SUMMARY
Protein Histidine Kinases (PHKs) function in Two Component Signaling pathways utilized extensively by bacteria and archaea. Many PHKs participate in three distinct, but interrelated signaling reactions: autophoshorylation, phosphotransfer (to a partner Response Regulator (RR) protein), and dephosphorylation of this RR. Detailed biochemical and structural characterization of several PHKs have revealed how the domains of these proteins can interact to assemble the three active sites that promote the necessary chemistry and how these domain interactions might be regulated in response to sensory input: the relative orientation of helices in the PHK dimerization domain can reorient, via cogwheeling (rotation) and kinking (bending), to effect changes in PHK activities that likely involve sequestration/release of the PHK catalytic domain by the dimerization domain.
INTRODUCTION
Scope and Perspective of this Review
Protein histidine kinases (PHKs) that function in Two Component Signaling pathways (TCSs) are ubiquitous in the prokaryotic world. These systems allow bacterial and archaeal cells to sense and respond to a wide variety of stimuli ranging from physical conditions (temperature, osmolarity, light) to concentrations of specific chemicals (nutrients, chemical signals for quorum sensing) [1]. In many TCSs, PHKs serve as receptors for stimuli and as regulators that control the activity of downstream signaling components (Response Regulators) via phosphorylation. In each such system, the PHK autophosphorylates on a specific histidine side chain (hereafter referred to as the phospho-accepting His), and then this phosphoryl group is passed to a cognate Response Regulator (RR), a modification that alters the activity of the RR. Most RRs are DNA-binding proteins that function as activators or inhibitors of transcription in a phosphorylation-dependent manner [2]. In addition to >10,000 cataloged examples of PHKs in prokaryotes [3•], there are some that have been found in eukaryotes: mostly fungi , amoebae, and land plants, but not metazoans [4]. Defining how these enzymes function is important for understanding the machinery utilized by many organisms to perceive and respond to their worlds. Further interest in PHKs stems from observations that some regulate expression of cell components vital for survival and/or virulence in pathogenic microbes, and so they might be exploited as targets for new antimicrobial drugs [5–8]. Such efforts would benefit from a detailed understanding of PHK biochemistry and their structural/functional organization.
TCSs and PHKs have been the subject of many insightful reviews that have summarized various aspects of their common activities and sequences [9,10], their structures [3•], and their evolution [11–13]. This review adopts a different perspective, focusing on just two of the many interesting aspects of PHKs: their active site structures and the possible mechanisms underlying regulation of the activities of these active sites.
PHK activities
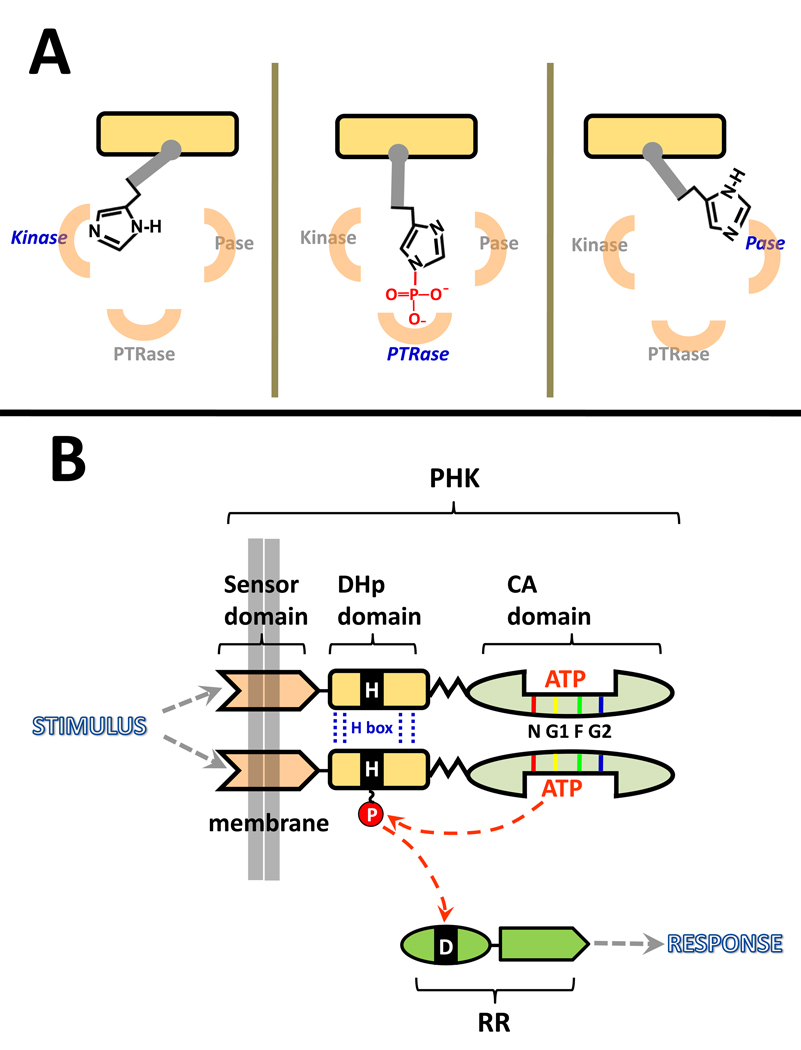
From an enzymology perspective, PHKs are interesting because many participate in three distinct, but related, phosphotransfer reactions: autophosphorylation (phosphotransfer from ATP to a histidine side chain), phosphorylation of a cognate response regulator (RR) protein (phosphotransfer from P~His to an aspartate side chain), and dephosphorylation of the P~RR (phosphotransfer from P~Asp to water). This review will consider the first two of these activities in some detail, but the third is the subject of a separate review in this issue [14], and so it will be described only briefly here. In all three of these reactions, the phospho-accepting His of the PHK is a central player. One can envision these enzymes functioning by toggling this His among three alternative positions, as depicted in Fig. 1A. This toggling would assemble three distinct active sites by: (i) positioning the His (or P~His) in close proximity to a phosphodonor or phosphoacceptor, and (ii) placing the His (P~His) in a mileau of functional groups that tune its reactivity in appropriate ways. What does this His encounter at each active site? How does it get from one site to another? Below, I will address these questions by first summarizing current understanding of the autokinase active site and the phosphotransfer active site, and then I will consider how toggling of the phospho-accepting His from one site to the other might be accomplished by PHKs and regulated in response to stimuli. To follow this discussion it is important to have a basic understanding of the structural organization of PHKs.
Figure 1.
Schematic diagram of the role played by PHKs in two-component signal transduction systems (TCSs). (A) Many PHKs have three distinct but interrelated enzymatic activities that involve positioning the phospho-accepting His in three active sites. The active sites for phosphotransferase (PTRase) and phosphatase(Pase) activities are likely to be very similar but are portrayed as being physically distinct for the purpose of illustration. Although the diagram depicts the phospho-accepting His rotating from site to site, this reorientation might also involve movement of the active sites relative to an essentially stationary phospho-accepting His. (B) PHKs function as homodimers that autophosphorylate then pass their phosphoryl group to an aspartate side chain located in the receiver domain of a cognate response regulator protein. Each PHK monomer has three distinct structural/functional domains: a transmembrane sensor, a DHp domain, and a CA domain. Sequence comparisons have defined a homology box within DHp (H box) that spans the phospho-accepting histidine. In addition, there are homology boxes (5–10 amino acids) located within the CA domain at/near the ATP binding pocket: N, G1 (sometimes called the D box), and G2 (sometimes called the G box) are conserved in all PHKs, while the F box is present in some, but not all, PHKs (note that DesK portrayed in Fig. 2 and Fig. 3 lacks the lacks an F box) [9,61,62]. The diagram depicts autophosphorylation via an intradimer cis mechanism; some PHKs utilize a trans mechanism.
PHK Domain Architecture
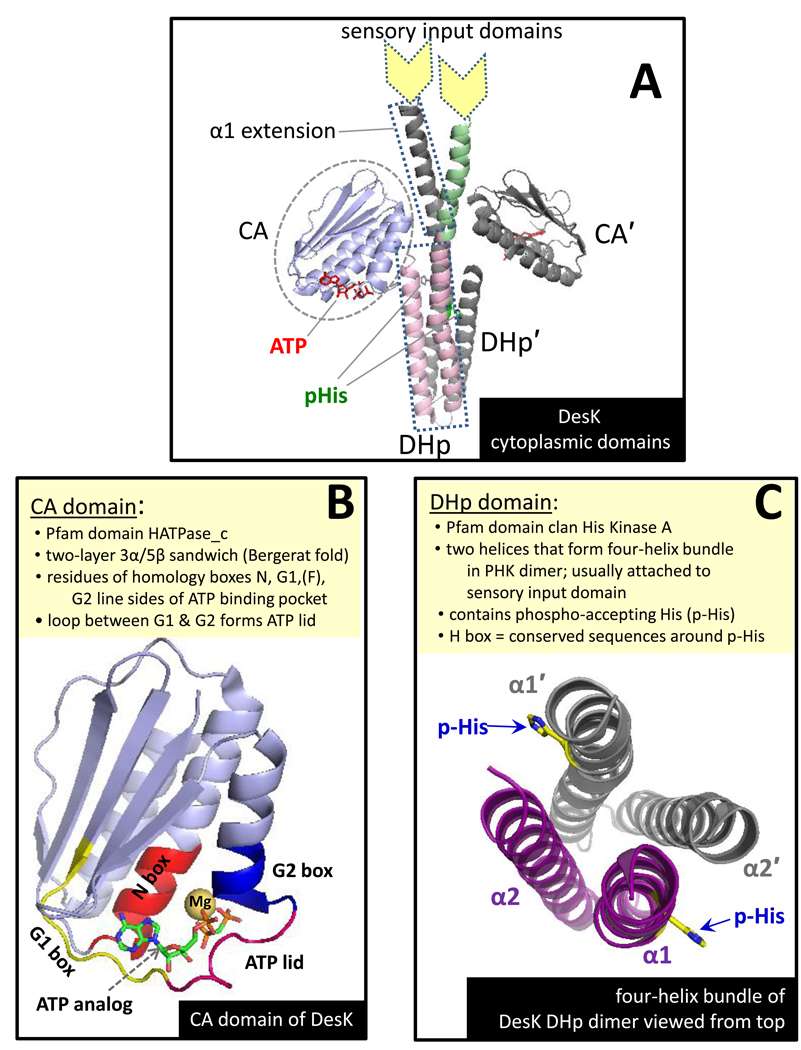
PHKs have a modular architecture with distinct structural domains playing different functional roles (Fig. 1B) [3•,9,15]. Most have an amino-terminal sensor domain (stimulus-specific, not conserved) that spans the membrane [16,17]. This domain connects to a conserved cytoplasmic domain called the DHp (dimerization and histidine phosphotransfer) domain which, in turn, is connected to the CA domain (catalytic and ATP-binding), another conserved component of all PHKs . The phospho-accepting His resides in the DHp domain. Several aspects of the structures of the DHp and CA domains are summarized in Fig. 2. At present, there are no available structures for full-length PHKs, so the exact relative orientations of the sensor, DHp, and CA domains aren’t known. However, clearly the DHp and CA domains need to associate, at least transiently, to allow transfer of the phosphoryl group from ATP to the phospho-accepting His, and the sensor domain must have some mechanism to manipulate DHp-CA interaction to achieve regulation of PHK activity. To generate versions of PHKs that are amenable to analysis by high resolution methods (xray crystallography and NMR methods), researchers have removed the sensory domains, generating structures that I will refer to as DHp+CA proteins, and there are also high resolution structures available for isolated DHp [18 ,19] and CA domains [20–25].
Figure 2.
Summary of key features of PHK structure as illustrated with B. subtilis DesK. (A) The crystal structure of the butterfly-shaped dimer formed by a DesK fragment that includes the CA and DHp domains as well as a helical extension of α1 of the DHp. The protomer in back is colored grey. Color coding for the front protomer: CA domain (blue), DHp helices (pink), extension of DHp helix (green). This panel was modeled after Fig. 1 of Jacques et al. [55•] (B) The CA domain of DesK with nonhydrolyzable ATP analog ADPCP bound. The location of the conserved homology box residues are shown in different colors as well as the ‘ATP-lid’ (magenta loop that folds over the polyphosphate groups of ATP). Note that DesK does not have an F box [9]. (C) A top-down view of the four-helix bundle formed by the dimerized DHp domain (with CA domain and helical extensions removed for purposes of illustration). α1 and α2 of one protomer are purple; helices of the second protomer are grey. The short connector linking α1 to α2 is at the bottom of the helices from this perspective, and the membrane/sensor input side would be the top. This figure was created using coordinates from PDB ID 3GIE (DesKDHp+CA H188E mutant) manipulated in PyMol to replace the mutant side chain with the phospho-accepting His (H188).
For some well characterized PHKs, autophosphorylation occurs in trans within the PHK dimer (the phospho-accepting His in one protomer receives is phosphoryl group from the ATP molecule bound to the CA domain of the partner protomer) [26,27], but recent results demonstrate that at least some PHKs utilize a cis mechanism within the PHK dimer [28••]. At present, it isn’t clear whether there are any inherent advantages or consequences resulting from a PHK utilizing the cis versus the trans mechanism.
There are some ‘nonconventional PHKs’ that have a more elaborate domain organization than that presented above. For example, CheA, one of the most extensively studied PHKs, mediates chemotaxis signaling events in many prokaryotes. In CheA, a cytoplasmic protein, there is no membrane-spanning sensory input domain, and the phospho-accepting His is located in an HPt domain rather than in its dimerization domain [22,29]. Considerable progress has been made toward understanding the structure and biochemical mechanism of the CheA HPt domain [30–32•] and several other HPt-utilizing proteins [33,34], including yeast Ypd1 [35,36•]. There are other PHKs (‘hybrid PHKs’) that have an even more elaborate nonconventional domain organization than CheA, for example by including a receiver domain. Although this group constitutes ~25% of known PHKs [3•] they are beyond the scope of this review.
A GLIMPSE OF THE AUTOKINASE ACTIVE SITE
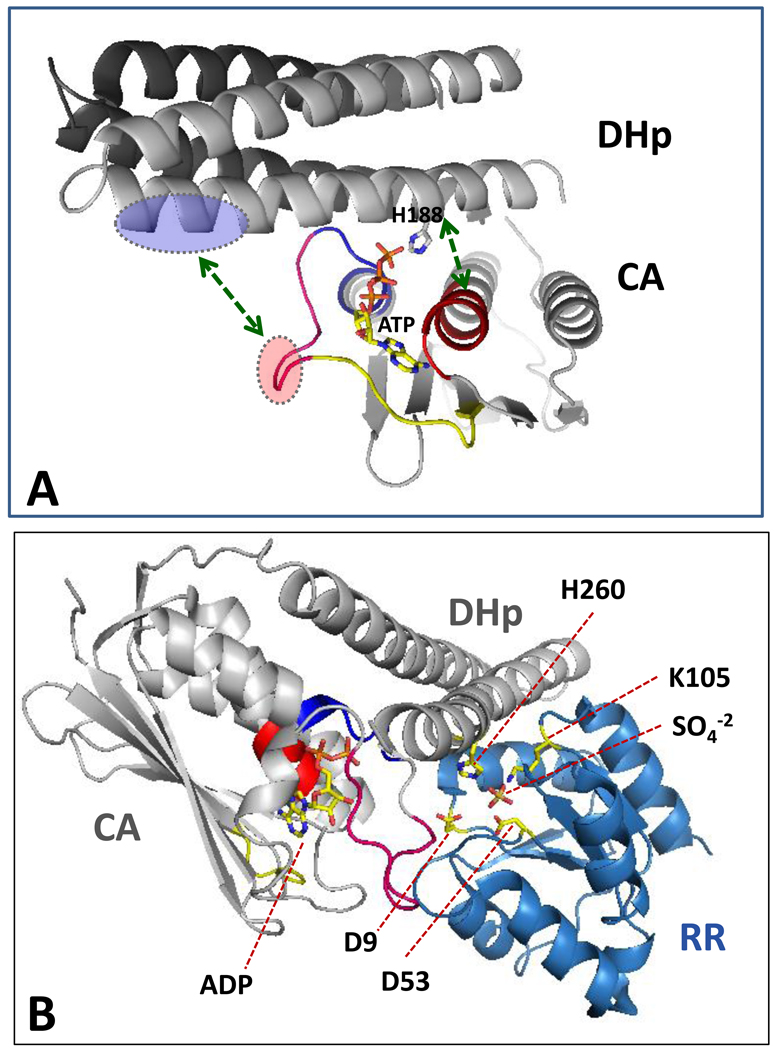
Visualizing a PHK active site with all of the expected components poised for catalysis is a goal that has eluded the PHK field, to date. The main obstacle here is the flexibility of the interdomain hinge connecting the DHp and CA domains. This flexibility appears to make it difficult to crystallize DHp+CA inclusive proteins, although the individual domains do readily crystallize. Recently, however, several groups have succeeded in crystallizing and characterizing nucleotide-bound DHp+CA versions of PHKs: HK853 (from Thermotoga maritima) [28••,37•]; DesK (a thermosensing PHK from Bacillus subtilis) [38••]; KinB (part of the sporulation activating pathway in Geobacillus stearothermophilus)[54]; and ThkA (from T. maritima) [39]. The HK853 and DesK DHp+CA proteins crystallize with their DHp and CA domains in different orientations that may provide key insights into how PHKs function. In some DHp+CA structures there is a clear and extensive binding interface between the two domains. However, these appear to represent kinase-incompetent conformations: the phospho-accepting His and the bound nucleotide are clearly not oriented in a manner appropriate for the autophosphorylation reaction. By contrast, in some of the DesK structures (e.g., Fig. 2A) there is no visible interaction between the DHp and CA domains. Using one of these structures as a starting point, Albanesi et al. [38••] found that by simply pivoting the CA domain (as a rigid body) on a Gly residue in the interdomain hinge region, they could bring the phospho-accepting His of the DesK DHp into close alignment with the γ-phosphate of ATP bound at the CA domain (Fig. 3A). This pivoting also aligned potential complementary binding surfaces located on DHp and the CA domains, and so it seems likely that this domain-pivoted structure provides an exciting first glimpse of an assembled PHK autokinase active site. The segments of the DHp and CA that mediate the interdomain contacts include the ATP lid of the CA and several clustered basic residues of the DHp, as well as salt bridges between DHp α1 side chains and residues in the N box of the CA domain (green arrows in Fig. 3A).
Figure 3.
Views of the PHK autokinase and phosphotransferase active sites. (A) A PHK poised for autophosphorylation: DesKDHp+CA with ATP bound. This figure was generated by manually docking the one DesK CA domain onto the four-helix bundle of a DHp dimer (as described by Albanesi et al. [38••] (docking by rigid body rotation about a pivot point in the hinge linking DHp to CA). Coordinates for this structure were kindly provided by Dr. Alejandro Buschiazzo and correspond to Fig. S4 in reference [38••]. This docking orients the phospho-accepting His near the γ –phosphoryl of bound ATP and allows some complementary interdomain electrostatic interactions (green arrows), for example between acidic side chains of the ATP-lid (red oval) and basic side chains of the DHp (blue oval). Color coding of homology boxes of the CA domain (N, G1, G2, and ATP-lid) is the same as in Fig. 2. (B) A PHK poised for phosphotransfer: HK853DHp+CA bound to RR468 (generated using PDB 3DGE). Two molecules of R468 bind to the dimeric HK853, but in this diagram only one molecule of each protein is shown to improve clarity (i.e., only two helices of the four-helix bundle are shown). The PHK:RR complex brings the phospho-accepting His (H260) of HK853 close to D53 of RR468 (the phosphorylation site of RR468) and close to RR side chains that catalyze phosphotransfer (e.g., D9, D10, M55, T83, and K105); a sulfate ion occupies a position that may mimic that of phosphate during phosphotransfer reactions. Two key RR side chains (M55 and K105) interact with the PHK (M55 with E348 in the CA domain; and K105 with R263 and T267 in the DHp domain); their abilities to contribute to catalysis of the phosphotransfer reaction might be influenced by these associations. However, RR468, like all response regulators, can catalyze its own phosphorylation using small molecule phosphodonors (such as acetyl phosphate) in the absence of any PHK [63], and so it is likely that, like other RRs, RR468 does the ‘heavy lifting’ in catalyzing the PHK→RR phosphotransfer reaction [64], while the PHK might make a comparatively small contribution by altering the positions or efficacy of the catalytic scaffold provided by the RR. The PHK might, in addition, contribute to the general hydrophobic environment in which this catalytic scaffold operates, an environment expected to enhance the strength of charge-charge and H-bonding interactions [36•,40].
A VIEW OF THE PHOSPHOTRANSFER ACTIVE SITE
Once a PHK has accomplished autophosphorylation, it needs to allow a cognate RR protein to grab its high-energy phosphoryl group. A structure that likely provides a representative view of the PHK phosphotransferase active site that mediates this exchange is shown in Fig. 3B. This view was generated using the first high resolution structure of a PHK:RR complex (T. maritima HK853DHp+CA in complex with RR468) recently reported by Casino et al. [28••]. Some aspects of this structure confirm expectations based on a low resolution PHK:RR structure(ThkA:TrrA) [25,39] and on high resolution structures of RRs with (nonkinase) phosphotransfer proteins [36•,40], but other aspects of the new structure have provided some surprises. For example, the RR interacts not only with the DHp domain (an expected interaction that buries 885 Å2 of surface area), but also with the CA domain (an unanticipated interaction that buries 150 Å2), as well as with the DHp-CA linker (140 Å2) [28••]. Although the RR-CA domain interaction is relatively small, it includes two contacts involving an interesting region of the CA, the ‘lid’ of the ATP-binding pocket. This interaction creates an intriguing situation: the nucleotide binding site of the CA domain is occupied by an ADP molecule that is trapped in position because the RR is effectively holding down the lid. In this conformation HK853 would not be able to able to catalyze autophosphorylation. This raises the possibility that RR-CA binding might provide a mechanism for shutting off PHK autokinase active site under certain circumstances, such as when the PHK is operating as a phosphotransferase or as a phosphatase [28••]. Previous work has emphasized the potential of the ATP-lid for promoting PHK autophosphorylation [29,41]; the idea that lid-closure could serve as a negative regulator of this activity provides an interesting new perspective.
SIGNALING EVENTS: HOW ARE PHK ACTIVITIES REGULATED BY SENSORY INPUT?
To be useful parts of sensory response systems, PHKs need to respond to specific input signals by modulating one or more of their three activities such that phosphorylation level of their cognate response regulators are dialed up or down in an appropriate manner. For some PHKs, signaling events control only the autokinase activity [42–45] or exclusively the phosphatase activity [46,47], while some PHKs modulate both autokinase and phosphatase activities in response to sensory input [48,49]. The discussion in the preceding paragraph suggests that simultaneous, reciprocal regulation of autokinase and phosphatase activities could result from the structure of PHK:RR complexes via mechanisms such as closing the ATP-lid. To date, there is no system in which PHK-RR phosphotransfer activity is known to serves as the primary control point, although not many systems have been analyzed from this perspective.
One long-standing and popular model for regulation of PHK autokinase activity involves control of DHp↔CA interaction: in short, PHKs would respond to sensory input by controlling access of their phospho-accepting His to the ATP bound to the CA domain [3•,37•,43,50,51]. How might this regulation be achieved? Here again, the recently reported structures of DesK and HK853 could provide some key insights [28••,38••]. In each of these proteins, the DHp domain is part of a coiled-coil structure that is attached to a membrane-spanning helix that is part of the sensory input domain of the PHK. It is easy to envision these PHKs using this helical connection to adjust the conformation of the DHp domain in response to stimulus-responsive conformational changes in the sensory domains. Although they lack the sensory input domains, the DHp+CA versions of DesK and HK853 are capable of adopting different conformations that could represent the signaling states that the full-length proteins adopt in response to stimuli. Switching from one conformation to another involves changing the orientations of the helices of the DHp domain via mechanisms that can be described as “cogwheeling” and “bending”.
Cogwheeling
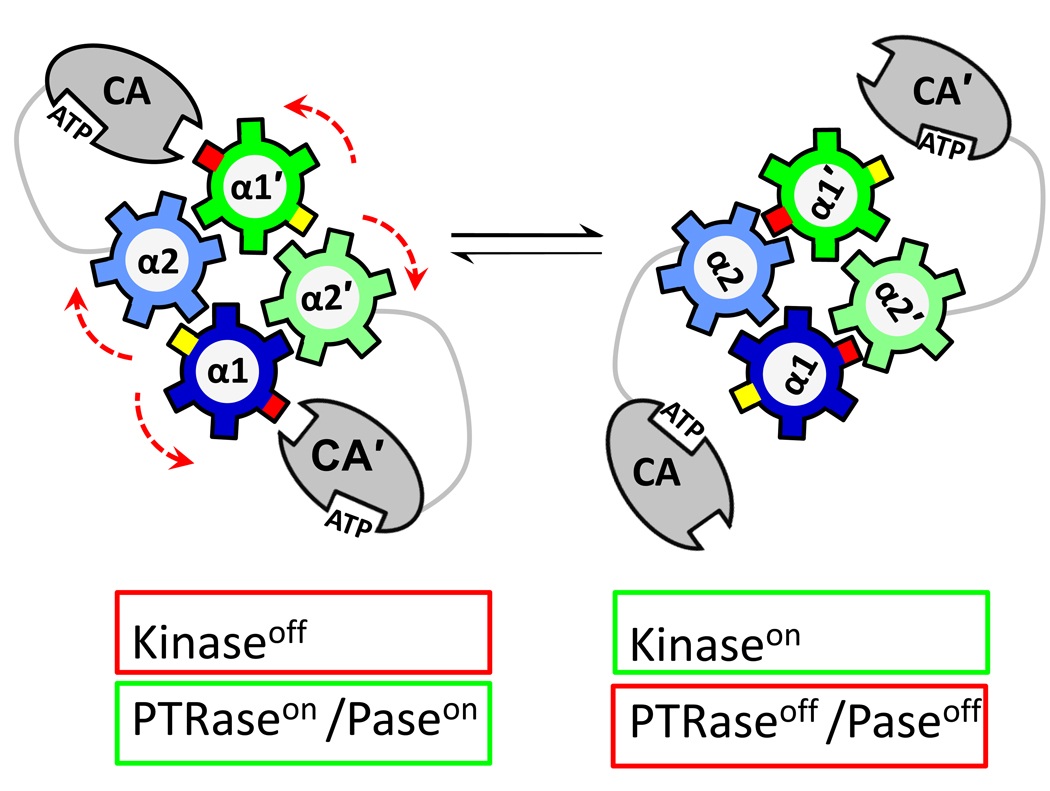
Comparisons of alternative conformations of DesK and HK853 indicate that the helices of the dimeric DHp four-helix bundle are rotated relative to one another as depicted schematically in Fig. 4. This shift has a significant effect on the accessibility of DHp surfaces for interactions with the CA domain of the PHK and for the ability of the PHK to interact with the RR. Thus, cogwheel rotation of DHp helices would affect autokinase activity as well as phosphotransfer and phosphatase activities. A similar cogwheel mechanism has been proposed as a mechanism underlying signaling events mediated by HAMP domains [52•] and by a distinct domain referred to as ‘the signaling helix’ [53]. These domains are frequently observed in signal transduction proteins (not only in PHKs), and so cogwheeling could be a general mechanism exploited by many signaling pathways, not just TCSs.
Figure 4.
Schematic diagram depicting how rearrangement of the helices within the DHp domain could alter PHK activities. The DHp helices of the four-helix bundle formed in the PHK dimer are depicted (top-down view) as cogwheels. In the starting structure (left), a surface (red cog) of the DHp is not buried within the bundle and is available to sequester the CA domain: CA cannot access the phospho-accepting His. In this conformation, the PHK is not active as an autokinase, but the DHp does have interaction surfaces for RR binding, so it can function as phosphotransferase (if it has been phosphorylated) or as a phosphatase (if it has not). Signaling events can cause cogwheel rotation of the helices (by 60° in this diagram to match that reported by Albanesi et al. [38••]). In this new orientation (right), the red cog of the DHp is no longer accessible, and the CA domain has been released: now it can access the DHp surface (yellow cog) to complete assembly of the kinase active site, including the phospho-accepting His: now the PHK is active as an autokinase. This reorientation also inhibits phosphotransferase and/or phosphatase activities of the PHK, either because it hides DHp surfaces important for RR binding or because the CA now competes effectively with the RR for binding surface on the DHp.
Bending
In addition to cogwheeling, the DHp domain of DesK appears to undergo a distinct kind of conformational shift (kinking or bending) when the phospho-accepting His becomes phosphorylated [38••]. The possible functional consequences of bending α1 of the DHp are qualitatively similar to those described above for cogwheeling (and indeed may be interrelated): different segments of the DHp would be exposed or sequestered depending on whether the helix is less bent (unphosphorylated) or more bent (phosphorylated), and this would have consequences for interaction of the DHp domain with the CA domain and also for RR binding.
Sequestering CA
With DesK and HK853, and perhaps with other PHKs, one important consequence of DHp conformational shifts appears to be influencing the ability of DHp to sequester the CA domain in a kinase-inactive conformation as depicted in Fig. 4 [28••,38••]. Basically, this involves the DHp domain having the ability to lock the CA domain into a position where its ATP binding site is well separated from the phospho-accepting His [37•]. Moreover, in its sequestered location, the CA cannot impede DHp↔RR interactions necessary for phosphotransfer or phosphatase activities and may even promote RR binding by providing an extra binding surface that the RR can use in addition to its contacts with the DHp itself [28••].
Antikinase Blockades
Another type of PHK regulation has also been revealed as a result of recently published crystal structures of two PHKs (KinA and KinB) that participate in the signaling cascade regulating sporulation in Bacillus and Geobacillus [54•,55•]. These structures highlight the ability of regulatory proteins to affect PHK activity by binding to strategic locations of the DHp domain. For example, the ‘antikinase’ protein Sda (a KinB inhibitor) binds to the base of the KinB DHp domain, and in this position Sda prevents or hinders DHp↔CA intraprotein, interdomain interactions within KinB as well as interprotein interactions between KinA and the RR protein Spo0F, effectively blocking KinB autophosphorylation and slowing KinB→Spo0F phosphotransfer [54•]. A distinct antikinase, KipI, can bind to the PHK KinA, and this involves a region of the DHp similar to (but more extensive than) that utilized by Sda [55•]. Interestingly, KipI appears to recognize a segment of the KinA DHp that includes a proline residue that is conserved in numerous other PHKs (families 1, 2, 3, and 4 in the Grebe-Stock scheme [9]). This proline introduces a bend into α1 of the DHp , and this bend affects how tightly the helices of the dimeric DHp four helix bundle can wind around one another [38••]. Jacques et al. [55•] have proposed the interesting idea that KipI and related proteins might promote cis-trans isomerization at this conserved proline, a change that would alter the conformation of the DHp domain in a manner that would affect PHK activities. Perhaps other PHKs also utilize regulatory mechanisms that exploit the conformation of this proline.
CONCLUSIONS
Structural information generated over the past decade has dramatically improved our understanding of how PHKs function as enzymes, as receptors, and as signaling components. In particular, in the past few months new structures of DHp+CA proteins have revealed specific conformational changes that may underlie regulation of PHK activities in response to stimuli. Box 1 summarizes some important specific questions that are likely to be addressed over the next five to ten years to follow up on and extend recent advances. Answers to some of these questions will help us to answer an important overarching question: is there a universal mechanism for PHK regulation or are there several (or even many) mechanisms that have evolved to meet the demands of specific signaling pathways? In this regard, it is interesting to note the success of domain-swap experiments in which heterologous sensory input domains were fused to DHp+CA domains [56,57] and the ingenious rewiring of PHK-RR specificity by Skerker et al. [58]. The abilities of such ‘synthetic proteins’ to signal properly suggest that there are indeed shared regulatory mechanisms controlling PHK activities and that this knowledge can be exploited to create novel signaling circuits [59,60].
Box 1. Key Questions for Future Work.
Do DesK and HK853 crystal structures tell us how other conventional PHKs work?
PHK regulation models discussed here are based on the assumption that the alternative conformations observed for DesK and HK853 DHp+CA fragments represent conformations that ‘normal versions’ of PHKs would adopt. Testing these models will require analysis of domain-domain orientations of full-length PHKs in solution, perhaps using cross-linking methods developed in the Falke [65•] and Inouye [66•] labs or the EPR methods developed by the Crane group [67].
How do nonconventional PHKs (like CheA and NtrB) achieve regulation?
Two of the most intensively studied PHKs, CheA and NtrB modulate their activities over impressive ranges in response to stimuli (>100-fold ). Is this achieved using the same types of conformational changes utilized by conventional PHKs even though CheA and NtrB don’t have the traditional architecture?
Is there another level of PHK regulation?
PHK regulation involves some relatively large-scale changes in domain orientations, and these control whether active sites can assemble. However, after assembly, these sites might be further regulated by tweaking the orientation of key functional groups, as suggested by the interacting network of active site groups identified in the CheA P1 domain[32•] and in the Ypd1:Sln1(RR) complex[36•].
How are transmembrane signaling events linked to changes in DHp conformation?
In some PHKs (including DesK and HK853), helical signaling domains (e.g., HAMP) provide a communication link between the sensory input domain and the cytoplasmic DHp and CA domains, but in many PHKs this job is carried out by some other kind of signaling domain (e.g., PAS, GAF, Cache, CHASE, etc.) [16,17]. Can they also drive cogwheeling and bending within the DHp domain, or do they trigger distinct conformational changes?
Does clustering affect the conformations and activities of PHKs?
In the chemotaxis system and some other TCSs, the signaling proteins can cluster[68] in arrays that may include numerous copies of the PHK (e.g., anywhere from from ~10 to ~1,500 CheA molecules[69••]). Modelers have heralded this clustering as a key design feature for enhancing sensitivity and signal amplification[70,71], but we don’t really have any idea how PHK conformations might be influenced by clustering or how this might affect interactions with RRs.
What can we learn from PHK imposters?
There are some proteins (e.g., NifL [72] and ETR1 [73]) that, based on sequence comparisons, look like PHKs, but when examined in closer detail, don’t function as kinases in TCSs. Defining how these proteins do function may generate new insights into the abilities of real PHKs as well as provide further examples of how the Bergerat fold of the CA domain has been adapted to accomplish different specific functions in different enzyme families [41,74].
ACKNOWLEDGEMENTS
I thank Dr. Alejandro Buschiazzo (Intsitut Pasteur de Montevideo) and Dr. Alberto Marina (IBV-CSIC and CIBERER, Instituto de Biomedicina de Valencia) for providing information in advance of publication. Dr. Buschiazzo also kindly provided the coordinate file necessary for Fig. 3A. Research on PHKs in my laboratory is supported by Public Health Service grant GM052853 from the NIH National Institute of General Medical Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Hoch JA, Silhavy TJ. Two-Component Signal Transduction. Washington, DC: ASM Press; 1995. [Google Scholar]
- 2.Galperin MY. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J Bacteriol. 2006;46:4169–4182. doi: 10.1128/JB.01887-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gao R, Stock AM. Biological Insights from Structures of Two-Component Proteins. Ann Rev Microbiol. 2009;63:133–154. doi: 10.1146/annurev.micro.091208.073214. Both PHK afficianados and newcomers will appreciate this well written and up-to-date review.
- 4.Saito H. Histidine phosphorylation and two-component signaling in eukaryotic cells. Chem Rev. 2001;101:2497–2509. doi: 10.1021/cr000243+. [DOI] [PubMed] [Google Scholar]
- 5.Chauhan N, Calderone R. Two-Component Signal Transduction Proteins as Potential Drug Targets in Medically Important Fungi. Infection and Immunity. 2008;76:4795–4803. doi: 10.1128/IAI.00834-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hubbard J, Burnham MKR, Throup JP. Pathogenicity and histidine kinases: approaches toward the development of a new generation of antibiotics. In: Inouye M, Dutta R, editors. Histidine Kinases in Signal Transduction. Academic Press; 2003. pp. 459–506. [Google Scholar]
- 7.Okada A, Gotoh Y, Watanabe T, Furuta E, Yamamoto K, Utsumi R. Targeting two-component signal transduction: A novel drug discovery system. Meth Enzymol. 2007;422:386–395. doi: 10.1016/S0076-6879(06)22019-6. [DOI] [PubMed] [Google Scholar]
- 8.Stephenson K, Hoch JA. Two-component and phosphorelay signal-transduction systems as therapeutic targets. Curr Opin Pharmacology. 2002;2:507–512. doi: 10.1016/s1471-4892(02)00194-7. [DOI] [PubMed] [Google Scholar]
- 9.Grebe T, Stock JB. The histidine protein kinase superfamily. Adv Microb Physiol. 1999;41:139–227. doi: 10.1016/s0065-2911(08)60167-8. [DOI] [PubMed] [Google Scholar]
- 10.Mascher T, Helmann JD, Unden G. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol Mol Biol Rev. 2006;70:910. doi: 10.1128/MMBR.00020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koretke KK, Volker C, Bower ML, Lupas AN. Molecular evolution of histidine kinases. In: Inouye M, Dutta R, editors. Histidine Kinases in Signal Transduction. Academic Press; 2003. pp. 483–506. [Google Scholar]
- 12.Alm E, Huang K, Arkin A. The evolution of two-component systems in bacteria reveals different strategies for niche adaptation. PLos Comp Biol. 2006;2:1329–1342. doi: 10.1371/journal.pcbi.0020143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whitworth DE, Cock PJA. Evolution of prokaryotic two-component systems: insights from comparative genomics. Amino Acids. 2009;37:459–466. doi: 10.1007/s00726-009-0259-2. [DOI] [PubMed] [Google Scholar]
- 14.Kenney L. Unknown title. Curr Opin Microbiology. 2010;13 doi: 10.1016/j.mib.2010.01.013. unknown pages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dutta R, Qin L, Inouye M. Histidine kinases: diversity of domain organization. Mol Microbiol. 1999;34:633–640. doi: 10.1046/j.1365-2958.1999.01646.x. [DOI] [PubMed] [Google Scholar]
- 16.Wuichet K, Alexander RP, Zhulin IB. Comparative genomic and protein sequence analyses of a complex system controlling bacterial chemotaxis. Meth Enzymol. 2007;422:3–31. doi: 10.1016/S0076-6879(06)22001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheung J, Hendrickson WA. Sensor domains of two-component regulatory systems. Cur Opin Microbiology. 2010;13 doi: 10.1016/j.mib.2010.01.016. unknown pages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomomori C, Tanaka T, Dutta R, Park H, Saha S. Solution structure of the homodimeric core domain of Escherichia coli histidine kinase EnvZ. Nat Struct Biol. 1999;6:729–734. doi: 10.1038/11495. [DOI] [PubMed] [Google Scholar]
- 19.Varughese KI, Madhusudan, Zhou XZ, Whiteley JM, Hoch JA. Formation of a novel four-helix bundle and molecular recognition sites by dimerization of a response regulator phosphotransferase. Mol Cell. 1998;2:485–493. doi: 10.1016/s1097-2765(00)80148-3. [DOI] [PubMed] [Google Scholar]
- 20.Marina A, Mott C, Auyenberg A, Hendrickson WA, Waldburger CD. Structural and mutational analysis of the PhoQ histidine kinase catalytic domain. J Biol Chem. 2001;276:41182–41190. doi: 10.1074/jbc.M106080200. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka T, Saha S, Tomomori C, Ishima R, Liu D. NMR structure of the histidine kinase domain of the E. coli osmosensor EnvZ. Nature. 1998;396:88–92. doi: 10.1038/23968. [DOI] [PubMed] [Google Scholar]
- 22.Bilwes AM, Alex LA, Crane BR, Simon MI. Structure of CheA, a signal-transducing histidine kinase. Cell. 1999;96:131–141. doi: 10.1016/s0092-8674(00)80966-6. [DOI] [PubMed] [Google Scholar]
- 23.Bilwes AM, Quezada CM, Croal LR, Crane BR, Simon MI. Nucleotide binding by the histidine kinase CheA. Nat Struct Biol. 2001;8:353–360. doi: 10.1038/86243. [DOI] [PubMed] [Google Scholar]
- 24.Song Y, Peisach D, Pioszak AA, Xu Z, Ninfa AJ. Crystal structure of the C-terminal domain of the two-component system transmitter protein nitrogen regulator II (NRII; NtrB), regulator of nitrogen assimilation in Escherichia coli. Biochemistry. 2004;43:6670–6678. doi: 10.1021/bi049474r. [DOI] [PubMed] [Google Scholar]
- 25.Yamada S, Sugimota H, Kobayashi M, Ohno A, Nakamura H, Shiro Y. Structure of PAS-linked histidine kinase and the response regulator complex. Structure. 2009;17:1333–1344. doi: 10.1016/j.str.2009.07.016. [DOI] [PubMed] [Google Scholar]
- 26.Ninfa EG, Atkinson MR, Kamberov ES, Ninfa AJ. Mechanism of autophosphorylation of Escherichia coli nitrogen regulator II (NRII or NtrB): trans-phosphorylation between subunits. J Bacteriol. 1993;175:7024–7032. doi: 10.1128/jb.175.21.7024-7032.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Y, Inouye M. Intermolecular complementation between two defective mutant signal-transducing receptors of Escherichia coli. Proc Natl Acad Sci USA. 1991;88:11057–11061. doi: 10.1073/pnas.88.24.11057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Casino P, Rubio V, Marina A. Structural insight into partner specificity and phosphoryl transfer in two-component signal transduction. Cell. 2009;139:325–336. doi: 10.1016/j.cell.2009.08.032. This is the first high-resolution structure of a PHK:RR complex. Analysis of this structure and comparison to the structure of the PHK by itself demonstrate how the conformation of the DHp domain can change and how this could regulate PHK activity.
- 29.Bilwes AM, Park SY, Quezada CM, Simon MI, Crane BR. Structure and function of CheA, the histidine kinase central to bacterial chemotaxis. In: Inouye M, Dutta R, editors. Histidine Kinases in Signal Transduction. Academic Press; 2003. pp. 47–72. [Google Scholar]
- 30.Mourey L, Da Re S, Pédelacq J-D, Tolstykh T, Faurie C, Guillet V, Stock JB, Samama J-P. Crystal structure of the CheA histidine phosphotransfer domain that mediates response regulator phosphorylation in bacterial chemotaxis. J Biol Chem. 2001;276:31074–31082. doi: 10.1074/jbc.M101943200. [DOI] [PubMed] [Google Scholar]
- 31.Quezada CM, Gradinaru C, Simon MI, Bilwes AM, Crane BR. Helical shifts generate two distinct conformers in the atomic resolution structure of the CheA phosphotransferase domain from Thermotoga maritima. J Mol Biol. 2004;341:1283–1294. doi: 10.1016/j.jmb.2004.06.061. [DOI] [PubMed] [Google Scholar]
- 32. Quezada CM, Hamel DJ, Gradinaru C, Bilwes AM, Dahlquist FW, Crane BR, Simon MI. Structural and chemical requirements for histidine phosphorylation by the chemotaxis kinase CheA. J Biol Chem. 2005;280:30581–30585. doi: 10.1074/jbc.M505316200. This paper demonstrate that the phospho-accepting His of CheA is part of hydrogen-bonding network (including a Glu-His dyad) that dramatically enhances autophosphorylation kinetics. This means that the CheA autokinase active includes essential contributions from both the DHp and CA domains and that this site is assembled only transiently. The authors argue that CheA may be a special case and that this catalytic dyad may not be present in other PHKs, but one wonders whether they may be other examples of catalytic dyads out there in the vast array of PHKs.
- 33.Ikegami T, Okada T, Ohki I, Hirayama J, Mizuno T, Shirakawa M. Solution structure and dynamic character of the histidine-containing phosphotransfer domain of anaerobic sensor kinase ArcB from Escherichia coli. Biochemistry. 2001;40:375–386. doi: 10.1021/bi001619g. [DOI] [PubMed] [Google Scholar]
- 34.Rogov VV, Bernhard F, Lohr F, Dotsch V. Solution structure of the Escherichia coli YojN histidine-phosphotransferase domain and its interaction with cognate phosphoryl receiver domains. J Mol Biol. 2004;343:1035–1048. doi: 10.1016/j.jmb.2004.08.096. [DOI] [PubMed] [Google Scholar]
- 35.Xu Q, West AH. Conservation of structure and function among histidine-containing phosphotransfer (HPt) domains as revealed by the crystal structure of YPD1. J Mol Biol. 1999;292:1039–1050. doi: 10.1006/jmbi.1999.3143. [DOI] [PubMed] [Google Scholar]
- 36. Zhao X, Copeland DM, Soares AS, West AH. Crystal structure of a complex between the phosphorelay protein YPD1 and the response regulator domain of SLN1 bound to a phosphoryl analog. J. Mol. Biol. 2008;375:1141–1151. doi: 10.1016/j.jmb.2007.11.045. This structure of a RR:Hpt complex provides a high resolution view of a phosphotransfer active site. The bond distances observed support an associative mechanism for the phosphotransfer reaction.
- 37. Marina A, Waldburger CD, Hendrickson WA. Structure of the entire cytoplasmic portion of a sensor histidine-kinase protein. EMBO J. 2005;24:4247–4259. doi: 10.1038/sj.emboj.7600886. This work provided the first view of how the DHp and CA domains are oriented in a conventional PHK. The authors provide an excellent summary of how the various homology boxes fit into the structure and an insightful discussion of the domain movements required for PHK activities.
- 38. Albanesi D, Martin M, Trajtenberg F, Mansilla MC, Haouz A, Alzari PM, de Mendoza D, Buschiazzo A. Structural plasticity and catalysis regulation of a thermosensor histidine kinase. Proc Natl Acad Sci USA. 2009;106:16185–16190. doi: 10.1073/pnas.0906699106. The authors crystallized theDHp+CA domains of DesK in several distinct conformations that provide new insights into the mechanisms underlying regulation of PHK activities. One of these structures provides the first glimpse of an assembled PHK kinase active site; other structures suggest that cogwheeling and bending in the DHp domain are key regulatory events.
- 39.Yamada S, Akiyama S, Sugimoto H, Kumita H, Iito K, Fujisawa T, Nakamura H, Shiro Y. The signaling pathway in histidine kinase and the response regulator complex revealed by x-ray crystallography and solution scattering. J Mol Biol. 2006;362:123–139. doi: 10.1016/j.jmb.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 40.Zapf J, Sen U, Madhusudan, Hoch JA, Varughese KI. A transient interaction between two phosphorelay proteins trapped in a crystal lattice reveals the mechanism of molecular recognition and phosphotransfer in signal transduction. Structure. 2000;8:851–862. doi: 10.1016/s0969-2126(00)00174-x. [DOI] [PubMed] [Google Scholar]
- 41.Dutta R, Inouye M. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem Sci. 2000;25:24–28. doi: 10.1016/s0968-0004(99)01503-0. [DOI] [PubMed] [Google Scholar]
- 42.Borkovich KA, Simon MI. The dynamics of protein-phosphorylation in bacterial chemotaxis. Cell. 1990;63:1339–1348. doi: 10.1016/0092-8674(90)90429-i. [DOI] [PubMed] [Google Scholar]
- 43.Levit MN, Liu Y, Stock JB. Mechanism of CheA protein kinase activation in receptor signaling complexes. Biochemistry. 1999;38:6651–6658. doi: 10.1021/bi982839l. [DOI] [PubMed] [Google Scholar]
- 44.Fleischer R, Heermann R, Jung K, Hunke S. Purification, reconstitution, and characterization of the CpxRAP envelope stress system of Escherichia coli. J Biol Chem. 2007;282:8583–8593. doi: 10.1074/jbc.M605785200. [DOI] [PubMed] [Google Scholar]
- 45.Timmen M, Bassler BL, Jung K. AI-1 influences the kinase activity but not the phosphatase activity of LuxN of Vibrio harveyi. J Biol Chem. 2006;281:24398–24404. doi: 10.1074/jbc.M604108200. [DOI] [PubMed] [Google Scholar]
- 46.Stock AM, West AH. Response regulator proteins and their interactions with histidine protein kinases. In: Inouye M DR, editor. Histidine Kinases in Signal Transduction. Academic Press; 2003. pp. 237–271. [Google Scholar]
- 47.Brandon L, Dorus S, Epstein W, Altendorf K, Jung K. Modulation of KdpD phosphatase implicated in the physiological expression of the kdp ATPase of Escherichia coli. Mol Microbiol. 2000;38:1086–1092. doi: 10.1046/j.1365-2958.2000.02219.x. [DOI] [PubMed] [Google Scholar]
- 48.Jiang P, Atkinson MR, Srisawat C, Sun Q, Ninfa AJ. Functional dissection of the dimerization and enzymatic activities of Escherichia coli nitrogen regulator II and their regulation by the PII protein. Biochemistry. 2000;39:13433–13449. doi: 10.1021/bi000794u. [DOI] [PubMed] [Google Scholar]
- 49.Chamnongpol S, Cromie M, Groisman EA. Mg2+ sensing by the Mg2+ sensor PhoQ of Salmonella enterica. J Mol Biol. 2003;325:795–807. doi: 10.1016/s0022-2836(02)01268-8. [DOI] [PubMed] [Google Scholar]
- 50.Wolanin PM, Stock JB. Transmembrane signaling and the regulation of histidine kinase activity. In: Inouye M DR, editor. Histidine kinases in Signal Transduction. Academic Press; 2003. pp. 73–122. [Google Scholar]
- 51.Yamada S, Shiro Y. Structural basis of the signal transduction in the two-component system. Adv Exp Med Biol. 2008;631:22–39. doi: 10.1007/978-0-387-78885-2_3. [DOI] [PubMed] [Google Scholar]
- 52. Hulko M, Berndt F, Gruber M, Linder JU, Truffault V. The HAMP domain structure implies helix rotation in transmembrane signaling. Cell. 2006;126:929–940. doi: 10.1016/j.cell.2006.06.058. The first detailed structural information about a ubiquitous signaling domain. The authors describe a model by which rotation (cogwheeling) of helices in a coiled-coil can accomplish transmembrane signaling.
- 53.Anantharaman V, Balaji S, Aravind L. The signaling helix: a common functional theme in diverse signaling proteins. Biol Direct. 2006;1:25. doi: 10.1186/1745-6150-1-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bick MJ, Lamour V, Rajashankar KR, Gordiyenko Y, Robinson CV, Darst SA. How to switch off a histidine kinase: crystal structure of Geobacillus stearothermophilus KinB with the inhibitor Sda. J Mol Biol. 2009;386:163–177. doi: 10.1016/j.jmb.2008.12.006. This paper details the structure of the KinB DHp+CA domains and shows how interaction between these domains is blocked by the antikinase Sda.
- 55. Jaques DA, Langley DB, Jeffries CM, Cunningham KA, Burkholder WF, Guss JM, Trewhella J. Histidine kinase regulation by a cyclophilin-like inhibitor. J Mol Biol. 2008;384:422–435. doi: 10.1016/j.jmb.2008.09.017. The structure of this PHK:antikinase complex suggests an important role for a kinked region of α1 in the DHp (where a conserved proline is positioned). This is particularly interested in view of the bend-inducing effect of α1 phosphorylation reported by Albanesi et al. (Ref. 38). Is cis-trans isomerization at this proline involved in PHK regulation?
- 56.Appleman JA, Chen LL, Stewart V. Probing conservation of HAMP linker structure and signal transduction mechanism through analysis of hybrid sensor kinases. J Bacteriol. 2003;185:4872–4882. doi: 10.1128/JB.185.16.4872-4882.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoshida T, Phadtare S, Inouye M. The design and development of Tar-EnvZ chimeric receptors. Meth Enzymol. 2007;423:166–183. doi: 10.1016/S0076-6879(07)23007-1. [DOI] [PubMed] [Google Scholar]
- 58.Skerker JM, Perchuk BS, Siryaporn A, Lubin EA, Ashenberg O. Rewiring the specificity of two-component signal transduction systems. Cell. 2008;133:1043–1054. doi: 10.1016/j.cell.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ninfa AJ. Use of two-component signal transduction systems in the construction of synthetic genetic networks. Curr Opin Microbiology. 2010;13 doi: 10.1016/j.mib.2010.01.003. unknown. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ninfa AJ, Selinsky S, Perry N, XAtkins S, Song QX, Mayo A, Arps D, Woolf P, Atkinson MR. Using two component systems and other bacterial regulatory factors for the fabrication of synthetic genetic devices. Meth Enzymol. 2007;422:488–512. doi: 10.1016/S0076-6879(06)22025-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parkinson JS. Communication modules in bacterial signaling proteins. Ann Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 62.Wolanin PM, Thomason PA, Stock JB. Histidine protein kinases: key signal transducers outside the animal kingdom. Genome Biology. 2002;3 doi: 10.1186/gb-2002-3-10-reviews3013. reviews3013.3011-3013.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lukat GS, McCleary WR, Stock AM, Stock JB. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc Natl Acad Sci USA. 1992;89:718–722. doi: 10.1073/pnas.89.2.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.DaRe SS, Deville-Bonne D, Tolstykh T, Veron M, Stock JB. Kinetics of CheY phosphorylation by small molecule phosphodonors. FEBS Lett. 1999;457:323–326. doi: 10.1016/s0014-5793(99)01057-1. [DOI] [PubMed] [Google Scholar]
- 65. Gloor SL, Falke JJ. Thermal domain motions of CheA kinase in solution: disulfide trapping reveals the motional constraints leading to trans-autophosphorylation. Biochemistry. 2009;48:3631–3644. doi: 10.1021/bi900033r. Analysis of domain orientations in full-length PHKs (i.e., with all the domains present) is tough, but these authors demonstrate how it can be done and provide some interesting information about the orientation of domains in CheA.
- 66. Cai SJ, Khorchid A, Ikura M, Inouye M. Probing catalytically essential domain orientation in histidine kinase EnvZ by targeted disulfide crosslinking. J Mol Biol. 2003;328:409–418. doi: 10.1016/s0022-2836(03)00275-4. Disulfide trapping combined with molelcular modeling to assess domain orientations in a well characterized PHK.
- 67.Park S-Y, Borbat PP, Gonzalez-Bonet G, Bhatnagar J, Pollard AM, Freed JH, Bilwes AM, Crane BR. Reconstruction of the chemotaxis receptor-kinase assembly. Nat Struct Biol. 2006;13:400–407. doi: 10.1038/nsmb1085. [DOI] [PubMed] [Google Scholar]
- 68.Vescovi EG, Sciara MI, Castelli ME. Two-component systems in the spatial program of bacteria. Curr Opin Microbiology. 2010;13 doi: 10.1016/j.mib.2009.12.012. unknown. [DOI] [PubMed] [Google Scholar]
- 69. Greenfield D, McEvoy AL, Shroff H, Crooks GE, Wingreen NS, Betzig E. Self-organization of the Escherichia coli chemotaxis network imaged with super-resolution light microscopy. PLoS Biol. 2009;7:e1000137. doi: 10.1371/journal.pbio.1000137. Photactivated localization microscopy provides high-resolution images of assembled complexes of chemotaxis signaling proteins, and these provide new perspectives of the sizes, locations, and assembly mechanism.
- 70.Bray D, Levin M, Morton-Firth C. Receptor clustering as a cellular mechanism to control sensitivity. Nature. 1998;393:85–88. doi: 10.1038/30018. [DOI] [PubMed] [Google Scholar]
- 71.Endres RG, Oleksiiuk O, Hansen CH, Meir Y, Sourjik V, Wingreen NS. Variable sizes of Escherichia coli chemoreceptor signaling teams. Mol Sys Biol. 2008;4:211. doi: 10.1038/msb.2008.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Little R, Martiniz-Argudo I, Dixon R. Role of the central region of NifL in conformational switches that regulate nitrogen fixation. Biochem Soc Trans. 2006;34:162–164. doi: 10.1042/BST0340162. [DOI] [PubMed] [Google Scholar]
- 73.Wang WY, Hall AE, O'Malley R, Bleecker AB. Canonical histidine kinase activity of the transmitter domain of the ETR1 ethylene receptor from Arabidopsis is not required for signal transmission. Proc Natl Acad Sci USA. 2003;100:352–357. doi: 10.1073/pnas.0237085100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang W. Histidine kinases: extended relationship with GHL ATPases. In: Inouye M, Dutta R, editors. Histidine Kinases in Signal Transduction. Academic Press; 2003. pp. 220–236. [Google Scholar]