Abstract
The small molecule 4EGI-1 was identified as an inhibitor of cap-dependent translation initiation owing to its disruption of the eIF4E/eIF4G association through binding to eIF4E. 4EGI-1 exhibits growth-inhibitory and apoptosis-inducing activity in cancer cells; thus, we were interested in its therapeutic efficacy in human lung cancer cells. 4EGI-1, as a single agent, inhibited the growth and induced apoptosis of human lung cancer cells.When combined with the death ligand tumor necrosis factor-related apoptosis-inducing ligand (TRAIL), enhanced apoptosis-induced activity was observed. As expected, 4EGI-1 inhibited eIF4E/eIF4G interaction and reduced the levels of cyclin D1 and hypoxia-inducing factor-1α (HIF-1α), both of which are regulated by a cap-dependent translation mechanism. Moreover, 4EGI-1 induced CCAAT/enhancer-binding protein homologous protein-dependent DR5 expression and ubiquitin/proteasome- mediated degradation of cellular FLICE-inhibitory protein (c-FLIP). Small interfering RNA-mediated blockade of DR5 induction or enforced expression of c-FLIP abrogated 4EGI-1's ability to enhance TRAIL-induced apoptosis, indicating that both DR5 induction and c-FLIP down-regulation contribute to enhancement of TRAIL-induced apoptosis by 4EGI-1. However, inhibition of eIF4E/eIF4G interaction by knockdown of eIF4E effectively reduced the levels of cyclin D1 and HIF-1α but failed to induce DR5 expression, downregulate c-FLIP levels, or augment TRAIL-induced apoptosis. These results collectively suggest that 4EGI-1 augments TRAIL-induced apoptosis through induction of DR5 and down-regulation of c-FLIP, independent of inhibition of cap-dependent protein translation.
Introduction
Protein translational control is an important strategy by which eukaryotic cells regulate gene expression. A prime target of translational control is eukaryotic translation initiation factor 4E (eIF4E), which recognizes and binds to the 7-methylguanosine cap structure present at the 5′ untranslated regions of cellular messenger RNA (mRNA) and delivers these mRNA to the eIF4F translation initiation complex, which is composed of the cap-binding protein eIF4E, the RNA helicase eIF4A, and the scaffolding protein eIF4G [1,2]. Assembly of the eIF4F complex is dependent on eIF4E availability. Given that eIF4E is the least abundant among the initiator factors involved in the eIF4F complex, eIF4E is the rate-limiting factor for cap-dependent translation initiation [2]. Consequently, changes in eIF4E levels profoundly affect translation rates of certain proteins, particularly those related to cell growth and survival involved in oncogenesis (e.g., c-Myc, vascular endothelial growth factor, ODC, cyclin D1, hypoxia-inducing factor-1 [HIF-1], andMcl-1), which, under normal cellular conditions, are translationally repressed [3,4]. Under normal cellular conditions, eIF4E is bound by the inhibitory 4E-BPs, which sequester eIF4E from interaction with eIF4G, preventing the formation of the eIF4F translation initiation complex. The activities of 4E-BPs are regulated by phosphorylation through themammalian target of rapamycin (mTOR) pathway. Hypophosphorylated 4E-BPs sequester eIF4E, inhibiting translation, whereas hyperphosphorylated 4E-BPs do not bind eIF4E and the eIF4E is free to participate in translation initiation. The 4E-BPs compete with eIF4G for binding to eIF4E because they share the same binding motif on eIF4E [5–7].
Abnormal regulation of cellular eIF4F caused by elevated levels of initiation factors or dysregulation of 4E-BP phosphorylation plays an important role in oncogenesis. eIF4E expression is frequently elevated in many types of cancers and is associated with malignant progression [5,8]. Enforced overexpression of eIF4E in experimental systems induces cell transformation and tumorigenesis both in cell cultures and in animal models [4,9–12]. Similar results were also observed when eIF4G was overexpressed [13]. Conversely, ectopic expression of 4EBPs can partially revert eIF4E-transformed cells to a nonmalignant phenotype [14] and induce apoptosis in cells transformed by other oncogenes such as Ras [9,15]. Thus, the eIF4F complex and eIF4E are considered to be promising cancer therapeutic targets. Accordingly, eIF4E-specific antisense oligonucleotides have been shown to effectively repress the expression of eIF4E-regulated proteins (e.g., vascular endothelial growth factor, cyclin D1, survivin, c-Myc, Bcl-2), induce apoptosis, inhibit tumor growth in vivo with minimal toxicity, and are now being tested in phase 1 clinical trials [16]. Several rapamycin analogs, which decrease eIF4F levels by inhibiting mTOR-dependent 4E-BP phosphorylation, have antitumor activity and have been approved for cancer treatment or are being evaluated as cancer drugs in clinical trials [17–19].
The death ligand tumor necrosis factor-related apoptosis-inducing ligand (TRAIL), which induces apoptosis on binding to its receptors death receptors 4 and 5 (DR4 and DR5), has recently received much attention because it preferentially induces apoptosis in transformed or malignant cells but not in most normal cells [20]. Currently, recombinant human TRAIL is being tested as an anticancer agent in phase 1 clinical trials [21]. In addition, agonistic antibodies against DR4 and DR5, which directly activate the extrinsic apoptotic pathway, have been developed and tested in phase 1 or 2 trials and found to be well tolerated [22,23]. Thus, death receptor-mediated apoptosis, particularly TRAIL death receptor-mediated apoptosis, has been subject to intense research as a cancer therapeutic target [23,24]. However, an important issue in this regard is the intrinsic resistance of certain cancer cells to TRAIL/death receptor-induced apoptosis [25]. Fortunately, certain cancer therapeutic agents can sensitize various types of cancer cells to TRAIL-induced apoptosis or overcome the intrinsic apoptotic resistance to TRAIL [26]. Accordingly, these agents are potentially useful in combination with TRAIL or TRAIL receptor agonistic antibodies to enhance TRAIL- or TRAIL receptor-based cancer therapy.
TRAIL/death receptor-mediated apoptosis is primarily inhibited by cellular FLICE-inhibitory protein (c-FLIP), which inhibits caspase-8 activation by preventing recruitment of caspase-8 to the death-inducing signaling complex called DISC [27,28]. c-FLIP has multiple splice variants, however, only two of them (i.e., FLIPL and FLIPS) have been well characterized at the protein levels [29,30]. The levels of c-FLIP, including both FLIPL and FLIPS, are subject to regulation by ubiquitin/proteasome-mediated degradation [31–33]. It has been well documented that elevated c-FLIP expression protects cells from death receptor-mediated apoptosis, whereas down-regulation of c-FLIP by chemicals or small interfering RNA (siRNA) sensitizes cells to death receptor-mediated apoptosis [29]. Moreover, overexpression of c-FLIP also protects cells from apoptosis induced by cancer therapeutic agents such as etoposide and cisplatin [34–38].
The small molecule, 4EGI-1, has been identified through highthroughput screens of compound libraries to bind eIF4E and disrupt eIF4E/eIF4G association. This compound selectively inhibits capdependent translation and exhibits activity against multiple cancer cell lines including induction of apoptosis [39]. We are interested in its therapeutic activity in lung cancer. Moreover, we are particularly interested in identifying small molecules that can sensitize cancer cells to TRAIL-induced apoptosis. Interestingly, we found that 4EGI-1 alone not only induced apoptosis of human lung cancer but also cooperated with TRAIL in augmenting apoptosis. Thus, we focused our study on revealing the mechanisms by which 4EGI-1 enhances TRAIL-induced apoptosis and the role of cap-dependent translation in this process.
Materials and Methods
Reagents
4EGI-1 was purchased from EMD Chemicals, Inc, or Calbiochem (Gibbstown, NJ). It was dissolved in DMSO at the concentration of 100 mM, and aliquots were stored at -80°C. Stock solutions were diluted to the appropriate concentrations with growth medium immediately before use. The soluble recombinant human TRAIL was purchased from PeproTech, Inc (Rocky Hill, NJ). The proteasome inhibitor MG132 and the protein synthesis inhibitor cycloheximide (CHX) were purchased from Sigma Chemical Co (St Louis, MO). Rabbit polyclonal anti-DR5 antibody was purchased from ProSci, Inc (Poway, CA).Mouse monoclonal anti-DR4 antibody (B-N28) was purchased from Diaclone (Stamford, CT). Mouse monoclonal anti-FLIP antibody (NF6) was purchased Alexis Biochemicals (San Diego, CA). Rabbit polyclonal anti-caspase-8, anti-poly(ADP-ribose) polymerase (PARP), and anti-phospho-elF2α (Ser51) antibodies were purchased from Cell Signaling Technology, Inc (Beverly, MA). Mouse monoclonal c-Myc (9E10), anti-CCAAT/enhancer-binding protein homologous protein (CHOP) (B-3) antibodies, and rabbit polyclonal ATF4 (CREB-2; C-20) were purchased from Santa Cruz Biotechnology, Inc (Santa Cruz, CA). Mouse monoclonal cyclin D1 antibody (clone DCS-6) was purchased from Dako (Carpinteria, CA). Mouse monoclonal anti-β-actin and rabbit polyclonal anti-GAPDH antibodies were purchased from Sigma Chemical Co and Trevigen, Inc (Gaithersburg, MD), respectively.
Cell Lines and Cell Culture
Human non-small cell lung cancer cell lines used in this study were purchased from the American Type Culture Collection (Manassas, VA). The H157-Lac Z-5, H157-FLIPL-21 and H157-FLIPS-1 stable transfectants that express control Lac Z, ectopic FLIPL, and ectopic FLIPS, respectively, were described previously [40]. These cell lines were cultured in RPMI 1640 containing 5% fetal bovine serum at 37°C in a humidified atmosphere of 5% CO2 and 95% air.
Cell Survival Assay
Cells were seeded in 96-well cell culture plates and treated the next day with the agents indicated. The viable cell number was determined using the sulforhodamine B (SRB) assay, as previously described [41].
Detection of Apoptosis
Apoptosis was evaluated by Annexin V staining using Annexin V-PE apoptosis detection kit purchased from BD Biosciences (San Jose, CA) following the manufacturer's instructions. We also detected PARP cleavage by Western blot analysis (as described below) as an additional indicator of apoptosis.
Western Blot Analysis
Whole-cell protein lysates were prepared and analyzed by Western blot analysis as described previously [42,43].
Detection of DR5 mRNA
DR5 mRNA levels were detected with reverse transcription- polymerase chain reaction (RT-PCR) as described previously [44].
7-Methyl GTP Pull-down for Analysis of eIF4F Complex Formation
eIF4F complex in cell extracts was detected using affinity chromatography 7-methyl GTP (m7GTP)-Sepharose as described previously [45]. Briefly, the given cells were lysed in buffer containing 20 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM β-glycerophosphate, 1 mM Na3VO4, 1% Triton X-100, 2.5 mM sodium pyrophosphate, and protease inhibitor cocktail, on ice for 15 minutes. After centrifugation at 12,000g for 15 minutes at 4°C, the supernatants were collected and incubated with m7GTP-Sepharose (GE Healthcare Bio-Sciences, Corp/Amersham, Piscataway, NJ) at 4°C for 2 hours with constant shaking. Beads were washed three times with lysis buffer and three times with 1 x PBS. The samples were then denatured, and the supernatants were then loaded to SDS-PAGE for Western blot analysis.
Immunoprecipitation for Detection of Ubiquitinated c-FLIP
H157-FLIPL-21 cells, which stably express FLIPL, were transfected with HA-ubiquitin plasmid using the FuGENE 6 transfection reagent (Roche Diagnostics Corp, Indianapolis, IN) following the manufacturer's instructions. After 24 hours, the cells were treated with 4EGI-1 or MG132 plus 4EGI-1 for the given time and were then lysed for immunoprecipitation of Flag-FLIPL using Flag M2 monoclonal antibody (Sigma) as previously described [46] followed by the detection of ubiquitinated FLIPL with Western blot analysis using anti-HA antibody (Abgent, San Diego, CA).
SiRNA-Mediated Gene Silencing
The siRNA duplexes for nonsilencing control, DR5, and CHOP were described previously [43,44]. eIF4E siRNA (5′-GGACGAUGGCUAAUUACAUdTdT-3′) was described previously [47]. Transfection of these siRNA duplexes was conducted in six-well plates using the HiPerFect transfection reagent (Qiagen, Valencia, CA) following the manufacturer's manual. The gene-silencing effect was evaluated by Western blot analysis.
Transient Transfection and Luciferase Activity Assay
The reporter constructs harboring different DR5 5′-flanking regions, transient transfection, and luciferase assay were all the same as described previously [44,48].
Results
4EGI-1 Inhibits the Growth of Lung Cancer Cells and Cooperates with TRAIL to Augment Apoptosis
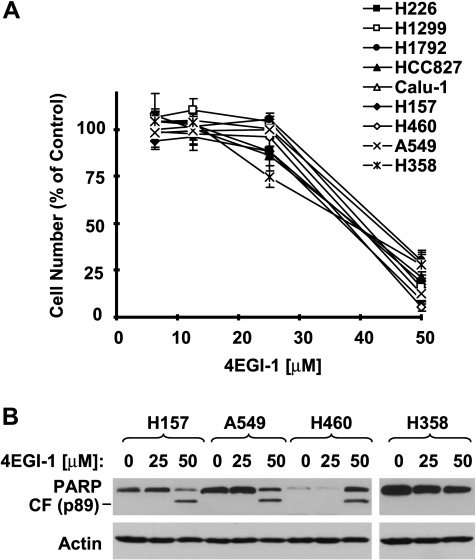
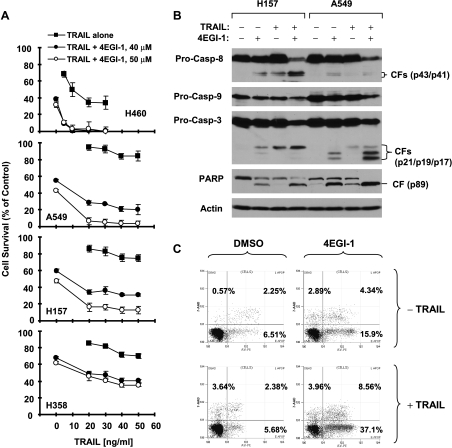
We first determined the effects of 4EGI-1 as a single agent on the growth of human lung cancer cells. As presented in Figure 1A, 4EGI-1 at a concentration range of 20 to 50 µM effectively inhibited the growth of the panel of lung cancer cell lines, with halfmaximal inhibitory concentration values, around 40 µM under the tested conditions. Moreover, it clearly induced the cleavage of PARP, a hallmarkof apoptosis, in the tested lung cancer cell lines evidenced by the appearance of cleaved band and/or reduction of intact PARP (Figure 1B), suggesting that 4EGI-1 also induces apoptosis. When combined with TRAIL, enhanced effects on decreasing cell survival were observed in every tested cell line because the combination of 4EGI-1 withTRAILwasmuchmore effective than either 4EGI-1 or TRAIL alone in decreasing the survival of the lung cancer cells (Figure 2A). ByWestern blot analysis, we detected more sever cleavage of caspases including caspase-3, caspase-8, and caspase-9 and PARP in cells treated with 4EGI-1 and TRAIL than those treated with 4EGI-1 or TRAIL alone (Figure 2B), evidenced by either reduction of proforms of the caspases or appearance of the cleaved bands. Also, direct detection of apoptosis with Annexin V staining showed that the combination of 4EGI-1 and TRAIL was more potent than either single agent alone in inducing apoptosis (Figure 2C). Specifically, the 4EGI-1 and TRAIL combination induced apoptosis in approximately 46% of cells, whereas 4EGI-1 and TRAIL alone induced apoptosis in approximately 20% and 8% of cells, respectively (Figure 2C). Collectively, these results indicate that 4EGI-1 cooperates with TRAIL to augment the induction of apoptosis.
Figure 1.
4EGI-1 inhibits the growth of human lung cancer cells (A) and induces PARP cleavage (B). (A) The indicated lung cancer cell lines were seeded in 96-well cell culture plates and treated the next day with the given concentrations of 4EGI-1. After 3 days, cell numbers were estimated using the SRB assay. Cell survival was expressed as the percent of control (DMSO-treated) cells. Data are the means of four replicate determinations. Bars, ±SDs. (B) The indicated cancer cell lines were treated with the given concentrations of 4EGI-1 for 24 hours. The cells were then harvested for preparation of whole-cell protein lysates and subsequent Western blot analysis for detecting PARP cleavage. CF indicates cleaved fragment.
Figure 2.
4EGI-1 enhances TRAIL-induced apoptosis as evaluated by cell survival (A), caspase activation (B), and Annexin V staining (C). (A) The indicated cell lines were seeded in 96-well cell culture plates and treated the next day with the given concentrations of 4EGI-1 alone, TRAIL alone, or their respective combinations. After 24 hours, cell numbers were estimated using the SRB assay. Data are the means of four replicate determinations. Bars, ±SDs. (B) The indicated cell lines were treated with 20 ng/ml TRAIL alone, 50 µM 4EGI-1 alone, and their combination. After 24 hours, the cells were harvested for preparation of whole-cell protein lysates and subsequent Western blot analysis. CF indicates cleaved fragment. (C) A549 cells were treated with 40 ng/ml TRAIL alone, 40 µM 4EGI-1 alone, and their combination. After 24 hours, the cells were subjected to measurement of apoptosis using Annexin V staining. The percent positive cells in the upper right and lower right quadrants were added to yield the total of apoptotic cells.
4EGI-1 Modulates DR5 and c-FLIP Levels That Contribute to Cooperative Induction of Apoptosis by the Combination of 4EGI-1 and TRAIL
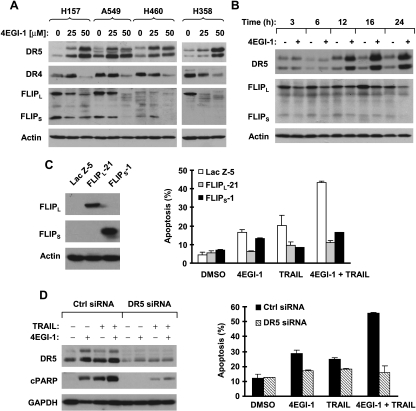
To determine the mechanism by which 4EGI-1 cooperates with TRAIL to augment apoptosis, we analyzed the modulatory effects of 4EGI-1 on DR4, DR5, and c-FLIP, which are often involved in drug-induced sensitization of TRAIL/death receptor-induced apoptosis [26]. As shown in Figure 3A, 4EGI-1 increased DR5 expression and reduced c-FLIP (both FLIPL and FLIPS) levels in every tested cell line; however, it decreased DR4 expression in these cell lines. Thus, we focused our further analyses on DR5 and c-FLIP. By time course analysis of the effects of 4EGI-1 on DR5 and c-FLIP, we observed that DR5 induction occurred at 12 hours and c-FLIP reduction happened at 3 hours after 4EGI-1 treatment. Both DR5 induction and c-FLIP reduction were sustained up to 24 hours (Figure 3B). It appears that c-FLIP reduction precedes DR5 induction in 4EGI-1-treated cells.
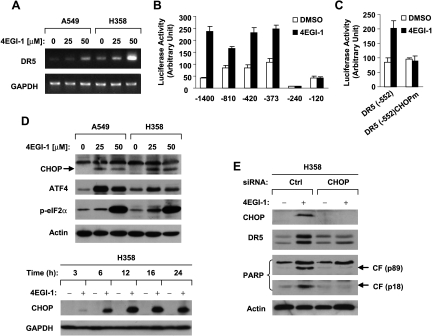
Figure 3.
4EGI-1 modulates the levels of DR5 and c-FLIP (A and B), which contribute to enhancement of TRAIL-induced apoptosis (C and D). (A and B) The given cell lines were treated with different concentrations of 4EGI-1 as indicated for 24 hours (A) or 50 µM 4EGI-1 for various times from 3 to 24 hours as indicated and then subjected to preparation of whole-cell protein lysates and subsequent Western blot analysis for the given proteins. (C) The H157 cell lines Lac Z-5, FLIPL-21 and FLIPS-1, which express different ectopic FLIPs, as detected by Western blot analysis (left panel), were treated with DMSO, 50 µM 4EGI-1 alone, 20 ng/ml TRAIL alone or 4EGI-1 plus TRAIL. After 24 hours, the cells were harvested and subjected to detection of apoptosis by Annexin V staining (right panel). Columns indicate means of duplicate experiments; bars, ±SE. (D) A549 cells were cultured in a six-well plate and, the next day, transfected with 60nMcontrol (Ctrl) or DR5 siRNA. Forty-eight hours after transfection, the cells were treated with 40 µM 4EGI-1 alone, 20 ng/ml TRAIL alone, or 4EGI-1 plus TRAIL for 24 hours and then harvested for detection of apoptosis by Annexin V staining. Columns indicate means of duplicate experiments; bars, ±SE.
We next asked whether c-FLIP reduction participated in cooperative induction of apoptosis by 4EGI-1 and TRAIL combination. To this end, we tested the apoptosis-inducing effects of 4EGI-1 and TRAIL combination in cell lines that overexpress ectopic c-FLIP (both FLIPL and FLIPS). We assumed that overexpression of ectopic c-FLIP prevents endogenous c-FLIP reduction and thus confers cell resistance to 4EGI-1/TRAIL-induced apoptosis if the c-FLIP is important for cooperative induction of apoptosis by 4EGI-1 and TRAIL combination. Indeed, both TRAIL alone and 4EGI-1 combined with TRAIL induced much less apoptosis in the cell lines that expressed either ectopic FLIPL or FLIPS than the control cell line that expressed the irrelevant protein Lac Z (Figure 3C), indicating that expression of ectopic c-FLIP clearly protects cells from 4EGI-1/TRAIL-induced apoptosis. Thus, we conclude that c-FLIP down-regulation contributes to cooperative induction of apoptosis by the 4EGI-1 and TRAIL combination.
Furthermore, we determined whether DR5 induction is important for enhancement of TRAIL-induced apoptosis by 4EGI-1 by knocking down DR5 expression and then examining its impact on 4EGI-1 and TRAIL-induced apoptosis. As shown in Figure 3D, transfection of DR5 siRNA decreased not only the basal levels of DR5 but also 4EGI-1-induced DR5 up-regulation (left panel), indicating successful knockdown of DR5 expression. Accordingly, the 4EGI-1 and TRAIL combination potently induced PARP cleavage (left panel) and increased the percentage of apoptotic cells (right panel) in control siRNA-transfected cells but only minimally in DR5 siRNA-transfected cells.Moreover, the effects of 4EGI-1 or TRAIL alone on PARP cleavage and induction of apoptosis were attenuated in DR5 siRNA-transfected cells compared with their effects in control siRNA-tranfected cells (Figure 3D). Thus, these results clearly indicate that DR5 induction also contributes to cooperative induction of apoptosis by the 4EGI-1 and TRAIL combination. It is likely that DR5 induction is also important for 4EGI-1-induced apoptosis.
4EGI-1 Enhances TRAIL-Induced Apoptosis Independent of Its Inhibitory Effect on Cap-Dependent Protein Translation
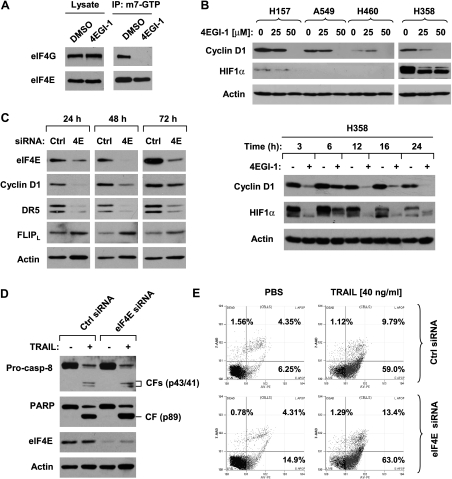
Given that 4EGI-1 was developed as an eIF4E/eIF4G interaction inhibitor [39], we asked whether the effects of 4EGI-1 on DR5 induction, c-FLIP reduction, and enhancement of TRAIL-induced apoptosis are secondary to its inhibition of cap-dependent protein translation. Thus, we first analyzed whether 4EGI-1 inhibits eIF4E and eIF4G interaction or eIF4F complex formation in our cell systems. In an m7GTP pull-down assay, we detected much lower amounts of eIF4G in 4EGI-1-treated cells than in DMSO-treated cells (Figure 4A), indicating that 4EGI-1 indeed disrupts the formation of eIF4F complex in our cell systems as reported [39]. Moreover, we examined the effects of 4EGI-1 on the expression of cyclin D1 and HIF-1α, which are known to be regulated through cap-dependent translation initiation mechanisms. As presented in Figure 4B, 4EGI-1 decreased the levels of both cyclin D1 and HIF-1α in concentration- and timedependent manners, suggesting that 4EGI-1 inhibits cap-dependent protein translation.We noted that HIF-1α expression was not detected in A549 and H460 cells; this is likely due to the normoxia condition in our experiment.
Figure 4.
4EGI-1 disrupts eIF4E and eIF4G interaction (A) and inhibits cyclin D1 and HIF-1α expression (B); however, eIF4E siRNA decreases cyclin D1 expression but does not induce DR5 expression, reduce c-FLIP levels (C) or enhance TRAIL-induced apoptosis (D and E). (A) H358 cells were treated with DMSO or 50 µM 4EGI-1 for 6 hours and then harvested for preparation of whole-cell protein lysates and subsequent m7GTP pull-down assay. (B) The given cell lines were treated with different concentrations of 4EGI-1 as indicated for 24 hours (upper panel) or 50 µM 4EGI-1 for the indicated times (lower panel) and then subjected to preparation of whole-cell protein lysates and subsequent Western blot analysis. (C) H358 cells were transfected with 20 nM control (Ctrl) or eIF4E siRNA for the indicated times and then subjected to preparation of whole-cell protein lysates and subsequent Western blot analysis. (D and E) A549 cells were transfected with 10 nM control (Ctrl) or eIF4E siRNA for 24 hours and then treated with 40 ng/ml TRAIL. After an additional 16 hours, the cells were harvested for either Western blot analysis of the given proteins (D) or Annexin V assay for detection of apoptotic cells (E). CF indicates cleaved form.
After these studies, we determined whether knockdown of eIF4E with eIF4E siRNA can generate similar effects as 4EGI-1 does in inducing DR5 expression, reducing c-FLIP levels and enhancing TRAILinduced apoptosis. Knockdown of eIF4E effectively decreased cyclin D1 levels but not c-FLIP levels. Interestingly, eIF4E knockdown actually decreased DR5 levels (Figure 4C). When treated with TRAIL, we detected comparable levels of the cleavage of caspase-8 and PARP (Figure 4D) and similar percentages of apoptotic cells (Figure 4E). These results indicate that inhibition of eIF4E with eIF4E siRNA does not increase DR5 expression, reduce c-FLIP levels, or sensitize cells to TRAIL-induced apoptosis.
Furthermore, we checked the effects of rapamycin, an mTOR inhibitor that is known to inhibit cap-dependent protein translation [49], on modulation of DR5, c-FLIP and TRAIL-induced apoptosis. Treatment of cells with rapamycin inhibited the phosphorylation of S6, a known readout of mTOR signaling activity, and decreased the levels of cyclin D1 and HIF-1α but did not increase DR5 or reduce c-FLIP levels (Figure W1A). Similarly, treatment of cells with rapamycin combined with TRAIL did not show any enhanced effects on decreasing cell survival (Figure W1B). Together, these results further our notion that 4EGI-1 cooperates with TRAIL to augment apoptosis independent of inhibition of cap-dependent protein translation.
4EGI-1 Induces DR5 Expression through a CHOP-Dependent Mechanism
To understand how 4EGI-1 upregulates DR5 expression, we then checked whether 4EGI-1 increases DR5 expression at the transcriptional level. By RT-PCR, we detected concentration-dependent increases of DR5 mRNA in cells exposed to 4EGI-1 (Figure 5A). Moreover, 4EGI-1 increased DR5 promoter activity (Figure 5B). These results indicate that 4EGI-1 indeed increases DR5 expression at the transcriptional level. Through deletion analysis of the DR5 promoter region, we noted that the region between -373 and -240 is critical for 4EGI-1 to transactivate the DR5 promoter because the DR5 -240 promoter fragment lost activity to respond to 4EGI-1, whereas the DR5 -373 fragment maintained its response to 4EGI-1 treatment (Figure 5B). It has been documented that the CHOP is a critical transcriptional factor that regulates DR5 expression [44,50,51]. We noted that the CHOP binding site locates at the region between -373 and -240 of the DR5 promoter. Thus, we questioned whether the CHOP binding site is responsible for 4EGI-1-mediated transactivation of the DR5 promoter. To this end, we compared the effects of 4EGI-1 on transcriptional activity of the DR5 promoter reporter construct with and without wild-type CHOP binding site. As expected, 4EGI-1 increased the luciferase activity in cells transfected with pGL3-DR5(-552) carrying the wild-type CHOP binding site but failed to do so in cells transfected with pGL3-DR5(-552)CHOPm, in which the CHOP binding site was inactivated (Figure 5C). Together, these results suggest that CHOP might be an important transcriptional factor responsible for 4EGI-1-induced DR5 up-regulation.
Figure 5.
4EGI-1 increases DR5 expression at the transcriptional level (A-C) through a CHOP-dependent mechanism (D and E). (A) The given cell lines were treated with the indicated concentrations of 4EGI-1 for 12 hours and then subjected to preparation of total cellular RNA and subsequent RT-PCR. (B and C) The given reporter constructs were cotransfected with pCH110 plasmid into H358 cells. After 24 hours, the cells were treated with DMSO or 50 µM 4EGI-1 for 14 hours and then subjected to luciferase assay. Columns indicate means of triplicate determinations; bars, ±SDs. (D) The indicated cell lines were treated with the given concentrations of 4EGI-1 for 24 hours (upper panel) or 50 µM 4EGI-1 for the indicated times (lower panel). The cells were then harvested for preparation of whole-cell protein lysates and subsequent Western blot analysis. (E) H358 cells were transfected with 60 nM control (Ctrl) or CHOP siRNA. After 48 hours, the cells were treated with 50 µM 4EGI-1 for 14 hours and then subjected to preparation of whole-cell protein lysates and subsequent Western blot analysis. CF indicates cleaved form.
Subsequently, we determined whether 4EGI-1 indeed alters CHOP expression. As presented in Figure 5D, 4EGI-1 increased CHOP levels in both A549 and H358 cells (top panel); this effect occurred at 6 hours after 4EGI-1 treatment, peaked at 12 hours and was sustained up to 24 hours (bottom panel). In addition, we found that 4EGI-1 increased the levels of ATF4 and p-eIF2α (Figure 5D), two proteins known to be increased during endoplasmic reticulum (ER) stress. Thus, it seems that 4EGI-1 induces CHOP expression with ER stress. To prove that CHOP is critical forDR5 induction by 4EGI-1, we usedCHOPsiRNA to knock down CHOP expression and then examined its impact on 4EGI-1-induced DR5 expression. As shown in Figure 5E, transfection of CHOP siRNA effectively blocked not only CHOP up-regulation by 4EGI-1 but also 4EGI-1-induced DR5 up-regulation, indicating that 4EGI-1 induces DR5 expression through a CHOP-dependent mechanism. Moreover, we detected increased cleavage of PARP in 4EGI-1-treated cells transfected with control siRNA but not in 4EGI-1-treated cells transfected with CHOP siRNA, suggesting that both CHOP and DR5 up-regulation are also important for 4EGI-1-induced apoptosis.
4EGI-1 Downregulates c-FLIP Levels through Ubiquitin/Proteasome-Mediated Degradation
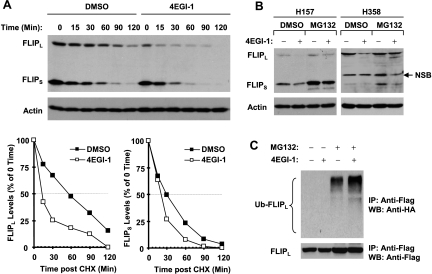
We also determined the mechanism by which 4EGI-1 reduces c-FLIP levels. Because c-FLIP proteins are known to be regulated by ubiquitin/proteasome-mediated degradation [31–33], we asked whether the observed down-regulation of c-FLIP by 4EGI-1 would be mediated through this process. Thus, we first determined whether 4EGI-1 affects c-FLIP stability. To this end, we treated H157 cells with either DMSO solvent control or 4EGI-1 for 5 hours and then washed away the drug followed by refilling the cells with fresh medium containing CHX. At the indicated times after addition of CHX, the cells were harvested for Western blot analysis for analyzing c-FLIP degradation rate. As presented in Figure 6A, the half-lives for FLIPL and FLIPS in control cells were approximately 60 and 30 minutes, respectively; however, they were approximately 15 and 20 minutes, respectively, in 4EGI-1-treated cells. Thus, it is clear that 4EGI-1 facilitates c-FLIP degradation. Moreover, we examined the effects of 4EGI-1 on c-FLIP in the absence and presence of the proteasome inhibitor MG132. We found that 4EGI-1-induced reduction of c-FLIP was abrogated by the presence of MG132 in both H157 and H358 cell lines (Figure 6B), suggesting that 4EGI-1-induced c-FLIP reduction is proteasome-dependent. In addition, we determined whether 4EGI-1 increased c-FLIP ubiquitination. By immunoprecipitation/Western blot analysis, we detected the highest levels of ubiquitinated FLIPL in cells treated with 4EGI-1 plus MG132 compared with cells exposed to 4EGI-1 alone or MG132 alone (Figure 6C), indicating that 4EGI-1 increases c-FLIP ubiquitination. Collectively, we conclude that 4EGI-1 facilitates ubiquitin/proteasome-mediated c-FLIP degradation, leading to down-regulation of c-FLIP.
Figure 6.
4EGI-1 decreases c-FLIP stability (A) and promotes ubiquitin/proteasome-mediated c-FLIP degradation (B and C). (A) H157 cells were treated with DMSO or 50 µM 4EGI-1 for 5 hours. The cells were then washed with PBS three times and refed with fresh medium containing 10 µg/ml CHX. At the indicated times, the cells were then harvested for preparation of whole-cell protein lysates and subsequent Western blot analysis. Protein levels were quantitated with NIH Image J software (Bethesda, MD) and were normalized to actin. The results were plotted as the relative c-FLIP levels compared with those at the time 0 of CHX treatment (lower panel). (B) The indicated cell lines were pretreated with 20 µM MG132 for 30 minutes and then cotreated with 50 µM 4EGI-1 for another 4 hours. The cells were then harvested for preparation of whole-cell protein lysates and subsequent Western blot analysis. NSB indicates nonspecific band. (C) H157-FLIPL-21 cells, which stably express ectopic flag-FLIPL, were transfected with HA-ubiquitin plasmid using FuGENE 6 transfection reagent for 24 hours. The cells were then pretreated with 20 µM MG132 for 30 minutes and then cotreated with 50 µM 4EGI-1 for 4 hours. Whole-cell protein lysates were then prepared for immunoprecipitation using anti-Flag antibody followed by Western blot (WB) analysis using anti-HA antibody for detection of ubiquitinated FLIPL (Ub-FLIPL) and anti-Flag antibody for detection of ectopic FLIPL.
Discussion
In the present study, we have demonstrated that the eIF4E and eIF4G interaction inhibitor 4EGI-1 effectively inhibited the growth of human lung cancer cells and induces apoptosis. These findings are consistent with previous work [39]. Moreover, we have shown that 4EGI-1 cooperates with TRAIL to augment induction of apoptosis in a panel of human lung cancer cells. To the best of our knowledge, this is the first study to show the interaction between 4EGI-1 and TRAIL/death receptor-induced apoptosis.
DR4, DR5, and c-FLIP are key components in the TRAIL-mediated apoptotic pathway [23]. Modulation of their expression levels alters cell sensitivity to TRAIL-induced apoptosis [26]. In this study, we found that 4EGI-1 increased DR5 expression and reduced c-FLIP levels inmultiple lung cancer cell lines. When DR5 induction was blocked by siRNA-mediated knockdown of DR5 expression or ectopic c-FLIP was expressed, the cooperative induction of apoptosis by the 4EGI-1 and TRAIL combination was abrogated (Figure 3). Thus, we conclude that both DR5 induction and c-FLIP down-regulation are critical events that mediate enhanced induction of apoptosis by the combination of 4EGI-1 and TRAIL.We noted that blockage of DR5 induction or overexpression of ectopic c-FLIP also attenuated apoptosis induced by 4EGI-1 alone (Figure 3), suggesting that DR5 induction and c-FLIP reduction also contribute to 4EGI-1-induced apoptosis. Unexpectedly, we also observed that DR4 expression was reduced by 4EGI-1 (Figure 3A). The mechanism underlying the down-regulation of DR4 by 4EGI-1 is unknown; however, this warrants our further investigation.
A previous study suggested that c-FLIP regulation in glioblastoma multiforme cells involves an mTOR-dependent translational mechanism [52]. Accordingly, rapamycin inhibited c-FLIP expression and sensitized glioblastoma multiforme cells to TRAIL-induced apoptosis [52]. In this study, 4EGI-1, as an eIF4E and eIF4G interaction inhibitor, disrupted the interaction between eIF4E and eIF4G and inhibited the expression of both cyclin D1 and HIF-1α, both of which are regulated by cap-dependent translation mechanisms, in our cell systems (i.e., lung cancer cells; Figure 4), indicating that 4EGI-1, as reported [39], indeed inhibits eIF4F formation and cap-dependent protein translation. However, neither eIF4E siRNA (which inhibits eIF4F formation) nor rapamycin (which inhibits mTOR-dependent protein translation) increased DR5 expression, reduced c-FLIP levels or enhanced TRAIL-induced apoptosis in our cell systems, although they did downregulate the levels of cyclin D1 and HIF-1α (Figures 4 and W1). These data suggest that 4EGI-1 is unlikely to modulate the levels of DR5 and c-FLIP and augment TRAIL-induced apoptosis through inhibition of eIF4F-mediated cap-dependent protein translation. Our data are in agreement with another recent study showing that inhibition of mTOR with either rapamycin or mTOR siRNA failed to enhance TRAIL-induced apoptosis in glioblastoma cells [53]. Thus, we suggest that 4EGI-1 modulates the levels of DR5 and c-FLIP and enhances TRAIL-induced apoptosis independent of inhibition of cap-dependent protein translation, for example, through “off-target” mechanisms.
ER stress andCHOP have been demonstrated to be involved inDR5 regulation [44,50,54]. In this study, 4EGI-1 increased DR5 mRNA and luciferase activity of the DR5 promoter (Figure 5), indicating that 4EGI-1 induces DR5 expression at the transcriptional level. Moreover, deletion and mutation analysis of the DR5 promoter revealed that the presence of the CHOP binding site is required for transactivation of the DR5 promoter by 4EGI-1 (Figure 5, B and C). In agreement, we found that 4EGI-1 increased CHOP levels, which could be detected even at 3 hours after 4EGI-1 treatment (Figure 5D), thus preceding DR5 up-regulation that occurred 12 hours after 4EGI-1 treatment (Figure 3B). Importantly, blockade of CHOP induction by silencing CHOP expression with CHOP siRNA abolished 4EGI-1's ability to induce DR5 expression (Figure 5E). Taken together, we conclude that 4EGI-1 induces a CHOP-dependent DR5 up-regulation. CHOP is known to be a typical protein associated with ER stress-induced apoptosis [55]. In addition to CHOP, we also detected increases in ATF4 and p-eIF2a, two other ER stress-associated proteins, in cells treated with 4EGI-1 (Figure 5D). Thus, it is possible that 4EGI-1 induces ER stress, leading to CHOP-dependent DR5 up-regulation. We noted that CHOP blockage abolished not only DR5 induction but also PARP cleavage by 4EGI-1 (Figure 5E), suggesting that CHOP induction is also important for 4EGI-1-induced apoptosis. This furthers our notion that DR5 induction is important for 4EGI-1-induced apoptosis as well.
c-FLIP proteins are known to be regulated by ubiquitin/proteasome-mediated degradation [31–33]. In this study, we found that 4EGI-1 treatment reduced c-FLIP stability and the presence of the proteasome inhibitor MG132 prevented c-FLIP from reduction by 4EGI-1. Moreover, we detected increased levels of ubiquitinated c-FLIP in cells cotreated with MG132 and 4EGI-1 using immunoprecipitation/Western blot analysis (Figure 6). All together, we conclude that 4EGI-1 downregulates c-FLIP levels by facilitating ubiquitin/proteasome- mediated degradation of c-FLIP.
In summary, the current study has revealed a novel biological function of 4EGI-1 that sensitizes human lung cancer cells to TRAIL-induced apoptosis. Cooperative augmentation of apoptosis by the 4EGI-1 and TRAIL combination involves CHOP-dependent DR5 induction and ubiquitin/proteasome-mediated c-FLIP degradation independent of inhibition of cap-dependent protein translation.
Supplementary Material
Acknowledgments
The authors thank H.-G. Wang (Penn State Hershey Cancer Institute, The Pennsylvania State University College of Medicine, Hershey, PA) for providing DR5 reporter constructs with a wild-type and mutant CHOP binding site and Anthea Hammond for editing the manuscript.
Abbreviations
- TRAIL
tumor necrosis factor-related apoptosis-inducing ligand
- c-FLIP
cellular FLICE-inhibitory protein
- CHX
cycloheximide
- siRNA
small interfering RNA
- CHOP
CCAAT/enhancer-binding protein homologous protein
- ER
endoplasmic reticulum
Footnotes
This study was supported by the Georgia Cancer Coalition Distinguished Cancer Scholar award (to S.-Y.S.), Department of Defense VITAL grant W81XWH-04-1-0142 (to S.-Y. Sun for Project 4) and National Cancer Institute R01 CA118450 and SPORE P50 grant CA128613. F.R.K. and S-Y.S. are Georgia Cancer Coalition Distinguished Cancer Scholars.
This article refers to supplementary material, which is designated by Figure W1 and is available online at www.neoplasia.com.
References
- 1.von der Haar T, Gross JD, Wagner G, McCarthy JE. The mRNA cap-binding protein eIF4E in post-transcriptional gene expression. Nat Struct Mol Biol. 2004;11:503–511. doi: 10.1038/nsmb779. [DOI] [PubMed] [Google Scholar]
- 2.Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- 3.Mamane Y, Petroulakis E, Rong L, Yoshida K, Ler LW, Sonenberg N. eIF4E—from translation to transformation. Oncogene. 2004;23:3172–3179. doi: 10.1038/sj.onc.1207549. [DOI] [PubMed] [Google Scholar]
- 4.Graff JR, Konicek BW, Carter JH, Marcusson EG. Targeting the eukaryotic translation initiation factor 4E for cancer therapy. Cancer Res. 2008;68:631–634. doi: 10.1158/0008-5472.CAN-07-5635. [DOI] [PubMed] [Google Scholar]
- 5.De Benedetti A, Graff JR. eIF-4E expression and its role in malignancies and metastases. Oncogene. 2004;23:3189–3199. doi: 10.1038/sj.onc.1207545. [DOI] [PubMed] [Google Scholar]
- 6.Goodfellow IG, Roberts LO. Eukaryotic initiation factor 4E. Int J Biochem Cell Biol. 2007;40:2675–2680. doi: 10.1016/j.biocel.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raught B, Gingras AC. eIF4E activity is regulated at multiple levels. Int J Biochem Cell Biol. 1999;31:43–57. doi: 10.1016/s1357-2725(98)00131-9. [DOI] [PubMed] [Google Scholar]
- 8.Thumma SC, Kratzke RA. Translational control: a target for cancer therapy. Cancer Lett. 2007;258:1–8. doi: 10.1016/j.canlet.2007.08.022. [DOI] [PubMed] [Google Scholar]
- 9.Avdulov S, Li S, Michalek V, Burrichter D, Peterson M, Perlman DM, Manivel JC, Sonenberg N, Yee D, Bitterman PB, et al. Activation of translation complex eIF4F is essential for the genesis and maintenance of the malignant phenotype in human mammary epithelial cells. Cancer Cell. 2004;5:553–563. doi: 10.1016/j.ccr.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 10.Wendel HG, De Stanchina E, Fridman JS, Malina A, Ray S, Kogan S, Cordon-Cardo C, Pelletier J, Lowe SW. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature. 2004;428:332–337. doi: 10.1038/nature02369. [DOI] [PubMed] [Google Scholar]
- 11.Mavrakis KJ, Zhu H, Silva RL, Mills JR, Teruya-Feldstein J, Lowe SW, Tam W, Pelletier J, Wendel HG. Tumorigenic activity and therapeutic inhibition of Rheb GTPase. Genes Dev. 2008;22:2178–2188. doi: 10.1101/gad.1690808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruggero D, Montanaro L, Ma L, Xu W, Londei P, Cordon-Cardo C, Pandolfi PP. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat Med. 2004;10:484–486. doi: 10.1038/nm1042. [DOI] [PubMed] [Google Scholar]
- 13.Fukuchi-Shimogori T, Ishii I, Kashiwagi K, Mashiba H, Ekimoto H, Igarashi K. Malignant transformation by overproduction of translation initiation factor eIF4G. Cancer Res. 1997;57:5041–5044. [PubMed] [Google Scholar]
- 14.Rousseau D, Gingras AC, Pause A, Sonenberg N. The eIF4E-binding proteins 1 and 2 are negative regulators of cell growth. Oncogene. 1996;13:2415–2420. [PubMed] [Google Scholar]
- 15.Li S, Sonenberg N, Gingras AC, Peterson M, Avdulov S, Polunovsky VA, Bitterman PB. Translational control of cell fate: availability of phosphorylation sites on translational repressor 4E-BP1 governs its proapoptotic potency. Mol Cell Biol. 2002;22:2853–2861. doi: 10.1128/MCB.22.8.2853-2861.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graff JR, Konicek BW, Vincent TM, Lynch RL, Monteith D, Weir SN, Schwier P, Capen A, Goode RL, Dowless MS, et al. Therapeutic suppression of translation initiation factor eIF4E expression reduces tumor growth without toxicity. J Clin Invest. 2007;117:2638–2648. doi: 10.1172/JCI32044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sawyers CL. Will mTOR inhibitors make it as cancer drugs? Cancer Cell. 2003;4:343–348. doi: 10.1016/s1535-6108(03)00275-7. [DOI] [PubMed] [Google Scholar]
- 18.Le Tourneau C, Faivre S, Serova M, Raymond E. mTORC1 inhibitors: is temsirolimus in renal cancer telling us how they really work? Br J Cancer. 2008;99:1197–1203. doi: 10.1038/sj.bjc.6604636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abraham RT, Gibbons JJ. The mammalian target of rapamycin signaling pathway: twists and turns in the road to cancer therapy. Clin Cancer Res. 2007;13:3109–3114. doi: 10.1158/1078-0432.CCR-06-2798. [DOI] [PubMed] [Google Scholar]
- 20.Kelley SK, Ashkenazi A. Targeting death receptors in cancer with Apo2L/TRAIL. Curr Opin Pharmacol. 2004;4:333–339. doi: 10.1016/j.coph.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 21.Bellail AC, Qi L, Mulligan P, Chhabra V, Hao C. TRAIL agonists on clinical trials for cancer therapy: the promises and the challenges. Rev Recent Clin Trials. 2009;4:34–41. doi: 10.2174/157488709787047530. [DOI] [PubMed] [Google Scholar]
- 22.Rowinsky EK. Targeted induction of apoptosis in cancer management: the emerging role of tumor necrosis factor-related apoptosis-inducing ligand receptor activating agents. J Clin Oncol. 2005;23:9394–9407. doi: 10.1200/JCO.2005.02.2889. [DOI] [PubMed] [Google Scholar]
- 23.Ashkenazi A. Targeting the extrinsic apoptosis pathway in cancer. Cytokine Growth Factor Rev. 2008;19:325–331. doi: 10.1016/j.cytogfr.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 24.Takeda K, Stagg J, Yagita H, Okumura K, Smyth MJ. Targeting death-inducing receptors in cancer therapy. Oncogene. 2007;26:3745–3757. doi: 10.1038/sj.onc.1210374. [DOI] [PubMed] [Google Scholar]
- 25.Zhang L, Fang B. Mechanisms of resistance to TRAIL-induced apoptosis in cancer. Cancer Gene Ther. 2005;12:228–237. doi: 10.1038/sj.cgt.7700792. [DOI] [PubMed] [Google Scholar]
- 26.Elrod HA, Sun SY. Modulation of death receptors by cancer therapeutic agents. Cancer Biol Ther. 2007;7:163–173. doi: 10.4161/cbt.7.2.5335. [DOI] [PubMed] [Google Scholar]
- 27.Krueger A, Baumann S, Krammer PH, Kirchhoff S. FLICE-inhibitory proteins: regulators of death receptor-mediated apoptosis. Mol Cell Biol. 2001;21:8247–8254. doi: 10.1128/MCB.21.24.8247-8254.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Budd RC, Yeh WC, Tschopp J. cFLIP regulation of lymphocyte activation and development. Nat Rev Immunol. 2006;6:196–204. doi: 10.1038/nri1787. [DOI] [PubMed] [Google Scholar]
- 29.Wajant H. Targeting the FLICE inhibitory protein (FLIP) in cancer therapy. Mol Interv. 2003;3:124–127. doi: 10.1124/mi.3.3.124. [DOI] [PubMed] [Google Scholar]
- 30.Kataoka T. The caspase-8 modulator c-FLIP. Crit Rev Immunol. 2005;25:31–58. doi: 10.1615/critrevimmunol.v25.i1.30. [DOI] [PubMed] [Google Scholar]
- 31.Kim Y, Suh N, Sporn M, Reed JC. An inducible pathway for degradation of FLIP protein sensitizes tumor cells to TRAIL-induced apoptosis. J Biol Chem. 2002;277:22320–22329. doi: 10.1074/jbc.M202458200. [DOI] [PubMed] [Google Scholar]
- 32.Poukkula M, Kaunisto A, Hietakangas V, Denessiouk K, Katajamaki T, Johnson MS, Sistonen L, Eriksson JE. Rapid turnover of c-FLIPshort is determined by its unique C-terminal tail. J Biol Chem. 2005;280:27345–27355. doi: 10.1074/jbc.M504019200. [DOI] [PubMed] [Google Scholar]
- 33.Chang L, Kamata H, Solinas G, Luo JL, Maeda S, Venuprasad K, Liu YC, Karin M. The E3 ubiquitin ligase itch couples JNK activation to TNFα-induced cell death by inducing c-FLIP(L) turnover. Cell. 2006;124:601–613. doi: 10.1016/j.cell.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 34.Kamarajan P, Sun NK, Chao CC. Up-regulation of FLIP in cisplatinselected HeLa cells causes cross-resistance to CD95/Fas death signalling. Biochem J. 2003;376:253–260. doi: 10.1042/BJ20030659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Longley DB, Wilson TR, McEwan M, Allen WL, McDermott U, Galligan L, Johnston PG. c-FLIP inhibits chemotherapy-induced colorectal cancer cell death. Oncogene. 2006;25:838–848. doi: 10.1038/sj.onc.1209122. [DOI] [PubMed] [Google Scholar]
- 36.Abedini MR, Qiu Q, Yan X, Tsang BK. Possible role of FLICE-like inhibitory protein (FLIP) in chemoresistant ovarian cancer cells in vitro. Oncogene. 2004;23:6997–7004. doi: 10.1038/sj.onc.1207925. [DOI] [PubMed] [Google Scholar]
- 37.Wilson TR, McLaughlin KM, McEwan M, Sakai H, Rogers KM, Redmond KM, Johnston PG, Longley DB. c-FLIP: a key regulator of colorectal cancer cell death. Cancer Res. 2007;67:5754–5762. doi: 10.1158/0008-5472.CAN-06-3585. [DOI] [PubMed] [Google Scholar]
- 38.Rogers KM, Thomas M, Galligan L, Wilson TR, Allen WL, Sakai H, Johnston PG, Longley DB. Cellular FLICE-inhibitory protein regulates chemotherapy-induced apoptosis in breast cancer cells. Mol Cancer Ther. 2007;6:1544–1551. doi: 10.1158/1535-7163.MCT-06-0673. [DOI] [PubMed] [Google Scholar]
- 39.Moerke NJ, Aktas H, Chen H, Cantel S, Reibarkh MY, Fahmy A, Gross JD, Degterev A, Yuan J, Chorev M, et al. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell. 2007;128:257–267. doi: 10.1016/j.cell.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 40.Raja SM, Chen S, Yue P, Acker TM, Lefkove B, Arbiser JL, Khuri FR, Sun SY. The natural product honokiol preferentially inhibits cellular FLICE-inhibitory protein and augments death receptor-induced apoptosis. Mol Cancer Ther. 2008;7:2212–2223. doi: 10.1158/1535-7163.MCT-07-2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun SY, Yue P, Dawson MI, Shroot B, Michel S, Lamph WW, Heyman RA, Teng M, Chandraratna RA, Shudo K, et al. Differential effects of synthetic nuclear retinoid receptor-selective retinoids on the growth of human non-small cell lung carcinoma cells. Cancer Res. 1997;57:4931–4939. [PubMed] [Google Scholar]
- 42.Sun SY, Yue P, Wu GS, El-Deiry WS, Shroot B, Hong WK, Lotan R. Mechanisms of apoptosis induced by the synthetic retinoid CD437 in human non-small cell lung carcinoma cells. Oncogene. 1999;18:2357–2365. doi: 10.1038/sj.onc.1202543. [DOI] [PubMed] [Google Scholar]
- 43.Liu X, Yue P, Zhou Z, Khuri FR, Sun SY. Death receptor regulation and celecoxib-induced apoptosis in human lung cancer cells. J Natl Cancer Inst. 2004;96:1769–1780. doi: 10.1093/jnci/djh322. [DOI] [PubMed] [Google Scholar]
- 44.Sun SY, Liu X, Zou W, Yue P, Marcus AI, Khuri FR. The farnesyl-transferase inhibitor lonafarnib induces CCAAT/enhancer-binding protein homologous protein-dependent expression of death receptor 5, leading to induction of apoptosis in human cancer cells. J Biol Chem. 2007;282:18800–18809. doi: 10.1074/jbc.M611438200. [DOI] [PubMed] [Google Scholar]
- 45.Wang X, Proud CG. Methods for studying signal-dependent regulation of translation factor activity. Methods Enzymol. 2007;431:113–142. doi: 10.1016/S0076-6879(07)31007-0. [DOI] [PubMed] [Google Scholar]
- 46.Chen C, Sun X, Ran Q, Wilkinson KD, Murphy TJ, Simons JW, Dong JT. Ubiquitin-proteasome degradation of KLF5 transcription factor in cancer and untransformed epithelial cells. Oncogene. 2005;24:3319–3327. doi: 10.1038/sj.onc.1208497. [DOI] [PubMed] [Google Scholar]
- 47.Svitkin YV, Herdy B, Costa-Mattioli M, Gingras AC, Raught B, Sonenberg N. Eukaryotic translation initiation factor 4E availability controls the switch between cap-dependent and internal ribosomal entry site-mediated translation. Mol Cell Biol. 2005;25:10556–10565. doi: 10.1128/MCB.25.23.10556-10565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin YD, Chen S, Yue P, Zou W, Benbrook D, Liu S, Le TC, Berlin KD, Khuri FR, Sun SY. CAAT/enhancer binding protein homologous proteindependent death receptor 5 induction is a major component of SHetA2-induced apoptosis in lung cancer cells. Cancer Res. 2008;68:5335–5344. doi: 10.1158/0008-5472.CAN-07-6209. [DOI] [PubMed] [Google Scholar]
- 49.Sabatini DM. mTOR and cancer: insights into a complex relationship. Nat Rev Cancer. 2006;6:729–734. doi: 10.1038/nrc1974. [DOI] [PubMed] [Google Scholar]
- 50.Yamaguchi H, Wang HG. CHOP is involved in endoplasmic reticulum stress-induced apoptosis by enhancing DR5 expression in human carcinoma cells. J Biol Chem. 2004;279:45495–45502. doi: 10.1074/jbc.M406933200. [DOI] [PubMed] [Google Scholar]
- 51.Shiraishi T, Yoshida T, Nakata S, Horinaka M, Wakada M, Mizutani Y, Miki T, Sakai T. Tunicamycin enhances tumor necrosis factor-related apoptosisinducing ligand-induced apoptosis in human prostate cancer cells. Cancer Res. 2005;65:6364–6370. doi: 10.1158/0008-5472.CAN-05-0312. [DOI] [PubMed] [Google Scholar]
- 52.Panner A, James CD, Berger MS, Pieper RO. mTOR controls FLIPS translation and TRAIL sensitivity in glioblastoma multiforme cells. Mol Cell Biol. 2005;25:8809–8823. doi: 10.1128/MCB.25.20.8809-8823.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Opel D, Westhoff MA, Bender A, Braun V, Debatin KM, Fulda S. Phosphatidylinositol 3-kinase inhibition broadly sensitizes glioblastoma cells to death receptor- and drug-induced apoptosis. Cancer Res. 2008;68:6271–6280. doi: 10.1158/0008-5472.CAN-07-6769. [DOI] [PubMed] [Google Scholar]
- 54.Yoshida T, Shiraishi T, Nakata S, Horinaka M, Wakada M, Mizutani Y, Miki T, Sakai T. Proteasome inhibitor MG132 induces death receptor 5 through CCAAT/enhancer-binding protein homologous protein. Cancer Res. 2005;65:5662–5667. doi: 10.1158/0008-5472.CAN-05-0693. [DOI] [PubMed] [Google Scholar]
- 55.Oyadomari S, Mori M. Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ. 2004;11:381–389. doi: 10.1038/sj.cdd.4401373. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.