Abstract
Purpose
To analyze transforming growth factor beta-induced (TGFBI) gene mutations in a Chinese pedigree with Reis-Bücklers dystrophy (RBCD).
Methods
In a four-generation Chinese family with Reis-Bücklers dystrophy, six members were patients and the rest were unaffected. All members of the family underwent complete ophthalmologic examinations. Exons of TGFBI were amplified by polymerase chain reaction, sequenced, and compared with a reference database. The sequencing results were reconfirmed by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP).
Results
A single heterozygous C>T (R124C) point mutation was found in exon 4 of TGFBI in all six members of the pedigree affected with RBCD, but not in the unaffected members.
Conclusions
Within this pedigree, RBCD segregates with the R124C variance, which is a known mutation for lattice corneal dystrophy type I. Therefore, along with G623D and R124L, the R124C mutation in TGFBI is also found to be responsible for RBCD.
Introduction
Inherited corneal dystrophy is mainly classified as lattice, granular, Avellino, Thiel-Behnke, and Reis-Bücklers corneal dystrophy (RBCD/CDB1, OMIM 608470). RBCD, a dominantly inherited dystrophy, is characterized by bilateral, progressive, and painful corneal erosions, and significant visual impairment [1]. Histopathologically, the involvement of the Bowman’s layer, the presence of band-shaped granular and subepithelial deposits that stained intensely red with Masson trichrome, and “rod shaped bodies” in cornea were confirmed in this disease [2,3]. The mechanisms remain unclear, but it is generally accepted that transforming growth factor beta-induced (TGFBI) is actively involved in the pathogenesis of RBCD.
Initially known as kerato-epithelin, TGFBI is an extracellular matrix protein induced by transforming growth factor-beta 1 and is highly expressed in the corneal epithelium. It contains a Arg-Gly-Asp (RGD) motif that acts as a ligand recognition sequence for several integrins, and thus is associated with cell-collagen interactions with a role in the regulation of cell-adhesion. Therefore, it is believed that TGFBI plays a role in corneal development and wound healing by mediating cell adhesion via its interaction with collagen, fibronectin, and integrins. The human TGFBI gene encodes a 682 amino acids protein (68 kDa) with four internal homologous repeats. Currently, more than 30 mutations in TGFBI have been demonstrated in four different types of corneal dystrophies [4].
Mutated TGFBI has been linked to the four types of corneal dystrophy, including RBCD [5-11]. Two mutations, R124L [12,13] and G623D [14], have previously been identified in patients with typical RBCD, including Chinese RBCD pedigrees. In this study, R124C mutation, rather than R124L or G623D, was identified— to the best of our knowledge—for the first time in a Chinese family with RBCD.
Methods
Patient recruitment
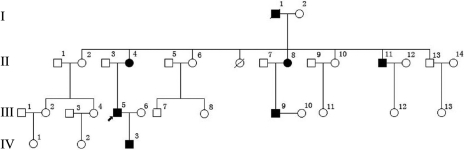
A four-generation Chinese family from the Sichuan province with RBCD was included in this study (Figure 1). This study includes six RBCD patients and six unaffected relatives. This study was approved by a local institutional medical ethics committee, and informed consent conforming to the tenets of the Declaration of Helsinki was obtained from each of participants.
Figure 1.
The pedigree of the family with Reis-Bücklers dystrophy (RBCD). The circle indicates female, the square indicates male, and the filled circle or square indicates the affected individual with RBCD. Arrow signifies the proband and slash through symbol indicates death.
Clinical examination
A Snellen best-corrected visual acuity test, a slit-lamp biomicroscopy, and a fundus examination were conducted by an experienced ophthalmologist for all subjects. Laser scanning in vivo confocal microscopy (Heidelberg Retina Tomograph III, Rostock Corneal Module [RCM]; Heidelberg Engineering GmbH, Heidelberg, Germany) were also performed on some of the affected and unaffected individuals.
DNA extraction and polymerase chain reaction
Peripheral blood samples were drawn from six RBCD patients and six unaffected members. Leukocyte DNA was extracted from 200 μl peripheral blood using a TIANamp Genomic DNA Kit (Tiangen Biotech Co. Ltd, Beijing, China), following the manufacturer’s instructions. DNA integrity was evaluated by 1% agarose gel electrophoresis. The intronic primers flanking the exons were designed based on genomic sequences of TGFBI (Consortium Human Build 37 NC_000005) and synthesized by Invitrogen Company (Carlsbad, CA). The sequences of the primers are listed in Table 1.
Table 1. Primers used in polymerase chain reaction for amplification of TGFBI.
| Exon | Primer direction | Sequence (5′→3′) | Annealing temperature (°C) |
|---|---|---|---|
| 1 |
Forward: |
GCTTGCCCGTCGGTCGCTA |
62 |
| |
Reverse: |
TCCGAGCCCCGACTACCTGA |
|
| 2 |
Forward: |
AGGCAAACACGATGGGAGTCA |
60 |
| |
Reverse: |
TAGCACGCAGGTCCCAGACA |
|
| 3 |
Forward: |
CCAGATGACCTGTGAGGAACAGTGA |
60 |
| |
Reverse: |
CCTTTTATGTGGGTACTCCTCTCT |
|
| 4 |
Forward: |
TCCTCGTCCTCTCCACCTGT |
58 |
| |
Reverse: |
CTCCCATTCATCATGCCCAC |
|
| 5 and 6 |
Forward: |
CCTGGGCTCACGAGGGCTGAGAACAT |
64 |
| |
Reverse: |
GCCCCTCTTGGGAGGCAATGTGTCCC |
|
| 7 |
Forward: |
GTGAGCTTGGGTTTGGCTTC |
63 |
| |
Reverse: |
ACCTCATGGCAGGTGGTATG |
|
| 8 |
Forward: |
TGAGGTTATCGTGGAGTG |
53 |
| |
Reverse: |
CACATCAGTCTGGTCACA |
|
| 9 |
Forward: |
ACTCACGAGATGACATTCCT |
60 |
| |
Reverse: |
TCCAGGGACAATCTAACAGG |
|
| 10 |
Forward: |
TAGAAGATACCAGATGTTAAGG |
56 |
| |
Reverse: |
TGTCAGCAACCAGTTCTCAT |
|
| 11 |
Forward: |
CCTGCTACATGCTCTGAACAA |
58 |
| |
Reverse: |
GAATCCCCAAGGTAGAAGAAAG |
|
| 12 |
Forward: |
GACTCTACTATCCTCAGTGGTG |
58 |
| |
Reverse: |
ATGTGCCAACTGTTTGCTGCT |
|
| 13 |
Forward: |
CATTAGACAGATTGTGGGTCA |
60 |
| |
Reverse: |
GGGCTGCAACTTGAAGGTT |
|
| 14 |
Forward: |
GCGACAAGATTGAAACTCCAT |
58 |
| |
Reverse: |
CTCTCCACCAACTGCCACAT |
|
| 15 |
Forward: |
CCCTCAGTCACGGTTGTT |
58 |
| |
Reverse: |
GGAGTTGCCTTGGTTCTT |
|
| 16 |
Forward: |
CTTGCACAACTTATGTCTGC |
58 |
| |
Reverse: |
TGCACCATGATGTTCTTATC |
|
| 17 |
Forward: |
AGTGAAGTTTCACAAACCAC |
58 |
| Reverse: | CCACATTTGGGATAGGTC |
Summary of the primers and annealing temperatures used for the amplification of the 17 exons of TGFBI.
DNA fragments were amplified by PCR using a MyCycler thermocycler (Bio-Rad, Hercules, CA). The 30 μl PCR reaction mixture included 1× PCR buffer (GC buffer I were used for several reactions, see Table 1), 30 ng DNA, 2.5 mM MgCl2, 0.3 mM of each of dNTPs, 1.5U Pfu DNA polymerase, and 1.0 μM of each of the forward and reverse primers. All reagents used in this procedure were purchased from TaKaRa Biotechnology Co., Ltd. (Dalian, China). The reactions were incubated at 94 °C for 3 min, followed by 35 cycles of 94 °C for 10 s, 53–64 °C for 30 s, and 72 ° for 1–2 min, followed by a final extension at 72 °C for 5 min.
Sequencing and PCR-RFLP analysis
The PCR products were purified with a TIANgel Midi Purification Kit (Tiangen Biltech Co. Ltd, Beijing, China) and sequenced using an ABI 377XL automated DNA sequencer (Applied Biosystems, Foster City, CA). Sequence data were compared pair-wise with the published TGFB1 sequences. To confirm the results of sequence analysis, a commonly used method of polymerase chain reaction-restriction fragment length polymorphism analysis (PCR-RFLP) [15] was performed, as the pathogenic mutation (R124C) resulted in a deprivation of a restriction endonuclease AciI recognition site. Exon 4 of TGFBI was amplified from each family member by PCR using the primers mentioned above, purified by a PCR purification kit (Qiagen, Hamburg, Germany) and digested with an AciI restriction enzyme (New England Biolabs, Inc., Beverly, MA) for 3 h at 37 °C. Fragments were analyzed by electrophoresis.
Results
Clinical presentation
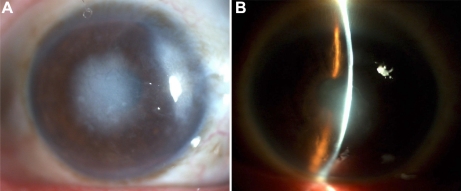
In the family, six individuals with RBCD and six unaffected individuals were examined. The proband (a 33-year-old male, patient III:3, Figure 1) experienced recurrent photophobia, progressive vision loss, and corneal erosion since the age of 10. At the time of the examination, the best corrected visual acuity was hand motion (OD) and 6/600 (OS). Slit lamp examination showed multiple annular grayish opacities at the subepithelial and anterior stroma of the central cornea of both eyes. Representative clinical photographs of the cornea of an affected family member are shown in Figure 2. Patient IV:3, the 4-year-old son of the proband, presented with no clinical symptoms, but manifested bilateral diffuse and small dot epithelial and subepithelial opacities in the central cornea (Figure 2).
Figure 2.
Slit-lamp examination showed an irregular corneal surface and several discrete, gray-white opacities in the subepithelial area and Bowman’s layer. Some of the opacities were associated with marked corneal scarring. No neovascularization and stromal lattice were observed.
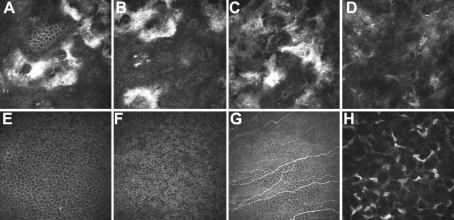
In vivo laser scanning confocal microscopy was performed on the proband. Focal depositions of homogeneous reflective materials with rounded and hyporeflective edges were observed (Figure 3).
Figure 3.
In vivo laser scanning confocal microscopy. A–D: Patient III:3 36 μm (A), 42 μm (B), 59 μm (C), and 110 μm (D) from the corneal surface. Focal deposition of homogeneous reflective materials with rounded and hyporeflective edges were observed (A–C). Amyloid-like deposits that were hyperreflective and with poorly demarcated margins were observed (D). E–H: Normal individual: 36 μm (A), 42 μm (B), 57 μm (C), and 113 μm (D) from the corneal surface. Epithelium cells (E, F) nerve fibers (G), and corneal stromal cells (H) were observed.
TGFBI mutation analysis
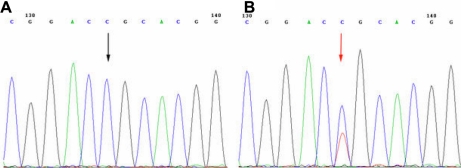
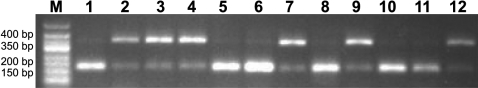
A single heterozygous C>T mutation was found (R124C) in exon 4 of TGFBI in all affected members of this pedigree (Figure 4). This R124C mutation co-segregated with the disorder within the family. This mutation causes an Arginine to Cysteine substitution at the protein level. The results of the sequencing analysis were confirmed by PCR-RFLP analysis (Figure 5). After digestion, wild type alleles were cut into two fragments, 181 bp and 172 bp, whereas the affected patients’ alleles were cut into three fragments, 353 bp, 181 bp, and 172 bp.
Figure 4.
TGFBI heterozygous mutation in the family. A: Unaffected individuals of the family. B: Patients. The sequence in an affected member shows a heterozygous C>T transversion (indicated by the arrow).
Figure 5.
PCR-RFLP. Analysis of R124C mutation by polymerase chain reaction restriction fragment length polymorphism (PCR-RFLP). Lane 1–12: II:2, II:4, II:8, II:11, II:13, III:4, III:5, III:6, III:9, III:13, IV:1, and IV:3 of the pedigree. Lane 2, 3, 4, 7, 9, and 12 are affected individuals, and the rest are unaffected individuals of the pedigree.
Discussion
Corneal dystrophy is a group of diseases with autosomal dominant inheritance. Until now, corneal dystrophies failed to be clearly classified because of the variability in phenotypic expression of the diseases. A proposed corneal dystrophy classification system, which is identical or similar to those in the current nomenclature, is anatomically based with dystrophies classified according to the layer mainly involved, such as the epithelial and subepithelial, Bowman’s layer, stroma, Descemet’s membrane, and endothelium [16]. Clinical characteristics such as the depth of the cornea affected, the morphology of the deposits, and the histopathological features are also important for classifying different corneal dystrophies [17]. However, dystrophies with overlapping and atypical characteristics are still too similar to be distinguished from one another. A Chinese family with atypical RBCD was recently reported [14]. This family presented with a unique corneal dystrophy within the Bowman’s layer and the corneal stroma. However, no lattice was noted in the proband or other affected members, and the deposits were located in the mid-stroma of the cornea, which was different from the phenotypes previously reported in lattice corneal dystrophy patients with R124C mutation. Some corneal dystrophies affect multiple corneal layers and therefore cannot be classified as a single type based on morphologic criteria.
Phenotypically, the pedigree we documented here exhibited typical features of RBCD. The affected individuals presented with a gray-white geographic opacity in the anterior to mid-stroma of both eyes. In addition, geometric and round opacities in the subepithelial layers and anterior to mid-stroma were found in all of the affected family members. The clinical features, including recurrent erosion and gradually developing opacities of the Bowman’s layer, were consistent with the characteristic of RBCD [1,18] and to those found in the families previously described by Afshari et al. [10] and Aldave et al. [19]. In vivo confocal microscopy images were also consistent with the reported findings in RBCD [20].
TGFBI has been closely involved with the inherited corneal dystrophies, as mutations in this gene were identified in at least 5 types of corneal dystrophies, including granular corneal dystrophy (R555W), Avellino corneal dystrophy (R124H), lattice corneal dystrophy type I (R124C), Thiel-Behnke corneal dystrophy (R555L), and RBCD (R124L, G623D) [10,19]. Among those mutations reported, R124 appeared to be a “hot-spot” point mutation in TGFBI [21,22] as the R124 mutation has been detected in three types of corneal dystrophies, including Avellino corneal dystrophy, lattice corneal dystrophy type I, and RBCD. In previous reports, R124C is associated with lattice corneal dystrophy [15,21,23]. Interestingly, R124 in exon 4 of TGFBI is conserved among several species, including Homo sapiens, Mus musculus (R124), Pan troglodytes (R124), Macaca mulatta (R124), and Rattus norvegicus (R124), and is incompletely conserved in Gallus gallus (R117) and Danio rerio (R118). It strongly suggests that this residue is an important—functional and structural—site of the protein.
To date, TGFBI was the only gene found to be associated with RBCD. Here, we demonstrated an unusual R124C mutation in TGFBI associated with the RBCD, which was a mutation known to be responsible for corneal lattice dystrophy type I. Therefore, along with G623D and R124L, the R124C mutation in TGFBI is also found to be responsible for RBCD.
In 1996, Small et al. [24] suggested that Reis-Bücklers, lattice type 1, Avillino, and granular corneal dystrophies are all the same disease as they stem from the same gene products. Recently, it was suggested by the International Committee for Classification of Corneal Dystrophy that the following corneal dystrophies be named as TGFBI corneal dystrophies, including granular corneal dystrophy type 3 (RBCD), Thiel–Behnke corneal dystrophy (TBCD), classic lattice corneal dystrophy (LCD1), Granular corneal dystrophy, type 1 (classic; GCD1), and Granular corneal dystrophy, type 2 (granular-lattice; GCD2) [16]. Corneal dystrophies caused by mutations in TGFBI are characterized by abnormal extracellular deposits of mutated TGFBI protein in the corneal stroma. Most of TGFBI mutations so far reported are located in the fourth Fas1 domain with two mutational hot spots in R124 and R555. There is a strong correlation between phenotype-genotype in most corneal dystrophies caused by TGFBI mutations. The corneal dystrophy with R124L mutation usually has a worse prognosis, whereas the clinical manifestations of corneal dystrophies resulting from R555T or R555C mutations are usually mild. Other mutations such as P501T and N622K are related to various subtypes of lattice-like dystrophies. However, as dystrophies have a known common genetic basis, they may be classified into a single category, TGFBI dystrophy [16].
Acknowledgments
The authors wish to thank Dr. Yi Mou for his technical help.
References
- 1.Wittebol-Post D, Pels E. The dystrophy described by Reis and Bücklers. Separate entity or variant of the granular dystrophy? Ophthalmologica. 1989;199:1–9. doi: 10.1159/000310006. [DOI] [PubMed] [Google Scholar]
- 2.Kuchle M, Green WR, Volcker HE, Barraquer J. Reevaluation of corneal dystrophies of Bowman's layer and the anterior stroma (Reis-Bucklers and Thiel-Behnke types): a light and electron microscopic study of eight corneas and a review of the literature. Cornea. 1995;14:333–54. doi: 10.1097/00003226-199507000-00001. [DOI] [PubMed] [Google Scholar]
- 3.Wheeldon CE, De Karolyi BH, Patel DV, Sherwin T, McGhee CN, Vincent AL. A novel phenotype-genotype relationship with a TGFBI exon 14 mutation in a pedigree with a unique corneal dystrophy of Bowman’s layer. Mol Vis. 2008;14:1503–12. [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Z, Wang YQ, Gong QH, Xie LX. An R124C mutation in TGFBI caused lattice corneal dystrophy type I with a variable phenotype in three Chinese families. Mol Vis. 2008;14:1234–9. [PMC free article] [PubMed] [Google Scholar]
- 5.Munier FL, Frueh BE, Othenin-Girard P, Uffer S, Cousin P, Wang MX, Heon E, Black GC, Blasi MA, Balestrazzi E, Lorenz B, Escoto R, Barraquer R, Hoeltzenbein M, Gloor B, Fossarello M, Singh AD, Arsenijevic Y, Zografos L, Schorderet DF. BIGH3 Mutation Spectrum in Corneal Dystrophies. Invest Ophthalmol Vis Sci. 2002;43:949–54. [PubMed] [Google Scholar]
- 6.Atchaneeyasakul LO, Appukuttan B, Pingsuthiwong S, Yenchitsomanus PT, Trinavarat A, Srisawat C. Study Group. A novel H572R mutation in the transforming growth factor-beta-induced gene in a Thai family with lattice corneal dystrophy type I. Jpn J Ophthalmol. 2006;50:403–8. doi: 10.1007/s10384-006-0357-6. [DOI] [PubMed] [Google Scholar]
- 7.Funayama T, Mashima Y, Kawashima M, Yamada M. Lattice corneal dystrophy type III in patients with a homozygous L527R mutation in the TGFBI gene. Jpn J Ophthalmol. 2006;50:62–4. doi: 10.1007/s10384-005-0260-6. [DOI] [PubMed] [Google Scholar]
- 8.Nakagawa Asahina S, Fujiki K, Enomoto Y, Murakami A, Kanai A. Case of late onset and isolated lattice corneal dystrophy with Asn544Ser (N544S) mutation of transforming growth factor beta-induced (TGFBI, BIGH3) gene. Nippon Ganka Gakkai Zasshi. 2004;108:618–20. [PubMed] [Google Scholar]
- 9.Ellies P, Renard G, Valleix S, Boelle PY, Dighiero P. Clinical outcome of eight BIGH3-linked corneal dystrophies. Ophthalmology. 2002;109:793–7. doi: 10.1016/s0161-6420(01)01025-9. [DOI] [PubMed] [Google Scholar]
- 10.Afshari NA, Mullally JE, Afshari MA, Steinert RF, Adamis AP, Azar DT, Talamo JH, Dohlman CH, Dryja TP. Survey of patients with granular, lattice, avellino, and Reis-Bücklers corneal dystrophies for mutations in the BIGH3 and gelsolin genes. Arch Ophthalmol. 2001;119:16–22. [PubMed] [Google Scholar]
- 11.Kannabiran C, Klintworth GK. TGFBI gene mutations in corneal dystrophies. Hum Mutat. 2006;27:615–25. doi: 10.1002/humu.20334. [DOI] [PubMed] [Google Scholar]
- 12.Tian X, Liu ZG, Li Q, Li B, Wang W, Xie PY, Fujiki K, Murakami A, Kanai A. Analysis of gene mutation in Chinese patients with Reis-Bücklers corneal dystrophy. Zhonghua Yan Ke Za Zhi. 2005;41:239–42. [PubMed] [Google Scholar]
- 13.Yu J, Zou LH, He JC, Liu NP, Zhang W, Lu L, Sun XG, Dong DS, Wu YY, Yin XT. Analysis of mutation of BIGH3 gene in Chinese patients with corneal dystrophies. Zhonghua Yan Ke Za Zhi. 2003;39:582–6. [PubMed] [Google Scholar]
- 14.Li D, Qi YH, Wang L, Li H, Zhou N, Zhao LM. An atypical phenotype of Reis-Bücklers corneal dystrophy caused by the G623D mutation in TGFBI. Mol Vis. 2008;14:1298–302. [PMC free article] [PubMed] [Google Scholar]
- 15.Romero P, Vogel M, Diaz JM, Romero MP, Herrera L. Anticipation in familial lattice corneal dystrophy type I with R124C mutation in the TGFBI (BIGH3) gene. Mol Vis. 2008;14:829–35. [PMC free article] [PubMed] [Google Scholar]
- 16.Weiss JS, Møller HU, Lisch W, Kinoshita S, Aldave AJ, Belin MW, Kivelä T, Busin M, Munier FL, Seitz B, Sutphin J, Bredrup C, Mannis MJ, Rapuano CJ, Van Rij G, Kim EK, Klintworth GK. The IC3D classification of the corneal dystrophies. Cornea. 2008;27:S1–83. doi: 10.1097/ICO.0b013e31817780fb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poulaki V, Colby K. Genetics of anterior and stromal corneal dystrophies. Semin Ophthalmol. 2008;23:9–17. doi: 10.1080/08820530701745173. [DOI] [PubMed] [Google Scholar]
- 18.Rice NS, Ashton N, Jay B, Blach RK. Reis-Bücklers' dystrophy: a clinico-pathological study. Br J Ophthalmol. 1968;52:577–603. doi: 10.1136/bjo.52.8.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aldave AJ, Rayner SA, King JA, Affeldt JA, Yellore VS. A unique corneal dystrophy of Bowman's layer and stroma associated with the Gly623Asp mutation in the transforming growth factor beta-induced (TGFBI) gene. Ophthalmology. 2005;112:1017–22. doi: 10.1016/j.ophtha.2004.12.044. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi A, Sugiyama K. In vivo laser confocal microscopy findings for Bowman's layer dystrophies, (Thiel-Behnke and Reis-Bücklers corneal dystrophies). Ophthalmology. 2007;114:69–75. doi: 10.1016/j.ophtha.2006.05.076. [DOI] [PubMed] [Google Scholar]
- 21.Korvatska E, Munier FL, Djemai A, Wang MX, Frueh B, Chiou AG, Uffer S, Ballestrazzi E, Braunstein RE, Forster RK, Culbertson WW, Boman H, Zografos L, Schorderet DF. Mutation hot spots in 5q31-linked corneal dystrophies. Am J Hum Genet. 1998;62:320–4. doi: 10.1086/301720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Korvatska E, Yamada M, Yamamoto S, Okada M, Munier FL, Schorderet DF, Mashima Y. Haplotye analysis of Jaanese families with a superficial variant of granular corneal dystrohy: evidence for multiple origins of R124L mutation of keratoepithelin. Ophthalmic Genet. 2000;21:63–5. [PubMed] [Google Scholar]
- 23.Huerva V, Velasco A, Sanchez MC, Matias-Guiu X. Role of BIGH3 R124H mutation in the diagnosis of Avellino corneal dystrophy. Eur J Ophthalmol. 2008;18:345–50. doi: 10.1177/112067210801800305. [DOI] [PubMed] [Google Scholar]
- 24.Small KW, Mullen L, Barletta J, Graham K, Glasgow B, Stern G, Yee R. Mapping of Reis-Bücklers' corneal dystrophy to chromosome 5q. Am J Ophthalmol. 1996;121:384–90. doi: 10.1016/s0002-9394(14)70434-9. [DOI] [PubMed] [Google Scholar]