Abstract
The Genetics of Kidneys in Diabetes (GoKinD) study was initiated to facilitate research aimed at identifying genes involved in diabetic nephropathy (DN) in type 1 diabetes (T1D). In this review, we present on overview of this study and the various reports that have utilized its collection. At the forefront of these efforts is the recent genome-wide association (GWA) scan implemented on the GoKinD collection. We highlight the results from our analysis of these data and describe compelling evidence from animal models that further support the potential role of associated loci in the susceptibility of DN. To enhance our analysis of genetic associations in GoKinD, using genome-wide imputation (GWI), we expanded our analysis of this collection to include genotype data from more than 2.4 million common SNPs. We illustrate the added utility of this enhanced dataset through the comprehensive fine-mapping of candidate genomic regions previously linked with DN and the targeted investigation of genes involved in candidate pathway implicated in its pathogenesis. Collectively, GWA and GWI data from the GoKinD collection will serve as a springboard for future investigations into the genetic basis of DN in T1D.
Keywords: genome-wide association, diabetic nephropathy, type 1 diabetes, imputation
The Genetics of Kidneys in Diabetes (GoKinD) Study
For 20 years, researchers have known that genetics susceptibility plays an important role in the pathogenesis of diabetic nephropathy (DN) among patients with type 1 diabetes (T1D).1 Reports of clustering of DN among T1D members of families, or familial aggregation, by Seaquist et al.1, Borch-Johnsen et al.2, and Quinn et al.3 supported the genetic basis of this complication of diabetes and provided the foundation for investigators to begin searching for the genetic components underlying its cause. Despite the early optimism generated by several studies (primarily using linkage-based approaches) efforts aimed at identifying genes involved in the pathogenesis of DN have yet to provide conclusive evidence to implicate the causative role of specific susceptibility genes.
Early efforts to investigate the genetic causes of DN were limited in their ability to adequately assess its impact on the risk of disease. Specifically, the study collections being investigated by individual researchers were largely underpowered; a feature that, in all likelihood, has contributed to the irreproducibility among various reports and challenged the interpretation of these findings. Recognizing this as a critical bottleneck in the study of DN genes, the Juvenile Diabetes Research Foundation (JDRF), the National Institutes of Diabetes and Digestive and Kidney Disease (NIDDK), and the Centers for Disease Control and Prevention (CDC) sponsored the Genetics of Kidneys in Diabetes (GoKinD) study, an initiative to assemble a large DNA collection to facilitate investigator-driven research into the genetic basis of DN and T1D.4
Through collaborative recruitment efforts at both the Joslin Diabetes Center (JDC) and the George Washington University Biostatistics Center (GWU), 1,889 T1D patients, primarily self-reported Caucasians, were enrolled to the GoKinD study.4 This collection includes a total of 943 cases with DN and 946 controls without DN. Among the DN cases, 328 patients had persistent proteinuria, defined by a urinary albumin to creatinine ratio (ACR) ≥300 μg/mg in two of the last three measurements taken at least one month apart, and 615 patients had end-stage renal disease (ESRD) (dialysis or renal transplant). Controls had T1D for at least 15 years and normoalbuminuria, defined by an ACR <20μg/mg in two of the last three measurements taken at least one month apart (if a third measurement was required, a value <40μg/mg was necessary for inclusion), without ever having been treated with angiotensin-converting enzyme inhibitors or angiotensin receptor blockers, and were not being treated with antihypertensive medication at the time of recruitment into the study.
In keeping with the primary goal of the GoKinD study, this DNA collection and its corresponding clinical data are available to all approved researchers (see www.jdrf.org/gokind for further details).
Genome-wide Association (GWA) Analysis of the GoKinD Collection
In 2006, the GoKinD collection was selected by the Foundation for the National Institutes of Health (FNIH) for whole-genome association analysis by the Genetic Association Information Network (GAIN).5 As part of this public-private partnership, DNA from 1,879 available participants of GoKinD were submitted for genotyping on the Affymetrix 5.0 500K SNP Array by the GAIN genotyping laboratory at the Eli and Edythe L. Broad Institute (Cambridge, MA). Internal quality control by the GAIN genotyping laboratory resulted in the release of data for 467,144 SNPs from this platform. Pre-computed association data have been made available through the GAIN Database within dbGaP (www.ncbi.nlm.nih.gov/sites/entrez?db=gap, accession number pha002852.1). Additionally, individual phenotypic and genotypic data can be accessed by investigators following approval by the GAIN Data Access Committee (see dbGaP for instructions on accessing GAIN data).
We recently reported our primary analysis of these data.6 Following the application of more stringent quality control metrics, including restricting our analysis to autosomal SNPs with minor allele frequency (MAF) > 0.01, the rejection of Hardy-Weinberg assumptions (P ≤ 10−5), and the application of differential rates of missingness (by case/control status and MAF), we decreased the number of high-quality SNPs to 359,193 variants. Similarly, additional measures of sample quality control were implemented to ensure genotyping quality and identify instances of cryptic relatedness and non-European ancestry. Among the GoKinD participants, 93.0% had self-reported a primary race of ‘White’, while the remainder had either reported a non-White race (Black, Hispanic, Asian/Pacific Islander, or Native American) or failed to provide their ethnicity. Using principal component analysis (PCA), we identified and removed 129 non-European outliers from among these individuals. Interestingly, approximately 20% of these individuals had reported their ethnicity as White. In total, our quality control criteria reduced the GoKinD population to 1,705 individuals (885 controls and 820 DN cases, including 284 with proteinuria and 536 with ESRD) of European ancestry.
As has been reported for other complex genetic disorders, no single major gene that contributes to an increased risk of DN emerged from our analysis of these data. In total, we identified 11 SNPs that were significantly associated (P <1×10−5) with DN in T1D. The strongest association occurred on chromosome 9q with rs10868025 (P=5.0×10−7), a SNP located near the 5′-end of the FERM domain containing 3 (FRMD3) gene. Three additional genomic regions located on chromosomes 7p, 11p, and 13q were also associated with DN. On chromosome 7p, rs39059 (P= 5.0×10−6) localizes to the first intron of CHN2 (beta chimerin) isoform 2 and upstream of an alternatively spliced CPVL (serine carboxypeptidase vitellogenic-like) transcript. On chromosome 11p, rs451041 (P=3.1×10−6) is located in an intronic region of the CARS (cysteinyl-tRNA synthetase) gene. And on chromosome 13q, the region bounded by rs1411766/rs1742858 (P=1.8×10−6) is located approximately 384 kilo-basepair (kb) telomeric to the myosin heavy chain Myr 8 (MYO16) gene and 120 kb centromeric to the insulin receptor substrate 2 (IRS2) gene. Interestingly, significant evidence of linkage was recently identified at this same locus in studies of patients with non-diabetic ESRD and all-cause ESRD.7, 8 The imputation of un-genotyped SNPs at each of these loci, using population haplotype data available from the HapMap Project (www.hapmap.org) and the MACH software program (www.sph.umich.edu/csg/abecasis/MACH), identified 11 additional SNPs that were highly correlated with the original associations (P<1×10−5), including three variants that were more strongly associated with DN than our lead genotyped SNPs at their respective loci (rs1888747 on chromosome 9q and rs39075 and rs39076 on chromosome 7p). Finally, and perhaps most importantly, associations at the FRMD3 and CARS loci were replicated with time to the onset of severe nephropathy in the DCCT/EDIC study (P=0.02 and P=0.01, respectively), thereby bolstering the significance of these findings and implicating the involvement of two previously unsuspected pathways in the pathogenesis of DN. Association data from these analyses are also available through the GAIN Database within dbGaP (accession number pha002864.1).
We recently used data from this analysis to examine associations between SNPs in the engulfment and cell motility 1 (ELMO1) gene, a locus previously shown to be associated with DN in two ethnically distinct type 2 diabetic (T2D) populations9,10, and the risk of nephropathy in the GoKinD collection.11 We examined a total of 118 SNPs across this locus, including 106 genotyped SNPs. Locus-specific imputation was performed to examine 12 additional SNPs not genotyped on the Affymetrix 5.0 platform but that had previously been reported to be associated with DN. The strongest associations in ELMO1 occurred at rs11769038 (P = 1.7×10−3) and rs1882080 (P = 3.2×10−3) located in intron 16. No evidence of association for variants previously reported in T2D was observed in our collection. This investigation, however, marks the third report of associations in ELMO1 with DN. While these data appear to suggest that allelic heterogeneity exists at this locus, they also further establish ELMO1’s role in the susceptibility of this disease.
In addition to our investigations of DN-associated loci using GWA data from GoKinD, these data have also recently been used to interrogate T1D risk loci.12 Data from 1,601 T1D cases from GoKinD were combined with data from 1,704 non-diabetic controls from the National Institute of Mental Health (NIHM). Associations were detected at several previously confirmed T1D-accociated loci.13 Interestingly, a meta-analysis of these data and that from 1,960 T1D cases and 2,942 non-diabetic controls from the British Wellcome Trust Case Control Consortium (WTCCC) identified support for several previously undetected T1D loci, with the strongest associations occurring at variants in the cathepsin H (CTSH) and BTB and CNC homology 1, basic leucine zipper transcription factor 2 (BACH2) genes.
Concordance of DN Loci from GoKinD in Animal Models
Insights to the causative genes involved in various human diseases have been gained from inter-disciplinary approaches that incorporate data from human studies with that from animal models.14-16 Because comparative genomic analysis, specifically the identification of concordant disease loci in human and mouse, offers a complementary approach to human studies alone, we applied this approach to loci associated with DN in our recent GWA analysis of the GoKinD collection
Although we did not find evidence of concordant loci in the mouse for the homologous loci on human chromosomes 7p (near CPVL/CHN2) and 11p (near CARS), through comparative analysis, we were able to identify concordance at the human chromosome 9q region (near FRMD3) and albuminuria quantitative trait loci (QTL) reported in two independent studies of nephropathy in mice.17,18 The 2 mega-basepair (Mb) interval flanking FRMD3 contains 10 annotated genes. Interestingly, while the distal portion of the human locus is homologous to mouse chromosome 4 and falls within the 95% confidence intervals of the QTL identified in these studies, the proximal portion is homologous to a region on mouse chromosome 13 in which no QTL have been reported. Under the assumption that the same underlying gene contributes to the association and linkage signals observed in human and mouse, respectively, this breakpoint in synteny excludes all genes contained in the interval homologous to mouse chromosome 13 as likely nephropathy candidate genes and, there by, narrows the disease locus of interest to a 0.4 Mb interval containing only the RASEF (RAS and EF-hand domains-containing gene) and FRMD3 genes.
Further narrowing of this reduced region is possible through interval specific haplotype analysis of the individual strains contributing to the QTL on mouse chromosome 4. The QTL at this locus was identified in two independent crosses: (C57BL/6J x DBA/2J)F218 and (C57BL/6J x NZM)F1 x NZM17. In comparing the haplotypes of the strains from the (C57BL/6J x DBA/2J)F2 cross, we see that both C57BL/6J and DBA/2J share a common haplotype in the region where Rasef is located. Because there is no genetic variation between either strain along this haplotype, this region does not contribute to the observed QTL identified in this cross, and thus, we can discount this region as one likely to contain the underlining disease gene. Similarly, this same region is identical by descent within the C57BL/6J and NZM strains. When coupled with the data from our GWA analysis of GoKinD, these data exclude RASEF as a likely candidate disease gene at this locus and further implicate FRMD3 as a common nephropathy disease gene between both species.
In addition to concordance at the FRMD3 region, we recently reported similar findings at human chromosomes 13q and 1p with albuminuria-associated loci identified in aging mice.19 Using haplotype association mapping (HAM), a recently developed approach which utilizes high-density SNP data from many inbred strains to identify chromosomal haplotypes that are associated with a phenotype of interest, Tsaih et al. identified 9 candidate chromosomal regions associated with increased albuminuria. To assess whether the syntenic human loci were also associated with nephropathy, we identified the human chromosomal regions corresponding to each albuminuria-associated locus and examined each region for genetic associations in GWA data from GoKinD. Remarkably, concordance with homologous human chromosomal regions was identified at 2 of these 9 loci. Among these concordant loci, the strongest association occurred at a variant identified in our original analysis of the GoKinD data; at rs1411766 located on human chromosome 13q, a region concordant with an albuminuria-associated locus located between Myo16 and Irs2 on mouse chromosome 8. A second concordant locus was identified at NEGR1 (neuronal growth regulator 1) on human chromosome 1p (syntenic albuminuria-associated locus located at Negr1 on mouse chromosome 3). Although this variant was just below the threshold reported in our analysis the GWA data from GoKinD (rs6671557, P=4.9×10−5), this finding is particularly interesting given the concordant albuminuria-associated mouse locus identified using HAM.
The identification of concordance at these loci, as well as at the 9q region, provides additional support that these loci play a role in the susceptibility of DN.
Enhanced GWA Analysis of the GoKinD Collection through SNP Imputation
Our analysis of GWA data from the GoKinD collection included 359,193 SNPs genotyped on the Affymetrix 5.0 500K SNP Array, a platform that offers approximately 76% coverage of common variation across the entire genome.5, 6 As described above, we utilized imputation to supplement these data as specific loci identified in our GWA analysis, as well as to closely examine genetic variation at the ELMO1 locus. To further enhance the coverage of these association data, we recently expanded our analysis of the GoKinD dataset by performing genome-wide imputation (GWI) of un-genotyped SNPs available from the HapMap Project.
GWI of the GoKinD collection was carried out using phased, forward strand Phase II data (release 22, build 36) for all autosomal chromosomes from the HapMap CEU population and the MACH software program. Imputed genotypes were filtered for minor allele frequency > 1% and r2 > 0.30, resulting in 2,043,202 high-quality imputed SNPs and in an augmented dataset containing genotypes for a total of 2,402,395 SNPs (359,193 SNPs genotyped plus 2,043,202 imputed SNPs). All SNPs were then analyzed using stratified additive tests of association (adjusting for both gender and JDC/GWU strata, as previously described6) using PLINK.20
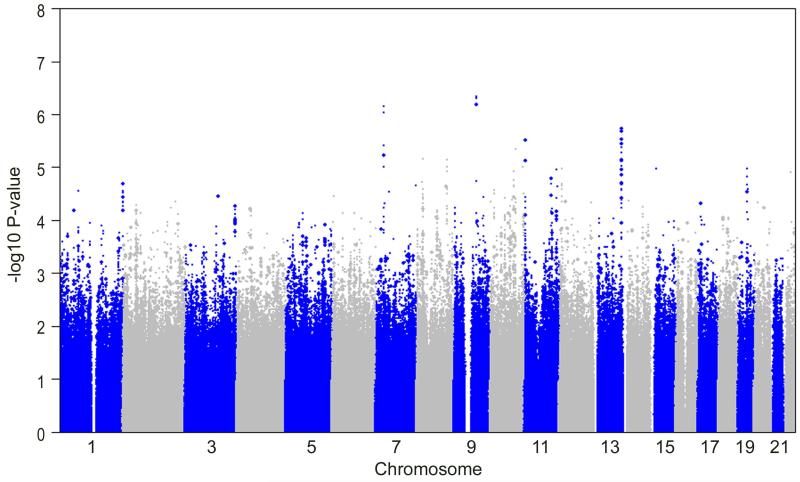
As shown in Figure 1, and as previously reported6, the strongest associations remain on chromosomes 7p, 9q, 11p, and 13q. As we have previously shown, imputed SNPs on both chromosomes 7p and 9q were more strongly associated with DN than the genotyped SNPs at these loci.6 In addition to these signals, the enhanced GoKinD dataset provided strong associations (P <1×10−5) at 4 additional loci not previously observed in the Affymetrix dataset. Among these signals, the strongest imputed association occurred on chromosome 10q with rs7071071 (P=4.5×10−6) located at the sorbin and SH3 domain containing 1 (SORBS1) gene. Four imputed SNPs clustered within a 27 kb region located 177 kb centromeric to the trichorhinophalangeal syndrome I (TRPS1) gene on chromosome 8p. Also on chromosome 8p, rs6986695 (P= 6.9×10−6) is located approximately 202 kb centromeric to the cell division cycle associated 2 (CDCA2) gene and 134 kb telomeric to the early B-cell factor 2 (EBF2) gene. And, on chromosome 10q, rs845086 (P= 9.8×10−6) localizes to an intergenic region flanked by the budding uninhibited by benzimidazoles 3 homolog (BUB3) and the G protein-coupled receptor 26 (GPR26) genes.
Figure 1.
Summary of enhanced GWA analysis of the GoKinD collection through SNP imputation. The −log10 P-values for GWI data were calculated using the Cochran-Mantel-Haenszel method (adjusting for gender and GoKinD sub-collection (JDC/GWU)) are shown for 359,193 genotyped (large circles) and 2,043,202 imputed SNPs (small circles) across the entire genome in the GoKinD collection.
Importantly, in addition to expanding the coverage of genotyped SNPs and uncovering additional loci of interest, GWI also allows researchers to effectively fine-map the entire genome. As we describe below, this augmented dataset also facilitates the comprehensive analysis of targeted regions, including the investigation of both candidate regions previously linked with DN and the investigation of candidate pathways implicated in DN.
Comprehensive Analysis of Candidate Regions in GoKinD
Attempts to identify the genetic determinants of DN in T1D have been made using various approaches, including linkage analysis.21-23 A pilot study performed at the Joslin Diabetes Center by our research group aimed to test 3 candidate loci comprised of genes from the angiotensin pathway: angiotensin-converting enzyme (ACE) on chromosome 17q, angiotensinogen (AGT) on chromosome 1q, and angiotensin II type 1 receptor (AGTR1) on chromosome 3q.21 The strongest linkage signal observed in this study occurred at the AGTR1 locus (LOD score = 3.1) and was centered on the ATCA marker, located approximately 15 kb downstream from ATGR1. A recent genome-wide linkage study in a Finnish T1D population revealed the highest linkage peak near this same locus (maximum LOD score, under a dominant model, = 2.67), located between microsatellite markers D3S3606 and D3S3694 on chromosome 3q.22 This region spans 39 cM and includes the AGTR1 locus identified at Joslin. To follow-up this finding, He et al. recently fine-mapped the 3q locus using a multi-stage case-control study approach involving T1D patients from Finland, Iceland, and the British Isles.24 In this report, the strongest association occurred in a non-coding region approximately 10 Mb centromeric of the original linkage peak at rs1866813 (P=7.1×10−6). Interestingly, a genome-wide linkage study in African-Americans with T2D identified evidence of suggestive linkage with ESRD at this same region on chromosome 3q.25
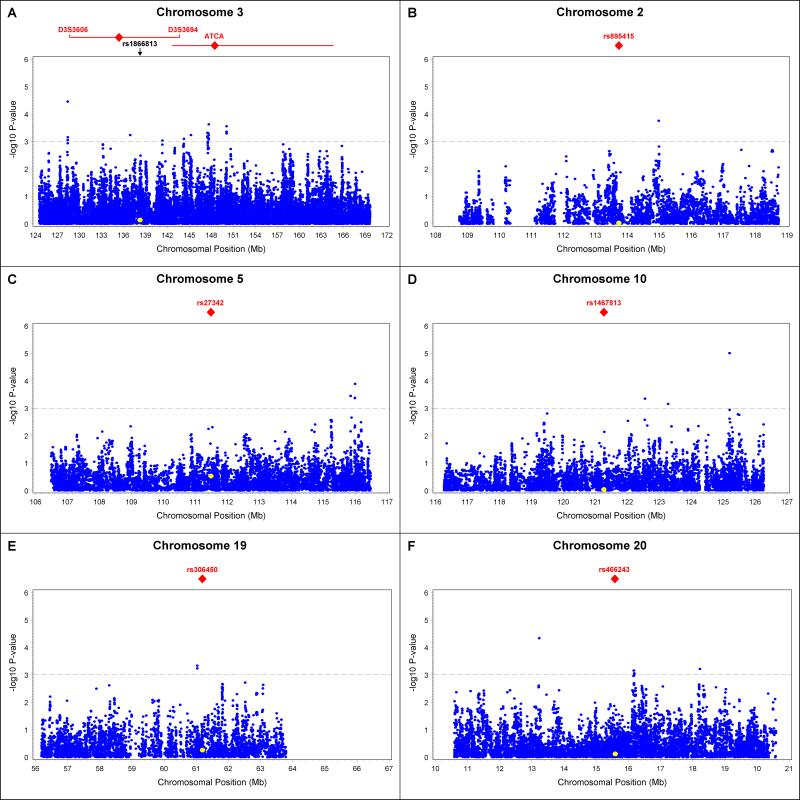
Because multiple groups have reported evidence of linkage and genetic associations at the 3q region, we chose to examine this candidate region more closely in GWI data from the GoKinD collection. Supportive evidence of associations was sought in this collection by analyzing all SNPs that mapped to the reported region on chromosome 3q (position 124.5 to 169.7 Mb), including a total of 5,444 genotyped and 32,277 imputed SNPs. The strongest association with DN in this region was observed at rs11915684 (P=3.5×10−5), a SNP located approximately 7 Mb centromeric from the ATCA marker located in the AGTR1 gene and approximately 20 Mb centromeric from the linkage peak reported in the Finnish population (Figure 2A).22 This SNP maps downstream of the C3orf56 gene and approximately 150 kb upstream of the plexin A1 (PLXNA1) gene. Interestingly, PLXNA1 encodes a protein which has been shown to be down-regulated in podocytes by advanced glycation end products (AGE).26 Additionally, several associated SNPs (at a level of P <10−3) were identified within the 2 Mb region flanking the linkage peak reported in the Finnish population, however, the variant rs1866813, reported by He et al., did not reach nominal statistical significance in the GoKinD collection.22,24
Figure 2.
Summary of comprehensive analysis of regions previously reported to be linkage with DN in T1D in GWI data from GoKinD. The −log10 P-values for GWI data were calculated using the Cochran-Mantel-Haenszel method (adjusting for gender and GoKinD sub-collection (JDC/GWU)) for SNPs (blue circles) mapping to regions previously reported to be linkage with DN in T1D. Panel A: Region of significant linkage on chromosome 3q. Red rhombi indicate the positions of the highest LOD-scores reported by Osterholm et al.22 (left) and Moczulski et al.21 (right, at ATCA marker). LOD-1 intervals are indicated by the red horizontal lines. Also indicated is position of SNP rs1866813 reported by He et al. to be associated with DN (P=7.1×10−6). In the GoKinD collection, this SNP is indicated by a yellow circle. Panels B to F: Regions flanking linkage markers for DN reported by Rogus et al. on chromosome 2q (Panel B), chromosome 5q (Panel C), chromosome 10q (Panel D), chromosome 19q (Panel E) and chromosome 20p (Panel F). Red symbols indicate the position of linked SNPs. These SNPs were imputed in the GoKinD collection and are indicated by yellow circles.
Our analysis of the chromosome 3q locus using GWI data from the GoKinD population demonstrates the potential value of this data in fine-mapping previously reported DN loci. In addition to this locus, evidence of linkage to DN has been reported at other regions of the genome and, similarly, these findings are worth examining in the context of the available GWI data from GoKinD as these data may be helpful in further exploring these regions. We recently reported results from a high-density SNP genome-wide linkage scan of 100 T1D sibpairs discordant for DN.23 In this study, evidence of linkage was observed at 5 distinct chromosomal regions. Near the region harboring the strongest signal (chromosome 19q at rs306450, LOD = 3.1), we identified a cluster of 4 highly-correlated SNPs (rs16986667, rs16986669, rs16986672 and rs2616944) associated (P<10−3) with DN in the GoKinD collection (Figure 2E). On chromosome 2q, significant linkage (LOD=2.1) was observed at rs895415. In the GoKinD collection, we identified one SNP associated with DN (rs10187644, P=1.7×10−4) approximately 1 Mb telomeric from this linkage signal (Figure 2B). On chromosome 5q, where we reported a LOD score of 2.7 for rs27342, there were several SNPs associated with DN (P<10−3) in the GoKinD collection that clustered approximately 5 Mb telomeric from the linkage marker (Figure 2C). At the chromosome 10q region (linkage marker rs1467813, LOD = 2.4), we identified a SNP (rs845086) highly associated with DN (P=9.8×10−6), which maps about 4 Mb telomeric from the linkage marker (Figure 2D). This SNP is located in a region that does not contain any known genes. Finally, on chromosome 20p, stratified linkage analysis among proteinuric patients identified a linkage peak at rs466243 (LOD = 2.8). At this locus, the strongest association with DN in the GoKinD collection was observed at rs17191018 (P=4.6×10−5) (Figure 2F), a marker located approximately 2 Mb telomeric from this linkage signal and within the fifth intron of isthmin 1 homolog (ISM1), a gene whose role in kidney physiology remains to be determined.
The GoKinD collection provides some confirmatory evidence for previous linkage findings. Our analysis suggests that within the large linkage region on chromosome 3q there may be additional interesting candidate genes for DN than those that have thus far been examined, including the PLXNA1 gene. Strong association signals were also observed on chromosomes 10q and 20p. The replication of these findings is critical to understanding whether these variants underlie the observed linkage signals and whether these associations can help to pinpoint the putative DN genes at these loci.
Candidate Pathway Analysis Using GWA Data from GoKinD
In addition to investigations spanning the genome or particular candidate loci, GWI data from the GoKinD collection can also be used in candidate gene-based approaches. There are number of possible candidate genes, physiological pathways, and mechanisms that are likely to play important roles in the development of DN. Renal fibrosis is one such mechanism.
It is well recognized that, irrespective of its initiating factors, the final stages of renal disease that contribute to its progression to ESRD, follow common pathway that ultimately result in glomerulosclerosis27 and tubulointerstitial fibrosis28,29. There is strong evidence that transforming growth factor beta (TGF-β) is a major mediator of this process.28-30 TGF-β induces fibrosis in the course of the development of DN31, as well as in other types of renal damage: during tissue repair32, immunoglobulin A (IgA) nephropathy, lupus nephritis, focal and segmental glomerulosclerosis33, and in chronic graft rejection34. Genes that constitute the TGF-β pathway are therefore well-justified candidates for genetic studies. Similarly, genes that encode the extracellular matrix (ECM) proteins and their cellular receptors may also be considered in this same context.
To illustrate a candidate gene/pathway approach to interrogate components of the TGF-β and ECM pathways, we identified all autosomal genes in each pathway and examined all available markers for genetic associations at these genes using GWI data from GoKinD. The genes constituting the TGF-β and ECM pathways were identified through the Kyoto Encyclopedia of Genes and Genomes (KEGG; www.genome.jp/kegg) and all genotyped and imputed SNPs located within these genes (including 50 kb of flanking sequence) were examined. The TGF-β pathway (KEGG: hsa04350) consists of 90 autosomal genes located in 85 distinct loci, while the ECM receptor-interaction pathway (KEGG: hsa04512) includes 86 autosomal genes across 79 distinct loci. Four related genes from the thrombospondin family (THBS1, THBS2, THBS3, and THBS4) are common to both the TGF-β pathway and the ECM pathway. Finally, because no genotyped or imputed SNPs mapped to the agrin (AGRN) gene region on chromosome 1, this ECM pathway gene was excluded from our analysis.
Our findings are summarized in Table 1, and additional details are provided in Online Supplementary Tables 1 and 2. Briefly, in the TGF-β pathway, there were 28 SNPs with P <10−3 from among a total of 9,979 SNPs. The most significant SNP (rs8135616, P=3.7×10−4) was located in the mitogen-activated protein kinase 1 (MAPK1) gene on chromosome 22. Several SNPs with the high statistical significance were located in the bone morphogenic protein receptor 1B (BMPR1B) locus on chromosome 4. Polymorphisms in BMPR1B were previously reported to be significantly associated with glomerular filtration rate (GFR) in a cohort of patients with sickle cell anemia.35 None of the 4 SNPs reported in this study were associated with DN in the GoKinD collection. Despite some evidence of association, given the number of SNPs examined in this pathway, our findings are not significant following adjustments for multiple testing.
Table 1.
Summary of Candidate Pathway Analysis in GWI Data from the GoKinD Collection
| Pathway | Number of Loci |
Average Locus Size (kb) |
Total Size of Loci (kb) |
Number of Genotyped SNPs |
Number of Imputed SNPs |
Number of Nominally Significant SNPs (P<0.05) |
Lowest P-value (SNP, gene) |
|---|---|---|---|---|---|---|---|
| TGF-β | 85 | 170.6 | 14,503.4 | 1,488 | 8,491 | 549 | 3.7×10−4 (rs8135616, MAPK1) |
| ECM | 74 | 214.2 | 15,847.5 | 2,498 | 13,040 | 770 | 3.1×10−4 (rs4703699, SV2C) |
TGF-β = transforming growth factor beta pathway
ECM = extracellular matrix receptor-interaction pathway
In the ECM pathway, the most significant SNP (rs4703699 on chromosome 5 in synaptic vesicle glycoprotein 2C [SV2C]) yielded a P-value of 3.1×10−4. In total, there were 22 SNPs with P <10−3 among the 15,538 tested SNPs (including 2,498 genotyped SNPs). Again, constrained by the number of comparisons included in this analysis, none of these variants reached a level indicative of statistical significance.
Taken together, our comprehensive analysis of genes involved in the TGF-β and ECM pathways using GWI data from the GoKinD collection suggests that common genetic variations at loci involved in fibrosis and ECM organization do not contribute to the genetic susceptibility of DN in T1D.
Previous Studies on the GoKinD Collection
Prior to the recent GWA study, the GoKinD collection had proven to be a valuable resource for genetic association studies, particularly those aimed at investigating specific candidate genes implicated in DN (Table 2). In a subset of the GoKinD collection that included 531 ESRD cases and 564 controls (with at least 20 years diabetes duration) of European ancestry, Millis et al. sought confirmatory evidence of associations in the plasmacytoma variant translocation (PVT1) gene that were previously identified with ESRD in Pima Indians with T2D.36 Interestingly, 2 SNPs (rs13447075; P =3.0×10−3 and rs2648862; P =8.0×10−3), including one located in the coding region of one of the gene’s transcript variants, were strongly associated with ESRD in GoKinD. Similarly, an association initially identified in T2D patients with both proliferative diabetic retinopathy and ESRD at a functional variant in the erythropoietin (EPO) gene was also replicated in 865 cases and 574 controls (without both DN and retinopathy) from the GoKinD collection.37 In a study of 2 non-synonymous SNPs in the intracellular adhesion molecule-1 (ICAM-1) gene in 662 DN cases and 620 controls, although no significant differences in allele frequencies were observed, Ma et al. found differences in the heterozygous frequencies of rs5498 (E469K).38 This association was only borderline among T1D patients with and without DN (P=0.05), however, a significant association was identified in comparisons among females patients (P=0.01), suggesting a potential gender-specific effect.
Table 2.
Summary of Studies Using the GoKinD DNA Collection
| Reference | Study Design | GoKinD Sample SubSets |
Chr. | SNP | Nearest Gene(s) |
P-value |
|---|---|---|---|---|---|---|
| Ma et al. 200742 | Case-control, candidate gene, 1 SNP |
Cases (N=577) and controls (N =597) |
7p15 | rs16139 | NPY | 0.48 |
| Millis et al. 200736 | Case-control, candidate gene, 24 SNPs |
Cases (N=531) and controls (N =564) |
8q24 | rs11993333 rs13447075 rs2648862 |
PVT1 | 0.02* 3.0×10−3* 8.0×10−3* |
| Ma et al. 200838 | Case-control, candidate gene, 2 SNPs |
Cases (N=662) and controls (N =620) |
19p13 | rs1799969 rs5498† |
ICAM-1 | 0.58 0.11 (genotypes: P=0.01 among females) |
| Tong et al. 200837 | Case-control, candidate gene, 1 SNP |
Cases (N=865) and controls (N =574)‡ |
7q22 | rs1617640 | EPO | 2.66×10−8 |
| Zhang et al. 200941 | Case-control, candidate gene, 3 SNPs |
Cases (N=578) and controls (N =599) |
3q27 | rs16861194 rs17300539 rs266729 |
ADIPOQ | 0.49 0.88 Males: 0.46, Females: 0.02 |
| Greene et al. 200839¶ |
Case-control, GWA scan, Affymetrix 10K |
DR3/4 cases (N=112) and controls (N =148) |
2p22 | rs1368086 rs725238 rs11886047 |
PLEKHH2 | 2.1×10−5 8.9×10−4 3.4×10−3 (genotypes: 601 cases and 577 controls) |
| Gu et al. 200843 | Case-control, candidate gene |
Cases (N=663) and controls (N =622) |
5p12 |
GHRd3 (genomic deletion of exon 3) |
GHR | 0.97 |
| Zhang et al. 200940 | Case-control, candidate gene, 7 SNPs |
Cases (N=578) and controls (N =599) |
3q22-24 | rs953239 rs7638459 rs17624218 rs7621642 rs2033912 rs3821647 rs7610200 |
TRPC1 | n.s n.s n.s Males: 0.66, Females: 0.05 Males: 0.69, Females: 0.05 Males: 0.95, Females: 0.10 n.s |
Chr. = chromosome, n.s = not significant
Utilized logistic regression under and additive model and adjusted for effects of age2 (age X age), age3 (age X age X age), diabetes duration, and cigarette smoking status.
Ma et al. reported that rs5498 had a high heterozygous index and note that the difference of heterozygous frequencies between DN cases and controls achieved borderline significance (P=0.052). Also observed a gender-specific effect of the genetic risk for DN.
Controls (N=574) were limited to those without nephropathy and retinopathy.
Both rs1368086 and rs725238, as well as 15 other SNPs, were genotyped in 246 GoKinD case trios. TdT analysis of these SNPs revealed significant over-transmission of rs11886047 to case probands. rs11886047 was subsequently genotyped in all GoKinD case-control singletons (N=601 cases, N=577 controls): no difference in allele frequencies were observed. However, there was a difference among genotype frequencies (P = 3.4×10−3).
It’s important to note that the study by Ma et al. included GoKinD patients of European (92%) and non-European (8%) ancestry. Difference in allele frequencies between subpopulations of different ancestry could result in biased estimates of these frequencies in a heterogeneous population. Among individuals of northern and western European (CEU), African (YRI), Japanese (JPT), or Chinese (CHB) descent from the HapMap populations, rs5498’s minor allele frequency differs vastly between ethnic groups (ranging from 0.18 in the African population to 0.46 in the Japanese population). Although it’s unclear whether such differences contribute to this finding, such considerations are important to ensure that genetic associations are due to the phenotype or trait of interest, rather than the underlying structure of the examined population.
Because the GoKinD collection does include samples of European and non-European ancestry, as we demonstrated in our analysis of GWA data from this collection, efforts to minimize potential biases due to population stratification are critical, particularly in analyses involving case-control comparisons. Realizing the importance of this, and to minimize its risk, Greene et al. selected patients from the GoKinD study with similar major histocompatibility complex (HLA) backgrounds and performed a GWA scan using 7,421 SNPs.39 Focusing on the subset of GoKinD study participants with the HLA Class II DR3/4 gene diplotype, including only 112 DN cases samples and 148 control samples, significant associations were identified at rs1368086 (P =2.1×10−5) and rs725238 (P =9.0×10−4) at the pleckstrin homology domain containing, family H (PLEKHH2) locus on chromosome 2p21. Both markers, and 15 additional SNPs at this locus, were subsequently examined in case trios from GoKinD using TdT analysis. Although neither rs1368086 nor rs725238 were over-transmitted to case probands, rs11886047, which is not in linkage disequilibrium (LD) with either of these markers, was significantly over-transmitted (P=0.03). Additional genotyping of rs11886047 in 601 DN cases and 577 controls from GoKinD revealed difference in its genotype frequencies (P=0.003) despite similar distributions of its allele frequencies.
Finally, the GoKinD collection has also been used to examine positional and biological candidate genes in the DN linkage region located on chromosome 3q.21,22,25 Two independent reports by Zhang and colleagues have investigated polymorphisms at the adiponectin (ADIPOQ) gene and the transient receptor potential channel 1 (TRPC1) gene within this region.40,41 Although both studies were largely negative, stratified analyses revealed that a variant located at an SP1 binding site in ADIPOQ’s promoter region was associated with DN among female patients from GoKinD, suggesting that gender-specific effects may contribute to the genetic risk of DN.
Conclusion
The GoKinD study was initiated to provide investigators with a large, well-characterized collection for research aimed at identifying genes involved in DN. In the 3 years since its assembly, through numerous genetic association studies, this collection has certainly achieved this goal. At the forefront of these accomplishments is the recent GWA scan of the GoKinD collection. In addition to contributing to our current understanding of the genetics of DN, through its availability at dbGaP, these data offer researchers an unprecedented opportunity to further investigate the underlying genetic basis of this disease by providing a high-quality dataset with comprehensive coverage of the entire genome. In this review, we discuss the initial findings from our analysis of these data, the added utility gained through GWI, and the usefulness of this expanded dataset. In keeping with the principal objectives of the GoKinD study, it is our belief that these research efforts will provide a springboard for future investigations of the genetic susceptibility of DN in T1D.
Supplementary Material
Acknowledgments
Supported in part by the National Institutes of Health (NIH): DK77532 (to ASK) and T32 DK007260-31 (to MGP), and Juvenile Diabetes Research Foundation (JDRF): 3-2009-397 (to JS)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Seaquist ER, Goetz FC, Rich S, Barbosa J. Familial clustering of diabetic kidney disease. Evidence for genetic susceptibility to diabetic nephropathy. N Engl J Med. 1989;320:1161–1165. doi: 10.1056/NEJM198905043201801. [DOI] [PubMed] [Google Scholar]
- 2.Borch-Johnsen K, Norgaard K, Hommel E, Mathiesen ER, Jensen JS, Deckert T, et al. Is diabetic nephropathy an inherited complication? Kidney Int. 1992;41:719–722. doi: 10.1038/ki.1992.112. [DOI] [PubMed] [Google Scholar]
- 3.Quinn M, Angelico MC, Warram JH, Krolewski AS. Familial factors determine the development of diabetic nephropathy in patients with IDDM. Diabetologia. 1996;39:940–945. doi: 10.1007/BF00403913. [DOI] [PubMed] [Google Scholar]
- 4.Mueller PW, Rogus JJ, Cleary PA, Zhao Y, Smiles AM, Steffes MW, et al. Genetics of Kidneys in Diabetes (GoKinD) study: a genetics collection available for identifying genetic susceptibility factors for diabetic nephropathy in type 1 diabetes. J Am Soc Nephrol. 2006;17:1782–1790. doi: 10.1681/ASN.2005080822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.GAIN Collaborative Research Group New models of collaboration in genome-wide association studies: the Genetic Association Information Network. Nat Genet. 2007;39:1045–1051. doi: 10.1038/ng2127. [DOI] [PubMed] [Google Scholar]
- 6.Pezzolesi MG, Poznik GD, Mychaleckyj JC, Paterson AD, Barati MT, Klein JB, et al. Genome-wide association scan for diabetic nephropathy susceptibility genes in type 1 diabetes. Diabetes. 2009;58:1403–1410. doi: 10.2337/db08-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freedman BI, Langefeld CD, Rich SS, Valis CJ, Sale MM, Williams AH, et al. A genome scan for ESRD in black families enriched for nondiabetic nephropathy. J Am Soc Nephrol. 2004;15:2719–2727. doi: 10.1097/01.ASN.0000141312.39483.4F. [DOI] [PubMed] [Google Scholar]
- 8.Freedman BI, Bowden DW, Rich SS, Valis CJ, Sale MM, Hicks PJ, et al. A genome scan for all-cause end-stage renal disease in African Americans. Nephrol Dial Transplant. 2005;20:712–718. doi: 10.1093/ndt/gfh704. [DOI] [PubMed] [Google Scholar]
- 9.Shimazaki A, Kawamura Y, Kanazawa A, Sekine A, Saito S, Tsunoda T, et al. Genetic variations in the gene encoding ELMO1 are associated with susceptibility to diabetic nephropathy. Diabetes. 2005;54:1171–1178. doi: 10.2337/diabetes.54.4.1171. [DOI] [PubMed] [Google Scholar]
- 10.Leak TS, Perlegas PS, Smith SG, Keene KL, Hicks PJ, Langefeld CD, et al. Variants in intron 13 of the ELMO1 gene are associated with diabetic nephropathy in African Americans. Ann Hum Genet. 2009;73:152–159. doi: 10.1111/j.1469-1809.2008.00498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pezzolesi MG, Katavetin P, Kure M, Poznik GD, Skupien J, Mychaleckyj JC, et al. Confirmation of Genetic Associations at ELMO1 in the GoKinD Collection Support its Role as a Susceptibility Gene in Diabetic Nephropathy. Diabetes. doi: 10.2337/db09-0641. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooper JD, Smyth DJ, Smiles AM, Plagnol V, Walker NM, Allen JE, et al. Meta-analysis of genome-wide association study data identifies additional type 1 diabetes risk loci. Nat Genet. 2008;40:1399–1401. doi: 10.1038/ng.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39:857–864. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korstanje R, DiPetrillo K. Unraveling the genetics of chronic kidney disease using animal models. Am J Physiol Renal Physiol. 2004;287:F347–352. doi: 10.1152/ajprenal.00159.2004. [DOI] [PubMed] [Google Scholar]
- 15.Garrett MR, Gunning WT, Radecki T, Richard A. Dissection of a genetic locus influencing renal function in the rat and its concordance with kidney disease loci on human chromosome 1q21. Physiol Genomics. 2007;30:322–334. doi: 10.1152/physiolgenomics.00001.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang A, Ziyadeh FN, Lee EY, Pyagay PE, Sung SH, Sheardown SA, et al. Interference with TGF-beta signaling by Smad3-knockout in mice limits diabetic glomerulosclerosis without affecting albuminuria. Am J Physiol Renal Physiol. 2007;293:F1657–1665. doi: 10.1152/ajprenal.00274.2007. [DOI] [PubMed] [Google Scholar]
- 17.Morel L, Rudofsky UH, Longmate JA, Schiffenbauer J, Wakeland EK. Polygenic control of susceptibility to murine systemic lupus erythematosus. Immunity. 1994;1:219–229. doi: 10.1016/1074-7613(94)90100-7. [DOI] [PubMed] [Google Scholar]
- 18.Sheehan S, Tsaih SW, King BL, Stanton C, Churchill GA, Paigen B, et al. Genetic analysis of albuminuria in a cross between C57BL/6J and DBA/2J mice. Am J Physiol Renal Physiol. 2007;293:F1649–1656. doi: 10.1152/ajprenal.00233.2007. [DOI] [PubMed] [Google Scholar]
- 19.Tsaih SW, Pezzolesi MG, Yuan R, Warram JH, Krolewski AS, Korstanje R. Genetic analysis of albuminuria in the aging mouse and concordance with loci for diabetic nephropathy found in a genome-wide association scan. Kidney Int. doi: 10.1038/ki.2009.434. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moczulski DK, Rogus JJ, Antonellis A, Warram JH, Krolewski AS. Major susceptibility locus for nephropathy in type 1 diabetes on chromosome 3q: results of novel discordant sib-pair analysis. Diabetes. 1998;47:1164–1169. doi: 10.2337/diabetes.47.7.1164. [DOI] [PubMed] [Google Scholar]
- 22.Osterholm AM, He B, Pitkaniemi J, Albinsson L, Berg T, Sarti C, et al. Genome-wide scan for type 1 diabetic nephropathy in the Finnish population reveals suggestive linkage to a single locus on chromosome 3q. Kidney Int. 2007;71:140–145. doi: 10.1038/sj.ki.5001933. [DOI] [PubMed] [Google Scholar]
- 23.Rogus JJ, Poznik GD, Pezzolesi MG, Smiles AM, Dunn J, Walker W, et al. High-density single nucleotide polymorphism genome-wide linkage scan for susceptibility genes for diabetic nephropathy in type 1 diabetes: discordant sibpair approach. Diabetes. 2008;57:2519–2526. doi: 10.2337/db07-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He B, Osterholm AM, Hoverfalt A, Forsblom C, Hjorleifsdottir EE, Nilsson AS, et al. Association of genetic variants at 3q22 with nephropathy in patients with type 1 diabetes mellitus. Am J Hum Genet. 2009;84:5–13. doi: 10.1016/j.ajhg.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bowden DW, Colicigno CJ, Langefeld CD, Sale MM, Williams A, Anderson PJ, et al. A genome scan for diabetic nephropathy in African Americans. Kidney Int. 2004;66:1517–1526. doi: 10.1111/j.1523-1755.2004.00915.x. [DOI] [PubMed] [Google Scholar]
- 26.Bondeva T, Ruster C, Franke S, Hammerschmid E, Klagsbrun M, Cohen CD, et al. Advanced glycation end-products suppress neuropilin-1 expression in podocytes. Kidney Int. 2009;75:605–616. doi: 10.1038/ki.2008.603. [DOI] [PubMed] [Google Scholar]
- 27.Pollak MR. Focal segmental glomerulosclerosis: recent advances. Curr Opin Nephrol Hypertens. 2008;17:138–142. doi: 10.1097/MNH.0b013e3282f5dbe4. [DOI] [PubMed] [Google Scholar]
- 28.Yang J, Liu Y. Dissection of key events in tubular epithelial to myofibroblast transition and its implications in renal interstitial fibrosis. Am J Pathol. 2001;159:1465–1475. doi: 10.1016/S0002-9440(10)62533-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeisberg M, Hanai J, Sugimoto H, Mammoto T, Charytan D, Strutz F, et al. BMP-7 counteracts TGF-beta1-induced epithelial-to-mesenchymal transition and reverses chronic renal injury. Nat Med. 2003;9:964–968. doi: 10.1038/nm888. [DOI] [PubMed] [Google Scholar]
- 30.Wang X, Ishimori N, Korstanje R, Rollins J, Paigen B. Identifying novel genes for atherosclerosis through mouse-human comparative genetics. Am J Hum Genet. 2005;77:1–15. doi: 10.1086/431656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ziyadeh FN. Mediators of diabetic renal disease: the case for tgf-Beta as the major mediator. J Am Soc Nephrol. 2004;15(Suppl 1):S55–57. doi: 10.1097/01.asn.0000093460.24823.5b. [DOI] [PubMed] [Google Scholar]
- 32.Deelman L, Sharma K. Mechanisms of kidney fibrosis and the role of antifibrotic therapies. Curr Opin Nephrol Hypertens. 2009;18:85–90. doi: 10.1097/MNH.0b013e32831c50a1. [DOI] [PubMed] [Google Scholar]
- 33.Yamamoto T, Noble NA, Cohen AH, Nast CC, Hishida A, Gold LI, et al. Expression of transforming growth factor-beta isoforms in human glomerular diseases. Kidney Int. 1996;49:461–469. doi: 10.1038/ki.1996.65. [DOI] [PubMed] [Google Scholar]
- 34.Sharma VK, Bologa RM, Xu GP, Li B, Mouradian J, Wang J, et al. Intragraft TGF-beta 1 mRNA: a correlate of interstitial fibrosis and chronic allograft nephropathy. Kidney Int. 1996;49:1297–1303. doi: 10.1038/ki.1996.185. [DOI] [PubMed] [Google Scholar]
- 35.Nolan VG, Ma Q, Cohen HT, Adewoye A, Rybicki AC, Baldwin C, et al. Estimated glomerular filtration rate in sickle cell anemia is associated with polymorphisms of bone morphogenetic protein receptor 1B. Am J Hematol. 2007;82:179–184. doi: 10.1002/ajh.20800. [DOI] [PubMed] [Google Scholar]
- 36.Millis MP, Bowen D, Kingsley C, Watanabe RM, Wolford JK. Variants in the plasmacytoma variant translocation gene (PVT1) are associated with end-stage renal disease attributed to type 1 diabetes. Diabetes. 2007;56:3027–3032. doi: 10.2337/db07-0675. [DOI] [PubMed] [Google Scholar]
- 37.Tong Z, Yang Z, Patel S, Chen H, Gibbs D, Yang X, et al. Promoter polymorphism of the erythropoietin gene in severe diabetic eye and kidney complications. Proc Natl Acad Sci. 2008;105:6998–7003. doi: 10.1073/pnas.0800454105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma J, Zhang D, Brismar K, Efendic S, Gu HF. Evaluation of the association between the common E469K polymorphism in the ICAM-1 gene and diabetic nephropathy among type 1 diabetic patients in GoKinD population. BMC Med Genet. 2008;9:47. doi: 10.1186/1471-2350-9-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greene CN, Keong LM, Cordovado SK, Mueller PW. Sequence variants in the PLEKHH2 region are associated with diabetic nephropathy in the GoKinD study population. Hum Genet. 2008;124:255–262. doi: 10.1007/s00439-008-0548-y. [DOI] [PubMed] [Google Scholar]
- 40.Zhang D, Freedman BI, Flekac M, Santos E, Hicks PJ, Bowden DW, et al. Evaluation of genetic association and expression reduction of TRPC1 in the development of diabetic nephropathy. Am J Nephrol. 2009;29:244–251. doi: 10.1159/000157627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang D, Ma J, Brismar K, Efendic S, Gu HF. A single nucleotide polymorphism alters the sequence of SP1 binding site in the adiponectin promoter region and is associated with diabetic nephropathy among type 1 diabetic patients in the Genetics of Kidneys in Diabetes Study. J Diabetes Complications. 2009;23:265–272. doi: 10.1016/j.jdiacomp.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 42.Ma J, Nordman S, Mollsten A, Falhammar H, Brismar K, Dahlquist G, et al. Distribution of neuropeptide Y Leu7Pro polymorphism in patients with type 1 diabetes and diabetic nephropathy among Swedish and American populations. Eur J Endocrinol. 2007;157:641–645. doi: 10.1530/EJE-07-0354. [DOI] [PubMed] [Google Scholar]
- 43.Gu HF, Efendic S, Brismar K. Lack of an association between GHR exon 3 polymorphism and diabetic nephropathy in the Genetics of Kidneys in Diabetes (GoKinD) population. Diabetologia. 2008;51:2333–2334. doi: 10.1007/s00125-008-1145-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.