In plants and mammals, the PEX7 PTS2 receptor relies on the PEX5 PTS1 receptor to shuttle proteins bearing peroxisome-targeting signals into peroxisomes. This study reveals that Arabidopsis pex7 mutants display defects not only in PTS2 import, but also in PEX5 accumulation and PTS1 import. Thus, PEX5 and PEX7 are interdependent in plants.
Abstract
Peroxisomes compartmentalize certain metabolic reactions critical to plant and animal development. The import of proteins from the cytosol into the organelle matrix depends on more than a dozen peroxin (PEX) proteins, with PEX5 and PEX7 serving as receptors that shuttle proteins bearing one of two peroxisome-targeting signals (PTSs) into the organelle. PEX5 is the PTS1 receptor; PEX7 is the PTS2 receptor. In plants and mammals, PEX7 depends on PEX5 binding to deliver PTS2 cargo into the peroxisome. In this study, we characterized a pex7 missense mutation, pex7-2, that disrupts both PEX7 cargo binding and PEX7-PEX5 interactions in yeast, as well as PEX7 protein accumulation in plants. We examined localization of peroxisomally targeted green fluorescent protein derivatives in light-grown pex7 mutants and observed not only the expected defects in PTS2 protein import but also defects in PTS1 import. These PTS1 import defects were accompanied by reduced PEX5 accumulation in light-grown pex7 seedlings. Our data suggest that PEX5 and PTS1 import depend on the PTS2 receptor PEX7 in Arabidopsis and that the environment may influence this dependence. These data advance our understanding of the biogenesis of these essential organelles and provide a possible rationale for the retention of the PTS2 pathway in some organisms.
INTRODUCTION
Because peroxisomes lack DNA, all necessary proteins are encoded in the nucleus and imported posttranslationally from the cytosol. There are two well-characterized signals that can direct proteins to enter the peroxisome matrix. The peroxisome-targeting signal (PTS) 1 consists of tripeptide variants at the extreme C terminus, and the PTS2 is a nonapeptide within the first ∼30 amino acids of the protein (reviewed in Brown and Baker, 2008). PTS2 proteins comprise approximately a quarter of plant peroxisomal proteins (Kamada et al., 2003; Reumann et al., 2004), including several enzymes necessary for fatty acid β-oxidation that are required for normal seedling development (Hayashi et al., 1998; Hooks et al., 1999; Eastmond et al., 2000; Adham et al., 2005; Pracharoenwattana et al., 2005, 2007).
Two PTS receptors bind and escort cargo proteins from the cytosol to the peroxisome for import: peroxin (PEX) 5 and PEX7, which recognize PTS1- (Brocard et al., 1994; Dodt et al., 1995; Fransen et al., 1995) and PTS2-containing proteins (Rehling et al., 1996), respectively. Receptors and cargo are translocated into the peroxisome, the cargo dissociates, and the receptors are recycled back to the cytosol where they can undergo further rounds of import (Dammai and Subramani, 2001; Nair et al., 2004; Brown and Baker, 2008). In plants and vertebrates, PTS2 proteins such as 3-ketoacyl-CoA thiolase (thiolase) can be processed into a mature form by removal of the N-terminal PTS2 recognition sequence upon entrance into the peroxisome (Brown and Baker, 2008).
Dissecting interactions among peroxins from different species has revealed differences in PEX5 and PEX7 docking with the PEX13 and PEX14 membrane peroxins (Brown and Baker, 2008). Some of the receptor docking differences between mammals and yeast reflect the existence in mammals of long (L) and short (S) isoforms of PEX5 that result from alternative splicing; both isoforms can bind the PTS1, but only PEX5L binds PEX7 and functions in PTS2 import (Braverman et al., 1998; Otera et al., 1998). Arabidopsis seems to only encode a PEX5L isoform, which is required for both PTS1 and PTS2 import (Hayashi et al., 2005; Woodward and Bartel, 2005). Like humans, rice has two PEX5 splice isoforms, and only the longer isoform binds PEX7 (Lee et al., 2006). Fungal PEX5 resembles PEX5S; PEX7 is bound by other fungal peroxins that serve the role in PTS2 import that is served by PEX5L in other organisms (Brown and Baker, 2008). Interestingly, the nematode Caenorhabditis elegans lacks PEX7 and the corresponding PTS2 import pathway; proteins that contain a PTS2 in other organisms are PTS1-targeted in C. elegans (Gurvitz et al., 2000; Motley et al., 2000).
PEX7 and PTS2 import are dependent on the PTS1 receptor PEX5 in many multicellular organisms. In this study, we provide evidence that the converse is also true in plants. We have characterized a new Arabidopsis pex7 allele, pex7-2, that displays defects not only in PEX7 accumulation and PTS2 import but also in PEX5 accumulation and PTS1 import. Our results support the hypothesis that PEX5 and PEX7 are interdependent in plants.
MATERIALS AND METHODS
Plant Material and Growth Conditions
All Arabidopsis thaliana plants were in the Columbia (Col-0) accession. pex7-2 was isolated as an indole-3-butyric acid (IBA)-response mutant from progeny of ethylmethanesulfonate-mutagenized Col-0 seeds (Lehle Seeds, Round Rock, TX) as described previously (Adham et al., 2005; Zolman et al., 2008) and was backcrossed once before phenotypic analyses. The pex7-1 mutant contains a T-DNA inserted 95 base pairs upstream of the start codon in the PEX7 5′-untranslated region (UTR) and has reduced PEX7 mRNA levels (Woodward and Bartel, 2005), pex5-10 contains a T-DNA inserted in PEX5 exon 5 (Zolman et al., 2005), and pex5-1 is a missense allele (Zolman et al., 2000).
For phenotypic assays, seeds were surface sterilized, stratified as indicated, and plated on plant nutrient media (Haughn and Somerville, 1986) supplemented with 0.5% sucrose or hormones (from ethanol-based stocks) as indicated. The volume of added ethanol was normalized for hormone-supplemented medium and unsupplemented controls. Seedlings were grown at 22°C in the dark or under continuous white or yellow-filtered light (Stasinopoulos and Hangarter, 1990), as indicated. Dark-grown plants were grown horizontally for 1 d under white light then transferred to the dark and incubated on vertically orientated plates; light-grown plants were incubated on horizontally oriented plates.
Map-based Cloning
DNA was isolated from IBA-resistant plants selected from F2 progeny of pex7-2 crossed to Ler tt4. Mapping with polymerase chain reaction (PCR)-based molecular markers (Supplemental Table 1) localized the lesion to chromosome 1 between F6N18 and F28N24. The At1g29260 gene was PCR amplified from genomic DNA prepared from the mutant and sequenced (LoneStar Labs, Houston, TX).
The PEX7 cDNA was driven by the cauliflower mosaic virus 35S promoter by subcloning the SalI/NotI insert of pCR4-PEX7 (see below) into XhoI/NotI-cut 35SpBARN (LeClere and Bartel, 2001) to give 35S-PEX7a, which was transformed into pex7-2 by using the floral dip method (Clough and Bent, 1998). T1 plants were selected on 7.5 μg/ml glufosinate ammonium, and homozygous plants were selected from subsequent generations by analyzing the pattern of seedling glufosinate ammonium resistance.
Immunoblot Analysis
Seedling protein was extracted by grinding tissue on dry ice and then mixing with an equal volume of NuPAGE 2× loading buffer (Invitrogen, Carlsbad, CA). After centrifugation, supernatants were moved to fresh tubes with 1/10 volume of 0.5 M dithiothreitol (DTT) and then boiled for 5 min. Yeast protein was extracted from 5-ml cultures grown at 30°C to an OD of 0.8–1.0 by resuspending cells in two volumes of NuPAGE 2× loading buffer and half a volume of glass beads, heating to 100°C for 2.5 min, vortexing on high for 1 min, and then boiling again for 2.5 min. After centrifugation, supernatants were moved to fresh tubes with 1/10 volume of 0.5 M DTT and then boiled for 5 min. Five microliters (yeast) or 10 μl (seedlings) of supernatant was loaded onto NuPAGE 10% Bis-Tris gels (Invitrogen) next to broad-range prestained protein markers (P7708S; New England Biolabs, Beverly, MA) and Cruz Markers (Santa Cruz Biotechnology, Santa Cruz, CA). After electrophoresis, proteins were transferred for 35 min at 24 V to Hybond ECL nitrocellulose membranes (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) by using NuPAGE transfer buffer (Invitrogen). Membranes were blocked with 8% (wt/vol) nonfat dry milk in Tween Tris-buffered saline (Ausubel et al., 1999) and incubated at 4°C with rabbit anti-PEX5 (1:100 dilution; Zolman and Bartel, 2004), rabbit anti-PEX7 (1:800; generated and affinity purified against a peptide corresponding to the C-terminal 17 amino acids of PEX7 by Bethyl Laboratories, Montgomery, TX), rabbit anti-PED1 (1:10,000; Lingard et al., 2009), or rabbit anti-PMDH2 (1:2000; Pracharoenwattana et al., 2007) primary antibodies followed by incubation with a horseradish peroxidase-linked goat anti-rabbit immunoglobulin (Ig)G secondary antibody (sc-2030; Santa Cruz Biotechnology). As a loading control, membranes were incubated with mouse anti-HSC70 (1:5000, SPA-817; Assay Designs, Ann Arbor, MI) followed by horseradish peroxidase-linked anti-mouse IgG secondary antibody (sc-2031; Santa Cruz Biotechnology). Horseradish peroxidase was visualized using LumiGLO (Cell Signaling Technology, Danvers, MA) according to the manufacturer's instructions.
Microscopy
Col-0 expressing 35S-GFP-PTS1 (Zolman and Bartel, 2004) or 35S-PTS2-GFP (Woodward and Bartel, 2005) were crossed to pex7-2 and pex7-1 pex5-1, and lines homozygous for the mutant allele(s) and transgene were selected in subsequent generations. pex5-1 and pex7-1 expressing 35S-GFP-PTS1 or 35S-PTS2-GFP were described previously (Woodward and Bartel, 2005). Confocal images of cotyledon cells were obtained using a multiphoton laser scanning microscope (510 META NLO; Carl Zeiss, Thornwood, NY) equipped with a 63× oil immersion lens. Green fluorescent protein (GFP) and propidium iodide were excited using a 488 nm argon laser. Bandpass emission filters of 500–530 nm and 650–710 nm were used to detect GFP and propidium iodide or chlorophyll fluorescence, respectively.
Yeast Strains and Methods
Inserts for the yeast two-hybrid constructs were fused to the Gal4 DNA-binding domain using the pBI770 “bait” vector or the Gal4 activation domain by using the pBI771 “prey” vector (Kohalmi et al., 1998). All inserts and junctions were verified by sequencing. A PEX5 cDNA with an in-frame SalI site 5′ of the ATG was made using QuikChange site-directed mutagenesis (Stratagene, La Jolla, CA) with the primer PEX5-SalIF (Supplemental Table 2) on a PEX5 cDNA in the NotI site of pBluescript KS (pKS-PEX5; Zolman et al., 2000) to make pKS-PEX5b3. The SalI-Not1 PEX5 fragment from pKS-PEX5b3 was ligated into SalI/Not1-cut pBI771 to make pBI771-PEX5.
PEX7 and pex7-2 cDNAs with an in-frame SalI site 5′ of the ATG and a NotI site immediately downstream of the stop was made by PCR amplifying PEX7 cDNA APZ50H10R (Asamizu et al., 2000) or pex7-2 mutant genomic DNA, respectively, by using Ex-Taq (Takara Bio USA, Madison, WI) with PEX7-SalI-F and PEX7-NotI-R. Amplicons were cloned into pCR4-TOPO (Invitrogen) to give pCR4-PEX7 and pCR4-pex7-2. Inserts from these plasmids were subcloned into SalI/NotI-cut pBI770 to make pBI770-PEX7 and pBI770-pex7-2, respectively.
A PED1 cDNA flanked by an in-frame SalI site 5′ of the ATG and a NotI site immediately 3′ of the stop codon was PCR amplified from PED1 cDNA U09045 (Yamada et al., 2003) by using Triple Master Taq DNA polymerase (Eppendorf North America, New York, NY) with PED1-SalI-F1 and PED1-NotI-R. A truncated PED1 cDNA lacking the PTS2 was similarly amplified using PED1-SalI-F2 and PED1-NotI-R. Amplicons were cloned into pCR4-TOPO to make pCR4-PED1 and pCR4-tPED1. Inserts from these plasmids were subcloned into SalI/NotI-cut pBI771 to make pBI771-PED1 and pBI771-tPED1.
Plasmids were transformed (Gietz and Schiestl, 1995) into the AH109 (MATa, trp1-901, leu2-3, 112, ura3-52, his3-200, gal4Δ, gal80Δ, LYS2::GAL1UAS- GAL1TATA-HIS3, GAL2UAS-GAL2TATA-ADE2, URA3::MEL1UAS-MEL1TATA-lacZ, MEL1; Clontech, Mountain View, CA) Saccharomyces cerevisiae yeast strain. Strains were grown on synthetic complete media lacking Leu and Trp or lacking Leu, Trp, and His and supplemented with 2 mM 3-aminotriazole.
RESULTS
Isolation of an IBA-Response Mutant Defective in PEX7
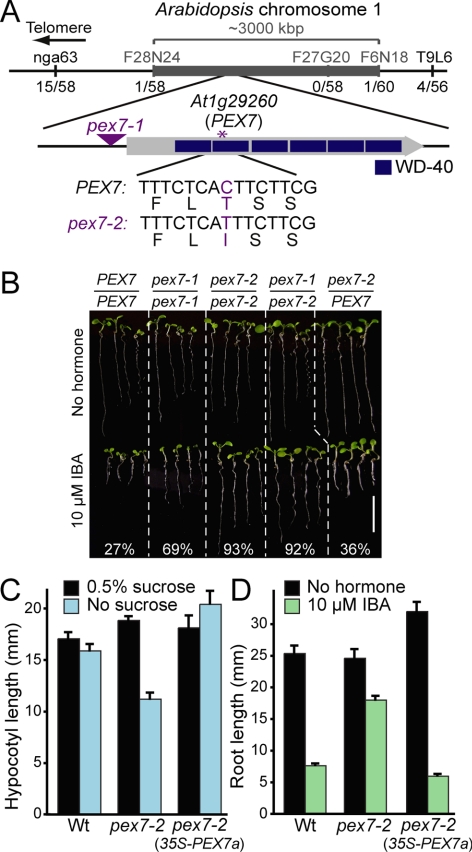
Because peroxisomes are the site of fatty acid β-oxidation and metabolism of the auxin precursor IBA, sucrose dependence and IBA resistance can be used to assess peroxisome function in Arabidopsis seedlings (Zolman et al., 2000). To identify peroxisome defects, we screened for mutants with IBA-resistant root elongation and used recombination mapping to localize the lesion in one such mutant to a region on chromosome 1 (Figure 1A) containing PEX7 (At1g29260), which encodes the Arabidopsis PTS2 receptor. Because the previously characterized pex7-1 mutant, which accumulates reduced PEX7 mRNA levels due to a T-DNA insertion in the PEX7 5′-UTR, also displays reduced IBA response (Woodward and Bartel, 2005), we sequenced the PEX7 coding region from mutant DNA. We found a C-to-T base change at position 371 (where the A of the ATG start codon is position 1) that results in a Thr124-to-Ile amino acid change in the second WD-40 repeat of PEX7 (Figure 1A). Although the PEX7 structure has not been solved from any organism, WD-40 proteins typically fold into β-propeller structures, with each propeller blade comprised of four β-strands. The identified mutation is predicted to fall near the C-terminal end of the second β-strand in the second propeller blade of PEX7. We named this new allele pex7-2.
Figure 1.
Positional cloning of the gene defective in pex7-2. (A) Recombination mapping localized the defect between F28N24 and F6N18. This interval includes the PEX7 gene (At1g29260), which encodes the PTS2 receptor. The pex7-2 C-to-T mutation causes a Thr-to-Ile missense mutation in the second WD-40 repeat; the triangle shows the position of the pex7-1 T-DNA insertion in the 5′-UTR (Woodward and Bartel, 2005). (B) pex7-2 fails to complement pex7-1. Four 8-d-old seedlings representing the range of root lengths after growth under yellow-filtered light on medium without or with 10 μM IBA are shown. Col-0 plants were used as wild type (PEX7/PEX7), and the mean percentage of root length on IBA versus unsupplemented medium is shown; n ≥ 10. Bar, 1 cm. (C) 35S-PEX7a rescues pex7-2 sucrose dependence. Seedlings were grown under white light for 1 d and transferred to the dark for five additional days on medium with or without 0.5% sucrose. Bars show mean hypocotyl lengths + SE; n ≥ 9. (D) 35S-PEX7a restores pex7-2 IBA responsiveness. Bars show mean root lengths (+SE; n ≥ 10) of 8-d-old seedlings grown under yellow-filtered light on 10 μM IBA or medium containing no hormone.
To determine whether the IBA-resistant root elongation of pex7-2 was caused by the mutation in the PEX7 gene, we conducted a complementation test with pex7-1. We found that pex7-2 was recessive and that pex7-2 failed to complement the pex7-1 resistance to IBA (Figure 1B). Moreover, we could fully restore pex7-2 mutant defects by expressing a wild-type PEX7 cDNA from the cauliflower mosaic virus 35S promoter in pex7-2 (Figure 1, C and D), confirming that the identified PEX7 lesion was responsible for the pex7-2 mutant phenotypes.
Peroxisome-defective Physiological Phenotypes in pex7-2
We used physiological assays that require peroxisomal metabolism to compare the extent of peroxisomal deficiency in the pex7-2 mutant to pex7-1 (Woodward and Bartel, 2005) and two mutants defective in the PTS1 receptor PEX5, the pex5-1 missense allele (Zolman et al., 2000) and the pex5-10 T-DNA insertion allele (Zolman and Bartel, 2004). pex7-2 displayed resistance to inhibition of root elongation by IBA similar to pex5-1 and pex7-1, responding to IBA at higher concentrations, whereas pex5-10 seemed unresponsive to IBA in this assay (Figure 2A and Supplemental Figure 1A). pex7-2 was more resistant to the promotion of lateral roots by IBA than pex7-1, but like other pex mutants, still responded robustly to the promotion of lateral roots by the synthetic auxin 1-naphthaleneacetic acid (NAA) (Figure 2B), implicating conversion of the IBA protoauxin to active indole-3-acetic acid rather than the ability to form lateral roots as the pex7-2 mutant defect.
Figure 2.
pex7-2 IBA resistance and sucrose dependence suggest IBA and fatty acid β-oxidation defects. (A) pex mutants are resistant to root elongation inhibition by IBA. Seeds were stratified 3 d at 4°C before 8 d of growth under yellow-filtered light on medium without hormone or supplemented with IBA. Bars show mean root lengths + SE; n ≥ 8. (B) pex mutants are resistant to lateral root promotion by IBA. Four-day-old seedlings grown under yellow-filtered light were transferred from hormone-free medium to medium containing IBA, the synthetic auxin NAA, or no hormone. After four additional days, root length was measured (top) and lateral roots emerged from the primary root were counted (bottom). Bars show means + SE; n ≥ 10. (C) Sucrose dependence of pex mutants. Seedlings were grown with or without sucrose for 1 d in light and 5 d in darkness (top) or for 8 d under white light (bottom). Bars show mean hypocotyl or root lengths + SE; n ≥ 9.
In addition to IBA response defects, peroxisome-defective mutants often fail to efficiently β-oxidize seed storage fatty acids following germination, resulting in developmental arrest or delay that can be restored by provision of sucrose (Hayashi et al., 1998; Zolman et al., 2000). We found that pex7-2, unlike pex7-1, was partially sucrose dependent in the dark, similar to pex5-1 but less severe than pex5-10 (Figure 2C and Supplemental Figure 1B). When grown in the light, however, pex7-2 was severely sucrose dependent, whereas pex5-1 and pex7-1 resembled wild type (Figure 2C and Supplemental Figure 1C). When grown on sucrose in the light, pex7-2 seedlings were smaller than pex7-1 and pex5-1, which resembled wild type, but not as small as pex5-10 (Figure 3A). Together, the pex7-2 phenotypes suggested a more severe defect in peroxisome function, and therefore PEX7 function, in pex7-2 than in pex7-1.
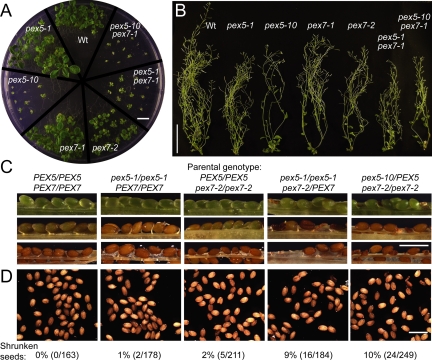
Figure 3.
Developmental defects of pex5 and pex7 mutant plants. (A) pex5-10, pex7-2, and pex5 pex7-1 double mutants are developmentally delayed. Seedlings were grown for 26 d on medium supplemented with 0.5% sucrose under white light. Bar, 1 cm. (B) pex5 and pex7 mutants produce relatively healthy and fertile adult plants. Plants from A were transferred to soil and grown under constant white light. Representative 55-d-old plants are shown. Bar, 10 cm. (C) pex7-2 confers embryo lethality in combination with pex5 mutants. Valves were removed from siliques of the indicated genotypes to reveal developing seeds. Images are arranged in order of increasing silique maturity (top to bottom). (D) Mature seed phenotypes of lines in panel C; the fraction of shrunken versus filled seeds is shown below the images. Bar, 1 mm.
Enhanced Peroxisome-defective Phenotypes in pex7 pex5 Double Mutants
The phenotypically weak pex7-1 mutation dramatically enhances pex5-1 phenotypes, consistent with the finding that both single mutants confer partial defects in PTS2 import (Woodward and Bartel, 2005). In spite of these defects, the pex5-1 pex7-1 double mutant survives to produce some viable seed (Woodward and Bartel, 2005). In contrast, we were unable to recover viable pex5-1 pex7-2 or pex5-10 pex7-2 double mutants. For example, we analyzed the progeny of pex5-1/pex5-1 pex7-2/PEX7 individuals and found 69% (18/26) pex5-1/pex5-1 pex7-2/PEX7 plants and 31% (8/26) pex5-1/pex5-1 PEX7/PEX7 plants, suggesting that the pex5-1 pex7-2 combination is lethal. Similarly, in the progeny of pex5-10/PEX5 pex7-2/pex7-2 plants, we observed 62% (8/13) pex5-10/PEX5 pex7-2/pex7-2 plants and 38% (5/13) PEX5/PEX5 pex7-2/pex7-2 plants, suggesting that pex5-10 pex7-2 is also lethal. Altogether, we analyzed 343 progeny of pex5-1 × pex7-2 crosses and 91 progeny of pex5-10 × pex7-2 crosses without recovering either double mutant. We analyzed developing seeds in pex5-1/pex5-1 pex7-2/PEX7 and pex5-10/PEX5 pex7-2/pex7-2 siliques and found shriveled seeds diagnostic of embryonic lethality (Figure 3C). At maturity, these plants generated ∼10% shrunken seeds (Figure 3D). We concluded that combining pex7-2 with either pex5-1 or pex5-10 confers embryonic lethality.
In contrast to the inviability of the pex5 pex7-2 double mutants, we were able to isolate the pex5-10 pex7-1 double mutant. The pex5-10 pex7-1 mutant resembled pex5-10; it did not germinate without sucrose (Figure 2C) and displayed similar IBA resistance as pex5-10 in root elongation (Figure 2A) and lateral root initiation (Figure 2B) assays. The general growth and development of pex5-10 pex7-1 also was similar to that of pex5-10; both mutants were slow to develop even when provided with sucrose in the light or the dark (Figures 2C and 3A) but eventually produced fertile adult plants (Figure 3B). The viability of pex5 pex7-1 double mutants and the inviability of pex5 pex7-2 double mutants are consistent with our phenotypic analyses of the relative defects in the pex7-1 and pex7-2 single mutants, which also suggest that pex7-2 more completely blocks PEX7 function than pex7-1.
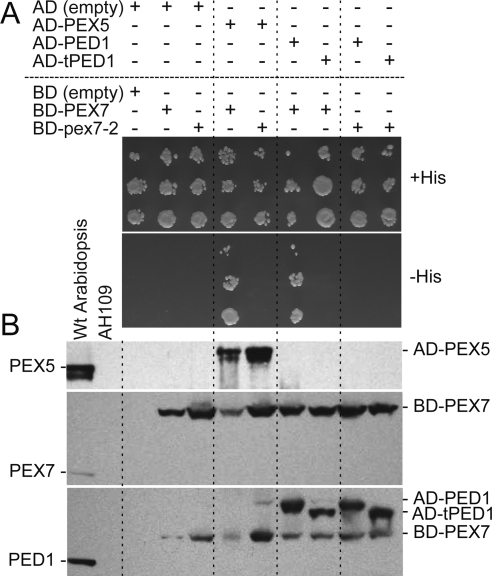
pex7-2 Disrupts Binding to PEX5 and PTS2-Cargo in Yeast Two-Hybrid Assays
In plants and mammals, PEX7 must bind both PTS2 cargo and the PTS1 receptor PEX5 to deliver PTS2 cargo to peroxisomes (Braverman et al., 1998; Otera et al., 1998; Nito et al., 2002; Hayashi et al., 2005; Woodward and Bartel, 2005). We used directed yeast two-hybrid assays to determine whether one or both of these functions was altered in pex7-2. As demonstrated previously (Nito et al., 2002), we found that Arabidopsis PEX7 and PEX5 interacted in the yeast two-hybrid assay. In contrast, pex7-2 did not interact with PEX5 in this assay (Figure 4A).
Figure 4.
The pex7-2 lesion disrupts PEX5 and PTS2-cargo binding in yeast. (A) Yeast two-hybrid assays were conducted with PEX7 and pex7-2 fused to the GAL4-DNA binding domain (BD) and PEX5, PED1 (a PTS2 protein), and tPED1 (without the PTS2 region) fused to the GAL4 activation domain (AD). Serial dilutions spotted onto permissive plates (+His) and plates on which growth requires interaction (−His) are shown after incubation at 30°C for 2 d. (B) Proteins extracted from yeast strains in A were separated using SDS-polyacrylamide gel electrophoresis and immunoblots were probed with the indicated antibodies. Extracts from 8-d-old wild-type Arabidopsis seedlings and the untransformed AH109 yeast were included as controls.
We also compared PEX7 and pex7-2 interactions with the PTS2 protein thiolase (PED1) to determine whether the pex7-2 mutation disrupted cargo binding. As expected, PEX7 interacted with PED1 but not with an N-terminally truncated PED1 version (tPED1) lacking the PTS2 region (Figure 4A). In contrast, pex7-2 paired with PED1 did not grow on selective medium (Figure 4A). The Gal4-pex7-2 and Gal4-PEX7 proteins accumulated similarly in yeast (Figure 4B), suggesting that the Gal4-pex7-2 and Gal4-PEX7 were similarly stable in yeast and that the pex7-2 lesion did not promote global pex7-2 unfolding. We concluded that the pex7-2 mutation disrupted both PEX5 and PTS2 cargo binding.
PTS Import Defects in pex7-2 Peroxisomes
We indirectly assessed PTS2 cargo import into pex7 peroxisomes by monitoring removal of the PTS2 signal from thiolase and peroxisomal malate dehydrogenase (PMDH). PTS2 processing occurs after import and is catalyzed by the peroxisomally localized PTS2-processing protease DEG15, a PTS1 protein (Helm et al., 2007; Schuhmann et al., 2008). Similar to previous reports of thiolase processing in pex7-1, pex5-10, and pex7-1 pex5-1 (Woodward and Bartel, 2005; Zolman et al., 2005), we found that in light-grown seedlings, thiolase and PMDH were fully processed in wild type, partially processed in the pex7-1, pex7-2, and pex5-1 single mutants, and only minimally processed in pex5-10 and the double mutants (Figure 5A). We found similar thiolase processing defects in dark-grown seedlings (Figure 5C), although pex7-2 defects seemed slightly less severe. Approximately half of the thiolase was processed in dark-grown pex7-2 seedlings (Figure 5C), whereas nearly all thiolase was unprocessed in light-grown pex7-2 seedlings (Figure 5A).
Figure 5.
Protein accumulation in pex7 and pex5 single and double mutants. (A and B) Light-grown pex7 mutants have PTS2 processing defects (A) and reduced PEX5 accumulation (B). Proteins extracted from 7-d-old seedlings grown on 0.5% sucrose under white light were analyzed by immunoblotting with the indicated antibodies. (C and D) Dark-grown pex7 mutants have PTS2 processing defects (C) and less severe receptor accumulation defects (D). Proteins were extracted from seedlings grown 1 d in light and 4 d in darkness on 0.5% sucrose and analyzed by immunoblotting. Protein was normalized to the same volume.
To directly analyze the effects of the pex7-2 mutation on matrix protein import, we crossed pex7-2 to transgenic lines containing peroxisome-targeted GFP and examined the resultant homozygous plants using fluorescence microscopy. Whereas wild-type cotyledon cells efficiently imported PTS2-GFP into punctate structures indicative of peroxisomes, we found that PTS2-GFP displayed mostly cytosolic fluorescence in cotyledons of both light- and dark-grown pex7-2 seedlings (Figure 6A). This localization confirmed that PTS2 import is impaired but not completely blocked in pex7-2. We also examined a PTS1 reporter and found that GFP-PTS1 displayed the expected punctate peroxisomal fluorescence pattern in cotyledons of dark-grown pex7 seedlings (Figure 6B). Surprisingly, however, GFP-PTS1 displayed primarily cytosolic fluorescence in light-grown pex7-2 seedlings (Figure 6B). This unanticipated PTS1 import defect was apparent in multiple cell types, including cotyledon epidermal cells and the underlying mesophyll cells (Figure 6B). To assess whether this defect in PTS1-GFP was unique to pex7-2, we also examined cotyledons of the weaker pex7-1 allele and in the pex5-1 pex7-1 double mutant. In both cases, we detected substantial cytosolic GFP-PTS1 in light-, but not dark-grown seedlings (Figure 6B), confirming that decreased PEX7 function can reduce PTS1 import, and demonstrating that this defect was not peculiar to the pex7-2 allele.
Figure 6.
pex7-2 defects in matrix protein import. (A) Cotyledons of 7-d-old light-grown seedlings or of seedlings grown 1 d in light and 4 d in darkness on 0.5% sucrose were analyzed using confocal fluorescence microscopy. PTS2 protein targeting was visualized by imaging fluorescence from a GFP derivative (PTS2-GFP) carrying the N-terminal peroxisome targeting signal from PED1 (Woodward and Bartel, 2005) in epidermal cells (first column) and the underlying mesophyll cells (second column). The mesophyll column shows merged images of GFP (green) and chlorophyll (chl; magenta) fluorescence. The large central vacuole of expanded plant cells consolidates most of the cytosol at the cell margins. Dark-grown cotyledons were briefly stained with propidium iodide, which stains cell walls, before visualization of epidermal cells; the last column shows merged image of GFP (green; third column) and propidium iodide (magenta) fluorescence. Bar, 20 μm. (B) Cotyledons of seedlings grown as described in A were analyzed as described in A. PTS1 protein targeting was visualized by imaging fluorescence from a GFP derivative (GFP-PTS1) carrying a C-terminal PTS1 (Zolman and Bartel, 2004). Bar, 20 μm.
Decreased PEX5 Levels in pex7 Mutants
The cytosolic fluorescence pattern of GFP-PTS1 in light-grown pex7 mutants suggested that PTS1 import, and by extension PEX5, depends on PEX7. To further understand the basis of the pex7 PTS1 import defects, we analyzed peroxin levels in mutant seedlings. We found that PEX7 levels were similar to wild type in both pex5 alleles but were reduced in both pex7 alleles (Figure 5, B and D), consistent with the reduced PEX7 mRNA level in pex7-1 (Woodward and Bartel, 2005) and suggesting that the pex7-2 lesion might impair PEX7 stability in seedlings in addition to disrupting PEX5 and cargo interactions (Figure 4A). As reported previously (Woodward and Bartel, 2005; Zolman et al., 2005), pex5-1 and pex5-10 mutants accumulated normal and undetectable levels of full-length PEX5, respectively. A smaller protein that cross-reacts with our PEX5 antibody, which was generated to a peptide corresponding to the PEX5 C terminus (Zolman and Bartel, 2004), was sometimes detected in pex5-10 extracts (Figure 5, B and D). This residual immunoreactivity, along with the small fraction of processed thiolase detected even in pex5-10 plants (Figure 5C; Zolman et al., 2005), is consistent with the possibility that the pex5-10 protein may retain partial function.
Although PEX5 levels were nearly normal in pex7 mutants grown in the dark (Figure 5D), light-grown seedlings of both pex7-1 and pex7-2 accumulated substantially less PEX5 protein than wild type (Figure 5B). This result suggested that the defects in GFP-PTS1 import displayed by the pex7 mutants could be explained by reduced PEX5 accumulation in these mutants and that PEX5 might depend on PEX7 for stability in light-grown seedlings.
DISCUSSION
Import of PTS2 cargo into mammalian and plant peroxisomes requires not only the PTS2 receptor PEX7 but also the PTS1 receptor PEX5 (Braverman et al., 1998; Otera et al., 1998; Hayashi et al., 2005; Woodward and Bartel, 2005; Lee et al., 2006). In this work, we used a new pex7 allele, pex7-2, to demonstrate that the reciprocal is also true: Arabidopsis PEX5 and PTS1 import depend on the PTS2 receptor PEX7.
The pex7-2 mutant displayed typical peroxisome-defective phenotypes such as IBA resistance and sucrose dependence during seedling development (Figure 2). The pex7-2 mutant defects were more severe than the previously characterized pex7-1 and pex5-1 mutants (Zolman et al., 2000; Woodward and Bartel, 2005) but less severe than the pex5-10 T-DNA insertion allele (Figures 2 and 3). We were unable to recover pex5-1 pex7-2 or pex5-10 pex7-2 double mutants (Figure 3), implying that peroxisomal matrix protein import is essential during Arabidopsis embryogenesis. Indeed, mutants defective in multiple isozymes of fatty acid β-oxidation enzymes display embryo lethality (Rylott et al., 2003, 2006), suggesting that one essential embryonic function for peroxisomes is β-oxidation. An essential role for peroxisome matrix protein import in embryogenesis is also suggested by the embryo lethality observed in null alleles of the ring-finger peroxins PEX2, PEX10, and PEX12 (Hu et al., 2002; Schumann et al., 2003; Sparkes et al., 2003; Fan et al., 2005). Our inability to recover the pex5-10 pex7-2 double mutant strongly suggests that the pex5-10 allele does not abolish PEX5 function, despite the presence of a T-DNA in exon five (Zolman and Bartel, 2004). Similarly, our inability to recover viable pex7-2 double mutants with even the pex5-1 missense allele indicates that pex7-2 is unlikely to be a null allele.
The pex7-2 Thr to Ile missense mutation disrupted interactions with both PTS2 cargo and the PTS1 receptor PEX5 in yeast two-hybrid assays (Figure 4). It remains formally possible that the reduced ability of pex7-2 to bind PEX5 was a secondary effect of reduced cargo binding, as yeast contains several PTS2 proteins that might interact with Arabidopsis PEX7. It seems less likely, however, that the reduced ability of pex7-2 to bind cargo in the yeast two-hybrid assay was a secondary effect of reduced PEX5 binding, as yeast PEX5 does not bind PEX7 (Brown and Baker, 2008).
Interestingly, the pex7-2 mutation resulted in reduced pex7-2 protein accumulation in plants, particularly in light-grown seedlings (Figure 5B). It is possible that the missense mutation impairs receptor folding, thereby rendering it more susceptible to proteolysis in vivo and unable to interact with binding partners. However, Gal4-pex7-2 accumulated normally in yeast (Figure 4B), suggesting that the pex7-2 Thr-to-Ile substitution does not lead to global pex7-2 unfolding. In addition, dark-grown pex7 seedlings accumulated more easily detected amounts of PEX7 protein (Figure 5). Alternatively, pex7-2 may fold correctly but may be subject to proteolysis in vivo because it is not protected by association with PEX5 or with cargo or because it is not efficiently recycled from the peroxisome, similar to what is seen with inefficiently recycled PEX5 (reviewed in Brown and Baker, 2008). Distinguishing among these possibilities will be aided by future studies that elucidate the PEX7 structure and define PEX7 binding interfaces.
We found that along with reduced PEX7 levels, both pex7-2 and pex7-1 displayed reduced PEX5 protein levels when grown in the light (Figure 5), suggesting that PEX5 depends on PEX7 for stability. Moreover, GFP-PTS1 was mislocalized to the cytosol in light-grown pex7 mutants (Figure 6), indicating that the reduced PEX5 levels that we observed in pex7 were accompanied by a reduced efficiency of PTS1 import. Because the pex7-1 mutation results in reduced accumulation of wild-type PEX7 (Woodward and Bartel, 2005), these defects are not a specific consequence of the pex7-2 lesion but likely result from reduced PEX7 function. To our knowledge, a dependence of PTS1 import on the PTS2 receptor has not previously been observed in plants. Interestingly, the possibility that PTS1 import can depend on the PEX7 pathway may not be unique to plants. A recent study in trypanosomes revealed that pex7 RNA interference lines had low PEX5 levels and mislocalized not only PTS2, but also PTS1 proteins (Galland et al., 2007). Moreover, H. polymorpha mutants lacking the PEX7 coreceptor PEX20 also display reduced PEX5 accumulation (Moscicka et al., 2007).
Other pex mutants also impact PEX5 levels. The Arabidopsis pex6 mutant has decreased PEX5 levels (Zolman and Bartel, 2004), consistent with the possibility that Arabidopsis PEX5 is degraded when not properly recycled, as has been demonstrated in nonplant systems (reviewed in Brown and Baker, 2008). Mammalian PEX7 interacts with the PEX2 RING-finger peroxin (Miyata et al., 2009) and Arabidopsis PEX7 interacts with PEX12, another RING-finger peroxin (Singh et al., 2009). In yeast, PEX12 catalyzes the PEX5 monoubiquitination that is essential for PEX5 recycling, whereas PEX2 catalyzes PEX5 polyubiquitination, which leads to PEX5 degradation (Platta et al., 2009). It is possible that the reduced PEX5 levels that we observed in pex7 mutants result from a PEX5 recycling defect, perhaps because PEX7 normally promotes PEX5 association with the recycling components through its association with PEX12.
Our analysis also revealed that the severity of peroxisomal import defects can be impacted by light conditions. The more substantial defects in PEX5 and PEX7 accumulation in the light (Figure 5) were accompanied by more severe pex7-2 sucrose dependence in the light compared with the dark (Figure 2C). Moreover, we observed GFP-PTS1 import defects only in light-grown pex7 seedlings (Figure 6). Other peroxisome-related mutants with light-enhanced sucrose dependence include acx4, a mutant defective in a PTS1-targeted short-chain acyl-CoA oxidase that is sucrose dependent in the light but not in the dark (Adham et al., 2005), and pex6-1, which displays more severe sucrose dependence in the light than in the dark (Zolman and Bartel, 2004). These light-exacerbated phenotypes suggest that particular PEX protein roles might have varied importance in different environmental conditions. It is intriguing that PTS2 proteins are overrepresented among fatty acid β-oxidation enzymes (Kamada et al., 2003), which act to metabolize fatty acids stored in seeds immediately after germination, whereas virtually all peroxisomal photorespiration enzymes are PTS1 proteins. Perhaps the role of PEX7 in promoting PEX5 accumulation becomes more important in the light when there is a reduced demand for PTS2 import.
The majority of peroxisomal matrix proteins are PTS1-containing proteins, and PEX5 has been at the forefront of peroxisome receptor research. Although the fact that C. elegans lacks PEX7 (Gurvitz et al., 2000; Motley et al., 2000) demonstrates that organisms can evolve peroxisomes that function without a PTS2 pathway, the observation that most examined eukaryotes maintain both PEX5 and PEX7 implies that the dual targeting system confers some selective advantage. This study has uncovered an importance for the PTS2 receptor PEX7 in facilitating PTS1 import. Such interdependence among targeting receptors may make it more difficult for one targeting system to be lost. For example, a role for PEX7 in PEX5 function provides a rationale for the continued existence of an apparent PEX7 homologue in Drosophila melanogaster (Motley et al., 2000), even though flies lack readily identifiable PTS2 proteins (Woodward, 2005). Further studies are needed to uncover the molecular mechanisms by which PEX7 promotes PEX5 accumulation and function.
Supplementary Material
ACKNOWLEDGMENTS
We thank Lucia Strader for microscopy assistance; Arthur Millius and A. Raquel Adham for initial pex7-2 isolation; Neda Nikravan for mapping assistance; Tyler Moss for assistance with PED1 constructs; Andrew Woodward for developing the F28N24 marker and for assistance in generating pex5-1 pex7-1 (GFP-PTS1); Steven Smith for the PMDH2 antibodies; and Sarah Christensen, Melanie Monroe-Augustus, Sarah Ratzel, Lucia Strader, and Andrew Woodward for critical comments on the manuscript. This research was supported by the National Science Foundation (MCB-0745122) and the Robert A. Welch Foundation (C-1309). N.M.R. was supported in part by a National Institutes of Health predoctoral fellowship (F31-GM081911) and the Rice-Houston Alliance for Graduate Education and the Professoriate Program (NSF HRD-0450363).
Abbreviations used:
- IBA
indole-3-butyric acid
- PEX
peroxin
- PTS
peroxisome-targeting signal.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-08-0672) on February 3, 2010.
REFERENCES
- Adham A. R., Zolman B. K., Millius A., Bartel B. Mutations in Arabidopsis acyl-CoA oxidase genes reveal distinct and overlapping roles in β-oxidation. Plant J. 2005;41:859–874. doi: 10.1111/j.1365-313X.2005.02343.x. [DOI] [PubMed] [Google Scholar]
- Asamizu E., Nakamura Y., Sato S., Tabata S. A large scale analysis of cDNA in Arabidopsis thaliana: generation of 12,028 non-redundant expressed sequence tags from normalized and size-selected cDNA libraries. DNA Res. 2000;7:175–180. doi: 10.1093/dnares/7.3.175. [DOI] [PubMed] [Google Scholar]
- Ausubel F., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K. Current Protocols in Molecular Biology. New York: Greene Publishing Associates and Wiley-Interscience; 1999. [Google Scholar]
- Braverman N., Dodt G., Gould S. J., Valle D. An isoform of Pex5p, the human PTS1 receptor, is required for the import of PTS2 proteins into peroxisomes. Hum. Mol. Genet. 1998;7:1195–1205. doi: 10.1093/hmg/7.8.1195. [DOI] [PubMed] [Google Scholar]
- Brocard C., Kragler F., Simon M. M., Schuster T., Hartig A. The tetratricopeptide repeat-domain of the PAS10 protein of Saccharomyces cerevisiae is essential for binding the peroxisomal targeting signal -SKL. Biochem. Biophys. Res. Commun. 1994;204:1016–1022. doi: 10.1006/bbrc.1994.2564. [DOI] [PubMed] [Google Scholar]
- Brown L.-A., Baker A. Shuttles and cycles: transport of proteins into the peroxisome matrix. Mol. Membr. Biol. 2008;25:363–375. doi: 10.1080/09687680802130583. [DOI] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Dammai V., Subramani S. The human peroxisomal targeting signal receptor, Pex5p, is translocated into the peroxisome matrix and recycled to the cytosol. Cell. 2001;105:187–196. doi: 10.1016/s0092-8674(01)00310-5. [DOI] [PubMed] [Google Scholar]
- Dodt G., Braverman N., Wong C., Moser A., Moser H. W., Watkins P., Valle D., Gould S. J. Mutations in the PTS1 receptor gene, PXR1, define complementation group 2 of the peroxisome biogenesis disorders. Nat. Genet. 1995;9:115–125. doi: 10.1038/ng0295-115. [DOI] [PubMed] [Google Scholar]
- Eastmond P. J., Hooks M. A., Williams D., Lange P., Bechtold N., Sarrobert C., Nussaume L., Graham I. A. Promoter trapping of a novel medium-chain acyl-CoA oxidase, which is induced transcriptionally during Arabidopsis seed germination. J. Biol. Chem. 2000;275:34375–34381. doi: 10.1074/jbc.M004945200. [DOI] [PubMed] [Google Scholar]
- Fan J., Quan S., Orth T., Awai C., Chory J., Hu J. The Arabidopsis PEX12 gene is required for peroxisome biogenesis and is essential for development. Plant Physiol. 2005;139:231–239. doi: 10.1104/pp.105.066811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransen M., Brees C., Baumgart E., Vanhooren J.C.T., Baes M., Mannaerts G. P., Veldhoven P. P. Identification and characterization of the putative human peroxisomal C-terminal targeting signal import receptor. J. Biol. Chem. 1995;270:7731–7736. doi: 10.1074/jbc.270.13.7731. [DOI] [PubMed] [Google Scholar]
- Galland N., Demeure F., Hannaert V., Verplaetse E., Vertommen D., Van der Smissen P., Courtoy P. J., Michels P. A.. Characterization of the role of the receptors PEX5 and PEX7 in the import of proteins into glycosomes of Trypanosoma brucei. Biochim. Biophys. Acta. 2007;1773:521–535. doi: 10.1016/j.bbamcr.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Gietz R., Schiestl R. Transforming yeast with DNA. Methods Mol. Cell. Biol. 1995;5:255–269. [Google Scholar]
- Gurvitz A., Langer S., Piskacek M., Hamilton B., Ruis H., Hartig A. Predicting the function and subcellular location of Caenorhabditis elegans proteins similar to Saccharomyces cerevisiae β-oxidation enzymes. Yeast. 2000;17:188–200. doi: 10.1002/1097-0061(20000930)17:3<188::AID-YEA27>3.0.CO;2-E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haughn G. W., Somerville C. Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol. Gen. Genet. 1986;204:430–434. [Google Scholar]
- Hayashi M., Toriyama K., Kondo M., Nishimura M. 2,4-dichlorophenoxybutyric acid-resistant mutants of Arabidopsis have defects in glyoxysomal fatty acid β-oxidation. Plant Cell. 1998;10:183–195. doi: 10.1105/tpc.10.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M., Yagi M., Nito K., Kamada T., Nishimura M. Differential contribution of two peroxisomal protein receptors to the maintenance of peroxisomal functions in Arabidopsis. J. Biol. Chem. 2005;280:14829–14835. doi: 10.1074/jbc.M411005200. [DOI] [PubMed] [Google Scholar]
- Helm M., et al. Dual specificities of the glyoxysomal/peroxisomal processing protease Deg15 in higher plants. Proc. Natl. Acad. Sci. USA. 2007;104:11501–11506. doi: 10.1073/pnas.0704733104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooks M. A., Kellas F., Graham I. A. Long-chain acyl-CoA oxidases of Arabidopsis. Plant J. 1999;20:1–13. doi: 10.1046/j.1365-313x.1999.00559.x. [DOI] [PubMed] [Google Scholar]
- Hu J., Aguirre M., Peto C., Alonso J., Ecker J., Chory J. A role for peroxisomes in photomorphogenesis and development of Arabidopsis. Science. 2002;297:405–409. doi: 10.1126/science.1073633. [DOI] [PubMed] [Google Scholar]
- Kamada T., Nito K., Hayashi H., Mano S., Hayashi M., Nishimura M. Functional differentiation of peroxisomes revealed by expression profiles of peroxisomal genes in Arabidopsis thaliana. Plant Cell Physiol. 2003;44:1275–1289. doi: 10.1093/pcp/pcg173. [DOI] [PubMed] [Google Scholar]
- Kohalmi S., Reader L.J.V., Samach A., Nowak J., Haughn G. W., Crosby W. L. Identification and characterization of protein interactions using the yeast 2-hybrid system. In: Gelvin S. B., Schiperoort R. A., editors. Plant Molecular Biology Manual. vol. M1. Dordrecht, The Netherlands: Kluwer; 1998. pp. 1–30. [Google Scholar]
- LeClere S., Bartel B. A library of Arabidopsis 35S-cDNA lines for identifying novel mutants. Plant Mol. Biol. 2001;46:695–703. doi: 10.1023/a:1011699722052. [DOI] [PubMed] [Google Scholar]
- Lee J. R., et al. Cloning of two splice variants of the rice PTS1 receptor, OsPex5pL and OsPex5pS, and their functional characterization using pex5-deficient yeast and Arabidopsis. Plant J. 2006;47:457–466. doi: 10.1111/j.1365-313X.2006.02797.x. [DOI] [PubMed] [Google Scholar]
- Lingard M., Monroe-Augustus M., Bartel B. Peroxisome-associated matrix protein degradation in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2009;106:4561–4566. doi: 10.1073/pnas.0811329106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata N., Hosoi K.-I., Mukai S., Fujiki Y. In vitro import of peroxisome-targeting signal type 2 (PTS2) receptor Pex7p into peroxisomes. Biochim. Biophys. Acta. 2009;1793:860–870. doi: 10.1016/j.bbamcr.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Moscicka K., Klompmaker S., Wang D., van der Klei I. J., Boekema E. J. The Hansenula polymorpha peroxisomal targeting signal 1 receptor, Pex5p, functions as a tetramer. FEBS Lett. 2007;581:1758–1762. doi: 10.1016/j.febslet.2007.03.061. [DOI] [PubMed] [Google Scholar]
- Motley A. M., Hettema E. H., Ketting R., Plasterk R., Tabak H. F. Caenorhabditis elegans has a single pathway to target matrix proteins to peroxisomes. EMBO Rep. 2000;1:40–46. doi: 10.1093/embo-reports/kvd010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair D. M., Purdue P. E., Lazarow P. B. Pex7p translocates in and out of peroxisomes in Saccharomyces cerevisiae. J. Cell Biol. 2004;167:599–604. doi: 10.1083/jcb.200407119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nito K., Hayashi M., Nishimura M. Direct interaction and determination of binding domains among peroxisomal import factors in Arabidopsis thaliana. Plant Cell Physiol. 2002;43:355–366. doi: 10.1093/pcp/pcf057. [DOI] [PubMed] [Google Scholar]
- Otera H., et al. Peroxisome targeting signal type 1 (PTS1) receptor is involved in import of both PTS1 and PTS 2, Studies with PEX5-defective CHO cell mutants. Mol. Cell. Biol. 1998;18:388–399. doi: 10.1128/mcb.18.1.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platta H. W., Magraoui F. E., Baumer B. E., Schlee D., Girzalsky W., Erdmann R. Pex2 and Pex12 function as protein-ubiquitin ligases in peroxisomal protein import. Mol. Cell. Biol. 2009;29:5505–5516. doi: 10.1128/MCB.00388-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pracharoenwattana I., Cornah J. E., Smith S. M. Arabidopsis peroxisomal citrate synthase is required for fatty acid respiration and seed germination. Plant Cell. 2005;17:2037–2048. doi: 10.1105/tpc.105.031856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pracharoenwattana I., Cornah J. E., Smith S. M. Arabidopsis peroxisomal malate dehydrogenase functions in β-oxidation but not in the glyoxylate cycle. Plant J. 2007;50:381–390. doi: 10.1111/j.1365-313X.2007.03055.x. [DOI] [PubMed] [Google Scholar]
- Rehling P., Marzioch M., Niesen F., Wittke E., Veenhuis M., Kunau W. H. The import receptor for the peroxisomal targeting signal 2 (PTS2) in Saccharomyces cerevisiae is encoded by the PAS7 gene. EMBO J. 1996;15:2901–2913. [PMC free article] [PubMed] [Google Scholar]
- Reumann S., Ma C., Lemke S., Babujee L. AraPerox. A database of putative Arabidopsis proteins from plant peroxisomes. Plant Physiol. 2004;136:2587–2608. doi: 10.1104/pp.104.043695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rylott E. L., Eastmond P. J., Gilday A. D., Slocombe S. P., Larson T. R., Baker A., Graham I. A. The Arabidopsis thaliana multifunctional protein gene (MFP2) of peroxisomal beta-oxidation is essential for seedling establishment. Plant J. 2006;45:930–941. doi: 10.1111/j.1365-313X.2005.02650.x. [DOI] [PubMed] [Google Scholar]
- Rylott E. L., Rogers C. A., Gilday A. D., Edgell T., Larson T. R., Graham I. A. Arabidopsis mutants in short- and medium-chain acyl-CoA oxidase activities accumulate acyl-CoAs and reveal that fatty acid β-oxidation is essential for embryo development. J. Biol. Chem. 2003;278:21370–21377. doi: 10.1074/jbc.M300826200. [DOI] [PubMed] [Google Scholar]
- Schuhmann H., Huesgen P. F., Gietl C., Adamska I. The DEG15 serine protease cleaves peroxisomal targeting signal 2-containing proteins in Arabidopsis. Plant J. 2008;148:1847–1856. doi: 10.1104/pp.108.125377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann U., Wanner G., Veenhuis M., Schmid M., Gietl C. AthPEX10, a nuclear gene essential for peroxisome and storage organelle formation during Arabidopsis embryogenesis. Proc. Natl. Acad. Sci. USA. 2003;100:9626–9631. doi: 10.1073/pnas.1633697100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh T., Hayashi M., Mano S., Arai Y., Goto S., Nishimura M. Molecular Components Required for the Targeting of PEX7 to Peroxisomes in Arabidopsis thaliana. Plant J. 2009;60:488–498. doi: 10.1111/j.1365-313X.2009.03970.x. [DOI] [PubMed] [Google Scholar]
- Sparkes I. A., Brandizzi F., Slocombe S. P., El-Shami M., Hawes C., Baker A. An Arabidopsis pex10 null mutant is embryo lethal, implicating peroxisomes in an essential role during plant embryogenesis. Plant Physiol. 2003;133:1809–1819. doi: 10.1104/pp.103.031252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasinopoulos T. C., Hangarter R. P. Preventing photochemistry in culture media by long-pass light filters alters growth of cultured tissues. Plant Physiol. 1990;93:1365–1369. doi: 10.1104/pp.93.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward A. W. Houston, TX: Rice University; 2005. Genes, Organelles, and Molecules That Influence Plant Development through Auxin Regulation. Ph.D. Dissertation. [Google Scholar]
- Woodward A. W., Bartel B. The Arabidopsis peroxisomal targeting signal type 2 receptor PEX7 is necessary for peroxisome function and dependent on PEX5. Mol. Biol. Cell. 2005;16:573–583. doi: 10.1091/mbc.E04-05-0422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., et al. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science. 2003;302:842–846. doi: 10.1126/science.1088305. [DOI] [PubMed] [Google Scholar]
- Zolman B. K., Bartel B. An Arabidopsis indole-3-butyric acid-response mutant defective in PEROXIN6, an apparent ATPase implicated in peroxisomal function. Proc. Natl. Acad. Sci. USA. 2004;101:1786–1791. doi: 10.1073/pnas.0304368101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman B. K., Martinez N., Millius A., Adham A. R., Bartel B. Identification and characterization of Arabidopsis indole-3-butyric acid response mutants defective in novel peroxisomal enzymes. Genetics. 2008;180:237–251. doi: 10.1534/genetics.108.090399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman B. K., Monroe-Augustus M., Silva I. D., Bartel B. Identification and functional characterization of Arabidopsis PEROXIN4 and the interacting protein PEROXIN22. Plant Cell. 2005;17:3422–3435. doi: 10.1105/tpc.105.035691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman B. K., Yoder A., Bartel B. Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics. 2000;156:1323–1337. doi: 10.1093/genetics/156.3.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.