Abstract
We performed integrated gene coexpression network analysis on two large microarray-based brain gene expression data sets generated from the prefrontal cortex obtained post-mortem from 101 subjects, 47 subjects with schizophrenia and 54 normal control subjects, ranging in age from 19 to 81 years. Twenty-eight modules of coexpressed genes with functional interpretations were detected in both normal subjects and those with schizophrenia. Significant overlap of “case” and “control” module composition was observed, indicating that extensive differences in underlying molecular connectivity are not likely driving pathology in schizophrenia. Modules of coexpressed genes were characterized according to disease association, cell type specificity, and the effects of aging. We find that genes with altered expression in schizophrenia clustered into distinct coexpression networks and that these were associated primarily with neurons. We further identified a robust effect of age on gene expression modules that differentiates normal subjects from those with schizophrenia. In particular, we report that normal age-related decreases in genes related to central nervous system developmental processes, including neurite outgrowth, neuronal differentiation, and dopamine-related cellular signaling, do not occur in subjects with schizophrenia during the aging process. Extrapolating these findings to earlier stages of development supports the concept that schizophrenia pathogenesis begins early in life and is associated with a failure of normal decreases in developmental-related gene expression. These findings provide a novel mechanism for the “developmental” hypothesis of schizophrenia on a molecular level.
Schizophrenia is a heterogeneous psychiatric disorder with a lifetime risk of ∼1%. Complex interactions between genetic and environmental factors are thought to result in abnormalities in central nervous system (CNS) gene expression leading to disease manifestation (Giegling et al. 2008). Accordingly, several global expression studies of schizophrenia have been published (for review, see Konradi 2005; Mirnics et al. 2006), with the expression of genes related to myelination, synaptic transmission, metabolism, and ubiquitination reported as being altered in brains of individuals with schizophrenia. However, not all of these differences have been replicated in every study, nor have they been integrated into a compelling and comprehensive biological context. While these standard analyses of differential expression in schizophrenia have resulted in the reporting of multiple lists of genes with altered expression in schizophrenia, most show mild fold-changes and nominal statistical significance after correcting for multiple hypothesis testing. Furthermore, standard analyses ignore the strong correlations that may exist between gene expression patterns. Consequently, interpreting the contribution(s) of individual genes to the pathophysiology of schizophrenia has been difficult, raising the need to search beyond simple differential expression of each gene in isolation.
Alternatively, gene coexpression network analysis can provide a more powerful approach for elucidating transcriptome patterns and dysfunction of gene expression at the systems level, digging further into the underlying molecular nature of this disease. This network approach organizes genes and their protein products into functional modules that are co-regulated and therefore are more likely to participate in similar cellular processes and pathways. Such analyses have been used to understand the molecular basis of other conditions, including cancer (Horvath et al. 2006; Hu et al. 2009), chronic fatigue syndrome (Presson et al. 2008), and body weight regulation (Fuller et al. 2007). Furthermore, network coexpression analysis greatly alleviates the multiple testing problems inherent in standard gene-centric methods of microarray data analysis by converting thousands of genes potentially related to the disease into a manageable number of gene coexpression modules (i.e., 10–200), and hence is a powerful data reduction strategy, allowing for the detection of subtle gene expression changes across groups of genes with statistically derived regulatory relationships.
In this study, we have applied network coexpression analysis to two large microarray data sets in order to characterize comprehensive molecular mechanisms in schizophrenia. We find similar fundamental gene co-regulation in both normal subjects and those with schizophrenia, suggesting that a major change in the underlying molecular connectivity is not a basis for pathology in this disease. Rather, the greatest molecular variation distinguishing subjects with schizophrenia from controls occurs at the level of collective changes in gene expression within functional networks and the differential effects of aging on key biological systems. The power to detect these changes is dramatically improved by network coexpression analysis, which can reveal small concerted gene expression changes that may not reach individual gene-level significance due to multiple testing issues. More specifically, we hypothesize that at least a proportion of disease pathogenesis results from a failure of normal age-related down-regulation of gene expression related to neuronal development and dopamine-related cellular signaling. These findings illuminate a novel molecular basis for schizophrenia that should facilitate diagnosis, prognosis, and therapeutic considerations.
Results
Generation of gene coexpression networks
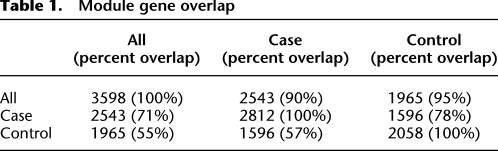
In order to form a framework for our systems-level analyses, we combined and analyzed two different brain gene expression data sets from individuals with schizophrenia and normal controls (Tang et al. 2009; http://www.brainbank.mclean.org/). These arrays, representing 13,012 genes in total, were used to reconstruct networks for 47 schizophrenic cases and 54 controls, separately and combined. In the combined network, 3598 genes were present, representing 90%–95% of the genes present when networks were reconstructed for cases and controls separately (Table 1). The case- and control-only networks contained significantly fewer genes (2812 for cases and 2058 for controls) and overlapped with one another at frequencies approximately expected by selecting two different subsets of genes from the overall network (Table 1). Formally, we compared the overlap of the schizophrenic case-only, control-only, and the combined networks with one another using the hypergeometric distribution to test for the probabilistic significance of module overlap (Fig. 1). All case modules and all control modules significantly overlapped with at least one of the combined network modules (Fig. 1A,B). When case- and control-only modules were compared to one another, again, all but one module from case modules, case Module 26, overlapped significantly with a control module and vice versa (control Module 16) (Fig. 1C). The excellent overlap of gene membership between the two networks and the combined network indicates that major differences in module composition do not differentiate subjects with schizophrenia from normal human subjects. This suggests that the underlying gene–gene connectivity of the CNS is not different in schizophrenia overall, although we cannot exclude perturbations in a limited number of gene regulatory relationships. Further functional analyses were considered using the higher confidence and completeness of the combined networks.
Table 1.
Module gene overlap
Figure 1.
Gene network overlaps. Heatmaps displaying the overlap between the combined case and control networks (“ALL1-28”) versus case networks (“Case 1-26”) (A), the combined case and control networks versus control networks (“Cntrl 1-23”) (B), the case versus control networks (C), and the control networks and those generated by Oldham et al. (2008) indicated by color names (D). Red indicates a significant overlap of gene composition in the designated modules according to the scale shown.
To gauge the accuracy of our reconstructed networks, we compared our control-only networks to the cortex networks generated from normal subjects (n = 67) in a recent study using similar methods (Fig. 1D; Oldham et al. 2008). The Oldham et al. network analysis, which maintained strict quality-control criteria for inclusion of microarray data sets, consisted of 4302 genes distributed over 20 different modules (Fig. 1D). Of the 2058 genes within our control network (out of 13,012 possible genes), 1387 matched genes within the Oldham et al. network (67.3%; P-value = 4.01 × 10−268). All of our control modules approached significance in overlap with at least one Oldham et al. module, with four modules, most of which were the smaller of our modules, showing marginal significance with an Oldham et al. module (Fig. 1D). These differences are likely explained by differences in sample size, sample composition, correlation measures, and tree-cutting methods used in our approach versus the Oldham et al. approach. Overall, the Oldham et al. network and our control network show excellent agreement, providing confidence in the accurate generation of our gene expression modules and demonstrating the reproducibility of similar global network analyses.
Network enrichment of differentially expressed genes in schizophrenia
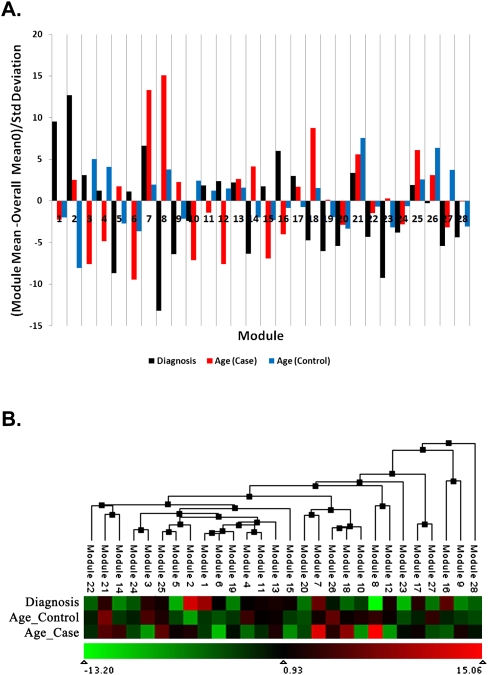
In order to determine which modules, and their corresponding processes, were associated with schizophrenia, we first identified genes showing altered expression in all schizophrenic subjects versus all controls. Multiple linear regressions were run for each gene individually, with age, sex, pH, and post-mortem interval as covariates in order to remove potential confounding effects of these factors on gene expression differences in schizophrenia. The resultant F-ratios were entered into a Wilcoxon rank-sum test by module in order to determine which modules contain an enrichment of genes associated with diagnosis. The results of this analysis are shown in Figure 2 and described further in Supplemental Table 1.
Figure 2.
(A) Module enrichment of differentially expressed and age-related genes. Enrichment of each module with differentially expressed genes in cases versus controls (black bars) and genes whose expression changes with age (red bars, schizophrenia; blue bars, controls) is depicted in terms of the number of standard deviations the Wilcoxon rank-sum score deviates from the overall average score. Positive scores correspond to a higher score than average and demonstrate an enrichment of differentially expressed or age-related genes, while negative scores correspond to a lower score than average and demonstrate a deficit of differentially expressed or age-related genes. The threshold for significance after correction for multiple tests is 2.91. (B) Eigengene clustering to display the relationships between modules. Each module is summarized by its first principal component (eigengene) and clustered on the basis of the absolute correlation between module eigengenes. The heatmap depicts module enrichment of differentially expressed and age-related genes as in A, where the color represents the number of standard deviations the Wilcoxon rank-sum score deviates from the overall average score.
Five modules (Modules 1, 2, 7, 16, and 21) were significantly enriched in genes differentially expressed between subjects with schizophrenia versus control subjects. (A list of all the genes present in all of the network modules is provided in Supplemental Table 2.) Heatmap depictions of these coexpression modules in all subjects are shown in Supplemental Figure 1. Eigengene clustering, to display the relationships between modules, revealed that these five modules do not cluster together, suggesting they represent gene expression perturbations in disparate biological systems (Fig. 2B). These distinct systems may give rise to, or be associated with, different clinical presentations of this disorder. We examined the functional roles encoded by these modules by Gene Ontology (GO) enrichment analysis. Top findings show that Module 1 is associated with oxidative phosphorylation, energy production, and metabolism (Supplemental Table 3), consistent with evidence indicating mitochondrial dysfunction and defects in brain metabolism leading to oxidative stress in schizophrenia (Prabakaran et al. 2004; Iwamoto et al. 2005). Module 2 is associated with angiogenesis, including a significant number of genes associated with diseases resulting from abnormalities in brain vasculature (Supplemental Tables 3, 4). These findings are consistent with evidence indicating abnormalities in cerebral vasculature in schizophrenia, potentially extenuating brain-metabolic defects through defects in nutrient delivery (Cohen et al. 1995; Schultz et al. 2002). Module 7 is associated with post-translational modification, neurogenesis, and neuron differentiation (Supplemental Table 3). Module 16 is associated with chromatin and nucleosome assembly and transcriptional regulation (Supplemental Table 3), consistent with the known perturbations of gene expression observed in numerous microarray studies (Lewis and Mirnics 2006; Mirnics et al. 2006). Finally, Module 21 is associated with inositol phosphate metabolism and metabotropic glutamate signaling (Supplemental Table 3), which have been widely implicated in schizophrenia etiology (Patil et al. 2007). The top five functions for each of these modules are highlighted in Supplemental Figure 2.
Module characterization according to cell type and disease association
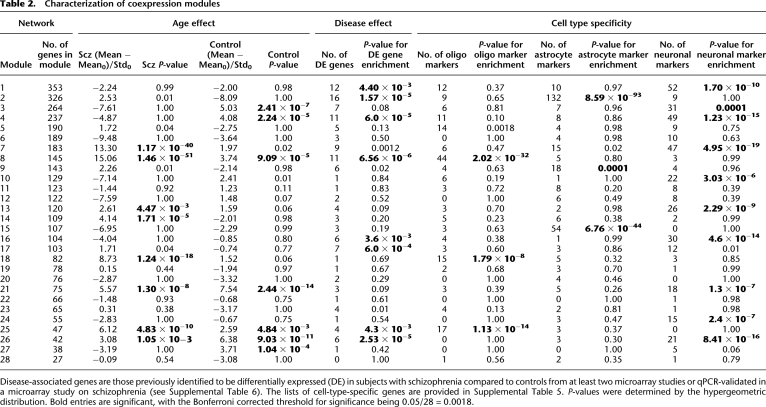
In order to characterize coexpression modules with regards to CNS cellular organization and their further association with schizophrenia, we cross-referenced the module gene lists to (1) published data of genes encoding markers of different CNS cell types (Supplemental Table 5; Cahoy et al. 2008) and (2) data of genes previously identified to be differentially expressed in schizophrenia from published microarray studies (Supplemental Table 6). It is important to note that our coexpression modules are derived from brain tissue, representing a heterogeneous collection of different cell types. Nonetheless, we found that certain modules consisted of genes preferentially associated with oligodendrocytes, astrocytes, and neurons. For example, Module 8 is enriched in genes encoding proteins essential for oligodendrocyte/myelin function (Table 2), including MAG, MOG, MAL, and PLLP (see Supplemental Table 2); Module 2 contained genes associated with astrocyte function, including SLC1A3 and VIM (see Supplemental Table 2); and Module 7 with neuronal function, including NTRK2, NRN1, and STX1A (see Supplemental Table 2). Heatmaps showing the strong correlations of these modules with CNS cell type are shown in Supplemental Figure 3. All modules containing genes differentially expressed between our case and control cohorts were significantly associated with a specific CNS cell type. Modules 1, 7, 16, and 21 were significantly associated with neurons (Table 2), and most of the processes encoded within these modules are neuron-specific. Additionally, we found that Module 2, associated with cerebral vasculature by GO, was significantly associated with astrocyte markers, which conforms to the role of astrocytes in vasomodulation and other brain vascular functions (Zonta et al. 2003).
Table 2.
Characterization of coexpression modules
Disease-associated genes are those previously identified to be differentially expressed (DE) in subjects with schizophrenia compared to controls from at least two microarray studies or qPCR-validated in a microarray study on schizophrenia (see Supplemental Table 6). The lists of cell-type-specific genes are provided in Supplemental Table 5. P-values were determined by the hypergeometric distribution. Bold entries are significant, with the Bonferroni corrected threshold for significance being 0.05/28 = 0.0018.
Next, we compared our module gene lists to previously identified disease-associated genes from the literature; these were defined from 18 previously published global gene expression studies performed on post-mortem brain from subjects with schizophrenia (the exact studies are referenced in Supplemental Table 6). We included (1) genes identified as significantly altered in their expression in schizophrenia in more than one microarray study, and (2) those genes whose expression changes were validated by qPCR in at least one study. We found that genes with altered expression in schizophrenia cluster into a subset of coexpression modules, rather than being evenly dispersed among the entire 28 modules. Accordingly, four of the five modules enriched in genes altered in their expression in schizophrenia in this study (Modules 1, 2, 7, and 16) showed a significant or marginally significant enrichment in genes previously validated for differential expression in schizophrenia. Module 8 also showed an overrepresentation of genes altered in their expression in schizophrenia based on literature findings (Table 2), which might be expected based on the wide range of reports of myelin dysfunction in this disease (for review, see Davis et al. 2003; Segal et al. 2007; Tkachev et al. 2007; Karoutzou et al. 2008). The clustering of genes differentially expressed in schizophrenia compared to controls, many of which were apparently unrelated in terms of known biological functions, into selected coexpression modules demonstrates the power of our approach in identifying systems-level dysfunction in the pathophysiology of schizophrenia in the face of mild expression changes overall.
The effects of aging on gene coexpression
We previously reported differences in age-related genes in schizophrenia versus control subjects (Tang et al. 2009). Here, we applied our network-based analysis to provide a more powerful approach to identify groups of functionally related genes showing significant relationships with aging. We ran multiple linear regressions for each gene individually against age for schizophrenics and normal subjects separately, with sex, pH, and postmortem interval (PMI) as covariates to remove any confounding effects. As with the analysis for differential expression with disease state, the resultant F-ratios were compared via a Wilcoxon rank-sum test according to module in order to determine which modules contained an enrichment of genes associated with age. The results of this analysis are shown in Figure 2A and in Table 2. As with differential expression due to disease, the modules displaying age effects do not cluster together, suggesting they represent aging perturbations in disparate biological systems (Fig. 2B). These different systems may give rise to distinct phenotypes associated with aging.
Four modules were associated with aging in both schizophrenics and normal subjects, but to varying degrees in the two populations (Table 2). Modules 8 and 21 are clearly and strongly associated with aging in both schizophrenics and normal subjects; however, there was a significantly stronger age association observed in schizophrenia compared to normal controls (P-value for the difference by rank sums = 6.28 × 10−24). As mentioned above, Module 8 is associated with oligodendrocyte function and myelination, processes known to change with aging, as well as in schizophrenia (Davis et al. 2003; Segal et al. 2007; Tkachev et al. 2007; Karoutzou et al. 2008). Similarly, while metabotropic glutamate signaling, the top GO function for Module 21, is altered in schizophrenia, age-related differences in the expression of glutamate-related genes in subjects with schizophrenia are likely to be part of the normal aging process (Supplemental Table 3; Simonyi et al. 1998). Modules 25 and 26 are more weakly associated with aging in both subjects with schizophrenia and normal subjects. These two are small modules containing a few aging genes of interest, such as SIRT2 and SREBF1, but did not give good GO results overall.
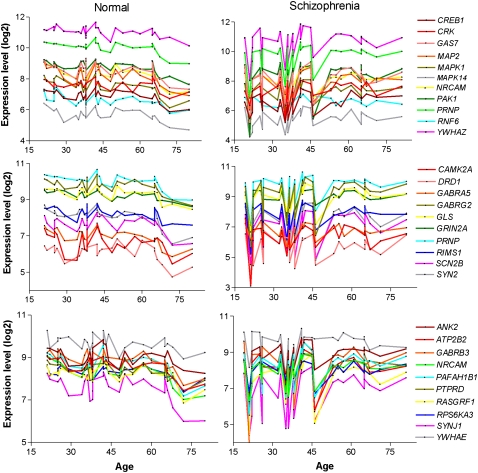
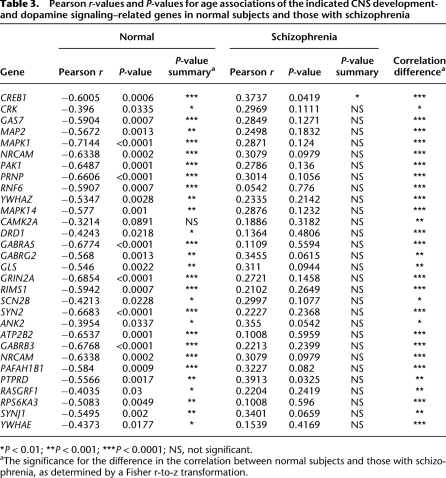
Next, we focused on those modules showing age associations in either case or control subjects. Three modules were associated with normal aging but did not exhibit an aging effect in subjects with schizophrenia. Modules 3, 4, and 27 were significantly associated with aging in normal subjects but show no age effect in subjects with schizophrenia. Functional network analysis, using the Ingenuity Systems Pathways Analysis knowledge base, was used to identify direct interactions between module genes and, thereby, assign broad biological functions to groups of interacting genes (Calvano et al. 2005). The top biological networks identified in each of the three modules were CNS development and function and cellular signaling (Supplemental Table 7). Additionally, Module 3 was associated with protein metabolism and Modules 4 and 27 with cellular assembly and organization. More detailed GO enrichment analysis of these module genes also revealed an association of Module 3 with protein metabolic processes, as well as regulation of neurotransmitter transport and glycolipid transport (Supplemental Table 8). Consistent with the pathways analysis, GO enrichment analysis of Module 4 genes revealed a strong association with CNS developmental processes, including neuronal differentiation and morphogenesis, neurite outgrowth, and synaptic transmission. Furthermore, Module 4 contained a large number of genes encoding protein kinases as well as neurotransmitter receptors, including the dopamine D1 receptor (DRD1) (see Supplemental Table 2). The expression of Module 4 genes related to these functions showed strong coregulation in all subjects, as well as a significant negative correlation with age in normal subjects (Fig. 3; Table 3). While strong coregulation of the expression of these 30 genes was evident in subjects with schizophrenia, no significant correlations with age were observed for any gene (Fig. 3; Table 3). This module also contained genes that were found to be significantly differentially expressed between controls and subjects with schizophrenia from the literature findings (P = 6.0 × 10−5) (Table 2), which could be explained by the differential aging effect, as most schizophrenic subjects previously examined were middle to late age (i.e., 40–80 yr of age). Finally, Module 27 contained many genes with unknown function and did not give good GO results overall.
Figure 3.
Expression coregulation of CNS developmental- and dopamine signaling–related genes and the effect of age on gene expression levels. The expression level of each gene (right of graphs) in individual subjects is depicted in chronological order by age. (Left) Normal subjects; (right) subjects with schizophrenia. (Top panels) Genes related to neurite outgrowth; (middle panels) genes related to dopamine synaptic transmission; (bottom panels) genes related to neuronal differentiation, morphogenesis, and migration. Pearson correlation coefficients (Pearson r-values) for the linear association with age in normal subjects and those with schizophrenia are provided in Table 3.
Table 3.
Pearson r-values and P-values for age associations of the indicated CNS development- and dopamine signaling–related genes in normal subjects and those with schizophrenia
*P < 0.01; **P < 0.001; ***P < 0.0001; NS, not significant.
aThe significance for the difference in the correlation between normal subjects and those with schizophrenia, as determined by a Fisher r-to-z transformation.
Three modules—Modules 7, 14, and 18—were found to be associated with age in schizophrenia but not in normal subjects. Top functional networks associated with these modules were distinct from those associated with aging in control subjects and included gene expression, lipid metabolism, and immunological disease (Supplemental Table 9). These functions may relate to features of disease progression in schizophrenia, as we have previously reported alterations in immune/inflammatory pathways and lipid metabolism in chronic illness but not in early-stage illness (Narayan et al. 2008). Module 7, previously identified to contain an enrichment of genes with altered expression in schizophrenia as mentioned above, showed Gene Ontologies related to post-translational modification, neurogenesis, and neuron differentiation (Supplemental Table 3), and interestingly, also showed an aging effect in this disease. GO results showed that Module 14 was specifically associated with chromosome organization, cell cycle, and response to DNA damage, while Module 18 was associated with intracellular transport, cell adhesion, and neuronal regeneration (Supplemental Table 8). Age-related gene expression changes detected in subjects with schizophrenia, but not in controls, might result from a progressive pathogenic process, a response to pathology, or, possibly, drug treatment, considering that a confounding factor in post-mortem research on schizophrenia is the unknown effect of antipsychotic drugs, which are known to alter gene expression (Thomas 2006). To address this issue, we selected eight genes from Modules 7, 14, and 18 related to different functional categories and tested whether their expression levels were altered in the brains of mice that were treated with haloperidol (2 mg/kg; 4 wk), the main drug with which most of the subjects with schizophrenia were treated. No changes in expression of any genes were found in response to drug treatment (Supplemental Table 10). While this argues against a drug effect in our subjects with schizophrenia, we cannot rule out the possibility that antipsychotic drug treatment might affect, to some extent, the observed transcriptome profiles.
Discussion
Our network-based approach constructs gene coexpression modules in an unbiased manner and generates gene sets that are related to one another due to gene coregulation. Application of this approach to two large microarray-based cortical gene expression data sets (n = 101 subjects in total) has revealed several important findings with regard to underlying molecular connectivity in schizophrenia, clustering of disease-related genes, and differential effects of the aging process on schizophrenia pathology.
First, we find that cortical transcriptome is organized into modules of coexpressed genes, and that significant preservation of coexpression relationships exists in normal and disease states, suggesting that major perturbations in gene connectivity do not distinguish schizophrenia from controls on a molecular level. Schizophrenia has been thought of as a disorder of reduced functional and structural cortical connectivity (Karlsgodt et al. 2008). Much evidence for this theory comes from magnetic resonance imaging studies showing abnormal functional connectivity between prefrontal cortex and multiple brain regions during certain cognitive tasks (Calhoun et al. 2009; Esslinger et al. 2009). Such effects might arise from neurotransmitter receptor–driven alterations in synaptic efficacy and modulation of plasticity. Furthermore, a breadth of evidence supports the existence of disrupted myelin/oligodendrocyte processes in schizophrenia (Davis et al. 2003; Segal et al. 2007; Tkachev et al. 2007; Karoutzou et al. 2008), which likely gives rise to structural abnormalities in the myelin sheath leading to altered neuronal communication. Although the root of dysfunctional integration among neuronal systems is not known, our findings suggest that the aberrant functional and structural connectivity observed in schizophrenia is not driven by differences in underlying molecular connectivity. Rather, we suggest that functional dysregulation of gene activity results from altered expression levels of genes within important processes, such as those listed in Supplemental Figure 2.
Standard analysis of differential gene expression comparing all 47 subjects with schizophrenia versus the 54 control subjects revealed that genes with altered expression in schizophrenia were significantly enriched in five module networks. These networks were associated with pathways previously implicated in the pathophysiology of schizophrenia, including oxidative phosphorylation, energy production, metabolism, synaptic neurotransmission, nervous system development, gene transcription, and metabotropic glutamate signaling, similar to previous findings (Prabakaran et al. 2004; Iwamoto et al. 2005; Lewis and Mirnics 2006; Mirnics et al. 2006; Patil et al. 2007; Paz et al. 2008; Maycox et al. 2009). Most importantly, four of the five networks were preferentially associated with neuronal function, strongly suggesting that neurons are primarily affected by the pathophysiology of schizophrenia. However, our results also provide evidence for dysfunction in astrocytes in schizophrenia. This evidence comes from the disease-linked coexpression module, Module 2, which not only contained an overrepresentation of astrocyte-specific genes, but also included genes involved in blood vessel development and brain vasculature (see Supplemental Table 3), which are roles fulfilled by astrocytes in the brain. Cross-referencing gene composition of all the modules to the published literature of altered gene expression in schizophrenia revealed consistent identification of our disease-associated modules and further revealed that disease-related genes cluster into coexpression modules. This suggests a common regulation of disease genes and/or the incorporation of disease genes into common biological groups supporting the concept of a systems-level pathological dysfunction in schizophrenia.
In our analysis, there was no link between Module 8, a module enriched in oligodendrocyte/myelin-related genes, and altered gene expression in schizophrenia in our cohorts; however, a strong association was found when compared to the literature microarray findings (Table 2). Both positive and negative findings regarding the role of oligodendrocyte-related gene expression in schizophrenia have been reported (Davis et al. 2003; Segal et al. 2007; Tkachev et al. 2007; Karoutzou et al. 2008; Mitkus et al. 2008; Maycox et al. 2009). Our lack of effect could be due to the broad age range of our subjects and the strong effect of aging to influence the expression of these genes, especially in subjects with schizophrenia.
Another advantage of our network analysis approach is to facilitate interpretation of disease-related genes by allowing assertion of a biological role for the module. The biological role of a module is based on the genes contained within the module that have known biological functions or are associated with a particular functional property. This approach can shed light on the function of new genes or genetic findings through “guilt-by-association.” For example, recent studies have identified a genetic variant (rs1344706) in the ZNF804A gene, a member of Module 21, which is associated with alterations in the coordinated activity of the dorsolateral prefrontal cortex and other brain regions (Esslinger et al. 2009). Biological processes associated with Module 21 include the metabotropic glutamate receptor signaling pathway, calcium ion transport, and phospholipase C activation (see Supplemental Table 3), thereby implicating these pathways in the functional coupling (correlated activity) of the dorsolateral prefrontal cortex. The strength of our approach is that we can now postulate that other genes in this module may represent novel targets for potential modifiers of this functional coupling.
Perhaps of highest relevance, our analyses revealed that age-related changes in gene expression differentiate control subjects from those with schizophrenia. These differences were extremely strong signals and reflected in modules being both significantly associated with aging in normal subjects and significantly unassociated with aging in schizophrenics (the mean F-stat for genes within this module are significantly lower than the average F-stat for genes with age). In particular, Modules 3 and 4 were found to be associated with aging in control subjects only. These modules were strongly associated with CNS developmental processes, including neuron formation and migration, synaptogenesis, synaptic pruning, and cell-to-cell signaling, especially involving the dopamine D1 receptor (see Fig. 3). Importantly, these processes are not observed when straightforward differential GO enrichment analysis is performed for the top age-related genes (Supplemental Fig. 4), which only captures more general processes such as axonogenesis, neuron differentiation, and synapse organization. Previous gene profiling studies performed on normal human prefrontal cortex during post-natal aging have reported a down-regulation of genes related to CNS development, including axon guidance, neuron development, and neurotransmitter signaling, from birth to an early twenties time window (Harris et al. 2009). This suggests that the down-regulation of similar genes we detected with aging in normal subjects in this study may reflect a continuum of developmental changes that occurs throughout the lifespan of the individual. We found these changes to be absent in the CNS of subjects with schizophrenia. It is worthwhile to note that gene expression measurements during post-natal stages of development in subjects with schizophrenia are not possible given that this disease is not typically diagnosed until the late teen years.
The neurodevelopmental model of schizophrenia posits that abnormalities during critical developmental periods increase the risk for the subsequent emergence of clinical symptoms (Marenco and Weinberger 2000; Rapoport et al. 2005). Hence, the lack of normal age-related down-regulation of important CNS developmental genes and pathways might trigger the onset of disease. The lack of the normal age-related decrease in dopamine-related cell signaling could be especially relevant. Previous studies have measured the expression of post-synaptic markers of the dopaminergic system in post-mortem brains of normal human individuals as early as 2 mo of age and continuing to 86 yr of age (Weickert et al. 2007). It was found that DRD2 and DRD1 mRNA levels were highest in the cortex of neonates and adolescents, respectively, and decreased progressively into adulthood and aging stages of life (Weickert et al. 2007). Extrapolating our findings from subjects with schizophrenia to developmental stages could suggest that the dopamine hyperactivity observed in this disease is associated with a lack of normal developmental-related decreases in dopamine metabolism/signaling. The continued molecular changes occurring throughout aging in schizophrenia could govern disease progression, as it has been argued that schizophrenia might be both a neurodevelopmental and a progressive disorder.
Our coexpression network analysis revealed a comprehensive description of transcriptome organization in schizophrenia. Our analyses indicate that genes showing altered expression in this disease cluster into coexpression networks associated primarily with neurons and, to a lesser extent, astrocytes. These coexpression networks allow us to identify the function of new genes based on membership in modules with known biological roles. Further, we find a strong differential aging effect between normal subjects and those with schizophrenia on key developmental processes. From these results, we conclude that at least part of the pathophysiology of schizophrenia results from a failure of normal age-related down-regulation of genes related to important developmental processes.
Methods
Post-mortem brain samples
This study incorporates post-mortem human brain samples from two different brain banks: The Victorian Brain Bank Network (VBBN) at the Mental Health Research Institute and the Harvard Brain Bank (HBB). The VBBN collection consists of prefrontal cortical (Brodmann Area [BA] 46) samples from 30 normal subjects and 30 subjects with schizophrenia. Demographic data for these individual subjects, including sex, age, post-mortem interval, recorded drug dose, and cause of death, are provided in our previous studies (Narayan et al. 2008). The HBB collection is comprised of prefrontal cortical (BA9) regions from 26 normal subjects and 19 subjects with schizophrenia. Demographic data for these individual subjects, including sex, age, post-mortem interval (PMI), handedness, ethnicity, and cause of death are provided on the National Brain Databank website (http://national_databank.mclean.harvard.edu/brainbank/Main). Ascertainment and diagnosis of all subjects were based on the Diagnostic and Statistical Manual of Mental Disorders (DSM-IV) criteria (American Psychiatric Association 1994). In the case of the VBBN collection, an additional validated instrument, the Diagnostic Instrument for Brain Studies, was used (Hill et al. 1996). The patients and controls from both banks were closely matched overall on gender (76.5% vs. 76.3% male; P = 0.98), age (49.85 ± 2.27 yr vs. 48.91 ± 2.66 yr; P = 0.78), pH (6.36 ± 0.034 vs. 6.318 ± 0.037; P = 0.44), and PMI (30.92 ± 2.0 vs. 32.30 ± 1.90 h; P = 0.19); however, the PMIs between the two brain banks were significantly different (40.4 ± 2.6 h vs. 21.6 ± 1.2 h; P < 0.0001). This difference could be due, in part, to different criteria for defining PMI.
Microarray expression data sets
Preparation of brain tissue, RNA extractions, microarray hybridizations, and quality-control procedures for our samples (VBBN) are described in Narayan et al. (2008). For the HBB samples, these procedures were performed at the Harvard Brain Tissue Resource Center by standard methods described on the National Brain Database website (http://www.brainbank.mclean.org/). Brain gene expression data from the VBBN cohort were generated on the Affymetrix Human Genome U133 Plus 2.0 Array, and those from the HBB cohort on the Affymetrix Human Genome U95a Array. The gene expression data from the HBB cohort were downloaded as cell intensity (cel) files from the National Brain Database website and merged with the VBBN data, also in cel format, for combined reanalysis.
The raw signal intensities for each individual from both cohorts (n = 105 subjects in total) were transformed using MAS5 normalization and subjected to Affymetrix quality-control measures. Arrays were filtered for GAPDH 3′/5′ ratios < 3 and Present call > 35% using the Affymetrix Expression Console, resulting in the removal of four samples in this manner, leaving a total of 54 control samples and 47 from subjects with schizophrenia. In the remaining samples, there were no significant differences in GAPDH 3′/5′ ratios (1.31 ± 0.04 vs. 1.40 ± 0.06; P = 0.244) or percent Present calls (46.10 ± 0.53 vs. 46.00 ± 0.58) between samples from controls and those with schizophrenia. The data from the VBBN subjects were generated on two separate dates, thus, batch effects for this data set were removed using the methods implemented in the ComBat algorithm (Johnson et al. 2007). Potential batch effects were removed from the Harvard data set by defining batches according to the creation date and removing any batch effects using ComBat. Probes common to the two Affymetrix Platforms were selected to merge the two data sets into a single data set (20,985 probes). Batch effects across the VBBN and HBB data sets were again removed by ComBat. Probes were assigned to gene symbols based on the annotation file provided by Affymetrix.
Network reconstruction
Networks were reconstructed for schizophrenic cases only, controls only, and combined expression data sets as follows. Mutual information scores quantifying relationships between the brain expression levels of probes were calculated using the ARACHNE algorithm with a P-value cutoff of 1 × 10−10 (Margolin et al. 2006). Gene mutual information scores were defined by selecting the highest mutual information value for probes mapping to each gene pair (13,012 genes total). These gene mutual information scores, I(x;y), were used to construct a symmetric, undirected, weighted, adjacency matrix, A, for which self-connections were not allowed (i.e., the diagonal of the matrix is set to 0), and the connection strength between genes x and y is simply equivalent to the mutual information score between genes x and y, such that the elements of the I are defined as: axy = ayx = I(x;y). The mutual information scores were standardized so that the maximum mutual information score in each network was set to one. The weighted adjacency matrix, A, was transformed to approximate an unweighted scale-free network topology of the type observed in other well-characterized biological systems (Zhang and Horvath 2005).
The transformed adjacency matrices, A, were converted to dissimilarity matrices, DISS, by replacing each value in the matrix by one minus the original value (i.e., the elements of the dissimilarity matrices were defined as: dissxy = dissyx = 1 − [I(x;y)/I(max)]s, where I(max) is the maximum mutual information score in the matrix (i.e., the standardization factor) and s is an integer used to transform the unweighted adjacency matrix to approximate the scale free criteria (s = 4 in all our matrices). These distance matrices were subjected to hierarchical clustering with average linkage using the R computational suite. Resulting trees from the cluster analyses were used to define gene networks using the Dynamic Tree Cut algorithm implemented in the cutreeHybrid approach implemented in R (Langfelder et al. 2008).
Acknowledgments
This work was supported, in part, by the Scripps Translational Science Institute Clinical Translational Science Award (NIH U54RR02504-01) (N.J.S. and A.T.), and, in part, by NIH grant MH069696 (E.A.T). A.T. is also supported in part by a Scripps Dickinson Fellowship. B.D. is a NHMRC senior research fellow (Level B; 400016).
Footnotes
[Supplemental material is available online at http://www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.101956.109.
References
- American Psychiatric Association. Diagnostic and statistical manual of mental disorders (DSM-IV) American Psychiatric Association; Washington, DC: 1994. [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: A new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Eichele T, Pearlson G. Functional brain networks in schizophrenia: A review. Front Hum Neurosci. 2009;3:17. doi: 10.3389/neuro.09.017.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, Chen RO, Brownstein BH, Cobb JP, Tschoeke SK, et al. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–1037. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- Cohen BM, Yurgelun-Todd D, English CD, Renshaw PF. Abnormalities of regional distribution of cerebral vasculature in schizophrenia detected by dynamic susceptibility contrast MRI. Am J Psychiatry. 1995;152:1801–1803. doi: 10.1176/ajp.152.12.1801. [DOI] [PubMed] [Google Scholar]
- Davis KL, Stewart DG, Friedman JI, Buchsbaum M, Harvey PD, Hof PR, Buxbaum J, Haroutunian V. White matter changes in schizophrenia: Evidence for myelin-related dysfunction. Arch Gen Psychiatry. 2003;60:443–456. doi: 10.1001/archpsyc.60.5.443. [DOI] [PubMed] [Google Scholar]
- Esslinger C, Walter H, Kirsch P, Erk S, Schnell K, Arnold C, Haddad L, Mier D, Opitz von Boberfeld C, Raab K. Neural mechanisms of a genome-wide supported psychosis variant. Science. 2009;324:605. doi: 10.1126/science.1167768. [DOI] [PubMed] [Google Scholar]
- Fuller TF, Ghazalpour A, Aten JE, Drake TA, Lusis AJ, Horvath S. Weighted gene coexpression network analysis strategies applied to mouse weight. Mamm Genome. 2007;18:463–472. doi: 10.1007/s00335-007-9043-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giegling I, Hartmann AM, Genius J, Benninghoff J, Möller HJ, Rujescu D. Systems biology and complex neurobehavioral traits. Pharmacopsychiatry. 2008;41:S32–S36. doi: 10.1055/s-2008-1081200. [DOI] [PubMed] [Google Scholar]
- Harris LW, Lockstone HE, Khaitovich P, Weickert CS, Webster MJ, Bahn S. Gene expression in the prefrontal cortex during adolescence: Implications for the onset of schizophrenia. BMC Med Genomics. 2009;2:28. doi: 10.1186/1755-8794-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C, Keks N, Roberts S, Opeskin K, Dean B, MacKinnon A, Copolov D. Postmortem brain studies in schizophrenia: The problems of diagnosis. Am J Psychiatry. 1996;153:533–537. doi: 10.1176/ajp.153.4.533. [DOI] [PubMed] [Google Scholar]
- Horvath S, Zhang B, Carlson M, Lu KV, Zhu S, Felciano RM, Laurance MF, Zhao W, Qi S, Chen Z, et al. Analysis of oncogenic signaling networks in glioblastoma identifies ASPM as a molecular target. Proc Natl Acad Sci. 2006;103:17402–17407. doi: 10.1073/pnas.0608396103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Zhou M, Jiang J, Wang J, Elashoff D, Gorr S, Michie SA, Spijkervet FK, Bootsma H, Kallenberg CG, et al. Systems biology analysis of Sjogren's syndrome and mucosa-associated lymphoid tissue lymphoma in parotid glands. Arthritis Rheum. 2009;60:81–92. doi: 10.1002/art.24150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto K, Bundo M, Kato T. Altered expression of mitochondria-related genes in postmortem brains of patients with bipolar disorder or schizophrenia, as revealed by large-scale DNA microarray analysis. Hum Mol Genet. 2005;14:241–253. doi: 10.1093/hmg/ddi022. [DOI] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Karlsgodt KH, Sun D, Jimenez AM, Lutkenhoff ES, Willhite R, van Erp TG, Cannon TD. Developmental disruptions in neural connectivity in the pathophysiology of schizophrenia. Dev Psychopathol. 2008;20:1297–1327. doi: 10.1017/S095457940800062X. [DOI] [PubMed] [Google Scholar]
- Karoutzou G, Emrich HM, Dietrich DE. The myelin-pathogenesis puzzle in schizophrenia: A literature review. Mol Psychiatry. 2008;13:245–260. doi: 10.1038/sj.mp.4002096. [DOI] [PubMed] [Google Scholar]
- Konradi C. Gene expression microarray studies in polygenic psychiatric disorders: Applications and data analysis. Brain Res Brain Res Rev. 2005;50:142–155. doi: 10.1016/j.brainresrev.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Langfelder P, Zhang B, Horvath S. Defining clusters from a hierarchical cluster tree: The Dynamic Tree Cut package for R. Bioinformatics. 2008;24:719–720. doi: 10.1093/bioinformatics/btm563. [DOI] [PubMed] [Google Scholar]
- Lewis DA, Mirnics K. Transcriptome alterations in schizophrenia: Disturbing the functional architecture of the dorsolateral prefrontal cortex. Prog Brain Res. 2006;158:141–152. doi: 10.1016/S0079-6123(06)58007-0. [DOI] [PubMed] [Google Scholar]
- Marenco S, Weinberger DR. The neurodevelopmental hypothesis of schizophrenia: Following a trail of evidence from cradle to grave. Dev Psychopathol. 2000;12:501–527. doi: 10.1017/s0954579400003138. [DOI] [PubMed] [Google Scholar]
- Margolin AA, Nemenman I, Basso K, Wiggins C, Stolovitzky G, Dalla Favera R, Califano A. ARACNE: An algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics. 2006;7:S7. doi: 10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maycox PR, Kelly F, Taylor A, Bates S, Reid J, Logendra R, Barnes MR, Larminie C, Jones N, Lennon M, et al. Analysis of gene expression in two large schizophrenia cohorts identifies multiple changes associated with nerve terminal function. Mol Psychiatry. 2009;14:1083–1094. doi: 10.1038/mp.2009.18. [DOI] [PubMed] [Google Scholar]
- Mirnics K, Levitt P, Lewis DA. Critical appraisal of DNA microarrays in psychiatric genomics. Biol Psychiatry. 2006;60:163–176. doi: 10.1016/j.biopsych.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Mitkus SN, Hyde TM, Vakkalanka R, Kolachana B, Weinberger DR, Kleinman JE, Lipska BK. Expression of oligodendrocyte-associated genes in dorsolateral prefrontal cortex of patients with schizophrenia. Schizophr Res. 2008;98:129–138. doi: 10.1016/j.schres.2007.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan S, Tang B, Head SR, Gilmartin TJ, Sutcliffe JG, Dean B, Thomas EA. Molecular profiles of schizophrenia in the CNS at different stages of illness. Brain Res. 2008;1239:235–248. doi: 10.1016/j.brainres.2008.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham MC, Konopka G, Iwamoto K, Langfelder P, Kato T, Horvath S, Geschwind DH. Functional organization of the transcriptome in human brain. Nat Neurosci. 2008;11:1271–1282. doi: 10.1038/nn.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil ST, Zhang L, Martenyi F, Lowe SL, Jackson KA, Andreev BV, Avedisova AS, Bardenstein LM, Gurovich IY, Morozova MA, et al. Activation of mGlu2/3 receptors as a new approach to treat schizophrenia: A randomized Phase 2 clinical trial. Nat Med. 2007;13:1102–1107. doi: 10.1038/nm1632. [DOI] [PubMed] [Google Scholar]
- Paz RD, Tardito S, Atzori M, Tseng KY. Glutamatergic dysfunction in schizophrenia: From basic neuroscience to clinical psychopharmacology. Eur Neuropsychopharmacol. 2008;18:773–786. doi: 10.1016/j.euroneuro.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabakaran S, Swatton JE, Ryan MM, Huffaker SJ, Huang JT, Griffin JL, Wayland M, Freeman T, Dudbridge F, Lilley KS, et al. Mitochondrial dysfunction in schizophrenia: Evidence for compromised brain metabolism and oxidative stress. Mol Psychiatry. 2004;9:684–697. doi: 10.1038/sj.mp.4001511. [DOI] [PubMed] [Google Scholar]
- Presson AP, Sobel EM, Papp JC, Suarez CJ, Whistler T, Rajeevan MS, Vernon SD, Horvath S. Integrated weighted gene co-expression network analysis with an application to chronic fatigue syndrome. BMC Syst Biol. 2008;2:95. doi: 10.1186/1752-0509-2-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport JL, Addington AM, Frangou S, Psych MR. The neurodevelopmental model of schizophrenia: Update 2005. Mol Psychiatry. 2005;10:434–449. doi: 10.1038/sj.mp.4001642. [DOI] [PubMed] [Google Scholar]
- Schultz SK, O'Leary DS, Boles Ponto LL, Arndt S, Magnotta V, Watkins GL, Hichwa RD, Andreasen NC. Age and regional cerebral blood flow in schizophrenia: Age effects in anterior cingulate, frontal, and parietal cortex. J Neuropsychiatry Clin Neurosci. 2002;14:19–24. doi: 10.1176/jnp.14.1.19. [DOI] [PubMed] [Google Scholar]
- Segal D, Koschnick JR, Slegers LH, Hof PR. Oligodendrocyte pathophysiology: A new view of schizophrenia. Int J Neuropsychopharmacol. 2007;10:503–511. doi: 10.1017/S146114570600722X. [DOI] [PubMed] [Google Scholar]
- Simonyi A, Xia J, Igbavboa U, Wood WG, Sun GY. Age differences in the expression of metabotropic glutamate receptor 1 and inositol 1,4,5-trisphosphate receptor in mouse cerebellum. Neurosci Lett. 1998;244:29–32. doi: 10.1016/s0304-3940(98)00127-x. [DOI] [PubMed] [Google Scholar]
- Tang B, Chang WL, Lanigan CM, Dean B, Sutcliffe JG, Thomas EA. Normal human aging and early-stage schizophrenia share common molecular profiles. Aging Cell. 2009;8:339–342. doi: 10.1111/j.1474-9726.2009.00468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas EA. Molecular profiling of antipsychotic drug function: Convergent mechanisms in the pathology and treatment of psychiatric disorders. Mol Neurobiol. 2006;34:109–128. doi: 10.1385/MN:34:2:109. [DOI] [PubMed] [Google Scholar]
- Tkachev D, Mimmack ML, Huffaker SJ, Ryan M, Bahn S. Further evidence for altered myelin biosynthesis and glutamatergic dysfunction in schizophrenia. Int J Neuropsychopharmacol. 2007;10:557–563. doi: 10.1017/S1461145706007334. [DOI] [PubMed] [Google Scholar]
- Weickert CS, Webster MJ, Gondipalli P, Rothmond D, Fatula RJ, Herman MM, Kleinman JE, Akil M. Postnatal alterations in dopaminergic markers in the human prefrontal cortex. Neuroscience. 2007;144:1109–1119. doi: 10.1016/j.neuroscience.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:Article 17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- Zonta M, Angulo MC, Gobbo S, Rosengarten B, Hossmann KA, Pozzan T, Carmignoto G. Neuron-to-astrocyte signaling is central to the dynamic control of brain microcirculation. Nat Neurosci. 2003;6:43–50. doi: 10.1038/nn980. [DOI] [PubMed] [Google Scholar]