Abstract
Objective
To review published methods for transcervical collection of fetal cells and to assess the potential of this approach for application in prenatal diagnosis.
Design
Retrospective analysis of efforts at prenatal diagnosis using trophoblast cells shed into the lower uterine pole that accumulate within the cervical mucus at the level of the internal os.
Results
Minimally invasive techniques that include cervical mucus aspiration, cervical swabbing, and cervical or intrauterine lavage can be used to retrieve trophoblast cells during the first trimester for diagnostic purposes, including prenatal genetic analysis. Fetal cells have been identified in these specimens with success rates that vary from 40 to 90%. The disparity in reported success rates can be a function of gestational age, collection method, operator variability, detection sensitivity, or pregnancy status. Molecular approaches have been devised to determine fetal sex and identify aneuploidies. Antibody markers have proven useful to select trophoblast cells for genetic analysis and to demonstrate that the abundance of recoverable fetal cells diminishes in abnormal gestations, such as ectopic pregnancy and blighted ovum.
Conclusions
Transcervical collection of fetal cells offers several avenues for prenatal diagnosis that with further refinement could one day provide valuable information for the management of ongoing pregnancies.
Keywords: Prenatal diagnosis, fetal cells, transcervical cell collection, cervical mucus, intrauterine lavage, trophoblast, genetic analysis, immunological markers, ectopic pregnancy
INTRODUCTION
It is thought that due to changing demographics, increased exposure to environmental toxins and intervention in the reproductive process, developmental abnormalities may be on the rise (1). The risk to any pregnant couple of having a live born infant with a chromosomal abnormality or structural defect has been previously estimated to be between 3% and 5% (2). Because of this considerable risk, much effort has been expended in recent decades to identify pregnancies at risk of chromosomal anomalies and genetic disorders at an early gestational age. The current standard of care involves screening maternal analytes and ultrasound markers, each alone or in combination, to identify at risk pregnancies, followed by referral for definitive diagnostic tests that include amniocentesis and chorionic villous sampling. While the former screening modalities have considerable rates of false positives and false negatives, the latter diagnostic tests are invasive and carry significant risk of fetal loss. Indeed, Mujezinovic et al. conducted a systematic analysis of 45 studies and reported a fetal loss rate of 1.9% for amniocentesis and 2% for chorionic villous sampling (3). Therefore, the search to develop safer methods to obtain genetic material from the fetus is ongoing and desperately needed.
Another alternative for prenatal diagnosis is preimplantation genetic diagnosis (PGD), which involves screening for chromosome abnormalities or single gene disorders in an embryo prior to implantation (4). The main advantage is avoidance of elective pregnancy termination, while offering a high likelihood that the fetus will be free of a specific disorder. Although PGD is an attractive method for prenatal diagnosis, it is an adjunct of assisted reproductive technology that requires in vitro fertilization, which has its own risks and high costs. Thus, PGD is not feasible as a universal diagnostic tool for genetic abnormalities in the general population.
Identification of fetal cells in maternal serum has been attempted, but this approach has been hindered by the relative rarity of fetal cells in maternal blood (1 fetal cell per 106–107 maternal cells) and associated difficulties in their isolation and analysis. Overall, the projected clinical efficacy has been disappointing (5). Nevertheless, recent discovery of fetal nucleic acids in maternal plasma has introduced several new possibilities for noninvasive prenatal diagnosis of chromosomal aneuploidies (6). Anomalies are revealed after the first eleven weeks of gestation by measuring the allelic ratio of single nucleotide polymorphisms in the coding region of placental mRNA, analysis of DNA fragments with different patterns of DNA methylation between fetal and maternal DNA, enrichment of the fractional concentration of fetal DNA in maternal plasma using physical or chemical methods, and the development of more precise digital polymerase chain reaction (PCR)-based methods for fetal nucleic acid analysis (6–7). Specific inheritable diseases could also be diagnosed with fetal DNA (8). These new approaches for prenatal diagnosis using maternal plasma are challenging for practical application because they require sophisticated and expensive technology. Presumably, large scale clinical trials will soon be initiated to validate the accuracy and safety of these approaches for routine clinical practice.
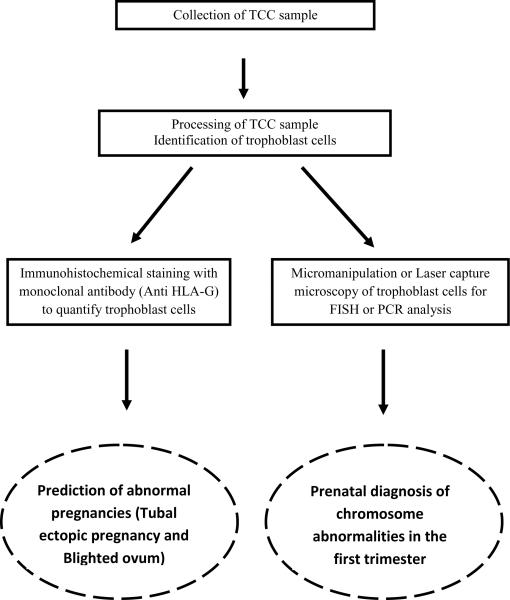
Prior to 13–15 weeks of gestation, it is believed that small areas of erosions allow trophoblast cells to cross the decidua capsularis and reach the uterine cavity (9). This process becomes less likely after the amniochorionic membrane seals the uterine cavity and the internal cervical os, which is thought to occur at three months of gestation. In 1971, Shettles (10) suggested that during early pregnancy, a similar shedding occurs into the uterine cavity, making chorionic cellular elements from the degenerating villi available in the endocervical canal. The possibility of capturing fetal cells from accessible regions of the reproductive tract suggests new approaches for early prenatal diagnosis. The isolation of fetal cells from the cervix and the endometrial cavity offers an attractive non-invasive alternative for very early (6–14 weeks, possibly as early as 5 weeks (11)) diagnosis. Since its first description, several investigators have reported the feasibility of isolating fetal cells from the cervical mucus or from fluid obtained by lavage of the endometrial cavity with varying degrees of success. The existing literature suggests that the present status of transcervical cell (TCC) sampling in prenatal diagnosis is experimental, but carries excellent potential for both genetic diagnosis and prediction of pregnancy outcome (Fig. 1) as laboratory methods are refined and standardized. The publication of both encouraging and discouraging reports of TCC sampling reliability warrants a reexamination of this topic, which has been previously surveyed (9;12;13). Here, we review the pertinent literature that describes TCC collection, the isolation of fetal cells, their subsequent analysis, and its applicability in the practice of modern clinical medicine.
Figure 1.
Transcervical cell analysis and potential clinical applications by quantification of trophoblast cells or genetic analysis using fluorescence in situ hybridization (FISH) or the polymerase chain reaction (PCR).
RETRIEVAL OF TROPHOBLAST CELLS FROM THE CERVIX
The ideal method that would reliably yield fetal cells in appreciable quantity should have no negative impact on the ongoing pregnancy and be free from infectious or traumatic complications. It should also be simple to perform and cost effective, with minimal inter-observer variability. A number of techniques have been devised to retrieve TCC samples from the endocervical canal and the endometrial cavity, including smears obtained with cotton swabs or a cytobrush (14–16), aspiration of cervical mucus with a catheter (17–19), endometrial biopsy with a Pipelle (20), and lavage of the endocervical canal (21–24) or the uterine cavity (14;22;25), all with variable success (Table 1).
Table 1.
Comparison of different methods of transcervical cell sample collection
| Method | Description | % of TCC sample with fetal cells | References |
|---|---|---|---|
| Antenatal cell extractor | Flushing of endocervical canal with saline to recover fetal cells | 50% |
18, 19 17 |
| Transcervical smears with cytobrush | Application of the cytobrush 2–3 cm into the endocervix and 360° rotation while the brush is being withdrawn to get cervical mucus. | 96.7% 23.9%* 95% |
15
14 16 |
| Aspiration with Pipelle De Cornier and Aspiracath | Introduction of the Pipelle or Aspiracath into the cervix to aspirate cervical canal mucus before elective termination of pregnancy | 50% of the aspirates | 20 |
| Endocervical Canal lavage | Application of a flexible plastic catheter connected to a 10 cc syringe into the endocervix which is flushed and immediately aspirated back to obtain cellular content. | 83.3% 50% 59.2% |
22
21 23 24 |
| Intrauterine lavage with or without ultrasound guidance | Utilization of a thin flexible plastic catheter connected to a 10 cc syringe, this is introduced passed the internal cervical os with or without transabdominal ultrasound guidance before 5 cc of saline is instilled and aspirated back under gentle suction. | 84.8% 82% 92.3% with ultrasound guidance |
25
14 22 |
TCC: Transcervical cells
Correct male sex prediction.
At present, the existing literature differs vastly and is often contradictory in projecting the relative efficacy of the various methods for retrieving fetal cells. Previously, emphasis has been placed on the feasibility of obtaining fetal cells and establishing their diagnostic utility, rather than a direct comparison of the relative efficacy of the various methods in randomized control trials, as recently reported (14;22). It has been noted that the post-collection processing of the TCC samples has tremendous variation from one study to another (26), which directly affects the yield of useful information. Techniques used to identify the fetal cells and the diagnostic end points (fetal sex vs gene disorders) have also differed, yielding heterogeneous groups for comparison with non-uniform results. Thus, there is a lack of information on well-described techniques for sample collection and analysis, resulting in considerable dependence on the technique and skill of individual operators.
In the landmark 1971 report by Shettles, identification of the Y chromosome was used to determine fetal sex from midcervical mucus samples obtained with cotton swabs (10). A limitation of using cotton swabs to retrieve TCC samples is the entrapment of cells within the cotton, which may reduce yield (9;27). The use of a cytobrush for cervical mucus retrieval or lavage of the endocervical canal with normal saline offers viable alternatives for TCC collection. A cytobrush inserted through the external os to a maximum depth of 2 cm and rotated a full turn during removal provides fetal cells in diagnostic quantities (15). However, other investigators failed to reproduce this success (26). Aspiration of the endocervical mucus with a single cannula also results in the detection of fetal cells in up to 70% of TCC samples from mothers with male fetuses (9). Furthermore, Kingdom et al. demonstrated that lavage of the endocervical canal retrieves more trophoblast cells than the cytobrush, and that cytobrush specimens may have a higher incidence of debris and maternal endocervical cells (27). A more effective method in terms of fetal cell yield is intrauterine lavage (IUL), in which a flexible catheter connected to a syringe filled with normal saline is used to flush the endometrial cavity. IUL and the other methods for TCC sampling are illustrated in an article by Adinolfi and Sherlock (12).
Reported rates of correct sex prediction are 25–70% for mucus collection (9;14) as compared to 75–90% for IUL samples (22;28). In particular, Cioni et al. (14) compared the outcomes of mucus collection to that of IUL in paired samples from a study population of 123 women between 6 to 12 weeks of gestation. Using quantitative fluorescence-PCR (QF-PCR) and fluorescence in situ hybridization (FISH), the correct sex prediction was obtained in 111 out of 123 cases with IUL (90.2%), and in 69 out of 123 cases with mucus collection (56%). Interestingly, Ergin et al. compared TCC retrieval by endocervical lavage to IUL (22). Although sufficient cell retrieval and correct prediction of fetal sex was more common by IUL, the difference in the two groups was not statistically significant due to the small sample size (n=25).
On a cautionary note, IUL may be considered more invasive than mucus collection, although a systemic investigation directed at the analysis of complications has never been performed. The majority of information on techniques of cervical cell retrieval derives from study populations composed primarily of pregnancies to be terminated. Chou et al. reported a 35 year old patient who underwent a uterine lavage procedure for prenatal sex determination at 7–8 weeks gestation and delivered a fetus with severe reduction defects of all four limbs at 38 weeks gestation (29).
Only one study attempted the retrieval of trophoblast cells from the cervix using a cytobrush in ongoing pregnancies, which resulted in the isolation of fetal cells in 35 of 37 normal intrauterine pregnancies sampled (95%), with no direct adverse outcome in the ongoing pregnancies (16). However, one patient with multiple previous episodes of first trimester bleeding that was sampled on her first prenatal visit had a spontaneous miscarriage one week later. In this study, cells were collected by rinsing the cytobrush in a fixative solution (ThinPrep kit, Hologic Corp., Marlborough, MA), which may have contributed to the high efficiency of trophoblast recovery. Because of the existence of very limited data, larger prospective studies incorporating normal ongoing pregnancies need to be conducted with long term follow up to detect possible adverse outcomes before it is considered safe to perform prenatal diagnosis by TCC sampling. This method for TCC sampling, as well as the others that have been proposed, can be performed in an office setting as an outpatient procedure. With the ThinPrep kit, cells in the fixative are stable for hours at room temperature and may be stored refrigerated for weeks.
POST-COLLECTION PROCESSING
The abundance of fetal cells acquired by TCC sampling depends on the method of collection and post-collection processing of the specimens. Along with fetal cytotrophoblast and syncitiotrophoblast cells, TCC specimens also contain maternal squamous cells, blood elements, spermatozoa and varying degrees of particulate contaminants. It is thought that gestational age can influence the proportion of fetal cells obtained (13). Reports indicate that cellular clumps within specimens often contain significant numbers of fetal cells. It is suggested that discarding the cellular clumps to obtain a single cell layer of cells for immunohistochemistry or FISH analysis can produce an underestimate of cell recovery (9). The cellular morphology should also be assessed, as spermatozoa and other contaminants can easily be identified.
Studies have evaluated the utility of mucolytics such as acetylcystine to digest cervical mucus in specimens for better identification of the cells. Fang et al. reported the presence of fetal cells in 60% of TCC samples retrieved by endocervical mucus aspiration using a cervical catheter made from an infant feeding tube (23). These investigators did not use any mucolytics and directly fixed their samples after analyzing the cellularity. Similarly, Fejgin et al. reported good results without using mucolytics (15). Others (21;30;31) reported using mucolytics only when the mucus was thick without affecting the specimen quality. In still another study, Cioni and colleagues (26) compared the TCC specimens collected by cytobrush that were processed using N-acetylcysteine, dithiothreitol, or no mucolytic, and reported neither to be superior. While these reports suggest that mucolytics may not be a major factor affecting the frequency of fetal cell detection, acidification of fixed samples collected by cytobrush disrupted mucus sufficiently for efficient immunohistochemical analysis after slide preparation by cytospin centrifugation (16). However, studies controlling for other confounding factors that influence fetal cell recovery are lacking.
METHODS OF ANALYSIS
Various types of useful diagnostic information can be obtained from TCC samples, depending on how the specimens are analyzed (Fig. 1). Most reported studies of fetal cells obtained from the cervical canal or by IUL are based on the identification of fetal sex and chromosome analysis using FISH or QF-PCR. A few studies have reported the use immunohistochemistry to identify fetal cells based on their expression of specific trophoblast marker proteins.
1. Genetic approach
Compared to standard karyotyping, FISH and QF-PCR can be exquisitely sensitive for the analysis of TCC and IUL samples. FISH directly labels specific genes or groups of genes with fluorescent dyes so that it is possible to visualize the number of copies of genes or chromosomes within individual cells, a perfect approach for analysis of the mixed cell populations found in cervical specimens. A wide variety of FISH probes are available that can be applied in the detection of aneuploidies on freshly fixed cells or fixed tissues stored in paraffin blocks. PCR amplification in the presence of DNA-binding dyes permits quantification of the relative number of copies of a gene in a small population of cells. The use of QF-PCR for prenatal diagnosis eliminates the need to culture and expand fetal cells and allows rapid diagnosis of some selected chromosomal anomalies (13, 18, 21, and X and Y)(32).
Table 2 summarizes results obtained by FISH and PCR to predict the sex of male fetuses using TCC samples collected by transcervical swab (33), cervical mucus (14;26), endocervical lavage (27;34–37), cytobrush (26) aspiration (38) or uterine lavage (14;25;39;40). FISH results were comparable to PCR in fetal sex determination. According to Bussani et al. (41), FISH revealed that 45/89 (51%) placental samples were derived from pregnancies with male fetuses. Correct sexing of the IUL samples from male pregnancies was achieved in 41/45 (91%) cases using the dual-FISH technique, and in 43/45 (95.5%) with PCR. All samples derived from male pregnancies tested for chromosome 21 were normal. From 57 samples subjected to micromanipulation, 51 (89.5%) showed discernible chorionic villous filaments or cell clumps of possible trophoblastic origin (41). Similarly, other authors have been able to show high rates of fetal cell detection in TCC and IUL samples using FISH (20;22;28;39).
Table 2.
Incidence of trophoblast cells in transcervical cell samples retrieved from mothers with male fetuses
| Method of sample collection | Method of sample analysis | References | |
|---|---|---|---|
| FISH | PCR | ||
| Transcervical swabs | - | 9/9 | 33 |
| Cervical mucus | 7/23 | 7/23 | 26 |
| 16/67 | 16/67 | 14 | |
| Endocervical lavage (10 ml) | 7/7 | 6/6 | 34 |
| 5/6 | 6/6 | 27 | |
| 6/6 | - | 35 | |
| 8/8 | - | 36 | |
| Endocervical lavage (5 ml) | 1/1 | 2/2 | 35 |
| Endocervical Lavage (3 ml) | - | 7/7 | 37 |
| Cytobrush | 4/6 | 6/6 | 26 |
| 20/66 | 20/66 | 38 | |
| Aspiration | 12/13 | 12/13 | 40 |
| Uterine Lavage (5–8 ml) | 10/12 | - | 14 |
| Uterine lavage (10 ml) | 13/13 | - | 25 |
| 55/67 | 55/67 | 39 | |
| - | 73/73 | 33 | |
| Uterine lavage (20 ml) | 5/5 | - | 26 |
FISH = fluorescence in-situ hybridization, PCR = polymerase chain reaction.
In addition to fetal sex determination and identification of major chromosomal abnormalities, TCC sampling has been applied successfully in the diagnosis of fetal Rh(D) blood group in an Rh(D)-negative mother (42) and hemoglobinopathy screens for thalasemia and sickle cell (43). With the technology of single cell DNA allelic profiling, TCC is feasible for more sophisticated genetic diagnostic procedures (20).
2. Immunohistochemical approaches
TCC samples have been examined by immunohistochemical staining with monoclonal antibodies (33;34;44). In 1995, Bulmer et al. used monoclonal antibodies (MAbs) FT 1.41.1, NDOG1 and 340 to identify syncytial fragments, and MAbs BC1 and NDOG5 to identify cytotrophoblasts (44). However, the detection rate depends on the method of sample collection; a 40% (4 of 10 cervical aspirate samples) detection rate was associated with cervical mucus aspiration, while a rate of 75% was obtained for IUL (9 of 12 lavage samples).
Human leukocyte antigen (HLA)-G is a class Ib major histocompatability complex protein that is expressed by human extravillous cytotrophoblast cells and is absent in all other uterine and placental cell populations (45–47). In 2003, Bulmer et al. employed MAbs against HLA-G to identify cytotrophoblasts cells in TCC samples retrieved by IUL (21). Cytotrophoblast cells characterized by their large, irregular hyperchromatic nuclei were HLA-G positive and were identified in 12 of 23 (52%) TCC samples. Interestingly, molecular examination of DNA by QF-PCR in HLA-G positive elements collected by laser capture micro-dissection from four of the patients revealed fetal markers, demonstrating the utility of this approach for prenatal genetic diagnosis (21). The combined immunohistochemical and molecular approach used in this study revealed considerable variation between the samples. The sensitivity of MAb labeling was relatively low even though HLA-G reactivity provides high specificity for identification of fetal-derived trophoblast cells. HLA-G is expressed by extravillous cytotrophoblast cellular elements, but not by syncytial fragments, limiting its ability to identify all fetal cells (46). The necessity for a set of MAbs reacting exclusively against antigens expressed on specific subpopulations of trophoblast cells will be crucial for an immunohistochemical approach to identify fetal cells comprehensively. More recently, it was demonstrated that extravillous cytotrophoblast cells could be consistently (>95% of specimens) identified using HLA-G as an antigenic marker in TCC specimens collected by cytobrush into a fixative rinse and prepared on microscope slides free of interfering mucus (16). Slides stained with the same antibody against HLA-G used by Bulmer et al. (21) and counterstained with hematoxylin reveal a small number of antibody-labeled cytotrophoblast cells on a dense background of cervical cell nuclei (Figure 2). Trophoblast frequency was approximately one in two thousand for all pregnancies successfully sampled between gestation weeks six and fourteen, while this value was reduced four to five-fold in specimens retrieved from women with ectopic pregnancy or blighted ovum (16). These findings suggest that, in addition to genetic testing, information can be gleaned from TCC analysis alerting clinicians to at-risk pregnancies.
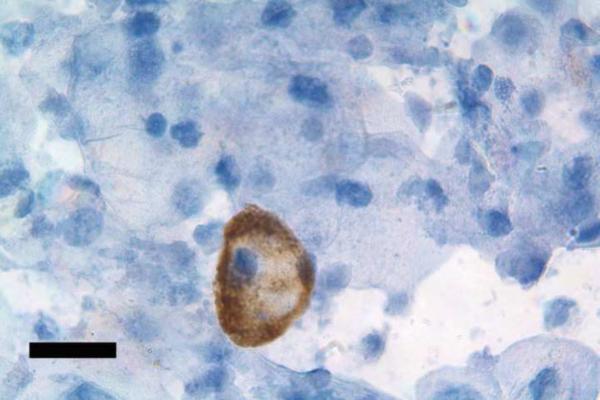
Figure 2.
HLA-G positive cytotrophoblast cell in a transcervical specimen. The slide was prepared as describe by Imudia et al. (18) from a specimen obtained during the first trimester of pregnancy. Antibody labeling was detected using monoclonal antibody G233 against HLA-G and a secondary antibody conjugated to peroxidase to produce a dark brown precipitate around the cytotrophoblast cell. Counterstaining with hematoxylin reveals nuclei (blue) of cervical cells, which are abundant in the specimen. Bar, 20 μm.
CONCLUSIONS
The recovery and analysis of fetal cells shed from the placenta into the cervical canal could provide wider availability of prenatal genetic diagnostics to the general patient population. With improvements in the efficacy and safety of trophoblast collection by TCC sampling using the cytobrush, and in the identification and isolation of those cells expressing trophoblast markers, small quantities of fetal DNA could be readily obtained for genetic testing. New sensitive technologies, such as those now under development for analysis of fetal DNA in maternal serum, could yield extensive information about the fetal genome from modest numbers of isolated cells. The ability to procure cytotrophoblast cells by TCC as early as six weeks of gestation could make this vital information available much earlier than current technologies, including the analysis of fetal DNA in maternal serum. Over the next few years, more studies using TCC sampling for prenatal diagnosis of chromosome abnormalities, paternity testing, screening for abnormal pregnancies in the first trimester and early diagnosis of obstetrical problems are expected. It is hoped that these advances will provide new and safer choices for pregnant women throughout the world who are in need of prenatal diagnosis, and a reliable tool for prediction of abnormal obstetric outcomes.
Capsule.
Transcervical sampling of trophoblast cells is a hopeful, non-invasive alternative for prenatal diagnosis of genetic disorders and pregnancy abnormalities in the first trimester.
ACKNOWLEDGEMENTS
This work was supported, in part, by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, DHHS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: There is no potential conflict of interest to be disclosed.
REFERENCES
- 1.Egan JF, Benn P, Borgida AF, Rodis JF, Campbell WA, Vintzileos AM. Efficacy of screening for fetal Down syndrome in the United States from 1974 to 1997. Obstet Gynecol. 2000;96:979–85. doi: 10.1016/s0029-7844(00)01044-9. [DOI] [PubMed] [Google Scholar]
- 2.O'Leary P, Breheny N, Dickinson JE, Bower C, Goldblatt J, Hewitt B, et al. First-trimester combined screening for Down syndrome and other fetal anomalies. Obstet Gynecol. 2006;107:869–76. doi: 10.1097/01.AOG.0000207562.09858.16. [DOI] [PubMed] [Google Scholar]
- 3.Mujezinovic F, Alfirevic Z. Procedure-related complications of amniocentesis and chorionic villous sampling: a systematic review. Obstet Gynecol. 2007;110:687–94. doi: 10.1097/01.AOG.0000278820.54029.e3. [DOI] [PubMed] [Google Scholar]
- 4.Handyside AH, Kontogianni EH, Hardy K, Winston RM. Pregnancies from biopsied human preimplantation embryos sexed by Y-specific DNA amplification. Nature. 1990;344:768–70. doi: 10.1038/344768a0. [DOI] [PubMed] [Google Scholar]
- 5.Steele CD, Wapner RJ, Smith JB, Haynes MK, Jackson LG. Prenatal diagnosis using fetal cells isolated from maternal peripheral blood: a review. Clin Obstet Gynecol. 1996;39:801–13. doi: 10.1097/00003081-199612000-00009. [DOI] [PubMed] [Google Scholar]
- 6.Lo YM. Noninvasive prenatal detection of fetal chromosomal aneuploidies by maternal plasma nucleic acid analysis: a review of the current state of the art. BJOG. 2009;116:152–7. doi: 10.1111/j.1471-0528.2008.02010.x. [DOI] [PubMed] [Google Scholar]
- 7.Chiu RW, Chan KC, Gao Y, Lau VY, Zheng W, Leung TY, et al. Noninvasive prenatal diagnosis of fetal chromosomal aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. Proc Natl Acad Sci U S A. 2008;105:20458–63. doi: 10.1073/pnas.0810641105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lun FM, Tsui NB, Chan KC, Leung TY, Lau TK, Charoenkwan P, et al. Noninvasive prenatal diagnosis of monogenic diseases by digital size selection and relative mutation dosage on DNA in maternal plasma. Proc Natl Acad Sci U S A. 2008;105:19920–5. doi: 10.1073/pnas.0810373105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adinolfi M, Sherlock J. First trimester prenatal diagnosis using transcervical cells: an evaluation. Hum Reprod Update. 1997;3:383–92. doi: 10.1093/humupd/3.4.383. [DOI] [PubMed] [Google Scholar]
- 10.Shettles LB. Use of the Y chromosome in prenatal sex determination. Nature. 1971;230:52–3. doi: 10.1038/230052b0. [DOI] [PubMed] [Google Scholar]
- 11.Cioni R, Bussani C, Bucciantini S, Scarselli G. Fetal cells in a transcervical cell sample collected at 5 weeks of gestation. J Matern Fetal Neonatal Med. 2005;18:271–3. doi: 10.1080/14767050500246391. [DOI] [PubMed] [Google Scholar]
- 12.Adinolfi M, Sherlock J. Fetal cells in transcervical samples at an early stage of gestation. J Hum Genet. 2001;46:99–104. doi: 10.1007/s100380170095. [DOI] [PubMed] [Google Scholar]
- 13.Bischoff FZ, Simpson JL. Endocervical fetal trophoblast for prenatal genetic diagnosis. Curr Opin Obstet Gynecol. 2006;18:216–20. doi: 10.1097/01.gco.0000192985.22718.17. [DOI] [PubMed] [Google Scholar]
- 14.Cioni R, Bussani C, Scarselli B, Bucciantini S, Marchionni M, Scarselli G. Comparison of two techniques for transcervical cell sampling performed in the same study population. Prenat Diagn. 2005;25:198–202. doi: 10.1002/pd.1104. [DOI] [PubMed] [Google Scholar]
- 15.Fejgin MD, Diukman R, Cotton Y, Weinstein G, Amiel A. Fetal cells in the uterine cervix: a source for early non-invasive prenatal diagnosis. Prenat Diagn. 2001;21:619–21. doi: 10.1002/pd.117. [DOI] [PubMed] [Google Scholar]
- 16.Imudia AN, Suzuki Y, Kilburn BA, Yelian FD, Diamond MP, Romero R, et al. Retrieval of trophoblast cells from the cervical canal for prediction of abnormal pregnancy: a pilot study. Hum Reprod. 2009;24:2086–92. doi: 10.1093/humrep/dep206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldberg MF, Chen AT, Ahn YW, Reidy JA. First-trimester fetal chromosomal diagnosis using endocervical lavage: a negative evaluation. Am J Obstet Gynecol. 1980;138:436–40. doi: 10.1016/0002-9378(80)90142-8. [DOI] [PubMed] [Google Scholar]
- 18.Rhine SA, Cain JL, Cleary RE, Palmer CG, Thompson JF. Prenatal sex detection with endocervical smears: successful results utilizing Y-body fluoroscence. Am J Obstet Gynecol. 1975;122:155–60. doi: 10.1016/s0002-9378(16)33486-x. [DOI] [PubMed] [Google Scholar]
- 19.Rhine SA, Palmer CG, Thompson JF. A simple alternative to amniocentesis for first trimester prenatal diagnosis. Birth Defects Orig Artic Ser. 1977;13:231–47. [PubMed] [Google Scholar]
- 20.Katz-Jaffe MG, Mantzaris D, Cram DS. DNA identification of fetal cells isolated from cervical mucus: potential for early non-invasive prenatal diagnosis. BJOG. 2005;112:595–600. doi: 10.1111/j.1471-0528.2004.00506.x. [DOI] [PubMed] [Google Scholar]
- 21.Bulmer JN, Cioni R, Bussani C, Cirigliano V, Sole F, Costa C, et al. HLA-G positive trophoblastic cells in transcervical samples and their isolation and analysis by laser microdissection and QF-PCR. Prenat Diagn. 2003;23:34–9. doi: 10.1002/pd.511. [DOI] [PubMed] [Google Scholar]
- 22.ErgIn T, Baltaci V, Zeyneloglu HB, Duran EH, ErgenelI MH, Batioglu S. Non-invasive early prenatal diagnosis using fluorescent in situ hybridization on transcervical cells: comparison of two different methods for retrieval. Eur J Obstet Gynecol Reprod Biol. 2001;95:37–41. doi: 10.1016/s0301-2115(00)00357-2. [DOI] [PubMed] [Google Scholar]
- 23.Fang CN, Kan YY, Hsiao CC. Detection of fetal cells from transcervical mucus plug before first-trimester termination of pregnancy by cytokeratin-7 immunohistochemistry. J Obstet Gynaecol Res. 2005;31:500–7. doi: 10.1111/j.1447-0756.2005.00329.x. [DOI] [PubMed] [Google Scholar]
- 24.Bahado-Singh RO, Kliman H, Feng TY, Hobbins J, Copel JA, Mahoney MJ. First-trimester endocervical irrigation: feasibility of obtaining trophoblast cells for prenatal diagnosis. Obstet Gynecol. 1995;85:461–4. doi: 10.1016/0029-7844(94)00416-b. [DOI] [PubMed] [Google Scholar]
- 25.Bussani C, Cioni R, Mattei A, Fambrini M, Marchionni M, Scarselli G. Prenatal diagnosis of common aneuploidies in transcervical samples using quantitative fluorescent-PCR analysis. Mol Diagn Ther. 2007;11:117–21. doi: 10.1007/BF03256231. [DOI] [PubMed] [Google Scholar]
- 26.Cioni R, Bussani C, Scarselli B, Bucciantini S, Barciulli F, Scarselli G. Fetal cells in cervical mucus in the first trimester of pregnancy. Prenat Diagn. 2003;23:168–71. doi: 10.1002/pd.562. [DOI] [PubMed] [Google Scholar]
- 27.Kingdom J, Sherlock J, Rodeck C, Adinolfi M. Detection of trophoblast cells in transcervical samples collected by lavage or cytobrush. Obstet Gynecol. 1995;86:283–8. doi: 10.1016/0029-7844(95)00127-d. [DOI] [PubMed] [Google Scholar]
- 28.Cioni R, Bussani C, Scarselli B, Barciulli F, Bucciantini S, Simi P, et al. Detection of fetal cells in intrauterine lavage samples collected in the first trimester of pregnancy. Prenat Diagn. 2002;22:52–5. doi: 10.1002/pd.236. [DOI] [PubMed] [Google Scholar]
- 29.Chou MM, Lin SK, Ho ES. Severe limb reduction defects after uterine lavage at 7–8 weeks' gestation. Prenat Diagn. 1997;17:77–80. [PubMed] [Google Scholar]
- 30.Rodeck C, Tutschek B, Sherlock J, Kingdom J. Methods for the transcervical collection of fetal cells during the first trimester of pregnancy. Prenat Diagn. 1995;15:933–42. doi: 10.1002/pd.1970151008. [DOI] [PubMed] [Google Scholar]
- 31.Tutschek B, Sherlock J, Halder A, Delhanty J, Rodeck C, Adinolfi M. Isolation of fetal cells from transcervical samples by micromanipulation: molecular confirmation of their fetal origin and diagnosis of fetal aneuploidy. Prenat Diagn. 1995;15:951–60. doi: 10.1002/pd.1970151010. [DOI] [PubMed] [Google Scholar]
- 32.Nicolini U, Lalatta F, Natacci F, Curcio C, Bui TH. The introduction of QF-PCR in prenatal diagnosis of fetal aneuploidies: time for reconsideration. Hum Reprod Update. 2004;10:541–8. doi: 10.1093/humupd/dmh046. [DOI] [PubMed] [Google Scholar]
- 33.Griffith-Jones MD, Miller D, Lilford RJ, Scott J, Bulmer J. Detection of fetal DNA in trans-cervical swabs from first trimester pregnancies by gene amplification: a new route to prenatal diagnosis? Br J Obstet Gynaecol. 1992;99:508–11. doi: 10.1111/j.1471-0528.1992.tb13792.x. [DOI] [PubMed] [Google Scholar]
- 34.Adinolfi M, Davies A, Sharif S, Soothill P, Rodeck C. Detection of trisomy 18 and Y-derived sequences in fetal nucleated cells obtained by transcervical flushing. Lancet. 1993;342:403–4. doi: 10.1016/0140-6736(93)92816-c. [DOI] [PubMed] [Google Scholar]
- 35.Adinolfi M, Sherlock J, Tutschek B, Halder A, Delhanty J, Rodeck C. Detection of fetal cells in transcervical samples and prenatal diagnosis of chromosomal abnormalities. Prenat Diagn. 1995;15:943–9. doi: 10.1002/pd.1970151009. [DOI] [PubMed] [Google Scholar]
- 36.Ishai D, Amiel A, Diukman R, Cogan O, Lichtenstein Z, Abramovici H, et al. Uterine cavity lavage: adding FISH to conventional cytogenetics for embryonic sexing and diagnosing common chromosomal aberrations. Prenat Diagn. 1995;15:961–5. doi: 10.1002/pd.1970151011. [DOI] [PubMed] [Google Scholar]
- 37.Massari A, Novelli G, Colosimo A, Sangiuolo F, Palka G, Calabrese G, et al. Non-invasive early prenatal molecular diagnosis using retrieved transcervical trophoblast cells. Hum Genet. 1996;97:150–5. doi: 10.1007/BF02265257. [DOI] [PubMed] [Google Scholar]
- 38.Briggs J, Miller D, Bulmer JN, Griffith-Jones M, Rame V, Lilford R. Non-syncytial sources of fetal DNA in transcervically recovered cell populations. Hum Reprod. 1995;10:749–54. doi: 10.1093/oxfordjournals.humrep.a136026. [DOI] [PubMed] [Google Scholar]
- 39.Chang SD, Lin SL, Chu KK, Hsi BL. Minimally-invasive early prenatal diagnosis using fluorescence in situ hybridization on samples from uterine lavage. Prenat Diagn. 1997;17:1019–25. [PubMed] [Google Scholar]
- 40.Daryani YP, Penna LK, Patton MA. Detection of cells of fetal origin from transcervical irrigations. Prenat Diagn. 1997;17:243–8. doi: 10.1002/(sici)1097-0223(199703)17:3<243::aid-pd44>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 41.Bussani C, Cioni R, Scarselli B, Barciulli F, Bucciantini S, Simi P, et al. Strategies for the isolation and detection of fetal cells in transcervical samples. Prenat Diagn. 2002;22:1098–101. doi: 10.1002/pd.469. [DOI] [PubMed] [Google Scholar]
- 42.Adinolfi M, Sherlock J, Kemp T, Carritt B, Soothill P, Kingdom J, et al. Prenatal detection of fetal RhD DNA sequences in transcervical samples. Lancet. 1995;345:318–9. doi: 10.1016/s0140-6736(95)90306-2. [DOI] [PubMed] [Google Scholar]
- 43.Adinolfi M, el-Hashemite N, Sherlock J, Ward RH, Petrou M, Rodeck C. Prenatal detection of Hb mutations using transcervical cells. Prenat Diagn. 1997;17:539–43. doi: 10.1002/(sici)1097-0223(199706)17:6<539::aid-pd106>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 44.Bulmer JN, Rodeck C, Adinolfi M. Immunohistochemical characterization of cells retrieved by transcervical sampling in early pregnancy. Prenat Diagn. 1995;15:1143–53. doi: 10.1002/pd.1970151211. [DOI] [PubMed] [Google Scholar]
- 45.Chumbley G, King A, Gardner L, Howlett S, Holmes N, Loke YW. Generation of an antibody to HLA-G in transgenic mice and demonstration of the tissue reactivity of this antibody. J Reprod Immunol. 1994;27:173–86. doi: 10.1016/0165-0378(94)90002-7. [DOI] [PubMed] [Google Scholar]
- 46.Loke YW, King A, Burrows T, Gardner L, Bowen M, Hiby S, et al. Evaluation of trophoblast HLA-G antigen with a specific monoclonal antibody. Tissue Antigens. 1997;50:135–46. doi: 10.1111/j.1399-0039.1997.tb02852.x. [DOI] [PubMed] [Google Scholar]
- 47.McMaster MT, Librach CL, Zhou Y, Lim KH, Janatpour MJ, DeMars R, et al. Human placental HLA-G expression is restricted to differentiated cytotrophoblasts. J Immunol. 1995;154:3771–8. [PubMed] [Google Scholar]