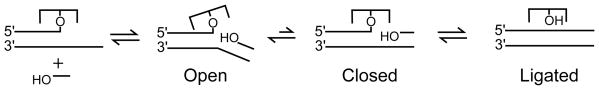Abstract
The type I DNA topoisomerase from vaccinia virus (vTopo) forms a reversible covalent 3′-phosphotyrosyl linkage with a single strand of duplex DNA at the preferred sequence 5′-(C/T)CCTTp↓N−1N−2N−3-3′. The enzyme-DNA covalent adduct is recombinogenic in cells, because the nicked strand downstream of the cleavage site can dissociate and be replaced by another DNA strand, potentially resulting in genome rearrangements if the enzyme executes strand ligation. Topo I could play an active role in strand exchange, either by altering the kinetics or thermodynamics of DNA strand binding, or by serving as a proofreading gate to prevent ligation of incoming DNA strands containing mismatches. To address these questions, we have measured the kinetic and thermodynamic parameters for strand annealing to a purified vaccinia Topo I-DNA (vTopo-DNA) covalent complex containing a single-strand overhang, and then compared them with the same overhang duplex in the absence of vTopo. We found that vTopo accelerates the strand association rate by two-fold, but has no effect on the rate of strand dissociation. vTopo has a similar small effect on the annealing parameters of a series of DNA strands containing single mismatches. In contrast, single base mismatches at the -1, -2 or -3 positions decreased the forward rate and equilibrium constant for reversible strand ligation by 10-fold. These data establish that while vTopo is a bystander during the annealing step of strand exchange, the enzyme strongly discriminates against mismatches close to the cleavage site during the subsequent events leading to strand ligation. A mechanism emerges where vTopo oscillates between an open state where the downstream DNA segment does not interact with the enzyme, and a closed state where catalytically important contacts are formed with this region. This oscillation between an open and closed state of the covalently bound enzyme is likely important for regulating the number of DNA superhelical turns that are removed during the lifetime of the covalent complex with supercoiled substrates.
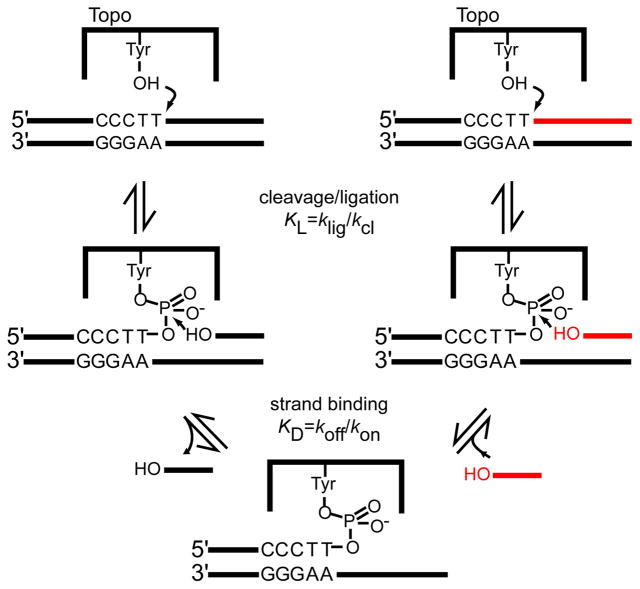
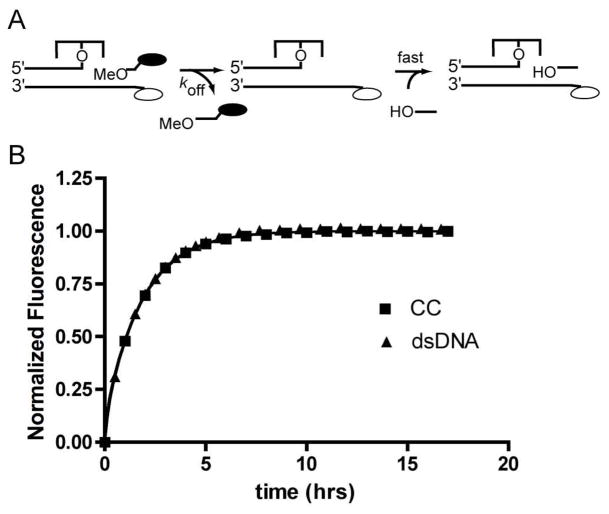
Type I DNA topoisomerases (Topo I) relieve supercoils generated during replication and other DNA manipulations in cells (1). The relaxation reaction is initiated by attack of a active site tyrosine at a phosphodiester bond, resulting in formation of a reversible covalent link between the enzyme and a single strand of the DNA (Figure 1) (2). Downstream of the covalent link, the nicked duplex is free to rotate, allowing the removal of multiple supercoils from the DNA before the strand is religated (3, 4). The generation of a reversible strand nick by Topo I is mechanistically identical to that of the related tyrosine recombinases, and is correspondingly recombinogenic (5, 6). In both systems, the DNA strand downstream of the nick can dissociate and be replaced with another strand prior to ligation, resulting in DNA rearrangements (Figure 1) (7, 8).
Figure 1.
Reversible DNA cleavage and ligation by topoisomerase IB and DNA strand exchange. The native strand (black) is cleaved during the first transesterification reaction, and can dissociate from the Topo-DNA covalent complex. Another DNA strand with a 5′-OH (red) can associate and attack the DNA-protein phosphotyrosyl bond, resulting in DNA rearrangements. The extent of binding and ligation of strands to the covalent complex is determined by the binding equilibrium (KD) and the reversible ligation equilibrium (klig/kcl = KL).
The recombinogenic potential of the Topo-DNA adduct has relevance for chemotherapy and cancer. The lifetime of the covalent DNA complex of human Topo I (hTopo) is increased by the widely used chemotherapy drug camptothecin (CPT) which intercalates at the site of the Topo I covalent attachment, thereby sterically hindering strand ligation (9, 10). The drug-stabilized complex results in DNA double strand breaks when a replication fork encounters the adduct (11). The increased persistence of the recombinogenic covalent complex in the presence of CPT can alternatively result in DNA translocations if DNA rearrangements occur prior to the encounter of a replication fork (12, 13). These translocations have been linked to CPT-induced secondary cancers (13, 14, 15).
The small type I Topo from vaccinia virus (vTopo) shares the same reaction mechanism and conserved active site structure with the hTopo, and has played an invaluable role in developing an understanding of Topo I reaction chemistry and supercoil release (3, 4, 16, 17, 18, 19, 20). A unique attribute of vTopo is its site-specific DNA cleavage and ligation chemistry that only occurs at DNA sites containing the pentapyrimidine sequence 5′-(C/T)CCTTp↓N−1N−2N−3-3′ (21). This high sequence selectivity allows for the formation of site-specific covalent complexes, and facilitates mechanistic investigations that are difficult or impossible with the largely nonspecific human enzyme. In addition, vTopo resolves pre-formed Holliday junctions that contain the consensus cleavage sequence, and when expressed in E. coli, promotes illegitimate recombination (5, 22, 23). Thus, vTopo is an experimentally tractable enzyme that exhibits many mechanistic aspects of the DNA recombination reactions catalyzed by both hTopo and the tyrosine recombinases, and is an excellent system to study Topo I-mediated DNA recombination in vitro (2).
One essential step of site specific DNA recombination is strand exchange within the Topo I or recombinase-DNA covalent complex, which is poorly characterized. In the absence of an enzyme, a nicked DNA strand can be replaced with a free homologous strand, but the rate of this approach-to-equilibrium reaction will be limited by the slow dissociation of the bound strand, as well as the slow association of the incoming strand, which involves a time consuming search for homology (24). Given these intrinsic kinetic barriers, a plausible mechanism for enzyme-mediated strand exchange can be envisioned where the covalently bound enzyme forms duplex destabilizing interactions that facilitate strand dissociation. Subsequently, the enzyme could reduce the time scale of the search for homology by restricting the conformational freedom of the incoming strand, and by orienting the bases with respect to the complementary target. Alternatively, Topo I could merely play a passive role in strand replacement, where downstream strand dissociation and annealing depends solely on the intrinsic kinetic properties of the nicked DNA.
A related aspect of DNA strand exchange and ligation by Topo I is the specificity of the overall process for perfectly matched as opposed to mismatched incoming DNA strands. Discrimination could occur at the strand annealing step, or at subsequent steps that lead to chemical ligation of the DNA backbone. In the absence of Topo I, annealing of a mismatched strand will be thermodynamically disfavored. However, if the lifetime of the bound mismatched strand is significant compared to the strand ligation rate, mismatched strands may become covalently integrated. Thus, Topo I may enhance specificity above and beyond the equilibrium discrimination provided by Watson-Crick base pairing by directly sensing mismatches during the annealing or chemical ligation step of strand exchange. Such a mechanism is plausible because the two nucleotides just downstream of the cleavage site are known to be critical for the DNA cleavage reaction, and thirteen downstream base pairs are within the footprint of the enzyme (25, 26).
In this study, we examine whether vTopo plays an active or passive role in DNA strand exchange, and also evaluate the energetic contributions of the ground-state and transition-state interactions of the enzyme that lead to discrimination against mismatched incoming strands during ligation.
MATERIALS AND METHODS
Protein purification
The cloning and purification of wild type vaccinia topoisomerase was performed as previously described (19).
DNA substrates
The sequences making up the 20/32FAM and 24Bio/32FAM DNA duplexes as well as the downstream 12 mer strands are:
20: 5′-AACATATCCGTGTCGCCCTT-3′
32FAM: 5′-FAM- CACTATCGGAAAAAGGGCGACACGGATATGTT-3′, FAM = 6-carboxyfluorescein linked through the 5′ phosphate.
24Bio: 5′-AACATATCCGTGTCGCCCTTTTTC-3′-BioTEG. The 3′-BioTEG substitution is a biotin connected to the DNA via a ~20 atom polyethyleneglycol linker.
Downstream 12 mer strands: Fully matched parent sequence, 5′-TTTCCGATAGTG-3′. Additional oligonucleotides derived from this sequence are: -1TC, -3TC, -6GA and -9AG which include a point mutation at the indicated position from the 5′-end. 12DAB, OMe12DAB, OMe-6GADAB and OMe-3TCDAB were also derived from this sequence. ‘OMe’ and ‘DAB’ indicate 5′-OMethyl and 3′-Dabcyl-dT substitutions, respectively.
5′-OMe and 3′-DAB doubly substituted oligonucleotides were synthesized on an ABI 394 Synthesizer with phosphoramidites from Glen Research. Dabcyl solid support was obtained from Biosearch Technologies. These oligonucleotides were purified on an anion exchange column and desalted with a C-18 reverse phase desalting column. The accuracy and purity of the synthesis was confirmed with MALDI-TOF and 20% polyacrylamide, 7M urea gel electrophoresis. The remainder of the oligonucleotides were ordered from Integrated DNA Technologies (IDT).
The 20/32FAM and 24Bio/32FAM duplexes were prepared in 15 mM Tris-HCl pH 8.0, 20 mM NaCl by heating to 90 °C, followed by slow cooling to 30 °C. The unlabeled 20 mer was in 10% excess over 32FAM, while 24Bio was in 50% excess over the 32FAM labeled strand.
Purification of Covalent Complex
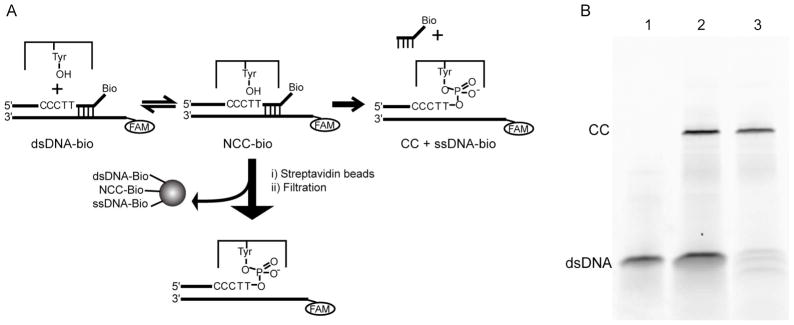
We have developed a robust method for easily purifying the vTopo-DNA covalent complex from noncovalently bound enzyme and unreacted DNA (Figure 2). 500 nM vTopo and 500 nM 24Bio/32FAM duplex in 30 mM NaCl, 20 mM Tris-HCl pH 8.0, 1 mM dithiothreitol (DTT) were incubated at room temperature (20 °C) for 30 minutes. 50 μl streptavidin agarose (Novagen) slurry was rinsed with reaction buffer and added to the protein-duplex reaction. The mixture was rotated at room temperature for 30 minutes, heated at 37 °C for 10 minutes and spun over a 0.22 μm filter to separate the biotin-bound streptavidin-agarose beads from the covalent complex (Figure 2). Attached to the beads and therefore purified away from the covalent complex is the unreacted 24Bio/32FAM duplex, any non-covalent protein-DNA complex, and the 4Bio reaction product. The flow through contains the FAM labeled purified covalent complex and any unreacted, unbound protein, which is estimated at less than 25% of the covalent complex by gel electrophoresis, and is spectroscopically silent in the measurements made herein. The covalent complex purity was estimated at 90% based on 10% sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoesis (PAGE) pre-run and run at 10W for 30 minutes. The FAM labeled free DNA and covalent complex was imaged on the gel using a Typhoon Fluorescence 520 nm filter. The concentration of the purified complex was estimated (usually 150 nM – 200 nM) by comparison to a FAM labeled DNA standard on the gel and in the fluorimeter. Each purification was stored at 4 °C and used within 5 days of preparation to prevent hydrolyzed DNA from contaminating the experiment.
Figure 2.
Purification of the vTopo I-DNA covalent complex. (A) Purification scheme. The enzyme is added to the biotinylated DNA duplex (dsDNA-bio) and following formation of the non-covalent complex (NCC-bio), the covalent complex is formed (CC) and the biotinylated single strand (ssDNA-bio) dissociates from the covalent complex. The reaction mixture is incubated with streptavidin beads and filtered, resulting in separation of the biotinylated products and the purified covalent complex. FAM label used in subsequent experiments depicted with an open circle. (B) SDS-PAGE gel of purification steps. Lane 1: Biotinylated DNA duplex (dsDNA) alone. Lane 2, Reaction mixture prior to strepdavidin-agarose purification. Lane 3: Purified covalent complex.
Equilibrium Binding Measurements
KD measurements for DNA 12mer strands were made by titrating 12DAB, OMe12DAB, OMe-3TCDAB or OMe-6GADAB into a solution containing 10–20 nM 20/32FAM overhang duplex or 10–30 nM Topo-20/32FAM covalent complex (CC) (30 mM NaCl, 20 mM TrisHCl pH 8.0, 1 mM DTT). The quenching of the FAM signal at each DAB-labeled 12mer strand addition was monitored at 25 °C on a Fluoromax-3 fluorimeter (excitation wavelength 495 nm, emission scan from 500–600 nm, excitation slit = 1 nm, emission slit = 5 nm). Each addition was allowed to reach equilibrium, which took up to 50 minutes at low strand concentrations, before an emission scan was collected and the fluorescence at the emission peak wavelength of 518 nm was recorded. The fluorescence quenching data were fitted to eq 1:
| (1) |
Where Fo and Ff are the initial and final fluorescence values, [D] is the total free overhang duplex or enzyme linked over hang duplex, [S] is the oligonucleotide concentration, and b = [D] + [S] + KD. A global fit of titrations to at least two duplex concentrations was made for the 12DAB-DNA duplex and OMe12DAB-CC measurements; the error reported is the error in the global fit. The error reported for the OMe12DAB-DNA duplex KD is the standard deviation of two independent measurements. For binding of the mismatched strands to the overhang DNA duplex and covalent complex, the errors are the deviations of the data from the nonlinear least-squares fit to eq 1.
DNA Strand Association Rate Constant Measurements (kon)
Increasing concentrations of 12mer strands (12DAB, OMe12DAB or 5′-OMe mismatched strands) were added to 10 nM 20/32FAM or 10 nM Topo-20/32FAM (CC) in the reaction buffer noted above. The oligonucleotides were always in >10-fold excess over the other binding partner to maintain pseudo-first order conditions. The quenching of the FAM signal was monitored on an Applied Photophysics stopped-flow with excitation wavelength 495 nm and a >510 nm cutoff optical emission filter. The association curve was fit to a single exponential function (eq 2), where F is the fluorescence intensity, Fo is the amplitude and kobs is the observed rate of association.
| (2) |
The observed rate constants were plotted as a function of 12 mer strand concentration and fit to eq 3, to obtain the association rate constant kon. In this experiment, the intercept is indistinguishable from zero, requiring that koff be independently measured using a different approach.
| (3) |
The reported errors are standard deviations from the theoretical best-fit curve obtained by nonlinear regression fitting. The NaCl dependence of the kobs (eq 2) for OMe12DAB strand association to the free 20/32FAM and covalent complex was studied using a single concentration of 500 nM OMe12DAB and NaCl concentrations in the range 30 to 400 mM.
DNA Strand Dissociation Rate Constant (koff)
Dissociation rate constants were obtained under irreversible conditions by rapidly trapping the free DNA overhang with a large excess of an unlabeled 12mer DNA strand in the reaction buffer noted above. In these experiments, 200 nM of the 20/32FAM duplex or covalent complex was pre-bound with 10% excess of the 12DAB or OMe12DAB strand, using the buffer conditions noted above. For the fully matched 12DAB strands, an unlabeled 12 mer strand was added in 20-fold excess to rapidly anneal to the free DNA overhang exposed after 12DAB dissociation. FAM fluorescence increases were monitored on short time scales (<1000s) using a stopped-flow device, and on longer timescales using a standard fluorimeter (see above). For the mismatched 12 mer strands, where it was not possible to obtain complete binding to the overhang DNA because of their weak binding constants, a 12mer trap complementary to the dissociating mismatched strand was used. As a control, both traps yielded identical the rate constants for dissociation of the fully matched strands (not shown). The dissociation curves required fitting to a double exponential function (eq 4), where F0 is the initial fluorescence and A1 and A2 are the amplitudes for the k1 and k2 observed rates, respectively.
| (4) |
The dissociation rate constants that we report correspond to a kinetic phase that accounts for 90% of the total fluorescence increase; the time dependence of this kinetic phase was found to vary with each mismatch. The small remaining 10% amplitude was invariant with each strand investigated, even as the rate associated with the main amplitude changed 104-fold between strands. This small amplitude phase has been previously observed attributed to sample impurities (24). Although this kinetic phase could in principle be included in a two step model for strand dissociation, the koff value obtained from the larger amplitude phase entirely accounts for the measured KD using the relationship KD = koff/kon. Thus, we only report the koff obtained from the major kinetic phase. The reported errors in koff are standard deviations of at least two or three measurements.
Strand Ligation Kinetics
Ligation experiments were performed in the 30 mM NaCl reaction conditions above with substrate concentrations set such that the incoming strand association was faster than ligation chemistry. 10 μM of 5′-OH 12 mer, -1TC, -3TC, -6GA or -9AG strands were added to 40 nM covalent complex and strand ligation was monitored over time by following the disappearance of the fluorescent covalent complex. The reaction was quenched at various times with addition of an equal volume of 10% SDS, and the covalent complex was resolved from free DNA using electrophoresis through a 10% polyacrylamide gel containing SDS. Because ligated DNA (32/32FAM) and hydrolyzed covalent complex DNA (20/32FAM) run similarly on the gel, the product bands were corrected by subtracting counts corresponding to the fraction of hydrolyzed DNA at time zero. The fraction covalent complex over time was fit to a single-exponential function (eq 2) to obtain the observed rate constant for ligation, klig,obs. The endpoint of the fully paired, -3TC and -6GA reactions were recorded and used along with the measured dissociation constants of the corresponding OMe/DAB substituted strands to calculate concentrations of the ligated DNA and unreacted Topo-DNA covalent complex at the reaction equilibrium (see supplementary information). The equilibrium constant of the ligation reaction, KL, the strand ligation rate, klig, and duplex cleavage rates, kcl, were then calculated for these strands using eqs 5 and 6:
| (5) |
| (6) |
RESULTS
General Approach
Topo I-mediated strand exchange can be divided into two distinct steps (Figure 1): (1) a strand binding step, which involves replacement of the nicked, base paired DNA strand with a new incoming strand, and (2) a strand ligation step, which results in covalent attachment of the new DNA strand at the vTopo cleavage site (Figure 1). To evaluate the role of vTopo in the strand binding step, we synthesized end-labeled DNA constructs with molecular beacon reporters that allow direct measurement of the DNA strand binding equilibria, as well as the strand association and dissociation rates. Through a kinetic and thermodynamic comparison of matched and mismatched strand binding to free DNA overhangs and analogous overhangs in the Topo-DNA covalent complex, the contribution of topoisomerase to the strand binding step was determined. In complementary experiments, the chemical step of DNA strand ligation was investigated in single-turnover reactions using purified Topo IB-overhang DNA covalent complex and a series of matched and mismatched incoming strands. These two approaches allow for complete dissection of the kinetic and thermodynamic contributions of the enzyme to strand binding and ligation with fully matched and mismatched DNA strands.
Thermodynamics of DNA Strand Binding (KD)
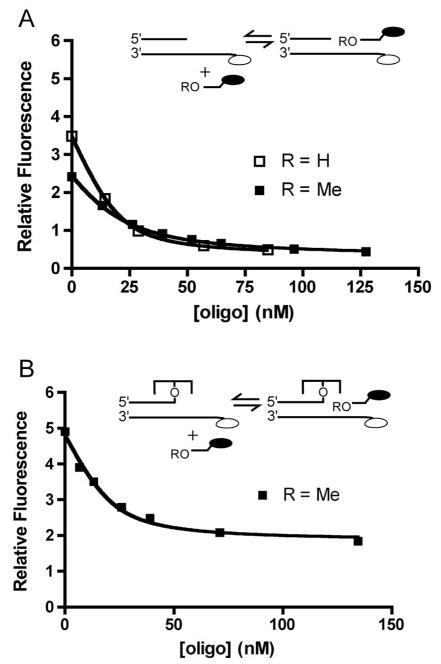
Strand binding to DNA overhangs was measured by following the decrease in fluorescence of a 5′-fluorescein probe on the overhang at increasing concentrations of a complementary 12 mer strand that contained a 3′-dabcyl quench (12DAB) (Figure 3). The experiment was performed on the free overhang DNA (Figure 3A), and the same DNA covalently attached to the enzyme (Figure 3B). To prevent strand ligation in the case of the Topo-DNA covalent complex, the 5′-OH nucleophile on the incoming strand was blocked by 5′-O-methyl group substitution (OMe12DAB). Structural modeling suggests that such a substitution is accommodated in the active site, and this is confirmed by the minor differences in the binding parameters for 5′-OH and 5′-O-methyl strands (Supplemental Figure S1, Table 1). The strand binding measurements were made using low salt concentrations (30 mM) and low free DNA or covalent complex concentrations, so that a reversible equilibrium between the free and bound DNA strand was achieved during the titration. The incoming DNA strand with a 5′-OH was found to bind to the overhang of the free DNA duplex with a KD OH, DNA = 3.8 ± 0.3 nM, which differs by only two-fold from the analogous 5′-O-methyl strand binding to free DNA (KDOMe, DNA = 9.2 ± 1.5 nM). In the context of the covalent vTopo-overhang duplex, the incoming 5′-O-methyl strand binds with a similar affinity to the free overhang duplex (KD, OMe, CC = 4.6 ± 1.8 nM). These thermodynamic measurements indicate that vTopo has only a two-fold effect on the equilibrium constant for strand binding as compared to free DNA.
Figure 3.
Determination of the equilibrium dissociation constants for incoming 12DAB strand binding to a free 20/32FAM overhang duplex. (A) Binding of the OH12DAB and OMe12DAB strands to the free 20/32FAM overhang duplex. (B) Binding of OMe12DAB strand to the 20/32FAM overhang duplex covalently attached to vTopo. In both panels, the dark and open ovals represent the 3′-dabcyl molecular quench and 5′-fluorescein label, respectively.
Table 1.
Thermodynamic and kinetic parameters of matched and mismatched strand binding to free dsDNA and the covalent complex (CC)
| KD (nM) | koff (s−1) | kon (× 104 M−1s−1) | ||||
|---|---|---|---|---|---|---|
| dsDNA | CC | dsDNA | CC | dsDNA | CC | |
| HOmatchedDAB | 3.8 ± 0.3 | (1.9 ± 0.2) × 10−4 | 6.3 ± .05 | 3.2 ± 0.3a | ||
| OMematchedDAB | 9.2 ± 1.5 | 4.6 ± 1.8 | (1.8 ± 0.2) × 10−4 | (2 ± 0.6) × 10−4 | 2.4 ± .07 | 4.7 ± 0.14 |
| OMe-3TCDAB | 900 ± 100 | 1500 ± 154 | 0.063 ± 0.002 | 0.065 ± 0.013 | 4.1 ± 0.4 | 7.8 ± 0.2 |
| OMe-6GADAB | (2.4 ± 0.15) × 104 | (1.5 ± 0.15) × 104 | 1.6 ± 0.01 | 1.9 ± 0.03 | 6.9 ± 1 | 11 ± 4 |
The on-rate of the 5′ OH strand was measured from the slope of a plot of kobs for strand ligation as a function of [HOmatchedDAB] using 30 mM NaCl where the rate limiting step is strand association.
Kinetics of Strand Association (kon) and Dissociation (koff)
The above thermodynamic measurements indicate that vTopo enhances the strand binding equilibrium by about two-fold. However, this measurement alone cannot exclude the possibility that vTopo facilitates strand exchange by increasing the dissociation and association rates to roughly equal extents. To address this question, we measured the second-order rate constant for strand annealing and the first-order rate constant for strand dissociation in the context of the free overhang duplex and in the covalent vTopo-DNA complex.
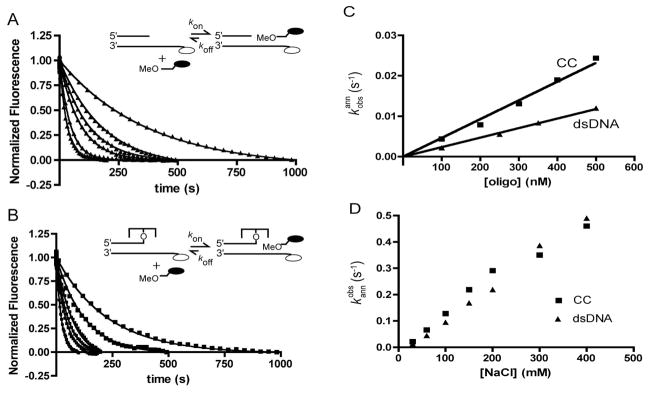
The second-order rate constants for strand annealing were determined from the slopes of the concentration dependence of the pseudo first-order rate constants for OMe12DAB association to the free overhang duplex and the covalent complex (Figure 4A, B, C). Although the data in Figure 2 are truncated at 0.5 μM [OMe12DAB], plots of kobs against [OMe12DAB] were linear over the entire range 0.1 μM to 10 μM with no downward curvature that would indicate a change in rate-limiting step. From these data, association constants of konOMe, DNA = (2.4 ± 0.1) × 104 M−1s−1 and konOMe, CC = (4.7 ± 0.15) ×104 M−1s−1 were determined. The strand association rate constants were found to be strongly dependent on the concentration of sodium chloride (Figure 4D). Increasing concentrations of NaCl from 30 to 400 mM led to a 20-fold increase in strand association rates to both the free DNA and covalent complex. These findings are significant because at low salt, strand association is much slower than the chemical step of strand ligation (see ligation studies below) (26), but at physiological salt concentrations (~150 mM), the strand association rate is comparable to the rate of strand ligation.
Figure 4.
Association rate constants for incoming 12 mer DNA strands. (A) Time dependence of OMe12DAB binding to free 20/32FAM overhang duplex (dsDNA, 10 nM) at 100 to 1000 nM [OMe12DAB] strand. (B) Time dependence of OMe12DAB strand binding to the covalent complex overhang (CC, 10 nM) at 100 to 1000 nM [OMe12DAB] strand. (C) Concentration dependences of the observed strand association rates to the free overhang DNA (dsDNA, ▲) and the covalent complex (CC■). The lines are the best fits to eq 3. (D) Salt concentration dependences of the observed strand association rates for the free overhang DNA and the covalent complex ([OMe12DAB] = 500 nM).
The dissociation rate constants of the OMe12DAB strand from the free overhang duplex and the covalent complex were determined in trapping experiments (Figure 5). Irreversible dissociation of the bound OMe12DAB strand was followed by blocking its re-association with a high concentration of an unlabeled 12 mer trap strand that was complementary to the overhang, or alternatively, by using a trap stand that was complementary to the departing strand. Both methods gave identical results2. These measurements showed that the strand dissociation rates in the absence and presence of covalently bound protein were identical within error: koffOMe, DNA = (1.8 ± 0.2) × 10−4 s−1, koffOMe, CC = (2 ± 0.6) × 10−4 s−1. When the strand KD is calculated from the ratio of the measured off- and on-rates (koff/kon), values within 20% of the measured KD are obtained. Thus the two-fold tighter binding affinity of the OMe12DAB strand for the covalent complex as compared to the free overhang DNA is quantitatively accounted for by the two-fold increase in the strand association rate constant in the presence of vTopo.
Figure 5.
Dissociation of downstream oligonucleotide. (A) Experimental scheme. The OMe12DAB strand is pre-bound to the covalent complex or the free dsDNA (not shown). Following spontaneous dissociation of the OMe12DAB strand, a fast association of an unlabeled trap oligonucleotide prevents re-annealing of the OMe12DAB strand. (B) Superposition of dissociation profiles of the OMe12DAB strand from the covalent complex (CC) and from the free (dsDNA).
The agreement between the koff value directly measured in trapping experiments and the value calculated from the equation koff = KD × kon provides strong validation that the on and off-rate measurements are following the same process as measured in the thermodynamic experiments. This is significant because the trapping experiment involves association of an incoming strand in the presence of a bound strand, while the KD and kon measurements involve simply association and dissociation of a single strand to the overhang. The internal agreement between the different measurements shows that the incoming strand does not alter the off-rate of the departing strand through the formation of a ternary complex2.
Binding of Mismatched Downstream Strands
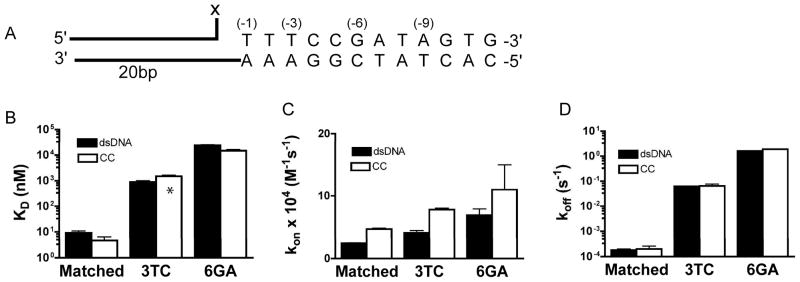
To determine if Topo I discriminates against mismatches during strand binding, single base mismatches were introduced into the OMe12DAB incoming strand at positions -3 and -6 from the cleavage site. At position -3, a T→C substitution was made resulting in a C/A mispair, and at position -6 a G→A substitution was made resulting in a A/C mispair. These constructs were used in KD, kon and koff measurements using the free overhang DNA and the covalent complex. The mismatches caused almost no change in the association rate to the free DNA or the covalent complex as compared to a fully matched strand, but mismatches increased koff by two to three orders of magnitude depending on the position and nature of the mismatch (Figure 6). As observed with the fully matched incoming strand, the presence of vTopo resulted in a modest two-fold increase in the association rate for each mismatch construct as compared to the free overhang DNA, but did not alter the dissociation rate (Figure 6, Table 1).
Figure 6.
Thermodynamics and kinetics of mismatched strand binding. The thermodynamics and kinetics of mismatched OMe-3TCDAB or OMe-6GADAB downstream strand binding to the covalent complex (open) and free dsDNA (black) are compared to fully matched OMe12DAB binding. (A) Downstream strand sequence used for substitution at the -3, or -6 position in these experiments. X is equal to OH for dsDNA overhangs and vTopo for the CC overhangs (B) Dissociation equilibrium constants for matched and mismatched strands. The KD for -3TC covalent complex is calculated from koff/kon, all other values are obtained from equilibrium measurements. (C) Association rates for matched and mismatched strands. (D) Dissociation rates for matched and mismatched strands.
DNA Strand Ligation
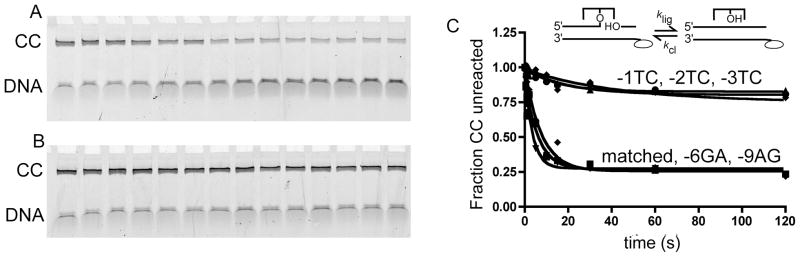
vTopo strand ligation and cleavage is a freely reversible process with a ligation equilibrium constant defined as KL = klig/kcl. Previous studies with fully matched substrates have found that in general the ligated state is favored by factor of ~4 to 10 over the covalent Topo-DNA complex, depending on conditions (27, 28) (Figure 1). To discover if vTopo discriminates against mismatches at the chemical step of ligation, we measured the KL, klig and kcl for paired and mispaired strands. For the fully matched sequence used in binding studies above1, the ligation equilibrium favored ligated DNA by 3-fold, with the ligation and cleavage rates equal klig = 0.12 s−1 and kcl = 0.04 s−1, respectively (Table 2). When strand ligation was evaluated with a series of single base mismatches at the -1, -2, -3, -6 or -9 positions 3′ to the DNA nick, significant decreases in the amount of strand ligation for -1, -2 and -3 substitutions were observed (Figure 7). Less than 20 % of the covalently bound DNA was ligated for the -1TC, -2TC and -3TC mismatched strands, compared with > 75% reacted for the fully paired and -6GA and -9AG mismatched strands.
Table 2.
Ligation rates and ligation equilibria of matched and mismatched strandsa
| Endpoint | klig,obs (s−1) | KL | klig (s−1) | kcl (s−1) | |
|---|---|---|---|---|---|
| matched | 0.75 ± 0.02 | 0.16 ± 0.003 | 3.0 | 0.12 | 0.04 |
| -1TC | 0.20 ± 0.02 | 0.05 ± 0.01 | ND | ND | ND |
| -2TC | 0.20 ± 0.02 | 0.023 ± 0.01 | ND | ND | ND |
| -3TC | 0.16 ± 0.02 | 0.08 ± 0.01 | 0.22 | 0.015 | 0.07 |
| -6GA | 0.74 ± 0.02 | 0.22 ± 0.10 | 7.1 | 0.19 | 0.027 |
| -9AG | 0.75 ± 0.02 | 0.13 ± 0.02 | ND | ND | ND |
Propagated relative errors in the calculated values of KL, klig and kcl for the matched and -3TC strands are less than 30 %; the errors for -6GA are less than 60%. ND, not determined.
Figure 7.
Matched and mismatched DNA strand ligation. (A) SDS-polyacrylamide gel electrophoresis of ligation products from 0 to 120 seconds with fully paired and, (B) -3TC mismatch incoming strand. (C) Plot of fraction covalent complex (CC) remaining as a function of time in strand ligation reactions using a perfectly matched incoming strand, and strands containing mismatches in the -1, -2, -3, -6 and -9 positions.
The ligation equilibrium, KL, was calculated for paired, -3TC and -6GA strands using the measured strand binding equilibrium constant (KD), and the relative amounts of ligated and unligated DNA from the experiment shown in Figure 7 (see Supplemental Materials). This analysis showed that the -3TC mismatch shifted KL towards the unligated state (KL−3TC = 0.2), due to a 10-fold decrease in the ligation rate and two-fold increase in the cleavage rate relative to the matched strand. In contrast, the same calculation for the strand with the -6GA mismatch indicated that the KL, klig and kcl were restored to the values observed for the fully matched strand (Figure 7, Table 1). These data reveal important vTopo interactions with the downstream strand that extend out to at least the -3 position, but not the -6 position (see discussion below).
DISCUSSION
In the past, vTopo catalysis has been studied predominantly in the direction of DNA cleavage using suicide cleavage substrates with short downstream strands that rapidly dissociate(18, 19), or by approach-to-equilibrium cleavage measurements using longer duplexes where the downstream strand remains annealed (17). Consequently, a much better understanding of the vTopo-DNA interactions has been achieved in the forward cleavage direction, as compared to the reverse process of strand annealing and ligation. The present findings provide unique insights into the interactions and mechanism of the reverse reaction and reflect on DNA supercoil release and Topo I and recombinase-mediated DNA recombination (5, 6, 22, 23, 29).
The most salient implication from this study is that vTopo samples an open and closed state during strand exchange and ligation (Scheme 1). The open state is defined as one where enzyme interactions with the downstream DNA are weak; this state need not involve a gross conformational rearrangement as compared to the closed state. The evidence for an open state is the small effect of the enzyme on the strand binding equilibrium and kinetics as compared to the intrinsic parameters observed in the absence of the enzyme (Figures 3–5). These measurements reveal a mechanism of strand annealing where vTopo is largely a bystander as the downstream strands dissociate and associate solely directed by DNA base pairing properties (Scheme 1). More complicated mechanisms involving enzymatic destabilization of the bound strand, a secondary DNA binding site for the incoming DNA strand, or a significant reduction in the time frame for the homology search between the strands are excluded by these data. The modest but reproducible 2-fold increase in the strand association rate could reflect a small contribution from electrostatic guidance by positively charged residues that are observed to neighbor the downstream duplex in the covalent complex (30). The indicated weak interactions of vTopo with the downstream DNA during strand annealing are also consistent with a previous study where it was suggested that the interactions with the downstream DNA region weakened after formation of the covalent complex, thereby allowing DNA supercoils to be released (see below) (17).
Scheme 1.
In contrast to the passive role of vTopo during strand annealing, the data suggest that the enzyme interacts more strongly with the downstream region during the subsequent step of strand ligation, and indicates that a transient closed state of the enzyme-DNA complex forms immediately preceding ligation. Such a transient intermediate is strongly suggested from the differential effects of mismatches on the annealing and ligation steps of strand exchange. All of the mismatches had similar destabilizing effects on the strand annealing parameters to the free DNA and vTopo-DNA overhangs (Figure 6, Table 1), confirming the absence of enzyme involvement at this step. In contrast, mismatches at the -1, -2 and -3 positions had significant effects on the strand ligation kinetics and equilibrium (Figure 7 and Table 2), indicating the presence of an additional state subsequent to annealing that allows the formation of important interactions between the enzyme and substrate within three base pairs of the cleavage site.
To further explore the effects of mismatches on strand ligation, a kinetic dissection of the reversible ligation reaction was carried out for the -3 and -6 mismatches (Table 2, Supplemental Materials). This analysis showed that the -3 mismatch decreased klig by 10-fold, but slightly enhanced kcl. In contrast, damaging effects on ligation were not observed for the -6 mismatch. Thus, in addition to destabilizing the annealed strand in the open complex by about +3.5 kcal/mol, the -3 mismatch further destabilizes vTopo-DNA interactions within the transient closed state by an additional +1.4 kcal/mol. Both effects serve to increase the overall activation barrier for ligation under conditions where strand binding is rate determining. An alternative explanation for the deleterious effect of the -3 mismatch involving selective destabilization of the transition state for ligation is not easily reconciled with the data because the cleavage rate was not decreased, and cleavage proceeds through the same transition state as ligation. Although a detailed analysis was not performed for the -1 or -2 mismatches, the similar effects of these mismatches on the observed ligation rate and extent suggests a similar mechanism (Table 2). Thus, with respect to the specificity of strand exchange, mismatches are disfavored at two steps relative to a fully matched strand. First, regardless of the position of the mismatch relative to the cleavage site, mismatches will be destabilizing towards strand annealing, and will be disfavored in competition with a fully matched strand. Second, mismatches located within three positions of the cleavage site will destabilize the closed state, shifting the equilibrium away from ligation of the mismatched strands.
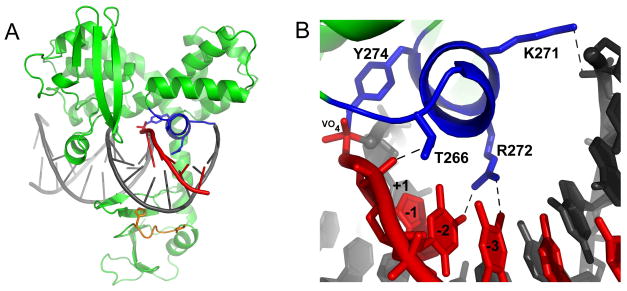
These mechanistic features for mismatch discrimination for vTopo appear to extend to the broader family of tyrosine recombinases. For lambda integrase, Cre recombinase and Flp recombinase, mispairs within two nucleotides of the tyrosyl-DNA linkage are prevented from ligation, but not strand association (31). In addition, for Flp recombinase, it has been shown that mispairs within three nucleotides downstream of the cleavage site inhibited ligation, while the reverse cleavage reaction proceeded uninhibited (32). Inspection of the non-covalent and covalent complex structures of hTopo and variola Topo, as well as the non-covalent Cre recombinase structure, reveals a conserved ~10 amino acid helix that is positioned appropriately to provide these selective interactions (30, 33, 34). A recent structure of the variola Topo in complex with DNA containing a covalent vanadate transition state mimic (Figure 8) (35)3 shows that the N-terminus of this helix approaches the downstream region of the scissile DNA strand, where several hydrogen bonds are made with the first few downstream nucleotides (Figure 8). Consistent with the lack effects of mismatches at the -6 position, no further contacts are observed with this strand. The selective effects of DNA mismatches at the -1 to -3 positions during ligation, but not cleavage, may indicate a greater role for correct base pairing in positioning the 5′-OH for nucleophilic attack during ligation. In contrast, correct base pairing may be less important during the cleavage reaction because the DNA backbone is covalently intact.
Figure 8.
Downstream strand binding by a poxvirus topoisomerase. (A) Overall structure of the variola poxvirus Topo in complex with DNA containing a covalent vanadate transition state mimic (PDB ID 3IGC) (35)3. The short helix (blue) that makes several contacts with the cleaved downstream strand (red) is shown. In addition, a loop (orange) from the N-terminal domain that hydrogen bonds with the complementary uncleaved strand is shown. (B) Detailed view of conserved helix (blue) which contacts the cleaved strand (red) out to three nucleotides 3′ to the cleavage site. At the base of this interacting helix is the catalytic tyrosine, Y274, which is coordinated along with 3′- and 5′-OH groups to the vanadate transition state mimic (VO4). Hydrogen bonds are shown as black dashes.
A related question is whether tyrosine recombinases can kinetically facilitate strand dissociation and annealing, or like vTopo, rely on the intrinsic dynamics of DNA base pairing. A conclusive answer to this question is not currently available, but the rates of strand dissociation in Table 1 (~2 × 10−4 s−1) for a 12 mer DNA strand measured at 22 °C) are not kinetically competent to account for the maximal observed rates of strand recombination at a LoxP site catalyzed by Cre recombinase at a similar temperature (~0.005 to 0.03 s−1) (29). However, the uncertainty in the sequence dependence of the strand dissociation rate, and the fact that the Cre recombination occurs through a shorter 8 bp spacer, still allows for the distinct possibility that intrinsic DNA strand unpairing dynamics may limit the overall process.
Implications for Topo-Catalyzed Supercoil Release and DNA Recombination
The open state, here characterized in terms of strand replacement during strand exchange, may be the same state populated during free rotation by the downstream strand during supercoil release. Previous ensemble and single-molecule studies of supercoil release by vTopo have indicated that the enzyme removes ~5 to 14 superhelical turns each time the DNA backbone is nicked (3, 4). This type of mechanism requires that the enzyme transiently releases its grasp on the downstream DNA region to allow rotation of the duplex around the uncleaved strand (3). Thus, in the context of the open to closed dynamic transition revealed here, the number of supercoils removed is determined by the frequency of strand rotation compared to the frequency of closing. The present data provide no information on the opening and closing transition rates, but closing must be competitive with duplex rotation (~20 s−1) or complete unwinding would occur at each nicking event, which was not observed (3).
The vTopo I studies described here suggest a model for camptothecin (CPT)-induced recombination events (15). In the covalent complex, where the dissociation rate of the nicked strand is ~104-fold slower than strand ligation, a drug such as CPT that hinders strand ligation could extend the lifetime of the covalent complex such that the probability of strand dissociation is significantly increased. After strand dissociation, CPT may also depart because its binding mode includes DNA intercalation (10) and there could then be competitive annealing between the departing strand and a new strand, which may contain mismatches. If the mismatched region is beyond three nucleotides from the cleavage site, association of the new strand could lead to ligation and illegitimate recombination (6, 36, 37). Although the measured effects with a short 12mer DNA strand may not fully replicate the features of all types of recombination events, especially those that occur during DNA replication, these measurements do, for the first time, quantify the role of the enzyme during the partial reaction of strand hybridization, which is a requisite step in all recombination reactions.
Supplementary Material
Abbreviations
- vTopo
vaccinia type I topoisomerase
- hTopo
human type I topoisomerases
- CC
covalent complex of vTopo with DNA
Footnotes
The matched DNA sequences used in this study contains a T at the -1 position relative to the cleavage site. Previous work has shown that an A at this position accelerates the cleavage and ligation rates by about 10-fold relative to T, but does not change the cleavage equilibrium significantly. The nature of the base at this position has no effect on interpretations in this work.
In addition to switching the trapping strand, and confirming that trapping was zero-order with respect to [trap], it was important to show that the trapping strand does not bind appreciably to vTopo and competitively disrupt the interactions between the enzyme and the bound duplex DNA. To exclude this possibility we performed two experiments. First, we used fluorescence anisotropy to confirm that a FAM-labeled 12 mer strand binds very weakly to free vTopo (KD12 mer = 2.5 ± 0.5 μM) (Supplemental Figure 1A). Second, using the same experimental approach, we confirmed that the catalytically inactive Y274F mutant enzyme binds noncovalently to the 32/32FAM duplex 25-fold more tightly than the 12 mer (KD = 100 ± 50 nM) (Supplemental Figure 1B). Moreover, if competitive binding were affecting the observed results, nonlinear plots of kobs against 12 mer concentration would have been observed in Figure 4C, which is not the case.
We thank Dr. Gregory D. Van Duyne for providing the crystallographic coordinates for the variola topoisomerase in complex with vanadate and DNA (PDB ID 3IGC) prior to their release.
This work was supported by grant GM068626 from the National Institutes of Health to J.T.S. and an Anticancer Drug Development Training Grant Fellowship from the National Cancer Institute to M.R.S.
References
- 1.Champoux JJ. DNA TOPOISOMERASES: Structure, Function, and Mechanism. Annu Rev Biochem. 2001;700:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 2.Shuman S. Vaccinia Virus DNA Topoisomerase: A Model Eukaryotic Type IB Enzyme. BBA-Gene Structure and Expression. 1998;1400:321–337. doi: 10.1016/s0167-4781(98)00144-4. [DOI] [PubMed] [Google Scholar]
- 3.Stivers JT, Harris TK, Mildvan AS. Vaccinia DNA Topoisomerase I: Evidence Supporting a Free Rotation Mechanism for DNA Supercoil Relaxation. Biochemistry. 1997;36:5212–5222. doi: 10.1021/bi962880t. [DOI] [PubMed] [Google Scholar]
- 4.Koster DA, Croquette V, Dekker C, Shuman S, Dekker NH. Friction and Torque Govern the Relaxation of DNA Supercoils by Eukaryotic Topoisomerase IB. Nature. 2005;434:671–674. doi: 10.1038/nature03395. [DOI] [PubMed] [Google Scholar]
- 5.Shuman S. Recombination Mediated by Vaccinia Virus DNA Topoisomerase I in Escherichia Coli is Sequence Specific. Proc Natl Acad Sci USA. 1991;88:10104–10108. doi: 10.1073/pnas.88.22.10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu J, Schiestl RH. Topoisomerase I Involvement in Illegitimate Recombination in Saccharomyces Cerevisiae. Mol Cell Biol. 1996;16:1805–1812. doi: 10.1128/mcb.16.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shuman S. DNA Strand Transfer Reactions Catalyzed by Vaccinia Topoisomerase I. J Biol Chem. 1992;267:8620–8627. [PubMed] [Google Scholar]
- 8.Grindley ND, Whiteson KL, Rice PA. Mechanisms of Site-Specific Recombination. Annu Rev Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- 9.Hsiang YH, Hertzberg R, Hecht S, Liu LF. Camptothecin Induces Protein-Linked DNA Breaks Via Mammalian DNA Topoisomerase I. J Biol Chem. 1985;260:14873–14878. [PubMed] [Google Scholar]
- 10.Staker BL, Feese MD, Cushman M, Pommier Y, Zembower D, Stewart L, Burgin AB. Structures of Three Classes of Anticancer Agents Bound to the Human Topoisomerase I-DNA Covalent Complex. J Med Chem. 2005;48:2336–2345. doi: 10.1021/jm049146p. [DOI] [PubMed] [Google Scholar]
- 11.Hsiang YH, Lihou MG, Liu LF. Arrest of Replication Forks by Drug-Stabilized Topoisomerase I-DNA Cleavable Complexes as a Mechanism of Cell Killing by Camptothecin. Cancer Res. 1989;49:5077–5082. [PubMed] [Google Scholar]
- 12.Wethington SL, Wright JD, Herzog TJ. Key Role of Topoisomerase I Inhibitors in the Treatment of Recurrent and Refractory Epithelial Ovarian Carcinoma. Expert Rev Anticancer Ther. 2008;8:819–831. doi: 10.1586/14737140.8.5.819. [DOI] [PubMed] [Google Scholar]
- 13.Larsen AK, Gobert C. DNA Topoisomerase I in Oncology: Dr Jekyll Or Mr Hyde? Pathol Oncol Res. 1999;5:171–178. doi: 10.1053/paor.1999.0209. [DOI] [PubMed] [Google Scholar]
- 14.Pourquier P, Pommier Y. Topoisomerase I-Mediated DNA Damage. Adv Cancer Res. 2001;8020487357:189–216. doi: 10.1016/s0065-230x(01)80016-6. [DOI] [PubMed] [Google Scholar]
- 15.Pourquier P, Pilon AA, Kohlhagen G, Mazumder A, Sharma A, Pommier Y. Trapping of Mammalian Topoisomerase I and Recombinations Induced by Damaged DNA Containing Nicks Or Gaps. Importance of DNA End Phosphorylation and Camptothecin Effects. J Biol Chem. 1997;272:26441–26447. doi: 10.1074/jbc.272.42.26441. [DOI] [PubMed] [Google Scholar]
- 16.Nagarajan R, Stivers JT. Major Groove Interactions of Vaccinia Topo I Provide Specificity by Optimally Positioning the Covalent Phosphotyrosine Linkage. Biochemistry. 2006;45:5775–5782. doi: 10.1021/bi060133i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagarajan R, Stivers JT. Unmasking Anticooperative DNA-Binding Interactions of Vaccinia DNA Topoisomerase I. Biochemistry. 2007;46:192–199. doi: 10.1021/bi061706u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwon K, Jiang YL, Song F, Stivers JT. 19F NMR Studies of Vaccinia Type IB Topoisomerase. Conformational Dynamics of the Bound DNA Substrate. J Biol Chem. 2002;277:353–358. doi: 10.1074/jbc.M109450200. [DOI] [PubMed] [Google Scholar]
- 19.Kwon K, Stivers JT. Fluorescence Spectroscopy Studies of Vaccinia Type IB DNA Topoisomerase. Closing of the Enzyme Clamp is Faster than DNA Cleavage. J Biol Chem. 2002;277:345–352. doi: 10.1074/jbc.M109449200. [DOI] [PubMed] [Google Scholar]
- 20.Stivers JT, Jagadeesh GJ, Nawrot B, Stec WJ, Shuman S. Stereochemical Outcome and Kinetic Effects of Rp- and Sp-Phosphorothioate Substitutions at the Cleavage Site of Vaccinia Type I DNA Topoisomerase. Biochemistry. 2000;39:5561–5572. doi: 10.1021/bi992429c. [DOI] [PubMed] [Google Scholar]
- 21.Shuman S, Prescott J. Specific DNA Cleavage and Binding by Vaccinia Virus DNA Topoisomerase I. J Biol Chem. 1990;265:17826–17836. [PubMed] [Google Scholar]
- 22.Shuman S. Vaccinia DNA Topoisomerase I Promotes Illegitimate Recombination in Escherichia Coli. Proc Natl Acad Sci USA. 1989;86:3489–3493. doi: 10.1073/pnas.86.10.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sekiguchi J, Seeman NC, Shuman S. Resolution of Holliday Junctions by Eukaryotic DNA Topoisomerase I. Proc Natl Acad Sci USA. 1996;93:785–789. doi: 10.1073/pnas.93.2.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reynaldo LP, Vologodskii AV, Neri BP, Lyamichev VI. The Kinetics of Oligonucleotide Replacements. J Mol Biol. 2000;297:511–520. doi: 10.1006/jmbi.2000.3573. [DOI] [PubMed] [Google Scholar]
- 25.Shuman S. Site-Specific Interaction of Vaccinia Virus Topoisomerase I with Duplex DNA. Minimal DNA Substrate for Strand Cleavage in Vitro. J Biol Chem. 1991;266:11372–11379. [PubMed] [Google Scholar]
- 26.Minkah N, Hwang Y, Perry K, Van Duyne G, Hendrickson R, Lefkowitz EJ, Hannenhalli S, Bushman FD. Variola Virus Topoisomerase: DNA Cleavage Specificity and Distribution of Sites in Poxvirus Genomes. Virology. 2007;365:60–69. doi: 10.1016/j.virol.2007.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stivers JT, Shuman S, Mildvan AS. Vaccinia DNA Topoisomerase I: Kinetic Evidence for General Acid-Base Catalysis and a Conformational Step. Biochemistry. 1994;33:15449–15458. doi: 10.1021/bi00255a027. [DOI] [PubMed] [Google Scholar]
- 28.Wittschieben J, Shuman S. Mechanism of DNA Transesterification by Vaccinia Topoisomerase: Catalytic Contributions of Essential Residues Arg-130, Gly-132, Tyr-136 and Lys-167. Nucleic Acids Res. 1997;25:3001–3008. doi: 10.1093/nar/25.15.3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin SS, Chu VC, Baldwin E. Modulation of the Active Complex Assembly and Turnover Rate by Protein-DNA Interactions in Cre-LoxP Recombination. Biochemistry. 2003;42:6814–6826. doi: 10.1021/bi0272306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perry K, Hwang Y, Bushman FD, Van Duyne G. Structural Basis for Specificity in the Poxvirus Topoisomerase. Mol Cell. 2006;23:343–354. doi: 10.1016/j.molcel.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 31.Rajeev L, Malanowska K, Gardner JF. Challenging a Paradigm: The Role of DNA Homology in Tyrosine Recombinase Reactions. Microbiol Mol Biol Rev. 2009;73:300–309. doi: 10.1128/MMBR.00038-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee J, Jayaram M. Role of Partner Homology in DNA Recombination. Complementary Base Pairing Orients the 5′-Hydroxyl for Strand Joining during Flp Site-Specific Recombination. J Biol Chem. 1995;270:4042–4052. doi: 10.1074/jbc.270.8.4042. [DOI] [PubMed] [Google Scholar]
- 33.Stewart L. A Model for the Mechanism of Human Topoisomerase I. Science. 1998;279:1534–1541. doi: 10.1126/science.279.5356.1534. [DOI] [PubMed] [Google Scholar]
- 34.Ghosh K, Guo F, Van Duyne G. Synapsis of loxP Sites by Cre Recombinase. J Biol Chem. 2007;282:24004–24016. doi: 10.1074/jbc.M703283200. [DOI] [PubMed] [Google Scholar]
- 35.Perry K, Hwang Y, Bushman FD, Duyne G. Insights from the Structure of a Smallpox Virus Topoisomerase-DNA Transition State Mimic. Structure. 2011;18:127–137. doi: 10.1016/j.str.2009.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bullock P, Champoux JJ, Botchan M. Association of Crossover Points with Topoisomerase I Cleavage Sites: A Model for Nonhomologous Recombination. Science. 1985;230:954–958. doi: 10.1126/science.2997924. [DOI] [PubMed] [Google Scholar]
- 37.Champoux JJ. Topoisomerase I is Preferentially Associated with Isolated Replicating Simian Virus 40 Molecules After Treatment of Infected Cells with Camptothecin. J Virol. 1988;62:3675–3683. doi: 10.1128/jvi.62.10.3675-3683.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.