Abstract
We recently found that normal human brain and low-grade astrocytomas express the receptor protein tyrosine phosphatase mu (PTPµ) and that the more invasive astrocytomas, glioblastoma multiforme (GBM), downregulate full-length PTPµ expression. Loss of PTPµ expression in GBMs is due to proteolytic cleavage that generates an intracellular and potentially a cleaved and released extracellular fragment of PTPµ. Here, we identify that a cleaved extracellular fragment containing the domains required for PTPµ-mediated adhesion remains associated with GBM tumor tissue. We hypothesized that detection of this fragment would make an excellent diagnostic tool for the localization of tumor tissue within the brain. To this end, we generated a series of fluorescently tagged peptide probes that bind the PTPµ fragment. The peptide probes specifically recognize GBM cells in tissue sections of surgically resected human tumors. To test whether the peptide probes are able to detect GBM tumors in vivo, the PTPµ peptide probes were tested in both mouse flank and intracranial xenograft human glioblastoma tumor model systems. The glial tumors were molecularly labeled with the PTPµ peptide probes within minutes of tail vein injection using the Maestro FLEX In Vivo Imaging System. The label was stable for at least 3 hours. Together, these results indicate that peptide recognition of the PTPµ extracellular fragment provides a novel molecular diagnostic tool for detection of human glioblastomas. Such a tool has clear translational applications and may lead to improved surgical resections and prognosis for patients with this devastating disease.
Introduction
The prognosis for high-grade brain tumors such as glioblastoma multiforme (GBM) is extremely poor, with a median survival of approximately 1 year from diagnosis [1,2]. Several biologic characteristics contribute to the lethality of GBM tumors, including their uncontrolled proliferation in the restricted cranial space and their highly dispersive nature [1,3]. Surgical resection remains the primary treatment of glial tumors [4], and a more complete resection has been linked to improved survival [5].However, by the time of diagnosis, GBM cells have usually dispersed extensively into the surrounding brain, making it difficult for the surgeon to precisely localize the tumor margin [6]. Magnetic resonance imaging (MRI)-guided stereotactic techniques are typically used to maximize resection. However, MRI is limited in its ability to detect sparse tumor cells invading surrounding normal brain [7]. Because nearly all glioblastomas recur locally, better detection and surgical resection would likely improve patient survival. The development of in vivo molecular imaging reagents could significantly improve glioblastoma detection in patients in a noninvasive fashion and facilitate more complete resection while minimizing surgical complications.
Receptor protein tyrosine phosphatases (RPTPs) are important regulators of adhesion-dependent signals [8–10]. Protein tyrosine phosphatase µ (PTPµ) protein is highly expressed in normal brain and plays a role in the stabilization of cell-cell contacts [11,12]. The extracellular segment of PTPµ contains motifs found in cell adhesion molecules, including a meprin, A5 (neuropilin), µ domain (MAM), an immunoglobulin (Ig) domain, and four fibronectin type III (FNIII) repeats [8]. PTPµ has a juxtamembrane domain and two tandem tyrosine phosphatase domains in its intracellular segment [8]. PTPµ likely transduces signals in response to adhesion that may regulate contact inhibition of movement. PTPµ binds homophilically (i.e., the “ligand” for PTPµ is an identical PTPµ molecule on an adjacent cell) [13–15].Cells expressing two very similar RPTPs, PTPκ and PTPµ, which share more than 60% amino acid sequence similarity, do not bind to one another in aggregation assays [15].
The Ig domain of PTPµ is responsible for promoting homophilic interactions [16] and proper cell surface localization [17]. The sorting of closely related molecules such as PTPµ and PTPκ is attributed to the MAM domain [15]. However, MAM, Ig, and the first two FNIII repeats are the minimal extracellular domains required for efficient cell-cell adhesion [15,16,18–20]. Crystallographic studies demonstrated that the MAM and Ig domains are tightly associated into what seems to be one functional entity [19]. Additional crystal structure analysis predicted that the adhesive interface between two PTPµ molecules is between the MAM and Ig domains of one PTPµ molecule with the first and second FNIII domains of the second PTPµ molecule [20]. Because of what was termed the extensive interaction interface observed in crystal packing [20], when PTPµ is involved in cell-cell adhesion, its binding sites are likely fully engaged.
We recently found that normal brain and primary rat astrocytes express PTPµ, but the most dispersive glial tumors, GBMs, downregulate full-length PTPµ expression [12]. The down-regulation of PTPµ occurs through proteolytic cleavage in human GBM tumors [21], which releases the cytoplasmic domain of PTPµ from the plasma membrane. We found that signaling through the released cytoplasmic domain was necessary for increased migration of GBM cancer cells in vitro. PTPµ cleavage may be an important event that deregulates normal contact inhibition of movement. In this article, we identify the presence of an extracellular fragment of PTPµ in human GBM tumors but not in normal brain tissue. This extracellular fragment contains the domains required for PTPµ-mediated homophilic cell-cell adhesion.We hypothesize that detection of this fragment may facilitate the identification of the tumor margin because it is present at the tumor edge. Therefore, we devised a strategy to detect this extracellular PTPµ fragment by exploiting what we know about PTPµ-mediated homophilic binding to generate a series of fluorescently tagged peptide probes to surface-exposed sites of PTPµ. The peptide probes specifically recognize primary human GBM cells in tissue sections of surgically resected tumor. The peptides recognize GBM tumors in vivo in two different human GBM xenografts in nude mice. Most importantly, the peptides cross the compromised blood-brain barrier to label intracranial GBM tumors. These results indicate that the cleaved PTPµ extracellular fragment is a novel biomarker of GBM cells that can be used as a molecular diagnostic imaging agent to noninvasively detect the tumor margin of human glioblastomas in vivo.
Materials and Methods
Peptide Synthesis and Conjugation
SBK1, SBK2, SBK3, and SBK4 peptides either were synthesized on a synthesizer (Model 433A; Applied Biosystems, Foster City, CA) by Dr. Marty Pagel and his laboratory in the Department of Biomedical Engineering, Case Western Reserve University or were purchased from Genscript Corporation (Piscataway, NJ). An N-terminal glycine residue was added to peptides SBK1, SBK2 and SBK3 during synthesis. After synthesis, the N-terminal glycine residue of each peptide was specifically coupled to Texas Red (TR)-X (single isomer) or Alexa-750 succinimidyl ester dye (Molecular Probes, Inc, Eugene, OR), which has a five-carbon spacer between the succinimide group that couples to the N-terminal amine and the fluorophore.
Human Brain Tissue Protein Extraction
Using a Food and Drug Administration-approved computer navigational device and software (BrainLab Vector Vision 2 and i-Plan 2.0, Westchester, IL), the neurosurgeon coregistered multiple scalp points with volumetric RAGE T1 ± gadolinium MRI (1.5-mm segments/0 skips) obtained the day before surgery using a standard technique (Z-Touch, BrainLab, Inc., Westchester, IL). After achieving precision of 1 mm or less and confirmation using various objective skull landmarks, surgery was performed using stereotactic techniques. After pathologic confirmation of GBM was obtained, the stereotactic device was used to identify the edge and center of the gadolinium-enhanced tumor mass, and paired specimens of approximately 100 mg each were preserved in liquid nitrogen or formalin-paraffin, respectively. In some cases, noneloquent brain was also identified and similarly preserved if it was part of the region to be resected. Similarly, discarded tissue from patients undergoing cortical resections for intractable epilepsy was collected for noncancerous “normal” tissue after the neuropathologists released the sample. Only discarded tissue was used, and approval was obtained from the University Hospitals Case Medical Center and Case Comprehensive Cancer Center's Institutional Review Board. After tissue resection, the samples were snap frozen and thawed on ice in lysis buffer, and protein was extracted as previously described [12]. Mouse flank Gli36ΔΔ5 or LN-229 tumors were excised, snap frozen, and then lysed as described [12] using a PRO 200 tissue tear or (PRO Scientific, Inc, Monroe, CT). All samples were separated by SDS-PAGE and transferred to nitrocellulose for immunoblot analysis with antibodies generated with a peptide corresponding to amino acids 42 to 60 within the MAM domain in the extracellular segment of PTPµ (BK9) [16].
Peptide Labeling of Human Brain Tissue
Human glioblastoma or noncancerous “normal” epilepsy tissue samples were immediately fixed in 10% neutral-buffered formalin (Sigma-Aldrich, St. Louis, MO). Tissues were ethanol-dehydrated and paraffin-embedded. Tissue sections were cut at 5-µm intervals and stored at room temperature (RT). Before staining, tissue sections were deparaffinized and blocked with 2% goat serum in PBS for 20 minutes at RT. PTPµ-TR peptides were diluted in block buffer and incubated with the tissue sections for 1 hour at RT in the dark. The sections were rinsed with PBS, coverslipped with Citifluor Antifadent Mounting Medium, AF1 (Electron Microscopy Sciences, Hatfield, PA) and imaged immediately at 40x on a Nikon TE-200 inverted microscope (Nikon Instruments Inc.,Melville, NY), using a SPOT-RT camera and SPOT software version 3.2 (Diagnostic Instruments, Inc, Sterling Heights, MI). High-magnification phase and fluorescent images were obtained using the same exposure settings between multiple experiments. The working concentrations for the peptides were determined empirically for tissue staining and are as follows: SBK1-TR, 40 µM; SBK2-TR, 10 µM; SBK3-TR, 10 µM; and SBK4-TR, 3.3 µM. The SBK4-TR peptide was the most effective in labeling human GBM tissue sections.
Antibody-Blocking Experiments
To block PTPµ binding sites, human tumor tissue sections were preincubated for 1 hour at RT with BK2 monoclonal antibody, raised against amino acids 42 to 60 within the MAM domain of PTPµ [16], before incubating with SBK2-TR (10 µM) or SBK4-TR (3.3 µM) peptide in block buffer for 1 hour at RT in the dark. Tissue sections were rinsed and imaged as described above.
Heterotopic Xenograft Flank Tumors
HumanGli36Δ5 glioblastoma cells constitutively overexpress the vIII mutant forms of the EGFR gene [22]. Human LN-229 glioblastoma cells were obtained from American Type Culture Collection (Manassas, VA). Gli36Δ5 or LN-229 cells were harvested for flank implantation by trypsinization. In some experiments, the cells were infected with lentivirus to express green fluorescent protein (GFP) [23] 48 hours before harvesting. The cells (2 x 106 cells for flank tumor implants) were resuspended in a 1:1 dilution of PBS and Matrigel (BD Biosciences, Franklin Lakes, NJ) for a total volume of 250 to 300 µl per flank tumor implant per animal.
NIH athymic nude female mice (5–8 weeks and 20–25 g on arrival; NCI-NIH) were maintained at the Athymic Animal Core Facility at Case Western Reserve University according to institutional policies. All animal protocols were approved by the Institutional Animal Care and Use Committee (IACUC). For flank tumor implants, mice were anesthetized with inhaled isofluorane-oxygen for immobilization. The Matrigel-cell mixture was loaded into a 1-ml syringe fitted with a 26-gauge needle and kept on ice. The mixture was injected subcutaneously in the right flank region of the mouse.
Peptide Labeling of Mouse Flank Tumor Sections
NIH athymic nude female mice implanted with Gli36Δ5 or LN-229 flank tumors for 2 to 3 weeks were anesthetized with inhaled isoflurane-oxygen and sacrificed by decapitation. Flank tumors were then excised, fixed in 4% paraformaldehyde in PEM buffer (80 mM Pipes, 5 mM EGTA, 1 mM magnesium chloride, 3% sucrose), pH 7.4, embedded in OCT, and cryosectioned at 5-µm intervals. Slides were stored at -20°C. For peptide labeling experiments, the slides were thawed and incubated with peptide diluted to 100 to 200 µM as described above.
In Vivo Imaging of Flank Tumors
Nude mice with Gli36Δ5 or LN-229 flank tumors were imaged at 9 to 28 days after cell injection. Fluorophore-conjugated PTPµ peptides were diluted to 100 µM (SBK2-Alexa) or 200 µM (SBK1-TR, SBK3-TR, SBK4-TR, and scrambled SBK2-TR control peptide) in PBS and injected (total volume, 150 µl) through a lateral tail vein using a 28-gauge insulin syringe. In the animals containing GFP-expressing tumor cells, Alexa-750-labeled peptide was used owing to its limited spectral overlap with GFP. Spectral fluorescence images were obtained using the Maestro FLEX In Vivo Imaging System using the appropriate filters for GFP (tumor; excitation = 445–490 nm, emission = 515 nm long-pass filter, acquisition settings = 500–720 in 10-nm steps), TR (peptide; excitation = 575–605 nm, emission = 645 nm; acquisition settings = 630–850 in 10-nm steps), or Alexa-750 (peptide; excitation = 671–705 nm, emission = 750 nm long-pass filter, acquisition settings = 730–950 in 10-nm steps). Acquisition settings were 53 milliseconds for GFP and 1000 milliseconds for either TR or Alexa-750-labeled peptide. Before peptide injection, background images were acquired through the skin to provide autofluorescence spectra. After peptide injection, fluorescence images were acquired at 5- to 15-minute intervals during 2 to 3 hours. The multispectral fluorescence images were background-subtracted and unmixed, using Maestro software (Cambridge Research & Instrumentation, Inc, Woburn, MA), to spectrally separate the autofluorescence animal signal from the peptide signals. Regions of interest (ROIs) were selected over the tumor or nontumor skin. Pixel values for the peptide signal, in photons measured at the surface of the animal, were determined within these ROIs. Higher pixel values corresponded to presence of tumor. Pixel values in the tumor ROIs were normalized to the nontumor ROI and peptide concentration and were then plotted. Each PTPµ peptide was tested on a minimum of three animals containing Gli36Δ5 and three animals containing LN-229 flank tumors. Statistical analyses were performed using Microsoft Excel and an unpaired Student's t test.
Orthotopic Xenograft Intracranial Tumors
Gli36Δ5-GFP or LN-229-GFP cells were harvested for intracranial implantation by trypsinization and concentrated to 1 x 105 cells per microliter of PBS. For brain tumor implants, NIH athymic nude female mice were anesthetized by intraperitoneal injection of 50 mg/kg ketamine/xylazine and fitted into a stereotaxic rodent frame (David Kopf Instruments, Tujunga, CA). A small incision was made just lateral to midline to expose the bregma suture. A small (0.7 mm) burr hole was drilled at AP = +1, ML = -2.5 from bregma. Glioblastoma cells were slowly deposited at a rate of 1 µl/min in the right striatum at a depth of -3 mm from dura with a 10-µl syringe (26-gauge needle; Hamilton Co, Reno, NV). The needle was slowly withdrawn, and the incision was closed with sutures.
In Vivo Labeling of Intracranial Tumors
Nude mice with Gli36Δ5-GFP or LN-229-GFP intracranial tumors were imaged at 9 to 12 days after GBM cell implant. Fluorophore-conjugated PTPµ peptides were injected through the tail vein as described above. After a 25-minute incubation for clearance of unbound PTPµ peptide, the animals were sacrificed, and the brains were removed and imaged either whole or sliced into coronal sections at 1-mm intervals. Individual brain sections containing tumor were placed on a black slide and examined using the Maestro FLEX In Vivo Imaging System as described above. Untreated brains containing Gli36Δ5-GFP intracranial tumors were used to provide autofluorescence spectra. ROIs were selected over the tumor region in each brain slice. Pixel values for the peptide signal, in photons measured from the slice, were determined within these ROIs. The multispectral fluorescence images were background-subtracted and analyzed using the Maestro software as described previously. Statistical analyses were performed using Microsoft Excel and an unpaired Student's t test.
Results
An Extracellular Fragment of PTPµ Is Detected in Human Glioblastomas
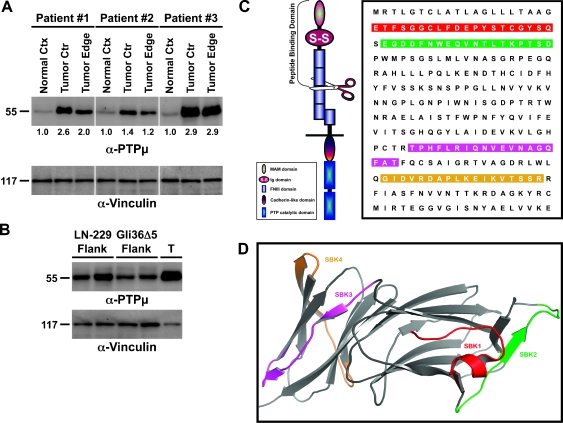
We recently demonstrated that full-length PTPµ is downregulated in human GBM tumors [12] through proteolytic processing [21]. We tested whether any extracellular fragments of PTPµ could be detected in human GBM tumor tissue lysates using the PTPµ-specific extracellular monoclonal antibody, BK9 [16]. BK9 was generated against the MAM domain of PTPµ [16]; it is specific to PTPµ and fails to cross-react even with closely related type IIb RPTPs (data not shown). Here, we show that a 55-kDa protein is recognized by BK9 (Figure 1A). Another PTPµ-specific polyclonal antibody, 494, also recognizes the 55-kDa protein (data not shown). The MAM domain of PTPµ is 160 amino acids in length, and the Ig and FNIII domains are both approximately 80 to 90 amino acids long; therefore, the 55-kDa fragment likely contains the MAM, Ig, and the first two FNIII repeats (Figure 1C). On the basis of these observations, the 55-kDa protein is an extracellular fragment of PTPµ.
Figure 1.
A 55-kDa extracellular fragment of PTPµ is detected in human glioblastoma tissue. (A) GBM tumors from three patients were divided into center (ctr) and edge samples, lysed, separated by SDS-PAGE, and immunoblotted using an antibody against the MAM domain of PTPµ. Noncancerous normal cortex (ctx) from the same patients was loaded for comparison. Equal protein load was verified by stripping and reprobing the immunoblot with an antibody to vinculin. Densitometry values for the 55-kDa extracellular fragment are shown under each GBM tumor center and edge lane and are normalized to the normal cortex from the same patient. (B) LN-229 or Gli36Δ5 xenograft flank tumor protein extracts were immunoblotted as described above. Human GBM tumor tissue (T) was loaded on the same blot for comparison. The numbers on the left side of each immunoblot correspond to the molecular weight in kilodaltons. (C) PTPµ structure and peptide probe sequences. PTPµ is a transmembrane protein that mediates efficient cell-cell adhesion through the MAM domain, Ig domain, and FNIII repeats within its extracellular segments. The scissors indicate the approximate site where PTPµ is cleaved to generate a 55-kDa N-terminal extracellular fragment. The sequence is shown for the PTPµ MAM and Ig domains. The highlighted regions indicate the sequences used to generate PTPµ peptide probes. (D) Crystal structure of the Ig and MAM domains of PTPµ (PDB ID:2V5Y). SBK1 (red) and SBK2 (green) were derived from the N-terminal MAM domain, whereas SBK3 (magenta) and SBK4 (camel) were from the Ig domain.
The 55-kDa extracellular fragment of PTPµ is detected in the center and edge of the resected GBM tumor at much higher levels than in normal cortical brain tissue from the same patient (Figures 1A and 4A). Densitometry indicates that the center and edge of GBM tumors contain approximately two-fold higher levels of the extracellular fragment (Figure 1A). The human edge samples were determined by pathologists to contain fewer tumor cells proportionally to normal cells, suggesting that the PTPµ extracellular fragment is concentrated at the tumor edge and may be exploitable as a biomarker of the tumor margin.
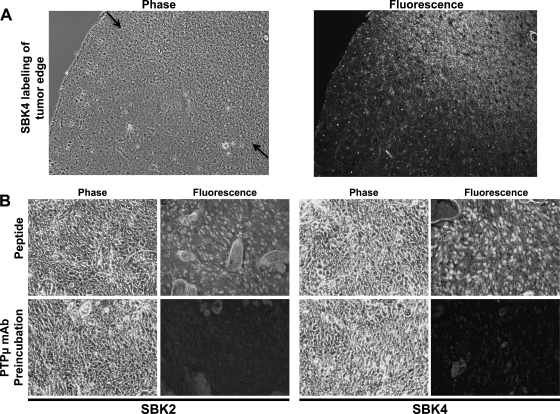
Figure 4.
PTPµ peptide SBK4 demarcates glioblastoma cells at the tumor edge. (A) Surgically resected human GBM tumor edge section was labeled with TR-conjugated SBK4 peptide. Arrows in the phase image indicate the approximate position of the margin between GBM and normal brain tissue. (B) PTPµ mAb blocks SBK2 and SBK4 peptide binding to human glioblastoma tissue. Preincubation of a PTPµ extracellular antibody raised against the MAM domain (BK2) specifically blocked SBK2-TR and SBK4-TR peptide binding to GBM tumor tissue sections.
Peptide Design and Optimization
PTPµ-dependent cell adhesion requires the MAM, Ig, and first two FNIII domains, all of which are predicted to be present in the 55-kDa cleaved fragment [15–20]. On the basis of crystallographic data indicating which residues are surface-exposed on PTPµ [19,20], and our binding studies [16], we designed peptide probes that would bind homophilically to the adhesive MAM and Ig domains contained within the cleaved fragment of the extracellular segment of PTPµ (Figure 1, C and D). Four peptides derived from the MAM (SBK1 and SBK2 peptides) or Ig (SBK3 and SBK4 peptides) domains of PTPµ were generated (Figure 1, C and D). As a control, a scrambled version of the SBK2 peptide was also synthesized. The peptides were fluorescently labeled with TR-X succinimidyl ester dye on the N-terminus.
Detection of Human Glioblastomas with the PTPµ Peptide Probes
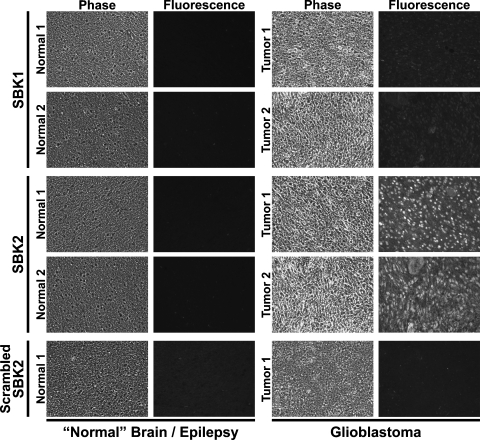
To examine the localization of the 55-kDa PTPµ fragment within GBM tumors, the TR-conjugated peptides were used as probes to label sections of GBM or noncancerous human brain obtained from epilepsy patients. The PTPµ-TR peptides each recognized GBM tissue to varying degrees, although the best labeling occurred with peptides SBK2 and SBK4 (Figures 2 and 3). Effective peptide concentrations for tissue labeling were determined by dilution histochemical analyses using human GBM tissue sections and indicated that the range of 3 to 40 µM was effective. The SBK1 peptide weakly labeled GBM tissue but did not label the noncancerous human brain (Figure 2). The SBK2, SBK3, and SBK4 peptides typically labeled the parenchyma above background with particularly bright labeling of the cell bodies (Figures 2 and 3). Endothelial cells express high levels of PTPµ [24–28] and were also brightly labeled within the tumor (Figures 2 and 3). Two representative GBM tumors are shown (tumors 1 and 2). Similar results were observed in six different GBM samples (data not shown). In contrast, scrambled control peptide (scrambled SBK2) did not label the GBM tissue (Figures 2 and 3). When PTPµ peptides were incubated with sections of noncancerous brain tissue obtained from epilepsy patients, no specific structures were labeled in either white or gray matter regions (Figures 2 and 3). Similar results were observed in six different samples of noncancerous “normal brain” (data not shown).
Figure 2.
SBK2 peptide probe specifically recognizes human glioblastoma tissue but not normal brain. Sections of noncancerous normal cortical brain tissue fromepilepsy patients or GBM tumor were histochemically labeled with TR-conjugated SBK1, SBK2, or scrambled SBK2 peptides. Two examples of normal and GBM tissue are shown, which are representative of six different samples examined.
Figure 3.
SBK3 and SBK4 peptide probes specifically recognize human glioblastoma tissue but not normal brain. Sections of noncancerous normal cortical brain tissue from epilepsy patients or GBM tumor were histochemically labeled with TR-conjugated SBK3, SBK4, or scrambled SBK2 peptides. Two examples of normal and GBM tissue are shown, which are representative of six different samples examined.
The correct determination of the tumor edge is of great importance to the neurosurgeon during tumor resection. In tissue sections from patients containing the edge of GBM tumor, the PTPµ peptide SBK4 clearly labeled the GBM tumor edge against the normal brain tissue background (Figure 4A). The scrambled control peptide did not label the tumor edge (data not shown). In normal brain tissue, when PTPµ is involved in cell-cell adhesion, its binding sites may be fully engaged and therefore unavailable for recognition by the PTPµ peptides. Alternatively, in GBM tissue, the extracellular domain of PTPµ may undergo a conformational change after cleavage that allows recognition by the PTPµ peptides.
The PTPµ Peptides Specifically Recognize PTPµ
To confirm that the PTPµ peptides were binding to an extracellular fragment of PTPµ in GBM tissue, we performed antibody-blocking experiments. To block peptide binding, we used the PTPµ-specific monoclonal antibody, BK2, which was raised against the MAM domain of PTPµ and recognized native protein [16]. The BK2 antibody does not cross-react with other closely related type IIb RPTPs (data not shown). GBM tissue sections were preincubated with BK2 before incubating with the SBK2 peptide, generated from the MAM domain, or SBK4 peptide, generated from the Ig domain of PTPµ. Because of the tightly associated structure of the MAM-Ig domain of PTPµ [19], we predicted that BK2 would sterically block access of both peptides to the extracellular fragment. Preincubation with BK2 caused a complete block of SBK2 and SBK4 peptide binding (Figure 4B). The detection of the extracellular PTPµ fragment on blots using both BK9 monoclonal and 494 polyclonal antibodies and the blocking of the peptide binding by the BK2 monoclonal antibody in tissue sections demonstrate that the SBK2 and SBK4 peptides recognize and bind to an extracellular fragment of PTPµ in human GBM tissues.
Peptide Recognition of Glioblastoma-Associated PTPµ Fragment as a Diagnostic Tool
Specific recognition of GBM cells in vivo through peptide binding would provide a powerful diagnostic tool. We developed a xenograft animal model system of GBM for in vivo labeling. Two GFP-expressing human glioblastoma cell lines, LN-229 and Gli36Δ5, were injected individually into the flanks of nude mice for the production of xenograft tumors. At 2 to 3 weeks after injection, the tumors were excised and homogenized for biochemical analysis. The extracellular fragment of PTPµ is present in both LN-229 and Gli36Δ5 cells when grown as flank tumors (Figure 1B).
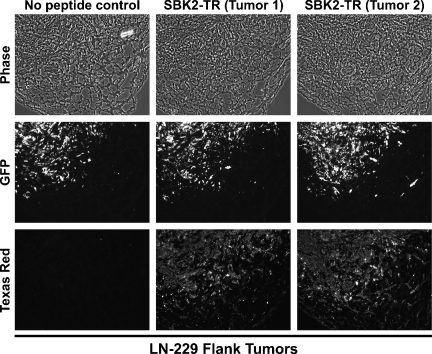
To observe localization of the PTPµ extracellular fragment in the xenograft tumors, sections of fixed tumors were incubated with either SBK2 or SBK4 peptides. The PTPµ peptides labeled both the Gli36Δ5 (Figure W1) and LN-229 (Figures 5 and W2) tumors, in a pattern that closely overlaid the tumor cells as evidenced by GFP fluorescence. Small clusters of labeled cells were clearly visible at the tumor edge (Figure 5); thus, the peptides may be useful for detection of the tumor margin. The recognition of the extracellular fragment by PTPµ peptides suggested that this animal model was a viable system for further study in vivo.
Figure 5.
Small cell clusters from LN-229 flank tumors label with PTPµ peptide SBK2. Flank tumors of LN-229 cells were excised, fixed, and sectioned. The LN-229 cells express GFP. Binding of the TR-conjugated SBK2 peptide is shown in two tumor samples. SBK2 peptide labels small clusters of cells in the tumor microenvironment.
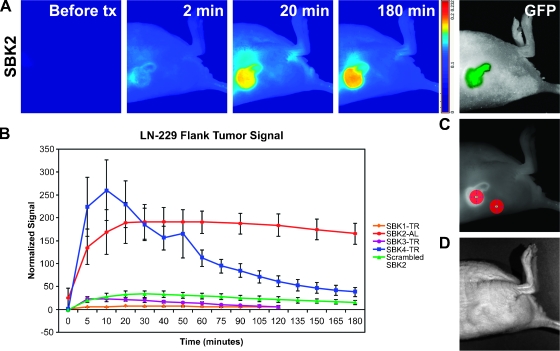
Flank tumors provide a useful model for molecular imaging studies owing to accessibility and ease of imaging using fluorescence detection methods. Nude mice with Gli36Δ5 or LN-229 flank xenografts were imaged through the skin with the Maestro FLEX In Vivo Imaging System using the appropriate filters for GFP, TR, or Alexa-750. Background images of the flank region containing the tumor were acquired, PTPµ peptides were administered through tail vein injection, and the animals were imaged at regular intervals during the course of 2 to 3 hours. Fluorescence was observed throughout the tumor within 1 minute after injection, but maximal tumor labeling occurred within 10 to 20 minutes after injection. Unbound circulating peptide was cleared from the animals quickly, resulting in a clear demarcation of tumor over the normal tissue background (Figures 6A and 7A). In most cases, the tumor remained labeled above background for at least 3 hours. The average signal in the tumor was normalized to the average signal in nontumor skin and plotted (Figures 6B and 7B). SBK2 peptide rapidly bound to the Gli36Δ5-GFP tumor and remained bound at a level greater than that in the surrounding skin during the course of the 3-hour experiment (Figure 6). Peak levels of Gli36Δ5-GFP tumor labeling were achieved in 10 to 20 minutes. Of note, similar results were obtained when the tumor was composed of LN-229-GFP human GBM cell line (Figure 7), suggesting that the peptide may be useful for labeling glioblastomas in general. The SBK4 peptide labeled both Gli36Δ5 and LN-229 flank tumors to a similar extent as SBK2, but SBK4 labeling intensity decreased at a faster rate over time (Figures 6B and 7B). The levels of Gli36Δ5 and LN-229 tumor labeling with SBK2 or SBK4 peptides were significantly different from the scrambled control peptide, as analyzed with an unpaired Student's t test. Two other peptides, SBK1 and SBK3, bound the GBM tumors poorly in vivo, with no significant difference from the scrambled control peptide (Figures 6B and 7B).
Figure 6.
PTPµ peptides SBK2 and SBK4 recognize Gli36Δ5 mouse flank tumors in vivo. TR- or Alexa-750 (AL)-conjugated PTPµ peptides were administered intravenously to mice with xenograft flank tumors of Gli36Δ5 cells. (A) Fluorescent images of SBK2-AL peptide labeling. Panel-labeled “before tx” (before time course) shows the animal autofluorescent background. The tumor cells are expressing GFP. (B) Time course of peptide binding to flank tumors (n = 3 animals tested per peptide). Average normalized signals acquired in the tumor region of interest were plotted. Error bars, SEM from the three animals. (C) ROIs shown over the tumor and nontumor skin. (D) Bright field image of the flank tumor labeled in (A).
Figure 7.
PTPµ peptides SBK2 and SBK4 recognize LN-229 mouse flank tumors in vivo. TR- or Alexa-750 (AL)-conjugated PTPµ peptides were administered intravenously to mice with xenograft flank tumors of LN-229 cells. (A) Fluorescent images of SBK2-AL peptide labeling. Panel-labeled “before tx” (before time course) shows the animal autofluorescent background. The tumor cells are expressing GFP. (B) Time course of peptide binding to flank tumors (n = 3 animals tested per peptide). Average normalized signals acquired in the tumor region of interest were plotted. Error bars, SEM from the three animals. (C) ROIs shown over the tumor and nontumor skin. (D) Bright field image of the flank tumor labeled in A.
Peptide Crossing of the Compromised Blood-Brain Barrier and Recognition of Glioblastoma Tumors
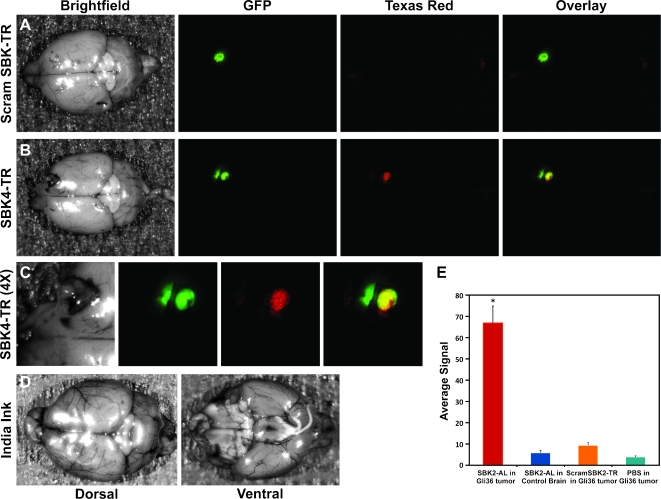
Whereas the results of flank tumor labeling demonstrate that the PTPµ peptides are capable of labeling GBM tumors in vivo, it was important to determine whether these peptides can cross the bloodbrain barrier, which is compromised in glioblastomas, to reach physiological targets in the brain. Athymic nude mice implanted with LN-229-GFP or Gli36Δ5-GFP intracranial tumors were used for PTPµ peptide labeling experiments. SBK2 or SBK4 peptide was injected through the tail vein and allowed to circulate in the mouse for 25 minutes to permit binding to the glioma brain tumor and initial clearance from nontumor tissue. The Maestro FLEX In Vivo Imaging System cannot image through the skull. Therefore, the animals were sacrificed, and the treated brain was removed and imaged whole or sectioned into 1-mm coronal slices and analyzed using the Maestro System as described above using filters appropriate for GFP, TR, or Alexa-750 fluorescence (Figure 8). The location of the tumor in the brain is indicated by GFP fluorescence. The TR scrambled control peptide did not label the tumor (Figure 8A). The TR SBK4 peptide crossed the compromised blood-brain barrier and bound to the main glioma tumor mass (Figure 8B). This particular tumor protruded into the skull, and a small piece of the skull is attached that contains GFP-labeled tumor cells (Figure 8, B and C, on the left side of each image). The tumor area was magnified four times for ease of visualization of the coincident labeling of GFP and SBK4 within the Gli36Δ5 tumor (Figure 8C, yellow-overlay image). India ink perfusion of the brain indicates the location of the vasculature in that area of the brain (Figure 8D), which is distinct from the labeling pattern observed with the SBK4 peptide. Quantitation of brain slice labeling in Gli36Δ5-GFP tumors showed the SBK2 peptide labeled at a significantly higher level than either scrambled peptide or PBS control injections (Figure 8E). Together, these studies suggest that the PTPµ peptide probes SBK2 and SBK4 specifically bind to the extracellular PTPµ fragment, label human GBM tumors, and can cross the compromised blood-brain barrier to identify the GBM tumor noninvasively in vivo.
Figure 8.
PTPµ peptides SBK2 and SBK4 label LN-229 and Gli36Δ5 intracranial tumors in vivo. TR-conjugated PTPµ peptides were administered intravenously to mice with xenograft intracranial tumors of LN-229 or Gli36Δ5 cells. (A–C) Bright field images of brains containing LN-229-GFP tumors are shown on the left after tail vein injection of the peptides. GFP fluorescence of the brains is shown second, indicating the location of the LN-229-GFP tumor. TR fluorescent images of the brains are shown third. The overlay image of the GFP and TR is shown on the far right. (A) Scrambled control peptide did not label the tumors. (B) The SBK4 peptide crossed the compromised blood-brain barrier to label LN-229 tumors. (C) The SBK4 peptide labeling is shown at four times magnification to enhance visualization of the overlay of green and red fluorescence, which appears as yellow. (D) India ink perfusion of the brains indicates the location of blood vessels in that region of the brain. (E) Fluorescence quantitation of peptide binding in brain slices containing Gli36Δ5-GFP tumor cells labeled with SBK2-AL peptide (n = 6), scrambled SBK2-TR peptide (n = 7), or PBS (n = 6). Quantitation of SBK2-AL labeling of control brain slices (n = 4) is also shown. Error bars, SE. Statistical significance from control calculated with the Student's t-test is shown (*P < .001).
Discussion
Two biologic characteristics contribute to the lethality of many human brain tumors: 1) their uncontrolled proliferation in the restricted cranial space and 2) the highly dispersive nature of tumors such as glioblastomas [1]. The rapid and extensive dispersal of GBMs results in the formation of secondary masses throughout the brain, some of which occur in inoperable regions. This, combined with an increased tumor load, leads to rapid mortality [6]. MRI is limited in its ability to detect these small clusters of glioblastoma cells that can form secondary masses [7]. Thus, MRI-guided surgical resections are likely to overlook GBM cells that have dispersed away from the main tumor mass, leading to poor prognosis. Clearly, the development of specific molecular diagnostics is critical for the detection and eradication of GBM. In this article, we report the identification of an extracellular fragment of PTPµ that can be molecularly imaged in vivo and used as a unique biomarker of GBM cells. Importantly, the peptides described in this article offer novel molecular diagnostic tools that are able to cross the compromised bloodbrain barrier to label GBM tumors.
GBM cell dispersal occurs along characteristic pathways of anatomic structures in the brain that are rich in cell adhesion molecules and extracellular matrix molecules that serve as permissive substrates for cell migration [29,30]. GBM cell dispersal requires the production of proteolytic enzymes [31], which gives the cell the ability to move through its environment [29]. GBM cells overexpress growth factor receptor protein tyrosine kinases and their ligands, an important prerequisite for tumor growth and dispersal [32]. The activity of the receptor tyrosine kinases is normally kept in check by the opposing activity of RPTPs such as PTPµ, which are important regulators of adhesion-dependent signals [8–10].
Human tissue samples of GBM and noncancerous “normal” brain from epileptic foci were examined by immunoblot analyses. These studies revealed a dramatic increase of a 55-kDa N-terminal extracellular fragment of PTPµ in dispersive GBMs when compared with normal brain. Using peptides that specifically recognize this PTPµ extracellular fragment, it was determined that the PTPµ fragment is common to high-grade glioblastomas. The PTPµ extracellular fragment is present in human tumor “edge” samples, and the peptides are able to demarcate tumor cells in tissue sections, suggesting that the peptides could be used diagnostically for molecular imaging of dispersive brain tumors or the tumor margin in vivo. To assess whether this is the case, experiments were performed using a rodent flank human GBM tumor model. The proof-of-principle experiments used fluorescently labeled peptide probes to image the tumor cells in live rodents using the Maestro FLEX In Vivo Imaging System. Systemic introduction of the peptide probes resulted in rapid and specific labeling of the flank tumors within minutes. Labeling occurred primarily within the tumor and at the tumor margin, indicating that the extracellular PTPµ fragment remains associated with the tumor. Most importantly, the PTPµ peptides crossed the compromised blood-brain barrier to specifically label GBM tumor cells in the brain. Together, these data demonstrate that the PTPµ peptide probes could be used as molecular indicators of highgrade glioblastoma.
The development of the PTPµ peptide probes was based on a large body of structural and functional data. The sites required for PTPµ-mediated homophilic adhesion have been well characterized by our laboratory and others [11,13–20,33]. The crystal structure of PTPµ provided valuable information regarding which regions of each functional domain are likely to be exposed to the extracellular environment and therefore available for homophilic binding and detection by a peptide probe. Of the four peptides generated, we found two that are compelling as specific markers of high-grade GBM (SBK2 and SBK4). A point of interest is that these peptides recognize two different domains of PTPµ (SBK2: the MAM domain; SBK4: the Ig domain) that are folded in a close conformation [19], and are both required for efficient cell-cell adhesion [34].
We previously determined that PTPµ is cleaved at three different sites to generate both a mature protein and, ultimately, an intracellular fragment that is found in both the cytoplasm and the nucleus [21]. In that study, we determined that cleavage of the extracellular domain of PTPµ through a metalloprotease (a matrix metalloproteinase or A Disintegrin And Metalloprotease [ADAM]) was necessary for further processing [21]. Although we cannot identify the site of cleavage because metalloproteases lack a specific amino acid recognition sequence, it may be that such an enzyme is functioning to yield the extracellular fragment of PTPµ. Another type IIb RPTP, PTPκ is also cleaved by an ADAM to release an extracellular fragment [35]; thus, the cleavage we observe here may be a common phenomenon. It is likely that cleavage of other cell surface receptors also occurs in GBMs and many other tumors. In support of this, metalloproteases [36] and metalloprotease cleavage of adhesion molecules are linked to tumor progression [37]. A similar molecular detection strategy could be used with any other homophilic or heterophilic-binding cell surface protein whose ligand-binding site is known. A large variety of cell surface proteins, including other phosphatases, are cleaved at the cell surface [35,38–40]. These proteins represent additional targets for the development of novel molecular diagnostics [41]. Furthermore, the PTPµ peptides could be used as a starting point to develop higher affinity small molecules with similar ligand-binding capabilities.
In the context of this study, a question remains: What is the biologic significance of the cleavage of PTPµ and shedding of the extracellular fragment? Recently, we examined whether the absence of PTPµ influences the migratory behavior of glioblastoma cells in the complex environment of the brain [12]. We found that full-length PTPµ influences contact-dependent signaling by negatively regulating migration of glial cells [12]. Therefore, the loss of PTPµ protein expression through proteolysis and fragment generation may be advantageous to GBM during tumor progression. It is unclear whether the association of the PTPµ fragment with dispersing GBM cells has an additional functional role to modulate cell-cell adhesion or interaction with endothelial cells, which also express PTPµ [24–28]. Alternatively, the extracellular fragment may activate intracellular signaling cascades by binding to cell surface PTPµ on other cells in the tumor microenvironment. Because the extracellular fragment contains all the domains of PTPµ required for efficient homophilic binding and it maintains adhesive activity, it is intriguing to speculate that the PTPµ shedding generates a “highway” for tumor cell migration in the microenvironment.
Neurosurgeons routinely use stereotactic techniques and intraoperative MRI in surgical resections. This allows them to identify and sample tissue from distinct regions of the tumor such as the tumor edge or center. Frequently, they also sample regions of brain on the tumor margin that are outside the tumor edge that appear to be grossly normal but are infiltrated by dispersing tumor cells on histologic examination. Such surgical techniques have been instrumental in characterizing various molecules associated with the invasive phenotype [42,43].With the discovery of molecular diagnostics for GBM cells, intraoperative imaging techniques could guide neurosurgical resection and eliminate the “educated guess” of the location of the tumor margin by the neurosurgeon. Previous studies have determined that more extensive surgical resection improves patient survival [5,44,45]. Thus, PTPµ peptide probes that function as diagnostic molecular imaging agents have the potential to increase patient survival. Currently available intraoperative imaging devices could be retrofitted to visualize these PTPµ peptide probes. Alternatively, the PTPµ peptides could be labeled with other agents to identify the best imaging modality for visualizing small numbers of dispersing GBM tumor cells. Such an approach has clear translational applications and may lead to improved outcomes for patients with this devastating disease.
Supplemental Data
Acknowledgments
The authors thank Marty Pagel, Meser Ali, and Richard Agnes for technical help with peptide production and labeling. Catherine Doller, Visual Sciences Research Center (PO-EY11373), CWRU, provided excellent histologic services. Scott Becka, Nathan Cohen, and Jing Wang provided technical assistance; and J.T. Kalnay encouraged the collaboration with the Case Center for Imaging Research. Lastly, the authors thank Sara Lou for production of figures and the members of the Brady-Kalnay laboratory for insightful discussions and critical reading of the manuscript. This work is dedicated to Tabitha Yee-May Lou who recently lost her battle with glioblastoma.
Abbreviations
- FNIII
fibronectin type III repeats
- GBM
glioblastoma multiforme
- GFP
green fluorescent protein
- Ig
immunoglobulin domain
- MRI
magnetic resonance imaging
- MAM
meprin, A5 (neuropilin), µ domain
- PTPµ
protein tyrosine phosphatase µ
- RT
room temperature
- ROI
region of interest
- TR
Texas Red
Footnotes
This study was supported by the Case Center for Imaging Research and a pilot grant from the Case Comprehensive Cancer Center. The research was also supported by National Institutes of Health grants R01-NS051520 (S.B.-K., S.R. and R.H.M.) and R01-NS063971 (S.B.-K., J.P.B., R.H.M. and A.E.S.). A.E.S. was supported by funding from National Cancer Institute grants K08-CA101954 and R01-CA116257, the Ivy Brain Tumor Foundation, and the Cancer Genome Atlas. D.T.L. was supported by a postdoctoral fellowship from Visual Sciences Training Grant T32-EY007157.
This article refers to supplementary materials, which are designated by Figures W1 and W2 and are available online at www.neoplasia.com.
References
- 1.Ichimura K, Ohgaki H, Kleihues P, Collins VP. Molecular pathogenesis of astrocytic tumours. J Neurooncol. 2004;70:137–160. doi: 10.1007/s11060-004-2747-2. [DOI] [PubMed] [Google Scholar]
- 2.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK. World Health Organization Classification of Tumours of the Nervous System. Lyon, France: IARC; 2007. [Google Scholar]
- 3.Louis DN. Molecular pathology of malignant gliomas. Annu Rev Pathol. 2006;1:97–117. doi: 10.1146/annurev.pathol.1.110304.100043. [DOI] [PubMed] [Google Scholar]
- 4.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 5.Sanai N, Berger MS. Glioma extent of resection and its impact on patient outcome. Neurosurgery. 2008;62:753–764. doi: 10.1227/01.neu.0000318159.21731.cf. discussion 264–756. [DOI] [PubMed] [Google Scholar]
- 6.Nakada M, Nakada S, Demuth T, Tran NL, Hoelzinger DB, Berens ME. Molecular targets of glioma invasion. Cell Mol Life Sci. 2007;64:458–478. doi: 10.1007/s00018-007-6342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sorensen AG, Batchelor TT, Wen PY, Zhang WT, Jain RK. Response criteria for glioma. Nat Clin Pract Oncol. 2008;5:634–644. doi: 10.1038/ncponc1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brady-Kalnay SM. Protein tyrosine phosphatases. In: Beckerle M, editor. Cell Adhesion: Frontiers in Molecular Biology. Vol. 39. Oxford, UK: Oxford University Press; 2001. pp. 217–258. [Google Scholar]
- 9.Ensslen-Craig SE, Brady-Kalnay SM. Receptor protein tyrosine phosphatases regulate neural development and axon guidance. Dev Biol. 2004;275:12–22. doi: 10.1016/j.ydbio.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 10.Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nat Rev Mol Cell Biol. 2006;7:833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 11.Burden-Gulley SM, Brady-Kalnay SM. PTPµ regulates N-cadherin-dependent neurite outgrowth. J Cell Biol. 1999;144:1323–1336. doi: 10.1083/jcb.144.6.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burgoyne AM, Palomo JM, Phillips-Mason PJ, Burden-Gulley SM, Major DL, Zaremba A, Robinson S, Sloan AE, Vogelbaum MA, Miller RH, et al. PTPµ suppresses glioblastoma cell migration and dispersal. J Neurooncol. 2009;11:767–778. doi: 10.1215/15228517-2009-019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brady-Kalnay SM, Flint AJ, Tonks NK. Homophilic binding of PTPµ, a receptor-type protein tyrosine phosphatase, can mediate cell-cell aggregation. J Cell Biol. 1993;122:961–972. doi: 10.1083/jcb.122.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gebbink MFBG, Zondag GCM, Wubbolts RW, Beijersbergen RL, van Etten I, Moolenaar WH. Cell-cell adhesion mediated by a receptor-like protein tyrosine phosphatase. J Biol Chem. 1993;268:16101–16104. [PubMed] [Google Scholar]
- 15.Zondag G, Koningstein G, Jiang YP, Sap J, Moolenaar WH, Gebbink M. Homophilic interactions mediated by receptor tyrosine phosphatases µ and κ. J Biol Chem. 1995;270:14247–14250. doi: 10.1074/jbc.270.24.14247. [DOI] [PubMed] [Google Scholar]
- 16.Brady-Kalnay SM, Tonks NK. Identification of the homophilic binding site of the receptor protein tyrosine phosphatase PTPµ. J Biol Chem. 1994;269:28472–28477. [PubMed] [Google Scholar]
- 17.Del Vecchio RL, Tonks NK. The conserved immunoglobulin domain controls the subcellular localization of the homophilic adhesion receptor proteintyrosine phosphatase µ. J Biol Chem. 2005;280:1603–1612. doi: 10.1074/jbc.M410181200. [DOI] [PubMed] [Google Scholar]
- 18.Cismasiu VB, Denes SA, Reilander H, Michel H, Szedlacsek SE. The MAM (meprin/A5-protein/PTPµ) domain is a homophilic binding site promoting the lateral dimerization of receptor-like protein-tyrosine phosphatase µ. J Biol Chem. 2004;279:26922–26931. doi: 10.1074/jbc.M313115200. [DOI] [PubMed] [Google Scholar]
- 19.Aricescu AR, Hon WC, Siebold C, Lu W, van der Merwe PA, Jones EY. Molecular analysis of receptor protein tyrosine phosphatase µ-mediated cell adhesion. EMBO J. 2006;25:701–712. doi: 10.1038/sj.emboj.7600974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aricescu AR, Siebold C, Choudhuri K, Chang VT, Lu W, Davis SJ, van der Merwe PA, Jones EY. Structure of a tyrosine phosphatase adhesive interaction reveals a spacer-clamp mechanism. Science. 2007;317:1217–1220. doi: 10.1126/science.1144646. [DOI] [PubMed] [Google Scholar]
- 21.Burgoyne AM, Phillips-Mason PJ, Burden-Gulley SM, Robinson S, Sloan AE, Miller RH, Brady-Kalnay SM. Proteolytic cleavage of PTPµ regulates glioblastoma cell migration. Cancer Res. 2009;69:6960–6968. doi: 10.1158/0008-5472.CAN-09-0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tyminski E, Leroy S, Terada K, Finkelstein DM, Hyatt JL, Danks MK, Potter PM, Saeki Y, Chiocca EA. Brain tumor oncolysis with replication-conditional herpes simplex virus type 1 expressing the prodrug-activating genes, CYP2B1 and secreted human intestinal carboxylesterase, in combination with cyclophosphamide and irinotecan. Cancer Res. 2005;65:6850–6857. doi: 10.1158/0008-5472.CAN-05-0154. [DOI] [PubMed] [Google Scholar]
- 23.Tyagi M, Karn J. CBF-1 promotes transcriptional silencing during the establishment of HIV-1 latency. EMBO J. 2007;26:4985–4995. doi: 10.1038/sj.emboj.7601928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Campman M, Yoshizumi M, Seidah NG, Lee ME, Bianchi C, Haber E. Increased proteolytic processing of protein tyrosine phosphatase µ in confluent vascular endothelial cells: the role of PC5, a member of the subtilisin family. Biochemistry. 1996;35:3797–3802. doi: 10.1021/bi952552d. [DOI] [PubMed] [Google Scholar]
- 25.Bianchi C, Sellke FW, Neel BG. Receptor-type protein-tyrosine phosphatase µ is expressed in specific vascular endothelial beds in vivo. Exp Cell Res. 1999;248:329. doi: 10.1006/excr.1999.4428. [DOI] [PubMed] [Google Scholar]
- 26.Koop EA, Lopes SM, Feiken E, Bluyssen HA, van der Valk M, Voest EE, Mummery CL, Moolenaar WH, Gebbink MF. Receptor protein tyrosine phosphatase µ expression as a marker for endothelial cell heterogeneity; analysis of RPTPµ gene expression using LacZ knock-in mice. Int J Dev Biol. 2003;47:345–354. [PubMed] [Google Scholar]
- 27.Koop EA, Gebbink MF, Sweeney TE, Mathy MJ, Heijnen HF, Spaan JA, Voest EE, VanBavel E, Peters SL. Impaired flow-induced dilation in mesenteric resistance arteries from receptor protein tyrosine phosphatase-µ-deficient mice. Am J Physiol Heart Circ Physiol. 2005;288:H1218–H1223. doi: 10.1152/ajpheart.00512.2004. [DOI] [PubMed] [Google Scholar]
- 28.Sui XF, Kiser TD, Hyun SW, Angelini DJ, Del Vecchio RL, Young BA, Hasday JD, Romer LH, Passaniti A, Tonks NK, et al. Receptor protein tyrosine phosphatase µ regulates the paracellular pathway in human lung microvascular endothelia. Am J Pathol. 2005;166:1247–1258. doi: 10.1016/s0002-9440(10)62343-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tysnes BB, Mahesparan R. Biological mechanisms of glioma invasion and potential therapeutic targets. J Neurooncol. 2001;53:129–147. doi: 10.1023/a:1012249216117. [DOI] [PubMed] [Google Scholar]
- 30.Demuth T, Berens ME. Molecular mechanisms of glioma cell migration and invasion. J Neurooncol. 2004;70:217–228. doi: 10.1007/s11060-004-2751-6. [DOI] [PubMed] [Google Scholar]
- 31.Rao JS. Molecular mechanisms of glioma invasiveness: the role of proteases. Nat Rev Cancer. 2003;3:489–501. doi: 10.1038/nrc1121. [DOI] [PubMed] [Google Scholar]
- 32.Sathornsumetee S, Rich JN. Designer therapies for glioblastoma multiforme. Ann N Y Acad Sci. 2008;1142:108–132. doi: 10.1196/annals.1444.009. [DOI] [PubMed] [Google Scholar]
- 33.Ensslen-Craig SE, Brady-Kalnay SM. PTP µ expression and catalytic activity are required for PTP µ-mediated neurite outgrowth and repulsion. Mol Cell Neurosci. 2005;28:177–188. doi: 10.1016/j.mcn.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 34.Aricescu AR, Siebold C, Jones EY. Receptor protein tyrosine phosphatase µ: measuring where to stick. Biochem Soc Trans. 2008;36:167–172. doi: 10.1042/BST0360167. [DOI] [PubMed] [Google Scholar]
- 35.Anders L, Mertins P, Lammich S, Murgia M, Hartmann D, Saftig P, Haass C, Ullrich A. Furin-, ADAM10-, and gamma-secretase-mediated cleavage of a receptor tyrosine phosphatase and regulation of β-catenin's transcriptional activity. Mol Cell Biol. 2006;26:3917–3934. doi: 10.1128/MCB.26.10.3917-3934.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roy R, Yang J, Moses MA. Matrix metalloproteinases as novel biomarkers and potential therapeutic targets in human cancer. J Clin Oncol. 2009;27:5287–5297. doi: 10.1200/JCO.2009.23.5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pruessmeyer J, Ludwig A. The good, the bad and the ugly substrates for ADAM10 and ADAM17 in brain pathology, inflammation and cancer. Semin Cell Dev Biol. 2009;20:164–174. doi: 10.1016/j.semcdb.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 38.Streuli M, Krueger N, Ariniello P, Tang M, Munro J, Blattler W, Adler D, Disteche C, Saito H. Expression of the receptor-linked protein tyrosine phosphatase LAR: proteolytic cleavage and shedding of the CAM-like extracellular region. EMBO J. 1992;11:897–907. doi: 10.1002/j.1460-2075.1992.tb05128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haapasalo A, Kim DY, Carey BW, Turunen MK, Pettingell WH, Kovacs DM. Presenilin/gamma-secretase-mediated cleavage regulates association of leukocyte—common antigen-related (LAR) receptor tyrosine phosphatase with β-catenin. J Biol Chem. 2007;282:9063–9072. doi: 10.1074/jbc.M611324200. [DOI] [PubMed] [Google Scholar]
- 40.Chow JP, Fujikawa A, Shimizu H, Noda M. Plasmin-mediated processing of protein tyrosine phosphatase receptor type ζ in the mouse brain. Neurosci Lett. 2008;442:208–212. doi: 10.1016/j.neulet.2008.07.028. [DOI] [PubMed] [Google Scholar]
- 41.Barr AJ, Ugochukwu E, Lee WH, King ON, Filippakopoulos P, Alfano I, Savitsky P, Burgess-Brown NA, Muller S, Knapp S. Large-scale structural analysis of the classical human protein tyrosine phosphatome. Cell. 2009;136:352–363. doi: 10.1016/j.cell.2008.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mariani L, Beaudry C, McDonough WS, Hoelzinger DB, Kaczmarek E, Ponce F, Coons SW, Giese A, Seiler RW, Berens ME. Death-associated protein 3 (Dap-3) is overexpressed in invasive glioblastoma cells in vivo and in glioma cell lines with induced motility phenotype in vitro. Clin Cancer Res. 2001;7:2480–2489. [PubMed] [Google Scholar]
- 43.Hoelzinger DB, Mariani L, Weis J, Woyke T, Berens TJ, McDonough WS, Sloan A, Coons SW, Berens ME. Gene expression profile of glioblastoma multiforme invasive phenotype points to new therapeutic targets. Neoplasia. 2005;7:7–16. doi: 10.1593/neo.04535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stummer W, Novotny A, Stepp H, Goetz C, Bise K, Reulen HJ. Fluorescence-guided resection of glioblastoma multiforme by using 5-aminole-vulinic acid-induced porphyrins: a prospective study in 52 consecutive patients. J Neurosurg. 2000;93:1003–1013. doi: 10.3171/jns.2000.93.6.1003. [DOI] [PubMed] [Google Scholar]
- 45.Lacroix M, Abi-Said D, Fourney DR, Gokaslan ZL, Shi W, DeMonte F, Lang FF, McCutcheon IE, Hassenbusch SJ, Holland E, et al. A multivariate analysis of 416 patients with glioblastoma multiforme: prognosis, extent of resection, and survival. J Neurosurg. 2001;95:190–198. doi: 10.3171/jns.2001.95.2.0190. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.