Spo13 is a sporulation-specific subunit of the Schizosaccharomyces pombe spindle pole body. We show that Spo13 acts specifically as an exchange factor for the Rab GTPase Ypt2 and that this activity is essential for spore formation. The meiotic spindle pole body may promote membrane assembly by stimulating the activity of a Rab GTPase.
Abstract
Spore morphogenesis in yeast is driven by the formation of membrane compartments that initiate growth at the spindle poles during meiosis II and grow to encapsulate daughter nuclei. Vesicle docking complexes, called meiosis II outer plaques (MOPs), form on each meiosis II spindle pole body (SPB) and serve as sites of membrane nucleation. How the MOP stimulates membrane assembly is not known. Here, we report that SpSpo13, a component of the MOP in Schizosaccharomyces pombe, shares homology with the guanine nucleotide exchange factor (GEF) domain of the Saccharomyces cerevisiae Sec2 protein. ScSec2 acts as a GEF for the small Rab GTPase ScSec4, which regulates vesicle trafficking from the late-Golgi to the plasma membrane. A chimeric protein in which the ScSec2-GEF domain is replaced with SpSpo13 is capable of supporting the growth of a sec2Δ mutant. SpSpo13 binds preferentially to the nucleotide-free form of ScSec4 and facilitates nucleotide exchange in vitro. In vivo, a Spspo13 mutant defective in GEF activity fails to support membrane assembly. In vitro specificity experiments suggest that SpYpt2 is the physiological substrate of SpSpo13. These results demonstrate that stimulation of Rab-GTPase activity is a property of the S. pombe MOP essential for the initiation of membrane formation.
INTRODUCTION
Formation of ascospores by yeast cells is an unusual cell division event in which daughter cells are formed by de novo synthesis of new plasma membranes around daughter nuclei, providing an excellent model system by which to study the generation of novel intracellular membrane compartments. (Neiman, 1998, 2005). In response to nitrogen starvation, the fission yeast Schizosaccharomyces pombe exits the mitotic cell cycle, mates to form diploid cells, and enters meiosis. During meiosis II, four newly formed membrane compartments, termed forespore membranes (FSMs), appear in the cytosol (Shimoda, 2004). As meiosis is completed each of the four haploid nuclei produced by meiosis is engulfed within a forespore membrane. Capture of a nucleus and associated cytoplasm by an FSM gives rise to a nascent spore, with the forespore membrane now serving as the plasma membrane of the spore.
Real-time videomicroscopy of FSM formation in S. pombe, and of the analogous prospore membrane in Saccharomyces cerevisiae, has revealed that membrane formation can be dissected into three distinct stages: 1) membrane initiation at the spindle pole body (SPB), 2) expansion to engulf a daughter nucleus, and 3) closure to complete cytokinesis (Nakamura et al., 2008; Diamond et al., 2009). Notably, assembly of FSMs always starts at spindle pole bodies. In yeast, spindle pole bodies, which are functional analogues of centrosomes in higher cells, have distinct nuclear and cytoplasmic faces, called the inner and outer plaques, respectively (Jaspersen and Winey, 2004). In dividing cells, the inner and outer plaques serve as microtubule-organizing centers. However, at the beginning of the meiosis II, there is a protein exchange on the outer plaques, which are now called meiosis II outer plaques (MOPs) (Shimoda, 2004; Neiman, 2005). In S. pombe, MOPs appear as multilayered disk-shaped structures in the electron microscope (Hirata and Shimoda, 1994). The assembly of the MOP changes the function of the outer plaque from microtubule nucleation to membrane nucleation (Shimoda, 2004; Neiman, 2005). Nascent FSMs are assembled on the outermost surface of the MOPs and the membranes remain in contact with the MOPs until very late in FSM development (Hirata and Shimoda, 1994; Nakamura et al., 2008).
In S. cerevisiae, genetic and cell biological studies have demonstrated that the prospore membrane compartment is derived from the coalescence of post-Golgi secretory vesicles (Neiman, 1998, 2005). This is likely true for the S. pombe FSM as well as mutations in genes encoding soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins that mediate fusion at the plasma membrane in vegetative cells disrupt FSM assembly (Nakase et al., 2001; Nakamura-Kubo et al., 2003; Nakamura et al., 2005). In both S. pombe and S. cerevisiae, deletion of the genes for individual MOP components leads to failure of the structure to assemble and a complete block to membrane formation, indicating that the MOP is essential for vesicle coalescence (Knop and Strasser, 2000; Bajgier et al., 2001; Nickas et al., 2003; Nakase et al., 2008). In mutants that fail to properly form MOPs, the precursor vesicles are dispersed from the spindle poles. This differentiates MOP mutants from mutants in proteins such as SNAREs that are involved directly in membrane fusion. In SNARE mutants, precursor vesicles accumulate on and around the MOP surface (Nakanishi et al., 2006). These results, and the direct association of precursor vesicles with the MOP, suggest that the MOP functions as a vesicle tethering complex and promotes SNARE-mediated membrane fusion. How the MOP stimulates fusion of the FSM precursor vesicles is not understood.
Docking or tethering complexes function upstream of SNARE-mediated fusion in a variety of other vesicle fusion events, and often function in association with Rab GTPases (Pfeffer, 1999). For example, association of secretory vesicles with the plasma membrane is mediated by a tethering complex termed the exocyst and the exocyst is an effector of the Rab GTPase ScSec4 (TerBush et al., 1996; Guo et al., 1999). Rab GTPases are important regulators of vesicle traffic (Stenmark, 2009). A Rab GTPase cycles between its guanosine triphosphate (GTP)-bound and guanosine diphosphate (GDP)-bound forms (Segev, 2001; Stenmark, 2009). A Rab protein in its GTP-bound form is switched on to bind to effectors. The exchange of GDP with GTP in a Rab GTPase is facilitated by a guanine nucleotide exchange factor (GEF), and the GEF activity directs where a Rab protein executes its function (Jones et al., 2000b; Ortiz et al., 2002; Cai et al., 2008).
In S. cerevisiae Sec4, the protein is activated by the GEF ScSec2. ScSec2 itself is localized to secretory vesicles, thus the GEF activity of ScSec2 serves to activate ScSec4 on the surface of the vesicles, promoting ScSec4-mediated interaction of the vesicle with the exocyst complex (Nair et al., 1990; Walch-Solimena et al., 1997; Guo et al., 1999). The exchange activity of ScSec2 resides in an NH2-terminal coiled-coil domain (amino acids 1-160), to which ScSec4 binds (Walch-Solimena et al., 1997; Dong et al., 2007). In addition to the GEF domain, the remaining 599 amino acids of the ScSec2 protein are required for its proper association with secretory vesicles (Elkind et al., 2000; Ortiz et al., 2002). Mutants impaired in ScSec2 localization display growth defects due to a failure of polarized delivery of ScSec4-containing vesicles (Walch-Solimena et al., 1997); however, overexpression of only the GEF domain is sufficient to complement the growth defect of a sec2 deletion (Nair et al., 1990; Dong et al., 2007).
In S. pombe, the MOP is composed of three sporulation-specific proteins: SpSpo15, SpSpo2, and SpSpo13 (Nakase et al., 2008). Localization of SpSpo13 is dependent on the other two MOP components, and SpSpo13 is thought to be located at the membrane-proximal surface of the MOP (Nakase et al., 2008). Here, we report that SpSpo13 has homology to the ScSec2 GEF domain. SpSpo13 can replace the GEF domain of Scsec2 in vivo and the SpSpo13 protein can bind to ScSec4 and stimulate GDP release in vitro. A point mutation that impairs SpSpo13 GEF activity leads to a failure of FSM assembly. GDP release assays using different S. pombe Ypt proteins suggest that SpYpt2 is the physiological substrate of SpSpo13. These results provide the first description of a biochemical activity associated with the MOP and provide insight into how this docking complex functions to regulate the initiation of FSM formation.
MATERIALS AND METHODS
Yeast Strains Construction
The strains used in this study are listed in Table 1. HJ79 was constructed as follows. First, a sec2Δ heterozygous diploid, HJ75, was constructed by deleting SEC2 in AN120 by polymerase chain reaction (PCR) (Longtine et al., 1998) using pFA6a-His3MX6 as a template and HJO105 and HJO106 as primers (oligonucleotides [oligos] are available upon requested). Second, pRS416-TEFpr-ScSEC2 was transformed into HJ75, and the resulting transformants were sporulated and dissected. An Ura+ His+ segregant was selected and named HJ79.
Table 1.
Strains used in this study
| Name | Genotype | Source |
|---|---|---|
| S. cerevisiae | ||
| AN120 | MATa/MATα ARG4/arg4-NspI his3SK/his3SK hoΔ::LYS2/hoΔ::LYS2 leu2/leu2 | Neiman et al. (2000) |
| lys2/lys2RME1/rme1Δ::LEU2 trp1::hisG/trp1::hisG ura3/ura3 | ||
| HJ75 | MATa/MATα ARG4/arg4-NspI his3SK/his3SK ho::LYS2/ho::LYS2 leu2/leu2 | This study |
| lys2/lys2RME1/rme1::LEU2 trp1::hisG/trp1::hisG ura3/ura3 sec2Δhis5+/SEC2 | ||
| HJ79 | MATa arg4-NspI his3SK ho::LYS2 leu2 lys2 rme1::LEU2 trp1::hisG ura3 sec2Δhis5+ | This study |
| HJ75-4 | MATα arg4-NspI his3SK ho::LYS2 leu2 lys2 RME1 trp1::hisG ura3 sec2Δhis5+pRS416-TEFpr-ScSEC2 | This study |
| S. pombe | ||
| GP46 | h+ ade6-M375 ura4-294 | Virgin et al. (1995) |
| B82 | h90 spo13-B82 ade6-M210 | Bresch et al. (1968) |
| ANP3 | h90 spo13-B82 ura4-294 | This study |
| GP1327 | h90 ade6-52 ura4-294 leu1-32 | Lin and Smith (1995) |
| HJP1 | h90 spo13-B82 ura4-294 leu1-32 | This study |
| FY12476a | h90 spo13::ura4+ura4 leu1 sid4GFP::kanR |
a This strain was obtained from the Yeast Genetic Resource Center of Japan supported by the National Bioresource Project (YGRC/NBRP) (http://yeast.lab.nig.ac.jp/nig/).
ANP3 is an S. pombe segregant from a cross between GP46 (a gift from Gerry Smith, Fred Hutchinson Cancer Research Center, Seattle, WA) and B82 (a gift from Chikashi Shimoda, Osaka City University, Osaka, Japan). HJP1 was obtained as a segregant from a cross of ANP3 and GP1327 (a gift from Gerry Smith).
Plasmids
The plasmids used in this study are listed in Table 2. To make plasmids pRS416TEF-ScSEC2 and pRS414TEF-ScSEC2, full-length ScSEC2 was amplified by PCR using S. cerevisiae genomic DNA from strain AN120 as a template and HJO97 and HJO98 as primers (sequences of all primers used are available upon request). The PCR products were purified, digested with EcoRI and XhoI, and cloned into similarly digested pRS416TEF or pRS414TEF (Mumberg et al., 1995). ScSEC21-160 and ScSEC2161-759 were similarly amplified by PCR from strain AN120 using oligos HJO97 and HJO99, and HJO104 and HJO98, respectively. The EcoRI- and XhoI-digested PCR products were cloned into similarly digested pRS414TEF to get pRS414TEF-ScSEC21-160 and pRS414TEF-ScSEC2161-759.
Table 2.
Plasmids used in this study
| Plasmid | Selected features | Source |
|---|---|---|
| pRS416TEF | CEN, ARSH4, URA3 | Mumberg et al. (1995) |
| pRS414TEF | CEN, ARSH4, TRP1 | Mumberg et al. (1995) |
| pRS416TEF-ScSEC2 | CEN, ARSH4, URA3 | This study |
| pRS414TEF-ScSEC2 | CEN, ARSH4, TRP1 | This study |
| pRS414TEF-ScSEC21-160 | CEN, ARSH4, TRP1 | This study |
| pRS414TEF-ScSEC2161-759 | CEN, ARSH4, TRP1 | This study |
| pRS414TEF-Spspo13+ | CEN, ARSH4, TRP1 | This study |
| pRS414TEF-Spspo13+-ScSEC2161-759 | CEN, ARSH4, TRP1 | This study |
| pRS425TEF-Spypt2+ | 2 micron, LEU2 | This study |
| pGP564-ScSEC4 | 2 micron, LEU2 | Jones et al. (2008) |
| pGP564-ScYPT1 | 2 micron, LEU2 | Jones et al. (2008) |
| pGP564-ScYPT31 | 2 micron, LEU2 | Jones et al. (2008) |
| pJRU-MCS2 | YRp, ars1, ura4+ | Moreno et al. (2000) |
| pJRU-MCS2-PSpSpo13-Spspo13+ | YRp, ars1, ura4+ | This study |
| pJRU-MCS2-PSpSpo13-Spspo13-F79A | YRp, ars1, ura4+ | This study |
| pJRU-MCS2-PSpSpo13-Spspo13+-mRFP | YRp, ars1, ura4+ | This study |
| pJRU-MCS2-PSpSpo13-Spspo13-F79A -mRFP | YRp, ars1, ura4+ | This study |
| FY532a (pREP41-GFP-psy1+) | YRp, ars1, LEU2 | Nakamura et al. (2001) |
| pET15b | His-tag, AmpR | Novagen |
| pET15b-ScSEC4 | His-tag, AmpR | This study |
| pET15b-ScYPT1 | His-tag, AmpR | This study |
| pET15b-Spypt1+ | His-tag, AmpR | This study |
| pET15b-Spypt2+ | His-tag, AmpR | This study |
| pET15b-Spypt3+ | His-tag, AmpR | This study |
| pGEX5X-1 | GST, AmpR | GE Healthcare |
| pGEX5X-1-ScSEC21-160 | GST, AmpR | This study |
| pGEX3X-Spspo13+ | GST, AmpR | This study |
| pGEX3X-Spspo13-F79A | GST, AmpR | This study |
a This plasmid was obtained from the Yeast Genetic Resource Center of Japan supported by the National Bioresource Project (YGRC/NBRP) (http://yeast.lab.nig.ac.jp/nig/).
The Spspo13+ coding region was amplified by PCR using S. pombe genomic DNA as a template and HJO116 and HJO134 as oligos. Because the genomic DNA of Spspo13+ contains an intron after the fifth codon, HJO116 was designed to add these codons to the 5′ end second exon and thereby remove the intron. The Spspo13+ coding region was then digested with EcoRI and XhoI, and cloned into similarly digested pRS414TEF to get pRS414TEF-Spspo13+.
To construct the chimeric plasmid, pRS414TEF-Spspo13+-ScSEC2161-759, the coding region of Spspo13+, without the stop codon, was PCR amplified from S. pombe genomic DNA using HJO116 and HJO117 as oligos. A BamHI-EcoRI–digested PCR fragment was then cloned into similarly digested pRS414TEF-ScSEC2161-759.
Plasmid pJRU-MCS2 (Moreno et al., 2000) was used as the vector backbone for plasmids used in S. pombe. The Spspo13+ promoter (−500 upstream of first ATG) was amplified from S. pombe genomic DNA by oligos HJO178 and HJO179. The PCR products was purified, digested with XhoI and PstI, and cloned into similarly digested pJRU-MCS2 to create pJRU-MCS2-PSpSpo13. To make pJRU-MCS2-PSpSpo13-Spspo13+, the Spspo13+ coding region was PCR amplified using S. pombe genomic DNA as a template and HJO176 and HJO145 as oligos. A KpnI-SacI–digested PCR fragment was cloned into similarly digested pJRU-MCS2-PSpSpo13.
The plasmid pJRU-MCS2-PSpSpo13-Spspo13+-mRFP was constructed in three steps. First, a PCR fragment containing the monomeric red fluorescent protein (mRFP) gene was amplified using pTi mRFP as a template (Gao et al., 2005) and YSO33 and HNO944 as primers and cloned as a HindIII-XhoI fragment into pRS424TEF (Mumberg et al., 1995). Next, the mRFP fragment from this construct was isolated as an EcoRI-XhoI fragment and used to replace the ScSEC2 coding region of pRS414TEF-Spspo13+-ScSEC2161-759, creating pRS414TEF-Spspo13+-mRFP. Finally, Spspo13-mRFP was PCR amplified from pRS414TEF-Spspo13+-mRFP by using HJO176 and HJO188 as oligos. This PCR product was KpnI-SacI digested and cloned into pJRU-MCS2-PSpSpo13.
pJRU-MCS2-SpSpo13pr-Spspo13-F79A was created by site-directed mutagenesis of the plasmid pJRU-MCS2-SpSpo13pr-Spspo13+ using oligos HJO182 and HJO183 (QuikChange kit; Stratagene, La Jolla, CA). The same oligos were used for site-directed mutagenesis of pJRU-MCS2-SpSpo13pr-Spspo13+-mRFP to make pJRU-MCS2-SpSpo13pr-Spspo13-F79A-mRFP. The Spspo13 gene was fully sequenced after mutagenesis to ensure that no other mutations were introduced during the procedure (sequencing performed at the Stony Brook DNA Sequencing Facility).
To construct plasmids expressing 6XHis-tagged ScSEC4 or ScYPT1, ScSEC4 and ScYPT1 were PCR amplified using S. cerevisiae genomic DNA as a template and using oligos HJO150 and HJO151, HJO152 and HJO153, respectively. The PCR products were purified, digested with XhoI and BamHI, and cloned into similarly digested plasmid pET15b (Novagen, Madison, WI) To make plasmids expressing glutathione transferase (GST)-tagged ScSEC21-160, an EcoRI-XhoI fragment of pRS414-TEFpr-ScSEC21-160 containing ScSEC21-160 was cloned into pGEX5X-1 (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). To make plasmids expressing GST-tagged Spspo13+, the Spspo13+ gene was first PCR amplified using pRS414-TEFpr-Spspo13+ as a template and HJO137 and HJO134 as oligos. The PCR product was then cloned into pGEX3X (GE Healthcare) as a BamHI-EcoRI fragment. The plasmid expressing GST-tagged Spspo13-F79A was constructed by site-directed mutagenesis of the plasmid pGEX3X-Spspo13+ using oligos HJO182 and HJO183.
S. pombe Sporulation Assays
Freshly transformed S. pombe cells were grown on selective medium at 32°C for 2 d. The cells were then patched onto an SPA plate (1%, wt/vol glucose, 7.3 mM KH2PO4, 1 ml of 1000× vitamin stock, and 3%, wt/vol Difco Bacto Agar [Difco, Detroit, MI]) to induce mating and sporulation. Sporulation was assayed by observation in the light microscope after 15–18 h incubation on the SPA plate at room temperature.
Fluorescence Microscopy
Freshly transformed S. pombe cells were cultured in 3 ml of EMM2 (3 g/l potassium hydrogen phthalate, 2.2 g/l sodium phosphate dibasic, 5 g/l ammonium chloride, 20 g/l dextrose, 2.1 g/l minimal salts, 0.2 g/l vitamins, and 3 mg/l trace elements), with selective supplements (75 μg/ml adenine and 225 μg/ml histidine) for 24 h. The cells were precipitated, washed twice with 1 ml EMM-N sporulation medium (3 g/l potassium hydrogen phthalate, 2.2 g/l sodium phosphate dibasic, 20 g/l dextrose, 2.1 g/l minimals, 0.2 g/l vitamins, and 3 mg/l trace elements). The cells were induced to enter meiosis by incubating in 3 ml of EMM-N at room temperature. After 9-h incubation in the EMM-N, aliquots of cells were examined in an Axioplane2 microscope (Carl Zeiss, Thornwood, NY). Fluorescence images were obtained by using Axiovision release 4.7.
Recombinant Protein Preparation
All recombinant proteins were expressed in Escherichia coli (BL21) and induced at room temperature by adding 0.5 mM isopropyl β-d-thiogalactoside for 4 h. The 6XHis-tagged Rab proteins were purified using nickel-nitrilotriacetic acid Superflow columns under native conditions (QIAGEN, Valencia, CA). To stabilize nucleotide-binding protein, 0.1 mM GDP was included in the lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 8.0), wash buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, pH 8.0) and elution buffer (50 mM NaH2PO4, 300 mM NaCl, and 250 mM imidazole, pH 8.0). After elution, samples were concentrated by centrifugation in a Microcon YM-10 centrifugal filter unit (Millipore, Billerica, MA) and kept in storage buffer (20 mM Tris, pH 8.0, 100 mM KCl, 1 mM dithiothreitol [DTT], 0.1 mM GDP, 100 μM phenylmethylsulfonyl fluoride [PMSF], and 20% glycerol) at −20°C.
GST fusion proteins were purified as described previously (Nakanishi et al., 2004). After elution and concentration, samples were kept in storage buffer (20 mM Tris, pH 8.0, 100 μM PMSF, and 20% glycerol) at −20°C.
GST Pull-Down Assay
GST pull-down assays were modified from Ortiz et al. (2002). For binding experiments with the nucleotide-bound form of Rab proteins, 0.8 μg of 6XHis-ScSec4 or 6XHis-ScYpt1 was preloaded with GDP or guanosine 5′-(β,γ-imido)triphosphate (GppNHp) in MgCl2 binding buffer (1× phosphate-buffered saline [PBS], 5 mM MgCl2, 1 mg/ml bovine serum albumin [BSA], and 1 mM DTT) for 30 min at room temperature. The GST fusion proteins immobilized on glutathione-Sepharose beads (30 μl of beads) were incubated with the preloaded Rab proteins (0.3 μg) in the MgCl2 binding buffer for 60 min at room temperature. After the binding reactions, the glutathione-Sepharose beads were washed five times with PBS buffer containing MgCl2 and resuspended in 2× SDS sample buffer (60 mM Tris-HCl, pH 6.8, 10% glycerol, 2% SDS, 0.05 mg/ml bromphenol blue, and 5% β-mercaptoethanol). Pull-down products were separated by SDS-polyacrylamide gel electrophoresis and analyzed by Western blot. 6XHis-ScSec4 and 6XHis-ScYpt1 were first detected by rabbit anti-histidine antibody (Santa Cruz Biotechnology, Santa Cruz, CA), and then goat anti-rabbit peroxidase-conjugated secondary antibody was used (GE Healthcare). GST fusion proteins were detected by goat horseradish peroxidase-conjugated anti-GST antibody (Abcam, Cambridge, MA).
For binding experiments with the nucleotide-free form of the Rab proteins, 6XHis-ScSec4 and 6XHis-ScYpt1 were not preloaded with nucleotides but were incubated with GST fusion proteins immobilized on beads in EDTA binding buffer (1× PBS, 5 mM EDTA, 1 mg/ml BSA, and 1 mM DTT). The resins were washed with PBS buffer containing EDTA.
Nucleotide Exchange Assays
Purified 6XHis-tagged Rab proteins were incubated in loading buffer (50 mM Tris, pH 8.0, 100 mM KCl, 1 mM EDTA, and 1 mM DTT) containing a twofold molar excess of GDP (Jena Bioscience, Jena, Germany) or N-methylanthraniloyl (mant)-GDP (Invitrogen, Carlsbad, CA) for 30 min at room temperature. To terminate the loading reaction, MgCl2 was added to a final concentration of 20 mM, and free GDP or free mant-GDP was removed by centrifugation in a Microcon YM-10 filter unit (Millipore). 6XHis-Rab bound to GDP or mant-GDP was then concentrated in buffers containing 50 mM Tris, pH 8.0, 100 mM KCl, and 6 mM MgCl2.
For the GTP loading assay, 100 nM mant-GppNHp (Invitrogen) was added to reaction buffer (10% glycerol, 50 μg/ml BSA, 50 mM Tris, pH 8.0, 100 mM KCl, 1 mM DTT, and 6 mM MgCl2) and allowed to equilibrate for 300 s in a thermostated cuvette (15°C). To initiate the reaction, 6XHis-ScSec4 preloaded with GDP was added to the mixture to a final concentration of 400 nM. To test GEF activity, purified GST, GST-ScSec2, or GST-SpSpo13 was premixed with 6XHis-ScSec4-GDP and then added to the mixture at the concentrations indicated. Fluorescence emission was monitored every 5 s for a total of 1500 s by using a fluorimeter (FP-6200; Jasco, Tokyo, Japan) with the following settings: λex = 290 nm, λem = 460 nm, and slits = 5/5.
For GDP release assay, 6XHis-ScSec4 was loaded with mant-GDP as describe above and added to the reaction buffer containing an excess of GppNHp (final concentration, 20 μM) (Jena Bioscience). The dissociation of mant-GDP was first monitored without a GEF protein by using the fluorimeter with the same settings as described in GTP loading studies. At the indicated time point, purified GST-ScSec2, GST-SpSpo13, or GST-SpSpo13F79A, or EDTA, was added to the mixture at desired concentrations, and the fluorescence emission was then monitored for another 1300 s.
RESULTS
The MOP Component SpSpo13 Has Homology to ScSec2 GEF Domain
Spspo13+ encodes a protein of 138 amino acids predicted to form a coiled-coil structure (Nakase et al., 2008). Iterative blast searches with the full-length SpSpo13 revealed a strong patch of conservation between SpSpo13 and the ScSec2 protein of budding yeast. Full-length SpSpo13 shares 24% identity and 43% similarity with the N-terminal 160 amino acids that contain the GEF activity of ScSec2 (Figure 1). The crystal structure of this domain of ScSec2 has been solved in complex with ScSec4 (Dong et al., 2007; Sato et al., 2007). The residues of ScSec2 that contact ScSec4 are highly conserved in SpSpo13 (Figure 1). This sequence homology suggests that SpSpo13 might have GEF activity similar to ScSec2.
Figure 1.
SpSpo13 is homologous to the ScSec2 GEF domain. (A) Sequence alignment of ScSec2 GEF domain with SpSpo13. Identical residues are highlighted in yellow, and conservative changes are shaded gray. ScSec2 residues that are found in a close contact with ScSec4 in the cocrystal structure are in red (Dong et al., 2007). Spspo13 residues that are conserved in the ScSec4 binding surface are in green. Phe79 of SpSpo13 that was mutated to alanine is indicated in green. (B) View of the cocrystal structure of ScSec2 GEF domain and ScSec4 (PDB 2OCY; Dong et al., 2007). ScSec4 is in gray, above and the ScSec2 dimer is below. The switch I and II regions of ScSec4 are labeled. The side chains of the ScSec2 residues highlighted in A are shown in red, except for Phe109 in green.
SpSpo13+ Can Substitute for the ScSEC2 GEF Domain
If SpSpo13 has GEF activity, it might be able to substitute for ScSec2 in activating ScSec4 in S. cerevisiae. ScSEC2 is an essential gene in budding yeast (Nair et al., 1990). The N-terminal GEF domain is sufficient to support the essential function of ScSec2, and the remaining C-terminal region seems to be required for proper polarized localization of ScSec2 (Elkind et al., 2000; Ortiz et al., 2002). To examine whether SpSpo13+ can provide GEF function, Spspo13+/ ScSEC2 chimeras were constructed and tested for ability to rescue the growth of a sec2Δ strain.
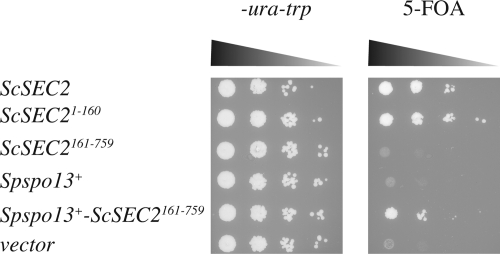
Using a plasmid shuffle approach, we showed that the ScSEC2 GEF domain alone (ScSEC21-160) was sufficient to restore the growth of the sec2Δ mutant at a level comparable with expression of full-length ScSEC2 gene, in agreement with an earlier report (Ortiz et al., 2002). In contrast, the ScSEC2 C-terminal region (ScSEC2161-759) failed to rescue the growth defect of the sec2Δ mutant (Figure 2).
Figure 2.
A Spspo13+-ScSec2 chimera rescues the growth defect of a sec2Δ. Strain HJ79 (sec2Δ; pCEN-URA3-ScSEC2) was transformed with CEN-TRP1-plasmids carrying the indicated genes. Numbers indicate amino acids. The transformed strains were grown in nonselective media overnight, and 10-fold serial dilutions of the overnight culture were spotted onto a synthetic plate lacking tryptophan and uracil or onto a plate containing 5-fluoroorotic acid (5-FOA). The plates were photographed after 2-d incubation at 30°C.
Although Spspo13+ alone did not suppress the sec2Δ growth defect, the Spspo13+-ScSEC2161-759 fusion did restore growth (Figure 2), indicating that this fusion gene encodes a functional protein that can substitute for ScSec2. This result indicates that SpSpo13 has GEF activity capable of activating ScSec4 in S. cerevisiae.
SpSpo13 Binds Preferentially to the Nucleotide-free Form of ScSec4
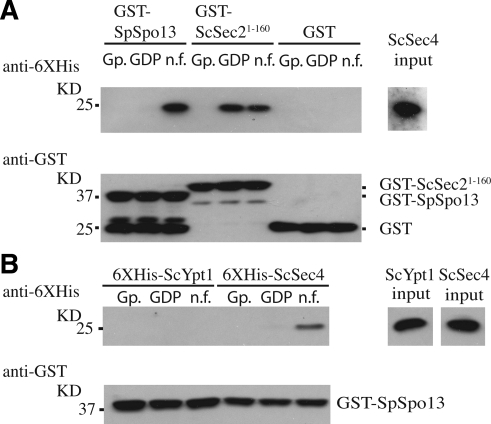
To test directly whether SpSpo13 can bind to ScSec4, glutathione transferase-tagged-SpSpo13 (GST-SpSpo13) was prepared from E. coli. The predicted molecular mass of the GST-SpSpo13 fusion protein is ∼44 kDa, and a band that ran a little above the 37-kDa marker was confirmed as GST-SpSpo13 by Western blot analysis using anti-GST antibodies (Figure 3A). GEFs preferentially bind to the nucleotide-free form of their target GTPase (Bos et al., 2007). GST-SpSpo13 attached to glutathione-Sepharose beads was therefore incubated with three different forms of recombinant hexahistidine-tagged ScSec4 (6XHis-ScSec4): the GDP-bound form, GppNHp (a nonhydrolysable analogue of GTP)-bound form, or the nucleotide-free form. Binding of ScSec2 GEF domain GST-ScSec21-160 was performed as a positive control in the binding assay.
Figure 3.
SpSpo13 binds preferentially to the nucleotide free form of ScSec4. GST-pull down assays. (A) 6XHis-ScSec4 bound to GppNHp (Gp.), GDP, or in its nucleotide-free (n.f.) form was mixed with the indicated GST fusion. After precipitation of the GST fusion proteins, the resulting pellets were analyzed by Western blot using anti-6XHis antibodies to detect precipitated 6XHis-ScSec4 (top) or with anti-GST antibodies to detect SpSpo13 and ScSec21-160. (B) GST-SpSpo13 immobilized on Sepharose beads was mixed with different derivatives of 6XHis-ScYpt1 or 6XHis-ScSec4 and analyzed as described in A. Input (right) represents 10% of Rab proteins used per binding reaction.
GST-SpSpo13 preferentially bound to the nucleotide-free form of ScSec4 compared with the GDP- or GppNHp-bound form, whereas GST-ScSec21-160 displayed binding to both the GDP-bound and nucleotide-free forms. The interaction of GST-SpSpo13 and ScSec4 was specific because GST alone did not bind to any conformations of ScSec4, and GST-SpSpo13 did not bind to another Rab GTPase, ScYpt1 (Figure 3B). These binding characteristics of GST-SpSpo13 support the idea that SpSpo13 is a guanine nucleotide exchange protein.
SpSpo13 Has Guanine Nucleotide Exchange Activity toward ScSec4 In Vitro
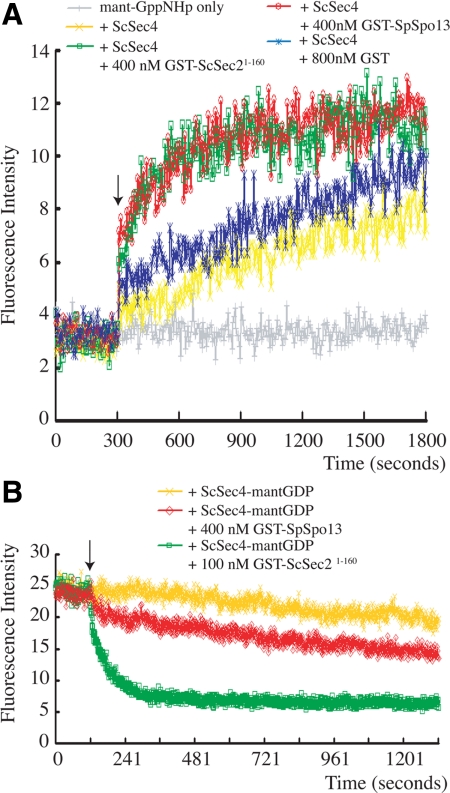
To confirm that SpSpo13 can act as a GEF, a real time fluorescent nucleotide exchange assay that monitored the level of ScSec4 activation was performed (Rojas et al., 2003). In this assay, ScSec4 was preloaded with GDP and added to buffer containing a limited amount of mant-GppNHp. Binding of the mant-GppNHp, a fluorescent nonhydrolysable analogue of GTP, to ScSec4 is detected by monitoring a fluorescence resonance energy transfer (FRET) interaction between a tryptophan residue on ScSec4 and the fluorophore. Although little FRET was detected with buffer containing only the fluorophore, an increase of fluorescence intensity was observed when ScSec4-GDP alone was added (Figure 4A). The fluorescence intensity increased linearly over the time course after ScSec4 was added, probably due to the high intrinsic GDP-release rate of ScSec4 (Ortiz et al., 2002). Notably, when ScSec4 was premixed with GST- SpSpo13 and added to the buffer containing mant-GppNHp, the increase in fluorescence intensity during first 200 s was dramatically higher, and the fluorescence intensity soon reached an asymptotic value (Figure 4A). Similar results were seen when ScSec4 was premixed with the known GEF, GST-ScSec21-160 (Figure 4A). In contrast, the initial rate of ScSec4 activation did not rapidly increase when ScSec4 premixed with GST.
Figure 4.
SpSpo13 facilitates nucleotide exchange on ScSec4. (A) SpSpo13 stimulates GTP binding. ScSec4 was preloaded with GDP. At t = 300 s (indicated by arrow), ScSec4 alone, ScSec4 mixed with GST, ScSec4 mixed with GST-ScSec21-160, or ScSec4 mixed with GST-SpSpo13 was added to buffer containing the fluorescent GTP analogue mant-GppNHp. We used 400 nM ScSec4-GDP per reaction. Binding of the analogue to ScSec4 was monitored by following the fluorescence signal created by a FRET interaction between ScSec4 and the mant-GppNHp. The gray line indicates the basal level of fluorescence signal from the fluorophore under these conditions. (B) SpSpo13 stimulates GDP release. 6XHis-ScSec4 preloaded with the fluorescent GDP analogue mant-GDP was incubated in buffer containing GppNHp. We used 400 nM ScSec4-mant-GDP per reaction. At the time indicated by the arrow, buffer, GST-ScSec21-160, or GST-SpSpo13 was added to the reaction mixture. Release of GDP was monitored by a loss of fluorescent signal from the ScSec4-mant-GDP FRET interaction. The gray line indicates the basal level of fluorescence signal from the fluorophore under these conditions.
This assay establishes that SpSpo13 can stimulate GTP binding by ScSec4 but, because of the low concentration of mant-GppNHp present in the buffer, is not informative about the kinetics of GDP release (Rojas et al., 2003). To examine these kinetics, the displacement of mant-GDP from ScSec4 in a solution containing a large excess of GppNHp was examined. In the reverse of the mant-GppNHp loading assay, release of mant-GDP from ScSec4 was monitored by a decrease of FRET (Figure 4B). Addition of GST-ScSec21-160 to buffer containing the mant-GDP loaded ScSec4 triggered a swift drop of fluorescence intensity to a basal level, indicating that GST-ScSec21-160 efficiently facilitates mant-GDP dissociation. Addition of GST-SpSpo13 to the preloaded ScSec4 also stimulated a drop in fluorescence intensity but at a much slower rate (Figure 4B), indicating that SpSpo13 facilitates mant-GDP release of ScSec4 with slower kinetics than GST-ScSec21-160. These data demonstrate that SpSpo13 can act as a GEF for ScSec4 in vitro, though not as efficiently as its natural GEF, ScSec2. This result is consistent with our finding that Spspo13+-ScSEC2161-759 rescues the growth of a sec2Δ mutant less well than the native ScSEC2 in vivo (Figure 2).
A Mutation in Spspo13 That Impairs GEF Activity Blocks FSM Formation
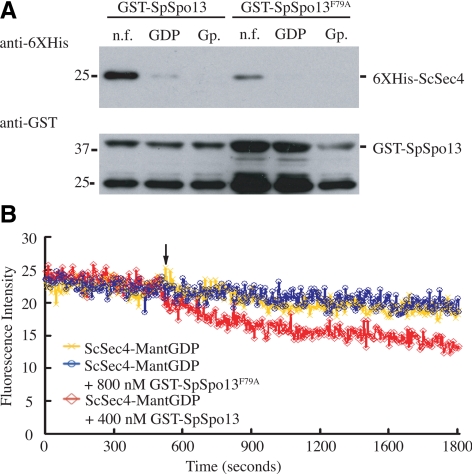
Within the ScSec4 binding site of ScSec2, Phe109 of ScSec2 is a critical residue for its GEF activity and binding affinity for ScSec4 (Sato et al., 2007; Figure 1). We reasoned that if SpSpo13 acts on an S. pombe Rab in a similar way to ScSec2 on ScSec4, then the corresponding residue of SpSpo13, Phe79, should be essential for SpSpo13 GEF activity. Therefore, Phe79 of SpSpo13 was mutated to alanine, and the resulting mutant protein was tested in the GST-pull-down and GEF assays. The pull-down assay revealed that GST-SpSpo13F79A still bound to the nucleotide-free form of ScSec4 but at a lower level than wild-type GST-SpSpo13 (Figure 5A). In the GEF assay, adding GST-SpSpo13F79A did not stimulate mant-GDP dissociation, indicating that the mutant protein has lost GEF activity in vitro (Figure 5B). Moreover, expression of a Spspo13-F79A-ScSEC2161-759 fusion gene in a sec2Δ mutant does not support cell growth in S. cerevisiae (data not shown), suggesting that the mutant protein lacks GEF activity in vivo as well.
Figure 5.
Mutation of conserved residue in SpSpo13 leads to the loss of GEF activity in vitro and a FSM assembly defect in vivo. (A) GST-SpSpo13 displays reduced binding to ScSec4. GST-SpSpo13 or GST-SpSpo13F79A immobilized on glutathione-Sepharose beads was mixed with the nucleotide-free (n.f.), GDP-bound, or GppNHp-bound (Gp.) forms of 6XHis-ScSec4. The mixtures were centrifuged, and the resulting pellets analyzed by Western blot. Top, blot probed with anti-6XHis antibodies. Bottom, same samples probed with anti-GST antibodies. (B) GDP release assay. 6XHis-ScSec4 preloaded with the fluorescent GDP analogue mant-GDP was incubated in buffer containing GppNHp. At the time indicated by the arrow, buffer (yellow line), 400 nm GST-SpSpo13 (red line), or 800 nM GST-SpSpo13F79A (blue line) was added to the reaction mixture. Release of GDP was monitored by a loss of fluorescent signal from the ScSec4-mant-GDP FRET interaction.
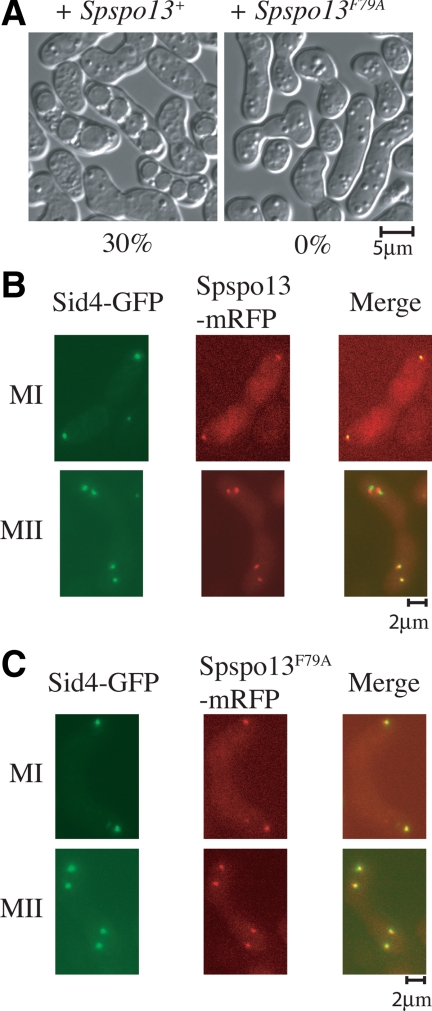
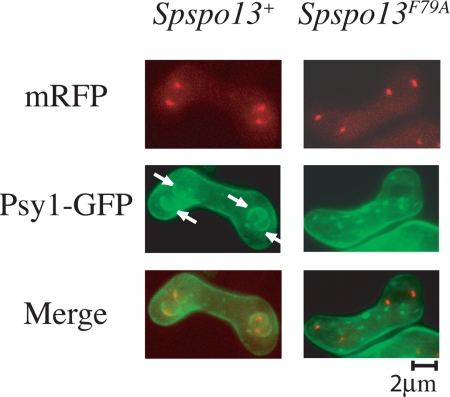
To examine the effect of the F79A mutant on sporulation and FSM formation in S. pombe, plasmids carrying Spspo13+ or Spspo13-F79A were expressed in a spo13-B82 mutant. The spo13-B82 allele has a nonsense mutation at the 53rd residue and fails to form FSMs or spores (Nakase et al., 2008) Introduction of the wild-type Spspo13+ restored sporulation to this strain; however expression of Spspo13-F79A did not (Figure 6A). SpSpo13 and SpSpo13F79A were next tagged with mRFP and expressed in sporulating cells under the native spo13 promoter. Sid4-GFP was used as a marker for the SPB (Chang and Gould, 2000). Spo13-mRFP was visible at the SPB, beginning with cells in meiosis I and persisted at the SPB throughout meiosis II, as reported previously (Nakase et al., 2008; Figure 6B). The localization of SpSpo13F79A-mRFP was indistinguishable from the wild-type protein (Figure 6C), indicating that the mutant protein is expressed and properly localized. To more closely examine the Spspo13-F79A defect, FSM assembly was visualized using GFP-Psy1, which encodes a SNARE protein and localizes at FSM during meiosis II (Maeda et al., 2009), and SpSpo13 was detected again by tagging with mRFP. Growing FSMs were observed adjacent to each of the SPBs in 100% of the cells expressing wild-type Spspo13+ (n = 20), but no FSMs were formed in cells expressing Spspo13-F79A (Figure 7) (n = 20). The GEF activity of SpSpo13 is therefore required for FSM assembly in S. pombe.
Figure 6.
SpSpo13F79A fails to support spore formation but localizes properly to the SPB. (A) spo13-F79A cells fail to sporulate. Strain ANP3 (h90, spo13-B82) was transformed with plasmids carrying Spspo13+ or Spspo13-F79A and incubated on sporulation medium. Sporulation was assessed by light microscopy. The sporulation frequency for each strain, measured as the percentages of asci out of 200 total zygotes, is shown. (B and C) SpSpo13F79A-mRFP and SpSpo13-mRFP localize to the meiotic SPB. Strain FY12476 (h90 spo13::ura4+ ura4 leu1 sid4GFP::kanR) was transformed with plasmids carrying Spspo13+-mRFP or SpSpo13F79A-mRFP, sporulated, and examined by fluorescence microscopy.
Figure 7.
Loss of GEF activity in SpSpo13 leads to an FSM assembly defect in vivo. Spspo13F79A cells do not form FSMs. Strain HJP1 (h90, spo13-B82) was cotransformed with plasmids carrying GFP-Psy1 and Spspo13+-mRFP or Spspo13F79A-mRFP, sporulated, and examined by fluorescence microscopy. Arrows indicate FSMs.
SpYpt2 Is the Probable In Vivo Target of Spspo13
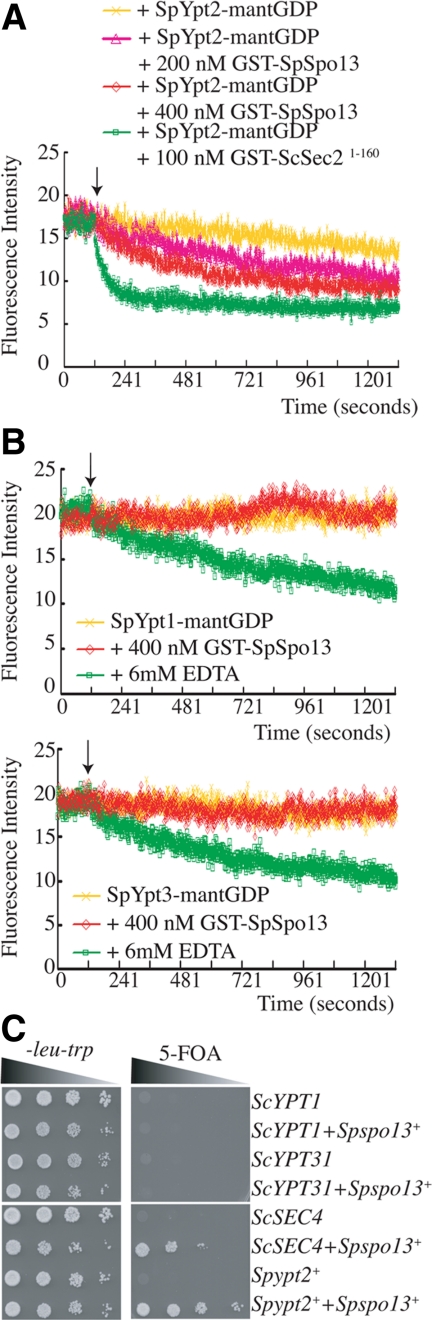
SpYpt2 is the S. pombe Rab most closely related to ScSec4 and, like ScSec4 has been shown to function in exocytosis (Craighead et al., 1993). Given the homology of ScSec4 and SpYpt2, the activity of SpSpo13 on ScSec4, and that ScSec4 has been shown to be required for prospore membrane formation during sporulation in S. cerevisiae (Neiman, 1998), SpYpt2 is a strong candidate to be the physiological target of SpSpo13. A 6XHis-tagged version of SpYpt2 was purified from E. coli, and the ability of SpSpo13 to stimulate GDP release was examined (Figure 8A). Similar to what was observed with ScSec4 as a substrate (Figure 4B), SpSpo13 stimulated GDP release, although not as efficiently as ScSec2. Thus, SpSpo13 can act on SpYpt2. To examine the specificity of SpSpo13, two other S. pombe Rab proteins, SpYpt1 and SpYpt3, were also purified and examined. Addition of 6 mM EDTA to the reaction was used as a control for GDP release in these experiments (Figure 8B). SpSpo13 failed to stimulate release of GDP from either SpYpt1 or SpYpt3, indicating specificity of SpYpt2 in vitro.
Figure 8.
SpSpo13 specifically facilitates nucleotide exchange by SpYpt2. (A) SpSpo13 stimulates GDP release by SpYpt2. 400 nM SpYpt2-mant-GDP was used per reaction. At the time point indicated by the arrow, GST-ScSec21-160 or GST-SpSpo13 was added to the reaction mixture at the indicated concentrations. (B) SpSpo13 cannot stimulate GDP release by SpYpt1 or SpYpt3. We used 400 nM SpYpt1-mant-GDP or 400 nM SpYpt3 per reaction. At the time point indicated by the arrow, 400 nM GST-SpSpo13 or 6 mM EDTA was added to the reaction mixture. (C) SpSpo13 genetically interacts with SpYpt2 in vivo. Strain HJ75-4 (sec2Δ; pCEN-URA3-ScSEC2) was cotransformed with a CEN-TRP1-plasmid carrying Spspo13+ and 2μ-LEU2-plasmids carrying indicated YPT genes. The transformed strains were grown in nonselective media overnight, and 10-fold serial dilutions of the overnight culture were spotted onto a synthetic plate lacking tryptophan and leucine or onto a plate containing 5-fluoroorotic acid (5-FOA). The plates were photographed after 2-d incubation at 30°C.
Deletion of SpYpt2 is lethal (Craighead et al., 1993), and overexpression of SpYpt2 did not rescue the spore formation defect of the spo13-F79A mutant (Yang, unpublished observations). Therefore, to look for evidence of in vivo interaction between SpSpo13 and SpYpt2, we used the sec2Δ rescue assay. SpSpo13 expression was unable to rescue the growth defect of a sec2Δ mutant (Figure 2). Similarly, overexpression of ScSec4, ScYpt1, SpYpt2, or ScYpt31 did not restore growth to the mutant. However, coexpression of SpSpo13 with SpYpt2 or with ScSec4 allowed rescue of the sec2Δ mutant (Figure 8C). In particular, coexpression of SpSpo13 and SpYpt2 completely rescued the growth defect. This result indicates that SpSpo13 can interact with SpYpt2 in vivo as well as in vitro.
DISCUSSION
We report here that a SPB component in S. pombe, SpSpo13, has homology to the GEF domain of ScSec2 and can function to stimulate nucleotide exchange both in vitro and in vivo. A mutation of SpSpo13 that impairs GEF activity blocks FSM assembly, indicating that the GEF activity is required for MOP-mediated membrane formation. Several lines of evidence suggest that SpYpt2 is the physiological Rab target of SpSpo13: 1) SpYpt2 is the homologue of ScSec4, which is required for prospore membrane formation in S. cerevisiae (Neiman, 1998); 2) SpSpo13 can act as a GEF for ScSec4 both in vivo and in vitro and 3) interact with SpYpt2 when coexpressed in S. cerevisiae; and 4) SpSpo13 specifically stimulates GDP release from SpYpt2 and not other S. pombe Rab proteins in vitro.
In S. cerevisiae, vesicles attach to the MOP before fusion (Nakanishi et al., 2006), suggesting that the MOP functions as a vesicle docking complex upstream of SNARE-mediated membrane fusion. In this light, our finding that the S. pombe MOP contains a GEF activity provides a strong parallel to other vesicle tethering complexes (Stenmark, 2009). For example, the TRAPP-I and TRAPP-II complexes involved in endoplasmic reticulum-to-Golgi trafficking and intra-Golgi trafficking, contain GEF activity directed toward Ypt1, and possibly Ypt31/Ypt32 (Jones et al., 2000a; Cai et al., 2008). Similarly, the Vps-C/HOPS complex involved in endosome–vacuole trafficking acts as a Ypt7-directed GEF (Wurmser et al., 2000). Stimulation of Rab activity may be a common mechanism by which tethering complexes promote the downstream events of vesicle fusion.
The fact that SpSpo13 is a component of the SPB also highlights a difference between the function of SpSpo13 and ScSec2. In addition to the GEF domain, ScSec2 has additional domains that target it to the vesicle and interact with the exocyst complex (Elkind et al., 2000; Ortiz et al., 2002). ScSec2 is therefore thought to associate with transport vesicles and travel with them to the site of membrane fusion (Walch-Solimena et al., 1997). By contrast, SpSpo13 is already localized to the future site of membrane fusion, the SPB, in meiosis I before precursor vesicles arrive. This leaves open the question of how the vesicles interact with SpSpo13. It may be that other components of the SPB initially interact with the vesicles and this serves to recruit them for SpSpo13 action.
In vitro, the GEF activity of SpSpo13 is not strong relative to the GEF domain of ScSec2, even when assayed on SpYpt2. It may be that the GST-SpSpo13 fusion protein does not fold properly in vitro. Alternatively, other SPB proteins missing from the in vitro reactions may be necessary to stimulate greater GEF activity of SpSpo13 in vivo.
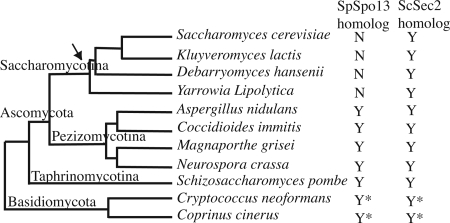
It should be noted that, in addition to SpSpo13, the S. pombe genome also contains an open reading frame (ORF) that seems to be a bona fide homologue of ScSec2, SPAC23C4.10. As with ScSec2, the predicted protein is large and contains an N-terminal GEF domain. Examination of other sequenced fungal genomes revealed the presence of two ScSec2/SpSpo13 related ORFs in many species. In the ascomycetes, such as Aspergillus, that contain both ORFs, one is shorter and more closely related to SpSpo13 and the other ORF is longer and more closely related to ScSec2. In the more distantly related basidiomycetes, such as Coprinus or Cryptococcus, one short and one long ORF are also present, although they are not obviously more closely related to SpSpo13 or ScSec2. Thus, in most fungi there seem to be two ScSec2/SpSpo13-related ORFs. However, in the Saccharomycotina, the lineage leading to budding yeasts, the shorter ORF has been lost and only the longer ScSec2-like ORF is present (Figure 9; Supplemental Figure S1).
Figure 9.
Phylogenetic distribution of SpSpo13/ScSec2 related proteins within representative fungi. Fungal tree is based on Fitzpatrick et al. (2006). Arrow indicates loss of an apparent SpSpo13 homologue in the Saccharomycotina lineage. Asterisks indicate that the two ORFs present in Cryptococcus cinerus and Cryptococcus neoformans, although one ORF is short and one ORF is long, do not obviously correspond to SpSpo13 and ScSec2, respectively.
Although no characterization of any of these genes has been reported, this observation raises the possibility that SPB-associated GEF activity is a conserved mechanism driving localized membrane formation during ascosporogenesis. In both S. pombe and S. cerevisiae, the MOP is essential for membrane formation. It is somewhat surprising therefore that there is no primary sequence homology between any of the S. pombe or S. cerevisiae MOP components so far identified. One interesting question is whether the S. cerevisiae MOP also contains a GEF activity, perhaps in a subunit structurally unrelated to SpSpo13/ScSec2. Alternatively, ScSec2 may provide the GEF activity at the MOP during S. cerevisiae meiosis. A temperature-sensitive sec2 allele, sec2-59, sporulates well at restrictive temperature (Neiman, unpublished observations). However, this allele carries a stop codon that leaves the GEF domain intact (Nair et al., 1990; Walch-Solimena et al., 1997). Testing the possible role of ScSec2 in sporulation in S. cerevisiae will require the isolation of an allele with conditional GEF activity.
The issue of how activation of ScSec4 leads to FSM precursor vesicle fusion remains to be determined. During exocytosis at the plasma membrane in S. cerevisiae, activated ScSec4 promotes fusion, in part, by mediating the interaction of the vesicle with the exocyst tethering complex (Guo et al., 1999). Presumably, SpYpt2 functions similarly in exocytosis in S. pombe. That activation of the SpYpt2 in FSM formation occurs in the context of a distinct tethering complex, the MOP, raises the question of whether or not the exocyst is required for coalescence of FSM precursor vesicles. If the MOP substitutes for the role of the exocyst in tethering the two membranes, then the Rab protein involved must promote fusion through some other route, perhaps by interaction with other factors that impinge on the assembly of SNARE complexes such as Sec1-family or tomosyn-family proteins (Aalto et al., 1997; Wiederkehr et al., 2004; Grosshans et al., 2006).
Finally, these results highlight an intriguing parallel between FSM formation and ciliogenesis in animal cells. During ciliogenesis a membrane cap initially forms on one of the centrioles. This membrane expands as the centriole migrates to the cell periphery and will eventually fuse with the plasma membrane and form the sheath of the cilium (Sorokin, 1962). Transport to this ciliary membrane requires Rab8, a member of the same Rab subfamily as Sec4 (Nachury et al., 2007; Yoshimura et al., 2007), and a centriole/basal body localized GEF, Rabin8, which is a Sec2/Spo13 family member (Nachury et al., 2007). Thus, in higher cells and in yeast, membrane organization by the microtubule organizing center uses orthologous Rabs and Rab-GEFs.
Supplementary Material
ACKNOWLEDGMENTS
We thank Janet Leatherwood (Stony Brook University) for plasmid pJRU-MCS2, Gerry Smith and Chikashi Shimoda for S. pombe strains, Rolf Sternglanz (Stony Brook University) for anti-histidine antibody, and Nancy Hollingsworth for critical comments on the manuscript. We thank the Yeast Genetic Resource Center of Japan supported by the National BioResource Project (NBRP/YGRC) for plasmid FY532 and strain FY12476.
Abbreviations used:
- FRET
fluorescence resonance energy transfer
- FSM
forespore membrane
- GEF
guanine nucleotide exchange factor
- GST
glutathione transferase
- MOP
meiosis II outer plaque
- SPB
spindle pole body.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-10-0842) on February 3, 2010.
REFERENCES
- Aalto M. K., Jantti J., Ostling J., Keranen S., Ronne H. Mso1p: a yeast protein that functions in secretion and interacts physically and genetically with Sec1p. Proc. Natl. Acad. Sci. USA. 1997;94:7331–7336. doi: 10.1073/pnas.94.14.7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajgier B. K., Malzone M., Nickas M., Neiman A. M. SPO21 is required for meiosis-specific modification of the spindle pole body in yeast. Mol. Biol. Cell. 2001;12:1611–1621. doi: 10.1091/mbc.12.6.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos J. L., Rehmann H., Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- Bresch C., Muller G., Egel R. Genes involved in meiosis and sporulation of a yeast. Mol. Gen. Genet. 1968;102:301–306. doi: 10.1007/BF00433721. [DOI] [PubMed] [Google Scholar]
- Cai Y., et al. The structural basis for activation of the Rab Ypt1p by the TRAPP membrane-tethering complexes. Cell. 2008;133:1202–1213. doi: 10.1016/j.cell.2008.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L., Gould K. L. Sid4p is required to localize components of the septation initiation pathway to the spindle pole body in fission yeast. Proc. Natl. Acad. Sci. USA. 2000;97:5249–5254. doi: 10.1073/pnas.97.10.5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craighead M. W., Bowden S., Watson R., Armstrong J. Function of the ypt2 gene in the exocytic pathway of Schizosaccharomyces pombe. Mol. Biol. Cell. 1993;4:1069–1076. doi: 10.1091/mbc.4.10.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond A. E., Park J. S., Inoue I., Tachikawa H., Neiman A. M. The anaphase promoting complex targeting subunit Ama1 links meiotic exit to cytokinesis during sporulation in Saccharomyces cerevisiae. Mol. Biol. Cell. 2009;20:134–145. doi: 10.1091/mbc.E08-06-0615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong G., Medkova M., Novick P., Reinisch K. M. A catalytic coiled coil: structural insights into the activation of the Rab GTPase Sec4p by Sec2p. Mol. Cell. 2007;25:455–462. doi: 10.1016/j.molcel.2007.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkind N. B., Walch-Solimena C., Novick P. J. The role of the COOH terminus of Sec2p in the transport of post-Golgi vesicles. J. Cell Biol. 2000;149:95–110. doi: 10.1083/jcb.149.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick D. A., Logue M. E., Stajich J. E., Butler G. A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol. Biol. 2006;6:99. doi: 10.1186/1471-2148-6-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X. D., Tachikawa H., Sato T., Jigami Y., Dean N. Alg14 recruits Alg13 to the cytoplasmic face of the endoplasmic reticulum to form a novel bipartite UDP-N-acetylglucosamine transferase required for the second step of N-linked glycosylation. J. Biol. Chem. 2005;280:36254–36262. doi: 10.1074/jbc.M507569200. [DOI] [PubMed] [Google Scholar]
- Grosshans B. L., Andreeva A., Gangar A., Niessen S., Yates J. R., 3rd, Brennwald P., Novick P. The yeast lgl family member Sro7p is an effector of the secretory Rab GTPase Sec4p. J. Cell Biol. 2006;172:55–66. doi: 10.1083/jcb.200510016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W., Roth D., Walch-Solimena C., Novick P. The exocyst is an effector for Sec4p, targeting secretory vesicles to sites of exocytosis. EMBO J. 1999;18:1071–1080. doi: 10.1093/emboj/18.4.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata A., Shimoda C. Structural modification of spindle pole bodies during meiosis II is essential for the normal formation of ascospores in Schizosaccharomyces pombe: ultrastructural analysis of spo mutants. Yeast. 1994;10:173–183. doi: 10.1002/yea.320100205. [DOI] [PubMed] [Google Scholar]
- Jaspersen S. L., Winey M. The budding yeast spindle pole body: structure, duplication, and function. Annu. Rev. Cell. Dev. Biol. 2004;20:1–28. doi: 10.1146/annurev.cellbio.20.022003.114106. [DOI] [PubMed] [Google Scholar]
- Jones G. M., Stalker J., Humphray S., West A., Cox T., Rogers J., Dunham I., Prelich G. A systematic library for comprehensive overexpression screens in Saccharomyces cerevisiae. Nat. Methods. 2008;5:239–241. doi: 10.1038/nmeth.1181. [DOI] [PubMed] [Google Scholar]
- Jones S., Newman C., Liu F., Segev N. The TRAPP complex is a nucleotide exchanger for Ypt1 and Ypt31/32. Mol. Biol. Cell. 2000a;11:4403–4411. doi: 10.1091/mbc.11.12.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S., Newman C., Liu F., Segev N. The TRAPP complex is a nucleotide exchanger for Ypt1 and Ypt31/32. Mol. Biol. Cell. 2000b;11:4403–4411. doi: 10.1091/mbc.11.12.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M., Strasser K. Role of the spindle pole body of yeast in mediating assembly of the prospore membrane during meiosis. EMBO J. 2000;19:3657–3667. doi: 10.1093/emboj/19.14.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Smith G. R. Molecular cloning of the meiosis-induced rec10 gene of Schizosaccharomyces pombe. Curr. Genet. 1995;27:440–446. doi: 10.1007/BF00311213. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Maeda Y., Kashiwazaki J., Shimoda C., Nakamura T. The Schizosaccharomyces pombe syntaxin 1 homolog, Psy1, is essential in the development of the forespore membrane. Biosci. Biotechnol. Biochem. 2009;73:339–345. doi: 10.1271/bbb.80575. [DOI] [PubMed] [Google Scholar]
- Moreno M. B., Duran A., Ribas J. C. A family of multifunctional thiamine-repressible expression vectors for fission yeast. Yeast. 2000;16:861–872. doi: 10.1002/1097-0061(20000630)16:9<861::AID-YEA577>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Mumberg D., Muller R., Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- Nachury M. V., et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- Nair J., Muller H., Peterson M., Novick P. Sec2 protein contains a coiled-coil domain essential for vesicular transport and a dispensable carboxy terminal domain. J. Cell Biol. 1990;110:1897–1909. doi: 10.1083/jcb.110.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Asakawa H., Nakase Y., Kashiwazaki J., Hiraoka Y., Shimoda C. Live observation of forespore membrane formation in fission yeast. Mol. Biol. Cell. 2008;19:3544–3553. doi: 10.1091/mbc.E08-04-0414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Kashiwazaki J., Shimoda C. A fission yeast SNAP-25 homologue, SpSec9, is essential for cytokinesis and sporulation. Cell Struct. Funct. 2005;30:15–24. doi: 10.1247/csf.30.15. [DOI] [PubMed] [Google Scholar]
- Nakamura T., Nakamura-Kubo M., Hirata A., Shimoda C. The Schizosaccharomyces pombe spo3+ gene is required for assembly of the forespore membrane and genetically interacts with psy1+-encoding syntaxin-like protein. Mol. Biol. Cell. 2001;12:3955–3972. doi: 10.1091/mbc.12.12.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura-Kubo M., Nakamura T., Hirata A., Shimoda C. The fission yeast spo14+ gene encoding a functional homologue of budding yeast Sec12 is required for the development of forespore membranes. Mol. Biol. Cell. 2003;14:1109–1124. doi: 10.1091/mbc.E02-08-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi H., de los Santos P., Neiman A. M. Positive and negative regulation of a SNARE protein by control of intracellular localization. Mol. Biol. Cell. 2004;15:1802–1815. doi: 10.1091/mbc.E03-11-0798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi H., Morishita M., Schwartz C. L., Coluccio A., Engebrecht J., Neiman A. M. Phospholipase D and the SNARE Sso1p are necessary for vesicle fusion during sporulation in yeast. J. Cell Sci. 2006;119:1406–1415. doi: 10.1242/jcs.02841. [DOI] [PubMed] [Google Scholar]
- Nakase Y., Nakamura T., Hirata A., Routt S. M., Skinner H. B., Bankaitis V. A., Shimoda C. The Schizosaccharomyces pombe spo20+ gene encoding a homologue of Saccharomyces cerevisiae Sec14 plays an important role in forespore membrane formation. Mol. Biol. Cell. 2001;12:901–917. doi: 10.1091/mbc.12.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakase Y., Nakamura-Kubo M., Ye Y., Hirata A., Shimoda C., Nakamura T. Meiotic spindle pole bodies acquire the ability to assemble the spore plasma membrane by sequential recruitment of sporulation-specific components in fission yeast. Mol. Biol. Cell. 2008;19:2476–2487. doi: 10.1091/mbc.E08-02-0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman A. M. Prospore membrane formation defines a developmentally regulated branch of the secretory pathway in yeast. J. Cell Biol. 1998;140:29–37. doi: 10.1083/jcb.140.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman A. M. Ascospore formation in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2005;69:565–584. doi: 10.1128/MMBR.69.4.565-584.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickas M. E., Schwartz C., Neiman A. M. Ady4p and Spo74p are components of the meiotic spindle pole body that promote growth of the prospore membrane in Saccharomyces cerevisiae. Eukaryot. Cell. 2003;2:431–445. doi: 10.1128/EC.2.3.431-445.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz D., Medkova M., Walch-Solimena C., Novick P. Ypt32 recruits the Sec4p guanine nucleotide exchange factor, Sec2p, to secretory vesicles; evidence for a Rab cascade in yeast. J. Cell Biol. 2002;157:1005–1015. doi: 10.1083/jcb.200201003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S. R. Transport-vesicle targeting: tethers before SNAREs. Nat. Cell Biol. 1999;1:E17–E22. doi: 10.1038/8967. [DOI] [PubMed] [Google Scholar]
- Rojas R. J., Kimple R. J., Rossman K. L., Siderovski D. P., Sondek J. Established and emerging fluorescence-based assays for G-protein function: Ras-superfamily GTPases. Comb. Chem. High Throughput Screen. 2003;6:409–418. doi: 10.2174/138620703106298509. [DOI] [PubMed] [Google Scholar]
- Sato Y., Shirakawa R., Horiuchi H., Dohmae N., Fukai S., Nureki O. Asymmetric coiled-coil structure with guanine nucleotide exchange activity. Structure. 2007;15:245–252. doi: 10.1016/j.str.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Segev N. Ypt and Rab GTPases: insight into functions through novel interactions. Curr. Opin. Cell Biol. 2001;13:500–511. doi: 10.1016/s0955-0674(00)00242-8. [DOI] [PubMed] [Google Scholar]
- Shimoda C. Forespore membrane assembly in yeast: coordinating SPBs and membrane trafficking. J. Cell Sci. 2004;117:389–396. doi: 10.1242/jcs.00980. [DOI] [PubMed] [Google Scholar]
- Sorokin S. Centrioles and the formation of rudimentary cilia by fibroblasts and smooth muscle cells. J. Cell Biol. 1962;15:363–377. doi: 10.1083/jcb.15.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- TerBush D. R., Maurice T., Roth D., Novick P. The Exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J. 1996;15:6483–6494. [PMC free article] [PubMed] [Google Scholar]
- Virgin J. B., Metzger J., Smith G. R. Active and inactive transplacement of the M26 recombination hotspot in Schizosaccharomyces pombe. Genetics. 1995;141:33–48. doi: 10.1093/genetics/141.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch-Solimena C., Collins R. N., Novick P. J. Sec2p mediates nucleotide exchange on Sec4p and is involved in polarized delivery of post-Golgi vesicles. J. Cell Biol. 1997;137:1495–1509. doi: 10.1083/jcb.137.7.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederkehr A., De Craene J. O., Ferro-Novick S., Novick P. Functional specialization within a vesicle tethering complex: bypass of a subset of exocyst deletion mutants by Sec1p or Sec4p. J. Cell Biol. 2004;167:875–887. doi: 10.1083/jcb.200408001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurmser A. E., Sato T. K., Emr S. D. New component of the vacuolar class C-Vps complex couples nucleotide exchange on the Ypt7 GTPase to SNARE-dependent docking and fusion. J. Cell Biol. 2000;151:551–562. doi: 10.1083/jcb.151.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura S., Egerer J., Fuchs E., Haas A. K., Barr F. A. Functional dissection of Rab GTPases involved in primary cilium formation. J. Cell Biol. 2007;178:363–369. doi: 10.1083/jcb.200703047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.