Abstract
The global diabetes epidemic poses a major challenge. Epigenetic events contribute to the etiology of diabetes; however, the lack of epigenomic analysis has limited the elucidation of the mechanistic basis for this link. To determine the epigenetic architecture of human pancreatic islets we mapped the genome-wide locations of four histone marks: three associated with gene activation—H3K4me1, H3K4me2, and H3K4me3—and one associated with gene repression, H3K27me3. Interestingly, the promoters of the highly transcribed insulin and glucagon genes are occupied only sparsely by H3K4me2 and H3K4me3. Globally, we identified important relationships between promoter structure, histone modification, and gene expression. We demonstrated co-occurrences of histone modifications including bivalent marks in mature islets. Furthermore, we found a set of promoters that is differentially modified between islets and other cell types. We also use our histone marks to determine which of the known diabetes-associated single-nucleotide polymorphisms are likely to be part of regulatory elements. Our global map of histone marks will serve as an important resource for understanding the epigenetic basis of type 2 diabetes.
Genetic and epigenetic factors determine cell fate and function. Recent breakthroughs in genotyping technology have led to the identification of more than 20 loci associated with the risk of type 2 diabetes (Sambuy 2007; Zhao et al. 2009). However, all together these loci explain <5% of the genetic risk for diabetes. Epigenetic events have been implicated as contributing factors for metabolic diseases (Barker 1988; Kaput et al. 2007). Unhealthy diet and a sedentary lifestyle likely lead to epigenetic changes that can, in turn, contribute to the onset of diabetes (Kaput et al. 2007). At present, the underlying molecular mechanisms for disease progression remain to be elucidated.
Epigenetic modifications encompass both DNA methylation and histone modifications (Cedar and Bergman 2009). In recent years, genome-wide maps of epigenetic marks have been generated for yeast (Pokholok et al. 2005) and several cell types in mice and humans (Bernstein et al. 2005; Roh et al. 2006; Barski et al. 2007; Mikkelsen et al. 2007; Pan et al. 2007; Zhao et al. 2007). However, no genome-wide map of histone modifications has been reported for the human pancreatic islet, a key player in the etiology of diabetes. In the present study, we have used chromatin immunoprecipitation with massively parallel sequencing (ChIP-seq) technology to create a genome-wide map of four histone modifications associated with gene activation or repression in human pancreatic islets.
Results and Discussion
Sample preparation and initial assessment
Chromatin from pancreatic islets was immunoprecipitated with antihistone antibodies to enrich for DNA fragments in regions of the genome associated with modified histones. Enrichment of occupancy by modified histone was validated using quantitative PCR (qPCR) (Supplemental Fig. 1; Supplemental Table 1). H3K4me1, H3K4me2, and H3K4me3 are frequently found near active gene promoters, while H3K4me1 is also often associated with enhancers, and inactive genes are associated with H3K27me3 (Strahl and Allis 2000; Bernstein et al. 2005; Mikkelsen et al. 2007; Roh et al. 2007). We found enrichment of H3K4me1 near the start site and the enhancer of the PDX1 gene, a transcription factor important for insulin gene regulation and beta-cell differentiation. H3K4me2 and very strong H3K4me3 occupancy was observed at the promoter of GAPDH (a housekeeping gene) and, surprisingly, at significantly lower levels at the insulin and glucagon promoters. The sparse methylation patterns at the highly active promoters of these major islet hormones suggest that epigenetic regulation of these genes in human islets is different from the regulation in mouse pancreatic islets and cell lines, which is dependent on hypermethylation of histone H3 at the insulin promoter (Deering et al. 2009).
Our qPCR results for H3K27me3 indicate a strong enrichment at the NEUROG3 promoter, a regulator of fetal islet development that is repressed during adult life. We also observed negligible enrichment of this repressive mark at the GAPDH promoter, which is consistent with the notion that H3K27me3 is absent in active genes. Surprisingly, we found that the promoter region of ALB, a liver-specific gene that is not expressed in the endocrine pancreas, exhibited very low levels of the H3K27me3 modification, indicating that a different mechanism must be responsible for its repression in the pancreas.
Genome-wide identification of enriched levels of histone modifications
Having assessed our ChIP results using gene-specific qPCR, we proceeded with the global location analysis for these four chromatin marks. Following next-generation sequencing and alignment of enriched DNA (see Supplemental Table 2 for numbers of sequence reads obtained), we computed the average profile of each modification with respect to the transcription start sites (TSSs) of known genes to verify that the results followed the expected patterns. We assigned a summary modification level to each gene based on the number of reads (per million sequence reads per kilobase pairs of DNA) that mapped to the first 2000 bp just downstream from the TSS (Supplemental Fig. 2). We verified summary modification levels of at least four genes for each mark using qPCR (Supplemental Fig. 3).
Next, we identified regions of statistically significant enrichment for H3K4me1 (189,103), H3K4me2 (31,906), H3K4me3 (36,763), and H3K27me3 (122,629) using the GLITR algorithm (Tuteja et al. 2009). The GLITR algorithm requires contiguous, overlapping, extended reads to identify a region of enrichment, which in many cases results in a locus of modification being split into a few pieces. We therefore merged nearby GLITR regions to create GLITR loci. We annotated these loci by proximity to RefSeqs, Known Genes, and miRNAs. We found 3826 (16.5%) of the H3K4me3 loci were >5 kbp from the nearest gene, indicating a large number of potentially novel transcriptional start sites active in islets (or possibly in contaminating exocrine tissue.) A larger fraction of H3K4me1 (34,994, 24.8%) and H3K4me2 (7762, 24.3%) loci were intergenic and represent potential regulatory regions.
The potential of H3K4me1 to identify regulatory regions enabled us to determine which of the previously identified noncoding diabetes-associated single nucleotide polymorphisms (SNPs) are most likely to be functional. We found that 18 of the 113 SNPs (15.9%) were within 500 bp of a GLITR H3K4me1 locus (Table 1). Fewer SNPs were near the H3K4me2 and H3K4me3 loci—four and two SNPs, respectively—which suggests that the H3K4me1-enriched SNPs are in distal or internal regulatory regions. Interestingly, four SNPs were associated with H3K27me3 regions, indicative of a repressed chromatin state in islets, and thus the corresponding genes may exert their effects on glucose homeostasis in other tissues or cause local disruption of the H3K27me3 distribution. We suggest that the 18 SNPs listed in Table 1, which are occupied by H3K4me1 in human islets and are therefore in potential enhancers, might be given priority in functional testing in the future.
Table 1.
Diabetes-associated SNPs marked by H3K4me1
We obtained 113 GWAS-identified SNPs associated with human diabetes from http://www.genome.gov/gwastoexcel.cfm. We then extracted chromosomal location and associated genes for the SNP from NCBI using eFetch. The 18 SNPs that are within 500 bp of an H3K4me1 GLITR region are listed. Because H3K4me1 is frequently associated with enhancers, the SNPs listed have the potential to affect gene expression of associated genes.
aKIAA0350 is currently known as CLEC16A.
The relationship of histone modifications and expression level depends on CpG island status
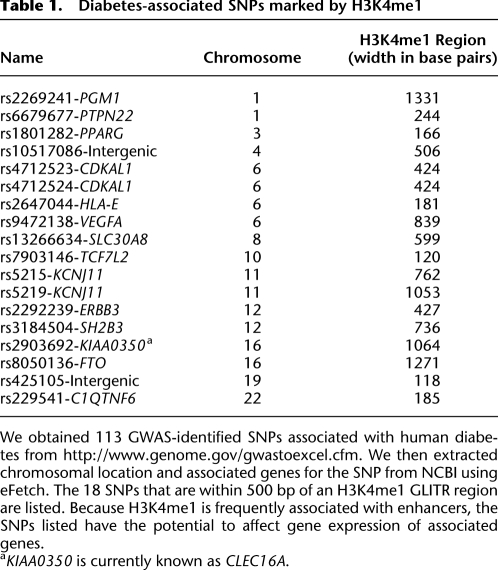
To explore the relationship between chromatin modifications and gene expression, we compared the expression levels of each gene to the input-normalized chromatin modification level of the gene as measured in a region 2 kb downstream from its TSS. We grouped all genes into five equally sized sets organized by expression level and computed histograms of the levels of histone modification for each of these gene sets. This analysis demonstrated that the behavior of H3K4me1 (Fig. 1A), H3K4me2 (Fig. 1B), H3K4me3 (Fig. 1C), and H3K27me3 (Fig. 1D) modifications is dependent on the presence (CpG+) or absence (CpG−) of a CpG island in the promoter (details in Supplemental Table 4.)
Figure 1.
Input-normalized modification levels of H3K4me1 (A), H3K4me2 (B), H3K4me3 (C), and H3K27me3 (D) depend on expression level and promoter type. Transcriptional start sites with a nearby CpG island (CpG+, yellow) show a relatively constant distribution of the H3K4me1 and H3K4me2 modifications levels that does not strongly correlate with gene expression. H3K4me3 levels are correlated with gene expression, even though they exhibit large variability. On the other hand, CpG− promoters (green) are usually unmodified except at high expression levels, where they begin to show higher levels of modifications. Thus, for the most part, the correlation of histone modifications and gene expression is due to a gradual increase in the proportion of CpG+ promoters at higher expression levels. The distribution of H3K27me3 is different, in accordance with its association with repression. At low expression levels, both CpG− and CpG+ promoters are occupied, and the overall levels of H3K27me3 modification decrease with increasing expression.
Within a given expression-level group, the median level of H3K4me1, H3K4me2, and H3K4me3 is dramatically higher on CpG+ genes than CpG− genes. The median level of H3K4me1 and H3K4me2 modification increases with expression, but the total shift from very low to very high expression is much less than the standard deviation of the distribution of the modification level. For example, the median modification level of H3K4me1 increased from 1.1-fold over input chromatin to 1.5 from very low to very highly expressed genes, but was accompanied by a standard deviation of about 0.8-fold. H3K4me3 showed a stronger correlation with gene expression, but was still quite modest in genes above the very low expression class, i.e., a total shift of 3.5-fold for CpG+ genes in a distribution with a 6.9 standard deviation. Interestingly, for all levels of H3K4 methylation, the shift in the median modification level of all genes (from very low expression to very high expression) is larger than either of the CpG+ or CpG− median shifts, indicating that a major component of the observed correlation between H3K4me* modification and expression is the shift from CpG− to CpG+ genes with increasing expression. For example, the shift of fold enrichment for very low to very highly expressed genes for CpG+ H3K4me3 was 6.4, whereas the total shift for all genes was 10.3. H3K27me3 does not appear to distinguish between the two promoter types to a significant degree (Fig. 1D). Overall, the level of H3K27me3 modification is inversely proportional to gene expression, which is consistent with its association with repression.
It is important to emphasize that the range of histone modifications is nearly constant from low to high expression. A promoter can be quite highly modified and yet be found in any of the expression classes. Interestingly, CpG− genes were generally unmodified, except at very high gene-expression levels. However, even at this level of expression, only about half of the genes have any evidence of H3K4me3 modification. Thus, the low H3K4me3 histone occupancy we had observed for insulin and glucagon by qPCR is not an exception, but rather follows a pattern found in many other highly expressed genes. It has been previously reported that CpG+ genes are generally housekeeping genes and are widely expressed, whereas CpG− genes are more tissue specific (Schug et al. 2005). Our current results indicate that the chromatin state of these two gene classes is different as well.
Co-occurrence of histone marks
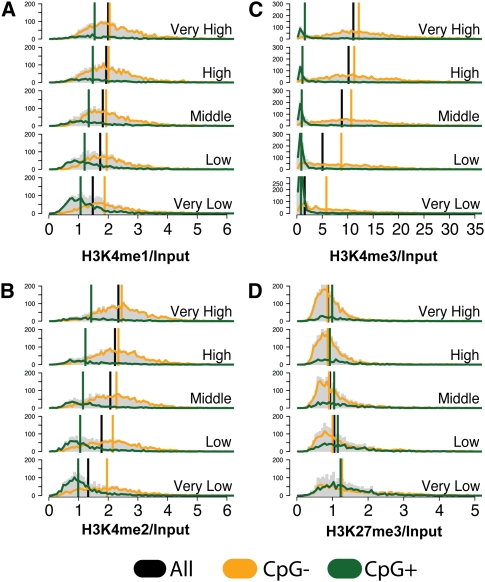
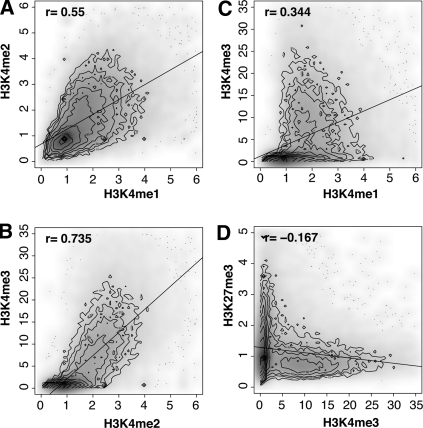
Histone modifications are known to act in a combinatorial fashion to determine the overall outcome of gene expression, so the colocalization of these marks may play an important role in regulating chromatin structure. We found that H3K4me1 and H3K4me2 are generally correlated (r = 0.55) (Fig. 2A), though there is considerable variability. The correlation is higher between H3K4me2 and H3K4me3 (r = 0.735) (Fig. 2B). However, correlation is limited between H3K4me1 and H3K4me3 (Fig. 2C). There exists a population of genes on which we observe occupancy by H3K4me1, but very little H3K4me3; another population demonstrates the opposite modification pattern. This likely reflects the total conversion of H3K4me1 (through H3K4me2) to H3K4me3. A clear example of the co-occurrence of H3K4me1, H3K4me2, and H3K4me3 marks can be seen at the ATF4 gene (Fig. 3A). A very sharp double peak of H3K4me3 is observed at the TSS, with weaker flanking peaks for H3K4me1 and H3K4me2 modifications.
Figure 2.
Co-occurrence of input-normalized histone modification near the promoters (0–2000 bp) in human islets: (A) H3K4me2 vs. H3K4me1, (B) H3K4me3 vs. H3K4me2, (C) H3K4me3 vs. H3K4me1, and (D) H3K27me3 vs. H3K4me3. (A–C) The H3K4me1,2,3 series tends to either be completely absent (values near 1), or to co-occur (values near the diagonal). (D) H3K27me3 and H4K4me3 tend not to co-occur, with a few exceptions (bivalent genes).
Figure 3.
Marks of active and repressed genes in islets. (A) Human islet chromatin enriched for H3K4me1, H3K4me2, H3K4me3, H3K27me3, and control input DNA from four samples was processed and sequenced. Reads were pooled and aligned to the NCBI Genome Build 36.1-hg18, to determine regions that were enriched for binding by modified histones. Note the strong double peak surrounding the transcriptional start site (left end) of ATF4, and the weaker peaks for H3K4me1 and H3K4me2. In B, the HOXB cluster contains significantly enriched regions for H3K27me3 as well as H3K4me3 at several transcription start sites.
Studies using embryonic stem (ES) cells have demonstrated the existence of “bivalent domains” in genes encoding developmental regulators (Boyer et al. 2006; Lee et al. 2006; Pan et al. 2007; Zhao et al. 2007). Recent evidence confirming the presence of bivalent domains in mouse embryonic fibroblasts cells, neural progenitor cells (Mikkelsen et al. 2007), and human T cells (Roh et al. 2006) has shown that these domains are not restricted to ES cells. A comparison of H3K4me3 and H3K27me3 in islets (Fig. 2D) indicates that these marks are usually mutually exclusive (or both absent), although there are a few genes with high levels of both marks. We found that developmental genes like the HOX gene clusters and neuronal transcription factors are bivalently marked in human pancreatic islets with both H3K4me3 at several transcriptional start sites and H3K27me3 covering the entire locus (Fig. 3B; data not shown). As these genes are important during early development and not expressed in adult islets, the presence of H3K27me3 is unsurprising. The role of H3K4me3 levels in these domains is unknown, but may be related to the retention of “memory” of earlier transcription or the reactivation of these genes under specific metabolic conditions.
The major hormone-encoding genes in the islet are not enriched for H3K4me3
Surprisingly, our ChIP-seq data indicate that H3K4me3 marks the insulin and glucagon promoters only very sparingly (Supplemental Figs. 4, 5), in contrast to the promoters of JUN, PDX1, and MAFB, which are heavily occupied by H3K4me3 despite their lower expression levels (Supplemental Figs. 6, 7A,B). Our initial hypothesis was that this lack of activating histone marks on the hormone-encoding gene promoters might be due to a complete absence of histones, perhaps due to the high transcription rate of these genes. To test this notion, we performed ChIP for total H3 histone at the promoters of JUN and GAPDH, which are occupied by H3K4me3, and insulin and glucagon, which are not (Supplemental Fig. 8). We found similar levels of histone H3 at all of these promoters; thus, the lack of H3K4me3 at the insulin and glucagon promoters is not due to a lack of histones. Therefore, alternative histone modifications or regulatory mechanisms must be responsible for the activation of these hormone-encoding genes in human islets.
Recent studies have demonstrated that the insulin and nearby genes in an extended 80-kb region are a part of a large, human islet-specific, open chromatin domain, and share a common control mechanism (Mutskov and Felsenfeld 2009). The presence of intergenic transcription in this region has been proposed to play a role in the maintenance of open chromatin structure, suggesting that a locus-specific control mechanism might be responsible for constitutive insulin gene expression in humans (Mutskov and Felsenfeld 2009). Our data also indicate a region (chr1:2,100,000–2,200,000 mm8) of high levels of H3K4me1, a mark associated with regulatory regions covering the insulin gene locus.
We also considered the possibility that the low levels of H3K4me3 modification at the insulin gene promoter might be due to a paucity of beta-cells in our samples. However, our ChIP-seq data suggest that this is not the case, as both PDX1 and MAFA (genes with expression restricted to beta-cells) are highly occupied by modified histones (Supplemental Fig. 7A; data not shown). Similarly, the promoter of MAFB (a gene expressed only in alpha-cells in the adult islet) is also occupied by H3K4me3 (Supplemental Fig. 7B). Furthermore, by qRT-PCR analysis, the expression of insulin and other pancreatic cell-specific genes is much higher than that of amylase, a marker for exocrine contamination (Supplemental Fig. 9). While we observed some variability in the levels of insulin mRNA expression between the individual donors, there was no significant difference in the H3K4me3 pattern at the insulin promoter between samples (Supplemental Fig. 10).
Tissue specificity of histone marks at promoters in human pancreatic islets
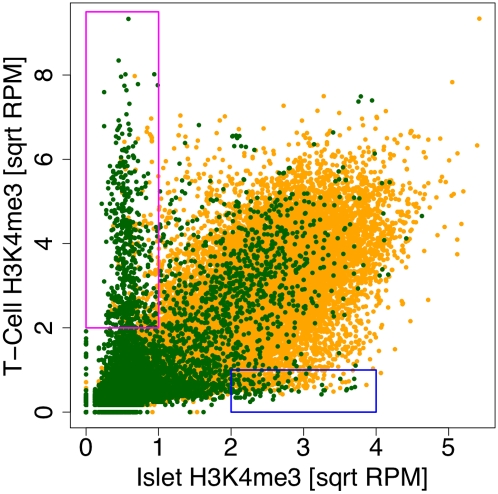
Previous efforts (Heintzman et al. 2009) have reported that chromatin structure at promoters is largely consistent between cells types and that the variation occurs at enhancers. To test the promoter part of this observation, we compared levels of H3K4me3 in islets with CD4+ T-cells (Barski et al. 2007). We found that while the levels of H3K4me3 on a majority of promoters were well correlated between islets and T-cells, there was a set of promoters that were differentially modified in the two tissues (Fig. 4). These genes include the small number of CpG− genes that showed higher H3K4me3 modification levels with higher expression levels in Figure 1. Thus, there are tissue-specific variations at the promoters of genes in terms of H3K4me3 occupancy. In addition, because most of the CpG− promoters that are expressed in islets are not marked with H3K4me3, it is possible that any unidentified marks correlated with CpG− gene expression exhibit tissue-specific variation at these promoters as well.
Figure 4.
Comparison of H3K4me3 at gene promoters in human islets and CD4+ T-cells. The square root of the summary input-normalized levels of H3K4me3 for CpG island-containing (yellow) and CpG island-less (green) genes are plotted. The CpG island-less genes in the magenta (395 genes) and blue (93 genes) boxes are modified only in one tissue or the other, indicating that this class of genes exhibits tissue-specific promoter modification.
Conclusion
This study represents the first description of the epigenome of the human pancreatic islet, a micro-organ of major importance in the regulation of metabolic disease and the etiology of diabetes. The pattern of histone modifications in the islet is complex. The correlation previously observed between H3K4me3 and gene expression levels, though upheld, was revealed to be dependent on the promoter architecture. Promoters containing a CpG island are likely to be methylated at H3K4 regardless of their expression level. On the other hand, many genes with CpG− promoters are not occupied by H3K4me3-modified histones even at the highest expression levels. This includes the two major hormone-producing genes. We also find, in contrast to previous reports, that there are tissue-specific differences in the histone methylation status of mammalian promoters. We have also demonstrated the presence of bivalent marks on many developmental regulators. Further studies are needed in order to establish the purpose of these marks and whether they are altered in disease states. This study will be useful to future researchers because of the fact that environment plays a large role in the etiology of diabetes, and is likely reflected in the epigenetic profile of the pancreatic islet.
Methods
Human islets
Islet isolation, viability, and purity was determined by the islet cell biology core of the DERC of the University of Pennsylvania as described previously (Deng et al. 2004). Purity of the samples used for ChIP-seq experiments and validation of ChIP-seq results were in a range of from 80% to 95%, and viability was in the range of from 84% to 97%, except for the sample A05 with the viability of 79% and the sample A10 with the purity of 70% (Supplemental Tables 2, 3).
RNA preparation and qPCR
Total RNA was prepared and its quality and quantity were assessed as described previously (Rieck et al. 2009). The cDNA preparation and qPCR were carried out as described previously (Rieck et al. 2009). GAPDH and beta-actin were used as housekeeping controls (Supplemental Fig. 9).
Chromatin preparation
Human pancreatic islets (10,000 islet equivalents from each donor) were sedimented by centrifugation at 13,000g for 30 sec and resuspended in 500 μL of PBS. Cells were processed and sonicated as described previously (Tuteja et al. 2008).
Chromatin immunoprecipitation (ChIP) and ChIP-seq
Immunoprecipitations were performed as described previously (Tuteja et al. 2008), except that 2 μg of chromatin and 2 μg of antibodies were used for each reaction. Chromatin was immunoprecipitated with antibodies for H3K4me3 (abcam catalog no. ab8580), H3K27me3 (Millipore catalog no. 07-449), H3K4me1 (abcam catalog no. ab8895), H3K4me2 (abcam catalog no. ab11946), and H3 (abcam catalog no. ab1791). qPCR was performed as described previously (Tuteja et al. 2008). Enrichment of target DNA regions was calculated using a control intergenic region as a reference and was quantified by comparing input DNA to the ChIP DNA. Primer sequences are listed in Supplemental Table 1.
The immunoprecipitated DNA was modified for sequencing following the Illumina protocol (Illumina, Inc.) (Supplemental Fig. 1). Modified DNA was assessed for size, purity, and quantity using an Agilent 2100 Bioanalyzer (Agilent Technologies) followed by 36-bp sequencing using an Illumina Genome Analyzer II according to the manufacturer's instructions. Sequenced DNA was aligned to the NCBI Genome Build 36.1 (hg18), allowing up to two mismatches, and without masking repeats. Reads for which the best alignment was unique were retained for further analysis. Reads for each modification were pooled to create a complete set for data analysis. Read quantities are summarized in Supplemental Table 2. Reads were loaded into a mysql database for further analysis.
Gene expression in human islets
mRNA from 31 samples of normal human islets was isolated following the protocol of Rieck et al. (2009), using either the Qiagen RNeasy or mirVana kits. Samples were then amplified and labeled using the Quick Amp Labeling Kit, One-Color (Agilent). Labeled DNA was hybridized for 17 h at 65°C on Agilent 4x44K Human Whole Genome chips. Slides were scanned with an Agilent Scanner (G2505C) and signals quantified with Agilent's Feature Extraction software (version 10.5). The gProcessedSignal was used for each spot and data were quantile normalized across all samples. The average intensity of each spot across replicates was used as the expression level. When a gene had multiple probes one was chosen arbitrarily.
Average modification profiles at gene transcription start sites.
As a basic validation of the data, we computed the average profile of each modification around the start site of known genes using locations downloaded from the UCSC Genome Browser (http://genome.ucsc.edu) for hg18. We extended the reads to a length of 100 bp and used bins of 20 bp across the region ±5 kb from each start site. Start sites were skipped if they were close to each other. The results are shown in Supplemental Figure 2. The H3K4me1, H3K4m2, and H3K4me3 profiles exhibit the strong bilateral peak with a central notch commonly observed. H3K27me3 is not focused near the TSS, but rather is a locus-wide mark and does not exhibit a peak.
Because the average profile exhibits significant upstream modification levels and because there are a number of divergently transcribed gene pairs, we refined this analysis so that we could determine a region to use for assigning a summary modification level to each gene. We created two groups of genes: those that are divergently transcribed (i.e., those with another gene's TSS within 1 kb) and those with no gene within 5 kb of the TSS (free genes). Using k-means clustering of the individual gene profiles (using the MeV program) in each group, we determined that there are a number of cases where the two genes in a divergently transcribed pair have different modification levels. Furthermore, about half of the free genes show a more asymmetric profile with very low upstream modification levels and much stronger downstream levels. Thus, we choose to use the region [0,2000] relative to the start site to assign an overall modification level to each gene.
Modification levels were quantified by counting the number of reads (extended to 100 bp) that overlapped with a target region. Read counts were normalized to reads per million per kilobase (RPMPK). When normalization to input was used, the modification level in RPMPK was divided by the input level in RPMPK. To summarize a gene's level of modification, we used the region of [0,2000] as it is least affected by nearby genes yet still captures the usual locus of modification.
Identifying significantly enriched regions
To identify peaks in modification data we used GLITR (Tuteja et al. 2009) and compared a sample of the reads for each histone modification and an equal number of input reads to a large group of input reads collected from a variety of human tissues. We choose regions that passed a false discovery rate cutoff of 1.5% as significant. As most of the modifications exhibit gaps in enrichment, e.g., the notch at the TSS, we group nearby GLITR regions into loci using distance cutoffs of 1000 bp for H3K4me3, 2435 bp for H3K4me2, 500 bp for H3K4me1, and 5000 bp for H3K27me3. The distances for H3K4me3 and H3K4me2 correspond to minima in the distribution in the log2 of the gaps between consecutive GLITR regions. The distributions of the other two modifications were unimodal, but kinks in the distribution suggested 500 bp for H3K4me1 and manual examination of the DLX5/6 region indicated that 5000 bp would create the appropriate locus.
External data
CpG island definitions and the locations of known genes were downloaded from the UCSC Genome Browser (Kuhn et al. 2009). Read alignments for Heintzman et al. (2009) were downloaded from http://dir.nhlbi.nih.gov/papers/lmi/epigenomes/data and processed in a similar fashion to our data.
Acknowledgments
We thank the organ donors and their families who made this study possible. We thank Ms. Amber Riblett for editing of the manuscript. This work was supported by the National Institutes of Health (USA) grants DK-R01-055342 (K.H.K.), as well as grants DK-U42RR016600 and U01 DK070430 (A.N.). This publication was made possible through core services provided by the DERC at the University of Pennsylvania from a grant sponsored by NIH DK-19525.
Footnotes
[Supplemental material is available online at http://www.genome.org. The ChIP-seq and gene expression data from this study have been submitted to ArrayExpress (http://www.ebi.ac.uk/microarray-as/ae/) under accession nos. E-MTAB-189 and E-MTAB-191, respectively.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.102038.109.
References
- Barker DJ. Childhood causes of adult diseases. Arch Dis Child. 1988;63:867–869. doi: 10.1136/adc.63.7.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, III, Gingeras TR, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. Linking DNA methylation and histone modification: Patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- Deering TG, Ogihara T, Trace AP, Maier B, Mirmira RG. Methyltransferase Set7/9 maintains transcription and euchromatin structure at islet-enriched genes. Diabetes. 2009;58:185–193. doi: 10.2337/db08-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng S, Vatamaniuk M, Huang X, Doliba N, Lian MM, Frank A, Velidedeoglu E, Desai NM, Koeberlein B, Wolf B, et al. Structural and functional abnormalities in the islets isolated from type 2 diabetic subjects. Diabetes. 2004;53:624–632. doi: 10.2337/diabetes.53.3.624. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaput J, Noble J, Hatipoglu B, Kohrs K, Dawson K, Bartholomew A. Application of nutrigenomic concepts to Type 2 diabetes mellitus. Nutr Metab Cardiovasc Dis. 2007;17:89–103. doi: 10.1016/j.numecd.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Kuhn RM, Karolchik D, Zweig AS, Wang T, Smith KE, Rosenbloom KR, Rhead B, Raney BJ, Pohl A, Pheasant M, et al. The UCSC Genome Browser Database: Update 2009. Nucleic Acids Res. 2009;37:D755–D761. doi: 10.1093/nar/gkn875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutskov V, Felsenfeld G. The human insulin gene is part of a large open chromatin domain specific for human islets. Proc Natl Acad Sci. 2009;106:17419–17424. doi: 10.1073/pnas.0909288106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, Jonsdottir GA, Stewart R, Thomson JA. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Rieck S, White P, Schug J, Fox AJ, Smirnova O, Gao N, Gupta RK, Wang ZV, Scherer PE, Keller MP, et al. The transcriptional response of the islet to pregnancy in mice. Mol Endocrinol. 2009;23:1702–1712. doi: 10.1210/me.2009-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh TY, Cuddapah S, Cui K, Zhao K. The genomic landscape of histone modifications in human T cells. Proc Natl Acad Sci. 2006;103:15782–15787. doi: 10.1073/pnas.0607617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh TY, Wei G, Farrell CM, Zhao K. Genome-wide prediction of conserved and nonconserved enhancers by histone acetylation patterns. Genome Res. 2007;17:74–81. doi: 10.1101/gr.5767907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambuy Y. Sideways Glance: Genome wide association studies for type 2 diabetes mellitus. Genes Nutr. 2007;2:245–248. doi: 10.1007/s12263-007-0061-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug J, Schuller WP, Kappen C, Salbaum JM, Bucan M, Stoeckert CJ., Jr Promoter features related to tissue specificity as measured by Shannon entropy. Genome Biol. 2005;6:R33. doi: 10.1186/gb/-2005-6-4-r33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Tuteja G, Jensen ST, White P, Kaestner KH. Cis-regulatory modules in the mammalian liver: Composition depends on strength of Foxa2 consensus site. Nucleic Acids Res. 2008;36:4149–4157. doi: 10.1093/nar/gkn366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja G, White P, Schug J, Kaestner KH. Extracting transcription factor targets from ChIP-Seq data. Nucleic Acids Res. 2009;37:e113. doi: 10.1093/nar/gkp536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XD, Han X, Chew JL, Liu J, Chiu KP, Choo A, Orlov YL, Sung WK, Shahab A, Kuznetsov VA, et al. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell. 2007;1:286–298. doi: 10.1016/j.stem.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Zhao J, Li M, Bradfield JP, Wang K, Zhang H, Sleiman P, Kim CE, Annaiah K, Glaberson W, Glessner JT, et al. Examination of type 2 diabetes loci implicates CDKAL1 as a birth weight gene. Diabetes. 2009;58:2414–2418. doi: 10.2337/db09-0506. [DOI] [PMC free article] [PubMed] [Google Scholar]