Abstract
Understanding the causes and consequences of variation in the rate of recombination is essential since this parameter is considered to affect levels of genetic variability, the efficacy of selection, and the design of association and linkage mapping studies. However, there is limited knowledge about the factors governing recombination rate variation. We genotyped 1920 single nucleotide polymorphisms in a multigeneration pedigree of more than 1000 zebra finches (Taeniopygia guttata) to develop a genetic linkage map, and then we used these map data together with the recently available draft genome sequence of the zebra finch to estimate recombination rates in 1 Mb intervals across the genome. The average zebra finch recombination rate (1.5 cM/Mb) is higher than in humans, but significantly lower than in chicken. The local rates of recombination in chicken and zebra finch were only weakly correlated, demonstrating evolutionary turnover of the recombination landscape in birds. The distribution of recombination events was heavily biased toward ends of chromosomes, with a stronger telomere effect than so far seen in any organism. In fact, the recombination rate was as low as 0.1 cM/Mb in intervals up to 100 Mb long in the middle of the larger chromosomes. We found a positive correlation between recombination rate and GC content, as well as GC-rich sequence motifs. Levels of linkage disequilibrium (LD) were significantly higher in regions of low recombination, showing that heterogeneity in recombination rates have left a footprint on the genomic landscape of LD in zebra finch populations.
Recombination acts to maintain and generate genetic variability in genomes. Specifically, recombination breaks up associations between alleles at linked loci, thereby creating novel haplotypes. This has implications for the effect of selection on molecular evolution as theoretical predictions suggest that recombination should be positively correlated with the efficiency of selection (Hill and Robertson 1966). The rate of recombination is therefore a critical parameter in population genetics and in evolutionary genomics.
Compared to our knowledge about the process behind the ultimate source of genetic diversity, de novo germ line mutations, our understanding of the patterns and causes of recombination rate variation is relatively limited (Jensen-Seaman et al. 2004). It is, however, clear that the rate of recombination is highly heterogeneous across the genome (Kong et al. 2002; Jensen-Seaman et al. 2004; Myers et al. 2006; Wahlberg et al. 2007). In at least some organisms, the majority of recombination events are concentrated in relatively narrow chromosomal regions, referred to as recombination hotspots (Arnheim et al. 2003). In addition, the recombination rate has been shown to co-vary with several sequence characteristics, in particular the GC-content (Birdsell 2002; International Chicken Genome Sequencing Consortium 2004). Moreover, some studies have shown that the recombination rate is higher in areas of the genome that show a higher degree of neutral sequence divergence, suggesting a link between the rates of recombination and point mutation (Filatov and Gerrard 2003; Hellmann et al. 2003; Hellmann et al. 2005; Huang et al. 2005; Bussell et al. 2006; Duret and Arndt 2008; but, see Noor 2008). The causal relationships between these parameters (recombination, base composition, and mutation) are not clear although there are indications that recombination may drive GC-content evolution (Meunier and Duret 2004). Specifically, this can be explained by the process of biased gene conversion, i.e., the tendency for the double-strand break repair mechanism to preferably insert C or G, rather than A or T, at mismatch positions (Marais 2003). On the other hand, data from higher primates indicate that recombination hotspots are transient (Ptak et al. 2005), and in order to explain currently observed base composition structures, hotspots would have to appear more frequently or persist for longer time periods in some regions than in others (Myers et al. 2005).
Besides the within-genome heterogeneity in the rate of recombination, the rate also varies among lineages (Nachman 2002; Jensen-Seaman et al. 2004; Hansson et al. 2005; Beye et al. 2006; Backström et al. 2008). However, there is little information on the phylogenetic signal of such variation (Jensen-Seaman et al. 2004). We currently do not know to what extent overall recombination rates are similar in related species and if homologous genomic regions show conserved rates of recombination. Answering such questions will require detailed information on recombination rates across species.
Understanding the basis of recombination rate variation is necessary to explain the patterns of genetic variation and the influence of natural selection on DNA sequence evolution (Maynard Smith and Haigh 1974; Jensen-Seaman et al. 2004). Knowledge of the recombination landscape is also essential for mapping efforts aimed at finding the genetic basis of phenotypes, in particular, for association mapping using population samples (Arnheim et al. 2003). Linkage disequilibrium (LD) is expected to be broken down more rapidly in regions of high recombination and the number of markers needed to efficiently cover such regions in an association scan should thus be larger than for other regions. In addition, the current distribution of recombination events along chromosomes affects the possibility to detect linkage between genotypes and phenotypes in traditional, pedigree-based genetic mapping efforts.
The zebra finch (Taeniopygia guttata) is an important model in neuroscience, in particular for studies of learning, memory, and behavior (Johnson and Whitney 2005). A draft sequence of the zebra finch genome has recently become available (Warren et al. 2010), making it the second bird species sequenced after the chicken (International Chicken Genome Sequencing Consortium 2004). Here we present a zebra finch genetic linkage map based on genotyping nearly 2000 evenly distributed single nucleotide polymorphisms (SNPs) from protein-coding regions in an extensive pedigree of more than 1000 individuals. By combining information on genetic and physical locations of the markers, we obtain estimates of rate of recombination at the megabase (Mb) scale. We use these data to study variation in recombination rate across the genome, and to analyze how this variation relates to different genomic parameters, how it affects levels of LD and to what extent recombination rates have remained conserved during avian evolution.
Results
Linkage map
We genotyped 1920 SNPs in a four-generation pedigree of 1079 zebra finches (Supplemental Fig. 1; Supplemental Table 1). In this pedigree, 1424 markers were informative (see Methods) of which 1404 showed significant linkage to at least one other marker in a two-point analysis. The resulting framework map consisted of 32 different linkage groups ranging from 0 to 118 cM. It contained 443 markers and covered 1341 cM in total. Linkage groups were equivalent to chromosomes 1–28 (except for chromosome 22 that was not covered in our analysis), plus the Z chromosome (Supplemental Fig. 2). Marker order in all linkage groups of the framework map was essentially identical to the order of markers in the physical assembly of the draft genome sequence. The only exceptions were TGU2, TGU26, and TGU28, which thus contain either assembly or linkage map errors; these chromosomes were excluded from the subsequent analysis of recombination rates. The female-specific autosomal map was marginally longer (1330 cM) than the male-specific map (1304 cM, a 2% difference).
The best-order map (1398 markers) was 1479 cM or 13% longer than the framework map (Supplemental Fig. 3). Based on the position of the most distal markers in the best-order map the coverage is ∼92% of the currently annotated zebra finch genome sequence (Supplemental Table 2).
Recombination rates
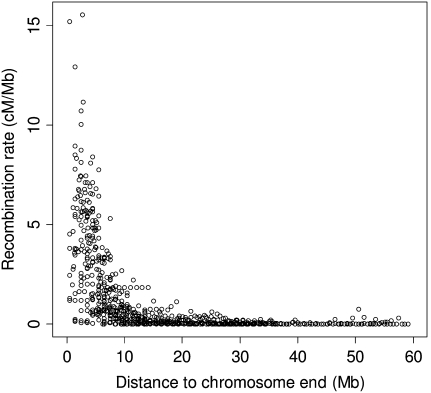
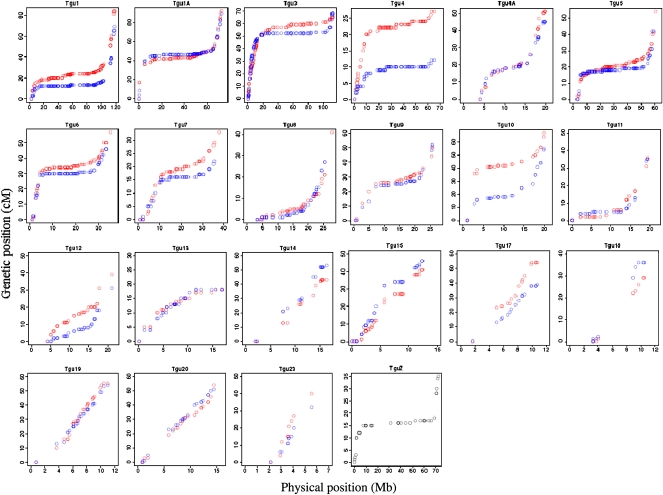
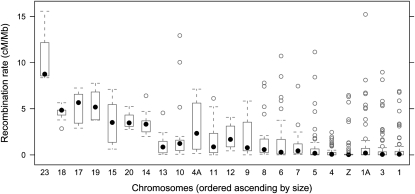
The recombination rate varied between 0 and 15.6 cM/Mb (mean ± SD 1.3 ± 2.2 cM/Mb) when measured in nonoverlapping 1 Mb windows across chromosomes. The distribution of recombination rates was heavily skewed with a large proportion of windows showing no recombination (Fig. 1). A closer inspection of the variation in recombination rate along chromosomes revealed a sharp increase toward telomeric regions. Moreover, there were extremely low rates of recombination in the central parts of the larger chromosomes, giving rise to a sigmoid relationship between the cumulative genetic length and physical position (Fig. 2; Supplemental Figs. 2, 3). As an example, the two distal 15 Mb of chromosome TGU1A measured 42 and 41 cM, while the genetic distance in the interior 44 Mb of this chromosome was only 6 cM. For microchromosomes (chromosomes <20 Mb) the rate of recombination was uniformly high over the entire chromosome (Fig. 2), and was similar to the rate in distal parts of larger chromosomes (see below). It is also clear from the distribution that male and female rates show good concordance across most of the genome, although there is a lack of concordance in a small proportion of regions. Notably, the sharp increase in recombination toward chromosome ends is seen for both male and female meiosis. There was a negative correlation between the mean recombination rate per chromosome and chromosome size (Spearman's rank correlation: rs = −0.91, N = 22, P < 10−8), and this seemed largely due to the low recombination rate in the central part of large chromosomes (Fig. 3). The Z chromosome showed the lowest median and the second lowest mean sex-average recombination rate of all chromosomes (Fig. 3).
Figure 1.
The zebra finch recombination rate (cM/Mb) in 1-Mb windows plotted against the distance (Mb) to the closest chromosome end.
Figure 2.
The relationship between the genetic (cM) and the physical (Mb) position of markers in the best-order map built based on the draft sequence of the zebra finch genome. Red and blue circles indicate the female- and male-specific genetic map, respectively. The length of the x-axis reflects the total physical size of the chromosome, as given by the genome assembly. Only chromosomes with >10 markers are included.
Figure 3.
Box-and-whisker plots for recombination rate in 1-Mb windows across chromosomes. Filled dots show the median and boxes the interquartile ranges. Outside the boxes, whiskers mark the largest and smallest values within 1.5 times the interquartile range, and open dots show data points outside the whisker range.
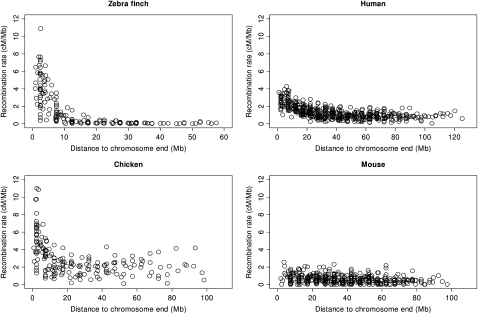
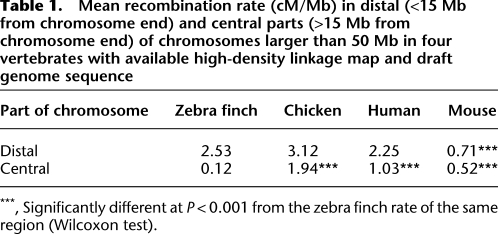
To illustrate the extreme heterogeneity and position dependence in rate of recombination along larger chromosomes we compared the relationship between recombination rate and distance to chromosome end in zebra finch, chicken (Groenen et al. 2009), mouse, and human (Jensen-Seaman et al. 2004). There was a general increase in recombination toward telomeres in all organisms, although the effect was weak in mouse (Fig. 4). The effect was by far the strongest in zebra finch, which was due to the combined effect of extremely low rates in central parts of chromosomes and highly elevated rates near telomeres (Table 1). The zebra finch recombination landscape on a regional scale can thus be characterized by well-defined recombination jungles and deserts.
Figure 4.
The relationship between the distance to the chromosome end (Mb) and the recombination rate (cM) measured in 5-Mb windows in zebra finch (this study), chicken (Groenen et al. 2009), mouse and human (Jensen-Seaman et al. 2004).
Table 1.
Mean recombination rate (cM/Mb) in distal (<15 Mb from chromosome end) and central parts (>15 Mb from chromosome end) of chromosomes larger than 50 Mb in four vertebrates with available high-density linkage map and draft genome sequence
***, Significantly different at P < 0.001 from the zebra finch rate of the same region (Wilcoxon test).
The relationship between recombination rate and other genomic parameters
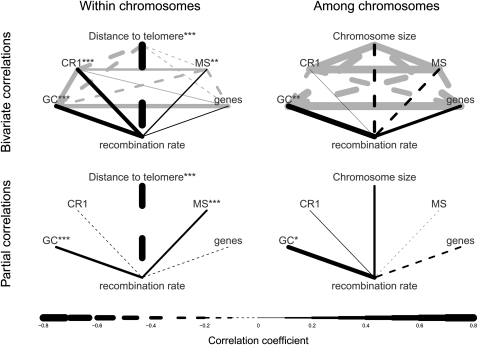
We explored the relationship between recombination rate and distance to chromosome end, base composition (GC content), repeat density (CR1 retrotransposons and microsatellites), gene density, and chromosome size on the megabase scale. Since recombination rate drops very close to zero further than 15 Mb away from chromosome ends, we initially limited the analysis to windows within each terminal 15 Mb (see Methods). Moreover, because recombination rate varied among chromosomes (variance component of chromosome identity standardized at 5 Mb from chromosome end: 0.27 ± 0.07, LRT: P < 10−13), we analyzed the effects separately for the among-chromosome and the within-chromosome variation.
The strongest association was seen between recombination rate and distance to chromosome end (r = −0.75). This effect was strong even when controlling for other genomic parameters (r = −0.67, Figure 5; Supplemental Table 3) and was about 7.5 times stronger than the second largest effect, which was GC content (r2 = 0.45 vs. 0.06). Within as well as among chromosomes, GC content correlated positively with recombination rate and this correlation was stable even after controlling for distance to chromosome end, CR1 repeat density, microsatellite density, gene density and chromosome size (within: r = 0.25, among: r = 0.43; Fig. 5; Supplemental Table 3). Furthermore, microsatellite density was positively associated with recombination rate within chromosomes (r = 0.19; Fig. 5). None of the other genomic parameters had similarly strong effects when controlling for the others. For example, chromosome size was not significantly related to recombination rate measured at 5 Mb distance from chromosome ends (Fig. 5). However, since the telomeric effect was so pronounced and chromosomes differ substantially in length, larger chromosomes had a lower average recombination rate than smaller chromosomes (Fig. 3).
Figure 5.
Correlations between recombination rate and genomic parameters. The upper two plots show raw pairwise correlations between genomic parameters and recombination rate, while the lower two plots show partial correlations controlling for all other genomic parameters in the plot. (Solid lines) Positive correlations; (dashed lines) negative correlations. Line width is proportional to the strength of the correlations. Gray lines show correlations among genomic parameters. Asterisks indicate significance levels for correlations with recombination rate (black lines; *P < 0.05; **P < 0.01; ***P < 0.001). CR1, chicken repeat 1 retrotransposon; MS, microsatellites.
We further analyzed the importance of specific GC-rich motifs that previously have been shown to influence recombination rate (Myers et al. 2008; Groenen et al. 2009). These included CpG, CCNCCNGGNGG, and CCTCCCT that were all highly correlated with GC content (r > 0.68). Within chromosomes, the motif CCTCCCT was significantly positively correlated with recombination rate after controlling for GC content and distance to telomere (r = 0.17, P = 0.0006; Supplemental Fig. 4), while controlling for the frequency of this motif rendered the effect of GC content negative and nonsignificant (Supplemental Table 3). After controlling for GC content, neither the frequency of the motif CCNCCNGGNGG nor the CpG density was significantly correlated to recombination rate, except for chromosome-average CpG density that was negatively related to recombination rate (r = −0.46, P = 0.032; Supplemental Fig. 4).
Since our initial analysis was limited to the telomeric 15 Mb, while we ignored the recombination deserts in the central parts of the chromosomes, we tested for differences in genomic parameters between the central parts and the telomeric ends. The GC content was significantly lower in central parts (LMM: t17 = −6.50, P = 2.9 × 10−5) and the same was true for the frequency of CCTCCCT motifs (t17 = −6.51, P = 2.9 × 10−5) and somewhat weaker also for CCNCCNGGNGG (t17 = −5.04, P = 2.9 × 10−4) and CpG frequency (t17 = −4.69, P = 5.3 × 10−4). This gives further evidence for the association between GC and CCTCCCT and recombination rate. Gene density did not differ between the central and terminal parts (t17 = −1.63, P = 0.13), microsatellite density tended to be higher (t17 = 2.05, P = 0.062) and CR1 density was lower in recombination deserts (t17 = −3.42, P = 0.0005).
To test if the pronounced telomere effect in recombination seen in zebra finch was due to a particularly pronounced increase in GC toward chromosome ends in this species, we compared the GC content of the terminal 15 Mb versus central parts of chromosomes (only chromosomes >50 Mb included) in zebra finch, chicken (also strong telomere effect), human (moderate effect), and mouse (limited effect). However, there was no obvious connection between the magnitude of the increase in recombination and the increase in GC: zebra finch, GC = (mean ± SD) 0.41 ± 0.031 vs. 0.39 ± 0.023 (Wilcoxon W = 55,095, P < 2.2 × 10−16); chicken, 0.41 ± 0.037 vs. 0.39 ± 0.035 (W = 49,816, P < 1.6 × 10−12); human, 0.44 ± 0.052 vs. 0.41 ± 0.043 (W = 505,426, P < 2.2 × 10−16); mouse, 0.41 ± 0.028 vs. 0.42 ± 0.036, (W = 275,900, P = 0.023).
Conservation of recombination rates during avian evolution
To address the degree of conservation of recombination rates in homologous regions of avian genomes we compared the rates in orthologous 1-Mb windows of chicken and zebra finch chromosomes (see Methods). There was a significant correlation (Spearman's rank correlation: rs = 0.50, N = 275, P < 2.2 × 10−16) between the rates in the two bird species (Supplemental Fig. 5). When analyzing large chromosomes (>50 Mb) and small chromosomes (<50 Mb) separately, the correlation was stronger for small (rs = 0.42, N = 107, P = 5.6 × 10−6) than for large chromosomes (rs = 0.29, N = 168, P = 1.27 × 10−4).
To investigate if intrachromosomal rearrangements affect the strength of the correlation we further divided the orthologous windows into two classes, “conserved windows” and rearranged windows (see Methods). Conserved windows represent orthologous chromosome segments that have remained in approximately the same position on chromosomes during avian evolution, while rearranged windows are orthologous chromosome segments that have changed location (relative to chromosome ends) during evolution. There was a significant positive correlation between recombination rates in the two species for conserved and rearranged windows within small chromosomes and for conserved windows within large chromosomes (Spearman's rank correlation: rs = 0.34–0.55, P = 9.45 × 10−13–2.40 × 10−2). However, for rearranged windows located on large chromosomes there was no longer a significant correlation (Spearman's rank correlation: rs = 0.14, N = 90, P = 0.19). One way to interpret these observations is that there is no strong signal of conserved recombination related to inherent characteristics of sequences contained within particular chromosomal regions. Rather, conserved recombination follows from strong karyotype conservation (Griffin et al. 2007) in combination with the fact that recombination occurs more frequently close to chromosome ends than in the center of chromosomes in both species, as well as in microchromosomes.
Recombination rate and linkage disequilibrium
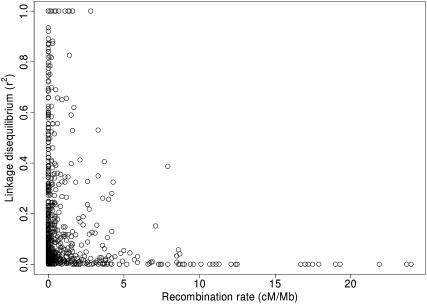
The heterogeneity in recombination rate observed within and among zebra finch chromosomes suggests that the pattern of LD is similarly heterogeneous. For example, selective sweeps may lead to longer blocks of LD in low, rather than in high, recombination rate regions. We measured LD between all pairs of loci within linkage groups using genotype data from parental birds of the pedigree. The extent of LD varied considerably within chromosomes (Supplemental Fig. 6). A very strong pattern of LD was observed for the middle part of the Z chromosome where significant LD (solid spine of LD; Barrett et al. 2005) extends over more than 50% of the chromosome (54.4 Mb). Strong signals of extensive LD were also seen on some of the autosomes (e.g., TGU7 and TGU10; Supplemental Fig. 6), in all those cases involving low recombination rate regions on larger chromosomes. LD on microchromosomes was consistently low. We tested if recombination rate and LD were correlated by analyzing all pairwise intermarker intervals of immediately adjacent SNPs throughout linkage groups. There was a negative correlation between the recombination rate and the level of LD (Spearman's rank correlation: rs = −0.19, P = 1.94 × 10−8), and high recombination intervals had exclusively low LD values, while intervals with low recombination displayed a higher variance (Fig. 6).
Figure 6.
The relationship between the recombination rate (cM/Mb) and the level of linkage disequilibrium (r2) for all adjacent marker pairs within linkage groups in the zebra finch genetic map.
LD decayed over physical distance although the variance in LD was large, especially for markers separated by short physical distances (Supplemental Fig. 7). To further investigate the relationship between decay of LD and chromosome class we grouped all distances into 100-kb intervals and calculated the mean LD for each interval for small chromosomes (<50 Mb) and large chromosomes (>50 Mb), respectively. When fitting a linear model to the logarithm of mean LD and physical distance, we observed the fastest decay of LD on small chromosomes (Supplemental Fig. 8). For both chromosomal classes, mean r2 is less than 0.25 already at the shortest interval of 100 kb, indicating that average LD decays rapidly in zebra finch chromosomes.
Discussion
The recombination landscape
Coupled with the recently obtained draft genome sequence (Warren et al. 2010), a genetic map of the zebra finch genome will be a great asset for dissection of the genetic basis of phenotypic traits of this model organism. For example, the observation that many behavioral traits are heritable and segregate in zebra finch populations (Forstmeier 2005; Evans et al. 2006; Forstmeier et al. 2009) calls for quantitative trait locus (QTL) mapping efforts of such traits. A zebra finch linkage map based on SNP markers was recently reported by Stapley et al. (2008). As judged from the location of framework markers common to the two maps, they show a high degree of consistency. The map presented here contains 67% more markers (1404 vs. 838) and is 38% longer (1479 vs. 1068 cM) than the Stapley et al. (2008) map. It is also based on data from a larger pedigree (1079 vs. 384 individuals). However, the most important difference between this study and that of Stapley et al. (2008) is that the access to a draft genome sequence of the zebra finch genome (Warren et al. 2010) means that the recombination landscape can now be studied for the first time in this model species. Accordingly, in most of the following we discuss findings based on the new ability to estimate the actual rates of recombination, by comparing genetic and physical distances in the zebra finch genome.
The mean recombination rate in the genomes of most eutherian mammals studied so far is in the range of 0.5–1.2 cM/Mb, with mouse and rat in the lower and human in the upper end (Jensen-Seaman et al. 2004; Dumont and Payseur 2007). Marsupials (as represented by opossum) have a much lower rate (≈0.2 cM/Mb; Samollow 2008), and this probably holds true for wallaby as well (Dumont and Payseur 2007). The high rate seen in chicken (3.1 cM/Mb; Groenen et al. 2009) led to the suggestion that, among vertebrates, birds have a relatively high rate of recombination (International Chicken Genome Sequencing Consortium 2004). However, there are several lines of evidence to suggest that recombination rates increased during domestication, as strong artificial selection in new environments will favor novel haplotypes (Burt and Bell 1987; Ross-Ibarra 2004). The rate observed in domestic chicken may therefore not be representative for other birds (Groenen et al. 2009), and partial linkage maps recently reported for several songbirds have indicated lower rates than in chicken (Hansson et al. 2005; Backström et al. 2008), although this is inferred in the absence of genome sequence data.
Genome size of birds is typically only 30%–50% of that of mammals, while chromosome number is usually much higher (mammals, median 2n = 46; birds, median 2n = 80; Gregory et al. 2007). The presence of numerous small microchromosomes, each contributing ≥50 cM to the genetic map assuming an obligate cross-over, will inevitably lead to elevated mean recombination rates in bird genomes. The average recombination rate that we observe for zebra finch, 1.5 cM/Mb (1439 cM in 933 Mb), is only slightly higher than that in humans. However, this is clearly an underestimate since 10 microchromosomes (≈10–20 Mb/chromosome) are not covered in the linkage map, nor included in the physical assembly. Assuming an obligate crossover per chromosome (Rodionov 1996), adding data from these microchromosomes adds >500 cM to the linkage map, while having relatively limited influence on the physical map. Together with the fact that the present map lacks terminal markers for several chromosomes and that recombination is strongly concentrated to telomeric regions should further contribute to an underestimate of the mean recombination rate. Taking these factors into account, we estimate by extrapolation that the mean overall zebra finch recombination rate is ≈2 cM/Mb (2400 cM in 1.2 Gb). This supports the idea that recombination is generally higher in birds than in mammals.
The rate of recombination has in some organisms been found to increase from the centromere to the telomere (Nachman 2002; Jensen-Seaman et al. 2004). However, to the best of our knowledge, in no case has such a pronounced telomere effect been observed as seen in zebra finch. The recombination landscape of zebra finch chromosomes is characterized by highly elevated rates in the distal 5–20 Mb regions, with rates of 5–10 cM/Mb in megabase windows closest to chromosome ends, and extremely low rates in central parts of chromosomes. The combined effect leads to the peculiar sigmoid relationship between cumulative genetic distance and physical position along chromosomes observed in the larger chromosomes (Fig. 2). About 90% of the total amount of recombination in zebra finch is concentrated to ≈23% of the genome. For the three largest chromosomes (TGU1–TGU3), 110–150 Mb in size, the interior “recombination desert” extends over ≈100 Mb with an average rate as low as ≈0.1 cM/Mb over these long intervals. The extent of recombination deserts decreases with decreasing chromosome size, to disappear for chromosomes <20 Mb; in essence, such microchromosomes can be seen to only contain telomeric regions when it comes to the recombination landscape.
At the level of resolution given by our marker set, with recombination estimated for 1-Mb intervals, we find no clear evidence for recombination hotspots outside the telomeric jungles. If such hotspots exist, they would be narrow and only make a limited contribution to the total amount of recombination given the absence of clear “steps” in the cumulative genetic distance in interior parts of chromosomes.
There is a sharp increase in recombination rate toward chromosome ends also in chicken (Groenen et al. 2009). The two bird genomes so far analyzed are thus similar in this respect, while the rate of recombination in interior parts of chromosomes is considerably higher in chicken than in zebra finch. One possible explanation for this difference is that low recombination rate regions are more responsive to selection for increased recombination during domestication. Moreover, while there is some evidence for a correlation in recombination rate between orthologous chromosome intervals of chicken and zebra finch, this correlation becomes very weak when controlling for chromosome identity. Given the high degree of chromosomal homology between these two species, with very few interchromosomal rearrangements, the raw correlation observed seems most easily explained by inherently large differences in recombination rate between large and small chromosomes. Jensen-Seaman et al. (2004) found only limited correlation in recombination rate among homologous chromosome intervals of human, mouse, and rat. Although there is some evidence for a global conservation of recombination rates among species (Dumont and Payseur 2007), it may very well be that regional and local recombination rates are poorly conserved given the strong effects of chromosome position and chromosome size on the rate of recombination.
Factors affecting recombination rates
GC content and presence of CCTCCCT motifs are strong predictors, with similar effect size, of the recombination rate after controlling for the effect of distance to chromosome end. Interestingly, while the effect of CCTCCCT remains significant after GC is controlled for, this is not the case for the effect of GC when CCTCCCT is controlled for. This illustrates that it may be premature to make conclusions based on bivariate regression analysis when motif abundance is likely to be associated with base composition. For example, like Groenen et al. (2009) reported for chicken, we found a raw correlation between the cohesin-binding site motif CCNCCNGGNGG and recombination rate; however, the effect is not significant after controlling for GC content. CCTCCCT, or the related degenerate motif CCNCCNTNNCCNC, have been reported to be overrepresented in highly recombining regions in humans (Myers et al. 2006, 2008), mice (Shifman et al. 2006), and chicken (Groenen et al. 2009). These motifs might have an impact on chromosome stability and it has been suggested that the CCNCCNTNNCCNC could promote double strand breakage at the replication stage in mtDNA (Myers et al. 2008).
It is difficult to reveal the causality of the observed relationships between GC content/GC-rich motifs and recombination rate, i.e., which parameter drives which. On the one hand, GC-rich sequence motifs could act as signals for recombination via, for example, preferential binding of cohesin complexes, which has been shown to mediate the efficiency of double strand break repair in yeast (Sjögren and Nasmyth 2001). On the other hand, the process of biased gene conversion could lead to increased GC content in regions with high recombination rates (Webster et al. 2003; Meunier and Duret 2004; Webster et al. 2005). It will be an important topic for further research to try to elucidate the causal relationship between base composition and recombination rate, especially since recent data indicate that biased gene conversion might be a potent evolutionary force also at functional sites (Galtier and Duret 2007; Berglund et al. 2009).
Linkage disequilibrium
While the buildup of LD can be a result of many different population genetic forces (genetic drift, selection, population structure, and admixture), recombination is the only major force that acts to break it down. This suggests that the local rate of recombination might strongly affect levels of LD (Slatkin 2008). This is confirmed in our data, in which we find that recombination and LD are negatively correlated. The overall extent of LD in out-crossing species with large effective population sizes has generally been found to be low (Conway et al. 1999; Remington et al. 2001; Edwards and Dillon 2004; Cutter et al. 2006; Heuertz et al. 2006). Not surprisingly, similarly low levels of LD, in most cases extending only a few hundred to a few thousands of base pairs, were observed in wild Australian populations of zebra finches, with an estimated population size in the range of 106 (Balakrishnan and Edwards 2009). Population bottlenecks result in loss of genetic diversity due to increased impact of drift and this has been observed in a comparison of wild and captive zebra finch populations (Forstmeier et al. 2007b). Since the loss of haplotypes occurs more rapidly than the loss of genotypes (genotypes cannot be lost without losing haplotypes, but haplotypes can be lost without losing genotypes), LD is expected to range over longer distances in bottlenecked populations. The fact that we observe significant LD, in some cases ranging over several megabases, in a laboratory zebra finch population is in line with predictions from a population bottleneck. Similar results have been obtained in studies of other bottlenecked populations, including derived human populations (Farnir et al. 2000; Reich et al. 2001; McRae et al. 2002; Nsengimana et al. 2004; Sutter et al. 2004; Nordborg et al. 2005; Backström et al. 2006; Aerts et al. 2007; Andreescu et al. 2007; Hernandez et al. 2007; McKay et al. 2007; Slate and Pemberton 2007; Amaral et al. 2008; Gray et al. 2009).
There are many captive zebra finch populations that are independently derived from wild birds and in which drift is likely to have resulted in significant allele frequency differences among populations (Forstmeier et al. 2007b). This provides an interesting framework for mapping traits of interest by admixture LD (Smith and O'Brien 2005). In addition, as exemplified for similarly very structured dog breeds (Sutter et al. 2004), a useful approach for trait mapping will be to use within-population LD to map candidate regions and then combine information from several populations for fine-tuned mapping of causative variants.
The significant heterogeneity in recombination rate observed in the zebra finch genome has practical implications for genetic mapping efforts aimed at finding trait loci. The conditions for detecting linkage and subsequently identifying candidate loci will depend on the genomic location of such loci. For loci located in the interior parts of larger chromosomes, it will be relatively straightforward to detect linkage between markers and trait loci, while it will be more demanding to detect such linkage for trait loci located close to telomeres. Conversely, when linkage has been established, it will be easier to narrow down intervals for candidate loci in the latter regions due to the higher likelihood of detecting recombination events between nearby markers and causative mutations. For both linkage and association mapping the development of SNP tags with a denser marker representation toward chromosome ends will provide a cost-effective means for upcoming mapping efforts in zebra finch.
Implications for the role of selection on genome evolution
The impact of natural and sexual selection on genome evolution is expected to be dependent on the rate of recombination. Indeed, theory suggests that the effect of selection at other loci on the same chromosome relates to the degree of linkage. A selective sweep can lead to the fixation of slightly deleterious alleles at linked loci and to a reduction in nucleotide diversity in linked regions (Maynard Smith and Haigh 1974; Barton 2000; Charlesworth 2007). Conversely, adaptive evolution may be hampered by linkage to deleterious alleles (Marais and Charlesworth 2003). Purifying selection can also lead to a loss of diversity, albeit less severe, in linked regions (background selection) (Charlesworth et al. 1997).
The remarkable heterogeneity in rate of recombination observed for the zebra finch genome suggests that the genomic footprints of selection may be more variable than so far seen for other genomes. If this pattern of recombination has persisted over a considerable amount of time, a number of predictions can be made. (1) When scaled by the underlying rate of mutation, polymorphism levels should be lower in the low-recombination central parts of the larger chromosome than in other parts of the genome (see, for example, Reich et al. 2002). (2) If protein evolution, at least in part, is affected by the accumulation of slightly deleterious mutations, this should be most pronounced in the low-recombination central parts of the larger chromosomes (Muller 1964), as manifested in high ratios of the rate of nonsynonymous substitution to the rate of synonymous substitution (dN/dS). (3) If adaptive protein evolution driven by positive selection is prevalent, this should be most pronounced in the high-recombination telomeric parts of the larger chromosomes and in microchromosomes (Marais and Charlesworth 2003; Presgraves 2005), again manifested in high dN/dS ratios. Whether high dN/dS ratios are caused by nearly neutral evolution or adaptive evolution can be assessed by contrasting levels of diversity and divergence in different regions (McDonald and Kreitman 1991). Testing these predictions will be important to further research aimed at elucidating the interaction between selection and recombination in molding genome evolution.
Methods
Pedigree structure and DNA preparation
The pedigree used in this study was raised at the Department of Behavioral Ecology and Evolutionary Genetics at the Max Planck Institute for Ornithology in Seewiesen, Germany. Previously, a total of 231 zebra finches had been brought from a laboratory population held at Sheffield University (Forstmeier et al. 2007b). Of those imported birds, 52 males and 54 females (parental generation) interbred successfully to form a four-generation pedigree of 1079 individuals (Supplemental Fig. 1; Supplemental Table 1).
About 40% of the individuals were bred by pairs housed long-term in separate cages so that parentage was known beyond any doubt. The other individuals were bred in aviaries holding up to 15 adults. For them parentage was assigned with the help of 10–18 microsatellite markers (Forstmeier et al. 2007a). This pedigree information was independently confirmed to be entirely correct during the course of the present study when checking the SNP genotypes for Mendelian inheritance.
For all individuals we extracted a minimum of 400 ng of DNA (measured by Nanodrop) from either blood or tissue samples (in case of deceased birds).
SNP selection and genotyping
Potential SNPs were identified as follows. A set of 2.1 million zebra finch 454 Life Sciences (Roche) GS transcriptome reads and their accompanying quality scores were retrieved from the NCBI Trace Archive (http://www.ncbi.nlm.nih.gov/Traces/home/) using a custom Perl script. We also obtained a set of 17,438 Ensembl-predicted chicken gene sequences using the BioMart tool (http://www.ensembl.org/biomart/martview). We then used the standalone version of MEGABLAST with default parameters to BLAST the trace reads against a database of chicken genes, followed by a reciprocal search using the trace reads as the subject and chicken genes as query. We parsed the MEGABLAST alignments to identify variable nucleotide positions between multiple zebra finch reads aligned to the same place in a chicken gene. Only variable positions with two states that occurred within best reciprocal BLAST hits were considered as potential SNPs. High-scoring segment pairs with E-values < 10−30 and all bases with quality scores < 20 were ignored. The zebra finch sequence flanking each potential SNP was also extracted from the MEGABLAST alignments. Potential SNPs were only retained for further analysis if there were fewer than 12 other variable sites in the flanking sequence within 200 bp, if we obtained at least three reads representing each allele and if the minor allele frequency >20%.
A set of 12.1 million zebra finch reads and their quality scores generated by Sanger sequencing were obtained from the NCBI Trace Archive using the same procedure as the 454 reads. We then performed reciprocal MEGABLAST searches with the Sanger reads and chicken gene sequences and parsed the alignments using the procedure described above. Due to the longer read length and larger number of sequences, different parameters were used to filter the resultant list. Here, potential SNPs needed to have fewer than eight other variable sites within 200 bp, at least three reads representing each allele, and minor allele frequency >30% to be retained for further analysis.
In total, 890 potential SNPs from the 454 reads, 1678 SNPs from the Sanger reads, 480 SNPs detected in both of the analyses, and 1693 SNPs from Stapley et al. (2008) were scored for primer design by Illumina. The final mapping panel was selected from this list using a custom algorithm to maximize genome coverage and design score. SNPs that were previously typed by Stapley et al. (2008), but had low heterozygosity or poor call rates were excluded. The final set of SNPs selected for genotyping consisted of 187 SNPs from the transcriptome sequencing, 917 SNPs from the raw reads of the zebra finch genome sequencing effort, 199 SNPs that were detected in both of these sources, and 617 SNPs from the panel used by Stapley et al. (2008). As much as possible, SNPs were selected to correspond to single-copy loci evenly distributed along all annotated chicken chromosomes. The mapping panel includes a total of 1920 SNPs, of which 1775 correspond to loci with known position in the chicken genome. Genotyping of these 1920 SNPs was attempted in all 1079 birds (≈2.1 million genotypes). All SNPs have been deposited in dbSNP (Supplemental Table 4).
SNP genotyping was done with the Illumina GoldenGate Assay (Fan et al. 2003) at the SNP Technology Platform in Uppsala, Uppsala University (http://www.genotyping.se). The average genotype call rate was 95.04% and the reproducibility was 99.98% according to duplicate analysis of 3.93% (81,563/2,073,600) of the genotypes. 108 of the 1920 SNPs failed to meet the quality criteria (more than three clusters observed or low sample call rate) and 380 SNPs showed no variation in the pedigree (minor allele frequency = 0.00%). An additional eight SNPs had to be rejected due to excessive inheritance failures; hence, 1424 markers could be used in linkage analysis.
Linkage mapping and comparative analyses
Linkage analyses were performed in the modified version of CRI-MAP v.2.4 (Green et al. 1990) as provided by Xuelu Liu (Monsanto). All possible marker pair combinations were tested for linkage with the option TWOPOINT at a threshold of LOD-score 3.0. Linked markers were assembled into linkage groups by the option AUTOGROUP. Due to the high rate of false positives expected when testing for linkage with ∼14002/2 combinations, markers were grouped if multipoint LOD-score exceeded 8. Markers were then ordered within linkage groups to generate a framework map using the option BUILD with a LOD-score threshold of 3.0. A best-order map was constructed by relaxing the threshold of BUILD until all markers were included in the map and the order was tested with recurrent runs of the option FLIPS5 until no better order could be found. Moreover, to be able to compare the genetic and physical positions, we constructed a best-order map by using the order indicated by the physical location of markers in the draft assembly of the zebra finch genome (see below) for all chromosomes with more than 10 markers in a single linkage group. There was complete agreement between the framework map and the best-order map based on physical location, except for TGU2, TGU26, and TGU28. All genetic distances were transformed to Kosambi cM and sex-specific, as well as sex-averaged distances, were computed for each linkage group. All genetic maps were drawn using MapChart (Voorrips 2002).
To compare recombination rates between chicken and zebra finch we used sex-averaged chicken recombination data from Groenen et al. (2009). Recombination rates were compared in all 1-Mb windows that did not include a chromosomal break point and that did not differ in length by more than 5%. Windows were classified as conserved if they were in the same direction and if the distance to chromosome end from the center of window was similar (<5% difference) between species, otherwise windows were classified as rearranged.
Linkage disequilibrium (LD) analysis
To assess the level of LD present in the mapping population we selected all individuals in the parental generation that did not share parents in the previous generation (Supplemental Fig. 1; Supplemental Table 1) and treated these as a population sample of individuals. This generated a set of 32 individuals (64 chromosomes) that could be used to assess the degree of linkage disequilibrium between markers. Intrachromosomal haplotypes were estimated with the ELB algorithm with default settings as implemented in the software Arlequin (Excoffier et al. 2005) and phased data was used to estimate pairwise LD (r2) in Haploview (Barrett et al. 2005). The physical position of SNPs was used to examine the decay of LD over physical distance. To investigate the relationship between the recombination rate and level of LD, we tested for correlations between the recombination rate and the degree of LD between adjacent markers within linkage groups.
Estimates of genomic parameters
The flanking sequences of SNP markers were used in BLAST searches against the zebra finch genome sequence (http://genome.ucsc.edu/cgi-bin/hgGateway?db=taeGut1) to determine the physical position of markers. Recombination rates (genetic distance in cM/physical distance in megabases) were estimated for a window size of 1 Mb. This was done by summing the amount of recombination of each marker pair with at least one marker within the limit of a window. For windows that lacked markers, the amount of recombination between the most closely located flanking markers was used. TGU2 was excluded from this analysis since we found discrepancies in gene order between our genetic framework map and the zebra finch genome sequence assembly. Chromosomes with less than five markers with known physical position were also excluded. For each 1 Mb window, we used in-house developed Perl-scripts to obtain data on genomic parameters: (1) GC content, (2) number of cohesion binding sites (CCNCCNGGNGG) (Groenen et al. 2009), (3) number of CCTCCCT motifs (Myers et al. 2005), (4) CpG fraction, (5) gene density, (6) density of chicken repeat 1 (CR1) elements, (7) density of microsatellites (both by RepeatMasker, http://repeatmasker.org), (8) distance to the closest chromosome end (from middle of window), and (9) chromosome size. To compare the distribution of recombination between species we used data presented in Jensen-Seaman et al. (2004) and Groenen et al. (2009). We calculated the recombination rate in 5-Mb windows for chicken and zebra finch for this specific analysis and included only chromosomes larger than 50 Mb.
Data analysis
Recombination rate was highly skewed and a transformation was necessary to be able to analyze it in a general linear model and correlation framework. Since the recombination rate dropped to almost zero at a distance of more than 15 Mb from the chromosome ends (Fig. 1), we analyzed the recombination rate only in the terminal 15 Mb of all chromosomes. This was justified because there was hardly any variation in recombination rate in the central part of the chromosomes. Excluding the central parts of the chromosomes effectively removed the large zero inflation in the data. We then used the Box-Cox method to find a normalizing transformation for recombination rate. Cube-root transformation was close to the optimal transformation and hence we used this in all further analyses of recombination rate. All predictors were log-transformed.
We used simple bivariate and partial correlations to estimate the strength of the relationships between recombination rate and genomic parameters. In a first step, we decomposed recombination rate in its within-chromosome and among-chromosome components by means of mixed models including chromosome identity as a random-effect. We also included distance-to-telomere as a fixed effect centered at 5 Mb from the telomeres, which is close to the average distance in the data set. Hence, between-chromosomes effects were estimated at a distance of 5 Mb for all chromosomes. We extracted best-linear unbiased predictors (BLUPs) for the chromosome effects as estimates of between-chromosome variation and the deviation from these as the within-chromosome variation. BLUPs are better suited for variance decomposition than averages, because they account for differences in sample sizes (that are large due to differences in chromosome size) and they enable the estimation of chromosome-wide effects at defined positions from the telomere (here 5 Mb).
We were interested in the correlations between recombination rate and genomic parameters, all of which were nonindependent on the same hierarchical levels as recombination rate and highly intercorrelated among each other. We first decomposed their variances into between- and within-chromosome effects as described for recombination rate. We then calculated bivariate between recombination rate and genomic parameters one-by-one. Furthermore, we calculated partial correlations for recombination rate with GC content, CR1 density, microsatellite density, gene density, and distance to telomere (or chromosome size) one-by-one, each time controlling for the effect of the others. Because of the very high correlations between GC content and GC-rich motifs (CpG, CCNCCNGGNGG, CCTCCCT), we did not include these parameters in this partial correlation analysis. Their effects are very similar to the effect of GC, if we replace GC by any one of these motifs. To elucidate if any of these motifs was overrepresented in regions of high recombination, we conducted partial correlations between the motifs controlling for GC content and distance to telomere, as well as for GC content controlling for motif frequency and distance to telomere.
The previous analysis was restricted to the terminal 15 Mb, because only these regions show elevated recombination rates, but it does not address why there is such low recombination in the central parts of the chromosomes. Therefore, we tested if genomic parameters differed between these “recombination jungles” in the telomeric ends and the “recombination deserts” in the central parts of the chromosomes (>15 Mb from telomeres). We used linear mixed models that included an indicator of center vs. telomeric ends and included chromosome identity as a random effect for the intercept and for the slope of the indicator variable. These random effects account for pseudoreplication within and for unequal sample sizes among chromosomes. Since all traits showed significant between-chromosomal variation, we used significance test with degrees of freedom equal to the number of chromosomes minus the number of parameters estimated. This yields conservative P-values compared to the alternative of using the number of data points minus the number of parameters estimated.
Acknowledgments
We thank S. Bauer, E. Bodendorfer, P. Brekke, A. Grötsch, J. Hacker, M. Halser, K. Martin, J. Minshull, P. Neubauer, J. Pellatt, A. Peter, N. Roddis, M. Ruhdofer, A. Türk, F. Weigel, B. Wörle, R. Yeates, and P. Young for providing animal care and help with breeding. T.R. Birkhead kindly provided the founder birds of this population. J. Slate provided information on SNPs. J. Müller commented on the manuscript. This study was funded by financial support from the German Research Foundation grants FO340/1-1, FO340/1-2, and FO340/1-3 (W.F.), the European Union grant HPMF-CT-2002-01871 (W.F.), the Max Planck Society (B.K.), the Swedish Research Council (H.E.), and the Knut and Alice Wallenberg Foundation (H.E.).
Footnotes
[Supplemental material is available online at http://www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.101410.109.
References
- Aerts J, Megens HJ, Veenendaal T, Ovcharenko I, Crooijmans R, Gordon L, Stubbs L, Groenen M. Extent of linkage disequilibrium in chicken. Cytogenet Genome Res. 2007;117:338–345. doi: 10.1159/000103196. [DOI] [PubMed] [Google Scholar]
- Amaral AJ, Megens H-J, Crooijmans RPMA, Heuven HCM, Groenen MAM. Linkage disequilibrium decay and haplotype block structure in the pig. Genetics. 2008;179:569–579. doi: 10.1534/genetics.107.084277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreescu C, Avendano S, Brown SR, Hassen A, Lamont SJ, Dekkers JCM. Linkage disequilibrium in related breeding lines of chickens. Genetics. 2007;177:2161–2169. doi: 10.1534/genetics.107.082206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnheim N, Calabrese P, Nordborg M. Hot and cold spots of recombination in the human genome: The reason we should find them and how this can be achieved. Am J Hum Genet. 2003;73:5–16. doi: 10.1086/376419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backström N, Qvarnström A, Gustafsson L, Ellegren H. Levels of linkage disequilibrium in a wild bird population. Biol Lett. 2006;2:435–438. doi: 10.1098/rsbl.2006.0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backström N, Karaiskou N, Leder EH, Gustafsson L, Primmer CR, Qvarnström A, Ellegren H. A gene-based genetic linkage map of the collared flycatcher (Ficedula albicollis) reveals extensive synteny and gene-order conservation during 100 million years of avian evolution. Genetics. 2008;179:1479–1495. doi: 10.1534/genetics.108.088195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan CN, Edwards SV. Nucleotide variation, linkage disequilibrium and founder-facilitated speciation in wild populations of the zebra finch (Taeniopygia guttata) Genetics. 2009;181:645–660. doi: 10.1534/genetics.108.094250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview, analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Barton N. Genetic hitchhiking. Philos Trans R Soc Lond B Biol Sci. 2000;355:1553–1562. doi: 10.1098/rstb.2000.0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund J, Pollard KS, Webster MT. Hotspots of biased nucleotide substitutions in human genes. PLoS Biol. 2009;7:e1000026. doi: 10.1371/image.pbio.v07.i01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beye M, Gattermeier I, Hasselmann M, Gempe T, Schioett M, Baines JF, Schlipalius D, Mougel F, Emore C, Rueppell O, et al. Exceptionally high levels of recombination across the honey bee genome. Genome Res. 2006;16:1339–1344. doi: 10.1101/gr.5680406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birdsell JA. Integrating genomics, bioinformatics, and classical genetics to study the effects of recombination on genome evolution. Mol Biol Evol. 2002;19:1181–1197. doi: 10.1093/oxfordjournals.molbev.a004176. [DOI] [PubMed] [Google Scholar]
- Burt A, Bell G. Mammalian chiasma frequencies as a test of two theories of recombination. Nature. 1987;326:803–805. doi: 10.1038/326803a0. [DOI] [PubMed] [Google Scholar]
- Bussell JJ, Pearson NM, Kanda R, Filatov DA, Lahn BT. Human polymorphism and human-chimpanzee divergence in pseudoautosomal region correlate with local recombination rate. Gene. 2006;368:94–100. doi: 10.1016/j.gene.2005.10.020. [DOI] [PubMed] [Google Scholar]
- Charlesworth B. A hitch-hiking guide to the genome: A commentary on “The hitch-hiking effect of a favourable gene” by John Maynard Smith and John Haigh. Genet Res. 2007;89:389–390. doi: 10.1017/S0016672308009580. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Nordborg M, Charlesworth D. The effects of local selection, balanced polymorphism and background selection on equilibrium patterns of genetic diversity in subdivided populations. Genet Res. 1997;70:155–174. doi: 10.1017/s0016672397002954. [DOI] [PubMed] [Google Scholar]
- Conway DJ, Roper C, Oduola AM, Arnot DE, Kremsner PG, Grobusch MP, Curtis CF, Greenwood BM. High recombination rate in natural populations of Plasmodium falciparum. Proc Natl Acad Sci. 1999;96:4506–4511. doi: 10.1073/pnas.96.8.4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter AD, Baird SE, Charlesworth D. High nucleotide polymorphism and rapid decay of linkage disequilibrium in wild populations of Caenorhabditis remanei. Genetics. 2006;174:901–913. doi: 10.1534/genetics.106.061879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont BL, Payseur BA. Evolution of the genomic rate of recombination in mammals. Evolution. 2007;62:276–294. doi: 10.1111/j.1558-5646.2007.00278.x. [DOI] [PubMed] [Google Scholar]
- Duret L, Arndt PF. The impact of recombination on nucleotide substitutions in the human genome. PLoS Genet. 2008;4:e1000071. doi: 10.1371/journal.pgen.1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards SV, Dillon M. Hitchhiking and recombination in birds: Evidence from Mhc-linked and unlinked loci in Red-Winged Blackbirds (Agelaius phoeniceus) Genet Res. 2004;84:175–192. doi: 10.1017/s0016672304007189. [DOI] [PubMed] [Google Scholar]
- Evans MR, Roberts ML, Buchanan KL, Goldsmith AR. Heritability of corticosterone response and changes in life history traits during selection in the zebra finch. J Evol Biol. 2006;19:343–352. doi: 10.1111/j.1420-9101.2005.01034.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Fan JB, Oliphant A, Shen R, Kermani BG, Garcia F, Gunderson KL, Hansen M, Steemers F, Butler SL, Deloukas P, et al. Highly parallel SNP genotyping. Cold Spring Harb Symp Quant Biol. 2003;68:69–78. doi: 10.1101/sqb.2003.68.69. [DOI] [PubMed] [Google Scholar]
- Farnir F, Coppieters W, Arranz J-J, Berzi P, Cambisano N, Grisart B, Karim L, Marcq F, Moreau L, Mni M, et al. Extensive genome-wide linkage disequilibrium in cattle. Genome Res. 2000;10:220–227. doi: 10.1101/gr.10.2.220. [DOI] [PubMed] [Google Scholar]
- Filatov DA, Gerrard DT. High mutation rates in human and ape pseudoautosomal genes. Gene. 2003;317:67–77. doi: 10.1016/s0378-1119(03)00697-8. [DOI] [PubMed] [Google Scholar]
- Forstmeier W. Quantitative genetics and behavioural correlates of digit ratio in the zebra finch. Proc R Soc Lond B Biol Sci. 2005;272:2641–2649. doi: 10.1098/rspb.2005.3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forstmeier W, Schielzeth H, Schneider M, Kempenaers B. Development of polymorphic microsatellite markers for the zebra finch (Taeniopygia guttata) Mol Ecol Notes. 2007a;7:1026–1028. [Google Scholar]
- Forstmeier W, Segelbacher G, Mueller JC, Kempenaers B. Genetic variation and differentiation in captive and wild zebra finches (Taeniopygia guttata) Mol Ecol. 2007b;16:4039–4050. doi: 10.1111/j.1365-294X.2007.03444.x. [DOI] [PubMed] [Google Scholar]
- Forstmeier W, Burger C, Temnow K, Derégnaucourt S. The genetic basis of zebra finch vocalizations. Evolution. 2009;63:2114–2130. doi: 10.1111/j.1558-5646.2009.00688.x. [DOI] [PubMed] [Google Scholar]
- Galtier N, Duret L. Adaptation or biased gene conversion? Extending the null hypothesis of molecular evolution. Trends Genet. 2007;23:273–277. doi: 10.1016/j.tig.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Gray MM, Granka JM, Bustamante CD, Sutter NB, Boyko AR, Zhu L, Ostrander EA, Wayne RK. Linkage disequilibrium and demographic history of wild and domesticated canids. Genetics. 2009;181:1493–1505. doi: 10.1534/genetics.108.098830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green P, Falls K, Crook S. Documentation for CRIMAP, version 2.4. Washington University School of Medicine; St. Louis, MO: 1990. [Google Scholar]
- Gregory TR, Nicol JA, Tamm H, Kullman B, Kullman K, Leitch IJ, Murray BG, Kapraun DF, Greilhuber J, Bennett MD. Eukaryotic genome size databases. Nucleic Acids Res. 2007;35:D332–D338. doi: 10.1093/nar/gkl828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin DK, Robertson LB, Tempest HG, Skinner BM. The evolution of the avian genome as revealed by comparative molecular cytogenetics. Cytogenet Genome Res. 2007;117:64–77. doi: 10.1159/000103166. [DOI] [PubMed] [Google Scholar]
- Groenen MAM, Wahlberg P, Foglio M, Cheng HH, Megens H-J, Crooijmans R, Besnier F, Lathrop M, Muir WM, Wong GK-S, et al. A high-density SNP-based linkage map of the chicken genome reveals sequence features correlated with recombination rate. Genome Res. 2009;19:510–519. doi: 10.1101/gr.086538.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson B, Akesson M, Slate J, Pemberton JM. Linkage mapping reveals sex-dimorphic map distances in a passerine bird. Proc R Soc Lond B Biol Sci. 2005;272:2289–2298. doi: 10.1098/rspb.2005.3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann I, Ebersberger I, Ptak SE, Paabo S, Przeworski M. A neutral explanation for the correlation of diversity with recombination rates in humans. Am J Hum Genet. 2003;72:1527–1535. doi: 10.1086/375657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann I, Prufer K, Ji H, Zody MC, Paabo S, Ptak SE. Why do human diversity levels vary at a megabase scale? Genome Res. 2005;15:1222–1231. doi: 10.1101/gr.3461105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez RD, Hubisz MJ, Wheeler DA, Smith DG, Ferguson B, Rogers J, Nazareth L, Indap A, Bourquin T, McPherson J, et al. Demographic histories and patterns of linkage disequilibrium in Chinese and Indian rhesus macaques. Science. 2007;316:240–243. doi: 10.1126/science.1140462. [DOI] [PubMed] [Google Scholar]
- Heuertz M, De Paoli E, Källman T, Larsson H, Jurman I, Morgante M, Lascoux M, Gyllenstrand N. Multilocus patterns of nucleotide diversity, linkage disequilibrium and demographic history of Norway Spruce [Picea abies (L.) Karst] Genetics. 2006;174:2095–2105. doi: 10.1534/genetics.106.065102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WG, Robertson A. The effect of linkage on limits to artificial selection. Genet Res. 1966;8:269–294. [PubMed] [Google Scholar]
- Huang S-W, Friedman R, Yu N, Yu A, Li W-H. How strong is the mutagenicity of recombination in mammals? Mol Biol Evol. 2005;22:426–431. doi: 10.1093/molbev/msi025. [DOI] [PubMed] [Google Scholar]
- International Chicken Genome Sequencing Consortium. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- Jensen-Seaman MI, Furey TS, Payseur BA, Lu Y, Roskin KM, Chen C-F, Thomas MA, Haussler D, Jacob HJ. Comparative recombination rates in the rat, mouse, and human genomes. Genome Res. 2004;14:528–538. doi: 10.1101/gr.1970304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson F, Whitney O. Singing-driven gene expression in the developing songbird brain. Physiol Behav. 2005;86:390–398. doi: 10.1016/j.physbeh.2005.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, et al. A high-resolution recombination map of the human genome. Nat Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- Marais G. Biased gene conversion: Implications for genome and sex evolution. Trends Genet. 2003;19:330–338. doi: 10.1016/S0168-9525(03)00116-1. [DOI] [PubMed] [Google Scholar]
- Marais G, Charlesworth B. Genome evolution: Recombination speeds up adaptive evolution. Curr Biol. 2003;13:R68–R70. doi: 10.1016/s0960-9822(02)01432-x. [DOI] [PubMed] [Google Scholar]
- Maynard Smith J, Haigh J. The hitch-hiking effect of a favourable gene. Genet Res. 1974;23:23–35. [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- McKay SD, Schnabel RD, Murdoch BM, Matukumalli LK, Aerts J, Coppieters W, Crews D, Neto ED, Gill CA, Gao C, et al. Whole genome linkage disequilibrium maps in cattle. BMC Genet. 2007;8:74. doi: 10.1186/1471-2156-8-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRae AF, McEwan JC, Dodds KG, Wilson T, Crawford AM, Slate J. Linkage disequilibrium in domestic sheep. Genetics. 2002;160:1113–1122. doi: 10.1093/genetics/160.3.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier J, Duret L. Recombination drives the evolution of GC-content in the human genome. Mol Biol Evol. 2004;21:984–990. doi: 10.1093/molbev/msh070. [DOI] [PubMed] [Google Scholar]
- Muller HJ. The relation of recombination to mutational advance. Mutat Res. 1964;1:2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- Myers S, Spencer CC, Auton A, Bottolo L, Freeman C, Donnelly P, McVean G. The distribution and causes of meiotic recombination in the human genome. Biochem Soc Trans. 2006;34:526–530. doi: 10.1042/BST0340526. [DOI] [PubMed] [Google Scholar]
- Myers S, Freeman C, Auton A, Donnelly P, McVean G. A common sequence motif associated with recombination hot spots and genome instability in humans. Nat Genet. 2008;40:1124–1129. doi: 10.1038/ng.213. [DOI] [PubMed] [Google Scholar]
- Nachman MW. Variation in recombination rate across the genome: Evidence and implications. Curr Opin Genet Dev. 2002;12:657–663. doi: 10.1016/s0959-437x(02)00358-1. [DOI] [PubMed] [Google Scholar]
- Noor MAF. Mutagenesis from meiotic recombination is not a primary driver of sequence divergence between Saccharomyces species. Mol Biol Evol. 2008;25:2439–2444. doi: 10.1093/molbev/msn186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M, Hu TT, Ishino Y, Jhaveri J, Toomajian C, Zheng H, Bakker E, Calabrese P, Gladstone J, Goyal R, et al. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 2005;3:e196. doi: 10.1371/journal.pbio.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nsengimana J, Baret P, Haley CS, Visscher PM. Linkage disequilibrium in the domesticated pig. Genetics. 2004;166:1395–1404. doi: 10.1534/genetics.166.3.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves DC. Recombination enhances protein adaptation in Drosophila melanogaster. Curr Biol. 2005;15:1651–1656. doi: 10.1016/j.cub.2005.07.065. [DOI] [PubMed] [Google Scholar]
- Ptak SE, Hinds DA, Koehler K, Nickel B, Patil N, Ballinger DG, Przeworski M, Frazer KA, Paabo S. Fine-scale recombination patterns differ between chimpanzees and humans. Nat Genet. 2005;37:429–434. doi: 10.1038/ng1529. [DOI] [PubMed] [Google Scholar]
- Reich DE, Cargill M, Bolk S, Ireland J, Sabeti PC, Richter DJ, Lavery T, Kouyoumjian R, Farhadian SF, Ward R, et al. Linkage disequilibrium in the human genome. Nature. 2001;411:199–204. doi: 10.1038/35075590. [DOI] [PubMed] [Google Scholar]
- Reich DE, Schaffner SF, Daly MJ, McVean G, Mullikin JC, Higgins JM, Richter DJ, Lander ES, Altshuler D. Human genome sequence variation and the influence of gene history, mutation and recombination. Nat Genet. 2002;32:135–142. doi: 10.1038/ng947. [DOI] [PubMed] [Google Scholar]
- Remington DL, Thornsberry JM, Matsuoka Y, Wilson LM, Whitt SR, Doebley J, Kresovich S, Goodman MM, Buckler ES., IV Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc Natl Acad Sci. 2001;98:11479–11484. doi: 10.1073/pnas.201394398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov AV. Micro versus macro: A review of structure and functions of avian micro- and macrochromosomes. Russ J Genet. 1996;32:517–527. [Google Scholar]
- Ross-Ibarra J. The evolution of recombination under domestication: A test of two hypotheses. Am Nat. 2004;163:105–112. doi: 10.1086/380606. [DOI] [PubMed] [Google Scholar]
- Samollow PB. The opossum genome: Insights and opportunities from an alternative mammal. Genome Res. 2008;18:1199–1215. doi: 10.1101/gr.065326.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shifman S, Bell JT, Copley RR, Taylor MS, Williams RW, Mott R, Flint J. A high-resolution single nucleotide polymorphism genetic map of the mouse genome. PLoS Biol. 2006;4:e395. doi: 10.1371/journal.pbio.0040395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjögren C, Nasmyth K. Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Curr Biol. 2001;11:991–995. doi: 10.1016/s0960-9822(01)00271-8. [DOI] [PubMed] [Google Scholar]
- Slate J, Pemberton JM. Admixture and patterns of linkage disequilibrium in a free-living vertebrate population. J Evol Biol. 2007;20:1415–1427. doi: 10.1111/j.1420-9101.2007.01339.x. [DOI] [PubMed] [Google Scholar]
- Slatkin M. Linkage disequilibrium—understanding the evolutionary past and mapping the medical future. Nat Rev Genet. 2008;9:477–485. doi: 10.1038/nrg2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MW, O'Brien SJ. Mapping by admixture linkage disequilibrium: Advances, limitations and guidelines. Nat Rev Genet. 2005;6:623–632. doi: 10.1038/nrg1657. [DOI] [PubMed] [Google Scholar]
- Stapley J, Birkhead TR, Burke T, Slate J. A linkage map of the zebra finch Taeniopygia guttata provides new insights into avian genome evolution. Genetics. 2008;179:651–667. doi: 10.1534/genetics.107.086264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter NB, Eberle MA, Parker HG, Pullar BJ, Kirkness EF, Kruglyak L, Ostrander EA. Extensive and breed-specific linkage disequilibrium in Canis familiaris. Genome Res. 2004;14:2388–2396. doi: 10.1101/gr.3147604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorrips RE. MapChart: Software for the graphical presentation of linkage maps and QTLs. J Hered. 2002;93:77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- Wahlberg P, Stromstedt L, Tordoir X, Foglio M, Heath S, Lechner D, Hellstrom AR, Tixier-Boichard M, Lathrop M, Gut IG, et al. A high-resolution linkage map for the Z chromosome in chicken reveals hot spots for recombination. Cytogenet Genome Res. 2007;117:22–29. doi: 10.1159/000103161. [DOI] [PubMed] [Google Scholar]
- Warren WC, Clayton DF, Ellegren H, Arnold AP, Hillier LW, Künstner A, Searle S, White S, Vilella A, Heger A, et al. The genome of the zebra finch: Special insights into vocal learning and communication. Nature. 2010 doi: 10.1038/nature08819. (in press). [DOI] [Google Scholar]
- Webster MT, Smith NG, Ellegren H. Compositional evolution of noncoding DNA in the human and chimpanzee genomes. Mol Biol Evol. 2003;20:278–286. doi: 10.1093/molbev/msg037. [DOI] [PubMed] [Google Scholar]
- Webster MT, Smith NG, Hultin-Rosenberg L, Arndt PF, Ellegren H. Male-driven biased gene conversion governs the evolution of base composition in human Alu repeats. Mol Biol Evol. 2005;22:1468–1474. doi: 10.1093/molbev/msi136. [DOI] [PubMed] [Google Scholar]