Abstract
Global studies of transcript structure and abundance in cancer cells enable the systematic discovery of aberrations that contribute to carcinogenesis, including gene fusions, alternative splice isoforms, and somatic mutations. We developed a systematic approach to characterize the spectrum of cancer-associated mRNA alterations through integration of transcriptomic and structural genomic data, and we applied this approach to generate new insights into melanoma biology. Using paired-end massively parallel sequencing of cDNA (RNA-seq) together with analyses of high-resolution chromosomal copy number data, we identified 11 novel melanoma gene fusions produced by underlying genomic rearrangements, as well as 12 novel readthrough transcripts. We mapped these chimeric transcripts to base-pair resolution and traced them to their genomic origins using matched chromosomal copy number information. We also used these data to discover and validate base-pair mutations that accumulated in these melanomas, revealing a surprisingly high rate of somatic mutation and lending support to the notion that point mutations constitute the major driver of melanoma progression. Taken together, these results may indicate new avenues for target discovery in melanoma, while also providing a template for large-scale transcriptome studies across many tumor types.
Cancers arise from the accumulation of genomic mutations and epigenetic changes that alter gene function and expression. In recent years, many new oncogenes and tumor suppressor genes have emerged from genome-wide analyses of human cancers (Stratton et al. 2009). Collectively, somatic base-pair mutations have been identified in many human genes through targeted DNA sequencing efforts (Forbes et al. 2008). DNA microarrays have been used to measure gains and losses in DNA copy number, as well as altered gene expression profiles. These efforts have provided key insights into the underlying biological mechanisms driving tumorigenesis. Additionally, they enable classification of patient subpopulations and the discovery of prognostic and predictive biomarkers (Sawyers 2008), inspiring the design of novel targeted approaches for clinical intervention (Stuart and Sellers 2009).
Despite their widespread utility, standard microarray and sequencing technologies have shown limited ability to interrogate key mRNA-based events such as gene fusions, alternative splicing, and base mutations involving the entire expressed fraction of the cancer genome. Gene fusions, in particular, have been recognized as a common and important feature of cancer since the discovery and characterization of the Philadelphia chromosome (Nowell and Hungerford 1960; Rowley 1973). Arising from translocations and other chromosomal abnormalities, gene fusions have been most commonly associated with hematological disorders and soft tissue sarcomas (Mitelman et al. 2007). Recent discoveries of prominent recurrent gene fusions in prostate cancer (Tomlins et al. 2005) and lung cancer (Soda et al. 2007) have revealed their prevalence in epithelial carcinomas as well (Prensner and Chinnaiyan 2009). That these recurrent events in epithelial carcinomas had gone undiscovered for so long is due to the fact that the technologies to systematically interrogate the genome for such alterations have emerged only recently. This raises the possibility that tumor types for which virtually no gene fusions are known, such as melanoma, may harbor important aberrations that can be identified by the application of these genomic tools.
Massively parallel sequencing technologies (Shendure and Ji 2008) offer the opportunity to characterize the cancer genome at unprecedented depth and sensitivity. Large-scale transcriptome sequencing (RNA-seq) using short paired-end reads has proved an effective means of precisely denoting exon structure and identifying novel transcription in a large variety of species (Cloonan et al. 2008; Lister et al. 2008; Morin et al. 2008; Mortazavi et al. 2008; Nagalakshmi et al. 2008; Sultan et al. 2008; Hillier et al. 2009; Yassour et al. 2009). Gene expression estimates from short-read sequencing have been shown to recapitulate microarray-based measurements (Marioni et al. 2008), while providing additional information regarding the existence and abundance of alternatively spliced variant transcripts (Pan et al. 2008; Wang et al. 2008). Gene fusions and other chimeric transcripts have been identified by RNA sequencing in selected prostate and breast cancer cell lines using technologies that generate single long reads spanning the fusion points (Maher et al. 2009a; Zhao et al. 2009). Paired-end short-read sequencing of RNA offers a particularly sensitive and efficient approach to gene fusion discovery due to the larger number of distinct reads and the increased physical coverage resulting from the long fragment length (Maher et al. 2009b). The millions of paired-end reads generated in a single lane of an Illumina Genome Analyzer II enable the additional characterization of gene expression levels, alternative splicing, sequence mutations, and allele-specific expression.
Here, we present a systematic framework for the integrative genomic analysis of cancer transcriptomic data. Using paired-end RNA-seq data from 10 patient-derived melanoma short-term cultures and cell lines, we discovered 11 novel expressed gene fusions produced by underlying genomic rearrangements, representing, to our knowledge, the first gene fusions reported in melanoma. We mapped these transcripts to base-pair resolution and interrogated their genomic origins using sample-matched high-density SNP array and chromosomal data. We also discovered 12 novel chimeric readthrough transcripts, including seven that were found in multiple melanoma samples. Further, we used the RNA-seq data to simultaneously interrogate sequence mutations, gene expression levels, alternative splicing, and allele-specific expression. These analyses have identified numerous novel genetic events in melanoma that offer several new biological insights into this malignancy and can be prioritized for further functional characterization. More generally, these results provide a template for the systematic analysis of transcriptomic data across broad collections of tumors.
Results
A framework for systematic cancer transcriptome analysis
To test the robustness of RNA-seq for transcriptome characterization in cancer, we performed paired-end RNA-seq using a cDNA library prepared from the well-characterized chronic myelogenous leukemia (CML) cell line K-562, which harbors the BCR-ABL1 gene fusion. Following cDNA library construction and shearing (Methods), fragments were size-selected to a fragment length range of 400–600 base pairs (bp) and loaded into separate flow-cell lanes on an Illumina Genome Analyzer II. We obtained 15.5 million pairs of 51-mer reads (8.8 million purity filtered pairs; Bentley et al. 2008), or 1.6 gigabases (Gb) of total sequence.
Next, we sought to implement an alignment strategy for RNA-seq read pairs that enabled identification of multiple types of mRNA alterations at optimal sensitivity and specificity (Supplemental Fig. S1). Here, all sequence reads were independently aligned to a reference consisting of the transcriptome (61,478 human transcripts in the Ensembl database) and the human genome (hg18), as described in Methods. This approach ensures that both annotated splice junctions and unannotated transcribed regions are accessible; reads encompassing splice junctions are split and then mapped to their genomic coordinates. For a read pair to be informative, we required a unique genomic placement for each read, and we eliminated duplicate pairs likely arising from PCR in library construction. As a result, we considered 4.0 million distinct read pairs informative for further analysis in K-562, achieving a mean sequence coverage of 4.4× for the annotated transcriptome (Supplemental Table S1). This two-tiered alignment and data filtering method markedly reduced the fraction of false-positive events detected by RNA-seq.
We reasoned that the gene fusions could be recognized by the presence of discordant read pairs in which each end mapped to a different chromosomal locus. However, we anticipated that artifactitious discordant read pairs could also arise from errors in sequence alignment, and that even a low misalignment rate would overwhelm our ability to discern bona fide gene fusions. Therefore, we limited our analysis to cases with (1) read pairs mapping uniquely in the human genome to opposite strands of separate protein-coding genes and (2) at least one 51-base read unambiguously spanning a junction between two exons of the genes.
As a positive control, we looked for reads connecting BCR on chromosome 22 and ABL1 on chromosome 9. We observed 37 distinct read pairs with ends mapping to the two genes, as well as 23 individual fusion-spanning reads, implicating a fusion between exon 14 of BCR and exon 2 of ABL1 (Supplemental Fig. S2). In addition to BCR-ABL1 we found two more gene fusions meeting these criteria (NUP214-XKR3 and BAT3-SLC44A4; Supplemental Fig. S3). We subsequently validated these gene fusions by RT-PCR and Sanger sequencing (Supplemental Fig. S4).
Having demonstrated the effectiveness of the technique for K-562, we then applied the method to 10 melanoma specimens (eight patient-derived short-term cultures and two cell lines, MeWo and 501 Mel). In addition to mRNA, we isolated genomic DNA from each melanoma, including matched normal DNA from three patients, to interrogate the genomic alterations underlying any events we observed from RNA-seq. We obtained one lane of mRNA sequence for each specimen, yielding an average of 14.2 million pairs of 51-mer reads, or nearly 1.5 Gb of total sequence per sample (Supplemental Table S1).
We analyzed the data to identify gene fusions, chimeric readthrough transcripts, and point mutations, as described below. We also determined gene expression levels and identified instances of alternative splicing and allele-specific expression for the entire melanoma transcriptome as a whole (described in Supplemental material). Altogether, these efforts provide a framework for the systematic integrated analysis of paired-end RNA-seq data.
Novel gene fusions in melanoma
Using the criteria above, we identified 11 novel gene fusions (Figure 1; Table 1), and successfully validated all 11 using RT-PCR followed by Sanger sequencing across the fusion point (Supplemental Fig. S5). Of these 11 fusions, four involve genes on separate chromosomes, and seven represent intrachromosomal events. All 11 gene fusions appear to be heterozygous based on the presence of reads consistent with the normal gene structure as well (Supplemental Table S2). To verify that these gene fusions arose from physical rearrangements in the genome, we tested 10 of 11 cases by performing long-range PCR with genomic DNA followed by end-sequencing of products. In nine of 10 cases, we confirmed the presence of a genomic rearrangement (Supplemental Table S3). The inability to detect a genomic rearrangement in the remaining case is likely due to inefficient long-range PCR; however, we cannot exclude the possibility that this gene fusion may have arisen from trans-splicing (Li et al. 2008).
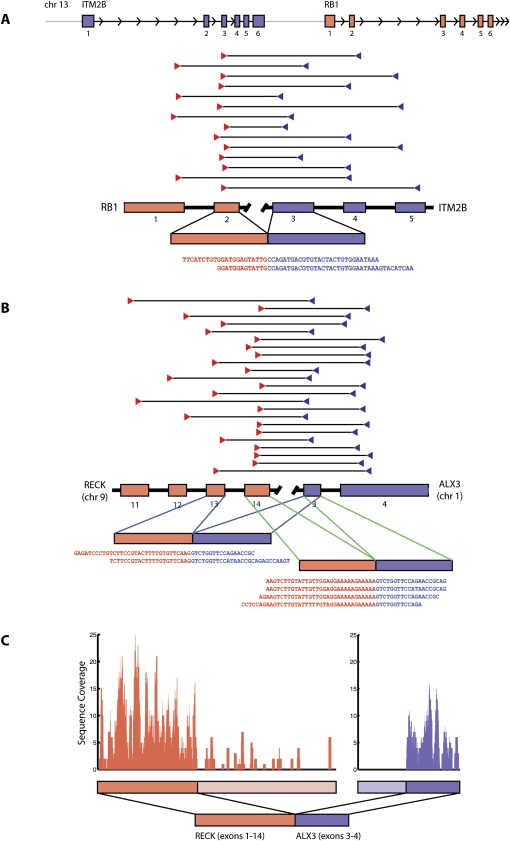
Figure 1.
Gene fusions in melanoma. (A) RB1-ITM2B gene fusion in short-term culture M990802. ITM2B is transcribed immediately upstream of RB1 on chromosome 13, yet in M990802, a fusion transcript beginning with the 5′ end of RB1 and ending with the 3′ end of ITM2B is implicated by 14 distinct read pairs and two individual fusion-spanning reads. (B) RECK-ALX3 gene fusion in short-term culture M000921. Two alternate transcripts are observed in the RNA-seq data: One joining exon 13 of RECK to exon 3 of ALX3, and one joining exon 14 of RECK to exon 3 of ALX3. (C) Spatial distribution of sequence reads for RECK and ALX3 are consistent with a fusion transcript containing the 5′ end of RECK and the 3′ end of ALX3.
Table 1.
Novel melanoma gene fusions
Novel gene fusions harboring at least two distinct discordant read pairs and at least one fusion-spanning individual 51-mer read are shown. Each gene fusion was validated by RT-PCR followed by Sanger sequencing of the product.
Based on current knowledge about mutations in cancer, it is expected that the majority of these gene fusions may be “passenger” mutations (Stratton et al. 2009). In the absence of directed functional follow-up experiments, one cannot distinguish “driver” from passenger mutations. Nonetheless, the genes implicated in these fusions include several genes with previous associations with cancer.
One gene fusion involves RB1, a well-characterized tumor suppressor gene. While deletions, truncations, and mutations of RB1 have been observed in many cancers, translocation is not a known mechanism for the disruption of RB1 function (Futreal et al. 2004). In this case (melanoma short-term culture M990802), the N terminus of RB1 is joined to the integral membrane protein ITM2B in an in-frame fusion. Of note, these genes are adjacent on chromosome 13 and transcribed in the same orientation, but RB1 normally lies downstream of ITM2B (Fig. 1A). The RNA-seq data also reveal an apparently full-length intact RB1 transcript at normal expression levels. Thus, if the fusion gene, in fact, is a driver mutation, it might therefore act in a dominant negative manner.
We also observed an in-frame fusion involving the DNA repair gene PARP1 (PARP1-MIXL1) in melanoma cell line 501 Mel, which may lead to genomic instability through loss of PARP1 function. The fusion product contains nearly the whole DNA-binding domain of PARP1, yet is missing the entire catalytic domain. If the fusion is a driver mutation, it thus might also act in a dominant negative manner.
Several other fusions also involved cancer-related genes. A RECK-ALX3 fusion is produced by a translocation between chromosomes 9 and 1 in melanoma short-term culture M000921 (Fig. 1B). ALX3 encodes a homeobox transcription factor, while RECK is an inhibitor of tumor invasion and metastasis (Takahashi et al. 1998). The TLN1 gene (present in the TMEM8B-TLN1 fusion) is present within a focal genomic amplification observed in oral squamous cell carcinoma (Snijders et al. 2005). CCT3 and GNA12 (implicated in the CCT3-C1orf61 and GNA12-SHANK2 fusions, respectively, both of which occur in the 501 Mel cell line) function in a cellular network predicted to play a role in colorectal cancer (Nibbe et al. 2009). On the other hand, several gene fusions involve poorly characterized genes or genes with no previous ties to cancer.
Despite the associations above, a causal role for these fusions in melanoma remains speculative until the appropriate functional experiments are performed. Nonetheless, the RNA-seq data directly pinpoint interesting candidate fusion events and thereby define functional experiments to interrogate their functional roles.
DNA and mRNA structural variations associated with melanoma gene fusions
The RB1-ITM2B and RECK-ALX3 gene fusions also exemplified interesting structural features observed repeatedly among the 11 events we discovered. RB1-ITM2B represents one of four cases where adjacent genes transcribed in the same orientation occur out of order in the fusion transcript. That is, the 5′ gene in the fusion transcript (RB1) actually lies 3′ of its partner (ITM2B) in the normal human genome. Such events may arise from tandem duplications, positioning the 3′ end of the first gene downstream of the 5′ end of the second gene (Fig. 1A). We examined this hypothesis using SNP microarrays to measure DNA copy number, as discussed below. The RECK-ALX3 fusion represents one of two cases where the individual fusion-spanning sequence reads indicate the presence of multiple alternatively spliced isoforms involving separate fusion points. We identified two sets of reads joining RECK and ALX3: Four reads joining exon 14 of RECK to exon 3 of ALX3, and two reads joining exon 13 of RECK to exon 3 of ALX3 (Fig. 1B). We also found reads implicating separate transcripts involving exon 4 of CCT3 fused to multiple exons of C1orf61 (data not shown). Our data are consistent with a single genomic breakpoint after exon 14 of RECK and before exon 3 of ALX3, leading to alternatively spliced fusion transcripts spanning this breakpoint. As shown in Figure 1C, the observed sequence coverage from RNA-seq confirms elevated expression of the 5′ end of RECK and the 3′ end of ALX3 in short-term culture M000921.
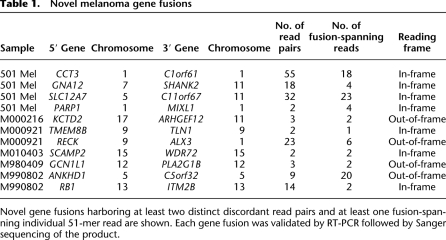
To study at higher resolution the underlying genomic changes producing these chimeric transcripts, we hybridized genomic DNA to Affymetrix SNP 6.0 microarrays for three melanomas harboring multiple gene fusions (eight gene fusions total). These SNP arrays enable the measurement of DNA copy number at more than 1.8 million markers across the genome (McCarroll et al. 2008). Although gene fusions are not necessarily expected to be accompanied by underlying chromosomal changes (for instance, balanced translocations are copy-neutral alterations) we nevertheless observed clear changes in DNA copy number (mostly amplifications) for seven of eight cases, with well-defined breakpoints evident within both genes for six fusions. As shown in Figure 2A, SNP arrays revealed copy number breakpoints within RECK and ALX3 at the exact positions expected from the observed fusion transcript. We confirmed the disruption of both loci using fluorescence in situ hybridization (FISH), as shown in Figure 2B. The SNP array results also support the hypothesis that intrachromosomal gene fusions may originate from tandem duplications (Campbell et al. 2008). For example, small amplified regions were observed for the loci producing RB1-ITM2B (chromosome 13) and ANKHD1-C5orf32 (chromosome 5). The boundaries of these regions are entirely consistent with the chimeric transcripts observed for each pair of genes (Fig. 2C,D).
Figure 2.
Genomic aberrations leading to gene fusions. (A) Evidence for copy number breakpoints inside RECK and ALX3 from Affymetrix SNP 6.0 microarrays. Raw probe signals were normalized (spots) and segmented (lines) as described in Methods. Exons in red represent those occurring in the RECK-ALX3 fusion transcript. (B) FISH in short-term culture M000921 at the RECK and ALX3 loci. Probes positioned 100–200 kb on both sides of each gene reveal amplifications 5′ of RECK and 3′ of ALX3. (C) Amplification involving ITM2B and RB1 on chromosome 13. A tandem duplication would position exon 2 of RB1 upstream of exon 3 of ITM2B (red), as observed in the fusion transcript. (D) Amplification involving C5orf32, ANKHD1, and intervening genes. A tandem duplication would position exon 1 of ANKHD1 upstream of exon 3 of C5orf32 (red), as observed in the fusion transcript.
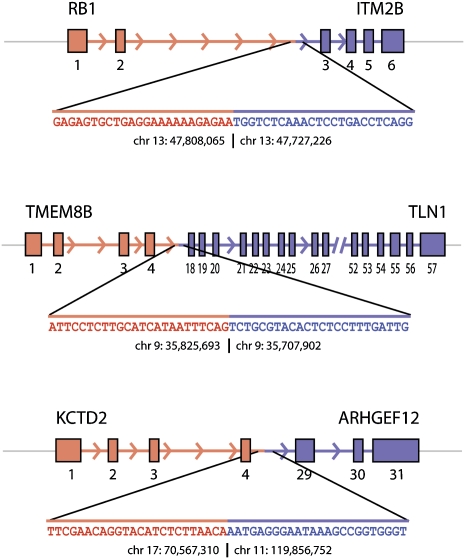
In three cases, we used Sanger sequencing to map the genomic breakpoints to base-pair resolution, as shown in Figure 3. We confirmed the precise location of the breakpoint producing RB1-ITM2B to occur within the second intron of RB1 and the second intron of ITM2B. Another potential tandem duplication producing TMEM8B-TLN1 was mapped to the expected introns. Finally, we identified the translocation site where chromosomes 17 and 11 are joined to produce KCTD2-ARHGEF12. In all three cases, the fused chromosomes involved uninterrupted stretches of native sequence from each locus joined at a single position, suggesting a clean break and reattachment for each chromosomal event.
Figure 3.
Genomic breakpoints mapped to base pair resolution. The precise locations of the fusion points in the genomic DNA were determined by Sanger sequencing for three gene fusions: RB1-ITM2B, TMEM8B-TLN1, and KCTD2-ARHGEF12.
Observed gene fusions appear to be private or low frequency events in melanoma
In principle, melanoma gene fusions may represent important driver events directing tumor progression. Such driver events might be expected to occur repeatedly in multiple independent melanomas. However, no fusion transcript resulting from an underlying chromosomal alteration was observed in more than one instance of the 10 samples analyzed by RNA-seq. We further screened for the presence of each fusion event by RT-PCR in 90 additional melanoma cell lines and short-term cultures derived from patients with metastatic disease. Each fusion product was detectable only in the original sample from which it was discovered and not in the additional 90 (data not shown). While these results do not exclude the possibility of gene fusions involving alternate partners, or even alternate exons within the same pairs of genes, we were unable to demonstrate the presence of any particular DNA rearrangement-driven fusion transcripts in more than one sample. Based on these results, the translocation-based gene fusions we discovered appear to be private events in melanomas (or else occur at very low frequencies). This suggests that many of these events may therefore constitute passenger mutations with little consequence for tumorigenesis. The apparent absence of any common, highly expressed gene fusions in melanoma is itself notable, suggesting that the underlying carcinogenic mechanisms pertinent to melanoma may differ from other tumor types with common gene fusions, such as prostate cancer and many hematologic malignancies.
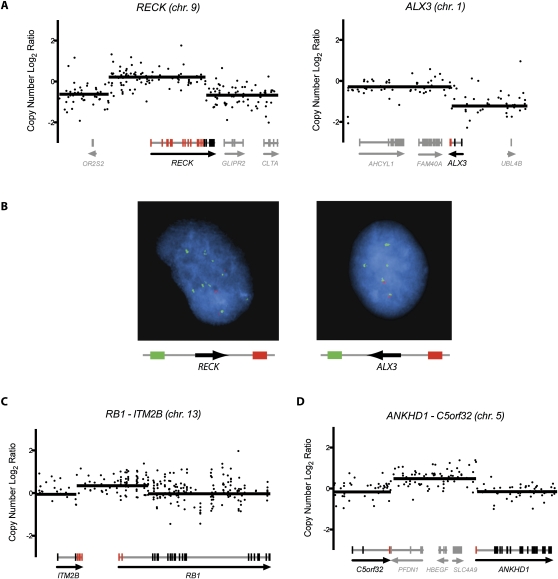
It is conceivable that for some gene fusions, one or both constituent partner genes may represent bona fide cancer genes that are more commonly altered by other genetic mechanisms; e.g., prevalent amplification or deletion events. To explore this possibility, we integrated our gene fusion results with existing melanoma chromosomal copy number data. Specifically, we wished to determine if any fusion partner genes identified here localized to genomic regions previously shown by our group to undergo statistically significant copy number alterations in melanoma (Lin et al. 2008). Of the 22 partner genes (Table 1), one gene (KCTD2) localized to a significant region of copy gain, and two genes (RB1 and ITM2B; comprising a single intrachromosomal fusion event) localized to a significant region of copy loss.
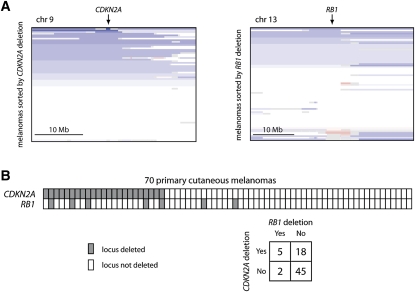
We then focused our attention on the RB1-ITM2B fusion transcript. In a previous analysis of chromosomal copy number data sets derived from 70 primary cutaneous melanomas (Curtin et al. 2005), a region on chromosome 13q encompassing the RB1/ITM2B locus was shown to be significantly deleted, second only to the ARF locus amongst melanoma chromosomal deletions (Fig. 4A; Lin et al. 2008). The ARF locus includes CDKN2A (encodes the p16 protein), a common melanoma tumor suppressor that up-regulates the retinoblastoma (RB1) pathway through suppression of cyclin/CDK function. In light of this functional overlap between CDKN2A and RB1 deletion, RB1 might seem to be an unlikely target gene of 13q deletion in melanoma. However, our data raise the possibility that dysregulation of RB1 is indeed a target of 13q deletion, representing a driver mechanism independent of CDKN2A deletion. Toward this end, the RB1-ITM2B fusion occurs together with CDKN2A deletion in the M990802 short-term culture (not shown). Furthermore, deletions involving the RB1 and CDKN2A loci co-occur at a statistically significant frequency in the aforementioned 70 primary cutaneous melanomas (P = 0.022; Fig. 4B). Conceivably, CDKN2A and RB1 may exert partially distinct functional roles as melanoma tumor suppressor genes. We emphasize that, alone, these results are insufficient to distinguish driver from passenger events; however, they illustrate how the integration of RNA-seq with orthogonal genomic data sets can identify candidate driver events for directed functional experiments.
Figure 4.
Co-occurrence of CDKN2A and RB1 deletions. (A) Array CGH copy number profiles for 70 primary cutaneous melanomas (Curtin et al. 2005). Blue segments correspond to deleted regions, and red segments correspond to amplified regions. (B) Deletions at both loci (CDKN2A and RB1) were found to significantly co-occur (P = 0.022).
Chimeric readthrough transcripts are recurrent in melanoma
In the analysis above, we specifically focused on gene fusions arising from chromosomal aberrations (i.e., involving genes on different chromosomes or in unexpected orientations on the same chromosome). However, we also observed evidence of “readthrough” transcripts joining nearby genes transcribed in the same orientation.
We identified 49 cases of transcripts involving distinct genes in the Ensembl database, based on the presence of at least two read pairs connecting the genes and at least one unambiguous individual fusion-spanning read. To eliminate possible errors in gene structure annotation or cases of incomplete transcriptional termination in normal cells, we eliminated 22 cases in which the genes were joined in other databases and 15 cases in which the genes were joined in reported EST sequences derived from nonmelanoma human tissues and cell lines.
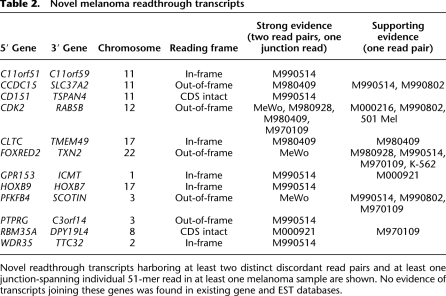
The remaining 12 cases appear to represent novel readthrough transcripts (Table 2). Five of these events are predicted to generate in-frame proteins; two leave the complete coding sequence of the initiating transcript intact, and the remaining five, though out-of-frame, add additional residues to the initiating transcript (average 21 amino acids). For seven of the 12 cases, we detected supporting RNA-seq read pairs in at least two of the samples studied, indicating that these chimeric transcripts may represent recurrent events in melanoma.
Table 2.
Novel melanoma readthrough transcripts
Novel readthrough transcripts harboring at least two distinct discordant read pairs and at least one junction-spanning individual 51-mer read in at least one melanoma sample are shown. No evidence of transcripts joining these genes was found in existing gene and EST databases.
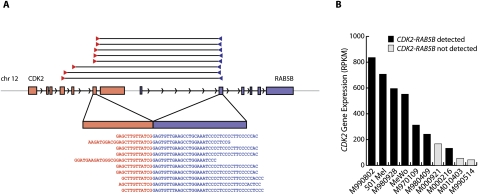
Tumor-specific readthrough transcripts have previously been linked to other cancers, such as prostate cancer (Maher et al. 2009a); thus, it is conceivable that at least some of the events discovered here may also contribute to tumorigenicity in melanoma. We note that one novel readthrough transcript, CDK2-RAB5B, was found in four of 10 melanoma RNA-seq samples (Fig. 5A; Table 2). CDK2 encodes a protein kinase that is a critical regulator of the G1/S phase transition of the cell cycle. The translated product of the readthrough transcript, which is identical in all four melanomas, consists of the first 264 of 298 amino acids of CDK2, followed by only two additional residues. This results in a premature truncation of the kinase domain by 22 amino acids, including deletion of the conserved arginine in subdomain XI, though the effect of this alteration on kinase activity is unclear. We found additional read pairs supporting the CDK2-RAB5B readthrough transcript in three more melanomas. Of note, CDK2 expression is elevated in the seven samples for which the readthrough was detected (Fig. 5B). The observation raises the possibility that elements in the newly added 3′ untranslated region (UTR) of the fusion transcript could contribute to mRNA stability and post-transcriptional regulation of CDK2 and, in turn, influence cellular proliferation.
Figure 5.
Novel recurrent readthrough transcript. (A) CDK2-RAB5B, shown here for the MeWo cell line, was independently discovered in four melanomas, with supporting evidence observed in three more. In MeWo, CDK2-RAB5B is implicated by eight distinct read pairs and 10 individual junction-spanning reads. (B) Expression level of CDK2 as measured by RNA-seq for 10 melanomas. RPKM, reads per kilobase of exon model per million mapped reads (Mortazavi et al. 2008).
Some readthrough transcripts may reflect low-level transcriptional readthrough in normal cells. However, we note that we did not detect read pairs connecting CDK2-RAB5B in the K-562 data set or RNA-seq data from primary glioblastoma and ovarian tumors sequenced through a separate effort (data not shown). These results raise the possibility that CDK2-RAB5B is both recurrent in and specific to the melanocyte lineage in general and melanoma in particular. They also underscore the future necessity of deploying RNA-seq across many additional tumors and cell types to characterize chimeric transcripts systematically and discern cancer-specific events. Such efforts will also lead to improved gene annotations and improve understanding of transcriptome regulation.
Somatic mutation discovery in melanoma by transcriptome analysis
In addition to mapping chimeric transcripts, RNA-seq provides sequence-level information for each individual transcript. Whereas several groups have developed methods for targeted high-throughput sequencing of genomic DNA by first capturing genomic regions of interest (typically exons) using molecular inversion probes (Porreca et al. 2007; Krishnakumar et al. 2008), microarray-based capture (Albert et al. 2007; Hodges et al. 2007; Okou et al. 2007), or solution-phase hybrid selection (Gnirke et al. 2009), RNA-seq offers a systematic means to determine sequence mutations in expressed protein-coding transcripts without the need for additional selection steps (Sugarbaker et al. 2008).
The ability to detect mutations from RNA-seq data depends heavily on the expression levels of the underlying genes. We found that the proportion of transcripts in each sample covered an average depth per nucleotide of at least 10-fold ranged from 5% to 17% (Supplemental Fig. S6). By analyzing these data, we generated sequence calls at an average of 4.7 million bases per sample in protein-coding regions (Methods). We then compared the sequence calls to the human reference sequence (hg18) to identify high quality variant calls. We estimated that the sensitivity for detecting variant sequences is ∼75% of that for calling reference genotypes.
These variants identified by comparison to the human reference sequence are expected to consist of (in descending order of frequency) inherited polymorphisms, somatic mutations, sequencing errors and bona fide differences between RNA and DNA (e.g., RNA editing and polyadenylation). The rate of single nucleotide polymorphisms (SNPs) in protein-coding regions of humans of European descent is ∼6 × 10−4 and, at present, roughly 94% of SNPs found in a sample are already present in the dbSNP database (Sherry et al. 2001; Ng et al. 2009). In contrast, the rate of somatic mutations in most cancer types is typically 1–2 × 10−6. By comparing the tumor sample to matched normal tissue from the same patient, one can eliminate the inherited polymorphisms that are found in both.
Based on the reported polymorphism rates in coding regions (∼6 × 10−4), the average amount of coding sequence called per sample (∼4.7 million bases), and the estimated difference in sensitivity (∼75%), we would expect to find ∼2100 variants per sample, with 94% in current databases. In fact, we found 1694 variants per sample, with 93% in current databases. This discrepancy likely results from regions of loss-of-heterozygosity in cancer. Approximately 130 variants per sample were novel, with most expected to be inherited SNPs.
To distinguish between novel inherited SNPs, somatic mutations, and false-positive calls, we selected 103 randomly chosen novel variants (including 95 nonsynonymous substitutions) in two of the melanoma samples and genotyped them in tumor and matched normal DNA. We observed that >95% were present in the tumor DNA, which is considerably higher than that obtained in previous RNA-seq studies (Sugarbaker et al. 2008) and that 67% of these were also present in the matched normal DNA, indicating that they are new inherited SNPs. The remainder is somatic mutations (Fig. 6A). This implies a somatic mutation rate of 1.1 × 10−5. We note that one cell line (MeWo) had more than twice as many novel variants as any other sample, raising the possibility that this melanoma harbors a “mutator” phenotype reflecting past exposure to UV radiation or possible treatment of the patient with an alkylating agent. The remaining samples exhibit a somatic mutation rate of 8 × 10−6, which is four- to eightfold higher than the rate in typical cancers.
Figure 6.
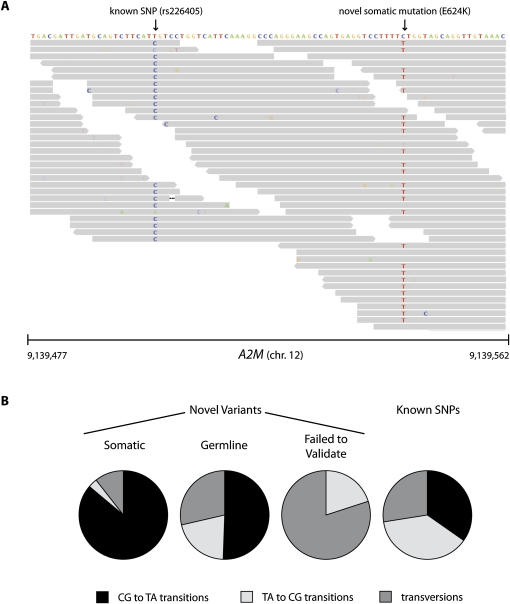
Validation of somatic mutations. (A) Sequence variants in exon 16 of A2M in melanoma short-term culture M970109. Illumina 51-mer reads are shown as gray boxes (arrowheads denote directionality of reads; nonreference bases are colored and shaded according to their quality scores). One variant (T → C) is homozygous and corresponds to a known SNP in dbSNP (Sherry et al. 2001). The other variant (C → T) is heterozygous and corresponds to a missense E624K mutation. This mutation was validated and confirmed to be somatic. (B) Distribution of transitions and transversions in sequence variants genotyped by Sequenom. Validated somatic mutations were largely CG to TA transitions (86%), representative of mutations induced by UV damage (Drobetsky et al. 1987). In contrast, only 53% of novel germline variants and 0% of variants that failed to validate were CG to TA transitions. Thirty six percent of all known SNPs detected by Illumina in the 10 melanoma samples were CG to TA transitions with respect to human genome reference hg18.
Previous sequencing efforts have also reported a markedly higher mutation rate for melanoma compared to other cancers (Greenman et al. 2007), with the excess explained by CG → TA transitions, which are typical of UV-induced mutations (Drobetsky et al. 1987). Among the validated somatic mutations identified here, 86% consist of CG → TA transitions, compared to 41% of somatic point mutations previously reported for all non-skin cancers (Fig. 6B; Forbes et al. 2008; P < 10−5; χ2 test of homogeneity). An increase in the proportion of CG→TA mutations from 41% to 86% would require a 8.5-fold increase in such mutations and would yield a fourfold increase in the total mutation rate. In short, the data support the idea that melanomas have an unusually high base-pair mutation rate, attributable to UV exposure. In addition, our data suggest a transcription-dependent mechanism of mutation and/or mismatch repair—in as much as novel C → T variants occur much less frequently on the transcribed than nontranscribed strand (72%:28%), whereas known C → T SNPs detected in these melanomas show no such bias (Vrieling et al. 1991).
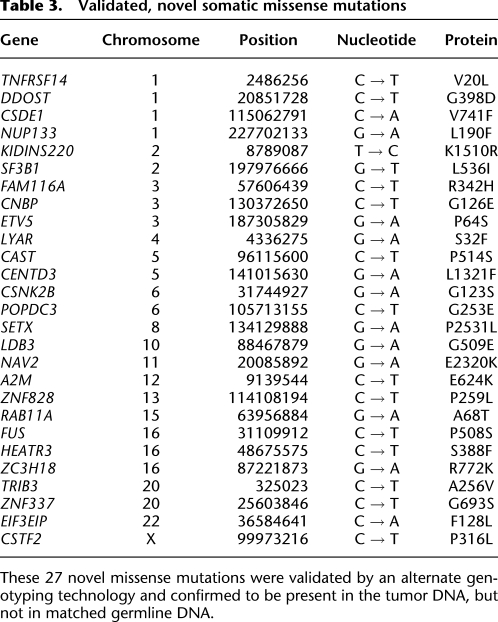
In all, we validated 27 novel somatic missense mutations involving genes listed in Table 3. Like the chimeric transcripts above, the majority of these somatic mutations are expected to be passenger events. However, several of these new mutations occur in genes previously shown to be mutated in cancer (Cancer Genome Atlas Research Network 2008; Forbes et al. 2008; Jones et al. 2008), namely A2M, CAST, CENTD3, FUS, NUP133, SF3B1, TNFRSF14, and TRIB3 (Fig. 6A). Additionally, three genes with missense mutations have previously been implicated in cancer-associated translocations: ETV5, CNBP, and FUS (Futreal et al. 2004). FUS (also named TLS) is a DNA/RNA-binding protein that is involved in a gene fusion with the CHOP transcription factor gene in some myxoid/round cell liposarcomas (Rabbitts et al. 1993). FUS has also been implicated in familial amyotrophic lateral sclerosis (ALS) through the identification of 14 separate germline mutations in the extreme C-terminal domain in ∼25 families (Kwiatkowski et al. 2009; Vance et al. 2009), and the somatic mutation that we observed (in melanoma short-term culture M970109) falls in the C terminus, very close to this cluster of familial mutations. Interestingly, we observed a validated mutation in another gene related to ALS: The M000921 line harbors a somatic mutation in SETX, a gene mutated in families with juvenile ALS and spinocerebellar ataxia (Chen et al. 2004; Moreira et al. 2004). It is unclear whether this finding indicates any connection between genes involved in melanoma and those involved in motor neuron degeneration.
Table 3.
Validated, novel somatic missense mutations
These 27 novel missense mutations were validated by an alternate genotyping technology and confirmed to be present in the tumor DNA, but not in matched germline DNA.
Altogether, we identified 721 novel, nonsynonymous coding variants in melanoma (Supplemental Table S5), though only a subset was subjected to validation to determine whether they are bona fide somatic mutations. Based on the results above, we expected that most are inherited SNPs, whereas ∼30% are somatic mutations. Nonetheless, the set of variants is interesting. One mutation observed in melanoma cell line 501 Mel (CTNNB1, chr3:41241117, C → T) is noted 135 times in the COSMIC database of somatic mutations in cancer (Forbes et al. 2008). This mutation changes serine 37 to phenylalanine in the beta-catenin protein, and has been observed in a wide range of tissues, including 21 skin cancer specimens. Another gene, SRRM2, harbors distinct missense variants in three out of 10 melanoma samples. Interestingly, the MeWo melanoma cell line exhibits novel nonsilent variants in two genes that are also implicated in the gene fusions above: A missense mutation in ANKHD1 (p.P1808S) and a nonsense mutation in SCAMP2 (p.Q301*). We confirmed the presence of both of these mutations by genotyping. Because MeWo is an established cell line with no matched normal DNA, we cannot determine whether the mutations are germline or somatic. Nevertheless, these observations lend support to the hypothesis that the genes involved in fusions may be altered recurrently by various mechanisms during the genesis or progression of melanoma.
Clearly, these melanomas also harbor many more somatic mutations than can be discerned from our data—which only reliably cover ∼12% of the protein-coding genes. For example, we note that BRAF and NRAS, two commonly mutated melanoma oncogenes, were not identified as mutated by this analysis because these genes are not among the top ∼12% of most abundant transcripts in this data set. Technological improvements since these data were generated have increased the sequencing output per Illumina lane more than threefold, significantly expanding the territory accessible to mutation discovery.
Gene expression-based alterations in melanoma by transcriptome analysis
Last, we estimated gene expression levels and identified instances of alternative splicing and allele-specific expression for the melanoma transcriptome as a whole. These results (for more detail, see Supplemental material) provide a framework for the systematic integrated analysis of paired-end RNA-seq data and further demonstrate the utility of RNA-seq in interrogating the full spectrum of RNA-based events relevant to cancer.
Briefly, we first quantified expression levels for all genes in each sample according to the RPKM measure (reads per kilobase of exon model per million mapped reads) (Mortazavi et al. 2008), reported in Supplemental Table S4, and observed high reproducibility among replicate library construction and Illumina sequencing experiments (Supplemental Fig. S7). Comparisons to microarray-based measurements of gene expression for these melanomas revealed good agreement between these technologies, with RNA-seq exhibiting a much wider dynamic range and greater precision for 97% of expressed genes (Supplemental Figs. S8, S9).
By counting individual sequence reads spanning exon junctions, we determined the relative abundance of different splice isoforms (Supplemental Fig. S10). Moreover, we discovered 4313 novel exon junctions in 2932 genes, including 12 of the 22 genes involved in the 11 gene fusions above.
We analyzed the data to look for instances of apparent allele-specific expression in these 22 genes; we found compelling evidence for allele-specific expression in one case: the gene SLC12A7 (Supplemental Fig. S11). A more complete description of this analysis is presented in Supplemental material.
Discussion
In this study, we have applied high-throughput paired-end sequencing of cDNA (RNA-seq) as a systematic means to identify chimeric transcripts and other genetic alterations that are expressed in tumors. In so doing, we discovered 11 novel melanoma gene fusions produced by underlying genomic rearrangements, representing, to our knowledge, the first such events in melanoma. These fusions involve common cancer-related genes, such as RB1 and PARP1, as well as several genes with no known cancer associations. We mapped each fusion point to base pair resolution and identified alternatively spliced fusion products in two cases. Fusion transcripts were also typically accompanied by genomic copy number alterations. Altogether, seven of the 11 fusions produced in-frame ORF derivatives, though the specific biological consequences of these events on melanoma genesis and maintenance remain to be determined.
We obtained an average of 14.5 million paired-reads from a single Illumina lane for each melanoma sample, which enabled us to estimate transcript abundance, identify alternatively spliced variants, and discover novel base pair mutations in addition to gene fusions. Because only one lane is needed, this strategy can readily be scaled up to interrogate multiple specimens of many diverse tumor types. This attribute differs importantly from whole-genome sequencing, which currently requires many dedicated whole flow cells per sample in order to detect mutations (Ley et al. 2008; Mardis et al. 2009; Shah et al. 2009). Significant improvements have been made in high-throughput sequencing technology since these melanoma RNA-seq data were generated, enabling even greater sequence coverage per lane of sequencing. In the future, the use of DNA barcoding may allow multiple samples to be pooled within a single lane for higher-throughput multiplexed sequencing (Craig et al. 2008).
The sensitivity for detecting gene fusions by paired-end, massively parallel sequencing depends on many factors, including expression level, transcript length, and cDNA library fragment length. With about 3 million distinct read pairs mapping uniquely to annotated protein-coding genes in the Ensembl database (as was obtained for a single Illumina lane of K-562 in this study), a heterozygous fusion point could theoretically be detected by our criteria in each of the top 50% of expressed genes with a probability of ≥70% (Supplemental Fig. S12). The most biologically meaningful translocations are likely to be those that produce fusion transcripts expressed at appreciable levels; thus, paired-end RNA-seq provides a robust means for identifying such events.
This study demonstrates for the first time that gene fusions and other chimeric transcripts occur frequently in melanoma. Although gene fusions were traditionally associated with hematological disorders and soft tissue sarcomas, several recent reports of recurrent gene fusions in prostate cancer (Tomlins et al. 2005) and lung cancer (Soda et al. 2007) reveal them to be important driver events of epithelial carcinomas as well. The discovery of a recurrent chimeric readthrough transcript involving CDK2 is noteworthy in this regard. Expression of this cyclin-dependent kinase in melanoma is regulated by MITF (Du et al. 2004), a master transcriptional regulator of melanocyte development that also functions as a melanoma oncogene (Garraway et al. 2005). In this context, CDK2 was shown to be required for melanoma cell proliferation (Du et al. 2004). The functional role(s) of truncated CDK2, when expressed as a readthrough transcript, represents an interesting area of further study.
All translocation-based gene fusions we discovered appear to be private events, although it is possible that some represent driver events. Overall, we observed that seven of 11 gene fusions produce in-frame products. The fact that the RB1-ITM2B gene fusion maps to a region commonly deleted in melanoma, raises the possibility that inactivation of the retinoblastoma tumor suppressor may contribute to melanoma pathology independent of CDKN2A/ARF deletion. If so, the observed co-occurrence of CDKN2A and RB1 deletions may imply that tumors harboring both events could prove refractory to pharmacologic CDK inhibition. Directed functional experiments will be necessary to determine the biological importance of each gene fusion and to delineate the possible mechanisms by which it might influence or alter gene function.
In addition to discovering gene fusion events, we used RNA-seq data to interrogate global properties of 10 melanoma transcriptomes. In doing so, we also directly examined many aspects of the 22 genes implicated in fusions. We discovered novel, unannotated splice variants for 12 of these genes (described in Supplemental materials). Additionally, we found that two of these genes harbor nonsilent mutations in the melanoma cell line MeWo. Although it is not possible to determine whether these particular mutations are germline or somatic in origin (due to the lack of a matched normal sample), they lend support to the hypothesis that some fusion partner genes could represent melanoma oncogenes that become altered by multiple genetic means during melanoma genesis.
We also discovered and validated 29 somatic mutations (27 missense) in the coding regions of additional genes. Although sequencing of many more samples is required to identify new genes that are significantly mutated in melanoma, we observed mutations in several known cancer genes (or orthologs), as well as a potential connection to neurodegenerative diseases. Regarding the latter, we confirmed somatic missense mutations in FUS and SETX in separate samples; both genes are mutated in familial ALS (Chen et al. 2004; Kwiatkowski et al. 2009; Vance et al. 2009). We also confirmed two somatic mutations (one missense, one synonymous) in the gene A2M (alpha-2-macroglobulin) in melanoma short-term culture M970109. Alpha-2-macroglobulin, a plasma proteinase inhibitor, mediates the degradation of A-beta in amyloid beta deposits, and polymorphisms in A2M have been associated with susceptibility to Alzheimer's disease (Blacker et al. 1998). Further, ITM2B, which is involved in a gene fusion, is a regulator of amyloid-beta production, and mutations in ITM2B have been previously associated with familial dementias similar to Alzherimer's disease (Matsuda et al. 2005). These data may provide a rationale for experimental approaches to explore a possible connectivity between melanoma genesis and mechanisms of neurodegeneration,
Extrapolating our empirical validation rates to all mutations called from RNA-seq, we estimate an average somatic mutation rate of 8 × 10−6 in melanoma. This is notably higher than other cancers, yet explained by the large excess of CG → TA transitions induced by UV exposure. Further, we observed a large variation in average mutation rates across these melanomas—most strikingly in the presence of one cell line with an especially high mutation rate (mutator phenotype). Interestingly, we also detected more gene fusions in one particular melanoma cell line (four fusions in 501 Mel) than in any of the other short-term cultures. This cell line also exhibits the largest number of copy number breakpoints deduced from SNP arrays, potentially indicative of excess genomic instability.
In any given sample, we obtained sufficient sequence coverage to call genotypes at ∼12% of bases contained in Ensembl transcripts. Undoubtedly, many more nonsilent mutations are present within the coding sequences of genes expressed at lower levels. Notable examples include mutation BRAF and NRAS, two known melanoma oncogenes that are not present in the top 12% of most highly expressed genes in this data set. Our data suggest that an additional fivefold to 10-fold more sequencing coverage would be necessary to reliably identify mutations in the vast majority of genes. Given the rapid increase in sequencing output per lane (threefold, just since these data were generated, with more in prospect), it should be possible to obtain such coverage with one to two lanes of sequence on an Illumina sequencer. On the other hand, targeted capture of genomic DNA or cDNA may be a more effective strategy to discover mutations in genes that may not be highly expressed (Levin et al. 2009). Thus, a mixed strategy of RNA-seq and targeted capture may prove optimal.
In conclusion, we have discovered 11 novel gene fusions and 12 novel readthrough chimeric transcripts in melanoma using a paired-end, massively parallel sequencing strategy. We also used these RNA-seq data to characterize the entire melanoma transcriptome represented herein, including sequence mutations, gene expression levels, alternative splicing, and allele-specific expression. In doing so, we demonstrate the capability of RNA-seq to interrogate the full spectrum of RNA-based alterations relevant to cancer through integrative analysis. This global approach can be readily applied across broad panels of additional tumor types to search for novel oncogenes and gene fusions. The therapeutic potential to target gene fusions, as demonstrated by the success of imatinib (Gleevec) in inhibiting the product of the BCR-ABL1 gene fusion in CML (Kantarjian et al. 2002), further underscores the benefit and importance of systematically deploying massively parallel sequencing technologies for comprehensive cancer genomic characterization.
Methods
cDNA library construction
cDNA libraries were constructed for 10 melanoma samples (two cell lines, eight patient-derived short-term cultures) and for the CML cell line, K-562. Patient-derived short-term cultures originated from stage 4 tumors and were passaged 10–18 times in vitro prior to RNA extraction (Supplemental Table S1). For each sample, we removed DNA (Turbo DNA-free treatment, Ambion) and isolated polyA+ RNA from 25 μg of total RNA. We synthesized cDNA from 65 to 101 ng of polyA+ RNA, with a peak length of ∼1500 bases using the SuperScript Double-Stranded cDNA Synthesis kit (Invitrogen), SuperScript III Reverse Transcriptase (Invitrogen), and 10 ng random hexamer primers (Invitrogen). Primer annealing was done at room temperature for 10 min followed by 1 h at 55°C for first strand synthesis and 2 h at 16°C for second strand synthesis. Following second strand synthesis and cDNA clean-up, each cDNA library was sheared by sonication with four alternating cycles between “high intensity” (30 sec; duty cycle, 5%; intensity, 5%; cycles per burst, 200) and “low intensity” (5 sec; duty cycle, 1%; intensity, 1%; cycles per burst, 200) in the Frequency Sweeping mode (Covaris S2 machine). Paired-end adapters for Illumina sequencing were added following Illumina protocols, except that five times less adapter mix was ligated to the cDNAs. For PCR amplification, between 6- and 12-μL gel size–selected DNA were mixed with 20 μL of 10× PCR buffer, 10 μL of DMSO, 1.6 μL of 25 mM dNTP mix, 2 μL each of 25 μM PCR primer PE 1.0 and 2.0, and 2 μL of Herculase Hotstart DNA polymerase (Stratagene) in a final volume of 200 μL. The samples were denatured for 5 min at 95°C; 18 cycles of 20 sec at 95°C, 30 sec at 65°C, and 30 sec at 72°C; and final 7 min at 72°C to polish ends before cooling to 4°C. PCR primers were removed by using 1.8× volume of Agencourt AMPure PCR Purification kit (Agencourt Bioscience Corporation). All melanoma cDNA libraries were prepared with a fragment length range of 400–600 bp; two libraries were prepared for K-562, exhibiting shorter (300–400 bp) and longer (400–600 bp) fragments. Each library was loaded into its own single Illumina flow cell lane, producing an average of 14.5 million pairs of 51-mer reads per lane (8.4 million purity filtered read pairs), or nearly 1.5 Gb of total sequence for each sample.
Alignment of reads to the transcriptome and genome
To estimate transcript abundances from these paired 51-mer reads, all reads were independently aligned to a single reference file consisting of all human transcripts in Ensembl build 52 and the human genome (hg18). Alignments were performed using the Burrows-Wheeler Alignment Tool (BWA), allowing up to four mismatches with the reference (Li and Durbin 2009). Reads aligning to the transcriptome were mapped to their genomic coordinates, and alignments were considered unique if all placements to the reference arise from a single genomic locus. 68% of reads mapped to the references with no more than four mismatches (81% of these corresponded to annotated genes in Ensembl).
Read pairs were considered informative if both reads independently aligned uniquely. Duplicate read pairs (originating from a single original template molecule) were removed to leave a single read pair per fragment. Both reads aligned to a single genomic locus in the expected orientation for >97% of informative read pairs. These read pairs were used to identify somatic base-pair mutations; the remaining read pairs were used to discover novel chimeric transcripts (gene fusions).
Identification of candidate gene fusions
Read pairs were selected as candidate fusion events if both reads mapped uniquely to different protein-coding genes, with one originating from the transcribed strand and the other originating from the antisense strand. (Reads mapping within 1 megabase (Mb) on the same chromosome in the expected orientation were temporarily set aside as possible unannotated transcripts or readthrough events.) Reads in discordant pairs were trimmed by 10 nucleotides (nt) at each end to 31 nt total and realigned to all Ensembl52 transcripts using BWA, allowing up to four mismatches. Discordant read pairs for which trimmed reads could be placed on a single gene were filtered out. To select the highest confidence candidates for validation, we searched the set of 51-mer reads that did not map to Ensembl transcripts or to the genome for the presence of at least one individual read containing a hypothetical fusion point between any two exons in the corresponding genes. (A reference file was created for each candidate gene fusion consisting of all hypothetical junctions between an exon of the 5′ gene and an exon of the 3′ gene. Unmapped reads were screened against this reference using BWA.) All events for which the positions of the read pairs and individual fusion-spanning reads were consistent with a gene fusion were prioritized for further study. This analysis revealed three high-confidence gene fusions in K-562 and 11 high-confidence gene fusions in melanoma.
To model the sensitivity for detecting gene fusions, we considered each gene in the Ensembl database (18,615 total) to harbor a single fusion point (Supplemental Fig. S12). Using the observed number of read pairs mapping to each gene in the K-562 sample, the length of each transcript, and the average cDNA fragment length amplified in an Illumina cluster, we calculated the probability of detecting this fusion point in each gene. We modeled the number of read pairs mapping to each transcript as a Poisson distribution, the mapped position of read pairs as a uniform distribution, and the cDNA fragment length as a Gaussian distribution (428 ± 105, as observed for K-562), and we postulated the breakpoint to occur in only one of two copies of the gene.
Validation of gene fusions by RT-PCR and sequencing
Fusion candidates from the Illumina paired-end cDNA sequencing data analysis were validated experimentally on the transcript level by reverse transcriptase PCR (RT-PCR). Total RNA was extracted from each tumor sample, and cDNA was synthesized from 5 μg of RNA (Qiagen QuantiTect Reverse Transcription kit). Gene-specific PCR primers were designed to flank the hypothesized fusion breakpoints. Following PCR and gel electrophoresis, all RT-PCR amplified bands were gel excised (Qiagen QIAquick Gel Extraction kit) and subjected to Sanger sequencing. All 11 candidate melanoma gene fusions (including two splice isoforms for two separate gene fusions) were confirmed using this approach. Primer sequences for RT-PCR are displayed in Supplemental Table S6.
Long-range genomic PCR
Gene fusions were validated on the genomic level using long-range PCR (LR-PCR). The breakpoints in genomic DNA producing the observed fusion transcripts were hypothesized to occur in the intron immediately following the upstream exon and the intron immediately preceding the downstream exon. For each gene of a given fusion pair, primers were designed along the genomic exon–intron boundaries. If either intron exceeded 15 kilobases (kb), primers were positioned in 15-kb increments. LR-PCR reactions (TakaRa LA PCR kit) were performed according to the manufacturers protocols. Following gel electrophoresis, all LR-PCR amplified bands were gel excised (Qiagen QIAquick Gel Extraction kit) and subjected to Sanger sequencing. Nine of 10 candidate fusions tested were confirmed at the genomic level based on the mapping of the end sequence products.
FISH
To assess for rearrangements of both the RECK and ALX3 loci, two unique break-apart FISH assays were designed using bacterial artificial chromosome (BAC) FISH probes to hybridize with the neighboring centromeric and telomeric regions of each gene. BAC clones were selected from the March 2006 build of the human genome using the University of California, Santa Cruz Genome Browser and were obtained from the BACPAC Resource Center (CHORI). For ALX3, the centromeric BAC clone was biotin-14-deoxycytidine triphosphate (dCTP)–labeled RP11-36L11 (eventually conjugated to produce a red signal), and the telomeric BAC clone was digoxigenin-dUTP–labeled RP11-195M16 (eventually conjugated to produce a green signal). For RECK, the centromeric BAC clone was biotin-14-dCTP–labeled RP11-58A20 (red), and the telomeric BAC clone was digoxigenin-dUTP–labeled RP11-112J3 (green). Probe preparation and hybridization procedure were performed as previously described (Perner et al. 2008). Probes were applied to preparations of M000921 preserved in 3:1 methanol:acetic acid. Assays were analyzed under a 100× oil immersion objective using a fluorescence microscope (Olympus) equipped with appropriate filters, a charge-coupled device camera, and the Cytovision FISH imaging and capturing software (Applied Imaging). For each case, at least 100 nuclei were analyzed.
Screening for recurrent translocations
After the candidate gene fusions were validated by RT-PCR, 90 additional melanoma samples were screened for the presence of each fusion. Of these samples, 39 were melanoma cell lines obtained from commercial sources, and 51 were patient-derived melanoma short-term cultures. The short-term cultures were kindly provided by M. Herlyn (Wistar Institute) or prepared as described previously (Hoek et al. 2008). RNA was extracted from each of these samples and cDNA was synthesized, as described above. RT-PCR was performed on each sample simultaneously in 96-well plate format using the primer pairs that were used to validate the fusions. cDNA prepared from the original samples served as positive controls. We were unable to detect recurrent fusions in new samples for any these chimeric transcripts.
SNP 6.0 arrays
DNA from melanomas M000921, M990802, and 501 Mel were hybridized to Affymetrix SNP Array 6.0 microarrays concurrently with 15 normal HapMap samples. SNP 6.0 data were processed from raw CEL files to segmented copy number data as described previously (Cancer Genome Atlas Research Network 2008). Briefly, raw Affymetrix CEL files were converted to a single value for each probe set representing a SNP allele or a copy number probe. Copy numbers were then inferred based upon estimating probe set-specific linear calibration curves, followed by normalization by the most similar HapMap normal samples. Segmentation of normalized log2 ratios was performed using the circular binary segmentation (CBS) algorithm.
Identification of read-through transcripts
Read pairs were selected as candidate readthrough events if (1) both reads mapped uniquely to different genes in Ensembl on the same chromosome and transcribed in a consistent orientation, (2) one read originated from the transcribed strand and the other read originated from the antisense strand, and (3) reads mapped within 1 Mb of each other. All candidate readthrough transcripts implicated by at least two distinct read pairs (244 total candidates) were screened for unmapped 51-mer reads containing a hypothetical fusion point between any two exons in the corresponding genes, using BWA as above. Forty-nine candidates harbored at least one fusion-spanning 51-mer read in a location consistent with the observed read pairs. These 49 readthrough transcripts were manually inspected in the UCSC Genome Browser for existing evidence of transcription. Twenty-two corresponded to annotated genes in other (non-Ensembl) databases and/or full-length human mRNAs, and 15 more were supported by spliced human ESTs in GenBank. The remaining 12 are novel readthrough transcripts.
Co-ocurrence of RB1 and CDKN2A deletions
Array CGH copy number data sets for 70 primary cutaneous melanomas (Curtin et al. 2005) were segmented using the GLAD method (gain and loss analysis of DNA) as described previously (Lin et al. 2008). For each sample, if the designated copy number status of the corresponding closest marker exceeded a log2-ratio threshold of −0.3, the locus was inferred to be deleted. Of the seven samples with a deletion at the RB1 locus, five also harbored deletions at CDKN2A (P = 0.022, χ2 test of homogeneity).
Mutation calling
Read pairs where both reads aligned uniquely to the same chromsome in the proper orientation (with duplicate pairs removed) were considered for sequence variant identification. Each position in an Ensembl transcript was assigned a LOD score indicating the likely accuracy of the call, according to the observed sequence coverage, allele distribution, base quality score, and reference genotype (hg18). Of 69,511,900 bases in Ensembl transcripts, an average of 8,145,636 total bases (and 4,687,155 CDS bases) exhibited LOD > 5. Bases that disagreed with the reference genome were classified as known SNPs if present in dbSNP (Sherry et al. 2001) (build 130), or novel variants. We estimated the sensitivity for calling nonreference variants at 75% by comparing the rate of polymorphism at all high-coverage positions (≥50×) to the rate of polymorphism at all positions where the genotype was called (LOD > 5). Novel variants were discarded if they occurred in more than one sample (melanoma or K-562). While it is possible that this filtering step may eliminate bona fide recurrent variants, in practice, most recurrent events represent technical or analytical artifacts in this small discovery set.
Mutation validation by Sequenom
Candidate base pair variants called by RNA-seq were interrogated using an independent mass spectrometric genotyping technology (Sequenom iPLEX genotyping). Variants were amplified in multiplex PCR reactions consisting of up to 24 loci each, using 10 ng of template DNA from a single melanoma sample. Single base extension was performed on the shrimp alkaline phosphatase treated PCR product using iPLEX enzyme and mass-modified terminators (Sequenom iPLEX-GOLD reagents kit). SpectroCHIPs with 384-wells were analyzed by a MassArray MALDI-TOF Compact system with a solid phase laser mass spectrometer (Bruker Daltonics Inc.). The resulting spectra were called and analyzed by the SpectroTyper v.4.0 software.
Assays were designed for 103 randomly chosen protein-coding variants in two melanoma samples (and their corresponding normals), including 95 putative nonsynonymous coding mutations. Additionally, two nonsynonymous variants in ANKHD1 and SCAMP2 were tested in the unmatched melanoma cell line MeWo. We observed an overall validation rate of 95% in the tumor DNA, with 67% of these true variants also present in the matched normal germline DNA. In all, we validated 29 novel somatic coding mutations (27 nonsynonymous). (Eight assays were technical failures.)
Acknowledgments
We thank Cory Johannessen, Gordon Saksena, Scott Carter, William Lin, Carsten Russ, Jim Bochicchio, and the staff of the Broad Institute Biological Samples Platform, Genome Analysis Platform, and Genome Sequencing Platform. We thank the Novartis Institute of BioMedical Research and the Genomics Institute of the Novartis Research Foundation for providing RNA from melanoma cell lines. This work was funded by grants from the Novartis Institutes of Biomedical Research, the Starr Cancer Consortium, and the Adelson Medical Research Foundation.
Footnotes
[Supplemental material is available online at http://www.genome.org. The sequencing and microarray data from this study have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under SuperSeries accession no. GSE17593.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.103697.109.
References
- Albert TJ, Molla MN, Muzny DM, Nazareth L, Wheeler D, Song X, Richmond TA, Middle CM, Rodesch MJ, Packard CJ, et al. Direct selection of human genomic loci by microarray hybridization. Nat Methods. 2007;4:903–905. doi: 10.1038/nmeth1111. [DOI] [PubMed] [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacker D, Wilcox MA, Laird NM, Rodes L, Horvath SM, Go RC, Perry R, Watson B, Jr, Bassett SS, McInnis MG, et al. Alpha-2 macroglobulin is genetically associated with Alzheimer disease. Nat Genet. 1998;19:357–360. doi: 10.1038/1243. [DOI] [PubMed] [Google Scholar]
- Campbell PJ, Stephens PJ, Pleasance ED, O'Meara S, Li H, Santarius T, Stebbings LA, Leroy C, Edkins S, Hardy C, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YZ, Bennett CL, Huynh HM, Blair IP, Puls I, Irobi J, Dierick I, Abel A, Kennerson ML, Rabin BA, et al. DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4) Am J Hum Genet. 2004;74:1128–1135. doi: 10.1086/421054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloonan N, Forrest AR, Kolle G, Gardiner BB, Faulkner GJ, Brown MK, Taylor DF, Steptoe AL, Wani S, Bethel G, et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods. 2008;5:613–619. doi: 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- Craig DW, Pearson JV, Szelinger S, Sekar A, Redman M, Corneveaux JJ, Pawlowski TL, Laub T, Nunn G, Stephan DA, et al. Identification of genetic variants using bar-coded multiplexed sequencing. Nat Methods. 2008;5:887–893. doi: 10.1038/nmeth.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin JA, Fridlyand J, Kageshita T, Patel HN, Busam KJ, Kutzner H, Cho KH, Aiba S, Brocker EB, LeBoit PE, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353:2135–2147. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- Drobetsky EA, Grosovsky AJ, Glickman BW. The specificity of UV-induced mutations at an endogenous locus in mammalian cells. Proc Natl Acad Sci. 1987;84:9103–9107. doi: 10.1073/pnas.84.24.9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Widlund HR, Horstmann MA, Ramaswamy S, Ross K, Huber WE, Nishimura EK, Golub TR, Fisher DE. Critical role of CDK2 for melanoma growth linked to its melanocyte-specific transcriptional regulation by MITF. Cancer Cell. 2004;6:565–576. doi: 10.1016/j.ccr.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Forbes SA, Bhamra G, Bamford S, Dawson E, Kok C, Clements J, Menzies A, Teague JW, Futreal PA, Stratton MR. The Catalogue of Somatic Mutations in Cancer (COSMIC) Curr Protoc Hum Genet. 2008;57:10.11.1–10.11.26. doi: 10.1002/0471142905.hg1011s57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, Beroukhim R, Milner DA, Granter SR, Du J, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, Fennell T, Giannoukos G, Fisher S, Russ C, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier LW, Reinke V, Green P, Hirst M, Marra MA, Waterston RH. Massively parallel sequencing of the polyadenylated transcriptome of C. elegans. Genome Res. 2009;19:657–666. doi: 10.1101/gr.088112.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges E, Xuan Z, Balija V, Kramer M, Molla MN, Smith SW, Middle CM, Rodesch MJ, Albert TJ, Hannon GJ, et al. Genome-wide in situ exon capture for selective resequencing. Nat Genet. 2007;39:1522–1527. doi: 10.1038/ng.2007.42. [DOI] [PubMed] [Google Scholar]
- Hoek KS, Schlegel NC, Eichhoff OM, Widmer DS, Praetorius C, Einarsson SO, Valgeirsdottir S, Bergsteinsdottir K, Schepsky A, Dummer R, et al. Novel MITF targets identified using a two-step DNA microarray strategy. Pigment Cell Melanoma Res. 2008;21:665–676. doi: 10.1111/j.1755-148X.2008.00505.x. [DOI] [PubMed] [Google Scholar]
- Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantarjian H, Sawyers C, Hochhaus A, Guilhot F, Schiffer C, Gambacorti-Passerini C, Niederwieser D, Resta D, Capdeville R, Zoellner U, et al. Hematologic and cytogenetic responses to imatinib mesylate in chronic myelogenous leukemia. N Engl J Med. 2002;346:645–652. doi: 10.1056/NEJMoa011573. [DOI] [PubMed] [Google Scholar]
- Krishnakumar S, Zheng J, Wilhelmy J, Faham M, Mindrinos M, Davis R. A comprehensive assay for targeted multiplex amplification of human DNA sequences. Proc Natl Acad Sci. 2008;105:9296–9301. doi: 10.1073/pnas.0803240105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski TJ, Jr, Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- Levin JZ, Berger MF, Adiconis X, Rogov P, Melnikov A, Fennell T, Nusbaum C, Garraway LA, Gnirke A. Targeted next-generation sequencing of a cancer transcriptome enhances detection of sequence variants and novel fusion transcripts. Genome Biol. 2009;10:R115. doi: 10.1186/gb-2009-10-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley TJ, Mardis ER, Ding L, Fulton B, McLellan MD, Chen K, Dooling D, Dunford-Shore BH, McGrath S, Hickenbotham M, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wang J, Mor G, Sklar J. A neoplastic gene fusion mimics trans-splicing of RNAs in normal human cells. Science. 2008;321:1357–1361. doi: 10.1126/science.1156725. [DOI] [PubMed] [Google Scholar]
- Lin WM, Baker AC, Beroukhim R, Winckler W, Feng W, Marmion JM, Laine E, Greulich H, Tseng H, Gates C, et al. Modeling genomic diversity and tumor dependency in malignant melanoma. Cancer Res. 2008;68:664–673. doi: 10.1158/0008-5472.CAN-07-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher CA, Kumar-Sinha C, Cao X, Kalyana-Sundaram S, Han B, Jing X, Sam L, Barrette T, Palanisamy N, Chinnaiyan AM. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009a;458:97–101. doi: 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher CA, Palanisamy N, Brenner JC, Cao X, Kalyana-Sundaram S, Luo S, Khrebtukova I, Barrette TR, Grasso C, Yu J, et al. Chimeric transcript discovery by paired-end transcriptome sequencing. Proc Natl Acad Sci. 2009b;106:12353–12358. doi: 10.1073/pnas.0904720106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda S, Giliberto L, Matsuda Y, Davies P, McGowan E, Pickford F, Ghiso J, Frangione B, D'Adamio L. The familial dementia BRI2 gene binds the Alzheimer gene amyloid-β precursor protein and inhibits amyloid-β production. J Biol Chem. 2005;280:28912–28916. doi: 10.1074/jbc.C500217200. [DOI] [PubMed] [Google Scholar]
- McCarroll SA, Kuruvilla FG, Korn JM, Cawley S, Nemesh J, Wysoker A, Shapero MH, de Bakker PI, Maller JB, Kirby A, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- Moreira MC, Klur S, Watanabe M, Nemeth AH, Le Ber I, Moniz JC, Tranchant C, Aubourg P, Tazir M, Schols L, et al. Senataxin, the ortholog of a yeast RNA helicase, is mutant in ataxia-ocular apraxia 2. Nat Genet. 2004;36:225–227. doi: 10.1038/ng1303. [DOI] [PubMed] [Google Scholar]
- Morin R, Bainbridge M, Fejes A, Hirst M, Krzywinski M, Pugh T, McDonald H, Varhol R, Jones S, Marra M. Profiling the HeLa S3 transcriptome using randomly primed cDNA and massively parallel short-read sequencing. Biotechniques. 2008;45:81–94. doi: 10.2144/000112900. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, Shaffer T, Wong M, Bhattacharjee A, Eichler EE, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibbe RK, Markowitz S, Myeroff L, Ewing R, Chance MR. Discovery and scoring of protein interaction subnetworks discriminative of late stage human colon cancer. Mol Cell Proteomics. 2009;8:827–845. doi: 10.1074/mcp.M800428-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowell PC, Hungerford DA. A minute chromosome in human chronic leukemia. Science. 1960;132:1497. [Google Scholar]
- Okou DT, Steinberg KM, Middle C, Cutler DJ, Albert TJ, Zwick ME. Microarray-based genomic selection for high-throughput resequencing. Nat Methods. 2007;4:907–909. doi: 10.1038/nmeth1109. [DOI] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- Perner S, Wagner PL, Demichelis F, Mehra R, Lafargue CJ, Moss BJ, Arbogast S, Soltermann A, Weder W, Giordano TJ, et al. EML4-ALK fusion lung cancer: A rare acquired event. Neoplasia. 2008;10:298–302. doi: 10.1593/neo.07878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porreca GJ, Zhang K, Li JB, Xie B, Austin D, Vassallo SL, LeProust EM, Peck BJ, Emig CJ, Dahl F, et al. Multiplex amplification of large sets of human exons. Nat Methods. 2007;4:931–936. doi: 10.1038/nmeth1110. [DOI] [PubMed] [Google Scholar]
- Prensner JR, Chinnaiyan AM. Oncogenic gene fusions in epithelial carcinomas. Curr Opin Genet Dev. 2009;19:82–91. doi: 10.1016/j.gde.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbitts TH, Forster A, Larson R, Nathan P. Fusion of the dominant negative transcription regulator CHOP with a novel gene FUS by translocation t(12;16) in malignant liposarcoma. Nat Genet. 1993;4:175–180. doi: 10.1038/ng0693-175. [DOI] [PubMed] [Google Scholar]
- Rowley JD. A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- Sawyers CL. The cancer biomarker problem. Nature. 2008;452:548–552. doi: 10.1038/nature06913. [DOI] [PubMed] [Google Scholar]
- Shah SP, Morin RD, Khattra J, Prentice L, Pugh T, Burleigh A, Delaney A, Gelmon K, Guliany R, Senz J, et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461:809–813. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijders AM, Schmidt BL, Fridlyand J, Dekker N, Pinkel D, Jordan RC, Albertson DG. Rare amplicons implicate frequent deregulation of cell fate specification pathways in oral squamous cell carcinoma. Oncogene. 2005;24:4232–4242. doi: 10.1038/sj.onc.1208601. [DOI] [PubMed] [Google Scholar]
- Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart D, Sellers WR. Linking somatic genetic alterations in cancer to therapeutics. Curr Opin Cell Biol. 2009;21:304–310. doi: 10.1016/j.ceb.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Sugarbaker DJ, Richards WG, Gordon GJ, Dong L, De Rienzo A, Maulik G, Glickman JN, Chirieac LR, Hartman ML, Taillon BE, et al. Transcriptome sequencing of malignant pleural mesothelioma tumors. Proc Natl Acad Sci. 2008;105:3521–3526. doi: 10.1073/pnas.0712399105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, Seifert M, Borodina T, Soldatov A, Parkhomchuk D, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- Takahashi C, Sheng Z, Horan TP, Kitayama H, Maki M, Hitomi K, Kitaura Y, Takai S, Sasahara RM, Horimoto A, et al. Regulation of matrix metalloproteinase-9 and inhibition of tumor invasion by the membrane-anchored glycoprotein RECK. Proc Natl Acad Sci. 1998;95:13221–13226. doi: 10.1073/pnas.95.22.13221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieling H, Venema J, van Rooyen ML, van Hoffen A, Menichini P, Zdzienicka MZ, Simons JW, Mullenders LH, van Zeeland AA. Strand specificity for UV-induced DNA repair and mutations in the Chinese hamster HPRT gene. Nucleic Acids Res. 1991;19:2411–2415. doi: 10.1093/nar/19.9.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassour M, Kaplan T, Fraser HB, Levin JZ, Pfiffner J, Adiconis X, Schroth G, Luo S, Khrebtukova I, Gnirke A, et al. Ab initio construction of a eukaryotic transcriptome by massively parallel mRNA sequencing. Proc Natl Acad Sci. 2009;106:3264–3269. doi: 10.1073/pnas.0812841106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Caballero OL, Levy S, Stevenson BJ, Iseli C, de Souza SJ, Galante PA, Busam D, Leversha MA, Chadalavada K, et al. Transcriptome-guided characterization of genomic rearrangements in a breast cancer cell line. Proc Natl Acad Sci. 2009;106:1886–1891. doi: 10.1073/pnas.0812945106. [DOI] [PMC free article] [PubMed] [Google Scholar]