Abstract
Non-coding RNAs (ncRNAs) that regulate gene expression in cis or in trans are a shared feature of prokaryotic and eukaryotic genomes. In mammals, cis-acting functions are associated with macro ncRNAs, which can be several hundred thousand nucleotides long. Imprinted ncRNAs are well-studied macro ncRNAs that have cis-regulatory effects on multiple flanking genes. Recent advances indicate that they employ different downstream mechanisms to regulate gene expression in embryonic and placental tissues. A better understanding of these downstream mechanisms will help to improve our general understanding of the function of ncRNAs throughout the genome.
Introduction
In recent years, tiling-array analyses (see Glossary, Box 1) and genome-wide cDNA sequencing have shown not only that most of the mammalian genome is transcribed, but also that the majority of the mammalian transcriptome consists of non-coding (nc) RNAs (Carninci et al., 2005; Engstrom et al., 2006; Kapranov et al., 2002; Katayama et al., 2005; Okazaki et al., 2002). Because a full classification system for ncRNAs is still outstanding, they are generally described according to their mature length, location and orientation with respect to the nearest protein-coding gene. For example, a new group called large intervening non-coding (linc) RNAs comprise ncRNAs that lie outside annotated genes in the mouse genome (Guttman et al., 2009). When their function is known, ncRNAs can also be classified by whether they act in cis or trans (see Glossary, Box 1). Trans-acting functions are associated with short ncRNAs, such as short interfering (si) RNAs (21 nt), micro (mi) RNAs (~22 nt), piwi-interacting RNAs (26-31 nt) and short nucleolar (sno) RNAs (60-300 nt). By contrast, cis-acting functions have so far only been associated with macro ncRNAs (see Glossary, Box 1), which can be up to several hundred thousand nucleotides long. Interestingly, whereas the number of protein-coding genes is no indication of an organism’s morphological complexity, macro ncRNA number increases with complexity, indicating a potential functional role in gene regulation (Amaral and Mattick, 2008). In support of this hypothesis, many ncRNAs show distinct cell-type-specific and developmental-stage-specific expression profiles (Dinger et al., 2008; Mercer et al., 2008). To date, however, only a few macro ncRNAs have been analysed in detail and shown to have functional gene-regulatory roles (Yazgan and Krebs, 2007; Prasanth and Spector, 2007).
Box 1. Glossary
- Cis-acting function
The ability of a DNA sequence or transcript to regulate the expression of one or more genes on the same chromosome. This contrasts with trans-acting function (see below).
- CTCF
A CCCTC-binding factor that is an 11-zinc-finger protein that binds insulator elements.
- Differentially methylated region (DMR)
A CG-dinucleotide-rich genomic region that, in diploid cells, is methylated on one parental chromosome and unmethylated on the other. Gametic DMRs acquire their parental-specific DNA methylation during gametogenesis, either in the developing haploid oocyte or sperm, whereas somatic DMRs acquire their parental-specific DNA methylation in somatic diploid cells.
- DNA/RNA FISH
A fluorescence in situ hybridisation technique that uses a complementary DNA or RNA strand to determine the localisation of DNA sequences or RNA transcripts in cell nuclei.
- Dosage compensation
An epigenetic regulatory mechanism present in mammals, flies and worms that equalises the expression of genes on the X chromosome between XY/X0 males and XX females.
- Epigenetics
Modifications of DNA or chromatin proteins that alter the ability of DNA to respond to external signals.
- Gene regulation in cis
See cis-acting function.
- Gene regulation in trans
See trans-acting function.
- Genomic imprinting
An epigenetic mechanism that induces parental-specific gene expression in diploid mammalian cells (see Box 2).
- Imprint control element (ICE)
A short DNA sequence [also known as an ICR (imprint control region) or IC (imprinting centre)] that controls the imprinted expression of multiple genes in cis. All known ICEs are also gametic DMRs; however, their identification requires the in vivo analysis of a deletion of the DMR.
- Insulator element
A genetic boundary element that binds insulator proteins to separate a promoter on one side of the insulator element from the activating effects of an enhancer located on the other side.
- Macro ncRNAs
ncRNAs that can be as short as a few hundred nucleotides or as long as several hundred thousand nucleotides, the function of which does not depend on processing into short or micro RNAs.
- Tiling-array analysis
Commercial chips containing 25-60 nt oligonucleotide probes designed to continuously cover a genomic region that are used to produce unbiased maps of histone modifications following chromatin immunoprecipitation (ChIP-chip), or of DNA methylation following methylated DNA immunoprecipitation (MeDIP-chip), or of gene expression following cDNA hybridisation (RNA-chip).
- Trans-acting function
The ability of a DNA sequence or transcript to regulate the expression of one or more genes on different chromosomes or to regulate mature RNAs in the cytoplasm.
The best-known functional mammalian macro ncRNAs are the inactive X-specific transcript (Xist) and X (inactive)-specific transcript, antisense (Tsix), which are overlapping transcripts required for X chromosome inactivation in female mammals – an epigenetic dosage-compensation mechanism (see Glossary, Box 1) that equalises X-linked gene expression between the sexes. Xist is expressed from, and localises to, the inactive X chromosome and, by an unknown mechanism, targets repressive chromatin modifications and gene silencing to this chromosome. Tsix overlaps with the entire Xist gene in an antisense orientation and silences Xist on the active X chromosome (reviewed by Wutz and Gribnau, 2007). The next-best-studied mammalian macro ncRNAs are those involved in genomic imprinting (see Glossary, Box 1; Box 2). To date, 90 genes show imprinted expression in the mouse (http://www.har.mrc.ac.uk/research/genomic_imprinting), and their imprinted status is mostly conserved in humans (www.otago.ac.nz/IGC). Imprinted genes mostly occur in clusters that contain 2-12 genes, and in most of these clusters at least one gene is a macro ncRNA. So far, two of the three tested imprinted macro ncRNAs have been shown to be required for the imprinted expression of the whole cluster (Barlow and Bartolomei, 2007). Thus, imprinted macro ncRNAs are able to regulate small clusters of autosomal genes in cis and offer an excellent model system not only to investigate how ncRNAs regulate genes epigenetically, but also to investigate the general biology of ncRNA transcripts.
Box 2. Genomic imprinting: basic biology, history and clinical implications.
Mammals are diploid organisms whose cells contain two matched sets of chromosomes, one inherited from the mother and one from the father. Thus, mammals have two copies of every gene with the same potential to be expressed in any cell. Genomic imprinting is an epigenetic mechanism that affects ~1% of genes and restricts their expression early in development to one of the two parental chromosomes. Genes that show parental-specific expression were hypothesised to exist in mammals following a series of landmark observations that began to accumulate thirty years ago. These included the failure of embryos to develop by parthenogenesis in the absence of fertilisation, the phenotype of embryos that had inherited two copies of one parental chromosome in the absence of the other parental copy, and the inability to generate viable embryos that contained two maternal or paternal pronuclei through oocyte nuclear transfer experiments. This hypothesis was corroborated in 1991 by the discovery of three imprinted genes: the maternally expressed Igf2r gene, the paternally expressed Igf2 gene and the maternally expressed H19 ncRNA. Whereas genomic imprinting now offers one of the best models in which to investigate epigenetic gene regulation in mammals, it also has considerable implications for modern molecular medicine in the management of genetic diseases that map to autosomes but are only inherited from one parent, and in the efforts to apply assisted reproductive or cloning technologies to human reproduction.
In this review, we focus on six well-studied mouse imprinted clusters and their associated macro ncRNAs (Fig. 1) and review three main areas: first, how imprinted macro ncRNAs are themselves epigenetically regulated by DNA methylation imprints, and their role in inducing imprinted expression and epigenetic modifications in imprinted clusters; second, what is currently known about the organisation and the transcriptional biology of imprinted macro ncRNAs; and third, why developmental and tissue-specific variation in imprinted expression indicates that multiple mechanisms might operate downstream of imprinted ncRNAs.
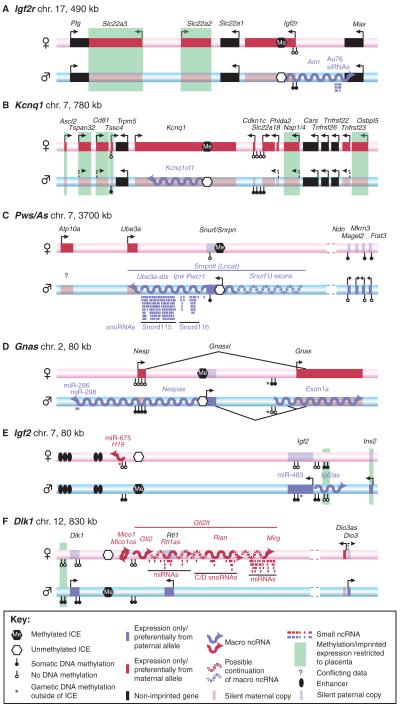
Fig. 1. Six well-studied imprinted clusters.
(A-F) The genomic organisation of six well-studied mouse imprinted clusters. The maternal chromosome is shown as a pink bar, the paternal chromosome as a blue bar. Protein-coding genes are shown as boxes: solid red box, maternally expressed gene on the expressed allele; transparent red box, maternally expressed gene on the repressed allele; solid blue box, paternally expressed gene on the expressed allele; transparent blue box, paternally expressed gene on the repressed allele. Macro ncRNAs are shown as wavy lines: red for maternally expressed, blue for paternally expressed. Arrows indicate transcriptional direction: solid arrows, strong transcription; dashed arrows, weak transcription. Note that many of the indicated genes show tissue- or temporal-restricted gene expression (not indicated). See key for further details. chr., chromosome; ICE, imprint control element; miRNA, microRNA; siRNA, short interfering RNA; snoRNA, short nucleolar RNA.
Imprinted macro ncRNAs are epigenetically regulated
A key feature of imprinted gene clusters is the presence of an imprint control element (ICE) (see Glossary, Box 1), which has been genetically defined by deletion experiments in mice or through the mapping of minimal naturally occurring deletions in humans (Table 1). The ICE is epigenetically modified on only one parental chromosome by a DNA methylation ‘imprint’, which is acquired during maternal or paternal gametogenesis and is maintained on the same parental chromosome in the diploid embryo. As the other parental chromosome lacks ICE DNA methylation, this region in a diploid cell is also known as a gametic differentially methylated region (gDMR). The unmethylated ICE controls the imprinted expression of the whole cluster; upon its deletion, imprinted genes are no longer expressed in a parental-specific pattern (Bielinska et al., 2000; Fitzpatrick et al., 2002; Lin et al., 2003; Thorvaldsen et al., 1998; Williamson et al., 2006; Wutz et al., 1997). Note that the term ‘imprinted’ refers to the presence of DNA methylation on the ICE and not to gene expression status and that the above-mentioned deletion experiments show that only the unmethylated ICE is active. Four of the six well-studied imprinted clusters in the mouse (Igf2r, Kcnq1, Pws/As, Gnas) are maternally imprinted and thus gain their ICE DNA methylation imprint during oogenesis. This imprint is maintained only on the maternal chromosome in diploid cells (Fig. 1A-D). The remaining two clusters (Igf2, Dlk1) are paternally imprinted and gain their ICE DNA methylation imprint during spermatogenesis. This imprint is maintained only on the paternal chromosome in diploid cells (Fig. 1E,F).
Table 1. Imprint control elements of six well-studied imprinted clusters.
| Imprinted cluster | ICE (a gametic DMR) Genomic position (UCSC mm9) |
Associated histone modifications | |
|---|---|---|---|
| DNA methylated ICE | Unmethylated ICE | ||
| Igf2r | Region 21,2 Chr17: 12934307-12935355 |
Maternal:3 H3K9me3, H4K20me3 |
Paternal:3 H3K4me3, H3K4me2, H3K9ac |
| Igf2 | H19 DMD4 Chr7: 149766154-149767791 |
Paternal:5,6 H3K9me3, H4K20me3 |
Maternal:5,6 H3K4me2, H3K4me3, H3ac |
| Kcnq1 | KvDMR17 Chr7: 150481310-150482463 |
Maternal:5,8 H3K9me, H3K27me |
Paternal:5,8 H3K4me, H3K9ac, H3K14ac, H3ac, H4ac |
| Pws/As | Part may be contained in 4.8 kb9, which is orthologous to the human PWS-IC Chr7: 67147668-67152741 |
Maternal:5,10 H3K9me3, H4K20me3, H3K27me3 (ES only) |
Paternal:5,10 H3K4me2, H3K4me3, H3ac, H4ac |
| Gnas | Nespas DMR11 Chr2: 174117482-174125568 |
Overlapping H3K4me3/H3K9me3 peaks in ES cells, H3K4me3 paternal bias (unmethylated ICE)12 |
|
| Dlk1 | IG-DMR13 Chr12: 110765047-110769203 |
Paternal:5,6 H3K9me, H4K20me3 |
Maternal:5,6 H3K4me3, H3K4me2, H3ac, H4ac |
ac, acetylation; Chr, mouse chromosome; DMR, differentially methylated region; H3, histone 3; H4, histone 4; ICE, imprint control element; K, lysine; me2, dimethylation; me3, trimethylation; PWS-IC, Prader-Willi imprint control element; UCSC, University of California Santa Cruz genome browser.
References:
In the Igf2r, Kcnq1 and Gnas clusters, the ICE contains a promoter for a macro ncRNA [Airn (108 kb), Kcnq1ot1 (91 kb) and Nespas (~30 kb), respectively] that has an overlap in the antisense direction with only one gene in each imprinted cluster (for references, see Table 2). In the Pws/As cluster, the provisionally named Snrpn-long-transcript (Snrpnlt, also known as Lncat) is an unusually long macro ncRNA that could cover 1000 kb of genomic sequence. The Snrpnlt ncRNA overlaps in antisense orientation with the Ube3a gene, which is located 720 kb downstream. The size of the ICE in this cluster has not been precisely determined in the mouse, as the smallest available ICE deletion only shows a partial or mosaic imprinting defect (Bressler et al., 2001). In the paternally imprinted Igf2 and Dlk1 clusters, the ICE is found 5-14 kb upstream of the H19 macro ncRNA (2.2 kb) and the provisionally named Gtl2-long-transcript (Gtl2lt; length unknown), respectively. H19 lacks any known transcriptional overlap with the other genes in the cluster, whereas Gtl2lt overlaps with Rtl1. In short, although the organisation of these six well-studied imprinted clusters appears to be complex, they generally follow two simple rules: (1) an unmethylated ICE is required for macro ncRNA expression; and (2) most imprinted mRNA genes are not expressed from the chromosome from which the macro ncRNA is expressed.
Table 2. Non-coding RNAs in imprinted clusters in the mouse genome.
| Imprinted cluster (mouse chromosome) |
Macro ncRNA |
Published size | Unspliced or spliced/subcellular localisation |
Cis-silencing function | Host for small RNAs |
|---|---|---|---|---|---|
|
Igf2r (17) |
Airn1,* | 108 kb2 | 95% unspliced (N) 5% spliced to ~1 kb (C)3 |
Yes4 | Au76 siRNA5 |
|
Kcnq1 (7) |
Kcnq1ot1 6 | ~90 kb7 | Unspliced7,8 (N)9,‡ | Yes8,10 | |
|
Pws/As (7) |
Snrpnlt† (Lncat)11 |
Numerous splice variants11-13 Genomic size: 1000 kb11 |
Spliced11-13 | n.d. | Snord64 MBII-1314,15 Snord115 MBII-5214,15 Snord116 MBII-8516,17 |
| Might contain all ncRNAs listed below | |||||
|
Snurf U exons11 |
n.d. | n.d. | n.d. | ||
| Pwcr1 18 | 2.2 kb18 | Unspliced18 | n.d. | ||
| Ipw 19 | 0.5-12 kb19 Genomic size: 60 kb |
Several splice variants19 mainly (N)13,§ |
n.d. | ||
|
Ube3a- ats20 |
n.d. | Spliced/unspliced n.d. mainly (N)13,§ |
n.d. | ||
|
Gnas (2) |
Nespas 21 | Unspliced at least 3.35 kb22 Spliced 1.4-15.8 kb22,23 Genomic size: at least 30 kb23 |
Several spliced and unspliced versions22-24 |
n.d. | mmu-mir-29625 mmu-mir-29825 |
| Exon1A 26 | Unspliced >1.1 kb23 Spliced >1.4 kb26 Genomic size 19 kb |
Spliced and unspliced23,26 |
n.d. | ||
|
Igf2 (7) |
H19 27 | 2.2 kb28 Genomic size: 2.5 kb |
Spliced28 (C)29 | no30 | mmu-mir-67531 |
| Igf2as 32 | 4.8 kb32 Genomic size: 10.7 kb |
Alternative promoters, spliced32 |
n.d. | ||
|
Dlk1 (12) |
Mico1 33 | 2 kb33 | Unspliced33 | n.d. | In silico predicted precursor miRNA33 |
| Mico1os 33 | 2 kb33 | Unspliced33 | n.d. | In silico predicted precursor miRNA33 |
|
| Gtl2lt † | Genomic size: 208 kb |
Hypothetical transcript; might contain all ncRNAs listed below34-36 |
mmu-mir: 770/673/493/337/540/665/431/433/1 27/434/136/341/1188/370/882/379/4 11/299/380/1197/323/758/329/494/6 79/1193/543/495/667/376c/654/376b /376a/300/381/487b/539/544/352/13 4/668/485/435/154/496/377/541/409/ 412/369/4134,37-40 MBII-48(5), MBII-49(4), ImsnoRNA1, MBII-426(1), ImsnoRNA2, MBII- 343(1), ImsnoRNA3, MBII-78(1), MBII-19(3)15,41,42 |
||
| Gtl2 43 | 1.9-7 kb43-45 Genomic size: 30.7 kb |
Several splice variants43 mainly (N)43,§ |
n.d. | ||
| Rtl1as 40 | n.d. | Processed40 | miRNAs involved in trans silencing of Rtl146 |
||
| Rian 47 | 5.4 kb47 Genomic size: 6.1 kb |
Spliced47 mainly (N)47,‡ |
n.d. | ||
| Mirg 40 | 1.3 kb40 Genomic size: 14.5 kb |
Spliced40 | n.d. | ||
The six well-studied imprinted clusters and their associated ncRNAs are listed with the macro ncRNA name, sizes of unspliced and spliced variants, genomic size if different from the longest unspliced version, and cellular localisation [note that nuclear localisation was sometimes inferred from RNA FISH (see Glossary, Box 1) data that did not exclude export to the cytoplasm]. Silencing function and hosted small ncRNAs are indicated.
References:
C, cytoplasmic; ImsnoRNA, snoRNAs at the Irm locus; MBII, mouse brain 2; mmu-mir, Mus musculus microRNA; N, nuclear; n.d., not determined.
Renamed from Air by HUGO nomenclature committee.
Provisional name.
By RNA FISH.
By whole-mount in situ hybridisation.
Imprinted macro ncRNAs can host short ncRNAs
The length of the macroRNAs of the imprinted clusters, i.e. Airn, Kcnq1ot1, Nespas, Snrpnlt, H19 and Gtl2lt, ranges from 2.2-1000 kb (Table 2). Intriguingly, as shown in Fig. 1, the majority of these imprinted macroRNAs, with the possible exception of Kcnq1ot, also serve as host transcripts for trans-acting short RNAs, such as siRNAs, which are involved in gene silencing by the RNA interference pathway (reviewed by Mattick and Makunin, 2006), miRNAs, which function as translational gene repressors (reviewed by Cannell et al., 2008), and snoRNAs, which are involved in rRNA processing (reviewed by Brown et al., 2008). For example, siRNAs encoded by the Au76 pseudogene region lie within the Airn ncRNA in the Igf2r cluster and are found in oocytes (Watanabe et al., 2008). In the Pws/As cluster, two snoRNA clusters (Snord115, Snord116) are located within the Snrpnlt macro ncRNA (Cavaille et al., 2000; Huttenhofer et al., 2001). In the Gnas cluster, two miRNAs are located within the Nespas macro ncRNA (Holmes et al., 2003). In the Igf2 cluster, both the H19 ncRNA and the protein-coding Igf2 gene contain a miRNA within their transcriptional unit (Cai and Cullen, 2007; Landgraf et al., 2007). In the Dlk1 cluster, four separate macro ncRNAs have been described [Gtl2 (Meg3), Rtl1as, Rian, Mirg] that might be contained within the Gtl2lt ncRNA. Gtl2, Rtl1as and Mirg contain multiple miRNAs, whereas Rian contains multiple snoRNAs (Cavaille et al., 2002; Houbaviy et al., 2003; Huttenhofer et al., 2001; Kim et al., 2004; Lagos-Quintana et al., 2002; Seitz et al., 2004; Seitz et al., 2003). Interestingly, miRNAs generated from Rtl1as have been shown to be involved in the trans-silencing of Rtl1 through an siRNA-mediated pathway (Davis et al., 2005). Few of the small ncRNAs in these clusters have been analysed in detail and most probably have trans-acting functions, which indicates that they are unlikely to be involved in regulating imprinted gene expression that depends on a cis-acting mechanism. However, their presence suggests a functional link between macro ncRNAs and short ncRNAs.
Imprinted ncRNAs – atypical mammalian transcripts?
An unusual feature of many imprinted ncRNAs is that they are unspliced or spliced with a low intron/exon ratio, in contrast to the majority of mammalian mRNA-encoding genes, which are intron rich. Notably, the export of ncRNAs to the cytoplasm correlates with splicing (Table 2). The H19 ncRNA is fully spliced and exported to the cytoplasm (Brannan et al., 1990; Pachnis et al., 1984), whereas the Kcnq1ot1 ncRNA is unspliced and retained in the nucleus (Pandey et al., 2008). The Airn ncRNA produces unspliced and spliced transcripts at a ratio of 19:1, and only the spliced transcripts are exported to the cytoplasm (Seidl et al., 2006). In the Gnas cluster, both Exon1A and Nespas ncRNAs are also found as spliced and unspliced forms, but their cellular localisation is unknown (Holmes et al., 2003; Li et al., 1993; Liu et al., 2000; Williamson et al., 2002). Both Airn and Kcnq1ot1 were shown by RNA fluorescence in situ hybridisation (RNA FISH; see Glossary, Box 1) to form RNA ‘clouds’ at their site of transcription (Braidotti et al., 2004; Nagano et al., 2008; Terranova et al., 2008). We do not yet know whether these ncRNA ‘clouds’ explain their ability to repress flanking genes or whether this ‘cloud-like’ appearance is a consequence of a lack of splicing, as mRNA genes mutated to inhibit splicing also show nuclear retention and intranuclear RNA focus formation (Custodio et al., 1999; Ryu and Mertz, 1989).
Although not all imprinted macro ncRNAs have been studied in sufficient detail, at least some have been shown to have unusual transcriptional features that are not generally associated with mammalian mRNA genes, such as reduced splicing potential or low intron/exon ratio, nuclear retention and accumulation at the site of transcription. Investigations into the control of these unusual transcriptional features, and into their role in the functional properties of imprinted ncRNAs, are only just beginning. An obvious genetic element that might account for unusual transcriptional features is the promoter. It was recently shown in fission yeast that splicing regulation is promoter driven (Moldon et al., 2008). It was therefore surprising to find that the endogenous Airn promoter does not control the low splicing capacity of the Airn ncRNA (Stricker et al., 2008), and further work is required to determine how these unusual macro ncRNA features arise.
Macro ncRNAs are functional in imprinted expression
The presence of macro ncRNAs in imprinted clusters raises the question of whether they play a functional role in imprinting. In the case of the two paternally expressed imprinted macro ncRNAs Airn and Kcnq1ot1, experiments in which the ncRNA was truncated by the homologous insertion of a polyadenylation cassette have demonstrated that these ncRNAs are indeed necessary for imprinted expression. Paternal chromosomes that carry a truncated Airn or Kcnq1ot1 ncRNA lose the repression of all protein-coding genes in the imprinted cluster in both embryonic and placental tissues, whereas maternal alleles are unaffected (Mancini-Dinardo et al., 2006; Shin et al., 2008; Sleutels et al., 2002). These experiments showed that these macro ncRNAs act by repressing multiple flanking genes in cis in both embryonic and placental tissues. Nespas is similar to Airn and Kcnq1ot1 in that it is transcribed from a promoter contained within the unmethylated ICE on the paternal allele and has an antisense orientation with respect to the imprinted protein-coding Nesp gene. However, it is not yet known whether Nespas has a cis-silencing role similar to Airn and Kcnq1ot1. By contrast, the maternally expressed H19 ncRNA is known to be dispensable for the imprinted expression of Igf2 (Schmidt et al., 1999). Instead, a methylation-sensitive insulator element (see Glossary, Box 1) contained in the ICE regulates the ability of enhancers that lie downstream of H19 to interact physically with the upstream H19 and Igf2 promoters (Fig. 1E). On the unmethylated maternal allele, CTCF (see Glossary, Box 1) binds the ICE and restricts the access of enhancers to the H19 promoter. On the methylated paternal allele, CTCF cannot bind, and the enhancers interact preferentially with the Igf2 promoter, facilitating its transcription (Bell and Felsenfeld, 2000; Hark et al., 2000). An additional mechanism to induce imprinted expression is present in the newly described H13 imprinted cluster on mouse chromosome 2 (Wood et al., 2007) (not shown in Fig. 1). The H13 (histocompatibility 13 antigen) (Graff et al., 1978) gene contains an intronic, maternally methylated gDMR. The transcription of full-length functional H13 from the maternal chromosome depends on the methylation of this gDMR. On the paternal allele, the unmethylated gDMR acts as a promoter for the Mcts2 retrogene, and Mcts2 expression correlates with the premature polyadenylation of H13 (Wood et al., 2008). To date, it is unknown whether the Mcts2 retrogene is coding or non-coding, nor whether Mcts2 expression or the unmethylated gDMR is required to block the production of full-length H13 transcripts.
Thus, two out of the three tested imprinted macro ncRNAs act in cis to induce the imprinted expression of flanking genes. These macro ncRNAs might also possess additional functions. For example, as their promoter is contained in the ICE, they could play a role in acquiring the gametic DNA methylation imprint. However, a recent publication indicates that the transcription of overlapping imprinted protein-coding genes, rather than of ncRNAs, is needed to acquire methylation imprints in the Gnas cluster (Chotalia et al., 2009). This work shows that a truncation of the Nesp mRNA transcript (Fig. 1D) by the insertion of a polyadenylation cassette, which abolishes transcription through the ICE, impairs acquisition of the ICE methylation mark in oocytes. This might represent a common theme for oocyte-specific DNA methylation imprints, given that an overlapping mRNA gene has been reported to be transcribed through five other maternal gDMRs in oocytes, but not in sperm. Perhaps surprisingly, ncRNA transcription might play a role in maintaining the unmethylated state of the ICE in the early embryo. This is indicated by experiments in which the Airn promoter was deleted from the paternal chromosome in embryonic stem (ES) cells, which resulted in the methylation of the normally unmethylated paternal ICE (Stricker et al., 2008).
In summary, macro ncRNAs have been shown to function in inducing parental-specific gene expression in the Igf2r and Kcnq1ot1 imprinted clusters. In the Igf2 imprinted cluster, the ncRNA is expressed from the parental chromosome, which silences the protein-coding genes but does not itself play a functional role in the silencing. The functions of ncRNAs in the other imprinted clusters shown in Fig. 1 are yet to be tested.
Developmental and tissue-specific imprinted expression
How might developmental or tissue-specific imprinted expression arise? In this section, we discuss mechanisms that might differentially regulate imprinted expression, and describe recently developed in vitro model systems that provide an excellent tool with which to study these mechanisms (Fig. 2). Genomic imprinting consists of a cycle of events that begins when the ICE DNA methylation imprint is established on one parental allele during gametogenesis. After fertilisation, when the embryo is diploid, the ICE methylation imprint is maintained on the same parental allele through the action of the DNA methyltransferase DNMT1 (Li et al., 1993). In subsequent developmental or tissue-specific regulated steps, imprinted expression can be maintained by additional epigenetic modifications or lost in the absence of such factors. In the examples already discussed, temporal- and tissue-specific imprinted expression could be achieved by regulating ncRNA expression and function (for the Igf2r and Kcnq1 clusters) or by regulating insulator formation (for the Igf2 cluster). To complete the genomic imprinting life cycle, the ICE methylation imprint and any secondary epigenetic modifications are erased during early germ cell development to allow the parental gametes to acquire a maternal or paternal DNA methylation imprint ready for the next generation (reviewed by Barlow and Bartolomei, 2007).
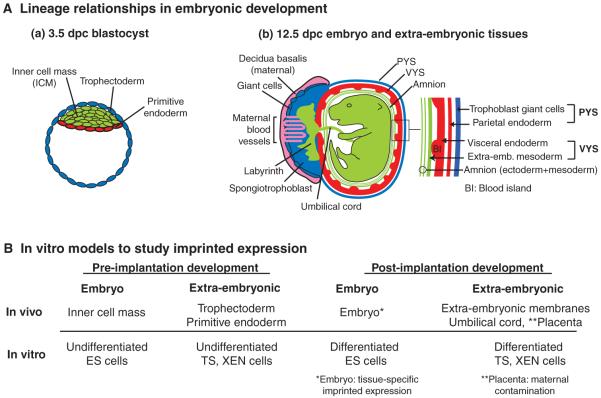
Fig. 2. Embryonic and extra-embryonic tissues in vivo and in vitro.
(A) Lineage relationships during embryonic development. (a) A blastocyst at 3.5 days post-coitum (dpc). The trophectoderm (blue) contributes to the parietal yolk sac (PYS) and the placenta of the 12.5 dpc embryo as depicted in b. The inner cell mass (ICM, green) gives rise to the embryo proper and contributes to the visceral yolk sac (VYS) and amnion extra-embryonic membranes. The primitive endoderm (red) differentiates into the endoderm layer of the PYS and VYS. (b) A 12.5 dpc embryo and its extra-embryonic tissues. The embryo (green) is surrounded by the amnion, which consists of ICM-derived ectoderm and mesoderm. The middle extra-embryonic membrane is the VYS, which consists of ICM-derived mesoderm (green) and endoderm (red). The outer membrane, the PYS, is lost after 13.5 dpc and consists of ICM-derived parietal endoderm (red) and trophoblast giant cells (blue), which are not ICM derived. The placenta consists of distinct layers: the inner labyrinth (green), the spongiotrophoblast (blue) and giant cells (blue). The outermost part of the placenta, the deciduas basalis (pink), is derived from maternal tissue. The intermingling of maternal blood vessels with the placenta is indicated. (B) In vitro model systems to study genomic imprinting. Undifferentiated embryonic stem (ES) cells are considered to mimic the blastocyst ICM, whereas undifferentiated trophoblast stem (TS) and extra-embryonic endoderm (XEN) cells mimic the blastocyst trophectoderm and primitive endoderm, respectively (Rossant, 2007). The differentiated derivatives of ES, TS and XEN cells might provide models for studying imprinting in post-implantation embryonic and extra-embryonic tissues, but care should be taken to ensure that the in vitro situations recapitulate what is observed in vivo.
There are several examples in which differential imprinted expression correlates with differential ncRNA expression. In the mouse brain, the imprinted expression of the Ube3a gene in the Pws/As cluster is seen in neurons that express the Ube3a-ats transcript, which might be continuous with Snrpnlt (Fig. 1C), but it is not seen in glial cells that lack this antisense RNA (Yamasaki et al., 2003). Glial cells show imprinted expression of Igf2r and express the Airn ncRNA, but neurons lack Airn and show non-imprinted Igf2r expression (Yamasaki et al., 2005). Imprinted expression of Igf2r also correlates with Airn expression in embryonic development. Preimplantation embryos lack Airn and express Igf2r from both parental chromosomes, whereas post-implantation embryos express Airn only from the paternal chromosome and Igf2r only from the maternal chromosome (Sleutels et al., 2002; Szabo and Mann, 1995). In terms of in vitro models for studying imprinting, it was shown recently for the Igf2r cluster that ES cell differentiation mimics the onset of imprinted expression and the gain of epigenetic modifications seen in the developing embryo (Latos et al., 2009). This work establishes the utility of ES cells to study the imprinted expression that is typical for embryonic tissue. However, as the imprinted expression of the Slc22a2 and Slc22a3 genes in the Igf2r cluster is restricted to the placenta labyrinth layer (Fig. 2A), this cannot be analysed in ES cells, which arise from a cell lineage that does not contribute to this tissue. Trophoblast stem (TS) cells are an obvious ES cell analogue for the study of genes that show imprinted expression only in placental tissues (Fig. 2B). However, differentiated TS cells appear to be an unsuitable model for the later stages of placental development, as the expression patterns and histone modifications detected in vivo are not recapitulated in vitro (Lewis et al., 2006).
The placenta provides a good example of tissue-specific variation in imprinted expression, as the majority of imprinted genes in the mouse only show imprinted expression in the placenta (Wagschal and Feil, 2006). This occurs because in many imprinted clusters a small number of centrally positioned genes show ‘ubiquitous’ imprinted expression (i.e. in embryo, placenta and adult), whereas additional genes in the cluster that extend upstream or downstream have imprinted expression only in the placenta (Fig. 1). As experiments that involve either ICE deletion or ncRNA truncation (as described above) show that imprinted expression in the embryo and the placenta are controlled by the same elements, there are two possible explanations for this phenomenon: either the ICE or the ncRNA acts differently in these two tissues to repress genes; or the placenta allows spreading of the basic mechanism that operates in embryonic tissue. The mouse placenta is a highly invasive organ, and a complete separation of embryonic and maternal tissue is not possible (Fig. 2A). This maternal contamination means that the placenta might not be a reliable tissue for analysing imprinted expression and epigenetic modifications. The placenta is an extra-embryonic tissue, which means that it is an embryonic tissue that does not contribute to the embryo itself. If ‘placental-specific’ imprinted expression were a general feature of extra-embryonic tissues, it might be more easily analysed in the extra-embryonic membranes (amnion, parietal and visceral yolk sac), which can be isolated without the presence of contaminating maternal tissue (Fig. 2A). The imprinted expression of some genes in the Igf2 and Kcnq1 imprinted clusters has been demonstrated in the visceral yolk sac (Davis et al., 1998; Frank et al., 1999); however, as the lineages of the extra-embryonic membranes and placenta differ, it remains to be determined whether the ‘placental-specific’ imprinted expression pattern is conserved in all extra-embryonic tissues.
DNA methylation represses a cis-acting repressor
As described above, deletion experiments show that only the unmethylated ICE is active in inducing the silencing of flanking protein-coding genes, either by activating a ncRNA promoter or by forming an insulator (see also Fig. 1). Thus, the ICE can be viewed as a cis-acting repressor and DNA methylation as a modification to repress this repressor. The analysis of ICE methylation can therefore offer insights into how this epigenetic modification is attracted to specific sequences and how it is used to inhibit ncRNA transcription and insulator function. In the maternal germline, the DNA methyltransferase-like protein DNMT3L, in concert with the DNA methyltransferase DNMT3A, are crucial players in the establishment of ICE germline DNA methylation (Bourc’his and Proudhon, 2008). The subsequent maintenance of ICE methylation requires the DNMT1 family of DNA methyltransferases (Hirasawa et al., 2008). Additional proteins, such as the Krüppel-associated box zinc-finger protein ZFP57, are also required for acquiring ICE methylation in the Pws/As cluster and for maintaining ICE methylation in Dlk1 (as well as in three other imprinted clusters not shown in Fig. 1), but play no role in the Igf2 and Igf2r clusters (Li et al., 2008). Although the exact mechanism by which ZFP57 acts is unknown, this finding raises the possibility that each ICE requires different additional factors for the acquisition and maintenance of germline DNA methylation. Exactly how de novo DNA methylation enzymes recognise ICE sequences is unclear. Many ICEs contain a run of tandem direct repeats that have been suggested to form a secondary structure that induces DNA methylation (Neumann and Barlow, 1996). Tandem direct repeat sequences from the Igf2r and Kcnq1 cluster ICEs are able to induce maternal germline methylation in a transgenic model (Reinhart et al., 2006). The role of these repeats in the endogenous Igf2r ICE is not yet known; however, the in vivo deletion of a subset of tandem repeats from the Kcnq1ot1 ICE or of direct repeats that flank the Igf2 ICE did not change ICE DNA methylation (Lewis et al., 2004; Mancini-Dinardo et al., 2006). By contrast, a mouse strain-specific loss of methylation was observed following the deletion of a repeat region in the paternally imprinted Rasgrf1 cluster located on mouse chromosome 9 (not shown in Fig. 1) (Yoon et al., 2002). Thus, further work is needed to determine exactly how the methylation machinery in oocytes targets ICE sequences. Although the analysis of imprinted genes highlights one of the few reported cases of a functional role for DNA methylation in gene silencing, it should be noted that its silencing effect is directed towards the ncRNA. As shown in Fig. 1, imprinted protein-coding genes are expressed from the parental chromosome that carries the methylated ICE. Thus, in imprinted clusters, DNA methylation acts by repressing a repressor of imprinted protein-coding genes.
Histone modifications associated with imprinted gene clusters
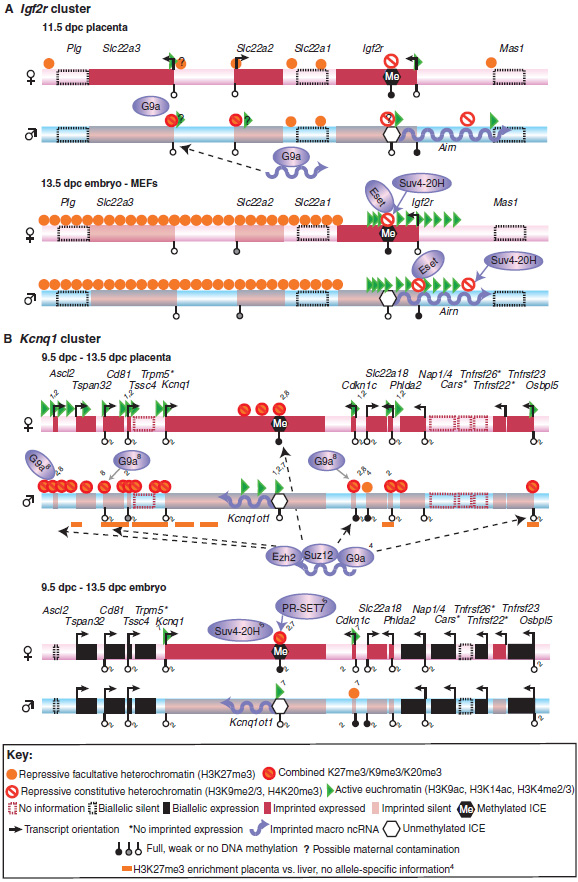
The previous section described how DNA methylation directly regulates ICE activity, but does not directly silence imprinted protein-coding genes. Here, we discuss current progress in understanding the potential roles played by histone modifications in restricting expression of macro ncRNAs to one parental allele and imprinted protein-coding genes to the other allele. Recent studies have shown that the ICE carries histone modifications that are specific to the DNA-methylated or the DNA-unmethylated allele (Fig. 3 and Table 1). Genome-wide sequencing and oligonucleotide tiling-array analyses have been used to show that the DNA-methylated ICE is marked by focal repressive histone modifications of the type found in constitutive centromeric and telomeric heterochromatin (Table 3), such as H3K9me3, H4K20me3 and the presence of HP1 (Mikkelsen et al., 2007; Regha et al., 2007). The discovery of focal repressive heterochromatin changed our general understanding of chromatin, which has traditionally been classified as either heterochromatin or euchromatin and considered to represent, respectively, transcriptionally repressed and active regions (Huisinga et al., 2006). As the ICE is often located within transcribed genes (Fig. 3), it is now clear that focal heterochromatin can exist inside actively transcribed regions without blocking RNA polymerase II (RNAPII) elongation. In the Igf2r and Kcnq1 clusters, the repressive H3K27me3 mark is present on the methylated ICE only in undifferentiated ES cells. In the Igf2r cluster, H3K27me3 is absent from embryonic fibroblasts (Fig. 3A), but in the Kcnq1 cluster it is present in both embryo and placenta (Fig. 3B) (Latos et al., 2009; Lewis et al., 2006; Lewis et al., 2004; Umlauf et al., 2004). The unmethylated ICE lacks repressive modifications but carries active histone modifications, such as H3K4me and H3/H4 acetylation. The presence of active and repressive histone modifications on the same DNA sequence in diploid cells that modify different parental chromosomes can be used to identify an ICE (Fig. 3). The usefulness of this approach was demonstrated in a genome-wide study of diploid ES cells that identified short regions that carry both repressive H3K9me3 and active H3K4me3 modifications on the ICE of the six imprinted clusters shown in Fig. 1 (Mikkelsen et al., 2007).
Fig. 3. Allele-specific histone modifications at the Igf2r and Kcnq1 clusters.
Active and repressive histone modifications on the maternal or paternal allele are shown for the (A) Igf2r and (B) Kcnq1 clusters for embryo and placenta. For simplicity, histone modifications are combined into three groups: repressive constitutive heterochromatin (H3K9me2/3, H4K20me3), repressive facultative heterochromatin (H3K27me3) and active euchromatin (H3K9ac, H3K4me2/3). In both clusters, only the indicated positions were assayed in the placenta, with the exception of one ChIP-chip mapping of H3K27me3 (orange lines). (A) In the Igf2r cluster, an unbiased continuous genome-wide location (ChIP-chip) analysis was performed in the embryo, with the indicated positions showing enrichment. (B) In the Kcnq1 cluster, all positions shown in the placenta were also assayed in the embryo, and only the indicated positions show allele-specific enrichment. Blue shaded ellipses indicate histone-modifying enzymes, and the solid blue arrows extending from the ellipses indicate the involvement of the respective enzyme in the setting or maintaining of a histone mark. Both Airn (A) and Kcnq1ot1 (B) ncRNAs bind to histone-modifying enzymes. Dashed arrows (B) indicate binding of Kcnq1ot1 ncRNA to chromatin. Gene expression marked by an asterisk is inferred from the presence of active histone modifications (Mikkelsen et al., 2007). References: Igf2r in the placenta (Nagano et al., 2008); Igf2r in the embryo (Regha et al., 2007); Kcnq1 in the placenta: all histone modifications without a numbered reference (Umlauf et al., 2004); others: 1(Green et al., 2007), 2(Lewis et al., 2004), 3(Mikkelsen et al., 2007), 4(Pandey et al., 2008), 5(Pannetier et al., 2008), 6(Regha et al., 2007), 7(Umlauf et al., 2004), 8(Wagschal et al., 2008). ac, acetylation; H3, histone 3; H4, histone 4; ICE, imprint control element; K, lysine; me2, dimethylation; me3, trimethylation.
Table 3. Epigenetic modifications associated with imprinted genes in the mouse genome.
| Gene expression | Modification | Modifying enzyme or complex |
|---|---|---|
| Modifications associated with repressed genes and/or pericentric and telomeric heterochromatin | De novo DNA methylation, i.e. CpG to meCpG |
DNMT3A, DNMT3B, DNMT3L |
| DNA methylation of hemi- methylated DNA |
DNMT1, UHRF1 | |
| HP1 | ||
| H3K27me3 | PRC2 containing EED, EZH2 and SUZ12 | |
| H3K9me2 | G9A (KMT1C, EHMT2); functions as a heterodimer with GLP (KMT1D, EHMT1) |
|
| H3K9me3 | SUV39H1 (KMT1A), SUV39H2 (KMT1B), ESET (KMT1E, SETDB1) |
|
| H4K20me1 | PR-SET7 (KMT5A, SETD8) | |
| H4K20me3 | SUV4-20H1 (KMT5B), SUV4-20H2 (KMT5C) | |
| Modifications associated with expressed genes | H3K4me2 and H3K4me3 | Multiple lysine methyltransferases (KMT2A-G) |
| Acetyl groups at several lysines in the tail of histone H3 (K9, K14, K18, K23) and H4 (K5, K8, K16) |
Multiple lysine acetyltransferases (KAT1-13) |
CpG, CG dinucleotide; DNMT, DNA methyltransferase; EED, embryonic ectoderm development; ESET, ERG-associated protein with a SET domain 7; EZH2, enhancer of zeste homolog 2; GLP, GATA-like protein 1; H3, histone 3; H4, histone 4; HP1, heterochromatin protein 1; K, lysine; KAT, lysine acetyltransferase; KMT, lysine methyltransferase; me2, dimethylation; me3, trimethylation; meCpG, methylated CG dinucleotide; PRC2, Polycomb repressor complex 2; PR-SET7, PR/SET domain-containing protein; SUV, suppressor of variegation; SUZ12, suppressor of zeste 12; UHRF1, ubiquitin-like containing PHD and RING finger domains 1. References: (Allis et al., 2007a; Allis et al., 2007b; Kouzarides, 2007).
The histone modification profiles established so far show that repressive marks are associated with the DNA-methylated ICE, whereas active histone marks are associated with the unmethylated ICE. Although it has proven to be relatively straightforward to assign a function to DNA methylation in regulating ICE activity, a general function for histone modifications has not yet been identified. Repressive H3K9me3 modifications are regulated by three known histone methyltransferases, SUV39H1, SUV39H2 (also known as KMT1A and KMT1B) and ESET (KMT1E, SETDB1), whereas the repressive H4K20me3 modification is regulated by SUV4-20H1 and SUV4-20H2 (KMT5B and KMT5C) (Table 3). The repressive H3K9me3 mark is maintained and even enhanced on the ICE in embryonic fibroblasts that lack SUV39H1 and SUV39H2, whereas the repressive H4K20me3 is reduced in embryonic cells that lack SUV4-20H1 and SUV4-20H2 without removing either the repressive H3K9me3 mark or DNA methylation (Pannetier et al., 2008; Regha et al., 2007). The ESET methyltransferase was found to bind to the Igf2r ICE; however, its role could not be tested directly because ESET-deficient cells are not viable at any developmental stage (Dodge et al., 2004; Regha et al., 2007). Thus, suitable genetic systems are not yet available to test the role of the repressive H3K9me3 and H4K20me3 modifications in regulating ICE activity.
In contrast to the lack of a defined role for histone-modifying enzymes in regulating ICE activity, several reports describe a role for these enzymes in regulating placental, but not embryonic, imprinted expression. The Polycomb group protein EED, which is required for repressive H3K27me3 modifications, has been shown to repress the paternal allele of four out of 18 tested imprinted genes in embryos 7.5 days post-coitus (dpc), which mainly consist of extra-embryonic tissue at this stage (Mager et al., 2003). The affected genes were located in three different imprinted clusters, in which the majority of genes maintained correct imprinted expression. This indicates that EED does not play a general role in regulating imprinted expression, but is attracted to specific genes. The G9A (KMT1C, EHMT2) histone lysine methyltransferase, which dimerises with G9A-like protein (GLP; KMT1D, EHMT1) to induce repressive H3K9me2 modifications (Table 3), is necessary for the paternal repression of some genes in the Kcnq1 and Igf2r clusters in the placenta, but not in the embryo (Nagano et al., 2008; Wagschal et al., 2008). As mentioned above, in embryonic tissue, repressive heterochromatin (H3K9me3, H4K20me3, HP1), but not repressive H3K27me3, modifies the DNA-methylated ICE in a focal manner. Not all of these repressive modifications have been mapped throughout the imprinted clusters in the placenta, but repressive H3K9me2/3 and H3K27me3 marks were found at the promoters of silenced mRNA genes in the Igf2r cluster (Nagano et al., 2008) (Fig. 3A). By contrast, these repressive marks were found to be more widespread in the placenta on the chromosome that carries the silenced mRNA genes in the Kcnq1 cluster (Fig. 3B). In one study in the placenta (Nagano et al., 2008), both active and repressive histone modifications were found on genes that showed placental-specific imprinted expression. Although this might indicate the existence of ‘bivalent’ domains (Bernstein et al., 2006), care should be taken in interpreting these results owing to the risk of maternal tissue contamination in placental samples (Fig. 2).
In summary, the analysis of histone modifications shows that the same active and repressive histone modifications that correlate with expressed and silent genes also modify imprinted genes in an allele-specific manner. Further work is needed to determine which modifications reflect the cause as opposed to the consequence of imprinted expression. Although there is currently no indication that histone modifications co-operate with DNA methylation to restrict macro ncRNA expression to one parental allele, there is emerging data that, in the placenta, histone modifications might play a role in repressing imprinted mRNA genes in cis. As discussed in the next section, there might even be a link between macro ncRNA function and the establishment of histone modifications.
In the placenta, ncRNAs might target repressive histone marks
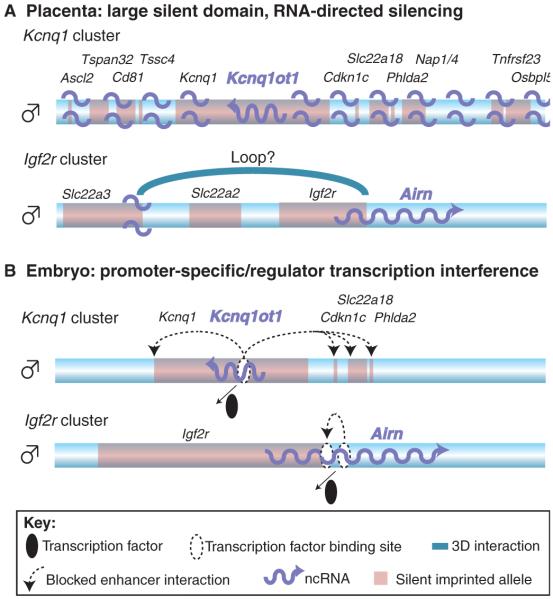
In both placental and embryonic tissue, the repression of multiple genes in the Igf2r and Kcnq1 clusters on the paternal chromosome depends on the Airn and Kcnq1ot1 macro ncRNAs (Mancini-Dinardo et al., 2006; Shin et al., 2008; Sleutels et al., 2002). However, the mechanism by which these ncRNAs induce repression is unknown. One significant open question is whether it is the ncRNA itself, or the act of its transcription, that is required for silencing (Pauler et al., 2007). Two recent studies indicate that the Airn and Kcnq1ot1 ncRNAs are themselves directly involved in silencing genes in the placenta. Kcnq1ot1 was found to localise physically to several silent genes on the paternal allele that lay hundreds of kb away from the Kcnq1ot1 promoter (Pandey et al., 2008). This finding is supported by RNA/DNA FISH, which showed partial overlap between the Kcnq1ot1 RNA and the flanking imprinted genes in the Kcnq1 cluster in the trophectoderm cells of early embryos, which contribute to the placenta (Terranova et al., 2008). Furthermore, Kcnq1ot1 also directly interacts with Polycomb group proteins, which are necessary for establishing the repressive H3K27me3 mark, and with G9A, which is involved in setting the repressive H3K9me2 mark (Pandey et al., 2008). Together, this indicates that in the placenta the Kcnq1ot1 ncRNA localises to chromatin and targets histone methyltransferases to the whole imprinted cluster. Notably, embryos that are deficient for G9A and for the Polycomb proteins EZH2 and RNF2 show a loss of paternal repression for some of the placental-specific imprinted genes in the Kcnq1 cluster (Terranova et al., 2008; Wagschal et al., 2008). Similarly, in the placenta, the Airn ncRNA in the Igf2r cluster lies in close proximity to the silent Slc22a3 promoter and has been shown to bind G9A. In addition, G9a-null embryos show a loss of placental imprinted expression for Slc22a3, but maintain Igf2r imprinted expression (Nagano et al., 2008). An RNA FISH study of the Airn and Kcnq1ot1 ncRNAs in TS cells and in preimplantation trophectoderm cells has also shown that both these ncRNAs are located in nuclear domains that are characterised by a high density of repressive H3K27me3 and by a lack of active histone modifications and RNAPII (Terranova et al., 2008). In summary, the evidence so far indicates that the Airn and Kcnq1ot1 ncRNAs induce imprinted expression by an RNA-directed targeting mechanism in the placenta that only affects genes that show placental-specific imprinted expression (Fig. 1). We present a model (Fig. 4A) according to which the ncRNA expressed from the unmethylated ICE is maintained at the site of transcription and associates with chromatin in cis. The ncRNA could localise throughout the cluster (Fig. 4A, Kcnq1ot1) or to specific promoters by looping (Fig. 4A, Airn), and might subsequently attract specific histone modifications that repress the transcription of multiple genes located at some distance from the ncRNA gene itself.
Fig. 4. Models of gene silencing by imprinted macro ncRNAs.
Imprinted macro ncRNAs might use different modes of silencing in embryonic and placental tissues. Only the paternal chromosome is shown (blue bar). The positions and names of silent imprinted genes are indicated in black font and by pale red boxes. Imprinted macro ncRNAs are indicated in blue font and by blue wavy arrows. (A) In the placenta, Kcnq1ot1 in the Kcnq1 cluster is transcribed from the paternal allele and localises to the whole imprinted domain by an unknown mechanism, inducing the recruitment of repressive histones, which leads to gene silencing. In the Igf2r cluster, Airn either locates to the silent Slc22a3 promoter, or the promoter forms a three-dimensional (3D) loop (indicated by a blue arch) to the Airn gene, similarly recruiting repressive histone modifications that silence Slc22a3. In both cases, the ncRNA itself is involved in the silencing process. (B) In embryonic tissues, only a few genes are silenced by the ncRNA in the Kcnq1 and Igf2r clusters. No localisation of the ncRNA to chromatin has been reported in the embryo. It is therefore possible that the transcription of the ncRNA is sufficient to silence all genes by interfering with the binding of essential transcription factors (black ellipse), thereby inducing gene silencing by interrupting enhancer interactions (dashed arrows).
Is ncRNA transcription more important in embryonic cells?
As described above, recent work suggests that in the placenta it is the ncRNA transcript itself that mediates gene silencing. However, imprinted ncRNAs exert different effects in the mouse embryo than in the placenta, as only a subset of the genes in the imprinted gene clusters show imprinted expression in embryonic and adult somatic tissue (Fig. 1, Fig. 4B). In the Igf2r cluster, the Airn ncRNA represses Igf2r in embryos, but represses Igf2r, Slc22a2 and Slc22a3 in the placenta. In the Kcnq1 cluster, the Kcnq1ot1 ncRNA silences Kcnq1, Cdkn1c, Slc22a18 and Phlda2 in the embryo, but an additional six genes in the placenta. A further indication of the differences between the embryo and the placenta is that G9A and EED, which are required for repressive H3K9me2 and H3K27me3 modifications, respectively, and for the imprinted expression of some placental genes, appear to play no role in the imprinted expression in the Igf2r and Kcnq1 clusters in embryonic tissues (Mager et al., 2003; Nagano et al., 2008; Wagschal and Feil, 2006). We have proposed previously that the Airn ncRNA might silence Igf2r because of transcriptional interference, and that Airn might act solely by transcription per se (Pauler et al., 2007). According to this model, ncRNA transcription either interferes directly with transcriptional initiation or with the activity of essential cis-regulatory elements (Fig. 4B). Several lines of evidence support a model in which Airn silences Igf2r by transcriptional interference in embryonic tissue. First, Airn has a short half-life of ~90 minutes, which argues against a function for the ncRNA in targeting repressive chromatin, as this would require it to be stable for at least one cell cycle (Seidl et al., 2006). Second, Airn does not induce widespread repressive chromatin in embryos (Regha et al., 2007). Third, the ability of Airn to silence Igf2r is dependent on promoter strength, a feature associated with transcriptional interference (Shearwin et al., 2005; Stricker et al., 2008). The Igf2r and Kcnq1 clusters differ in that the Kcnq1ot1 ncRNA represses multiple genes in the embryo, and as it is contained entirely within Kcnq1 (Pandey et al., 2008), it does not overlap with a promoter (Fig. 4B). However, it is possible to propose a transcriptional interference mode of silencing for this ncRNA by postulating the existence of crucial cis-regulatory elements that are overlapped by Kcnq1ot1. Although there is less evidence to support a transcriptional interference model for Kcnq1ot1, the lack of widespread repressive chromatin marks on genes in this cluster that show imprinted expression in the embryo (Pandey et al., 2008; Umlauf et al., 2004), as well as the absence of a role for G9A and EED (Mager et al., 2003; Wagschal et al., 2008), indicate that RNA-mediated targeting does not operate in embryonic tissues.
Conclusions
Mammalian macro ncRNAs, which comprise the majority of the transcriptome, have been suggested to play a role in the epigenetic regulation of gene expression, mainly on the basis of their expression patterns. In contrast to the uncertainty surrounding the function of most mammalian macro ncRNAs, imprinted macro ncRNAs have clearly been shown to regulate flanking genes epigenetically. Thus, imprinted genes offer a valuable in vivo and in vitro model not only to decipher the transcriptional biology of macro ncRNAs themselves and their regulation by DNA methylation, but also to shed light on the epigenetic mechanisms that underlie the macro ncRNA-mediated repression of flanking genes.
Acknowledgments
We thank Quanah Hudson and Stefan Stricker for comments on the manuscript. The authors are supported by grants from the Sixth European Union Framework Programme: the ‘HEROIC’ Integrated Project and the ‘EPIGENOME’ Network of Excellence, and the Austrian Science Fund (FWF).
References
- Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, et al. New nomenclature for chromatin-modifying enzymes. Cell. 2007a;131:633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Allis CD, Jenuwein T, Reinberg D. Overview and concepts. In: Allis CD, Jenuwein T, Reinberg D, Caparros M, editors. Epigenetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007b. pp. 23–61. [Google Scholar]
- Amaral PP, Mattick JS. Noncoding RNA in development. Mamm. Genome. 2008;19:454–492. doi: 10.1007/s00335-008-9136-7. [DOI] [PubMed] [Google Scholar]
- Barlow DP, Bartolomei MS. Genomic imprinting in mammals. In: Allis CD, Jenuwein T, Reinberg D, Caparros M, editors. Epigenetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. pp. 357–375. [Google Scholar]
- Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bielinska B, Blaydes SM, Buiting K, Yang T, Krajewska-Walasek M, Horsthemke B, Brannan CI. De novo deletions of SNRPN exon 1 in early human and mouse embryos result in a paternal to maternal imprint switch. Nat. Genet. 2000;25:74–78. doi: 10.1038/75629. [DOI] [PubMed] [Google Scholar]
- Bourc’his D, Proudhon C. Sexual dimorphism in parental imprint ontogeny and contribution to embryonic development. Mol. Cell. Endocrinol. 2008;282:87–94. doi: 10.1016/j.mce.2007.11.025. [DOI] [PubMed] [Google Scholar]
- Braidotti G, Baubec T, Pauler F, Seidl C, Smrzka O, Stricker S, Yotova I, Barlow DP. The Air noncoding RNA: an imprinted cis-silencing transcript. Cold Spring Harb. Symp. Quant. Biol. 2004;69:55–66. doi: 10.1101/sqb.2004.69.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannan CI, Dees EC, Ingram RS, Tilghman SM. The product of the H19 gene may function as an RNA. Mol. Cell. Biol. 1990;10:28–36. doi: 10.1128/mcb.10.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressler J, Tsai TF, Wu MY, Tsai SF, Ramirez MA, Armstrong D, Beaudet AL. The SNRPN promoter is not required for genomic imprinting of the Prader-Willi/Angelman domain in mice. Nat. Genet. 2001;28:232–240. doi: 10.1038/90067. [DOI] [PubMed] [Google Scholar]
- Brown JW, Marshall DF, Echeverria M. Intronic noncoding RNAs and splicing. Trends Plant Sci. 2008;13:335–342. doi: 10.1016/j.tplants.2008.04.010. [DOI] [PubMed] [Google Scholar]
- Cai X, Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA. 2007;13:313–316. doi: 10.1261/rna.351707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannell IG, Kong YW, Bushell M. How do microRNAs regulate gene expression? Biochem. Soc. Trans. 2008;36:1224–1231. doi: 10.1042/BST0361224. [DOI] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Cavaille J, Buiting K, Kiefmann M, Lalande M, Brannan CI, Horsthemke B, Bachellerie JP, Brosius J, Huttenhofer A. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl. Acad. Sci. USA. 2000;97:14311–14316. doi: 10.1073/pnas.250426397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaille J, Seitz H, Paulsen M, Ferguson-Smith AC, Bachellerie JP. Identification of tandemly-repeated C/D snoRNA genes at the imprinted human 14q32 domain reminiscent of those at the Prader-Willi/Angelman syndrome region. Hum. Mol. Genet. 2002;11:1527–1538. doi: 10.1093/hmg/11.13.1527. [DOI] [PubMed] [Google Scholar]
- Chamberlain SJ, Brannan CI. The Prader-Willi syndrome imprinting center activates the paternally expressed murine Ube3a antisense transcript but represses paternal Ube3a. Genomics. 2001;73:316–322. doi: 10.1006/geno.2001.6543. [DOI] [PubMed] [Google Scholar]
- Chotalia M, Smallwood SA, Ruf N, Dawson C, Lucifero D, Frontera M, James K, Dean W, Kelsey G. Transcription is required for establishment of germline methylation marks at imprinted genes. Genes Dev. 2009;23:105–117. doi: 10.1101/gad.495809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custodio N, Carmo-Fonseca M, Geraghty F, Pereira HS, Grosveld F, Antoniou M. Inefficient processing impairs release of RNA from the site of transcription. EMBO J. 1999;18:2855–2866. doi: 10.1093/emboj/18.10.2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Rocha ST, Edwards CA, Ito M, Ogata T, Ferguson-Smith AC. Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends Genet. 2008;24:306–316. doi: 10.1016/j.tig.2008.03.011. [DOI] [PubMed] [Google Scholar]
- Davis E, Caiment F, Tordoir X, Cavaille J, Ferguson-Smith A, Cockett N, Georges M, Charlier C. RNAi-mediated allelic trans-interaction at the imprinted Rtl1/Peg11 locus. Curr. Biol. 2005;15:743–749. doi: 10.1016/j.cub.2005.02.060. [DOI] [PubMed] [Google Scholar]
- Davis TL, Tremblay KD, Bartolomei MS. Imprinted expression and methylation of the mouse H19 gene are conserved in extraembryonic lineages. Dev. Genet. 1998;23:111–118. doi: 10.1002/(SICI)1520-6408(1998)23:2<111::AID-DVG3>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- de los Santos T, Schweizer J, Rees CA, Francke U. Small evolutionarily conserved RNA, resembling C/D box small nucleolar RNA, is transcribed from PWCR1, a novel imprinted gene in the Prader-Willi deletion region, which is highly expressed in brain. Am. J. Hum. Genet. 2000;67:1067–1082. doi: 10.1086/303106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaval K, Govin J, Cerqueira F, Rousseaux S, Khochbin S, Feil R. Differential histone modifications mark mouse imprinting control regions during spermatogenesis. EMBO J. 2007;26:720–729. doi: 10.1038/sj.emboj.7601513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F, Prints Y, Dhar MS, Johnson DK, Garnacho-Montero C, Nicholls RD, Francke U. Lack of Pwcr1/MBII-85 snoRNA is critical for neonatal lethality in Prader-Willi syndrome mouse models. Mamm. Genome. 2005;16:424–431. doi: 10.1007/s00335-005-2460-2. [DOI] [PubMed] [Google Scholar]
- Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Solda G, Simons C, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodge JE, Kang YK, Beppu H, Lei H, Li E. Histone H3-K9 methyltransferase ESET is essential for early development. Mol. Cell. Biol. 2004;24:2478–2486. doi: 10.1128/MCB.24.6.2478-2486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engstrom PG, Suzuki H, Ninomiya N, Akalin A, Sessa L, Lavorgna G, Brozzi A, Luzi L, Tan SL, Yang L, et al. Complex Loci in human and mouse genomes. PLoS Genet. 2006;2:e47. doi: 10.1371/journal.pgen.0020047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick GV, Soloway PD, Higgins MJ. Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat. Genet. 2002;32:426–431. doi: 10.1038/ng988. [DOI] [PubMed] [Google Scholar]
- Frank D, Mendelsohn CL, Ciccone E, Svensson K, Ohlsson R, Tycko B. A novel pleckstrin homology-related gene family defined by Ipl/Tssc3, TDAG51, and Tih1: tissue-specific expression, chromosomal location, and parental imprinting. Mamm. Genome. 1999;10:1150–1159. doi: 10.1007/s003359901182. [DOI] [PubMed] [Google Scholar]
- Gerard M, Hernandez L, Wevrick R, Stewart CL. Disruption of the mouse necdin gene results in early post-natal lethality. Nat. Genet. 1999;23:199–202. doi: 10.1038/13828. [DOI] [PubMed] [Google Scholar]
- Graff RJ, Brown DH, Snell GD. The alleles of the H-13 locus. Immunogenetics. 1978;7:413–423. doi: 10.1007/BF01844032. [DOI] [PubMed] [Google Scholar]
- Green K, Lewis A, Dawson C, Dean W, Reinhart B, Chaillet JR, Reik W. A developmental window of opportunity for imprinted gene silencing mediated by DNA methylation and the Kcnq1ot1 noncoding RNA. Mamm. Genome. 2007;18:32–42. doi: 10.1007/s00335-006-0092-9. [DOI] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- Hatada I, Morita S, Obata Y, Sotomaru Y, Shimoda M, Kono T. Identification of a new imprinted gene, Rian, on mouse chromosome 12 by fluorescent differential display screening. J. Biochem. 2001;130:187–190. doi: 10.1093/oxfordjournals.jbchem.a002971. [DOI] [PubMed] [Google Scholar]
- Hirasawa R, Chiba H, Kaneda M, Tajima S, Li E, Jaenisch R, Sasaki H. Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Dev. 2008;22:1607–1616. doi: 10.1101/gad.1667008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes R, Williamson C, Peters J, Denny P, Wells C. A comprehensive transcript map of the mouse Gnas imprinted complex. Genome Res. 2003;13:1410–1415. doi: 10.1101/gr.955503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell specific MicroRNAs. Dev. Cell. 2003;5:351–358. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- Huisinga KL, Brower-Toland B, Elgin SC. The contradictory definitions of heterochromatin: transcription and silencing. Chromosoma. 2006;115:110–122. doi: 10.1007/s00412-006-0052-x. [DOI] [PubMed] [Google Scholar]
- Huttenhofer A, Kiefmann M, Meier-Ewert S, O’Brien J, Lehrach H, Bachellerie JP, Brosius J. RNomics: an experimental approach that identifies 201 candidates for novel, small, non-messenger RNAs in mouse. EMBO J. 2001;20:2943–2953. doi: 10.1093/emboj/20.11.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cawley SE, Drenkow J, Bekiranov S, Strausberg RL, Fodor SP, Gingeras TR. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002;296:916–919. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- Kim J, Krichevsky A, Grad Y, Hayes GD, Kosik KS, Church GM, Ruvkun G. Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc. Natl. Acad. Sci. USA. 2004;101:360–365. doi: 10.1073/pnas.2333854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Labialle S, Croteau S, Belanger V, McMurray EN, Ruan X, Moussette S, Jonnaert M, Schmidt JV, Cermakian N, Naumova AK. Novel imprinted transcripts from the Dlk1-Gtl2 intergenic region, Mico1 and Mico1os, show circadian oscillations. Epigenetics. 2008;3:322–329. doi: 10.4161/epi.3.6.7109. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Landers M, Bancescu DL, Le Meur E, Rougeulle C, Glatt-Deeley H, Brannan C, Muscatelli F, Lalande M. Regulation of the large (approximately 1000 kb) imprinted murine Ube3a antisense transcript by alternative exons upstream of Snurf/Snrpn. Nucleic Acids Res. 2004;32:3480–3492. doi: 10.1093/nar/gkh670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latos PA, Stricker SH, Steenpass L, Pauler FM, Huang R, Senergin BH, Regha K, Koerner MV, Warczok KE, Unger C, et al. An in vitro ES cell imprinting model shows that imprinted expression of the Igf2r gene arises from an allele-specific expression bias. Development. 2009;136:437–448. doi: 10.1242/dev.032060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Meur E, Watrin F, Landers M, Sturny R, Lalande M, Muscatelli F. Dynamic developmental regulation of the large non-coding RNA associated with the mouse 7C imprinted chromosomal region. Dev. Biol. 2005;286:587–600. doi: 10.1016/j.ydbio.2005.07.030. [DOI] [PubMed] [Google Scholar]
- Lewis A, Mitsuya K, Umlauf D, Smith P, Dean W, Walter J, Higgins M, Feil R, Reik W. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat. Genet. 2004;36:1291–1295. doi: 10.1038/ng1468. [DOI] [PubMed] [Google Scholar]
- Lewis A, Green K, Dawson C, Redrup L, Huynh KD, Lee JT, Hemberger M, Reik W. Epigenetic dynamics of the Kcnq1 imprinted domain in the early embryo. Development. 2006;133:4203–4210. doi: 10.1242/dev.02612. [DOI] [PubMed] [Google Scholar]
- Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- Li T, Vu TH, Zeng ZL, Nguyen BT, Hayward BE, Bonthron DT, Hu JF, Hoffman AR. Tissue-specific expression of antisense and sense transcripts at the imprinted Gnas locus. Genomics. 2000;69:295–304. doi: 10.1006/geno.2000.6337. [DOI] [PubMed] [Google Scholar]
- Li X, Ito M, Zhou F, Youngson N, Zuo X, Leder P, Ferguson-Smith AC. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev. Cell. 2008;15:547–557. doi: 10.1016/j.devcel.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SP, Youngson N, Takada S, Seitz H, Reik W, Paulsen M, Cavaille J, Ferguson-Smith AC. Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat. Genet. 2003;35:97–102. doi: 10.1038/ng1233. [DOI] [PubMed] [Google Scholar]
- Liu J, Yu S, Litman D, Chen W, Weinstein LS. Identification of a methylation imprint mark within the mouse Gnas locus. Mol. Cell. Biol. 2000;20:5808–5817. doi: 10.1128/mcb.20.16.5808-5817.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyle R, Watanabe D, te Vruchte D, Lerchner W, Smrzka OW, Wutz A, Schageman J, Hahner L, Davies C, Barlow DP. The imprinted antisense RNA at the Igf2r locus overlaps but does not imprint Mas1. Nat. Genet. 2000;25:19–21. doi: 10.1038/75546. [DOI] [PubMed] [Google Scholar]
- Mager J, Montgomery ND, de Villena FP, Magnuson T. Genome imprinting regulated by the mouse Polycomb group protein Eed. Nat. Genet. 2003;33:502–507. doi: 10.1038/ng1125. [DOI] [PubMed] [Google Scholar]
- Mancini-Dinardo D, Steele SJ, Levorse JM, Ingram RS, Tilghman SM. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev. 2006;20:1268–1282. doi: 10.1101/gad.1416906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS, Makunin IV. Non-coding RNA. Hum. Mol. Genet. 2006;15(1):R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. USA. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi N, Wagatsuma H, Wakana S, Shiroishi T, Nomura M, Aisaka K, Kohda T, Surani MA, Kaneko-Ishino T, Ishino F. Identification of an imprinted gene, Meg3/Gtl2 and its human homologue MEG3, first mapped on mouse distal chromosome 12 and human chromosome 14q. Genes Cells. 2000;5:211–220. doi: 10.1046/j.1365-2443.2000.00320.x. [DOI] [PubMed] [Google Scholar]
- Moldon A, Malapeira J, Gabrielli N, Gogol M, Gomez-Escoda B, Ivanova T, Seidel C, Ayte J. Promoter-driven splicing regulation in fission yeast. Nature. 2008;455:997–1000. doi: 10.1038/nature07325. [DOI] [PubMed] [Google Scholar]
- Moore T, Constancia M, Zubair M, Bailleul B, Feil R, Sasaki H, Reik W. Multiple imprinted sense and antisense transcripts, differential methylation and tandem repeats in a putative imprinting control region upstream of mouse Igf2. Proc. Natl. Acad. Sci. USA. 1997;94:12509–12514. doi: 10.1073/pnas.94.23.12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P. The Air Noncoding RNA Epigenetically Silences Transcription by Targeting G9a to Chromatin. Science. 2008;322:1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- Neumann B, Barlow DP. Multiple roles for DNA methylation in gametic imprinting. Curr. Opin. Genet. Dev. 1996;6:159–163. doi: 10.1016/s0959-437x(96)80045-1. [DOI] [PubMed] [Google Scholar]
- Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- Pachnis V, Belayew A, Tilghman SM. Locus unlinked to alphafetoprotein under the control of the murine raf and Rif genes. Proc. Natl. Acad. Sci. USA. 1984;81:5523–5527. doi: 10.1073/pnas.81.17.5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachnis V, Brannan CI, Tilghman SM. The structure and expression of a novel gene activated in early mouse embryogenesis. EMBO J. 1988;7:673–681. doi: 10.1002/j.1460-2075.1988.tb02862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin level regulation. Mol. Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- Pannetier M, Julien E, Schotta G, Tardat M, Sardet C, Jenuwein T, Feil R. PR-SET7 and SUV4-20H regulate H4 lysine-20 methylation at imprinting control regions in the mouse. EMBO Rep. 2008;9:998–1005. doi: 10.1038/embor.2008.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauler FM, Koerner MV, Barlow DP. Silencing by imprinted noncoding RNAs: is transcription the answer? Trends Genet. 2007;23:284–292. doi: 10.1016/j.tig.2007.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth KV, Spector DL. Eukaryotic regulatory RNAs: an answer to the ‘genome complexity’ conundrum. Genes Dev. 2007;21:11–42. doi: 10.1101/gad.1484207. [DOI] [PubMed] [Google Scholar]
- Regha K, Sloane MA, Huang R, Pauler FM, Warczok KE, Melikant B, Radolf M, Martens JH, Schotta G, Jenuwein T, et al. Active and repressive chromatin are interspersed without spreading in an imprinted gene cluster in the mammalian genome. Mol. Cell. 2007;27:353–366. doi: 10.1016/j.molcel.2007.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart B, Paoloni-Giacobino A, Chaillet JR. Specific differentially methylated domain sequences direct the maintenance of methylation at imprinted genes. Mol. Cell. Biol. 2006;26:8347–8356. doi: 10.1128/MCB.00981-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossant J. Stem cells and lineage development in the mammalian blastocyst. Reprod. Fertil. Dev. 2007;19:111–118. doi: 10.1071/rd06125. [DOI] [PubMed] [Google Scholar]
- Royo H, Bortolin ML, Seitz H, Cavaille J. Small non-coding RNAs and genomic imprinting. Cytogenet. Genome Res. 2006;113:99–108. doi: 10.1159/000090820. [DOI] [PubMed] [Google Scholar]
- Ryu WS, Mertz JE. Simian virus 40 late transcripts lacking excisable intervening sequences are defective in both stability in the nucleus and transport to the cytoplasm. J. Virol. 1989;63:4386–4394. doi: 10.1128/jvi.63.10.4386-4394.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JV, Levorse JM, Tilghman SM. Enhancer competition between H19 and Igf2 does not mediate their imprinting. Proc. Natl. Acad. Sci. USA. 1999;96:9733–9738. doi: 10.1073/pnas.96.17.9733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JV, Matteson PG, Jones BK, Guan XJ, Tilghman SM. The Dlk1 and Gtl2 genes are linked and reciprocally imprinted. Genes Dev. 2000;14:1997–2002. [PMC free article] [PubMed] [Google Scholar]
- Schuster-Gossler K, Bilinski P, Sado T, Ferguson-Smith A, Gossler A. The mouse Gtl2 gene is differentially expressed during embryonic development, encodes multiple alternatively spliced transcripts, and may act as an RNA. Dev. Dyn. 1998;212:214–228. doi: 10.1002/(SICI)1097-0177(199806)212:2<214::AID-AJA6>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Seidl CI, Stricker SH, Barlow DP. The imprinted Air ncRNA is an atypical RNAPII transcript that evades splicing and escapes nuclear export. EMBO J. 2006;25:3565–3575. doi: 10.1038/sj.emboj.7601245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz H, Youngson N, Lin SP, Dalbert S, Paulsen M, Bachellerie JP, Ferguson-Smith AC, Cavaille J. Imprinted microRNA genes transcribed antisense to a reciprocally imprinted retrotransposon-like gene. Nat. Genet. 2003;34:261–262. doi: 10.1038/ng1171. [DOI] [PubMed] [Google Scholar]
- Seitz H, Royo H, Bortolin ML, Lin SP, Ferguson-Smith AC, Cavaille J. A large imprinted microRNA gene cluster at the mouse Dlk1-Gtl2 domain. Genome Res. 2004;14:1741–1748. doi: 10.1101/gr.2743304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearwin KE, Callen BP, Egan JB. Transcriptional interference – a crash course. Trends Genet. 2005;21:339–345. doi: 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JY, Fitzpatrick GV, Higgins MJ. Two distinct mechanisms of silencing by the KvDMR1 imprinting control region. EMBO J. 2008;27:168–178. doi: 10.1038/sj.emboj.7601960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skryabin BV, Gubar LV, Seeger B, Pfeiffer J, Handel S, Robeck T, Karpova E, Rozhdestvensky TS, Brosius J. Deletion of the MBII- 85 snoRNA gene cluster in mice results in postnatal growth retardation. PLoS Genet. 2007;3:e235. doi: 10.1371/journal.pgen.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- Smilinich NJ, Day CD, Fitzpatrick GV, Caldwell GM, Lossie AC, Cooper PR, Smallwood AC, Joyce JA, Schofield PN, Reik W, et al. A maternally methylated CpG island in KvLQT1 is associated with an antisense paternal transcript and loss of imprinting in Beckwith-Wiedemann syndrome. Proc. Natl. Acad. Sci. USA. 1999;96:8064–8069. doi: 10.1073/pnas.96.14.8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stricker SH, Steenpass L, Pauler FM, Santoro F, Latos PA, Huang R, Koerner MV, Sloane MA, Warczok KE, Barlow DP. Silencing and transcriptional properties of the imprinted Airn ncRNA are independent of the endogenous promoter. EMBO J. 2008;27:3116–3128. doi: 10.1038/emboj.2008.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo PE, Mann JR. Allele-specific expression and total expression levels of imprinted genes during early mouse development: implications for imprinting mechanisms. Genes Dev. 1995;9:3097–3108. doi: 10.1101/gad.9.24.3097. [DOI] [PubMed] [Google Scholar]
- Terranova R, Yokobayashi S, Stadler MB, Otte AP, van Lohuizen M, Orkin SH, Peters AH. Polycomb group proteins Ezh2 and Rnf2 direct genomic contraction and imprinted repression in early mouse embryos. Dev. Cell. 2008;15:668–679. doi: 10.1016/j.devcel.2008.08.015. [DOI] [PubMed] [Google Scholar]
- Thorvaldsen JL, Duran KL, Bartolomei MS. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 1998;12:3693–3702. doi: 10.1101/gad.12.23.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierling S, Dalbert S, Schoppenhorst S, Tsai CE, Oliger S, Ferguson-Smith AC, Paulsen M, Walter J. High-resolution map and imprinting analysis of the Gtl2-Dnchc1 domain on mouse chromosome 12. Genomics. 2006;87:225–235. doi: 10.1016/j.ygeno.2005.09.018. [DOI] [PubMed] [Google Scholar]
- Umlauf D, Goto Y, Cao R, Cerqueira F, Wagschal A, Zhang Y, Feil R. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat. Genet. 2004;36:1296–1300. doi: 10.1038/ng1467. [DOI] [PubMed] [Google Scholar]
- Verona RI, Thorvaldsen JL, Reese KJ, Bartolomei MS. The transcriptional status but not the imprinting control region determines allele-specific histone modifications at the imprinted H19 locus. Mol. Cell. Biol. 2008;28:71–82. doi: 10.1128/MCB.01534-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagschal A, Feil R. Genomic imprinting in the placenta. Cytogenet. Genome Res. 2006;113:90–98. doi: 10.1159/000090819. [DOI] [PubMed] [Google Scholar]
- Wagschal A, Sutherland HG, Woodfine K, Henckel A, Chebli K, Schulz R, Oakey RJ, Bickmore WA, Feil R. G9a histone methyltransferase contributes to imprinting in the mouse placenta. Mol. Cell. Biol. 2008;28:1104–1113. doi: 10.1128/MCB.01111-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- Wevrick R, Francke U. An imprinted mouse transcript homologous to the human imprinted in Prader-Willi syndrome (IPW) gene. Hum. Mol. Genet. 1997;6:325–332. doi: 10.1093/hmg/6.2.325. [DOI] [PubMed] [Google Scholar]
- Williamson CM, Skinner JA, Kelsey G, Peters J. Alternative non-coding splice variants of Nespas, an imprinted gene antisense to Nesp in the Gnas imprinting cluster. Mamm. Genome. 2002;13:74–79. doi: 10.1007/s00335-001-2102-2. [DOI] [PubMed] [Google Scholar]
- Williamson CM, Turner MD, Ball ST, Nottingham WT, Glenister P, Fray M, Tymowska-Lalanne Z, Plagge A, Powles-Glover N, Kelsey G, et al. Identification of an imprinting control region affecting the expression of all transcripts in the Gnas cluster. Nat. Genet. 2006;38:350–355. doi: 10.1038/ng1731. [DOI] [PubMed] [Google Scholar]
- Wood AJ, Roberts RG, Monk D, Moore GE, Schulz R, Oakey RJ. A screen for retrotransposed imprinted genes reveals an association between X chromosome homology and maternal germ-line methylation. PLoS Genet. 2007;3:e20. doi: 10.1371/journal.pgen.0030020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AJ, Schulz R, Woodfine K, Koltowska K, Beechey CV, Peters J, Bourc’his D, Oakey RJ. Regulation of alternative polyadenylation by genomic imprinting. Genes Dev. 2008;22:1141–1146. doi: 10.1101/gad.473408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wroe SF, Kelsey G, Skinner JA, Bodle D, Ball ST, Beechey CV, Peters J, Williamson CM. An imprinted transcript, antisense to Nesp, adds complexity to the cluster of imprinted genes at the mouse Gnas locus. Proc. Natl. Acad. Sci. USA. 2000;97:3342–3346. doi: 10.1073/pnas.050015397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MY, Tsai TF, Beaudet AL. Deficiency of Rbbp1/Arid4a and Rbbp1l1/Arid4b alters epigenetic modifications and suppresses an imprinting defect in the PWS/AS domain. Genes Dev. 2006;20:2859–2870. doi: 10.1101/gad.1452206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz A, Gribnau J. X inactivation Xplained. Curr. Opin. Genet. Dev. 2007;17:387–393. doi: 10.1016/j.gde.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Wutz A, Smrzka OW, Schweifer N, Schellander K, Wagner EF, Barlow DP. Imprinted expression of the Igf2r gene depends on an intronic CpG island. Nature. 1997;389:745–749. doi: 10.1038/39631. [DOI] [PubMed] [Google Scholar]
- Wutz A, Theussl HC, Dausman J, Jaenisch R, Barlow DP, Wagner EF. Non-imprinted Igf2r expression decreases growth and rescues the Tme mutation in mice. Development. 2001;128:1881–1887. doi: 10.1242/dev.128.10.1881. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Zhou H, Qu LH. Characterization of three novel imprinted snoRNAs from mouse Irm gene. Biochem. Biophys. Res. Commun. 2006;340:1217–1223. doi: 10.1016/j.bbrc.2005.12.128. [DOI] [PubMed] [Google Scholar]
- Yamasaki K, Joh K, Ohta T, Masuzaki H, Ishimaru T, Mukai T, Niikawa N, Ogawa M, Wagstaff J, Kishino T. Neurons but not glial cells show reciprocal imprinting of sense and antisense transcripts of Ube3a. Hum. Mol. Genet. 2003;12:837–847. doi: 10.1093/hmg/ddg106. [DOI] [PubMed] [Google Scholar]
- Yamasaki Y, Kayashima T, Soejima H, Kinoshita A, Yoshiura K, Matsumoto N, Ohta T, Urano T, Masuzaki H, Ishimaru T, et al. Neuron-specific relaxation of Igf2r imprinting is associated with neuron-specific histone modifications and lack of its antisense transcript Air. Hum. Mol. Genet. 2005;14:2511–2520. doi: 10.1093/hmg/ddi255. [DOI] [PubMed] [Google Scholar]
- Yazgan O, Krebs JE. Noncoding but nonexpendable: transcriptional regulation by large noncoding RNA in eukaryotes. Biochem. Cell Biol. 2007;85:484–496. doi: 10.1139/O07-061. [DOI] [PubMed] [Google Scholar]
- Yoon BJ, Herman H, Sikora A, Smith LT, Plass C, Soloway PD. Regulation of DNA methylation of Rasgrf1. Nat. Genet. 2002;30:92–96. doi: 10.1038/ng795. [DOI] [PMC free article] [PubMed] [Google Scholar]