Abstract
The objectives of this study were to investigate the feasibility of imaging green fluorescent protein (GFP)-expressing cells labeled with iron oxide nanoparticles with the fast low-angle positive contrast steady-state free precession (FLAPS) method and to compare them with the traditional negative contrast technique. The GFP-R3230Ac cell line (GFP cell) was incubated for 24 hours using 20 μg Fe/mL concentration of superparamagnetic iron oxide (SPIO) and ultrasmall superparamagnetic iron oxide (USPIO) nanoparticles. Cell samples were prepared for iron content analysis and cell function evaluation. The labeled cells were imaged using positive contrast with FLAPS imaging, and FLAPS images were compared with negative contrast T2*-weighted images. The results demonstrated that SPIO and USPIO labeling of GFP cells had no effect on cell function or GFP expression. Labeled cells were successfully imaged with both positive and negative contrast magnetic resonance imaging (MRI). The labeled cells were observed as a narrow band of signal enhancement surrounding signal voids in FLAPS images and were visible as signal voids in T2*-weighted images. Positive contrast and negative contrast imaging were both valuable for visualizing labeled GFP cells. MRI of labeled cells with GFP expression holds potential promise for monitoring the temporal and spatial migration of gene markers and cells, thereby enhancing the understanding of cell- and gene-based therapeutic strategies.
A standard method for tracking a reporter gene within a cell of interest is to link it to a second gene encoding a fluorescent protein. The activity of the gene can then be monitored by the magnitude of the fluorescent signal.1 However, the method is mainly used for an end-point assessment in animal studies.
More recently, it has become possible to depict biologic processes at the cellular and molecular levels using magnetic resonance imaging (MRI).2,3 Of primary interest is the development of MRI methods to noninvasively monitor the progress of targeted therapies.3,4 It is highly beneficial to image cells because MRI has the advantage of very high spatial resolution and the ability to measure multiple physiologic parameters with different pulse sequences. These physiologic parameters include oxygenation, blood flow, perfusion, and permeability.5–7 Super paramagnetic iron oxide (SPIO) and ultrasmall super-paramagnetic iron oxide (USPIO) MRI contrast agents can be used as compartmental, targeted, or smart probes during MRI.8,9 The SPIO and USPIO sequestered within cells can generate a large magnetic moment that creates substantial disturbances in the local magnetic field. This leads to rapid dephasing of protons, including those surrounding the targeted cell.8 MRI techniques provide good sensitivity for the detection of labeled cells with iron oxide nanoparticles, leading to apparent signal voids within gradient echo (GRE) images.10–13 More recently, off-resonance MRI techniques have provided the means for visualizing labeled cells and interventional devices with positive contrast.14–18 Among these methods, fast low-angle positive contrast steady-state free precession (FLAPS) imaging has been proposed for time-efficient acquisition of off-resonance positive contrast images.18 The FLAPS technique takes advantage of the unique spectral response of the steady-state free precession (SSFP) signal to generate signal enhancement from off-resonant spins while suppressing signal from on-resonant spins and using relatively small flip angles.14
The ability to noninvasively map gene markers has tremendous implications for biomedical research and is crucial in gene medicine.4 The success of gene therapy depends on precise and reproducible delivery and dynamic monitoring of the gene of interest. However, dynamically monitoring therapeutic genes in tissues is challenging, and the distribution of the genes typically remains uncertain.19 Real-time and direct MRI monitoring of genes within tissues is hindered by the target techniques.20 The purpose of this study was (1) to label green fluorescent protein (GFP)-expressing cells with USPIO and compare the labeling efficiency and GFP expression with previously reported SPIO-labeled GFP-expressing cells21 and (2) to track the GFP-expressing cells using positive contrast methods and compare the resulting images with those obtained using more conventional negative contrast MRI techniques.
Materials and Methods
Iron Oxide Magnetic Resonance Contrast Agents
Feridex (Advanced Magnetics, Cambridge, MA) is a commercially available SPIO magnetic resonance contrast agent. These SPIO nanoparticles are crystalline nanoparticles with a major crystal structure of Fe3O4 coated with carboxydextran. The core diameter of these SPIO nanoparticles is approximately 6.5 nm, and the total particle diameter is approximately 70 to 140 nm. SHU 555C or Supravist, the USPIO used for our study, is a ferucarbotran compound produced by Bayer Schering Pharma AG (Leverkusen, Germany). USPIO nanoparticles consist of a polycrystalline Fe2O3 magnetic core with a diameter of 3 to 4 nm embedded within a polymer coating with a hydrodynamic total particle diameter of 30 nm. Coatings of the cores of both nanoparticles are necessary to prevent aggregation of the nanoscale particles and improve biocompatibility. The mechanism of intracellular contrast enhancement with SPIO and USPIO is based on the difference in magnetic susceptibility between regions of labeled cells and background tissue. The change in susceptibility results in a field inhomogeneity in the vicinity of the labeled cells, causing spin dephasing in that region, which can be detected with MRI. The sensitivity for MRI detection of the labeled cells is related to the degree of intracellular iron loading.
Cell Culture and Labeling
A GFP-R3230Ac cell line, stably expressing GFP on R3230Ac cells and obtained from Dr. Mark W. Dewhirst’s group (Department of Radiation Oncology, Duke University Medical Center, Durham, NC), was used. The biologic background of this GFP-expressing cell line was previously described.22,23
The GFP cells were cultured in Dulbecco’s Modified Eagle’s Medium (Invitrogen, Grand Island, NY) with 10% fetal bovine serum (Biosource, Logen, UT) and penicillin-streptomycin at 37°C in air with 5% CO2 atmosphere in a 75 cm2 culture flask. The medium was replaced every 3 to 4 days. Both SPIO and USPIO groups were then incubated 24 hours in the culture medium with suspensions of SPIO and USPIO. The incubation iron concentrations for SPIO and USPIO were 20 μg/mL for all labeled cells, 10 mL of SPIO or USPIO culture medium for labeling 4.5 × 106 cells. The control group consisted of cells incubated with culture medium. Unincorporated iron particles were removed by repeated washing with phosphate-buffered saline (PBS). Samples of the cells were set aside for evaluation of cell labeling efficiency (n = 6, 1 × 104 cells per experiment), iron particle uptake (n = 3, 2 × 106 cells per experiment), viability (n = 6, 1 × 104 cells per experiment), and GFP expression assessment (n = 6, 2 × 104 cells per experiment). The remaining cells (7 × 106 per experiment) were used as MRI sample preparations (n = 6).
Histochemical Detection of USPIO and SPIO Labeling Efficiency
After incubation of cells within the SPIO and USPIO suspension and control groups within the culture medium for 24 hours, cells were washed three times with PBS to remove extracellular SPIO and USPIO. Samples of labeled and control cells were set aside for evaluation.
Labeling efficiency was determined through histologic staining with Prussian blue. In brief, the cells were fixed for 10 minutes with 4% glutaraldehyde. Cells were then incubated with a 1:1 mixture of 6% HCl (Sigma, St. Louis, MO) and 2% potassium ferrocyanide (Sigma) for another 30 minutes. After washing with PBS, cells were observed under a light microscope. Cells were considered Prussian blue positive if intracytoplasmic blue shading or granules could be detected. Labeling efficiency was determined by manual counting of Prussian blue–stained and unstained cells using a Zeiss microscope at 40 times magnification using Axiovision 4 software (Carl Zeiss Microscopy, Gottingen, Germany).
Iron content per cell was determined using an inductively coupled atomic emission spectrometer (ICP-AES; Varian, Palo Alto, CA). Cells were labeled, counted, and dissolved in 1% sodium dodecyl sulfate buffer. The spectrometer detection wavelength was set to 238 nm for iron and calibrated with three different standard samples.
GFP expression was quantified by confocal microscopy. The cells were grown on alcohol-cleaned glass coverslips in six-well plates in culture medium. Next, the medium was removed, and the cells were fixed using a fresh solution of 4% (w/v) paraformaldehyde (Sigma) for 15 minutes and rinsed thoroughly with PBS before mounting. The cells were observed using a Zeiss confocal laser microscope (LSM-510, Carl Zeiss Microscopy) with a 40× lens. GFP was excited using the 488 nm line of a krypton-argon laser, and the emitted fluorescence was detected with a 515 to 540 nm bandpass filter. During imaging of different groups of cells, the same system configuration was used, keeping all parameters fixed, including laser power, laser line, dichroic beam splitter for separating excitation and emission, pinhole size, and scanning speed. All images were acquired as multichannel TIFF. Offline image analysis was performed with MetaMorph software (Universal Imaging, Downingtown, PA). Color separation within the TIFF images was used to produce green-colored images. The signal intensity (SI) of each cell and background noise was measured by operator-defined regions of interests (ROI). The size of the ROI depended on the cell size. Average signal intensity over all of the cells (40 per group) was computed as the mean of all maximum intensity values from each cell measured using MetaMorph. The GFP signal intensity was normalized by background integrated SI for each cell. The signal to noise ratio (SNR) was computed as SNR = SIcell/SIbkgd, where SIcell is the signal intensity of the cell and SIbkgd is the average signal intensity of the background.
Cell Viability and Proliferation
Cell viability was determined using trypan blue assays. The cells were washed three times with PBS and harvested through trypsinization. These cells were resuspended with PBS and transferred into Eppendorf tubes: 0.5 mL of 0.4% trypan blue solution (Gibco, Grand Island, NY), 0.3 mL of PBS, and 0.2 mL of cell suspension in PBS (= dilution 1:5), which was allowed to stand for 5 to 15 minutes. A 10 μL sample of this mixture was dropped into a Neubauer’s counting chamber (EMS, Hatfield, PA). Cells stained blue were considered dead. The proliferative capacity of cells was determined by plating 5 × 103 cells in standard culture dishes, followed by counting the number of cells on days 0, 3, and 5 using a Neubauer’s counting chamber.
Sample Preparation for MRI
For MRI studies, cells were transferred to 1.5 mL test tubes. There were six samples of labeled cells: SPIO groups: 2 × 106, 1 × 106, and 0.5 × 106 cell samples and USPIO groups: 2 × 106, 1 × 106, and 0.5 × 106 samples. A seventh sample containing 2 × 106 unlabeled cells served as a control.
In Vitro MRI Protocol
MRI was performed using a clinical 3.0 T whole-body magnetic resonance system (Siemens Trio, Erlangen, Germany) using an eight-channel head coil for signal reception. To avoid susceptibility artifacts from the surrounding air in the scans, all samples were placed in a water bath and were manually shimmed. A two-dimensional T2*-weighted GRE sequence was prescribed with the following scan parameters to generate negative contrast: repetition time (TR)/echo time (TE) = 1,000/22.4 ms, flip angle = 90°, field of view (FOV) = 200 × 200 mm2, matrix = 384 × 384, imaging bandwidth (BW) = 620 Hz/pixel, slice thickness = 0.5 mm. These parameters yielded a voxel size ≈0.5 × 0.5 × 0.5 mm3. A two-dimensional FLAPS sequence23 was used to generate off-resonance positive contrast weighting with the following scan parameters: TR/TE = 5/2.5 ms, flip angle = 10°, FOV = 200 × 200 mm2, matrix = 384 × 384, BW = 610 Hz/pixel, slice thickness = 0.5 mm, voxel size ≈ 0.5 × 0.5 × 0.5 mm3.
Statistics
Quantitative data are presented as mean ± standard deviation. Comparisons between unlabeled (n = 6) and labeled (n = 6) GFP cell preparations were made using a two-tailed unpaired Student t-test. Repeated measures analysis of variance was used to assess differences between and within groups. The significance of statistical tests was set at p < .05.
Results
Cell Labeling Verification and Quantification
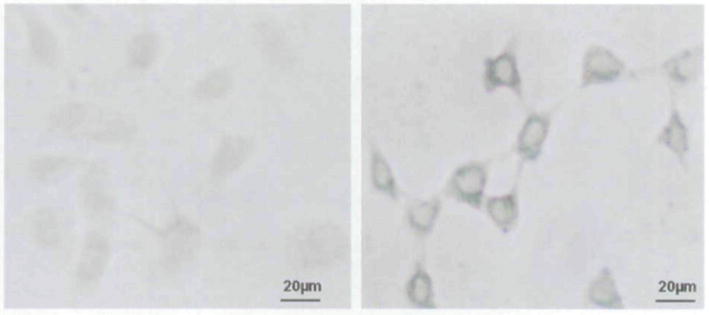
Prussian blue staining and inductively coupled atomic emission spectrometer (ICP-AES) analysis provided confirmation of cell labeling. Light microscopy revealed an abundant intracellular uptake of both SPIO and USPIO into cytoplasm using Prussian blue staining of labeled cells. However, no stainable iron was detected in the control cells. Reproducibility of labeling efficiency was approximately 100% (Figure 1). The results from ICP-AES showed that both SPIO and USPIO groups had a significantly higher uptake than the control group, with a higher amount of uptake for SPIO than for USPIO (mean ± SD: SPIO, 4.75 ± 0.11 pg Fe/cell; USPIO, 1.88 ± 0.04 pg Fe/cell; control, 0.19 ± 0.01 pg Fe/cell). These differences were significant when comparing SPIO with USPIO, SPIO with control, and USPIO with control (all p < .05).
Figure 1.
Light microscope images of Prussian blue staining of unlabeled (left) and labeled (right) green fluorescent protein cells. Left, No stainable iron is detected within the unlabeled cells. Right, Intracellular iron accumulation is demonstrated by blue coloring (×8 original magnification).
Cell Function Following Iron Oxide Nanoparticle Loading
Cell viability was determined following the labeling procedure. There was no observable increase in the amount of floating cells in the supernatant under the microscope, and the number of adherent cells was not significantly different between the control and both SPIO and USPIO groups. At least 92% of cells were deemed viable after the cell labeling experiment in each group. No significant differences in death rates between SPIO- and USPIO-labeled cells and control cells were observed.
Cell proliferation was not affected after SPIO and USPIO labeling when comparing with control cells, and the mean doubling times (hours) were 21.3 ± 0.4 (control group), 21.6 ± 0.5 (SPIO), and 21.8 ± 0.3 (USPIO). There were no statistically significant differences in cell proliferation times between these groups (p > .05 for each paired comparison).
GFP Expression Measurement
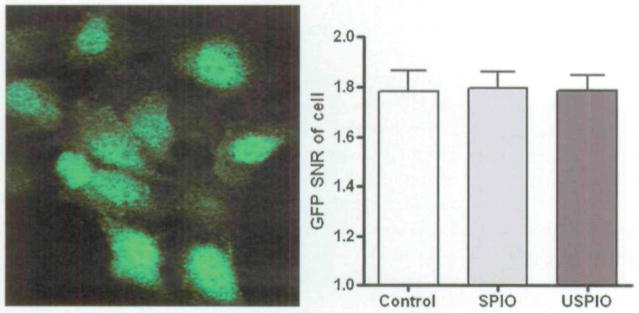
Confocal fluorescence microscopy revealed the GFP distribution within the USPIO-labeled cells. Fluorescence was detected in exposed cells and was contained within the cellular membrane (Figure 2). GFP signal intensity within each cell was subjected to quantitative image analysis with MetaMorph software. GFP SNR measurements demonstrated that the SPIO and USPIO labeling process did not affect GFP expression of the cells on Figure 2 (control, 1.78 ± 0.08; SPIO, 1.79 ± 0.07; USPIO. 1.78 ± 0.06). Comparison of GFP expression between control and both SPIO- and USPIO-labeled cells revealed no significant differences (p > .05 for each paired comparison). It was also observed that the labeling process (iron-loading dose) did not affect GFP signal intensity (expression).
Figure 2.
Confocal microscopic view of labeled cells. The left panel shows green fluorescent protein (GFP) expression of labeled cells, and the right panel shows quantitative signal to noise ratio (SNR) analysis with MetaMorph software; no difference was observed in GFP expression between superparamagnetic iron oxide (SPIO)- and ultrasmall superparamagnetic iron oxide (USPIO)-labeled and unlabeled groups, p > .05.
MRI of the Cell Pellets
Negative Contrast Imaging with GRE
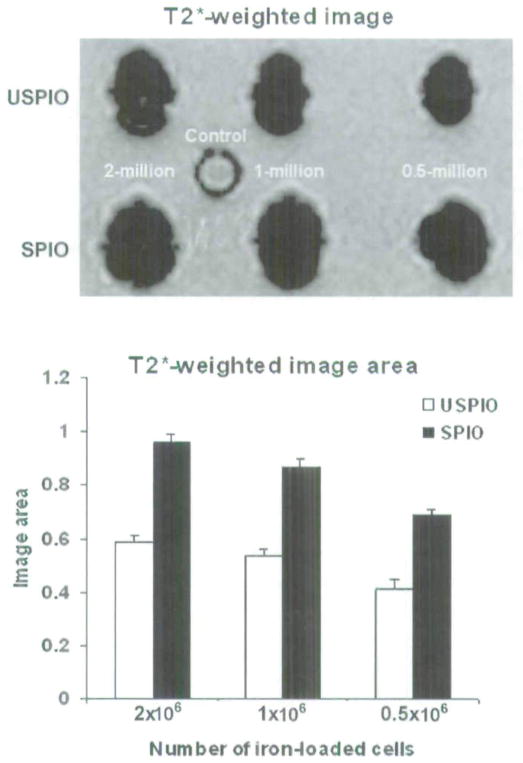
A strong signal decline surrounding all six test samples was observed in comparison with the control sample. All labeled cell samples were visible on the T2*-weighted MRIs as signal voids. The T2* effect of the SPIO-labeled cell pellets was markedly stronger than that of corresponding USPIO-labeled cell pellets. Representative images demonstrating these results are shown on the top panel of Figure 3. For a given cell pellet, the size of the signal void from SPIO-labeled cells was larger than cells labeled with USPIO (p < .05), whereas the size of the signal void generated by USPIO-labeled cells was larger than that of the control group (p < .05, bottom panel of Figure 3). In addition, it was also observed that the size of the signal void and the magnitude of negative enhancement depended on the number of labeled cells included within the sample. Although the total number of labeled cells was kept constant, the size of the signal void and signal enhancement depended on the diameter of iron oxide particles (ie, whether these cells were labeled with SPIO as opposed to USPIO; see Figure 3).
Figure 3.
Representative gradient echo T2*-weighted images of the ultrasmall superparamagnetic iron oxide (USPIO), superparamagnetic iron oxide (SPIO), green fluorescent protein cell pellets in the top panel: 2, 1, and 0.5 million labeled cells and 2 million unlabeled cells as a control group. All labeled cell pellets showed a marked susceptibility effect compared with the control group. Measurement of susceptibility-induced signal void area for each group in the T2*-weighted images is shown in the lower panel.
Positive Contrast Imaging with FLAPS
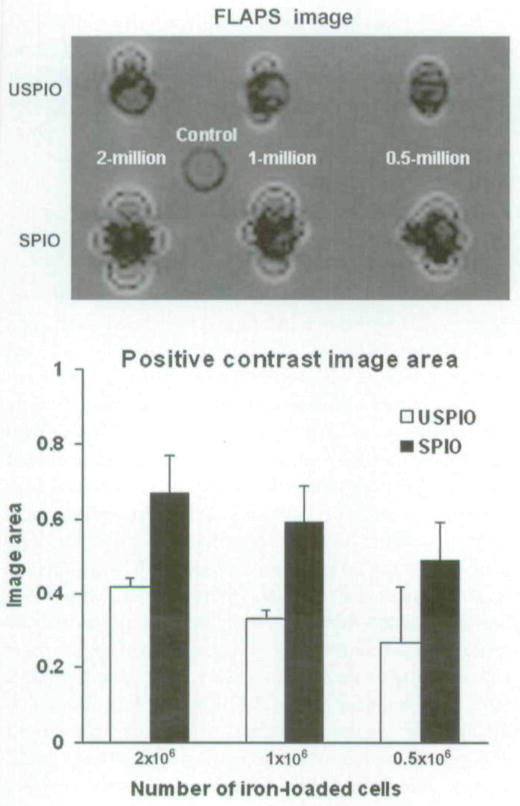
Positive contrast imaging of the cell pellets was observed as a narrow band of signal enhancement surrounding the signal void for six test samples, and no positive contrast was seen for the control group. Two-dimensional FLAPS MRIs of the same samples in Figure 3 are shown in the top panel of Figure 4. These results were consistent with T2*-weighted MRI measurements, with the extent of the positive enhancement (area of positive signal enhancement) clearly dependent on the number of labeled cells. As before, for a given cell pellet, the size of positive contrast enhancement was larger for SPIO-labeled cells than for USPIO-labeled cells. In contrast, however, FLAPS MRI showed a “dual” signal characteristic (both positive and negative contrast) of regions surrounding the labeled pellets (top panel of Figure 4). The results from quantitative evaluation of the relative area of positive contrast for each cell are shown at the bottom panel of Figure 4. Note that as with T2*-weighted results, this area also depends on the number of cells and the iron content within the cells.
Figure 4.
FLAPS images of the same clusters of ultrasmall superparamagnetic iron oxide (USPIO)- and superparamagnetic iron oxide (SPIO)-labeled cells as shown in Figure 3. There was a notable signal loss compared with the control with unlabeled cells. Positive signal contrast was observed as narrow bands of signal enhancement surrounding the signal voids (top panel) at the USPIO- and SPIO-labeled cell positions. Banding areas (size of regions exhibiting positive contrast) for each group are shown within the lower panel.
The hyperintense ring on T2*-weighted GRE images was likely due to secondary phase shift, but the rings from FLAPS images are from off-resonance enhancement. The hyperintense signals on the FLAPS images have a unique signature (similar to a dipole) that was previously described elsewhere.14 Note that in the FLAPS images, the intensity patterns are fluctuating (eg, dark, bright, dark, bright) and that this pattern coincides with the spectral response function of the SSFP signals in the presence of the magnetic perturber.
Discussion
The ability to image cells in vivo may be very useful for studying the effects of stem cell therapy, inflammation, tumors, immune response, etc. A number of investigators have shown that cells labeled with a commercially available iron oxide contrast medium can be imaged for weeks after such labeling and that the label does not damage the cells in vitro.24,25 In vivo cell tracking by MRI can provide a means to observe biologic processes and monitor cell therapy directly.26,27 Here, we demonstrated that iron oxide magnetic resonance contrast agents do not affect the marker gene expression within GFP cells, cell viability, or function. We have characterized an assay that combines MRI and microscopy to evaluate SPIO- and USPIO-labeled living GFP cells. Furthermore, the presence of iron-oxide labeled GFP cells can be detected with intravital microscopy and MRI, with a clinical scanner operating at 3.0 T permitting the possibility of dual-mode imaging for studying physiologic processes.
Reporter gene expression studies constitute a valuable and necessary step to further our understanding of gene marking for potential gene transfer opportunities. GFP has been a marker gene of choice for many studies. Its performance has been extensively characterized in many species, in part owing to its high intrinsic fluorescence, which allows for its easy and convenient detection in living cells via optical or confocal microscopy. Our experiments embarked on the characterization of bimodal imaging of SPIO- and USPIO-labeled GFP cells with confocal microscopy and MRI. In this study, (1) the efficiency of USPIO labeling was compared with SPIO labeling and (2) the applicability of positive contrast imaging techniques for visualization of those USPIO- and SPIO-labeled cells was evaluated.
A low-molecular-weight dextran coating around the iron core that stabilizes the particulate ensures the biocompatibility of Feridex and SHU 555 C. The use of two agents and others in other cell lines has been reported and has proven nontoxic both in vitro and in vivo.24–27 However, this has yet to be extended to GFP-expressing cells that have the capacity to allow for bimodal imaging.
We compared USPIO and SPIO for labeling the GFP cells. Determining the labeling efficiency by ICP-AES revealed that internalization of iron particles depends on the size of these particles (SPIO vs USPIO uptake: 4.75 ± 0.11 vs 1.88 ± 0.04 pg Fe/cell; p < .05). Given that the size of the perturbation caused by these susceptibility shifting agents is linked to the amount of iron internalized within the cells, it is SPIO labeling that is likely to be most suited for labeling GFP cells for visualization or tracking with MRI.
The changes in GFP cell viability were analyzed by trypan blue with a standard protocol. These results confirmed that no changes in GFP cell viability occurred after labeling procedures. A similar cell viability profile has been observed by other investigators who labeled cells without GFP expression using SPIO or USPIO.25,28 The growth characteristics of labeled GFP cells were assessed using a cell proliferation assay. Labeled GFP cells were not affected after 24 hours of incubation with 20 μg/mL of SPIO and USPIO suspension.
Our MRI studies were carried out using cell pellets ranging in diameter between 2.5 and 3.0 mm using a clinical scanner, which makes our data relevant to cell transplantation and tissue engineering applications.29,30 The results are expected effects of SPIO- and USPIO-labeled cells as hypointensity (negative contrast) within GRE T2*-weighted images and hyperintensity (positive contrast) within FLAPS images. Positive contrast with FLAPS imaging was comparable to standard GRE imaging with negative contrast.
With FLAPS imaging, an advantage is the ability to visualize the regions of “blooming” associated with magnetic susceptibility shifting agents as dual-contrast regions of signal void surrounded by bright signal; a disadvantage, however, with off-resonance positive contrast methods is increased sensitivity to unwanted off-resonance frequency shifts.18 Another advantage with off-resonance positive contrast, such as FLAPS, is that with purely negative contrast methods, it may be difficult to differentiate between signal loss owing to true regions of labeled cells and regions of signal voids owing to spin density variations.18 Furthermore, the signal void is better delineated with FLAPS because of the sharp contrast between the signal void and the band of signal enhancement. Finally, the dramatic contrast between the band of signal enhancement and the signal void area with FLAPS may improve identification of a region of labeled cells.14,18 For example, myocardial regeneration with stem cell transplantation is a possible treatment option to reverse the deleterious effects that occur after myocardial infarction. Iron-labeled stem cells have been used for in vivo tracking using MRI but produce negative contrast images that are difficult to interpret because acute myocardial infarction is treated by reperfusion therapy, aggravating microvascular obstruction and causing hemorrhage, which may mimic the signal voids caused by iron-labeled cells. With magnetic resonance–positive contrast sequences (such as FLAPS), it may be possible to track the therapeutic stem cells within the heart.18
Both negative and positive contrast effects act to extend the signal change well beyond the particle or cell. This form of signal amplification increases sensitivity for detecting the labeled GFP cells within a complex image background. With the use of signal amplification, potential future applications of SPIO and USPIO include “doping” of therapeutic cell preparations with a small fraction of labeled cells, to allow cell tracking without altering the majority of cells. It could potentially allow for better delineation and identification of labeled cells. The challenge with both positive and negative contrast methods is the difficulty of quantifying the number of labeled cells in vivo because of the susceptibility artifact produced by these nanoparticles.
In this study, we demonstrated that SPIO and USPIO can be used to effectively label GFP cells with no effects on cell viability, proliferation, or GFP expression. The present study also compared the characteristics of GFP cells labeled with different sizes of iron oxide nanoparticles. These permitted both cellular MRI and optical imaging. It is envisioned that bimodal labeling of cells with iron oxides and GFP expression offers the potential to visualize and track the cells in vivo and hence enhance our understanding and implementation of gene therapies.
Although this study demonstrates the feasibility of FLAPS MRI of GFP-expressing cells labeled with iron oxide nanoparticles, specific application with in vivo and in clinical settings will require further investigations. First, positive contrast imaging methods do not provide sufficient anatomic information; therefore, it is necessary to combine positive contrast techniques with conventional GRE or spin echo images for anatomic localization. Iron content will decrease following cell division. This may limit the ability to monitor cellular activity for long periods. Second, future research studies will be required to measure dynamic R2* changes following cell division as a function of cell number. Third, improvements in MRI spatial resolution may be necessary for use of these techniques within clinical scanners in a patient setting. Finally, at the present time, labeling cells with iron oxide MRI contrast agents cannot be used to interrogate the cell and gene function or viability during in vivo clinical studies. However, MRI of labeled GFP cells provides potential promise for tracking gene markers and cells.
In the summary, both SPIO and USPIO were used to effectively label GFP cells with no effects on cell viability, proliferation, and GFP expression. There was a greater uptake of iron oxide with SPIO rather than USPIO incubation as measured with ICP-AES. FLAPS-based off-resonance positive contrast and GRE-based negative contrast imaging may both be valuable for visualizing and potentially tracking SPIO- and USPIO-labeled GFP cells.
Acknowledgments
We would like to thank Dr. Mark W. Dewhirst and his group at the Department of Radiation Oncology, Duke University Medical Center, for providing the cell line used in this study. We would also like to acknowledge Dr. A.C. Larson for helping to revise the manuscript.
Footnotes
Financial disclosure of authors and reviewers: None reported.
References
- 1.Tam JM, Upadhyay R, Pittet MJ, et al. Improved in vivo whole-animal detection limits of green fluorescent protein-expressing tumor lines by spectral fluorescence imaging. Mol Imaging. 2007;6:269–76. [PubMed] [Google Scholar]
- 2.Glickson JD, Lund-Katz S, Zhou R, et al. Lipoprotein nanoplatform for targeted delivery of diagnostic and therapeutic agents. Mol Imaging. 2008;7:101–10. [PubMed] [Google Scholar]
- 3.Ly HQ, Frangioni JV, Hajjar RJ. Imaging in cardiac cell-based therapy: in vivo tracking of the biological fate of therapeutic cells. Nat Clin Pract Cardiovasc Med. 2008;5:S96–102. doi: 10.1038/ncpcardio1159. [DOI] [PubMed] [Google Scholar]
- 4.Morgul MH, Raschzok N, Schwartlander R, et al. Tracking of primary human hepatocytes with clinical MRI: initial results with Tat-peptide modified superparamagnetic iron oxide particles. Int J Artif Organs. 2008;31:252–7. doi: 10.1177/039139880803100309. [DOI] [PubMed] [Google Scholar]
- 5.Bachmann R, Conrad R, Kreft B, et al. Evaluation of a new ultrasmall superparamagnetic iron oxide contrast agent Clariscan (NC100150) for MRI of renal perfusion: experimental study in an animal model. J Magn Reson Imaging. 2002;16:190–5. doi: 10.1002/jmri.10149. [DOI] [PubMed] [Google Scholar]
- 6.Dennie J, Mandeville JB, Boxerman JL, et al. NMR imaging of changes in vascular morphology due to tumor angiogenesis. Magn Reson Med. 1998;40:793–9. doi: 10.1002/mrm.1910400602. [DOI] [PubMed] [Google Scholar]
- 7.Kim T, Hendrich K, Kim SG. Functional MRI with magnetization transfer effects: determination of BOLD and arterial blood volume changes. Magn Reson Med. 2008;60:1518–23. doi: 10.1002/mrm.21766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thorek DL, Tsourkas A. Size, charge and concentration dependent uptake of iron oxide particles by non-phagocytic cells. Biomaterials. 2008;29:3583–90. doi: 10.1016/j.biomaterials.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corot C, Robert P, Idée JM, et al. Recent advances in iron oxide nanocrystal technology for medical imaging. Adv Drug Deliv Rev. 2006;58:1471–504. doi: 10.1016/j.addr.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 10.Bos C, Delmas Y, Desmouliere A, et al. In vivo MR imaging of intravascularly injected magnetically labeled mesenchymal stem cells in rat kidney and liver. Radiology. 2004;233:781–9. doi: 10.1148/radiol.2333031714. [DOI] [PubMed] [Google Scholar]
- 11.Bulte JW, Douglas T, Witwer B, et al. Magnetodendrimers allow endosomal magnetic labeling and in vivo tracking of stem cells. Nat Biotechnol. 2001;19:1141–7. doi: 10.1038/nbt1201-1141. [DOI] [PubMed] [Google Scholar]
- 12.Daldrup-Link HE, Rudelius M, Piontek G, et al. Migration of iron oxide-labeled human hematopoietic progenitor cells in a mouse model: in vivo monitoring with 1.5-T MR imaging equipment. Radiology. 2005;234:197–205. doi: 10.1148/radiol.2341031236. [DOI] [PubMed] [Google Scholar]
- 13.de Vries IJ, Lesterhuis WJ, Barentsz JO, et al. Magnetic resonance tracking of dendritic cells in melanoma patients for monitoring of cellular therapy. Nat Biotechnol. 2005;23:1407–13. doi: 10.1038/nbt1154. [DOI] [PubMed] [Google Scholar]
- 14.Dharmakumar R, Koktzoglou I, Li D. Generating positive contrast from off-resonant spins with steady-state free precession magnetic resonance imaging: theory and proof-of principle experiments. Phys Med Biol. 2006;51:4201–15. doi: 10.1088/0031-9155/51/17/006. [DOI] [PubMed] [Google Scholar]
- 15.Suzuki Y, Cunningham CH, Noguchi K, et al. In vivo serial evaluation of superparamagnetic iron-oxide labeled stem cells by off-resonance positive contrast. Magn Reson Med. 2008;60:1269–75. doi: 10.1002/mrm.21816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dahnke H, Liu W, Herzka D, et al. Susceptibility gradient mapping (SGM): a new postprocessing method for positive contrast generation applied to superparamagnetic iron oxide particle (SPIO)-labeled cells. Magn Reson Med. 2008;60:595–603. doi: 10.1002/mrm.21478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilad AA, Walczak P, McMahon MT, et al. MR tracking of transplanted cells with “positive contrast” using manganese oxide nanoparticles. Magn Reson Med. 2008;60:1–7. doi: 10.1002/mrm.21622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dharmakumar R, Koktzoglou I, Li D. Factors influencing fast low-angle positive-contrast steady state free precession (FLAPS) magnetic resonance imaging. Phys Med Biol. 2007;52:3261–73. doi: 10.1088/0031-9155/52/11/021. [DOI] [PubMed] [Google Scholar]
- 19.Serganova I, Mayer-Kukuck P, Huang R, et al. Molecular imaging: reporter gene imaging. Handb Exp Pharmacol. 2008;185:167–223. doi: 10.1007/978-3-540-77496-9_8. [DOI] [PubMed] [Google Scholar]
- 20.Dirks RW, Tanke HJ. Advances in fluorescent tracking of nucleic acids in living cells. Biotechniques. 2006;40:489–96. doi: 10.2144/000112121. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Z, Mascheri N, Dharmakumar R, et al. Superparamagnetic iron oxide nanoparticle-labeled cells as an effective vehicle for tracking the GFP gene marker using magnetic resonance imaging. Cytotherapy. 2008;10:1–9. doi: 10.1080/14653240802420243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, Kon T, Li C, et al. High intensity focused ultrasound-induced gene activation in sublethally injured tumor cells in vitro. J Acoust Soc Am. 2005;118:3328–36. doi: 10.1121/1.2041247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shan S, Sorg B, Dewhirst MW. A novel rodent mammary window of orthotopic breast cancer for intravital microscopy. Microvasc Res. 2003;65:109–17. doi: 10.1016/s0026-2862(02)00017-1. [DOI] [PubMed] [Google Scholar]
- 24.Bulte JW, Kraitchman DL. Iron oxide MR contrast agents for molecular and cellular imaging. NMR Biomed. 2004;17:484–99. doi: 10.1002/nbm.924. [DOI] [PubMed] [Google Scholar]
- 25.Oude Engberink RD, van der Pol SM, Döpp EA, et al. Comparison of SPIO and USPIO for in vitro labeling of human monocytes: MR detection and cell function. Radiology. 2007;243:467–74. doi: 10.1148/radiol.2432060120. [DOI] [PubMed] [Google Scholar]
- 26.Frank JA, Anderson SA, Kalsih H, et al. Methods for magnetically labeling stem and other cells for detection by in vivo magnetic resonance imaging. Cytotherapy. 2004;6:621–5. doi: 10.1080/14653240410005267-1. [DOI] [PubMed] [Google Scholar]
- 27.Politi LS, Bacigaluppi M, Brambilla E, et al. Magnetic-resonance-based tracking and quantification of intravenously injected neural stem cell accumulation in the brains of mice with experimental multiple sclerosis. Stem Cells. 2007;25:2583–92. doi: 10.1634/stemcells.2007-0037. [DOI] [PubMed] [Google Scholar]
- 28.Sun R, Dittrich J, Le-Huu M, et al. Physical and biological characterization of superparamagnetic iron oxide- and ultrasmall superparamagnetic iron oxide-labeled cells: a comparison. Invest Radiol. 2005;40:504–13. doi: 10.1097/01.rli.0000162925.26703.3a. [DOI] [PubMed] [Google Scholar]
- 29.Kraitchman DL, Tatsumi M, Gilson WD, et al. Dynamic imaging of allogeneic mesenchymal stem cells trafficking to myocardial infarction. Circulation. 2005;112:1451–61. doi: 10.1161/CIRCULATIONAHA.105.537480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bulte JW, Duncan ID, Frank JA. In vivo magnetic resonance tracking of magnetically labeled cells after transplantation. J Cereb Blood Flow Metab. 2002;22:899–907. doi: 10.1097/00004647-200208000-00001. [DOI] [PubMed] [Google Scholar]