplementary Figure 1). These results suggest that reduction in Gfer causes an overall decrease in ESC function.
Down-Regulation of Gfer in Mouse ESCs Results in Reduced Proliferation and Enhanced Apoptosis
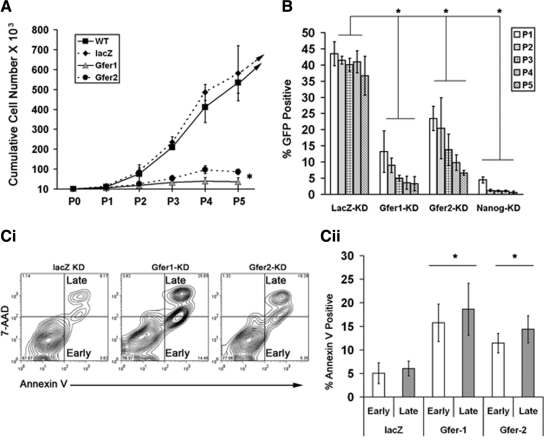
Next, we evaluated whether depletion of Gfer affects ESC proliferation. ESCs down-regulated for Gfer expression showed significantly diminished ability to proliferate in vitro, such that the cumulative cell yield at passage 4 of Gfer-KD ESCs was fourfold lower than that of WT controls (Figure 2A), indicating that Gfer KD ESCs lose their capacity to proliferate in culture. To assess whether down-regulation of Gfer also affects the capacity of ESCs to self-renew (Burdon et al., 2002), we mixed GFP-positive (GFP+) shRNA-infected ESCs with uninfected GFP-negative (GFP−) WT ESCs in a 3:1 ratio and evaluated the colonogenic capacity of the GFP+ stem cells by monitoring the GFP+/GFP− ratio for five passages (Ivanova et al., 2006). As down-regulation of Nanog has previously been shown to compromise ESC self-renewal (Ivanova et al., 2006; Wang et al., 2007), we included Nanog-KD ESCs as a positive control (Figure 2B). Our results confirmed that Nanog dramatically compromised the colonogenic ability of ESCs (Figure 2B). Further, KD of Gfer1 resulted in a significant 3.5-fold reduction in the number of GFP+ ESCs by the first passage and in a eightfold reduction by passage 3, suggesting that the presence of Gfer is important for ESC self-renewal (Figure 2B). Gfer2-KD also caused a reduction in ESC self-renewal such that there was a twofold reduction by passage 1 and fourfold reduction by passage 3, in the number of GFP+ ESCs. These data suggest that diminished pluripotency due to reduced Gfer expression significantly impairs the ability of ESCs to proliferate and self-renew in culture.
Figure 2.
Down-regulation of Gfer diminishes proliferation and cell survival in WT ESCs. (A) Average cumulative cell yield from n = 3, when WT-lacZ-KD, Gfer1-KD, and Gfer2-KD ESCs were cultured for seven passages (data from five passages are shown); *p = 0.001. (B) Self-renewal by lacZ, Gfer1, Gfer2, and Nanog KD ESCs, initially mixed (P0) at 3:1 ratio with WT ESCs, measured as percentage (%) of GFP-positive cells at all five passages by flow cytometry. Average percentage of GFP-positive ESCs from three experiments is shown; *p = 0.003. (C) Representative histogram (i) and graphs (ii) showing average (n = 4) percentage of apoptotic cells, measured by annexin V/7-AAD reactivity in WT-lacZ-KD, Gfer1-KD, and Gfer2-KD ESCs, at 72 h after cell sorting for lenti-GFP virus expression. Error bars, SD; *p = 0.0002.
Inability of ESCs to proliferate in culture could be indicative of a cell survival defect associated with depletion of Gfer. To investigate whether Gfer depletion results in loss of cell viability, we analyzed lacZ-KD and Gfer1/2-KD ESCs for annexin V/7-amino actinomycin D (7AAD) reactivity by single cell flow cytometry at 72 h after depleting Gfer (Figure 2Ci). There were three times more early (annexin V+/7AAD−) and late (annexin V+/7AAD+) apoptotic cells in the Gfer1/2-KD ESC population, compared with control lacZ-KD ESCs (Figure 2Cii), indicating that depletion of Gfer from ESCs results in a significant loss of cell viability through apoptosis.
Gfer KD Results in Loss of ESC Mitochondrial Function
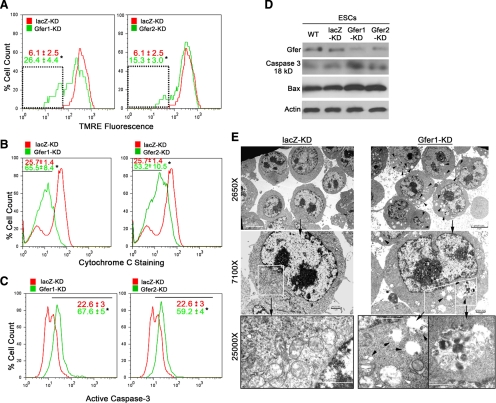
Permeabilization of the mitochondrial outer membrane and loss of ΔΨm are central events in the initiation of apoptosis. A major site of localization of Gfer is the IMS of the mitochondria (Klissenbauer et al., 2002). Additionally, Gfer is important for mitochondrial biogenesis and transport of proteins into mitochondrial IMS. Hence we hypothesized that decreased Gfer expression would result in permeabilization of the mitochondrial outer membrane, resulting in a loss of ΔΨm. To investigate this, we loaded WT, lacZ-KD, and Gfer1/2-KD ESCs with the red fluorescent ΔΨm indicator, TMRE, and analyzed fluorescence in single cells by flow cytometry. Intact mitochondria retain TMRE, whereas membrane depolarization and a loss of ΔΨm result in diminished fluorescence (Akao et al., 2003). We did not observe any difference in TMRE fluorescence intensities between WT and lacZ KD ESCs at 72 h in culture (Supplementary Figure 2). However, KD of Gfer from the ESCs resulted in a significant, 3-4-fold loss of TMRE fluorescence (Figure 3A), consistent with a loss of ESC ΔΨm. These results suggest that Gfer is required for the maintenance of ESC mitochondrial outer membrane integrity.
Figure 3.
KD of Gfer initiates mitochondrial dysfunction triggering mitophagy in ESCs. (A) Representative histograms depicting TMRE fluorescence (PE channel) in lacZ and Gfer1/2 KD ESCs. Numbers in red (lacZ-KD) and green (Gfer1/2-KD) are average (n = 3) percentage (%) of cells with TMRE fluorescence intensities within the gated region (dotted box) ± SD; *p = 0.0001. Mitochondrial membrane depolarization was assessed by loss of TMRE retention/fluorescence. (B) Histographic representation of the assay measuring the release of cytochrome c in the indicated ESC genotypes. Red (lacZ-KD) and green (Gfer1/2-KD) numbers are average (n = 3) percentage of cells within the gated region (black line) ± SD; *p = 0.001, measuring loss of PE-conjugated anti-cytochrome c antibody staining. (C) Percentage of cells showing positive staining (gated with a black line) for a PE-conjugated active caspase-3 antibody. Results represent average (n = 3) percentage of cells showing positive staining in the PE channel ± SD, *p = 0.01, for lacZ (red) and Gfer1/2-KD (green). (D) Representative immunoblot (n = 3) analyses of active caspase-3 and Bax in indicated ESC genotype. Gfer and actin levels are also shown. (E) Digital TEM images depicting ultrastructural details in lacZ (left) and Gfer-1 KD (right) ESCs. Scale bars, 5 μm at ×2650 and 1 μm at ×7100 and ×25,000 magnifications. Black arrowheads, representative autophagosomes; white arrowheads, degenerating mitochondria.
Loss of ΔΨm is usually accompanied by mitochondrial permeability transition (MPT) and the release of cytochrome c from mitochondria into the cytoplasm (Liu et al., 1996). We used a previously described flow cytometry–based quantitative assay to determine whether down-regulation of Gfer results in the release of cytochrome c (Waterhouse and Trapani, 2003). This assay utilizes plasma membrane permeabilization to enable cytosolic cytochrome c to diffuse out of the cells before fixation and intracellular staining with cytochrome c antibody. Thus, apoptotic cells will have less fluorescence than cells with intact mitochondria. In fact, KD of Gfer caused a significant (2–2.5-fold) increase in the percentage of ESCs showing a loss of cytochrome c staining, compared with lacZ-KD ESCs (Figure 3B). These data indicate enhanced cytochrome c release from the mitochondria after KD of Gfer in ESCs.
Cytoplasmic cytochrome c, in concert with Apaf1 and ATP, forms the apoptosome complex that activates the effector caspases-3, -6, and -7 (Wang, 2001). To confirm that enhanced cytochrome c release from the mitochondria after Gfer-KD in the ESCs also resulted in elevated caspase activity, we performed intracellular staining of fixed lacZ-KD and Gfer1/2-KD ESCs with a fluorescence-conjugated antibody that specifically recognizes the activated form of caspase-3. KD of Gfer from ESCs resulted in significantly elevated levels (threefold) of activated caspase-3, compared with lacZ-KD ESCs (Figure 3C). Immunoblots from WT, lacZ-KD, and Gfer1/2-KD ESCs also show higher activated caspase-3 (18 kDa) levels after KD of Gfer (Figure 3D). Further, as the Bcl-2/Bax family of apoptosis-related proteins have been implicated in mitochondria-associated cell survival and/or death decisions (Levine et al., 2008); we assessed the levels of Bcl-2, Bax, and Bad in WT, lacZ and Gfer1/2-KD ESCs and observed that the levels of Bax were higher in Gfer1/2-KD cells compared with WT and lacZ-KD ESCs (Figure 3D). Collectively, these data indicate that down-regulation of Gfer results in the failure of ESC mitochondria characterized by loss of ΔΨm and cytochrome c release, which triggers caspase-mediated cell-intrinsic apoptosis.
Gfer Depletion Triggers Mitochondrial Autophagy or Mitophagy in Mouse ESCs
The deletion of Gfer in yeast is lethal and is preceded by aberrant mitochondrial morphology (Becher et al., 1999). Because Gfer down-regulation results in a loss of ΔΨm and apoptosis in ESCs, we used transmission electron microscopy (TEM) to determine if mitochondria in Gfer-KD ESCs showed abnormal morphology that could be indicative of their failure. Previous ultrastructural studies have reported that undifferentiated WT human and mouse ESCs possess relatively few, round or oval, electron-translucent mitochondria with a perinuclear arrangement (Sathananthan et al., 2002; Baharvand and Matthaei, 2003). Mouse ESC mitochondria contain immature cristae that appear enlarged and translucent, giving the intracristal spaces a swollen appearance (Baharvand and Matthaei, 2003). Loss of “stemness” and onset of differentiation is characterized by cells possessing numerous, larger mitochondria with increasing morphological complexity (distinct cristae and denser matrix) that are scattered throughout the cytoplasm and by increased ATP production (Lonergan et al., 2007; Nesti et al., 2007).
Consistent with previous studies (Baharvand and Matthaei, 2003), our lacZ-KD (Figure 3E) and WT (Supplementary Figure 4B) ESCs possessed relatively few, round-to-oval, electron-translucent mitochondria with poorly defined cristae. As there were no significant ultrastructural differences between WT and lacZ-KD ESCs, we surmised that lentivirus transduction of ESCs per se did not affect their mitochondrial morphology. On the other hand, Gfer-KD ESCs contained few discernable mitochondria, and those present appeared swollen with abnormal morphology and at various stages of degeneration (Figure 3E; Supplementary Figure 3A) consistent with mitochondrial failure. Moreover, the cytoplasm of Gfer-KD ESCs contained numerous autophagosomes containing lysosomes and cytoplasmic organelles including mitochondria (Figure 3E, Supplementary Figure 3A), suggesting that the degenerated mitochondria in these cells are being eliminated by autophagy/mitophagy. Further, levels of autophagy-associated proteins light chain (LC) 3b, Beclin-1, and Atg5/12 complexes were higher in Gfer1-KD and Gfer2-KD ESCs compared with either WT or lacZ-KD ESCs (Supplementary Figure 3B). Taken together, our data reveal that the down-regulation of Gfer in the ESCs results in mitochondrial dysfunction; the ensuing MPT triggers the elimination of damaged mitochondria by mitophagy (Elmore et al., 2001), and massive degradation of such an essential organelle initiates apoptosis.
Our observations suggest that one mechanism by which Gfer maintains ESC stemness is by preserving the integrity of their mitochondria. We surmised that any assault to the very few mitochondria that the ESCs possess would also trigger mitophagy in these primitive cells. Complex I, a multisubunit redox machine, is the first enzyme in mitochondrial electron transport chain (ETC) that plays a central role in mitochondrial energy metabolism and can be pharmacologically inhibited by rotenone. Inhibition of ETC complex I with 10 μM rotenone for 24 h resulted in enhanced early (fivefold) and late (twofold) apoptotic populations in WT ESCs, indicating a marked reduction in cell viability (Supplementary Figure 4A). Further, treatment of ESCs with rotenone triggered mitochondrial autophagy or mitophagy, characterized by large autophagosomes containing several mitochondria (Supplementary Figure 4B). We also observed classic apoptotic features such as cell fragmentation within the rotenone-treated ESCs (Supplementary Figure 4B). Thus, inhibition of ETC complex I or Gfer down-regulation severely damages ESC mitochondria triggering extensive mitophagy and apoptosis.
Gfer Depletion Does Not Impair Mitochondrial Function and/or Cell Viability in Differentiated Cells
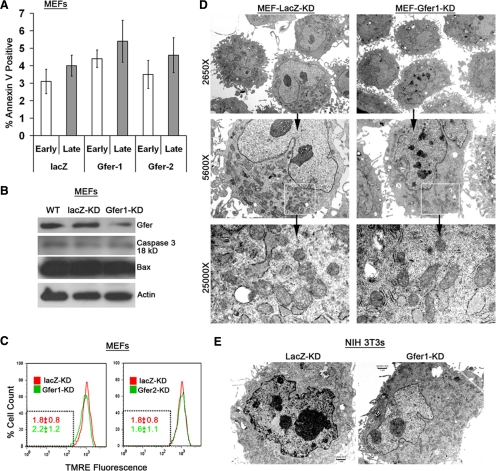
Gfer is an evolutionarily conserved, ubiquitous eukaryotic protein with essential roles in cell survival, maintenance of the mitochondrial genome, and organogenesis in higher eukaryotes (Polimeno et al., 1999; Klissenbauer et al., 2002). Our data indicate that Gfer promotes ESC survival by preserving mitochondrial structural integrity and function. These results raise the question of whether Gfer is a ubiquitous house-keeping gene essential for preservation of mitochondrial function and viability of all cells in which it is expressed. To investigate this, we performed lentivirus-mediated KD of Gfer in MEFs, a differentiated cell type derived from day 12.5 murine embryos. We achieved an 80% KD of Gfer using Gfer1 shRNA (Figure 4B) and a 70% KD using Gfer2 shRNA (not shown). We then evaluated whether KD of Gfer resulted in a loss of cell viability in MEFs by monitoring annexin V/7-AAD reactivity. Depletion of Gfer did not initiate any significant increases in the number of either early or late apoptotic cells (Figure 4A). Additionally, as shown in Figure 4B, KD of Gfer did not alter levels of activated form of caspase-3 or the proapoptotic Bax. Furthermore, reduction in Gfer did not alter the ΔΨm in primary MEFs (Figure 4C). These results indicate that down-regulation of Gfer does not initiate apoptosis or affect mitochondrial function in a differentiated cell type such as the MEF.
Figure 4.
Gfer KD does not affect cell viability or mitochondria in differentiated cells. (A) Graphs showing average (n = 3) percentage of apoptotic cells, measured by annexin V/7-AAD reactivity in indicated MEF genotypes, at 72 h after lentivirus infection. Error bars, SD; *p < 0.05. (B) Immunoblots (n = 2) showing Gfer, caspase 3 (18 kDa), Bax, and actin levels in indicated MEFs. (C) Representative histograms depicting TMRE fluorescence (PE channel) in LacZ and Gfer1/2 KD MEFs. Numbers in red (LacZ-KD) and green (Gfer1/2-KD) are the average (n = 3) percentage of cells with TMRE fluorescence intensities within the gated region (dotted box) ± SD; *p < 0.05. (D) Digital TEM images depicting ultrastructural details in LacZ (left) and Gfer-1 KD (right) MEFs. Magnifications, ×2650, ×5600, and ×25,000. (E) Digital TEM images depicting ultrastructural details at ×5600 magnification in LacZ (left) and Gfer-1 KD (right) NIH 3T3 cells.
We also performed TEM studies to determine whether Gfer-KD affected mitochondrial morphology in primary MEFs. Compared with ESCs, MEFs are larger cells that contain numerous mitochondria with well-distinct cristae, rough endoplasmic reticulum (ER), Golgi apparatus, and a few autophagic vesicles (Figure 4D). Although the mitochondrial cristae in Gfer-KD MEFs appeared slightly swollen compared with those in LacZ-KD MEFs, there was no alteration in the overall mitochondrial morphology or reduction in the number of mitochondria in these cells (Figure 4D). Additionally, KD of Gfer in NIH 3T3 cells, an immortalized MEF cell line, did not alter mitochondrial morphology (Figure 4E) or cell viability (not shown). Collectively, these data suggest that Gfer may largely be dispensable for mitochondrial function and/or survival of differentiated cells.
Overexpression of Gfer Results in Elongated ESC Mitochondria
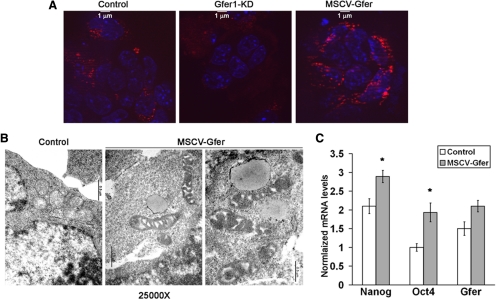
To test the hypothesis that manipulated up-regulation of Gfer would alter mitochondrial morphology, we overexpressed Gfer in ESCs, using the MSCV-IRES-GFP retrovirus system and observed the DsRed-labeled mitochondria in control, Gfer-KD, and MSCV-Gfer ESC colonies. Consistent with our previous TEM data, control ESCs possessed a few DsRed-positive mitochondria, whereas Gfer-KD ESCs contained very few, if any, DsRed-positive mitochondria. In contrast, MSCV-Gfer ESCs possessed numerous bright, DsRed-positive mitochondria that appeared slightly elongated (Figure 5A). Because they are very small cells with an average diameter of 4 μm, it was extremely difficult to evaluate mitochondrial morphology of ESC colonies using standard immunocytochemistry, even at high (×1000) magnification. We therefore performed TEM to determine whether Gfer overexpression alters mitochondrial morphology in ESCs. Up-regulation of Gfer results in enlarged, elongated mitochondria that contained prominent cristae (Figure 5B), suggesting that Gfer overexpression may block mitochondrial fission or promote mitochondrial fusion in ESCs. We also observed that overexpression of Gfer in WT ESCs conferred significant protection from apoptosis after a 24-h treatment with 10 μM rotenone (Supplementary Figure 4C). Moreover, Gfer overexpression resulted in significantly higher Nanog and Oct4 expression in ESCs (Figure 5C). Altogether, these data suggest that Gfer acts to promote ESC pluripotency by preserving the mitochondrial structural integrity and function.
Figure 5.
Overexpression of Gfer alters mitochondrial morphology and elevates pluripotent marker expression. (A) Representative digital ApoTome images (magnification, ×100) showing DsRed-positive mitochondria in ESCs infected with MSCV-IRES-GFP-control (control), FG12-GFP-Gfer1 shRNA, or MSCV-IRES-GFP-Gfer (MSCV-GFER) viruses. Scale bar, 1 μm; n = 3. (B) Digital TEM images at ×25,000 magnification (scale bar, 0.5 μm) depicting ultrastructural details of mitochondria in control (left) and MSCV-Gfer (both images on the right) ESCs. (C) Average mRNA levels of Nanog, Oct-4, and Gfer in day 6 control and MSCV-Gfer ESCs. The mRNA levels measured by qRT-PCR were normalized to actin; (n = 3); *p = 0.03 for Nanog and 0.006 for Oct4.
Gfer Modulates the Levels of Mitochondrial Fission GTPase Drp1 in Mouse ESCs
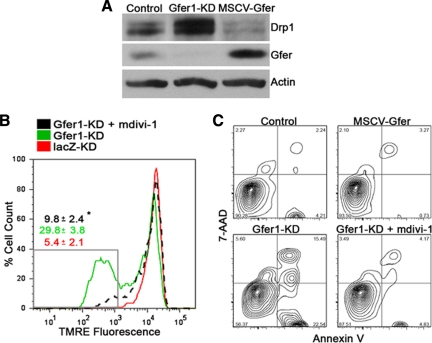
Overexpression of Gfer in ESCs results in large, elongated mitochondria, whereas depletion of this sulfhydryl oxidase enhances mitochondrial fragmentation. These data led us to hypothesize that Gfer affects ESC mitochondrial integrity by playing a role in the maintenance of mitochondrial fission–fusion dynamics. Mitochondria are dynamic organelles that undergo fusion, fission, and migration within cells. Because mitochondria constantly fuse and divide, any imbalance of these two processes alters the overall mitochondrial morphology, number, turnover, and function. Mitochondrial fusion is facilitated by the GTPases, mitofusins Mfn1/2 and optic atrophy protein 1 (OPA1). In contrast, the highly conserved Drp1 is the main effector of mitochondrial fission and overall mitochondrial dynamics. Enhanced cellular levels of Drp1 cause increased mitochondrial fragmentation and mitophagy. Enhanced mitochondrial division and fragmentation triggers cytochrome c release, caspase activation, and apoptosis (Frank et al., 2001; Smirnova et al., 2001; Tanaka and Youle, 2008; Benard and Karbowski, 2009; Ishihara et al., 2009). To further characterize the effect of Gfer on mitochondrial dynamics, we evaluated the levels of Drp1, OPA1, Mfn1, and Mfn2 in control, Gfer-KD, and MSCV-Gfer ESCs. As shown in Figure 6A, Drp1 was highly elevated upon Gfer down-regulation. Moreover, overexpression of Gfer resulted in a reduction in Drp1 levels (Figure 6A). In contrast, there were no significant changes in OPA1 or Mfn levels in Gfer-KD or Gfer overexpressing ESCs (Supplementary Figure 5A).
Figure 6.
Inhibition of Drp1 rescues mitochondrial dysfunction and apoptosis in Gfer-KD ESCs. (A) Representative immunoblots (n = 3) depicting Drp1, Gfer, and actin levels in control, Gfer-KD, or MSCV-Gfer ESCs. (B) TMRE fluorescence in lacZ-KD ESCs (red), Gfer1-KD ESCs (green), and Gfer1-KD ESCs treated for 24 h with 25 μM mdivi-1(dotted black). Numbers in red (lacZ-KD), green (Gfer1-KD), and black (Gfer1-KD + 25 μM mdivi-1) are average percentage of cells with TMRE fluorescence intensities within the gated region (gray box) ± SD; (n = 3); *p = 0.005. (C) Representative histograms (n = 3) showing percentage of apoptotic cells, in control or MSCV-Gfer ESCs (top panel) and in Gfer-KD ESCs or Gfer-KD ESCs treated for 24 h with vehicle or 25 μM mdivi-1 (bottom panel).
Inhibition of Drp1 Rescues Mitochondrial Dysfunction and Apoptosis in Gfer-KD ESCs
These data suggest that although elevated Drp1 facilitates enhanced mitochondrial fragmentation followed by mitophagy and apoptosis in Gfer-KD cells, lower Drp1 in Gfer overexpressing ESCs results in diminished fission, as evidenced by the presence of elongated mitochondria in these cells, and enhanced survival (Figure 6C). If this is true, then inhibition of Drp1 function should reverse the loss of mitochondrial function and cell viability in Gfer-KD ESCs. To test this idea, we treated Gfer-KD ESCs with mdivi-1, a recently identified specific, small-molecule inhibitor of Drp1 (Cassidy-Stone et al., 2008). Treatment of Gfer-KD ESCs with 25 μM mdivi-1 significantly reversed the loss of ΔΨm (Figure 6B) and rescued apoptosis (Figure 6C) in these cells. Thus, enhanced mitochondrial division triggered by high Drp1 levels is responsible for the loss of mitochondrial ΔΨm, mitophagy, and apoptosis in Gfer-depleted ESCs.
Expression of Dominant Negative Drp1 Restores Pluripotency in Gfer-KD ESCs
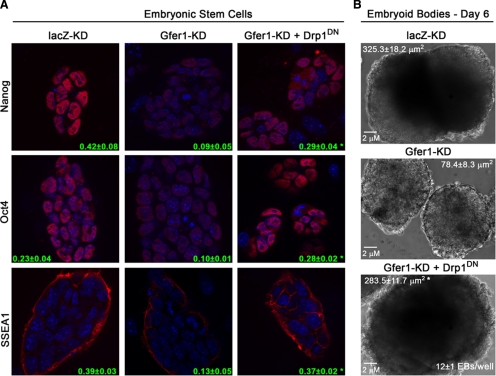
We hypothesized that enhanced mitochondrial fission triggered by elevated Drp1 in the absence of Gfer also compromised ESC pluripotency or “stemness.” To test this, we expressed the dominant-negative K38A mutant of Drp1 (Drp1DN) in WT, lacZ-KD, and Gfer-KD ESCs. The mutation of this critical lysine within the GTPase domain of Drp1 leads to the inhibition of its binding to GTP (Smirnova et al., 2001). Although the overall colony size was smaller in all ESCs expressing Drp1DN, inhibition of Drp1 did not alter pluripotency marker expression in lacZ-KD (Supplementary Figure 5B) or WT (data not shown) ESCs. In contrast, expression of Drp1DN resulted in a significant 2–3-fold increase of Nanog, Oct-4, and SSEA1 levels in Gfer-KD ESCs (Figure 7A). Thus, inhibition of Drp1 in ESCs depleted for Gfer resulted in restoration of Oct-4 and SSEA1 expression levels to those observed in control (lacZ-KD) ESCs. Moreover, Gfer-KD ESCs expressing Drp1DN formed embryoid bodies that were comparable in size and numbers to those formed by lacZ-KD ESCs. As shown in Figure 7B, day 6 Drp1DN-Gfer-KD EBs were similar in surface area (283 ± 11 μm2) compared with age-matched lacZ EBs (325 ± 18 μm2). Additionally, Drp1DN-Gfer-KD ESCs also yielded similar numbers of EBs (12 ± 1 per well) as lacZ-KD ESCs (Figure 1D). Taken together, our data indicate that Gfer modulates Drp1 in ESCs to maintain mitochondrial dynamics and consequently, may preserve pluripotency or “stemness.”
Figure 7.
Expression of Drp1DN restores pluripotency in Gfer-KD ESCs. (A) Digital ApoTome (optical section) images (magnification, ×630) depicting Nanog, Oct-4, and SSEA1 expression (all in red) in lacZ-KD and Gfer-KD ESCs and in Gfer-KD ESCs expressing Drp1DN. Blue, DAPI; green, numbers representing average signal intensities ± SD quantified from 35 colonies (n = 3); *p = 0.0001. (B) Day 6 embryoid bodies formed by lacZ-KD and Gfer-KD ESCs and by Gfer-KD ESCs expressing Drp1DN. Magnification, ×200; scale bar, 2 μm. Average surface area in μm2 ± SD from each genotype is shown (n = 3); *p = 0.001.
DISCUSSION
To maintain their pluripotency during embryogenesis and in culture, ESCs must employ highly efficient mechanisms to avoid genetic and/or mitochondrial damage. Understanding the nature of genes that preserve ESC stemness is therefore vital to realize the full therapeutic potential of ESCs in regenerative medicine. Here we identify the FAD-linked mitochondrial sulfhydryl oxidase Gfer as one such gene. Gfer enforces ESC mitochondrial fission–fusion balance by modulating Drp1 levels, and through this mechanism it maintains mitochondrial structural integrity and function and may thus promote ESC pluripotency. Overexpression of Gfer diminishes Drp1 levels, impedes mitochondrial fission, enhances ESC survival, and elevates pluripotency marker expression. Conversely, depletion of Gfer elevates Drp1 levels, which then orchestrates enhanced mitochondrial fragmentation, mitophagy, and apoptosis. Inhibition of Drp1 with a small molecule inhibitor, or expression of a dominant/negative mutant, rescues mitochondrial loss of function, apoptosis, and pluripotency in Gfer-depleted ESCs. Therefore, the effect of Gfer on mitochondrial function, and consequently pluripotency of ESCs, is mediated primarily by its regulation of Drp1 levels and/or activity.
Drp1 levels and activities are regulated by a combination of posttranslational modifications such as phosphorylation (Chang and Blackstone, 2007; Taguchi et al., 2007; Han et al., 2008), ubiquitination (Santel and Frank, 2008), and small ubiquitin-like modifier (SUMO)ylation (Zunino et al., 2007). How does Gfer modulate Drp1 levels and/or activity? The evolutionarily conserved FAD-dependent sulfhydryl oxidase activity of Gfer (Lisowsky et al., 2001) is conceivably important for this activity. Drp1 contains three domains: a highly conserved N-terminal GTPase domain, a middle dynamin/Vps1p homology (DVH) 1 domain, and a C-terminal DVH2 or GTPase effector domain (GED; Shin et al., 1999; Hoppins et al., 2007). The DVH1 and DVH2 or GED domains favor inter- and intramolecular interactions. Under physiological conditions, Drp1 exists as a dimer or tetramer and its self-assembly into higher order structures facilitates GTP hydrolysis and mitochondrial fission (Smirnova et al., 2001; Hoppins et al., 2007). Efficacy of the chemical inhibitor of Drp1, mdivi-1, is highly dependent on an unblocked sulfhydryl moiety that forms disulfide bonds on cysteine residues within regions outside of Drp1 GTPase domain (Cassidy-Stone et al., 2008). Thus, it is conceivable that the sulfhydryl oxidase Gfer, predominantly localized in the IMS but also present in the cytoplasm, catalyzes the formation of disulfide bonds on the same cysteine residues of Drp1 to inactivate the GTPase and/or facilitate structural changes that favor Drp1 turnover. In fact, Drp1 contains four evolutionarily conserved cysteine residues outside the GTPase domain that could serve as potential Gfer oxidization sites. The fact that Drp1 migrates as a double band during reducing SDS-PAGE (Figure 6A; Zunino et al., 2009), suggests partial oxidization of one or more of its cysteine pairs.
On the other hand, the regulation of Drp1 by Gfer could be indirect, with Gfer affecting the activity of a factor that regulates Drp1 activity. For example, overexpression of the SUMO protease SENP5 rescues SUMOylation-induced mitochondrial fragmentation by causing the down-regulation of DRP1; and deletion of SENP5 favors increased mitochondrial fragmentation (Zunino et al., 2007). SENP5 possesses six highly conserved cysteine residues, at least one of which, Cys713, is important for its catalytic activity. Hence it is possible that Gfer oxidizes disulfide bonds on one or more of the cysteine pairs on SENP5, thereby enhancing the protein's activity and stability, akin to its function in the import of proteins to the mitochondrial IMS. In yeast, Gfer catalyzes the formation of disulfide bridges between Mia40 and IMS proteins such as Cox17 and Tim13 to form a disulfide relay system that enables the import and retention of proteins in the mitochondrial IMS, a process critical for normal mitochondrial function (Mesecke et al., 2005). Perhaps Gfer also catalyzes inhibitory disulfide bridges between Drp1 and other protein(s) involved in the formation of higher-order structures that mediate mitochondrial fission.
Pluripotent ESCs contain a small number of mitochondria with poorly defined (immature) cristae and consequently, possess low ATP levels (Baharvand and Matthaei, 2003; Lonergan et al., 2007; Nesti et al., 2007). Paradoxically, ESCs also possess an unlimited proliferative potential, compared with differentiated cells such as the MEFs, and hence may depend on a relatively few proteins such as Gfer to maintain their mitochondrial morphology, dynamics, function, and therefore stemness. Even with 80% KD of Gfer (Figure 4B), we did not observe any associated loss of cell viability and/or mitochondrial morphology or function in MEFs. Although we interpreted these results to corroborate a selective role for Gfer in ESCs, it is also possible that the residual (20%) Gfer in these MEFs is able to support the maintenance of their mitochondria and therefore preserve cell viability. It is also possible that diminished pluripotency in Gfer-KD ESCS is consequential to a more general loss of its mitochondrial housekeeping function, resulting in an overall loss of cell viability. Exactly how mitochondrial stability and function contributes to stemness or pluripotency in ESCs is a critical but poorly understood aspect of stem cell biology.
Mitochondrial malfunction and mitophagy are associated with degenerative disorders such as Parkinson's and Huntington's. Elevated Drp1, enhanced mitochondrial fragmentation and pathological mitophagy is reported in familial and sporadic forms of Parkinson's disease (Dagda et al., 2009). Mutations in the human Gfer gene are associated with an infantile autosomal recessive form of myopathy, characterized at the cellular level by abnormal mitochondrial morphology, loss of mitochondrial function, and enhanced mutations of mtDNA (Di Fonzo et al., 2009). Moreover, reversal of cell death with a concomitant increase in Gfer expression was observed in a murine model of Huntington's treated with inhibitors of histone deacetylation (Gardian et al., 2005). These studies underscore a cytoprotective role for Gfer in the preservation of mitochondrial dynamics, structural integrity, and function during homeostasis.
In conclusion, the data presented herein support an essential prosurvival role for Gfer in the maintenance of murine ESC pluripotency by preserving the structural and functional integrity of their mitochondria, through modulation of the key mitochondrial fission factor Drp1. Our findings have therapeutic implications as they support a central role for a novel Gfer-Drp1 molecular link in preserving the stemness of ESCs, a primitive cell type with immense curative potential.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Drs. Sally Kornbluth, Jeffrey Rathmell, and Tso-Pang Yao (Department of Pharmacology and Cancer Biology, Duke University Medical Center) and Dr. Keith R. Davis (Owensboro Cancer Research Program [OCRP] for critical reading of the manuscript and helpful discussions. We also thank Cheryl Bock (Duke University Transgenic Core Facility) for the generous gift of B6 ESCs; Dr. Ferrell C. Campbell and Cathie G. Caple (University of Louisville Electron Microscopy Core Facility) for help with electron microscopy; and Sarah W. Grant and Cassandra Lockard for technical assistance. This work was supported by funds from the James Graham Brown Cancer Center and OCRP to U.S. and National Institutes of Health Grant R01 DK074701 to A.R.M.
Abbreviations used:
- SC
stem cell
- Drp1
Dynamin-related protein 1
- EB
embryoid body
- ESC
embryonic stem cell
- Gfer
growth factor erv1-like
- IMS
intermembrane space
- KD
knockdown
- MEF
mouse embryonic fibroblast
- ΔΨm
mitochondrial membrane potential.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-11-0937) on February 10, 2010.
REFERENCES
- Akao M., O'Rourke B., Teshima Y., Seharaseyon J., Marban E. Mechanistically distinct steps in the mitochondrial death pathway triggered by oxidative stress in cardiac myocytes. Circ. Res. 2003;92:186–194. doi: 10.1161/01.res.0000051861.21316.e9. [DOI] [PubMed] [Google Scholar]
- Baharvand H., Matthaei K. I. The ultrastructure of mouse embryonic stem cells. Reprod. Biomed. Online. 2003;7:330–335. doi: 10.1016/s1472-6483(10)61873-1. [DOI] [PubMed] [Google Scholar]
- Becher D., Kricke J., Stein G., Lisowsky T. A mutant for the yeast scERV1 gene displays a new defect in mitochondrial morphology and distribution. Yeast. 1999;15:1171–1181. doi: 10.1002/(SICI)1097-0061(19990915)15:12<1171::AID-YEA443>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Benard G., Karbowski M. Mitochondrial fusion and division: regulation and role in cell viability. Semin. Cell Dev. Biol. 2009;20:365–374. doi: 10.1016/j.semcdb.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdon T., Smith A., Savatier P. Signalling, cell cycle and pluripotency in embryonic stem cells. Trends Cell Biol. 2002;12:432–438. doi: 10.1016/s0962-8924(02)02352-8. [DOI] [PubMed] [Google Scholar]
- Cassidy-Stone A., Chipuk J. E., Ingerman E., Song C., Yoo C., Kuwana T., Kurth M. J., Shaw J. T., Hinshaw J. E., Green D. R., Nunnari J. Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization. Dev. Cell. 2008;14:193–204. doi: 10.1016/j.devcel.2007.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. R., Blackstone C. Cyclic AMP-dependent protein kinase phosphorylation of Drp1 regulates its GTPase activity and mitochondrial morphology. J. Biol. Chem. 2007;282:21583–21587. doi: 10.1074/jbc.C700083200. [DOI] [PubMed] [Google Scholar]
- Dagda R. K., Cherra S. J., 3rd, Kulich S. M., Tandon A., Park D., Chu C. T. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J. Biol. Chem. 2009;284:13843–13855. doi: 10.1074/jbc.M808515200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fonzo A., et al. The mitochondrial disulfide relay system protein GFER is mutated in autosomal-recessive myopathy with cataract and combined respiratory-chain deficiency. Am. J. Hum. Genet. 2009;84:594–604. doi: 10.1016/j.ajhg.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore S. P., Qian T., Grissom S. F., Lemasters J. J. The mitochondrial permeability transition initiates autophagy in rat hepatocytes. FASEB J. 2001;15:2286–2287. doi: 10.1096/fj.01-0206fje. [DOI] [PubMed] [Google Scholar]
- Frank S., Gaume B., Bergmann-Leitner E. S., Leitner W. W., Robert E. G., Catez F., Smith C. L., Youle R. J. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev. Cell. 2001;1:515–525. doi: 10.1016/s1534-5807(01)00055-7. [DOI] [PubMed] [Google Scholar]
- Gardian G., et al. Neuroprotective effects of phenylbutyrate in the N171–82Q transgenic mouse model of Huntington's disease. J. Biol. Chem. 2005;280:556–563. doi: 10.1074/jbc.M410210200. [DOI] [PubMed] [Google Scholar]
- Gatzidou E., Kouraklis G., Theocharis S. Insights on augmenter of liver regeneration cloning and function. World J. Gastroenterol. 2006;12:4951–4958. doi: 10.3748/wjg.v12.i31.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X.-J., Lu Y.-F., Li S.-A., Kaitsuka T., Sato Y., Tomizawa K., Nairn A. C., Takei K., Matsui H., Matsushita M. CaM kinase Iα-induced phosphorylation of Drp1 regulates mitochondrial morphology. J. Cell Biol. 2008;182:573–585. doi: 10.1083/jcb.200802164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S., Lackner L., Nunnari J. The machines that divide and fuse mitochondria. Annu. Rev. Biochem. 2007;76:751–780. doi: 10.1146/annurev.biochem.76.071905.090048. [DOI] [PubMed] [Google Scholar]
- Ishihara N., et al. Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat. Cell Biol. 2009;11:958–966. doi: 10.1038/ncb1907. [DOI] [PubMed] [Google Scholar]
- Ivanova N., Dobrin R., Lu R., Kotenko I., Levorse J., DeCoste C., Schafer X., Lun Y., Lemischka I. R. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- Ivanova N. B., Dimos J. T., Schaniel C., Hackney J. A., Moore K. A., Lemischka I. R. A stem cell molecular signature. Science. 2002;298:601–604. doi: 10.1126/science.1073823. [DOI] [PubMed] [Google Scholar]
- Kitsos C. M., Sankar U., Illario M., Colomer-Font J. M., Duncan A. W., Ribar T. J., Reya T., Means A. R. Calmodulin-dependent protein kinase IV regulates hematopoietic stem cell maintenance. J. Biol. Chem. 2005;280:33101–33108. doi: 10.1074/jbc.M505208200. [DOI] [PubMed] [Google Scholar]
- Klebes A., Sustar A., Kechris K., Li H., Schubiger G., Kornberg T. B. Regulation of cellular plasticity in Drosophila imaginal disc cells by the Polycomb group, trithorax group and lama genes. Development. 2005;132:3753–3765. doi: 10.1242/dev.01927. [DOI] [PubMed] [Google Scholar]
- Klissenbauer M., Winters S., Heinlein U. A., Lisowsky T. Accumulation of the mitochondrial form of the sulphydryl oxidase Erv1p/Alrp during the early stages of spermatogenesis. J. Exp. Biol. 2002;205:1979–1986. doi: 10.1242/jeb.205.14.1979. [DOI] [PubMed] [Google Scholar]
- Lange H., Lisowsky T., Gerber J., Muhlenhoff U., Kispal G., Lill R. An essential function of the mitochondrial sulfhydryl oxidase Erv1p/ALR in the maturation of cytosolic Fe/S proteins. EMBO Rep. 2001;2:715–720. doi: 10.1093/embo-reports/kve161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Sinha S., Kroemer G. Bcl-2 family members: dual regulators of apoptosis and autophagy. Autophagy. 2008;4:600–606. doi: 10.4161/auto.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisowsky T., Lee J. E., Polimeno L., Francavilla A., Hofhaus G. Mammalian augmenter of liver regeneration protein is a sulfhydryl oxidase. Dig. Liver Dis. 2001;33:173–180. doi: 10.1016/s1590-8658(01)80074-8. [DOI] [PubMed] [Google Scholar]
- Liu X., Kim C. N., Yang J., Jemmerson R., Wang X. Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell. 1996;86:147–157. doi: 10.1016/s0092-8674(00)80085-9. [DOI] [PubMed] [Google Scholar]
- Lonergan T., Bavister B., Brenner C. Mitochondria in stem cells. Mitochondrion. 2007;7:289–296. doi: 10.1016/j.mito.2007.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesecke N., Terziyska N., Kozany C., Baumann F., Neupert W., Hell K., Herrmann J. M. A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell. 2005;121:1059–1069. doi: 10.1016/j.cell.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Nesti C., Pasquali L., Vaglini F., Siciliano G., Murri L. The role of mitochondria in stem cell biology. Biosci. Rep. 2007;27:165–171. doi: 10.1007/s10540-007-9044-1. [DOI] [PubMed] [Google Scholar]
- Pasini D., Bracken A. P., Hansen J. B., Capillo M., Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol. Cell. Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polimeno L., Lisowsky T., Francavilla A. From yeast to man–from mitochondria to liver regeneration: a new essential gene family. Ital. J. Gastroenterol. Hepatol. 1999;31:494–500. [PubMed] [Google Scholar]
- Polimeno L., Margiotta M., Marangi L., Lisowsky T., Azzarone A., Ierardi E., Frassanito M. A., Francavilla R., Francavilla A. Molecular mechanisms of augmenter of liver regeneration as immunoregulator: its effect on interferon-gamma expression in rat liver. Dig. Liver Dis. 2000;32:217–225. doi: 10.1016/s1590-8658(00)80824-5. [DOI] [PubMed] [Google Scholar]
- Qin X. F., An D. S., Chen I. S., Baltimore D. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc. Natl. Acad. Sci. USA. 2003;100:183–188. doi: 10.1073/pnas.232688199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalho-Santos M., Yoon S., Matsuzaki Y., Mulligan R. C., Melton D. A. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- Santel A., Frank S. Shaping mitochondria: The complex posttranslational regulation of the mitochondrial fission protein DRP1. IUBMB Life. 2008;60:448–455. doi: 10.1002/iub.71. [DOI] [PubMed] [Google Scholar]
- Sathananthan H., Pera M., Trounson A. The fine structure of human embryonic stem cells. Reprod. Biomed. Online. 2002;4:56–61. doi: 10.1016/s1472-6483(10)61916-5. [DOI] [PubMed] [Google Scholar]
- Shin H. W., Takatsu H., Mukai H., Munekata E., Murakami K., Nakayama K. Intermolecular and interdomain interactions of a dynamin-related GTP-binding protein, Dnm1p/Vps1p-like protein. J. Biol. Chem. 1999;274:2780–2785. doi: 10.1074/jbc.274.5.2780. [DOI] [PubMed] [Google Scholar]
- Silver L. M. The peculiar journey of a selfish chromosome: mouse t haplotypes and meiotic drive. Trends Genet. 1993;9:250–254. doi: 10.1016/0168-9525(93)90090-5. [DOI] [PubMed] [Google Scholar]
- Smirnova E., Griparic L., Shurland D. L., van der Bliek A. M. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi N., Ishihara N., Jofuku A., Oka T., Mihara K. Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J. Biol. Chem. 2007;282:11521–11529. doi: 10.1074/jbc.M607279200. [DOI] [PubMed] [Google Scholar]
- Tanaka A., Youle R. J. A chemical inhibitor of DRP1 uncouples mitochondrial fission and apoptosis. Mol. Cell. 2008;29:409–410. doi: 10.1016/j.molcel.2008.02.005. [DOI] [PubMed] [Google Scholar]
- Wang J., Theunissen T. W., Orkin S. H. Site-directed, virus-free, and inducible RNAi in embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2007;104:20850–20855. doi: 10.1073/pnas.0710565105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. The expanding role of mitochondria in apoptosis. Genes Dev. 2001;15:2922–2933. [PubMed] [Google Scholar]
- Waterhouse N. J., Trapani J. A. A new quantitative assay for cytochrome c release in apoptotic cells. Cell Death Differ. 2003;10:853–855. doi: 10.1038/sj.cdd.4401263. [DOI] [PubMed] [Google Scholar]
- Zeng X., Rao M. S. Human embryonic stem cells: long term stability, absence of senescence and a potential cell source for neural replacement. Neuroscience. 2007;145:1348–1358. doi: 10.1016/j.neuroscience.2006.09.017. [DOI] [PubMed] [Google Scholar]
- Zunino R., Braschi E., Xu L., McBride H. M. Translocation of SenP5 from the nucleoli to the mitochondria modulates DRP1-dependent fission during mitosis. J. Biol. Chem. 2009;284:17783–17795. doi: 10.1074/jbc.M901902200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zunino R., Schauss A., Rippstein P., Andrade-Navarro M., McBride H. M. The SUMO protease SENP5 is required to maintain mitochondrial morphology and function. J. Cell Sci. 2007;120:1178–1188. doi: 10.1242/jcs.03418. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.