SUMMARY
In the largest E3 ligase subfamily, Cul3 binds a BTB domain, and an associated protein-interaction domain such as MATH recruits substrates for ubiquitination. Here we present biochemical and structural analyses of the MATH-BTB protein, SPOP. We define a SPOP-binding consensus (SBC), and determine structures revealing recognition of SBCs from the phosphatase Puc, the transcriptional regulator Ci, and the chromatin component MacroH2A. We identify a dimeric SPOP-Cul3 assembly involving a conserved helical structure C-terminal of BTB domains, which we call “3-box” due to its facilitating Cul3-binding and its resemblance to F-/SOCS-boxes in other cullin-based E3s. Structural flexibility between the substrate-binding MATH and Cul3-binding BTB/3-box domains potentially allows a SPOP dimer to engage multiple SBCs found within a single substrate, such as Puc. These studies provide a molecular understanding of how MATH-BTB proteins recruit substrates to Cul3, and how their dimerization and conformational variability may facilitate avid interactions with diverse substrates.
INTRODUCTION
Covalent attachment of ubiquitin via E1-E2-E3 enzyme cascades constitutes a primary mechanism for regulating protein function. Ubiquitin-protein ligases, or E3s, select substrates for ubiquitin conjugation. The largest E3 subclass consists of Cullin-RING Ligases (CRLs), which are modular, multisubunit enzymes, comprised of exchangeable substrate adaptors dedicated to particular cullin-based catalytic cores (Bosu and Kipreos, 2008; Cardozo and Pagano, 2004; Jackson and Eldridge, 2002; Petroski and Deshaies, 2005; Willems et al., 2004). The best understood CRL is the SCF ubiquitin ligase, composed of Skp1, Cul1, and a member of the F-box family of proteins (Bai et al., 1996; Feldman et al., 1997; Patton et al., 1998; Seol et al., 1999; Skowyra et al., 1997; Willems et al., 1996). Skp1 serves as an adaptor that simultaneously binds sequences near the N-terminus of Cul1 and the F-box motif of an F-box protein. In turn, F-box proteins contain additional protein interaction domains that recruit the substrate into a Cul1-Skp1-F-box protein complex, thereby facilitating ubiquitination of the target via the catalytic core assembled on the C-terminus of Cul1. Cullins-2 and 5 share a similar organization, wherein the Skp1-like protein EloC recruits members of the SOCS-box family to Cul2/5. Like an F-box, the SOCS-box links the substrate-binding domain to EloC, and also plays a role in cullin binding (Schulman et al., 2000; Stebbins et al., 1999). In contrast, Cul3 employs BTB proteins as substrate specific adaptors (Cardozo and Pagano, 2004; Furukawa et al., 2003; Geyer et al., 2003; Krek, 2003; Pintard et al., 2004; Pintard et al., 2003; Willems et al., 2004; Xu et al., 2003). “BTB” is a protein interaction/dimerization domain that is structurally homologous to the cullin-binding region of Skp1, and that binds Cul3 via motifs analogous to those in the Skp1-Cul1 complex (Bardwell and Treisman, 1994; Furukawa et al., 2003; Pintard et al., 2003; Stogios et al., 2005; Xu et al., 2003; Zollman et al., 1994). Many BTB-domain proteins also contain additional protein interaction domains, some of which have been shown to recruit ubiquitination targets (Petroski and Deshaies, 2005; Pintard et al., 2004; Willems et al., 2004). Thus, BTB proteins are thought to merge the functional properties of Skp1or EloC and their F-box or SOCS-box partners into a single polypeptide chain, without an intervening F- or SOCS- box.
The human genome encodes more than 150 proteins with recognizable BTB domains, often in combination with MATH, Kelch, or other interaction domains (Stogios et al., 2005). BTB proteins containing MATH and Kelch domains have been linked to substrate targeting by Cul3, although it is unclear precisely how many BTB proteins engage Cul3 in vivo. The best understood Cul3 adaptor is the BTB-Kelch protein Keap1, which mediates turnover of the Nrf2 transcription factor in oxidative stress-response paths (Cullinan et al., 2004; McMahon et al., 2003; Zhang and Hannink, 2003; Zhang et al., 2004). Structural studies of Keap1-Nrf2 identified a basic patch on the Kelch propeller that interacts with acidic motifs in Nrf2 (Lo et al., 2006; Padmanabhan et al., 2006; Tong et al., 2007).
In contrast with Keap1, little is known about how MATH domains select targets for ubiquitination by CRLs. The MATH domain, present in numerous diverse proteins, is most frequently found linked to a C-terminal BTB domain. Indeed, the MATH-BTB module is the 10th most abundant of 2-domain combinations encoded by 131 genomes (Vogel et al., 2004). Evidence that MATH-BTB proteins function in protein degradation came from the finding of C. elegans Mel-26 as a component of a Cul3-dependent E3 responsible for turnover of the Mei-1 meiosis-specific microtubule severing protein (Pintard et al., 2003; Xu et al., 2003). In mammals, the MATH-BTB protein SPOP has been linked to ubiquitination of MacroH2A, to regulate its deposition on the inactive X-chromosome (Hernandez-Munoz et al., 2005), and DAXX, to regulate transcriptional repression of pro-apoptotic proteins such as p53 (Kwon et al., 2006). More recently, the Drosophila SPOP ortholog HIB/Roadkill was found to regulate Hedgehog signaling during development by promoting degradation of the Ci transcription factor (Kent et al., 2006; Zhang et al., 2006). This process is independent of SCFSlimb-mediated processing of Ci to its repressive form (Jiang and Struhl, 1998). HIB/Roadkill also regulates TNF signaling in the fly eye via turnover of the MAPK phosphatase Puc (Liu et al., 2009).
Despite progress in understanding biological pathways in which SPOP functions, little is known about how target specificity is achieved. Known targets of the SPOP family lack obvious domains linking them to a common degradation mechanism, and unlike most SCF substrates, post-translational modifications are not known to control turnover of SPOP substrates. In spite of the numerous important pathways regulated by SPOP, and the abundance of MATH-BTB proteins, the molecular basis for substrate and Cul3 recognition remain poorly characterized. To address this problem, we report here biochemical, biophysical and crystallographic characterization of SPOP, its interactions with substrates, and with Cul3. Our results provide a framework for understanding how a dimeric MATH-BTB functions to assemble a dimeric Cul3 complex to promote recruitment and ubiquitination of substrates.
RESULTS AND DISCUSSION
SPOP domain structure
The 374-residue SPOP contains three domains: an N-terminal MATH domain (residues 28-166) that recruits substrates, an internal BTB domain (residues 190-297) that binds Cul3, and a C-terminal nuclear localization sequence (residues 365-374). We purified wild-type and mutant versions of four components: an N-terminal part containing residues 28-166 (SPOPMATH), a C-terminal part containing residues 172-329 (SPOPBTB+), and parts encompassing both a MATH and BTB (residues 28-329 - SPOPMATH-BTB+; residues 1-329 – SPOPN-MATH-BTB+). Below we describe dissection of SPOP’s molecular interactions by structural and functional characterization of SPOPMATH, SPOPBTB+, and SPOPMATH-BTB+.
A SPOP Binding Consensus in multiple SPOP substrates
To understand SPOPMATH-substrate interactions, we identified interacting regions on Puc and MacroH2A. We mapped a SPOP binding element on Puc by limited proteolysis. After treating a complex between a SPOP MATH domain and Puc1-390 with trypsin, a proteolytically-resistant complex containing a short Puc peptide (residues 91-113) was isolated by gel filtration (Fig 1A).
Figure 1. Identification of a SPOP Binding Consensus (SBC) sequence in multiple SPOP substrates.
(A) Identification of a SPOP-binding peptide in Puc. Left, Coommassie-stained SDS-PAGE gel showing products after trypsin digestion of a complex between a SPOP MATH domain and Puc1-390 (333:1 trypsin, 3 hr, rt). Right, Coommassie-stained SDS-PAGE gel of fractions from gel filtration (SD200) of trypsin digest products. Bottom, peptide co-purifying with SPOPMATH in fractions 33–35, identified by mass spectrometry.
(B) Identification of a MacroH2A sequence required for binding to SPOP. Top, Schematic view of MacroH2A deletions, highlighting residues 166-179. Bottom, Coommassie-stained SDS-PAGE gel of GST-pull-downs of GST, GST-MacroH2A 1, and GST-MacroH2A 2 coexpressed in E. coli with a HisMBP-tagged SPOP MATH domain.
(C) SBCs (red) in SPOP substrates Puc, MacroH2A, Ci and Daxx.
(D) Binding constants for SPOPMATH interactions with SBC peptides, measured by Surface Plasmon Resonance (BIACORE3000).
(E) Roles of individual Puc SBCs in in vitro ubiquitination. Western blots detecting His-Puc ubiquitination for wild-type (WT) and mutant Puc substrates. SBCm1, SBCm2 and SBCm3 refer to mutation at the 3 SBC sites.
Efforts to identify a SPOP binding element on MacroH2A focused on residues 166-372, which are sufficient for the interaction (Takahashi et al., 2002). Deletion mapping by pulling down a SPOP MATH domain demonstrated that MacroH2A residues 166-179 are required for the interaction (Fig 1B). For both Puc and MacroH2A, peptides corresponding to these regions (Puc93-107 and MacroH2A166-179) are sufficient for binding (Fig 1C–D).
Do Puc, MacroH2A and other substrates have common features mediating SPOP binding? We identified a 5-residue φ-π-S-S/T-S/T (φ-nonpolar; π-polar) SPOP Binding Consensus (SBC) motif in the sequences of Puc93-107, MacroH2A166-179, and two other SPOP substrates Ci and Daxx (Fig 1C). Notably, Puc, Ci, and Daxx all harbor multiple SBCs. Synthetic peptides with the SBC motif bind SPOPMATH as detected using Surface Plasmon Resonance (BIACORE). Different SBC peptides display a range of affinities similar to those observed for some other MATH domain-peptide interactions, as for TRAF6 (Ye et al., 2002). This agrees with the paradigm established by Keap1 that at least some BTB-Cul3 ligase substrates harbor multiple adaptor binding sites (Tong et al., 2007). The peptide corresponding to the Puc SBC that co-purifies with a SPOP MATH domain by gel filtration displays the highest affinity of ~4 μM, which is similar to the affinity of an SCF-substrate interaction (Hao et al., 2007).
We sought to examine roles of SPOPMATH-Puc SBC interactions in in vitro ubiquitination. As with other CRLs, SPOP-Cul3 mediates ubiquitination of Puc in the presence of E1 and E2 (Fig S3). Results with Puc mutants indicate that the contributions of Puc SBCs are proportional to their abilities to bind SPOP: mutation of SBC1 is highly deleterious even at the highest concentration we can assay, mutation of SBC3 is further detrimental in the context of the SBC1 mutant, and the mutant retaining only SBC1 is less active at low concentration (Fig 1E, Fig S4).
Structural Basis for SPOPMATH-SBC Interactions
To understand the molecular basis for SPOP MATH domain-SBC interactions, we determined structures of SPOPMATH complexed with peptides corresponding to three substrate SBCs (PucSBC1, MacroH2ASBC, CiSBC2) in numerous crystal forms (Table 1). SPOPMATH comprises an anti-parallel β-sandwich, and the structures superimpose moderately well with other MATH domains, such as from TRAF2 and TRAF6, with r.m.s. deviation (RMSD) values ranging between 3.0Å to 3.4Å (Fig S5). The SBC motifs adopt extended conformations, and bind the MATH central shallow groove (Fig 2). The SPOP binding sites for SBCs from different substrates are virtually identical. The SBCs also superimpose well, whereas sequences outside the conserved motif, even from the same peptides in different crystal forms, do not. This suggests that the primary sites of interaction are mediated by the SBC sequence. Indeed, binding affinities are similar for a range of peptides encompassing a given SBC motif (Fig S2).
Table 1.
Crystallographic and Refinement Statistics
| SPOPMATH-Puc SBC1_pep1 | SPOPMATHx-Puc SBC1_pep2 | SPOPMATH-MacroH2A_pep1 | SPOPMATHx-MacroH2A_pep1 | SPOPMATHx-MacroH2A_pep2 | SPOPMATH-Ci SBC2 | SPOPMATHx-Ci SBC2 | SPOPBTB/3-box | GigBTB/3-box+ | SPOPMATHx-BTB/3-box - Puc SBC1_pep3 (form 1) | SPOPMATHx-BTB/3-box - Puc SBC1_pep3 (form 2) | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SBC Peptide sequence | DEVTSTTSSS | ENLACDEVTSTTSSST | KAASADSTTEGTPAD | KAASADSTTEGTPAD | DSTTEGTPADGFTVL | NTLFPDVSSSTH | NTLFPDVSSSTH | n/a | n/a | DEVTSTT | DEVTSTT |
| Accession code | 3IVV.pdb | 3HQL.pdb | 3IVB.pdb | 3HQH.pdb | 3HSV.pdb | 3IQV.pdb | 3HQM.pdb | 3HTM.pdb | 3HVE.pdb | 3HQI.pdb | 3HU6.pdb |
| DATA COLLECTION | |||||||||||
| Beamline | APS 24-ID-C | ALS 8.2.2 | APS 24-ID-C | APS 22-BM | APS 22-BM | ALS 8.2.1 | APS 22-BM | NSLS X25 | ALS 8.2.2 | APS 24-ID-C | APS 24-ID-C |
| Wavelength (Å) | 0.9794 | 1 | 0.9794 | 1 | 1 | 1 | 1 | 0.9792 | 0.9794 | 0.9795 | 0.9795 |
| Space Group | C2 | C2 | P6522 | P6522 | C2 | C2221 | P1 | P1 | P21 | P212121 | P212121 |
| Cell Dimensions | |||||||||||
| a, b, c (Å) | 60.744, 56.492, 42.133 | 90.795, 43.690, 86.819 | 44.021, 44.021, 266.840 | 44.7, 44.7, 268.0 | 88.953, 43.086, 87.453 | 57.703, 59.829, 151.164 | 44.281, 48.100, 49.860 | 36.8, 88.7, 88.7 | 46.48, 55.6, 120.6 | 57.23, 106.87, 130.28 | 63.74, 107.74, 130.79 |
| α, β, γ (deg) | 90, 96.11, 90 | 90, 107.03, 90 | 90, 90, 120 | 90, 90, 120 | 90, 118.15, 90 | 90, 90, 90 | 63.00, 64.04, 62.89 | 90.8, 89.3, 89.9 | 90, 91.1, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 50-1.25 (1.29-1.25) | 20-1.66 (1.72-1.66) | 50-1.75 (1.81-1.75) | 50-2.29 (2.37-2.29) | 50-1.43 (1.48-1.43) | 50-2.1 (2.18-2.1) | 50-1.74(1.80-1.74) | 50-2.42(2.51-2.42) | 100-2.8(2.9-2.8) | 50-2.62(2.71-2.62) | 50-2.7(2.8-2.7) |
| Total Reflections | 358726 | 362,270 | 791,770 | 333,421 | 450,833 | 281,810 | 216,527 | 278,434 | 104,074 | 215,492 | 543,183 |
| Unique Reflections | 39304 (3926) | 38,876 (3796) | 16,750 (1564) | 8,052 (735) | 54,191 (5266) | 15678 (1558) | 31,963 (2996) | 42,413 (6731) | 15,314 (903) | 24,800 (2213) | 25,4401877 |
| Completeness (%) | 100.0 (99.9) | 99.5 (99.0) | 98.0 (97.4) | 98.9 (96.8) | 99.2 (97.7) | 100.0 (100.0) | 97.0(94.6) | 96.2(79.7) | 91.0(59.1) | 98.1(91.3) | 94.5(75.1) |
| Overall Rsym (%) | 3.8 (27.6) | 7.3 (39.0) | 8.8 (49.0) | 9.0 (18.2) | 5.7 (11.0) | 7.7 (36.5) | 3.1(10.2) | 6.5(33.5) | 5.7(17.1) | 10.6(29.6) | 17.4(59.0) |
| Overall I/σI | 21.8 (3.9) | 28.9 (3.1) | 23.0 (4.9) | 31.7 (8.3) | 31.7 (14.8) | 26.4 (4.4) | 30.3(9.5) | 25.5(4.9) | 20.8(5.3) | 20.0(3.7) | 20.1(2.4) |
| Mean redundancy | 3.7 (3.3) | 3.1 (2.8) | 4.8 (4.6) | 7.0 (4.5) | 4.3 (4.2) | 7.2 (7.3) | 2.0(1.9) | 1.9(1.7) | 3.2(2.6) | 4.6(3.8) | 9.4(5.9) |
| REFINEMENT | |||||||||||
| Rwork/Rfree | 0.199/0.177 | 0.247/0.258 | 0.247/0.218 | 0.238/0.276 | 0.175/0.206 | 0.279/0.233 | 0.177/0.218 | 0.219/0.259 | 0.303/0.333 | 0.226/0.275 | 0.246/0.295 |
| Reflections (working set) | 35,345 | 36,636 | 14,738 | 7,445 | 51,030 | 14,875 | 29,424 | 35,675 | 12,958 | 23,046 | 22,223 |
| Reflections (test set) | 1976 | 1,934 | 828 | 360 | 2,740 | 754 | 1,582 | 1,872 | 615 | 1,235 | 1,188 |
| overall B-factors (Å^2) | 14.9 | 18 | 16.8 | 45.4 | 14.9 | 53.9 | 14.5 | 37.3 | 56.3 | 18.7 | 19.6 |
| R.m.s deviations | |||||||||||
| Bond lengths (Å) | 0.008 | 0.013 | 0.009 | 0.01 | 0.009 | 0.008 | 0.009 | 0.007 | 0.011 | 0.009 | 0.009 |
| Bond angles (deg) | 1.2 | 1.36 | 1.25 | 1.3 | 1.2 | 1.69 | 1.19 | 1.3 | 1.5 | 1.4 | 1.4 |
| Ramachandran statistics | |||||||||||
| Most Favored | 90.40% | 98.05% | 93.90% | 92.10% | 97.96% | 87.30% | 98.43% | 94.85% | 88.77% | 94.89% | 83.70% |
| Additional allowed | 9.60% | 1.95% | 6.10% | 7.90% | 2.04% | 12.70% | 1.57% | 5.15% | 11.23% | 5.11% | 16.30% |
| Dissallowed | 0.00% | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Highest resolution shell is shown in parenthesis. Rwork = Σ|Fo−Fc|/ΣFo.
Rfree is the cross-validation of Rfactor with 5% of the total reflections omitted in model refinement.
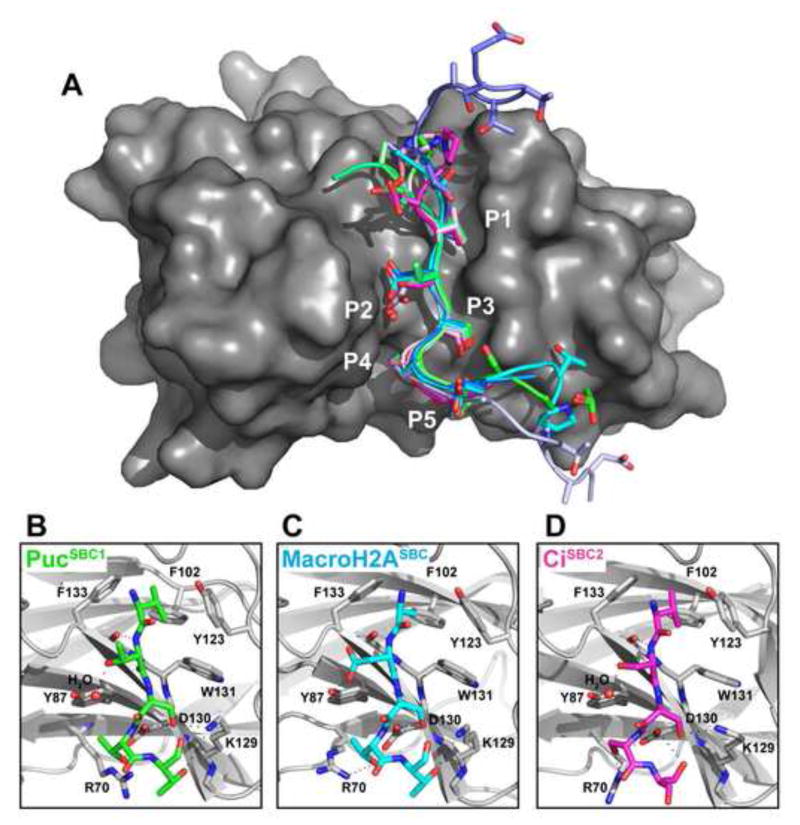
Figure 2. Structural Basis for SPOPMATH-SBC Interactions.
(A) Comparison of 11 independent structures of isolated SPOPMATH complexed with SBC peptides (3 from Puc – greens; 4 from MacroH2A – cyans/blues; 4 from Ci – pinks/magentas). After superposition over SPOPMATH mainchain, SBC peptides were displayed with backbones as cartoons and sidechains as sticks, docked in the structure of SPOPMATH (grey surface) from the complex with PucSBC1. The 5 φ-π-S-S/T-S/T motif positions are indicated P1–P5.
(B–D) Close-up views of SPOPMATH (grey) complexes with (B) PucSBC1 (green), (C) MacroH2ASBC (cyan), and (D) CiSBC2 (magenta), oriented as in panel A. Dashed lines - hydrogen bonds; red – oxygen; blue – nitrogen; red sphere – water.
SPOPMATH-SBC interactions are anchored by both hydrophobic and polar interactions, described here with SBC residues φ-π-S-S/T-S/T as P1-P5 (Position1 φ-, etc.). SPOP residues Y87, F102, Y123, W131 and F133 encircle the P1 hydrophobic side-chain (Puc V98, MacroH2A A170, Ci V1362). Many hydrogen bonds are also observed, mainly involving SPOP Y87 and D130 and SBC P2-P5 residues (Table S1, Fig 2). Sequence flexibility at the P2 position is explained by the ability of the Y87 hydroxyl to directly contact an Asp (MacroH2A), or form water-mediated hydrogen bonds to Thr or Ser (Puc or Ci, respectively). The structures also explain the apparent requirement for a Ser at P3: this sidechain both makes hydrogen bonds and is constrained by size within a small pocket formed by K129, D130, and W131 (Fig 2).
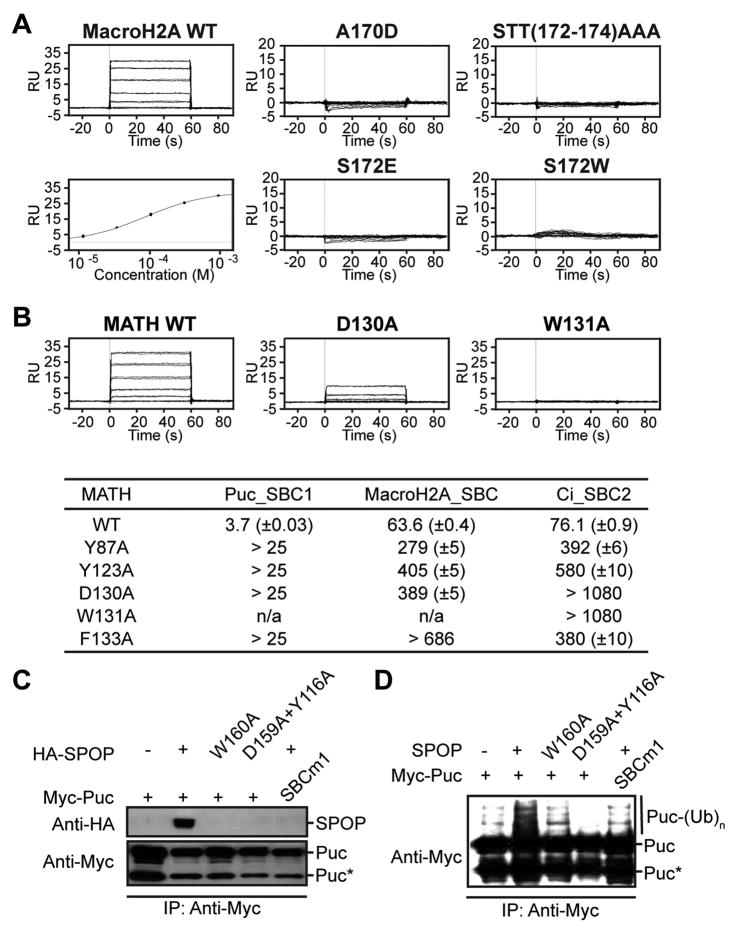
To test the significance of the observed interactions, we examined effects of mutations. First, we used BIACORE to test binding to a series of mutated peptides derived from MacroH2A. Abolishing the P1 hydrophobic interaction with an A170D substitution essentially eliminated binding. Furthermore, substitution of the P3 Ser with bulkier Glu or Trp residues also abolished the interaction, as did substituting the P3-5 STT residues with alanines (Fig 3A). Singly mutated SPOP MATH domains were also assayed (Fig 3B). D130A and W131A substitutions were highly deleterious with SBC peptides from all three substrates, supporting the view that these are key residues for substrate recognition. Mutants were further examined in cellular binding and ubiquitination assays. SPOP D130A and W131A and a Puc SBC1 mutant display parallel effects, disrupting the SPOP-Puc interaction and Puc ubiquitination in cells (Fig 3C, 3D).
Figure 3. Mutational Analysis of SPOPMATH-Substrate Interaction.
(A) BIACORE sensograms showing SPOPMATH binding to wildtype or indicated mutant versions of a MacroH2ASBC peptide. Bottom left - Fit used to calculate KD for SPOPMATH-MacroH2ASBC.
(B) Representative BIACORE sensograms examining binding between wildtype or indicated mutant versions of SPOPMATH and a PucSBC1 peptide. BIACORE binding data for SPOPMATH mutants and SBC peptides is summarized below.
(C) Western blots showing association of HA-SPOP (top) with Myc-Puc (bottom) or mutants after anti-Myc immunoprecipitation from Drosophila S2 cells co-transfected with the indicated constructs. D130 of human SPOP corresponds to D159 in Drosophila SPOP, W131 corresponds to W160, and Y87 corresponds to Y116. *Processed form of Puc (Liu et al., 2009).
(D) Anti-Myc western blot detecting ubiquitination of Puc from S2 cells co-transfected with wild-type or mutant versions of SPOP.
Previous genetic analysis of another MATH-BTB protein, MEL-26 from C. elegans, is also consistent with the functional importance of the crystallographically-observed MATH-substrate interactions. The SPOP and MEL-26 MATH domains share 30% sequence identity. MEL-26 controls degradation of the katanin mei-1 during the first mitotic division in C. elegans development. The MEL-26 residue Cys94, which corresponds to Tyr87 in the SPOPMATH substrate-binding cleft, was shown to be important for substrate targeting (Dow and Mains, 1998; Xu et al., 2003).
Interestingly, the Drosophila SPOP substrate Ci is subject to ubiquitination via multiple pathways. In the absence of Hedgehog stimulation, Ci is ubiquitinated by SCFSlimb in a phosphorylation-dependent manner (Jiang and Struhl, 1998). While many SCF substrates require phosphorylation, there is no evidence that this is the case with Cul3 substrates. In fact, phosphorylated residues within the SBC motif block binding to SPOP (Fig S6), raising the possibility that SBC phosphorylation negatively regulates turnover via SPOP. Future studies will be required to dissect the complex interplay between phosphorylation at perhaps a range of sites, and the biochemical decisions between different ubiquitination pathways.
Implications for substrate recognition by other MATH-BTB proteins
Unlike the situation with SPOP, but as with the majority of E3s, the identities of substrates for most MATH-BTB-Cul3 ubiquitin ligases remain unknown. The structural studies suggest that isolated MATH domains may be used to identify peptide sequences that serve as recognition sites for substrates of the MATH-BTB subfamily of Cul3 adaptors.
SPOPBTB+ forms a 2:2 complex with the Cul3 N terminal adaptor-binding domain
In addition to binding substrates via its MATH domain, SPOP dimerizes and binds Cul3 via its BTB domain. To better understand these features, we determined a crystal structure of SPOPBTB+. Residues 177-297 dimerize via a hydrophobic interface as in the published structures of BTB domains from the BTB-ZF family of transcription factors (Fig 4, S7), and display overall similarity to the Cul1, Cul2, and Cul5 adaptors Skp1 and Elongin C (Fig 5A). Indeed, BTB domains bind the Cul3 N-terminal domain in a manner resembling Skp1 interactions with Cul1 (Xu et al., 2003). To understand whether SPOPBTB+ maintains its dimeric structure in complex with Cul3, we examined binding with the Cul3 N-terminal domain by analytical ultracentrifugation (AUC). Both SPOPBTB+, and a version encompassing both a MATH and BTB domain form a 2:2 complex with the Cul3 N terminal domain (Fig 4B, S8), consistent with prior data indicating that BTB-Cul3 complexes in vivo are oligomeric (Chew et al., 2007). To address whether dimerization is required for Cul3 binding, we generated monomeric versions of SPOP by replacing four key hydrophobic BTB dimerization residues: L186D, L190D, L193D and I217K (Fig 4). The mutations (1) remove hydrophobic contacts that favor dimerization, (2) place negative charges in such a way as to repel each other in a dimer, and (3) allow for solubility and prevent aggregation by having the new exposed surface charged. The dimerization-defective SPOP copurifies with Cul3 N-terminal domain by gel filtration (not shown) and displays the same affinity as measured by AUC (Fig 4C, S8). However, the dimerization-defective SPOP is significantly impaired for ubiquitination activity (Fig 4D).
Figure 4. SPOPBTB+ forms a 2:2 dimer with Cul3 N-terminal domain.
(A) Left, overall view of the SPOPBTB+ dimer, with protomers in cyan (A) and red (B). Right, close-up view of dimer interface rotated 90° in x.
(B) Equilibrium AUC of SPOPBTB++Cul3ntd. Samples at 1.0 to 8.8 μM centrifuged at 8 (red), 12 (blue), and 16 (black) krpm 4 C. Lines show global nonlinear least-squares best-fit of all datasets/concentrations/speeds to a heterogeneous association model describing a 2:2 SPOPBTB+:Cul3ntd complex (MW 127.1 kDa) with indicated KD value. For clarity, only the 3 μM sample is shown.
(C) AUC of L186D, L190D, L193D, I217K mutant SPOPBTB++Cul3ntd performed as in B. Lines show global nonlinear least-squares best-fit of all datasets/concentrations/speeds to a heterogeneous association model describing a 1:1 SPOPBTB+ (mutant):Cul3ntd complex (MW 63.6 kDa) with the indicated KD value. For clarity, only the 2.0 μM sample is shown.
(D) Western blots of SPOPMATH-BTB+-mediated ubiquitination detecting His-Puc, for wild-type and L186D, L190D, L193D, I217K (dimer-defective) mutant SPOP.
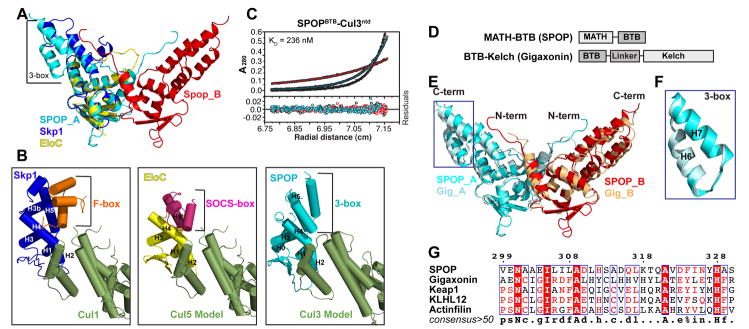
Figure 5. A conserved Cul3-Interacting box (3-box).
(A) Structural alignment of SPOPBTB+, Skp1 (blue) and EloC (yellow). One molecule of the SPOPBTB+ dimer is cyan (SPOP_A) and one is red (SPOP_B). The SPOP helix-pair (above “+”) not shared in Skp1 and EloC is labeled “3-box”.
(B) Structural comparison of the Skp1 (blue) -F-boxSkp2 (orange) -Cul1 (green) structure (1LDK.pdb), the Elongin C (yellow) – SOCS-boxVHL (magenta) structure (1VCB.pdb) docked onto a structural model of Cul5 (green), and the SPOPBTB/3-box (cyan) docked on a structural model of Cul3 (green). The relative locations of the SPOP 3-box, F-box, and SOCS-box are indicated.
(C) Equilibrium AUC of SPOPBTB (i.e., lacking the 3-box)+Cul3ntd. Samples at 1.2 to 8.0 μM centrifuged at 8 (red), 12 (blue), and 15 (black) krpm 4 C. Lines represent the global nonlinear least-squares best-fit of all datasets/concentrations/speeds to a heterogeneous association model describing a 2:2 SPOPBTB:Cul3ntd complex (MW 118.6 kDa) with the indicated KD value. For clarity, only the 3.0 μM sample is shown.
(D) Schematic of SPOP and Gigaxonin domain arrangements, which represent two distinct BTB subfamilies, MATH-BTB and BTB-Kelch, respectively.
(E) Overall structural alignment of SPOPBTB/3-box and GigaxoninBTB/3-box. Residues C-terminal of the Gigaxonin 3-box were omitted for clarity.
(F) Superposition of the SPOP and Gigaxonin 3-boxes. SPOP3-box –cyan; Gigaxonin3-box - light blue.
(G) Structure based sequence alignment (ESPript) of 32 amino acids corresponding to 3-boxes from 5 Cul3-interacting BTB proteins. SPOP 3-box residue numbers are shown on top.
The “3-box”: a conserved Cul3-binding element
Unexpectedly, SPOPBTB+ displays an additional structural feature: a pair of C-terminal α-helices (Fig 5A). To gain insight into potential functions of these helices, we compared SPOPBTB+ with other cullin-binding complexes, Skp1-F-box and ElonginC-SOCS-box. The SPOP helices are located at the position corresponding to an F-box or SOCS-box in other CRLs (Fig 5B). Given the major roles of F-/SOCS-boxes in physically linking BTB-like-fold adaptors to substrate-binding domains, the location of the SPOPBTB+ helix pair - at the extreme C-terminus distal from the site of BTB domain connection to the MATH domain - raises the question as to why a BTB protein would have an F-/SOCS-box-like structure. We considered that the F-box also has a secondary role, contacting Cul1 in the prior Cul1-Skp1-F-boxSkp2 crystal structure (Zheng et al., 2002). SOCS-boxes have also been implicated in Cul2 and Cul5 binding (Mahrour et al., 2008). Thus, we tested whether the SPOP helices might share this secondary function of F-/SOCS-boxes in cullin binding. Deletion of the helix pair decreased interactions between SPOP and Cul3, without diminishing the global stability of the isolated domain (Fig 5C, S9). Thus, due to functional parallels with F- or SOCS-box proteins and their interactions with cullins, we refer to this structure as “3-box”, for Cul3-interacting box, and we redefine our BTB+ construct as BTB/3-box.
To determine whether the 3-box is conserved, we examined the corresponding structure of another Cul3-binding BTB protein: a fragment corresponding to residues 1-258 of the human BTB-Kelch, Gigaxonin (Gig) (Fig S10). As with other BTB-Kelch proteins, the Gig BTB domain is located N-terminal of the substrate-binding domain, and thus displays a divergent domain arrangement from SPOP (Fig 5D). The overall GigBTB/3-box superimposed well with SPOPBTB/3-box (Cα RMSD 2.06Å). The 3-boxes of both proteins display virtually identical backbone structures (Cα RMSD 0.39 Å) despite distinct sequences (Fig 5F). Alignment of the corresponding 32 amino acids following BTB domains from five known Cul3 adaptors revealed conservation (Fig 5G), suggesting that the 3-box may also exist in Keap1, KLHL12 and Actinfilin. As most of the conserved residues are buried and mediate inter-helical packing, it is possible that Cul3 recognizes a range of sequences with a common 3-box tertiary structure.
Predicted 3-boxes in a subset of BTB proteins
Only a handful of mammalian BTB proteins have been explicitly shown to mediate ubiquitination (Allen et al., 2005; Angers et al., 2006b; Berthold et al., 2008; Cullinan et al., 2004; Itoh et al., 1999; Salinas et al., 2006). As we found two of these to contain a 3-box, we attempted to identify potential 3-boxes in other BTBs. A bioinformatics search for a paired helix structure within 40 amino acids beyond 183 human BTB domain C-termini predicts that like SPOP and Gig, all MATH-BTB and BTB-Kelch protein have a helix pair following the BTB domain, which may function as a 3-box (Fig S11). By contrast, we identified potential 3-box motifs in only a subset of the BTB-ZF, T1-like BTB, and other BTB proteins. It is possible that BTB proteins lacking an obvious 3-box may not function as Cul3 adaptors. One such example might be BTBD12, which lacks a 3-box and unlike 3-box containing BTB proteins does not copurify with Cul3 from cells (Cullinan et al., 2004; Svendsen et al., 2009). Alternatively, BTB proteins lacking a 3-box may utilize alternative mechanisms to strengthen interaction with Cul3.
It is tempting to speculate about relationships between F-box, SOCS-box, and BTB-proteins, although at this point it is unclear whether the functional similarities are a consequence of convergent evolution, or divergence from a common ancestor. In the case of divergent evolution, a BTB may represent an ancestral CRL adaptor, encompassing cullin- and substrate-binding in a single polypeptide, with a 3-box also contributing to cullin-binding.
Putting MATH and BTB domains together – an asymmetric dimer
To gain insights into how substrate, Cul3-binding, and dimerization are encompassed in one polypeptide, we determined the structure of a complex containing a dimeric SPOP spanning the MATH, BTB, and 3-box domains, with the MATH domains bound to a peptide corresponding to PucSBC1 (Fig 6). Structures were obtained from two slightly different crystal forms growing in the same condition (Table 1). In both, the MATH-SBCs and the BTB/3-box each resemble the structures of the individual domains. This includes the roughly 2-fold symmetry of the dimeric BTB/3-box domain.
Figure 6. Crystal Structures of dimeric SPOPMATHx-BTB/3-box-PucSBC1.
(A) Overall architecture of SPOPMATHx-BTB/3-box dimer. One molecule
(A) is colored cyan and the other (B) is red. Each MATH domain binds one PucSBC1 (green) peptide. Disordered regions not visible in electron density are represented with dotted lines to show connectivity.
(B) Superposition over a BTB domain for SPOPMATHx-BTB/3-box structures determined from crystals with slightly different unit cells. MATH_A1 (cyan) and MATH_B1 (red) are from crystal form 1, and correspond to the structure in (A). MATH_A2 (orange) and MATH_B2 (blue) are from crystal form 2.
(C) Coommassie-stained SDS-PAGE gel showing products of proteolysis of SPOPMATH-BTB/3-box by Endoproteinase Glu-C at room temperature for 30 min. The identities of products determined by Mass Spectrometry are shown.
(D) SPOPMATH and SPOPBTB/3-box domains do not cofractionate by sizing. A thrombin cleavage site was engineered in the interdomain linker in SPOPMATH-BTB/3-box (−T), and after treatment with thrombin (+T), the product was subject to gel filtration chromatography. Individual fractions were analyzed by Coommassie-stained SDS-PAGE gel, below.
The most striking feature of the structures is the asymmetric arrangement of the two substrate-binding MATH domains, which differ in orientation by 55° with respect to the BTB/3-box dimer. One MATH domain packs in the middle of the V-shaped groove created by the two protomers in the BTB domain. The other MATH is distal, and extends away from its associated BTB domain, as if to lengthen one side of the “V” (Fig 6A).
In addition to the significantly different MATH-BTB orientations, three other features of the crystal structures suggest that the MATH and BTB domains are flexibly tethered. First, in one crystal form, neither linker between the MATH and BTB is visible in the electron density. In the other, the backbone of one linker is visible due to crystal packing with the opposite MATH domain, although some side-chains are not visible. Second, comparing the complexes in the two crystal forms reveals a slight (~3Å) offset in the positions of the MATH domains relative to each other. Thus, a total of 3 BTB-MATH orientations are observed (Fig 6B). The particular domain arrangements in the structures may be the product of interdomain flexibility combined with orientations compatible with crystallization (Fig S12). Third, contacts between the MATH and BTB/3-box domains are minimal, with no interactions between side-chains preserved in both crystal forms for any domain arrangement.
We examined the linkage between the MATH and BTB domains in solution, using versions of the two longest forms of SPOP that we could express. First, we performed limited proteolysis. With Endoproteinase Glu C, the interdomain linker is preferentially susceptible to initial cleavage (Fig 6C, S13). Because extended reactions with Glu C lead to further breakdown (not shown), we engineered a thrombin site in the linker based on similarity to a distal SPOP relative (Fig S14). After elimination of their covalent linkage, the MATH and BTB domains separate during gel filtration (Fig 6D). Second, we performed Small Angle X-Ray Scattering analyses (SAXS). For both the longer and shorter constructs, the data do not fit to any single conformation. Instead, the data fit well to a population of multiple conformations, although the presence of the N-terminus appears to restrict the overall compactness of the complex (Fig S15). Overall, the data are consistent with the notion that the orientations between the BTB and MATH domains of SPOP are variable in our constructs.
Notably, there is also asymmetry beyond the BTB/3-box in the Gig1-258 crystal structure. Although poor electron density of one molecule in the asymmetric unit precludes building of side chains, the helices following the 3-box extend away from the BTB domain in architectures that clearly differs between the two protomers. This is further supported by different relative locations of selenomethionines (Fig S10). Thus, the linker between Cul3- and substrate-binding domains may be generally flexible in BTB proteins.
2:1 SPOP:Puc stoichiometry
Given the orientational flexibility of the two substrate-binding domains, we examined the stoichiometry of binding to Puc, a substrate with multiple SBCs. Both velocity and equilibrium AUC analyses of mixtures of 1:1–3 molar ratios of SPOPMATH-BTB-3-box and Puc1-390, which contains all three SBC motifs, reveal two species with molecular weights of 110 kDa and 40 kDa. These correspond to a 2 SPOP: 1 Puc complex and free Puc (Fig 7A, B), respectively. Thus, the SPOP dimer binds only one molecule of the substrate Puc in vitro, even in the presence of extra Puc. Although we cannot rule out the unlikely possibility that binding of one Puc molecule to SPOP sterically blocks association with a second Puc molecule, the 2 SPOP: 1 Puc stoichiometry may provide an explanation for the concentration-dependence of in vitro ubiquitination of a Puc mutant harboring only the single, high-affinity SBC1 (“m2m3”). At high concentrations, this mutant shows wild-type levels of ubiquitination (Fig 1). However, at low concentrations, this mutant is a poor substrate for SPOP-mediated ubiquitination (Fig S4). The results raise the possibility that a dimeric SPOP engages multiple Puc SBCs for the ubiquitination reaction.
Figure 7. A 1:2 substrate complex with the SPOP-Cul3 ubiquitin ligase.
(A) Velocity AUC of SPOPMATH-BTB/3-box + Puc1-390 at 20 °C, 60 krpm fit to a continuous distribution model c(s). Two peaks indicate molecular weights of 110 kDa and 39 kDa corresponding to the 1:2 Puc:SPOPMATH-BTB/3-box complex (MWcalc 112.5 kDa) and excess free Puc (MWcalc of 42.1 kDa).
(B) Equilibrium AUC of a sample as in (A). Samples at 1 to 6 μM centrifuged at 8 (red), 12 (blue), and 16 (black) krpm 4 C. Lines show global nonlinear least squares best-fit of all datasets/concentrations/speeds to a heterogeneous association model with 2 species, 2:1 SPOPMATH-BTB/3-box:Puc + Puc. For clarity, only the 1.1 μM sample is shown.
(C) Overall structure of SPOPMATHx-MacroH2ASBC (pep2). Two isolated MATH domains (chain A, cyan; chain B, pink) bind a single substrate peptide (green) at two suboptimal SBC sites.
(D) Schematic view of a SPOP-Cul3 ubiquitin ligase bound to a single substrate. Substrate is shown in grey, with SBCs in green, and ubiquitin-acceptor lysines as Ks. The two protomers of the dimeric SPOP complex are shown in cyan and red, with each BTB/3-box bound near the N-terminus of an elongated Cul3 (olive) activated with NEDD8 (orange) near the C-terminus. E2-bound Rbx1 RING domains are shown flexibly tethered to the Cul3 C-terminal domains. The high degree of conformational flexibility may allow substrates with a range of SBC configurations to be polyubiquitinated at multiple sites.
Potential insights into how a SPOP dimer might engage two SBCs from one target molecule may come from our structure of two isolated MATH domains bound to a single suboptimal peptide derived from MacroH2A (pep 2) that lacks the P1 Ala (Fig 7C, S2). One MATH domain binds the partial MacroH2ASBC, with the methyl group from the peptide’s N-terminal acetyl occupying the hydrophobic P1 position. Another MATH binds a downstream Ala as a P1 site, and the following Asp as a P2 residue. Subsequent residues in the peptide, which do not match the SBC motif sequence, are not visible in the electron density. Notably, this peptide (MacroH2A pep2) shows negligible binding to the SPOPMATH domain, presumably due to lack of a P1 hydrophobic residue (Fig S2). Thus, although this particular structure may be an artifact of crystallization, it nonetheless provides a “snapshot” of how an asymmetric arrangement of two SPOP MATH domains could achieve binding to multiple sites on a single polypeptide.
Flexibility and dimerization in CRL function
We found that the dimeric SPOP BTB domain can assemble with 2 molecules of Cul3, thereby generating a dimeric ubiquitin ligase containing two substrate-binding sites and two catalytic cores. Notably, several other ubiquitin E3s have been shown to dimerize. These include several CRLs, such as human SCFFbw7 and its yeast counterpart SCFCdc4 (Hao et al., 2007; Tang et al., 2007), and the MATH-domain containing simple RING E3s, Siah and TRAF6 (Polekhina et al., 2002; Yin et al., 2009).
To understand the function of SPOP, we generated models based on our multiple SPOP structures, and available cullin crystal structures. Based on modeling of the dimeric SPOP-SBC complex with Cul3, the substrate binding site is directed toward the catalytic core of the same cullin molecule to which the BTB domain is associated (Fig S16). However, our crystallographic and SAXS data indicate that the MATH domain adopts multiple orientations relative to the BTB domain through the flexible linker between the two. This feature of Cul3SPOP parallels that found with some other CRLs. The Cul4 adaptor DDB1 adopts multiple conformations (Angers et al., 2006a; Li et al., 2006; Scrima et al., 2008). Moreover, superposition of two molecules in one asymmetric unit of Cdc4 within the SCFCdc4 crystal structure indicates different positions of the substrate binding domains (Orlicky et al., 2003), reminiscent of our findings with SPOPMATH-BTB (Fig S17).
Recent structural studies indicate the ligation of NEDD8 to a conserved cullin Lys can serve as an additional source of conformational flexibility. Crystal structures revealed striking conformational differences between unmodified and NEDD8ylated versions of the Cul5 C-terminal domain in complex with Rbx1 (Duda et al., 2008). Taken together with enzymological and biophysical analyses, the data indicate that NEDD8 causes a switch in CRL structure from a closed autoinhibited form to a more dynamic active form (Duda et al., 2008; Saha and Deshaies, 2008; Yamoah et al., 2008). Thus, structural flexibility appears in many ways, for many CRLs.
The combination of dimerization and flexibility has many implications for substrate ubiquitination. First, the flexibility within SPOP may allow for SBCs in structurally diverse substrates to engage one or both MATH domains. Indeed, we found that Puc, which contains multiple functional SBCs, binds preferentially to SPOP in a 1:2 ratio, even when Puc is in excess (Fig 7), and that multiple SBCs contribute to SPOP ubiquitination, particularly at low concentration (Fig S4). This raises the possibility that one substrate molecule engages both SBC binding sites in the SPOP dimer (Fig 7). Furthermore, we obtained a crystallographic view of how two SPOPMATH domains could in principle adopt an asymmetric arrangement to bind a single target at multiple weak binding sites. Notably, multiple E3-binding sites are also found in several other dimeric CRL substrates. For example, the Fbw7 substrate Cyclin E contains one optimal and one suboptimal degron (Hao et al., 2007). Efficient turnover of a Cyclin E mutant in which the optimal degron is made suboptimal requires Fbw7 dimerization (Welcker and Clurman, 2007). Thus, the use of dimeric substrate binding domains may increase avidity for substrates, thereby facilitating binding to substrates with multiple degrons, or increasing binding efficiency for substrates containing multiple suboptimal degron sequences.
Dimerization may have additional roles beyond substrate recruitment, however, because the dimerization-defective SPOP is impaired for ubiquitination under all conditions tested (Fig 4). Dimerization may allow the substrate and elongating ubiquitin chain to sample a greater variety of orientations, as for dimeric SCFCdc4 (Tang et al., 2007). Although the precise manifestations of dimerization are still under debate, it seems that the use of two flexibly oriented substrate binding sites, combined with a flexible cullin-RING core, would allow a single E3 to recognize numerous substrates, potentially with high avidity, and with a range of conformational options for mediating their ubiquitination.
EXPERIMENTAL PROCEDURES
Protein expression
SPOPMATH, SPOPBTB, SPOPBTB+, Gig1-258, Cul3NTD (residues 1-384) and Puc1-390 and variants were expressed as GST-fusions (pGEX4T1), SPOPMATH-BTB+ and SPOPN-MATH-BTB+ as His-MBP fusions (pRSFDuet), and for ubiquitination assays His-Puc1-390 from pRSFDuet, in BL21(DE3) GOLD, and purified by combinations of affinity, ion exchange, and gel filtration chromatography (Supplemental Experimental Procedures).
X-ray diffraction
Crystals were grown in hanging drops, soaked in cryoprotectants, and flash-frozen (liquid N2) for data collection at APS, ALS, and NSLS. Structures were determined by molecular replacement, except for SPOPBTB/3-box and GigBTB/3-box for which phases were obtained by SAD. Details are in Supplemental Experimental Procedures and Table 1.
Biochemical and biophysical studies
Protein interactions were examined by co-immunoprecipitation from Drosophila S2 cells, GST-pulldowns, size exclusion chromatography, surface plasmon resonance (BIACORE), and analytical ultracentrifugation. Solution conformational analyses were performed by examining resistance to proteolysis, Small Angle X-ray Scattering, and Circular Dichroism. Puc ubiquitination was assayed from cells and from purified components in vitro by western blotting. Details are in Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
We are grateful to D. King, V. Pagala, K. Kodali, and X. Ding for mass spectrometry, to R. Cassell and P. Rodrigues for peptide synthesis and purification, to C. Ross, A. Fereira, and J. Bollinger for computational support, to L. Borg, and A. Voss for technical assistance, to S. Otieno and R. Kriwacki for assistance with CD, J. Wang and J. Obenauer at the Hartwell Center for bioinformatics, to D.W. Miller and S. Bozeman for administrative support, to D. Scott for advice, and to K.R. Rajashankar and I. Kurinov for assistance at synchrotron. This work was supported in part by ALSAC (American Syrian Lebanese Associated Charities), grants from the NIH (R01GM069530 to BAS, GM070565 to JWH, P30CA021765 to St. Jude), Beckman Young Investigator Awards to BAS and KPW, awards from the W. M. Keck Foundation, and the Searle Funds at The Chicago Community Trust from the Chicago Biomedical Consortium to KPW. BAS is an Investigator of the Howard Hughes Medical Institute (HHMI). MFC is an HHMI fellow of the Damon Runyon Cancer Research Foundation (DRG 2021-9). NECAT (APS) is funded by RR-15301 from the NCRR at the NIH. Support for SERCAT (APS) may be found at www.ser-cat.org/members.html. APS is funded by U.S. DOE, Office of Basic Energy Sciences, contract W-31-109-ENG-38. 8.2.2 (ALS) is supported by HHMI. SIBYLS/12.3.1 (ALS) is funded by NCI CA92584 and DOE DE-AC03-76SF00098. ALS is funded by DOE Contract DE-AC02-05CH11231. X25 (NSLS) is funded by DOE Contract DE-AC02-98CH10886.
Footnotes
ACCESSION CODES
SPOPMATH-PucSBC1_pep1 3IVV.pdb; SPOPMATHx-PucSBC1_pep2 - 3HQL.pdb; SPOPMATH-MacroH2ASBCpep1 3IVB.pdb; SPOPMATHx-MacroH2ASBCpep1 - 3HQH.pdb; SPOPMATHx-MacroH2ASBCpep2 - 3HSV.pdb; SPOPMATH-CiSBC2 3IVQ.pdb; SPOPMATHx-CiSBC2 - 3HQM.pdb; SPOPBTB/3-box - 3HTM.pdb; GigBTB/3-box+ - 3HVE.pdb; SPOPMATHx-BTB/3-box- PucSBC1pep3 (form 1) - 3HQI.pdb; SPOPMATHx-BTB/3-box-PucSBC1pep3 (form 2) - 3HU6.pdb
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allen E, Ding J, Wang W, Pramanik S, Chou J, Yau V, Yang Y. Gigaxonin-controlled degradation of MAP1B light chain is critical to neuronal survival. Nature. 2005;438:224–228. doi: 10.1038/nature04256. [DOI] [PubMed] [Google Scholar]
- Angers S, Li T, Yi X, MacCoss MJ, Moon RT, Zheng N. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature. 2006a;443:590–593. doi: 10.1038/nature05175. [DOI] [PubMed] [Google Scholar]
- Angers S, Thorpe CJ, Biechele TL, Goldenberg SJ, Zheng N, MacCoss MJ, Moon RT. The KLHL12-Cullin-3 ubiquitin ligase negatively regulates the Wnt-beta-catenin pathway by targeting Dishevelled for degradation. Nat Cell Biol. 2006b;8:348–357. doi: 10.1038/ncb1381. [DOI] [PubMed] [Google Scholar]
- Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- Bardwell VJ, Treisman R. The POZ domain: a conserved protein-protein interaction motif. Genes Dev. 1994;8:1664–1677. doi: 10.1101/gad.8.14.1664. [DOI] [PubMed] [Google Scholar]
- Berthold J, Schenkova K, Ramos S, Miura Y, Furukawa M, Aspenstrom P, Rivero F. Characterization of RhoBTB-dependent Cul3 ubiquitin ligase complexes--evidence for an autoregulatory mechanism. Exp Cell Res. 2008;314:3453–3465. doi: 10.1016/j.yexcr.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosu DR, Kipreos ET. Cullin-RING ubiquitin ligases: global regulation and activation cycles. Cell Div. 2008;3:7. doi: 10.1186/1747-1028-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- Chew EH, Poobalasingam T, Hawkey CJ, Hagen T. Characterization of cullin-based E3 ubiquitin ligases in intact mammalian cells--evidence for cullin dimerization. Cell Signal. 2007;19:1071–1080. doi: 10.1016/j.cellsig.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol. 2004;24:8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow MR, Mains PE. Genetic and molecular characterization of the caenorhabditis elegans gene, mel-26, a postmeiotic negative regulator of mei-1, a meiotic-specific spindle component. Genetics. 1998;150:119–128. doi: 10.1093/genetics/150.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman RM, Correll CC, Kaplan KB, Deshaies RJ. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- Furukawa M, He YJ, Borchers C, Xiong Y. Targeting of protein ubiquitination by BTB-Cullin 3-Roc1 ubiquitin ligases. Nat Cell Biol. 2003;5:1001–1007. doi: 10.1038/ncb1056. [DOI] [PubMed] [Google Scholar]
- Geyer R, Wee S, Anderson S, Yates J, Wolf DA. BTB/POZ domain proteins are putative substrate adaptors for cullin 3 ubiquitin ligases. Mol Cell. 2003;12:783–790. doi: 10.1016/s1097-2765(03)00341-1. [DOI] [PubMed] [Google Scholar]
- Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP. Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol Cell. 2007;26:131–143. doi: 10.1016/j.molcel.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Hernandez-Munoz I, Lund AH, van der Stoop P, Boutsma E, Muijrers I, Verhoeven E, Nusinow DA, Panning B, Marahrens Y, van Lohuizen M. Stable X chromosome inactivation involves the PRC1 Polycomb complex and requires histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase. Proc Natl Acad Sci U S A. 2005;102:7635–7640. doi: 10.1073/pnas.0408918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson PK, Eldridge AG. The SCF ubiquitin ligase: an extended look. Mol Cell. 2002;9:923–925. doi: 10.1016/s1097-2765(02)00538-5. [DOI] [PubMed] [Google Scholar]
- Jiang J, Struhl G. Regulation of the Hedgehog and Wingless signalling pathways by the F-box/WD40-repeat protein Slimb. Nature. 1998;391:493–496. doi: 10.1038/35154. [DOI] [PubMed] [Google Scholar]
- Kent D, Bush EW, Hooper JE. Roadkill attenuates Hedgehog responses through degradation of Cubitus interruptus. Development. 2006;133:2001–2010. doi: 10.1242/dev.02370. [DOI] [PubMed] [Google Scholar]
- Krek W. BTB proteins as henchmen of Cul3-based ubiquitin ligases. Nat Cell Biol. 2003;5:950–951. doi: 10.1038/ncb1103-950. [DOI] [PubMed] [Google Scholar]
- Kwon JE, La M, Oh KH, Oh YM, Kim GR, Seol JH, Baek SH, Chiba T, Tanaka K, Bang OS, et al. BTB domain-containing speckle-type POZ protein (SPOP) serves as an adaptor of Daxx for ubiquitination by Cul3-based ubiquitin ligase. J Biol Chem. 2006;281:12664–12672. doi: 10.1074/jbc.M600204200. [DOI] [PubMed] [Google Scholar]
- Li T, Chen X, Garbutt KC, Zhou P, Zheng N. Structure of DDB1 in complex with a paramyxovirus V protein: viral hijack of a propeller cluster in ubiquitin ligase. Cell. 2006;124:105–117. doi: 10.1016/j.cell.2005.10.033. [DOI] [PubMed] [Google Scholar]
- Liu J, Ghanim M, Xue L, Brown CD, Iossifov I, Angeletti C, Hua S, Negre N, Ludwig M, Stricker T, et al. Analysis of Drosophila Segmentation Network Identifies a JNK Pathway Factor Overexpressed in Kidney Cancer. Science. 2009;323:1218–1222. doi: 10.1126/science.1157669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo SC, Li X, Henzl MT, Beamer LJ, Hannink M. Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling. Embo J. 2006;25:3605–3617. doi: 10.1038/sj.emboj.7601243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahrour N, Redwine WB, Florens L, Swanson SK, Martin-Brown S, Bradford WD, Staehling-Hampton K, Washburn MP, Conaway RC, Conaway JW. Characterization of cullin-box sequences that direct recruitment of Cul2-Rbx1 and Cul5-Rbx2 modules to elongin BC-based ubiquitin ligases. J Biol Chem. 2008;283:8005–8013. doi: 10.1074/jbc.M706987200. [DOI] [PubMed] [Google Scholar]
- McMahon M, Itoh K, Yamamoto M, Hayes JD. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J Biol Chem. 2003;278:21592–21600. doi: 10.1074/jbc.M300931200. [DOI] [PubMed] [Google Scholar]
- Orlicky S, Tang X, Willems A, Tyers M, Sicheri F. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell. 2003;112:243–256. doi: 10.1016/s0092-8674(03)00034-5. [DOI] [PubMed] [Google Scholar]
- Padmanabhan B, Tong KI, Ohta T, Nakamura Y, Scharlock M, Ohtsuji M, Kang MI, Kobayashi A, Yokoyama S, Yamamoto M. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol Cell. 2006;21:689–700. doi: 10.1016/j.molcel.2006.01.013. [DOI] [PubMed] [Google Scholar]
- Patton EE, Willems AR, Sa D, Kuras L, Thomas D, Craig KL, Tyers M. Cdc53 is a scaffold protein for multiple Cdc34/Skp1/F-box proteincomplexes that regulate cell division and methionine biosynthesis in yeast. Genes Dev. 1998;12:692–705. doi: 10.1101/gad.12.5.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Pintard L, Willems A, Peter M. Cullin-based ubiquitin ligases: Cul3-BTB complexes join the family. Embo J. 2004;23:1681–1687. doi: 10.1038/sj.emboj.7600186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintard L, Willis JH, Willems A, Johnson JL, Srayko M, Kurz T, Glaser S, Mains PE, Tyers M, Bowerman B, Peter M. The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature. 2003;425:311–316. doi: 10.1038/nature01959. [DOI] [PubMed] [Google Scholar]
- Polekhina G, House CM, Traficante N, Mackay JP, Relaix F, Sassoon DA, Parker MW, Bowtell DD. Siah ubiquitin ligase is structurally related to TRAF and modulates TNF-alpha signaling. Nat Struct Biol. 2002;9:68–75. doi: 10.1038/nsb743. [DOI] [PubMed] [Google Scholar]
- Saha A, Deshaies RJ. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol Cell. 2008;32:21–31. doi: 10.1016/j.molcel.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas GD, Blair LA, Needleman LA, Gonzales JD, Chen Y, Li M, Singer JD, Marshall J. Actinfilin is a Cul3 substrate adaptor, linking GluR6 kainate receptor subunits to the ubiquitin-proteasome pathway. J Biol Chem. 2006;281:40164–40173. doi: 10.1074/jbc.M608194200. [DOI] [PubMed] [Google Scholar]
- Schulman BA, Carrano AC, Jeffrey PD, Bowen Z, Kinnucan ER, Finnin MS, Elledge SJ, Harper JW, Pagano M, Pavletich NP. Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature. 2000;408:381–386. doi: 10.1038/35042620. [DOI] [PubMed] [Google Scholar]
- Scrima A, Konickova R, Czyzewski BK, Kawasaki Y, Jeffrey PD, Groisman R, Nakatani Y, Iwai S, Pavletich NP, Thoma NH. Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex. Cell. 2008;135:1213–1223. doi: 10.1016/j.cell.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seol JH, Feldman RM, Zachariae W, Shevchenko A, Correll CC, Lyapina S, Chi Y, Galova M, Claypool J, Sandmeyer S, et al. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 1999;13:1614–1626. doi: 10.1101/gad.13.12.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- Stebbins CE, Kaelin WG, Jr, Pavletich NP. Structure of the VHL-ElonginC-ElonginB complex: implications for VHL tumor suppressor function. Science. 1999;284:455–461. doi: 10.1126/science.284.5413.455. [DOI] [PubMed] [Google Scholar]
- Stogios PJ, Downs GS, Jauhal JJ, Nandra SK, Prive GG. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005;6:R82. doi: 10.1186/gb-2005-6-10-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svendsen JM, Smogorzewska A, Sowa ME, O’Connell BC, Gygi SP, Elledge SJ, Harper JW. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell. 2009;138:63–77. doi: 10.1016/j.cell.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi I, Kameoka Y, Hashimoto K. MacroH2A1.2 binds the nuclear protein Spop. Biochim Biophys Acta. 2002;1591:63–68. doi: 10.1016/s0167-4889(02)00249-5. [DOI] [PubMed] [Google Scholar]
- Tang X, Orlicky S, Lin Z, Willems A, Neculai D, Ceccarelli D, Mercurio F, Shilton BH, Sicheri F, Tyers M. Suprafacial orientation of the SCFCdc4 dimer accommodates multiple geometries for substrate ubiquitination. Cell. 2007;129:1165–1176. doi: 10.1016/j.cell.2007.04.042. [DOI] [PubMed] [Google Scholar]
- Tong KI, Padmanabhan B, Kobayashi A, Shang C, Hirotsu Y, Yokoyama S, Yamamoto M. Different Electrostatic Potentials Define ETGE and DLG Motifs as Hinge and Latch in Oxidative Stress Response. Mol Cell Biol. 2007;27:7511–7521. doi: 10.1128/MCB.00753-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C, Bashton M, Kerrison ND, Chothia C, Teichmann SA. Structure, function and evolution of multidomain proteins. Curr Opin Struct Biol. 2004;14:208–216. doi: 10.1016/j.sbi.2004.03.011. [DOI] [PubMed] [Google Scholar]
- Welcker M, Clurman BE. Fbw7/hCDC4 dimerization regulates its substrate interactions. Cell Div. 2007;2:7. doi: 10.1186/1747-1028-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems AR, Lanker S, Patton EE, Craig KL, Nason TF, Mathias N, Kobayashi R, Wittenberg C, Tyers M. Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell. 1996;86:453–463. doi: 10.1016/s0092-8674(00)80118-x. [DOI] [PubMed] [Google Scholar]
- Willems AR, Schwab M, Tyers M. A hitchhiker’s guide to the cullin ubiquitin ligases: SCF and its kin. Biochim Biophys Acta. 2004;1695:133–170. doi: 10.1016/j.bbamcr.2004.09.027. [DOI] [PubMed] [Google Scholar]
- Xu L, Wei Y, Reboul J, Vaglio P, Shin TH, Vidal M, Elledge SJ, Harper JW. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature. 2003;425:316–321. doi: 10.1038/nature01985. [DOI] [PubMed] [Google Scholar]
- Yamoah K, Oashi T, Sarikas A, Gazdoiu S, Osman R, Pan ZQ. Autoinhibitory regulation of SCF-mediated ubiquitination by human cullin 1’s C-terminal tail. Proc Natl Acad Sci U S A. 2008;105:12230–12235. doi: 10.1073/pnas.0806155105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Arron JR, Lamothe B, Cirilli M, Kobayashi T, Shevde NK, Segal D, Dzivenu OK, Vologodskaia M, Yim M, et al. Distinct molecular mechanism for initiating TRAF6 signalling. Nature. 2002;418:443–447. doi: 10.1038/nature00888. [DOI] [PubMed] [Google Scholar]
- Yin Q, Lin SC, Lamothe B, Lu M, Lo YC, Hura G, Zheng L, Rich RL, Campos AD, Myszka DG, et al. E2 interaction and dimerization in the crystal structure of TRAF6. Nat Struct Mol Biol. 2009;16:658–666. doi: 10.1038/nsmb.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DD, Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol. 2003;23:8137–8151. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Zhang L, Wang B, Ou CY, Chien CT, Jiang J. A hedgehog-induced BTB protein modulates hedgehog signaling by degrading Ci/Gli transcription factor. Dev Cell. 2006;10:719–729. doi: 10.1016/j.devcel.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- Zollman S, Godt D, Prive GG, Couderc JL, Laski FA. The BTB domain, found primarily in zinc finger proteins, defines an evolutionarily conserved family that includes several developmentally regulated genes in Drosophila. Proc Natl Acad Sci USA. 1994;91:10717–10721. doi: 10.1073/pnas.91.22.10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.