Formins are conserved eukaryotic proteins that direct the nucleation and elongation of unbranched actin filaments. We define two nonoverlapping regions, Bnr1p-L1 (1-466) and Bnr1p-L2 (466-733) that can each localize to the bud neck independently of endogenous Bnr1p.
Abstract
Formins are conserved eukaryotic proteins that direct the nucleation and elongation of unbranched actin filaments. The yeast formins, Bni1p and Bnr1p, assemble actin cables from the bud cortex and bud neck, respectively, to guide overall cell polarity. Here we examine the regions of Bnr1p responsible for bud neck localization. We define two non-overlapping regions, Bnr1p-L1 (1-466) and Bnr1p-L2 (466-733), that can each localize to the bud neck independently of endogenous Bnr1p. Bnr1p-L1 and Bnr1p-L2 localize with septins at the bud neck, but show slightly differently spatial and temporal localization, reflecting the localization (Bnr1p-L1) or cell cycle timing (Bnr1p-L2) of full-length Bnr1p. Bnr1p is known to be very stably localized at the bud neck, and both Bnr1p-L1 and Bnr1p-L2 also show relatively stable localization there. Overexpression of Bnr1p-L1, but not Bnr1p-L2, disrupts septin organization at the bud neck. Thus Bnr1p has two separable regions that each contribute to its bud neck localization.
INTRODUCTION
The shape and internal organization of eukaryotic cells is determined by pathways that drive the assembly, spatial arrangement, and dynamics of the cytoskeleton. The actin-based cytoskeleton is especially important for the organization and morphology of the plasma membrane and processes associated with it, such as cytokinesis, endocytosis, and exocytosis. Cells have evolved mechanisms to regulate the rate-limiting nucleating step of actin filament assembly to control where and when actin filaments are assembled. The formins are a major and ubiquitous family of actin-nucleating proteins that regulate multiple actin-dependent processes, including cytokinesis (Chang et al., 1997), the formation of filopodia (Pellegrin and Mellor, 2005), and assembly of adherens junctions (Kobielak et al., 2004; Goode and Eck, 2007).
The defining feature of formins is the presence of adjacent FH1 and FH2 domains (formin homology domains 1 and 2), which generally reside in the C-terminal half of the protein (Higgs and Peterson, 2005; Goode and Eck, 2007). The FH1 region is rich in proline residues and recruits the profilin-actin complex, whereas the dimeric FH2 domain contains the actin-nucleating, processive capping and elongation activities. Together, FH1 and FH2 nucleate actin assembly and remain attached at the plus end while allowing for filament growth there (Pruyne et al., 2002; Sagot et al., 2002). Based on initial studies with mDia1, many formin proteins have a GTPase-binding domain (GBD) near the N-terminus (Watanabe et al., 1999). The GBD is followed by the DID (diaphanous inhibitory domain) that has been shown in mDia1 to bind a region downstream of the FH2 called the DAD (diaphanous autoregulatory domain) to inhibit the function of the formin (Alberts, 2001). The binding of a GTP-bound Rho protein to the GBD disrupts the DID–DAD interaction and relieves the inhibition in at least some cases (Watanabe et al., 1999; Li and Higgs, 2005).
Structural analysis has provided considerable insight into the organization of formins. The DID region of mDia1 consists of a series of armadillo repeats that is followed by a dimerization (DD) domain. The GTP-bound Rho protein binds the GBD and part of the DID in such a way that the DAD is displaced (Otomo et al., 2005; Rose et al., 2005; Nezami et al., 2006). In addition to providing the regulatory region, the N-terminal half of formins are believed to be involved in their localization, which is important for the assembly of actin filaments at the right place (Fujiwara et al., 1998). In this study, we examine the localization determinants in a yeast formin that specify where it will function.
Budding yeast has two formins, Bni1p and Bnr1p, that exhibit different localizations (Pruyne et al., 2004b). Bni1p is localized to growth sites, namely at the future bud site in unbudded cells, at the bud tip or cortex in small- to medium-budded cells, and at the bud neck in dividing cells. Bnr1p is localized to the bud neck after bud emergence and stays there until cytokinesis (Fujiwara et al., 1998; Kikyo et al., 1999; Ozaki-Kuroda et al., 2001; Pruyne et al., 2004a). Moreover, the localization of Bni1p is dynamic, whereas Bnr1p is stably associated with the bud neck (Buttery et al., 2007).
The localization of Bnr1p depends on bud neck-localized septins, GTP-binding proteins that function in cytokinesis in fungal and animal cells (Kikyo et al., 1999; Pruyne et al., 2004a). In budding yeast, five septins are expressed in vegetative cells: Cdc3p, Cdc10p, Cdc11p, Cdc12p, and Shs1p. Recent structural studies on the essential septin subunits show how they self-assemble into nonpolar filaments (Bertin et al., 2008). Septins show a cell cycle–regulated distribution. They are first assembled at the presumptive bud site and then form a ring through which the bud emerges, and so they remain at the bud neck. As the bud grows, the septins remain at the bud neck and extend into an hourglass shape, often called the septin collar, which then splits into two rings at cytokinesis (Gladfelter et al., 2001). A characteristic feature of conditional septin mutants is that they grow elongated buds when septin function is compromised, due to triggering of the morphological checkpoint (Lew, 2003).
Here we show that Bnr1p has two adjacent localization domains that can form independent, but slightly different, stable associations with septins at the bud neck, and when overexpressed they affect the function of septins differentially.
MATERIALS AND METHODS
Construction of Yeast Strains and Plasmids
Yeast strains and plasmids used in this study are described in Tables 1 and 2.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| ABY1599 | MATa his3Δ1 leu2Δ0 ura3Δ0 bni1Δ::KanR bnr1Δ::NatR pRS316-BNI1 | Lab collection |
| ABY1680 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 bnr1Δ::KanR | Invitrogen (Carlsbad, CA) |
| ABY1801 | MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 met15/met15 lys2/lys2 ura3Δ0/ura3Δ0 | Lab collection |
| ABY1825 | MATa his3Δ1 met15Δ0 leu2Δ0 ura3Δ0 bnr1Δ::KanR | Evangelista et al. (2002) |
| ABY1881 | MATα ura3 his3 leu2 lys2 trp1 BNR1-GFP::KanR | Pruyne et al. (2004) |
| ABY2097 | MATα ura3 his3 leu2 lys2 trp1 CDC3-mCherry::SpHIS5 BNR1-GFP::KanR | This study |
| ABY2807 | MATa ura3Δ0 his3Δ0 leu2Δ0 met15Δ0 CDC3-mCherry::KanR bnr1Δ::KanR | This study |
| ABY3089 | MATa his3Δ1 leu2Δ0 metΔ15 ura3Δ0 shs1Δ::KanR | Invitrogen |
| JGY1326 | MATa ura3 his3 leu2 lys2 trp1 CDC3-mCherry::SpHIS5 | Erfei Bi |
| M195 | MATa his3 leu2 ura3 cdc10-1 | John Pringle |
| MY10174 | MATa ura3Δ0 his3Δ200 leu2Δ0 met15Δ0 CDC3-mCherry::Kan fus2Δ::HIS3 | Mark Rose |
| Y1239 | MATa his3Δ1 leu2Δ0 metΔ15 ura3Δ0 | Evangelista et al. (2002) |
Table 2.
Plasmids used in this study
| Plasmid | Backbone | Genotype |
|---|---|---|
| pLG104 | pRS316 | PGAL1-10-3HA-BNR1(1-1473) |
| pLG106 | pRS316 | PGAL1-10-3HA-BNR1(1141-2199) |
| pLG130 | pRS316 | PGAL1-10-3HA-BNR1(1141-1473) |
| pLG164 | pRS316 | PGAL1-10-3HA-BNR1(1141-1818) |
| pLG186 | pRS316 | PGAL1-10-3HA-BNR1(1-1353) |
| pLG187 | pRS316 | PGAL1-10-3HA-BNR1(1-1368) |
| pLG188 | pRS316 | PGAL1-10-3HA-BNR1(1-1383) |
| pLG189 | pRS316 | PGAL1-10-3HA-BNR1(1-1398), 3HA-Bnr1-L1 |
| pLG190 | pRS316 | PGAL1-10-3HA-BNR1(1-1413) |
| pLG191 | pRS316 | PGAL1-10-3HA-BNR1(1-1428) |
| pLG192 | pRS316 | PGAL1-10-3HA-BNR1(1-1443) |
| pLG193 | pRS316 | PGAL1-10-3HA-BNR1(1-1458) |
| pLG194 | pRS316 | PGAL1-10-3HA-BNR1(1351-2199) |
| pLG195 | pRS316 | PGAL1-10-3HA-BNR1(1366-2199) |
| pLG196 | pRS316 | PGAL1-10-3HA-BNR1(1381-2199) |
| pLG197 | pRS316 | PGAL1-10-3HA-BNR1(1396-2199), 3HA-Bnr1-L2 |
| pLG198 | pRS316 | PGAL1-10-3HA-BNR1(1411-2199) |
| pLG199 | pRS316 | PGAL1-10-3HA-BNR1(1426-2199) |
| pLG200 | pRS316 | PGAL1-10-3HA-BNR1(1441-2199) |
| pLG201 | pRS316 | PGAL1-10-3HA-BNR1(1456-2199) |
| pLG202 | pRS316 | PGAL1-10-3HA-BNR1(1471-2199) |
| pLG311 | pRS316 | PBNR1-3GFP-BNR1(1-2199), 3GFP-Bnr1-L1-L2 |
| pLG314 | pRS316 | PBNR1-3GFP-BNR1(1396-2199), 3GFP-Bnr1-L2 |
| pLG320 | pRS316 | PGAL1-10-3GFP-BNR1(1-4128), 3GFP-Bnr1 |
| pLG325 | pRS316 | PGAL1-10-GFP-BNR1(1-1398), GFP-Bnr1-L1 |
| pLG328 | pRS316 | PGAL1-10-GFP-BNR1(1396-2199), GFP-Bnr1-L2 |
| pLG341 | pRS315 | PGAL1-10-mCherry-BNR1(1-1398), mCherry-Bnr1-L1 |
| pLG345 | pRS316 | PBNR1-3GFP-BNR1(1396-4128), 3GFP-Bnr1ΔL1 |
| pLG354 | pRS313 | PBNR1-3GFP-BNR1(1396-4128), 3GFP-Bnr1ΔL1 |
| pLG361 | pRS316 | PBNR1-3GFP-BNR1(1-1398, 2200-4128), 3GFP-Bnr1ΔL2 |
| pLG367 | pRS313 | PBNR1-3GFP-BNR1(1-1398, 2200-4128), 3GFP-Bnr1ΔL2 |
All plasmids were generated in this study.
To make ABY2097, ABY1881 (BNR1-GFP::Kan; Pruyne et al., 2004a) and JGY1326 (CDC3-mCherry::SpHIS5; Iwase et al., 2006) were crossed and sporulated. Segregants were then replica-plated onto SC plates lacking histidine and YPD plates containing G418 to select for ones that have both CDC3-mCherry::SpHIS5 and BNR1-GFP::Kan.
To make ABY2807, ABY1680 (bnr1Δ::Kan, lab collection) and MY10174 (CDC3-mCherry::Kan; Ydenberg and Rose, 2009) were crossed and sporulated. Segregants were then replica-plated onto YPD plates containing G418. Tetrads that produced four live spores, among which two were G418 resistant, were picked. The G418-resistant spores of such tetrads were selected as having both CDC3-mCherry::Kan and bnr1Δ::Kan.
To make the hemagglutinin (HA)-tagged Bnr1p constructs, we modified the pRS316-GAL1-10 promoter plasmid (Liu et al., 1992) by inserting a 3HA coding sequence with an in-frame NheI site in between the MluI and NotI sites and 500-bp BNR1 3′UTR between the NotI and SacI sites. Plasmids for localization experiments were made as follows: the respective sequences described in Table 2 were first amplified by PCR with added NheI and NotI sites and a stop codon 5′ to the NotI site. The products and the modified pRS316-GAL1-10-3HA plasmid were then digested with NheI and NotI and ligated.
The pRS316-GAL1-10-3HA plasmid was further modified to make green fluorescent protein (GFP)-tagged constructs. To make pRS316-GAL1-10-GFP or mCherry plasmid, the 3HA tag in pRS316-GAL1-10-3HA plasmid was replaced by a GFP or mCherry coding sequence flanked by MluI and NheI sites. To make pRS316-BNR1 promoter-3GFP plasmid, the GAL1-10 promoter (flanked by ApaI and BamHI sites) and a 3HA tag in pRS316-GAL1-10-3HA plasmid was replaced by 500-bp BNR1 promoter (flanked by ApaI and BamHI sites) and 3GFP coding sequence (flanked by MluI and NheI sites), respectively. Plasmids for localization experiments were made as follows: the respective sequences described in Table 2 were first amplified by PCR with added NheI and NotI sites and a stop codon 5′ to the NotI site. The products and the modified pRS316-GAL1-10-3HA plasmid were then digested with NheI and NotI and ligated.
Light Microscopy
Immunofluorescence microscopy with mouse anti-HA mAb (Roche, Indianapolis, IN) was performed as described (Evangelista et al., 2002). Live cell imaging samples were prepared by spotting cells onto slides covered with 2% agarose media.
Images were acquired with a Nikon Eclipse TE-2000U microscope (Melville, NY) on a spinning disk confocal imaging system (UltraView LCI, Perkin Elmer, Norwalk, CT) using a Nikon 100× 1.4 NA lens and digital camera (C4742-95–12ERG; Hamamatsu, Bridgewater, NJ) or with a Intelligent Imaging Innovations (3I, Denver, CO) spinning disk confocal imaging system (CSU-X;Yokogawa, Tokyo, Japan) on a Leica DMI6000B microscope (Deerfield, IL) using a 63× 1.4 NA lens and QuantEM EMCCD digital camera. Differential interference contrast (DIC) pictures were acquired with a Zeiss Axiovert 100TV microscope using a Fluar 100× 1.30 NA oil lens and digital camera (C4742-95-12ERG; Hamamatsu). Pictures were processed with ImageJ (NIH; http://rsb.info.nih.gov/ij/).
For the fluorescence recovery after photobleaching (FRAP) measurements, cells were immobilized on concanavalin A (ConA)-coated coverslips (0.5 mg/ml ConA for 15 min, washed with growth media three times and air-dried) and examined in the Leica spinning disk confocal microscope described above. Four prebleach images were acquired 5 s apart, and then half of the bud neck–localized GFP-tagged formin was bleached for 500 ms using a Photonic Instruments (St. Charles, IL) Mosaic FRAP system powered by 488-nm light provided by an argon laser. Fluorescence recovery and fluorescence loss from the unbleached part of the neck-localized GFP-formin in the same cell were documented every 5 s by a 100-ms exposure for 10 min. Background subtraction was done automatically by the SlideBook imaging program (Intelligent Imaging Innovations), and data were adjusted for bleaching based on images of an adjacent unbleached cell.
RESULTS
Bnr1p Has Two Distinct Localization Regions
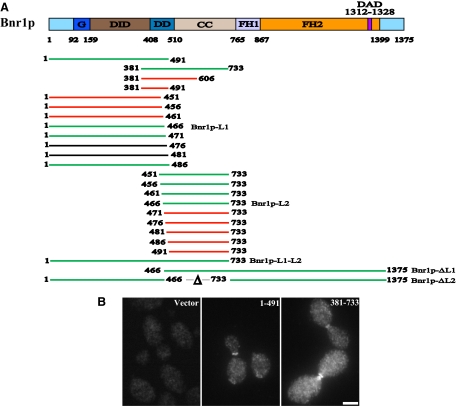
Earlier studies have shown that Bnr1p residues 35-500 are necessary and sufficient to localize Bnr1p to the bud neck (Kikyo et al., 1999). Consistent with this result, we found that overexpressed 3HA-Bnr1p-1-491 localizes to the bud neck (Figure 1, A and B). In previous studies, we reported that Bnr1p-388-1375 also localizes at the bud neck (Gao and Bretscher, 2008), whereas the C-terminal half, Bnr1p-746-1375 containing just the formin homology domains 1 and 2 (FH1 and FH2, respectively) to the C-terminus does not (Gao and Bretscher, 2009). Results from these two C-terminal constructs suggest that Bnr1p-388-746 should contain a localization signal. We made a construct to overexpress 3HA-tagged Bnr1p residues 381-733, and as expected, it exhibited bud neck localization (Figure 1, A and B). Therefore Bnr1p is localized by regions in its N-terminal half. Considering that both Bnr1p-1-491 and 381-733 localize to the bud neck, two possibilities arise: either 1) the localization signal resides in a small region, i.e., residues 381-491 or 2) there are two localization signals in the N-terminal half of Bnr1p. First we found that 3HA-tagged Bnr1p residues 381-491 expressed at high levels does not show bud neck localization (Figure 2), suggesting that the first possibility may not be correct. To test the second possibility, we made a series of deletion mutants progressively removing five amino acids from the C-terminus of the Bnr1p-1-491 construct down to Bnr1p-1-451 and progressively from the N-terminus of the Bnr1p- 451-733 construct down to Bnr1p 491-733. Analysis of the localization of all these constructs revealed that those containing either Bnr1p residues 1-466 or 466-733 localized to the bud neck, whereas Bnr1p-1-461 and 471-733 did not (Figure 2), although proteins of the correct size were expressed (Figure S1). Bnr1p therefore contains two separable localization regions, one we designate as Bnr1p-L1 residing within residues 1-466 and a second Bnr1p-L2 residing within residues 466-733. We attempted to shorten Bnr1p-L2 at the C-terminal end and found that a construct containing residues 381-606 does not localize to the bud neck (Figure 2). Thus Bnr1p-L1 and Bnr1p-L2 require quite long stretches of primary sequence, either for stability of the fragments or folding of domains. Based on limited sequence homology together with structural predictions based on mouse mDia1, the Bnr1p-L1 region is predicted to encompass the GBD and the DID region together with part of the dimerization domain. Bnr1p-L2 encompasses the dimerization domain and a region predicted to form a coiled-coil (Figure 1A).
Figure 1.
Bnr1p localization domains lie in the N-terminal 1-733 region. (A) Domain structures of Bnr1p and diagrams of Bnr1p constructs used in this study. G, GTPase binding domain; CC, coiled-coil domain; DAD, diaphanous autoregulatory domain; DD, dimerization domain; DID, diaphanous inhibitory domain; FH1 and FH2, formin homology domains 1 and 2; L1, Bnr1p-L1, localization region 1; Bnr1p-L2, Bnr1p localization region 2. Constructs that are localized at the bud neck are in green, constructs that are not localized at the bud neck are in red, and constructs that are not detected by immunoblotting are in black. (B) The N-terminal domain localizes Bnr1p to the bud neck. Localization of 3HA-Bnr1p-1-491 and 381-733 (under GAL1 promoter and induced with galactose for 2–3 h), as visualized by immunofluorescence with antibody to the HA-tag. Bar, 2 μm.
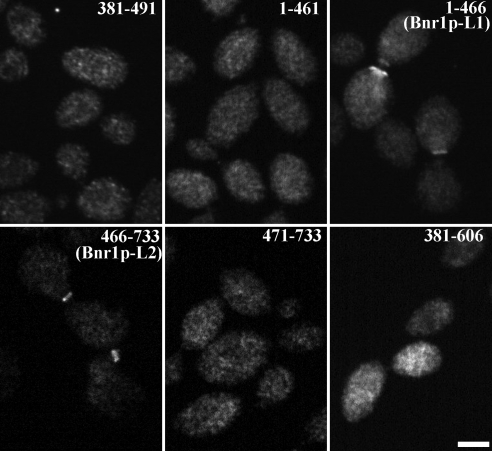
Figure 2.
Bnr1p has two distinct localization domains: Bnr1p-L1 and Bnr1p-L2. Localization of HA-Bnr1p-381-491, 1-461, 1-466 (Bnr1p-L1), 466-733 (Bnr1p-L2), 471-733 and 381-606 (under GAL1 promoter and induced with galactose for 2–3 h), as visualized by immunofluorescence with antibody to the HA-tag. Bar, 2 μm.
The Localizations of Bnr1p-L1 and Bnr1p-L2 Are Independent of Endogenous Bnr1p, But Depend on Septins
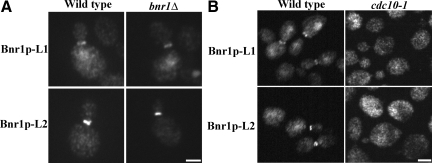
Using two-hybrid assays, we found that Bnr1p residues 1-491 and 446–773 self-interact, but did not interact with each other (not shown), suggesting that the N-terminal half of Bnr1p has two regions that self-interact in a parallel way. To see if the localization of Bnr1p-L1 or Bnr1p-L2 required dimerization with endogenous Bnr1p, we examined their localization in a bnr1Δ background. Their localization to the bud neck was unaffected (Figure 3A), thereby demonstrating that they can each localize independent of endogenous Bnr1p.
Figure 3.
The localization domains are not dependent on endogenous Bnr1p, but do depend on septins. (A) Localization of 3HA-Bnr1p-L1 and 3HA-Bnr1p-L2 (under GAL1 promoter and induced with galactose for 2–3 h) in wild-type and bnr1Δ cells, as visualized by immunofluorescence with antibody to the HA-tag. Bar, 2 μm. (B) Localization of 3HA-Bnr1p-L1 and 3HA-Bnr1p-L2 (under GAL1 promoter and induced with galactose for 2–3 h) in wild type and cdc10–1 cells (1 h at 35°C), as visualized by immunofluorescence with antibody to the HA-tag. Bar, 2 μm.
It has been shown that the bud neck localization of Bnr1p depends on septins (Kikyo et al., 1999; Pruyne et al., 2004a). We therefore examined their localization in cells harboring the conditional septin cdc10-1 mutation after shifting to the restrictive temperature for one hour. Both Bnr1p-L1 and Bnr1p-L2 showed a cytoplasmic distribution under these conditions, and so their localization is dependent on septins (Figure 3B). Bnr1p-L1 and Bnr1p-L2 both localized with septins when the nonessential septin subunit Shs1p was deleted (Figure S2).
Bnr1p-L1 and Bnr1p-L2 Exhibit Different Localization Patterns at the Bud Neck
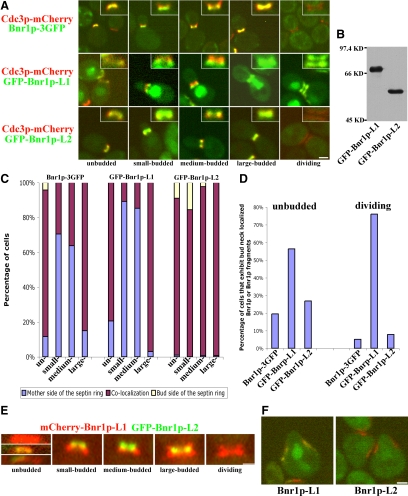
Septins recruit many factors to the bud neck that associate with specific regions of the septin ring or collar. Bnr1p has been reported to localize at the mother side of the septin ring and leaves the bud neck when the septin ring splits before cytokinesis (Gladfelter et al., 2001; Buttery et al., 2007). On close examination utilizing cells that express Cdc3p-mCherry and Bnr1p-GFP, we found that Bnr1p's localization at the bud neck has additional features during the cell cycle. In most unbudded cells, Bnr1p is not localized. When it is localized, Bnr1p colocalizes with septins at the future bud site. In small- to medium-budded cells, Bnr1p is present on the mother side of the septin ring. In large-budded cells, Bnr1p colocalizes with septins again and then, as reported, leaves the bud neck and is not localized when the septin ring splits (Figure 4, A and C).
Figure 4.
Overexpressed GFP-Bnr1p-L1 and GFP-Bnr1p-L2 localize differently at the bud neck and in shmooing cells. (A) Localization of Bnr1p-GFP expressed from the BNR1 promoter and GFP-Bnr1p-L1 or GFP-Bnr1p-L2 expressed from the GAL1 promoter (after 2–3-h galactose induction) in cells expressing Cdc3p-mCherry through the cell cycle. On the top-right corner of each micrograph shows the enlargement of the neck region. (B) Equal amount of total lysates of strains overexpressing GFP-Bnr1p-L1 or GFP-Bnr1p-L2 were blotted with antibodies to GFP. (C) Quantitation of the localization of endogenous Bnr1p-GFP or overexpressed GFP-Bnr1p-L1 or GFP-Bnr1p-L2 with respect to septins through the cell cycle. (D) In unbudded or dividing cells where septins are present, percentage of cells that show colocalized Bnr1p-GFP or overexpressed Bnr1p-L1 or Bnr1p-L2. The data for C and D were obtained from one representative experiment with n ≥ 700 for each strain. (E) Localization of mCherry-Bnr1p-L1 and GFP-Bnr1p-L2 expressed from the GAL1 promoter, after a 2–3-h galactose induction. (F) Localization of GFP-Bnr1p-L1 or GFP-Bnr1p-L2 expressed from the GAL1 promoter in shmooing cells expressing Cdc3p-mCherry. Cells were induced with galactose for 2–3 h and treated with α-factor for 2 h.
In cells prepared for immunofluorescence microscopy, fixation very often causes an artificial breakdown of the hourglass-shaped structure of the septins. Therefore we made constructs to visualize GFP-Bnr1p-L1 and GFP-Bnr1p-L2 in living cells also expressing the septin reporter Cdc3p-mCherry (Figure 4B). Overexpressed GFP-Bnr1p-L1 shows a nuclear staining that does not interfere with its bud neck localization. GFP-Bnr1p-L1 colocalizes with septins in unbudded cells, localizes to the mother side of the septin ring in small- to medium- budded cells, colocalizes with septins again in large budded cells and with the split septin rings (Figure 4, A and C). Thus, GFP-Bnr1p-L1 differs from full-length Bnr1p-GFP in that it is much better localized in unbudded cells and, unlike Bnr1p-GFP, localizes with the split septin rings in dividing cells (Figure 4D). When localized, GFP-Bnr1p-L2 colocalizes with septin rings at essentially all points of the cell cycle, although it is sometimes enriched on the bud side of the septin ring (Figure 4, A and C). However, like Bnr1p-GFP, GFP-Bnr1p-L2 was mainly delocalized in unbudded cells and disappeared from the bud neck in dividing cells (Figure 4D). Overall, GFP-Bnr1p-L1 reflects the selective localization pattern of full-length Bnr1p, but not its cell cycle timing, whereas GFP-Bnr1p-L2 associates with septins in a seemingly unrestricted manner, but with timing reflective of Bnr1p-GFP (Figure 4, C and D).
The different localization patterns of Bnr1p-L1 and Bnr1p-L2 are especially evident in cells expressing both mCherry-Bnr1p-L1 and GFP-Bnr1p-L2. In many unbudded cells, GFP-Bnr1p-L2 was not localized, whereas mCherry-Bnr1p-L1 was. When they are both present, GFP-Bnr1p-L1 and mCherry-Bnr1p-L2 either colocalized at the neck or localized next to each other. In small- to medium-budded cells, GFP-Bnr1p-L1 was clearly closer to the mother cell than mCherry-Bnr1p-L2. In large-budded cells, they colocalized in the middle of the bud neck. In dividing cells, although GFP-Bnr1p-L1 was present as two rings, mCherry-Bnr1p-L2 disappeared from the bud neck (Figure 4E).
Septins reorganize during mating factor–induced shmoo formation to a collar at the shmoo base, so they provide another situation in which to examine the localization of GFP-Bnr1p-L1 and GFP-Bnr1p-L2. Overexpressed GFP-Bnr1p-L1 localized to the septin collar, whereas GFP-Bnr1p-L2 did not (Figure 4F). The combined results show that Bnr1p-L1 shows a spatial localization similar to Bnr1p, whereas Bnr1p-L2 shows a distinctly different, but nevertheless septin-dependent, localization.
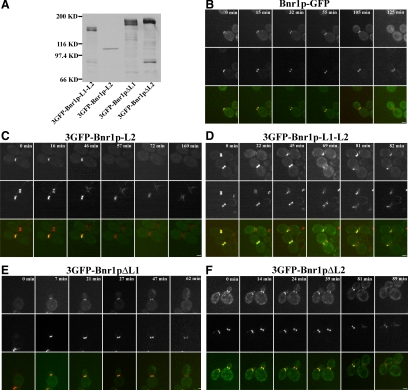
Our results reveal that short-term overexpressed Bnr1p-L1 and Bnr1p-L2 show differences in localization and timing. To explore whether expression of these localization domains is the same when present at endogenous levels, we made constructs expressing 3GFP-tagged Bnr1p constructs expressed from the BNR1 promoter (Figure 5A). Unfortunately, 3GFP-Bnr1p-L1 expressed at the endogenous level could not be visualized and 3GFP-Bnr1p-L2 or 3GFP-Bnr1p-L1-L2 could only be seen in a small percentage of cells. Nevertheless, we were able to use live cell imaging to determine where the two constructs localized early and late in the cell cycle with respect to the septin marker Cdc3-mCherry. In control cells, Bnr1p-3GFP disappears from the bud neck when the septin ring splits (Figure 5B, 125 min). Like the overexpressed construct, lower level expression of 3GFP-Bnr1p-L2 disappears from the bud neck when the septins form two rings (Figure 5C, 72 min), just like Bnr1p-GFP. By contrast, 3GFP-Bnr1p-L1-L2 not only colocalizes with septin double rings (Figure 5D, top cell, 22 min; and bottom cell, 69 min), but also stays at the bud neck with septins into the next cell cycle (Figure 5D, top cell, 45 min; and bottom cell, 81 min). Therefore additional regions of Bnr1p outside the Bnr1p-L1-L2 region also normally contribute to the timing of its correct localization.
Figure 5.
Bnr1p domains expressed from the endogenous promoter exhibit similar localization patterns. (A) Western blot with antibodies to GFP to show relative levels of expression of the indicated Bnr1p constructs. (B–F) Localization of Bnr1p-GFP (B), 3GFP-Bnr1p-L2 (C), 3GFP-Bnr1p-L1-L2 (D), 3GFP-Bnr1pΔL1 (E), and 3GFP-Bnr1pΔL2 (F) during the cell cycle. All constructs were expressed from the BNR1 promoter in cells also expressing Cdc3-mCherry. In each panel, the top row shows Bnr1p or Bnr1p constructs, the middle row shows Cdc3p-mCherry and the bottom row shows the merged images. The numbers indicate time points in minutes.
In addition to examining the localization of Bnr1p-L1 and Bnr1p-L2, we also investigated the localization of full-length Bnr1p constructs expressed from the endogeneous BNR1 promoter but lacking either one of the two localization domains. 3GFP-Bnr1pΔL1, retaining the L2 domain, colocalized with the septin marker, and disappearing when the septin ring split; i.e., it exhibited the same localization pattern as Bnr1p-L2 (Figure 5E). 3GFP-Bnr1pΔL2, retaining the L1 domain, was enriched on the mother side of the bud and therefore exhibited a localization pattern very similar to Bnr1p-L1, except that it did not localize very well to double septin rings (Figure 5F, left cell, 89 min). Thus, combining the results of the isolated localization domains with full-length protein lacking the specific domains provides a generally consistent view of how they contribute to localization of full-length Bnr1p.
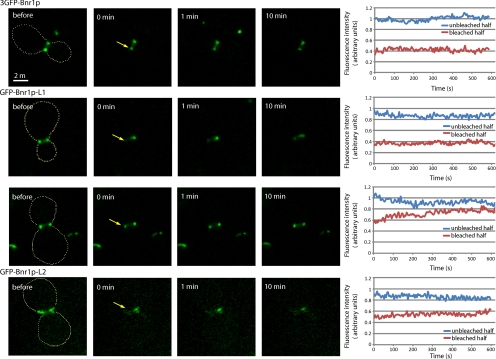
Bnr1p-L1 and Bnr1p-L2 Each Provide a Stable Association with the Bud Neck
Bnr1p-GFP has been shown by photobleaching studies to be stably associated with the bud neck (Buttery et al., 2007). We therefore examined the dynamics of 3GFP-Bnr1p and constructs lacking the specific localization domains, namely, GFP-Bnr1pΔL1 and GFP-Bnr1pΔL2 expressed in a bnr1Δ strain. To obtain sufficient signal, the constructs were expressed from the GAL1 promoter, and the cells were induced by growth in galactose for 4 hours. Cells expressing the different constructs were subject to a FRAP protocol where one-half of the bud neck–localized protein was bleached, and then the fluorescence of the unbleached half and the recovery of the bleached half was followed. Using this approach, bleached full-length 3GFP-Bnr1p fails to recover, and the unbleached region is stable, over a period of 10 min, consistent with previous studies (Figure 6A). Both GFP-Bnr1pΔL1 (localized by the L2 domain) and GFP-Bnr1pΔL2 (localized by the L1 domain) were also relatively stable. However, subtle differences were seen in GFP-Bnr1pΔL1 between small- and large-budded cells. For medium- to large-budded cells, a slow recovery was generally seen, whereas for small-budded cells, essentially no recovery was seen (Figure 6, B and C). In cells expressing GFP-Bnr1pΔL2, no significant recovery was ever seen (Figure 6D). Therefore, although some modest recovery can sometimes be seen for Bnr1pΔL1, overall, both the Bnr1p-L1 and Bnr1p-L2 regions provide stable association with the bud neck.
Figure 6.
Both Bnr1p-L1 and Bnr1p-L2 confer stable association with the bud neck. Cells expressing 3GFP-Bnr1p from the GAL1 promoter (top row) or GFP-Bnr1pΔL1 (second and third rows) expressed from the GAL1 promoter in a small-budded cell (second row) and a large-budded cell (third row) or GFP-Bnr1pΔL2 (bottom row) expressed from the GAL1 promoter were subjected to a FRAP protocol where one side of the bud neck–localized protein was partially bleached by a 500-ms pulse of 488-nm light, and the recovery, or loss from the unbleached side, was recorded every 5 s for 10 min. The panels on the left show time points before and after bleaching, and the graphs on the right show the results of a typical experiment. The arrows point to the region that was bleached. The results were adjusted by subtracting background fluorescence and corrected for fluorescence loss during imaging using a reference unbleached cell in the same field of view. Each photobleaching experiment was repeated at least five times on different cells with similar results.
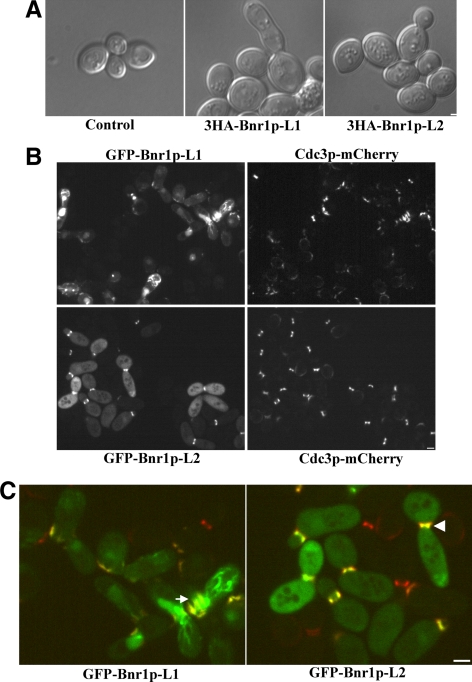
Overexpression of Bnr1p-L1, But Not Bnr1p-L2, Perturbs Septin Organization
Prolonged overexpression of full-length Bnr1p or Bnr1p residues 35-500, the region encompassing Bnr1p-L1, induces a septin mutant-like phenotype with elongated, sometimes unseparated buds (Kikyo et al., 1999). To explore if this phenotype is associated the defined localization domains, we examined the phenotype of cells overexpressing 3HA-Bnr1p-L1 and 3HA-Bnr1p-L2. Consistent with the earlier results, the elongated bud phenotype was seen after prolonged overexpression of 3HA-Bnr1p-L1. However, no morphological changes were seen upon 3HA-Bnr1p-L2 expression (Figure 7A). To investigate if this difference is caused by different effects on septins, we overexpressed GFP-Bnr1p-L1 or GFP-Bnr1p-L2 in cells expressing Cdc3p-mCherry. Prolonged overexpression of GFP-Bnr1p-L1 induced a severe cytokinesis defect in ∼10% of the cells, with elongated and unseparated buds, and the septin structure was greatly disrupted (Figure 7B). Interestingly, the septins were sometimes delocalized to a random location in the cell, and GFP-Bnr1p-L1 mostly colocalized with them (Figure 7C, arrow). By contrast, in cells overexpressing GFP-Bnr1p-L2, both septins and GFP-Bnr1p-L2 are localized at the bud neck normally (Figure 7C, arrowhead). However, overexpressed GFP-Bnr1p-L2, unlike 3HA-Bnr1p-L2, caused slightly elongated buds, for reasons that are not clear. We conclude that overexpression of Bnr1p-L1 has the potential to disrupt septin structure, whereas overexpression of Bnr1p-L2 does not, which explains why Bnr1p-L1 induces a cytokinesis defect.
Figure 7.
Prolonged overexpression of Bnr1p-L1 and Bnr1p-L2 has different phenotypes. (A) DIC micrographs of cells expressing 3HA-Bnr1p-L1 or 3HA-Bnr1p-L2 expressed from the GAL1 promoter after overnight induction with galactose. (B) Cells expressing Cdc3-mCherry and GFP-Bnr1p-L1 or GFP-Bnr1p-L2 expressed from the GAL1 promoter after overnight induction with galactose. (C) Enlargement of a region of the merged pictures in B. White arrow indicates disrupted septin structures, and white arrowhead indicates normal septin structures.
To begin to understand how Bnr1p-L1 and Bnr1p-L2 localize with septins, we tested their interaction with individual septins by two-hybrid analysis. However, we could not detect any interactions (data not shown). It is possible that Bnr1p-L1 or Bnr1p-L2 only interact with assembled septins or through one of the many other proteins found at the bud neck (Gladfelter et al., 2001).
DISCUSSION
Formins play important roles in regulating actin dynamics. Much has been learned about their actin nucleation/elongation activities, but their regulation and especially how they are localized is largely unknown. The existence of multiple localization domains, residing both in the N- and C-terminal halves of the protein, has recently been suggested for the association of formin Cdc12p in fission yeast with the contractile ring, although the N-terminal localization domain has only been examined in the context of endogenous Cdc12p (Yonetani et al., 2008).
We have identified two separate localization regions in the N-terminal half of Bnr1p, Bnr1p-L1 (residues 1-466), and Bnr1p-L2 (residues 466-733), each of which can independently localize to the bud neck in the absence of endogenous Bnr1p. Based on a comparison with the structure of mDia1, Bnr1p-L1 is composed of the GBD, DID, and part of the DD. Bnr1p-L2 consists of the rest of DD and the predicted coiled-coil domain between the DD and the FH1. Based on two-hybrid results, we found that both Bnr1p-1-491 and Bnr1p-446-733 self-interact, suggesting that the N-terminal half of Bnr1p exhibits parallel self-interactions. This is consistent with the suggested domain boundaries based on the mDia1 structure.
The association of Bnr1p-L1 and Bnr1p-L2 with the bud neck depends on septins, but their localizations are slightly different. Bnr1p-L1 shows a spatial distribution similar to full-length Bnr1p by selectively associating with the mother side of the septin ring in small- to medium-budded cells, whereas Bnr1p-L2 appears to associate with septins in a less restricted manner. To investigate whether the defined domains might play a role in the full-length protein, we also examined the localization properties of Bnr1p lacking either the Bnr1p-L1 or Bnr1p-L2 domain. In terms of spatial localization, Bnr1pΔL1 behaved similarly to Bnr1p-L2, and Bnr1pΔL2 behaved similarly to Bnr1p-L1. Therefore, the results examining the spatial localization of the isolated domains seem to reflect their independent roles in full-length Bnr1p and suggest that both domains probably contribute to the bud neck localization of full-length Bnr1p.
The temporal regulation through the cell cycle of Bnr1p-L1 and Bnr1p-L2 localization is different. Although Bnr1p-L2 reflects the temporal regulation of full-length Bnr1p, Bnr1p-L1 arrives early at the site of bud emergence and localizes to the split septin ring, whereas full-length Bnr1p does not. Bnr1p-L1-L2 has also lost temporal regulation, remaining with the split septin ring, reflecting the temporal pattern of Bnr1p-L1. The finding that Bnr1p-L1 and Bnr1p-L1-L2 associate with the split septin ring, whereas Bnr1ΔL2 (retaining L1) and full-length Bnr1p (retaining both L1 and L2) do not shows that the C-terminal region of Bnr1p confers temporal regulation. This part of Bnr1p, consisting of the FH1-FH2 to the C-terminus and containing the DAD sequence, does not localize to the bud neck itself and so must regulate the accessibility of the Bnr1p-L1 region. It is very likely that before bud emergence and at cytokinesis, full-length Bnr1p is in its dormant form, which not only inhibits its activity, but also its localization. A similar conformational regulation pattern has been shown for FRLα where Cdc42 regulates both nucleation activity and plasma membrane association (Seth et al., 2006). How this timing is achieved—either through structural changes in Bnr1p and septins or some other regulatory processes—remains a fascinating question.
Septins are involved in multiple cellular processes, most of which are achieved through serving as a scaffold to recruit proteins that function in these processes (Gladfelter et al., 2001). Clearly, septins perform their role in polarized growth by recruiting Bnr1p, which nucleates actin cable assembly. The transition of septin structures from a ring, to an hourglass, and then to split rings correlates with cell cycle progression. The localization of Bnr1p is consistent with its function. In small- to medium-budded cells, Bnr1p assembles actin cables that extend into the mother cell, facilitated by its localization at the mother side of the septin ring. Later it might move to the center of the septin ring to contribute to actomyosin ring formation. Based on our results, it is possible that Bnr1p is first recruited by septins through Bnr1p-L1 to localize to the mother side of septins, and then through the interaction of Bnr1p-L2 with unknown septin-interacting proteins, Bnr1p is gradually shifted to the middle of the septins.
The interactions of Bnr1p-L1 and Bnr1p-L2 with the bud neck are clearly different. Not only do they show differences in localization, they induce different phenotypes after prolonged overexpression. Cells overexpressing Bnr1p-L1 are dramatically elongated and have a cytokinesis defect. More importantly, septin structure is disrupted in such cells and sometimes localizes to a random location in the cell. In contrast, cells overexpressing Bnr1p-L2 have normal bud neck–localized septins. Although there are more than 20 bud neck–localized proteins whose localization depends on septins (Gladfelter et al., 2001), only a few of them have been reported to affect septin organization. Mutations in the Gin4p and Elm1p kinases lead to septin bars at the bud neck (Longtine et al., 1998; Bouquin et al., 2000), and overexpression of Bud3p or Cdc5p results in ectopic septin structures (Lord et al., 2000; Song et al., 2000). Prolonged overexpression of Bnr1p-L1 induces septin bars, suggesting that the function of one of the kinases may be disrupted. However, it also leads to severely mislocalized septins, which has only been reported in actin-overexpressing cells (Norden et al., 2004). The connection here is not clear, because Bnr1p-L1 does not contain the actin-nucleating FH1-FH2 domains.
We did not see any evident differences in the actin cytoskeleton between cells expressing Bnr1p-ΔL1 or Bnr1p-ΔL2 as their sole formin (Figure S3). This is not unexpected as cells with only a nonlocalized formin grow well and have a partially polarized actin cytoskeleton (Gao and Bretscher, 2009). It would be beneficial to identify synthetic lethal interactions with BNR1 other than BNI1 and to examine which one of the two localization domains is needed.
The localization of Bnr1p-L1 and Bnr1p-L2 at the bud neck both depend on septins, and both seem to contribute to the stable association of Bnr1p with the bud neck (Buttery et al., 2007), as FRAP analysis on constructs lacking either Bnr1p-L1 or Bnr1p-L2 reveal that they remain stably associated there. However, using two-hybrid approaches no direct interaction has been detected between Bnr1p and septins (Kikyo et al., 1999) or between Bnr1p-L1 or Bnr1p-L2 and the septin subunits (data not shown). This may be because Bnr1p-L1 and Bnr1p-L2 might only interact with assembled septin structures. With the identification of two distinct localization domains in Bnr1p, the next challenge will be to determine whether the interaction with septins is direct or indirect and exactly how this association is mediated and regulated.
Supplementary Material
ACKNOWLEDGMENTS
We thank Erfei Bi (University of Pennsylvania), Mark Rose (Princeton University), and John Pringle (Stanford University) for providing strains and plasmids that made this study possible. This work was supported by Public Health Service Grant GM39066.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-10-0861) on February 10, 2010.
REFERENCES
- Alberts A. S. Identification of a carboxyl-terminal Diaphanous-related formin homology protein autoregulatory domain. J. Biol. Chem. 2001;276:2824–2830. doi: 10.1074/jbc.M006205200. [DOI] [PubMed] [Google Scholar]
- Bertin A., McMurray M. A., Grob P., Park S. S., Garcia G., 3rd, Patanwala I., Ng H. L., Alber T., Thorner J., Nogales E. Saccharomyces cerevisiae septins: supramolecular organization of heterooligomers and the mechanism of filament assembly. Proc. Natl. Acad. Sci. USA. 2008;105:8274–8279. doi: 10.1073/pnas.0803330105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouquin N., Barral Y., Courbeyrette R., Blondel M., Snyder M., Mann C. Regulation of cytokinesis by the Elm1 protein kinase in Saccharomyces cerevisiae. J. Cell Sci. 2000;113(Pt 8):1435–1445. doi: 10.1242/jcs.113.8.1435. [DOI] [PubMed] [Google Scholar]
- Buttery S. M., Yoshida S., Pellman D. Yeast formins Bni1 and Bnr1 utilize different modes of cortical interaction during the assembly of actin cables. Mol. Biol. Cell. 2007;18:1826–1838. doi: 10.1091/mbc.E06-09-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F., Drubin D., Nurse P. cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. J. Cell Biol. 1997;137:169–182. doi: 10.1083/jcb.137.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista M., Pruyne D., Amberg D. C., Boone C., Bretscher A. Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat. Cell Biol. 2002;4:32–41. doi: 10.1038/ncb718. [DOI] [PubMed] [Google Scholar]
- Fujiwara T., Tanaka K., Mino A., Kikyo M., Takahashi K., Shimizu K., Takai Y. Rho1p-Bni1p-Spa2p interactions: implication in localization of Bni1p at the bud site and regulation of the actin cytoskeleton in Saccharomyces cerevisiae. Mol. Biol. Cell. 1998;9:1221–1233. doi: 10.1091/mbc.9.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L., Bretscher A. Analysis of unregulated formin activity reveals how yeast can balance F-actin assembly between different microfilament-based organizations. Mol. Biol. Cell. 2008;19:1474–1484. doi: 10.1091/mbc.E07-05-0520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L., Bretscher A. Polarized growth in budding yeast in the absence of a localized formin. Mol. Biol. Cell. 2009;20:2540–2548. doi: 10.1091/mbc.E09-03-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladfelter A. S., Pringle J. R., Lew D. J. The septin cortex at the yeast mother-bud neck. Curr. Opin. Microbiol. 2001;4:681–689. doi: 10.1016/s1369-5274(01)00269-7. [DOI] [PubMed] [Google Scholar]
- Goode B. L., Eck M. J. Mechanism and function of formins in the control of actin assembly. Annu. Rev. Biochem. 2007;76:593–627. doi: 10.1146/annurev.biochem.75.103004.142647. [DOI] [PubMed] [Google Scholar]
- Higgs H. N., Peterson K. J. Phylogenetic analysis of the formin homology 2 domain. Mol. Biol. Cell. 2005;16:1–13. doi: 10.1091/mbc.E04-07-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase M., Luo J., Nagaraj S., Longtine M., Kim H. B., Haarer B. K., Caruso C., Tong Z., Pringle J. R., Bi E. Role of a Cdc42p effector pathway in recruitment of the yeast septins to the presumptive bud site. Mol. Biol. Cell. 2006;17:1110–1125. doi: 10.1091/mbc.E05-08-0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikyo M., Tanaka K., Kamei T., Ozaki K., Fujiwara T., Inoue E., Takita Y., Ohya Y., Takai Y. An FH domain-containing Bnr1p is a multifunctional protein interacting with a variety of cytoskeletal proteins in Saccharomyces cerevisiae. Oncogene. 1999;18:7046–7054. doi: 10.1038/sj.onc.1203184. [DOI] [PubMed] [Google Scholar]
- Kobielak A., Pasolli H. A., Fuchs E. Mammalian formin-1 participates in adherens junctions and polymerization of linear actin cables. Nat. Cell Biol. 2004;6:21–30. doi: 10.1038/ncb1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew D. J. The morphogenesis checkpoint: how yeast cells watch their figures. Curr. Opin. Cell Biol. 2003;15:648–653. doi: 10.1016/j.ceb.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Li F., Higgs H. N. Dissecting requirements for auto-inhibition of actin nucleation by the formin, mDia1. J. Biol. Chem. 2005;280:6986–6992. doi: 10.1074/jbc.M411605200. [DOI] [PubMed] [Google Scholar]
- Liu H., Krizek J., Bretscher A. Construction of a GAL1-regulated yeast cDNA expression library and its application to the identification of genes whose overexpression causes lethality in yeast. Genetics. 1992;132:665–673. doi: 10.1093/genetics/132.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., Fares H., Pringle J. R. Role of the yeast Gin4p protein kinase in septin assembly and the relationship between septin assembly and septin function. J. Cell Biol. 1998;143:719–736. doi: 10.1083/jcb.143.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord M., Yang M. C., Mischke M., Chant J. Cell cycle programs of gene expression control morphogenetic protein localization. J. Cell Biol. 2000;151:1501–1512. doi: 10.1083/jcb.151.7.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezami A. G., Poy F., Eck M. J. Structure of the autoinhibitory switch in formin mDia1. Structure. 2006;14:257–263. doi: 10.1016/j.str.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Norden C., Liakopoulos D., Barral Y. Dissection of septin actin interactions using actin overexpression in Saccharomyces cerevisiae. Mol. Microbiol. 2004;53:469–483. doi: 10.1111/j.1365-2958.2004.04148.x. [DOI] [PubMed] [Google Scholar]
- Otomo T., Otomo C., Tomchick D. R., Machius M., Rosen M. K. Structural basis of Rho GTPase-mediated activation of the formin mDia1. Mol. Cell. 2005;18:273–281. doi: 10.1016/j.molcel.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Ozaki-Kuroda K., Yamamoto Y., Nohara H., Kinoshita M., Fujiwara T., Irie K., Takai Y. Dynamic localization and function of Bni1p at the sites of directed growth in Saccharomyces cerevisiae. Mol. Cell. Biol. 2001;21:827–839. doi: 10.1128/MCB.21.3.827-839.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrin S., Mellor H. The Rho family GTPase Rif induces filopodia through mDia2. Curr. Biol. 2005;15:129–133. doi: 10.1016/j.cub.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Pruyne D., Evangelista M., Yang C., Bi E., Zigmond S., Bretscher A., Boone C. Role of formins in actin assembly: nucleation and barbed end association. Science. 2002;297:612–615. doi: 10.1126/science.1072309. [DOI] [PubMed] [Google Scholar]
- Pruyne D., Gao L., Bi E., Bretscher A. Stable and dynamic axes of polarity use distinct formin isoforms in budding yeast. Mol. Biol. Cell. 2004a;15:4971–4989. doi: 10.1091/mbc.E04-04-0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne D., Legesse-Miller A., Gao L., Dong Y., Bretscher A. Mechanisms of polarized growth and organelle segregation in yeast. Annu. Rev. Cell Dev. Biol. 2004b;20:559–591. doi: 10.1146/annurev.cellbio.20.010403.103108. [DOI] [PubMed] [Google Scholar]
- Rose R., Weyand M., Lammers M., Ishizaki T., Ahmadian M. R., Wittinghofer A. Structural and mechanistic insights into the interaction between Rho and mammalian Dia. Nature. 2005;435:513–518. doi: 10.1038/nature03604. [DOI] [PubMed] [Google Scholar]
- Sagot I., Rodal A. A., Moseley J., Goode B. L., Pellman D. An actin nucleation mechanism mediated by Bni1 and profilin. Nat. Cell Biol. 2002;4:626–631. doi: 10.1038/ncb834. [DOI] [PubMed] [Google Scholar]
- Seth A., Otomo C., Rosen M. K. Autoinhibition regulates cellular localization and actin assembly activity of the diaphanous-related formins FRLalpha and mDia1. J. Cell Biol. 2006;174:701–713. doi: 10.1083/jcb.200605006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Grenfell T. Z., Garfield S., Erikson R. L., Lee K. S. Essential function of the polo box of Cdc5 in subcellular localization and induction of cytokinetic structures. Mol. Cell. Biol. 2000;20:286–298. doi: 10.1128/mcb.20.1.286-298.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N., Kato T., Fujita A., Ishizaki T., Narumiya S. Cooperation between mDia1 and ROCK in Rho-induced actin reorganization. Nat. Cell Biol. 1999;1:136–143. doi: 10.1038/11056. [DOI] [PubMed] [Google Scholar]
- Ydenberg C. A., Rose M. D. Antagonistic regulation of Fus2p nuclear localization by pheromone signaling and the cell cycle. J. Cell Biol. 2009;184:409–422. doi: 10.1083/jcb.200809066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonetani A., Lustig R. J., Moseley J. B., Takeda T., Goode B. L., Chang F. Regulation and targeting of the fission yeast formin cdc12p in cytokinesis. Mol. Biol. Cell. 2008;19:2208–2219. doi: 10.1091/mbc.E07-07-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.