Abstract
Accurate profiling of minute quantities of RNA in a global manner can enable key advances in many scientific and clinical disciplines. Here, we present low-quantity RNA sequencing (LQ-RNAseq), a high-throughput sequencing-based technique allowing whole transcriptome surveys from subnanogram RNA quantities in an amplification/ligation-free manner. LQ-RNAseq involves first-strand cDNA synthesis from RNA templates, followed by 3′ polyA tailing of the single-stranded cDNA products and direct single molecule sequencing. We applied LQ-RNAseq to profile S. cerevisiae polyA+ transcripts, demonstrate the reproducibility of the approach across different sample preparations and independent instrument runs, and establish the absolute quantitative power of this method through comparisons with other reported transcript profiling techniques and through utilization of RNA spike-in experiments. We demonstrate the practical application of this approach to define the transcriptional landscape of mouse embryonic and induced pluripotent stem cells, observing transcriptional differences, including over 100 genes exhibiting differential expression between these otherwise very similar stem cell populations. This amplification-independent technology, which utilizes small quantities of nucleic acid and provides quantitative measurements of cellular transcripts, enables global gene expression measurements from minute amounts of materials and offers broad utility in both basic research and translational biology for characterization of rare cells.
The widespread application of microarray technologies, and, most recently, high-throughput DNA sequencing technologies, to understand biological processes and human disease has resolved numerous “mysteries” in genomics and transcriptomics and has revolutionized the way we perform biomedical research. DNA sequencing technologies have eliminated several technical challenges posed by hybridization-based microarray strategies, such as limited dynamic range of detection and background due to cross-hybridization. However, several fundamental shortcomings still remain. These include (1) the lack of an absolute measurement making cross study comparisons challenging and (2) the requirement for high-quantities of valuable input material, namely, DNA/cDNA. Progress in many research areas, including stem cell biology, microbiology, cancer, paleoarcheology, forensics, and clinical diagnostics, is severely impeded by our inability to perform comprehensive and reliable molecular profiling analyses on low-quantity cell and nucleic acid samples. This is best exemplified by the challenges experienced in the oncology community, where often acquiring sufficient amounts of high-quality tissue specimens necessary for genomic characterization of tumors is difficult. If we are to successfully translate our research knowledge of genome biology to better diagnose and treat human disease, we must make progress on our ability to use subnanogram quantities of nucleic acid derived from patient samples, and explore methods that enable absolute measurements of these small quantities.
Various strategies have been explored since the late 1980s to enable molecular profiling analyses from as few as single cells in a genome-wide manner (Pfeifer et al. 1989; Van Gelder et al. 1990; Eberwine et al. 1992; Telenius et al. 1992; Zhang et al. 1992; Dean et al. 2002; Che and Ginsberg 2004). Much effort has been devoted to characterize the behaviors of these methods to better understand and address the biases and artifacts they introduce in various quantitative and qualitative applications (Pinard et al. 2006; Subkhankulova and Livesey 2006). These approaches generally rely on multiple sample manipulation steps such as restriction digestion, ligation, and amplification that may introduce artifacts/errors, such as the production of artifactual chimeric DNA/cDNA molecules (Murthy et al. 2005; Iwamoto et al. 2007; Talseth-Palmer et al. 2008). These manipulations also skew the original structure of the nucleic acid population and often yield unequal and unreproducible representation of the transcript molecules (Pinard et al. 2006; Subkhankulova and Livesey 2006; Linsen et al. 2009; Taniguchi et al. 2009). These difficulties render these methods problematic especially for “counting” applications where accurate quantitation and high fidelity are required.
Here, we present a low-quantity RNA sequencing (LQ-RNAseq) approach, which enables novel digital transcriptome profiling capable of generating whole transcriptome profiles in a highly quantitative manner from as few as 100 pg of RNA material. Unlike other reported RNA sequencing approaches (Cloonan et al. 2008; Mortazavi et al. 2008; Sultan et al. 2008), LQ-RNAseq benefits from the advantages of high-throughput single molecule sequencing (SMS) by synthesis (Harris et al. 2008; Lipson et al. 2009; Pushkarev et al. 2009), eliminating the need for bias-introducing manipulations such as amplification and ligation and dramatically reducing the amount of input RNA needed. We demonstrate the quantitative power, high reproducibility, and other aspects of the approach by profiling the well-studied Saccharomyces cerevisiae polyA+ transcriptome. We then extended the approach to profile mouse embryonic stem cells (ESs) and induced pluripotent stem cells (iPSs), identifying similarities and, importantly, differences in transcriptional activity between these otherwise very similar pluripotent stem cell populations. This is the first report of such minute cDNA quantities being sequenced in a massively parallel manner without potentially biasing manipulations such as ligation and amplification. LQ-RNAseq promises to be an efficient and easy-to-use strategy for attomole level RNA applications and offers researchers the opportunity to obtain reliable transcriptome profiles from extremely small nucleic acid quantities, including from rare cell or tissue types of biological importance.
Results
Overview of LQ-RNAseq
To facilitate the quantitative and in-depth analysis of attomole-level RNA materials, we developed LQ-RNAseq, an approach relying on high-throughput single molecule cDNA sequencing. LQ-RNAseq involves first-strand cDNA synthesis primed with random, not-so-random (NSR) (Armour et al. 2009) or other primers of choice, followed by terminal transferase (TdT)-mediated polyA-tailing of the single-stranded cDNA and sequencing on the Helicos Genetic Analysis System (Fig. 1; Lipson et al. 2009; Pushkarev et al. 2009). The simplicity of the sample preparation process, the requirement for only single-stranded cDNA, and the lack of potentially biasing nucleic acid manipulation steps such as ligation and amplification allow LQ-RNAseq to limit the input RNA quantity, minimize artifacts, and generate reliable transcriptome profiles. LQ-RNAseq is compatible with subnanogram RNA quantities, producing 5–6 million usable reads (between 25 and 55 nucleotides [nt] in length) per channel of a 50-channel HeliScope DNA sequencing run.
Figure 1.
Attomole-level LQ-RNAseq methodology. Picogram-level RNA is reverse-transcribed with random primers and treated with RNase. Purified single-stranded first-strand cDNA is poly-A-tailed and 3′ blocked with terminal transferase. The poly-A-tailed cDNA is sequenced with the Helicos Genetic Analysis System.
Characteristics and quantification capability of LQ-RNAseq
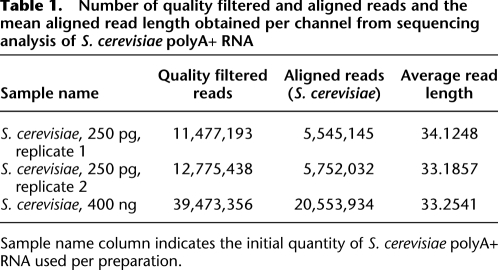
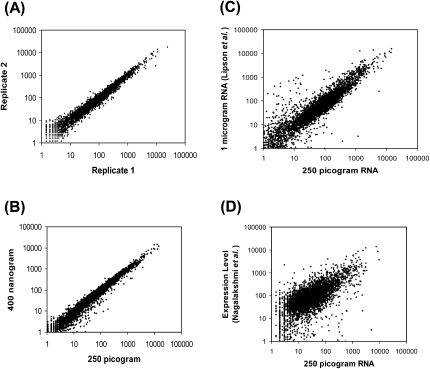
To evaluate the performance of this low-quantity approach, we first used it to sequence polyA+ RNA from the well-studied S. cerevisiae (strain DBY746). Using a starting amount of 250 pg of poly A+ RNA and random hexamer-priming for cDNA synthesis, we obtained 5 million usable sequence reads per channel of a 50-channel HeliScope sequencing run, each preparation being sufficient for loading up two to three channels (Table 1). The method yielded highly reproducible quantitative transcript counts between replicates prepared at different times and across independent sequencing runs (Pearson correlation, r = 0.9912) (Fig. 2A). A comparison of expression profiles obtained from a moderate 400-ng RNA quantity compared with a low 250-pg RNA quantity was highly correlated (r = 0.9763) (Fig. 2B). In comparing the data generated with this approach and with that of the published digital gene expression (DGE) method relying on oligo(dT) priming using ∼1 μg of a different RNA batch of the same yeast strain (Lipson et al. 2009), again data were shown to be closely correlated (r = 0.915) (Fig. 2C). This high agreement between the absolute transcript counts obtained with the published DGE results to LQ-RNAseq indicate the robustness and reliability of the expression profiles obtained from subnanogram RNA levels. Further comparison of the published expression levels obtained with an amplification-based RNA-seq approach (Nagalakshmi et al. 2008) from another S. cerevisiae strain to our data also revealed positive correlation (r = 0.661) (Fig. 2D), although lower than when comparing to the single molecule DGE data. The differences observed may be due to various factors, such as the different S. cerevisiae strains used in both studies, the ambiguities in yeast transcript 5′ and 3′ end annotations, different sample preparation steps, and “sampling effects” due to lower RNA quantities used in this study. In addition, we sequenced 5 ng of S. cerevisiae and human liver polyA+ RNAs and included 25 pg and 5 pg of two synthetic RNA spikes, observing accurate quantification of the spikes (Supplemental Table S1A). Collectively these analyses demonstrate the quantitative power and accuracy of LQ-RNAseq.
Table 1.
Number of quality filtered and aligned reads and the mean aligned read length obtained per channel from sequencing analysis of S. cerevisiae polyA+ RNA
Sample name column indicates the initial quantity of S. cerevisiae polyA+ RNA used per preparation.
Figure 2.
Reproducibility and quantitative ability of LQ-RNAseq. Reproducibility of S. cerevisiae polyA+ RNA (strain DBY746) expression profiles generated in two independent preparations of 250 pg of RNA run on different sequencers (A) and between 250-pg and 400-ng RNA preparations (B). (C,D) Comparison of LQ-RNAseq methodology to the published expression profile generated by oligo(dT) priming of 1 μg of polyA+ RNA from the same yeast strain used in this study (Lipson et al. 2009) (r = 0.915), and by random hexamer priming of 200 ng of RNA from another yeast strain (BY4741), adaptor ligation, and PCR amplification (Nagalakshmi et al. 2008) (r = 0.661). Log10 abundance is expressed using the number of unique reads aligned to each transcript in each sequencing experiment and expressed as reads number per kilobase per 1 million reads.
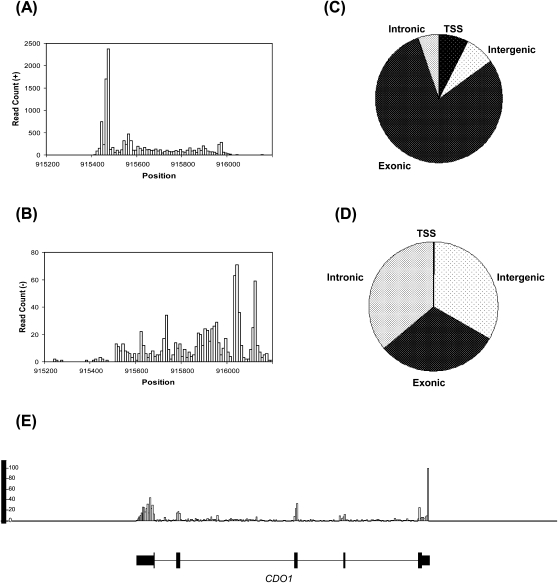
A qualitative analysis of the yeast sequence reads suggested that while the read coverage was relatively uniform across the transcripts, we did observe a 5′ bias, resulting in an accumulation of reads at or near the 5′ transcription start sites (Fig. 3). This is potentially due to the intact nature of the templates used and the relatively short length of yeast transcripts, allowing cDNA synthesis to reach 5′ transcript ends. Introduction of an RNA and/or cDNA fragmentation step for fresh RNA samples may reduce this bias without dramatically affecting the transcriptional profiles obtained (Supplemental Text; Supplemental Fig. S3) but could be problematic when dealing with minute quantities of RNA as it may be difficult to reproducibly control the RNA fragmentation step. This bias may also be lower in higher eukaryotes as transcript sizes are longer. This is exemplified by our human liver study where we see the proportion of reads mapping to transcription start regions to drop to 0.2% from the 7.4% level observed in S. cerevisiae, while the proportion of human LQ-RNAseq reads mapping to intronic and intergenic regions increase (Fig. 3D,E). Furthermore, for applications involving quantification of transcript levels, this bias should not have a negative impact.
Figure 3.
Coverage of reads aligned in the + direction (A) and − direction (B) binned in 10-nt intervals is exemplified across the HTA1 gene (location: chr4 915,521–915,919; transcription direction: +). (C) Assignment of uniquely aligned S. cerevisiae polyA+ RNA reads to the categories shown. TSS (transcription start site) category refers to regions 200 nt upstream of annotated coding region start sites; 7.4% of reads map to the yeast TSS regions. Reads in the S. cerevisiae intergenic regions include reads aligning mostly to the potentially transcriptionally active transposon repeats. (D) Assignment of uniquely aligned human liver polyA+ RNA reads to the categories shown. TSS category refers to regions 200 nt upstream and downstream of the annotated RefSeq transcription start sites; 0.2% of the reads map to the human TSS regions. (E) Human liver polyA+ RNA reads uniquely aligning to human genome was binned at 50-nt intervals and visualized using the Integrated Genome Browser. Panel exemplifies the distribution of reads across the human CDO1 (location: chr5 115,168,329–115,180,304; transcription direction: −) gene's exonic (thick bars) and intronic (thin lines) regions. Y-axis indicates the number of reads per bin.
While LQ-RNAseq offers an alternative RNA quantitation approach that is especially advantageous for analysis of minute cell quantities, this method may still suffer from common cDNA synthesis artifacts, such as spurious second-strand formation events and reverse transcriptase-related biases due to sequence context and/or higher order RNA structure. For instance, even though only first-strand cDNA is made and sequenced, examination of the strandedness of the procedure revealed 6.2%–8.9% of the sequences mapping to annotated exons aligned opposite to the known transcription direction (Fig. 3A,B). This proportion is consistent with the levels obtained from the 400-ng preparation, suggesting it is not due to subnanogram RNA levels used. While this may indicate potential antisense transcription events (Rosok and Sioud 2004; Johnson et al. 2005; Perocchi et al. 2007; He et al. 2008; Faghihi and Wahlestedt 2009), the primary contributor of these antisense reads is likely to be the reverse transcriptase tendency to generate spurious second-strand cDNA products (Spiegelman et al. 1970; Gubler 1987) and/or binding of excess random hexamers to first-strand cDNA during the reverse transcription step leading to priming of a second-strand cDNA.
Characterization of ESs and iPSs
We then used this method to examine genome-wide transcriptional states of mouse ES and iPS cells derived from mature B lymphocytes (B-iPS) (Hanna et al. 2008). Despite the rapid advances in cellular reprogramming, global gene expression profiles of iPS cells have only recently begun to be examined (Chin et al. 2009; Marchetto et al. 2009). In an effort to obtain whole-transcriptome views while minimizing reads emerging from undesirable RNA species such as ribosomal RNAs (rRNAs), we designed 408 mouse not-so-random (mNSR) primers in a manner similar to the approach described for human (Armour et al. 2009). Three nanograms of total RNA from FACS-sorted ES and B-iPS cells was converted to cDNAs with random hexamers or mNSR primers in biological duplicates, followed by 3′ poly-A tailing and SMS (Supplemental Table S1B). The percentage of reads emanating from rRNAs was 90%–94% with random hexamer priming, while this proportion dropped to 64%–75% with mNSR primers. Although the proportion of rRNA reads is still considerable in the mNSR-primed data set, the approximately threefold enrichment of non-rRNA reads in the mNSR-primed data set compared to the random-hexamer-primed case approaches the approximately fourfold level reported previously (Armour et al. 2009). Furthermore, we observed differences in the portion of rRNA reads we obtained with random hexamer-primed cDNA synthesis from ES and B-iPS cells (90%–94%), compared with data Armour et al. (2009) obtained from human reference RNA (67%). This difference may be due to potential cell type specific differences in the rRNA content. Alternatively, biases in the sample preparation or sequencing procedures of the LQ-RNAseq or the amplification-based approach used by Armour et al. (2009) might have contributed to under- or over-counting of generally GC-rich and highly structured rRNA species.
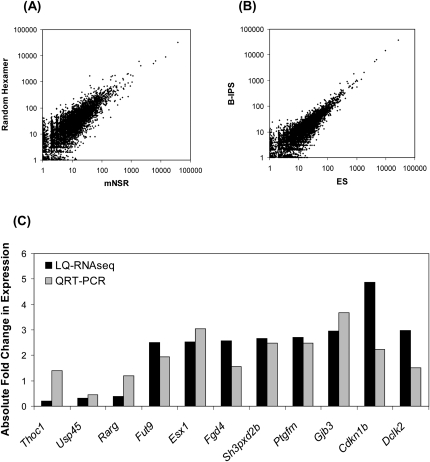
Gene expression profiles acquired with the mNSR primers were highly similar to the profiles with random hexamers (r = 0.971) (Fig. 4A). We successfully detected the expression of several known pluripotent stem cell markers (Pou5f1 [also known as Oct4], Nanog, and Lin28) in the B-iPS cells. Comparison of ES and B-iPS cell profiles revealed a high correlation (r = 0.974) (Fig. 4B) as previously reported for other iPS lines (Chin et al. 2009; Marchetto et al. 2009). Despite the global resemblance, 156 genes exhibiting differential expression were also detected (Supplemental Table S2), 11 of which were examined with the qRT-PCR assay to validate the measurements (Fig. 4C). These results indicate that there may be molecular differences in these otherwise very similar pluripotent stem cell populations. Further studies are needed to examine the significance and the cause of these differences and to determine whether these differences are related to cell origin and/or reprogramming procedures employed and whether they are biologically important.
Figure 4.
Transcriptome profiling of B-iPS and ES cells. (A) Random hexamer and mNSR primer approaches using B-iPS RNA yield similar gene expression profiles. (B) Induced pluripotent stem cells derived from B lymphocytes exhibit expression profiles similar to embryonic stem cells. Log10 abundance is expressed using the number of unique reads aligned to each transcript in each sequencing experiment and is expressed as reads number per kilobase per 1 million reads. (C) Validation of differentially expressed genes identified by LQ-RNAseq with the qRT-PCR assay. (Black bars) Fold differences observed with LQ-RNAseq; (gray bars) fold differences observed with qRT-PCR. The two genes (Thoc1 and Rarg) exhibiting discrepant expression level changes may be due to the difference in the way expression levels are calculated with the two assays and the potential presence of alternative transcript isoforms.
Discussion
LQ-RNAseq is a transcriptome analysis method that benefits from the recent advances in high-throughput single molecule DNA sequencing technology and enables subnanogram level RNA samples to be profiled in a genome-wide manner. We here demonstrated the performance and reproducibility of LQ-RNAseq by profiling the S. cerevisiae polyA+ transcriptome from 250 pg of RNA material. We also showed its quantitative power with spike-in experiments and comparison of the LQ-RNAseq data to previously generated yeast data sets using different sample preparation and sequencing strategies. We then examined the transcriptional states of mouse ES and B-iPS cells, observing differences that may have implications in the efficiency of the reprogramming process.
Unlike other reported RNA-seq approaches (Cloonan et al. 2008; Mortazavi et al. 2008; Sultan et al. 2008), LQ-RNAseq lacks amplification, ligation, or restriction digestion steps and thereby reduces biases relating to sample preparation steps. Furthermore, it requires only minute RNA quantities (250–500 pg). RNA quantities lower than this level may be used with LQ-RNAseq, but usable read yield obtained per channel may decrease. Second-strand cDNA is not generated; thus, this approach appears “mostly” strand-specific. Relatively simple alterations in the cDNA synthesis step, such as those proposed by Perocchi et al. (2007) may improve the strand-specificity of the approach. While the coverage of reads is relatively uniform across the transcription units, there is an accumulation of reads at the 5′ transcription start sites due to the lack of RNA or cDNA fragmentation steps in the standard LQ-RNAseq procedure. Fragmented RNA samples could also be profiled with LQ-RNAseq. However, accurate and reproducible fragmentation of RNA or cDNA without sample loss is problematic, especially when dealing with subnanogram RNA or cDNA samples. Differences of RNA fragmentation levels across samples may translate into difficulties particularly in comparative quantitative analyses. Therefore, at least for applications involving quantification of transcript levels, it may be preferable not to introduce an RNA or cDNA fragmentation step with this approach.
The approach presented here opens the path to new avenues of research in understanding the heterogeneity and cell dynamics of complex tissues and cell populations. It also represents an important step toward the ultimate goal of affordable, quantitative and bias-free molecular profiling capabilities from attomole-level RNA material obtained from as low as few/single cells. Future advances may further reduce the input RNA quantity requirements of LQ-RNAseq and reach single-cell levels. For example, sample loss during the RNA isolation step and the inefficiencies of cDNA synthesis reaction can be minimized by allowing cell lysis, cDNA synthesis, and other required modifications to take place in a single container, ideally in the flow cell itself. Coupling of microfluidic systems to sequencing flow cells may be necessary to load single cells to channels and enable subsequent nucleic acid manipulations. At present, the current efficiency of the poly-A-tailed cDNA template hybridization to the poly(dT)-coated sequencing flow cells is 10%–20%. Therefore, improvements in the flow cell design, nucleic acid hybridization, and chemistry to facilitate more efficient template capture within the flow cells may further reduce input template requirements. Furthermore, SMS DNA sequencing chemistry is not completely efficient, and only 15%–25% of the templates hybridized on the flow cell surfaces give rise to useable sequence reads, many never reaching the required minimum 25-nt length. Continuing optimizations in SMS DNA sequencing chemistry enabling longer reads and more efficient sequencing reactions will likely increase SMS yields and allow a higher percentage of templates on the flow cells to result in useable sequence reads. Perhaps other technologies in early stages of development such as direct RNA sequencing (Ozsolak et al. 2009) may offer an alternative route to further reduce input RNA levels if they achieve the satisfactory throughput and sequencing performance levels.
LQ-RNAseq is the first method allowing subnanogram RNA quantities to be sequenced in a massively parallel amplification-free manner. Our initial studies of transcriptome profiling from small quantity mouse stem cells provides an initial view of the biological potential for this application. Further, the simplicity and effectiveness of the method offers great opportunity for high-throughput transcriptome measurements and will likely enable the analysis of various low-quantity archival and clinical samples that are otherwise challenging with the existing approaches.
Methods
RNA preparation
mRNA from S. cerevisiae strain DBY746 grown under standard conditions, and human liver mRNA were obtained from Clontech. We diluted the RNAs to 50 pg/μL final stock concentration and used 5 μL/250 pg RNA cDNA preparation. Replicates were prepared independently from the same diluted RNA stock for reproducibility studies. RNA concentration measurements were done with the RiboGreen RNA quantitation kit (Invitrogen). The mouse Nanog-GFP iPS cell line derived from mature B lymphocytes (Hanna et al. 2008) and Oct4-GFP ESs were cultured on MEF feeder layer as described (Hanna et al. 2008). The top 5% of the GFP expressing ES or iPS cells were isolated by FACS (Aria, BD Biosciences), and aliquoted to 10,000 cells per tube. RNA was extracted from each sample using the miRNeasy kit (Qiagen). DNase I treatment was done using 2 units of DNase I (Ambion) in a 100-μL volume for 10 min at 37°C, followed by phenol/chloroform extraction and precipitation.
mNSR primer design
To reduce the number of reads originating from rRNA sequences, we used a selective hexamer approach as described previously (Armour et al. 2009). Briefly, we designed pools of DNA primers by enumerating all possible DNA hexamer sequences and removing all of those corresponding to the 28S, 18S, 16S, and 12S mouse rRNA sequences. After removing these ribosomal derived hexamers, we were left with 408 hexamers (Supplemental Table S3). To ensure that these 408 hexamers were well represented in the transcriptome, we enumerated all possible locations of these hexamers within known RefSeq transcripts. We computed the distance between hexamer locations and found the distribution to be similar to that previously described for human NSR primers (Armour et al. 2009).
cDNA preparation
First-strand cDNA was prepared from 250 pg of RNA using the SuperScript III first-strand cDNA synthesis kit (Invitrogen) using manufacturer's recommendations, except that 1 ng of random primers was used, and the incubation steps were modified as follows: 5 min at 70°C, 2 min at 4°C, 10 min at 25°C, 10 min at 37°C, and 45 min at 50°C. RNA was subsequently removed by RNase H (Invitrogen) and RNase If (NEB) digestion. The cDNA was purified with the QIAquick Nucleotide Removal Kit (Qiagen) following manufacturer recommendations. A subsequent ethanol precipitation of the cDNA was performed, and cDNA was dissolved in 10 μL of water. The synthetically produced RCA and LTP6 Arabidopsis thaliana spikes were obtained from Stratagene. The cDNA preparation for the 400 ng of RNA experiment was performed as described above, except that 50 ng of random primers were used. Three nanograms of B-iPS and ES cell RNA was combined with 1 ng of mNSR primers. The cDNA synthesis was performed in a 40-μL reaction volume following manufacturer recommendations, except that the incubation steps were modified as follows: 5 min at 65°C, 2 min at 4°C, and 90 min at 42°C. cDNA purification was done with the QIAquick Nucleotide Removal Kit (Qiagen).
Poly-A tailing and sequencing
cDNA was heat denatured at 95°C for 5 min followed by rapid snap-cooling on cooled aluminum block. Five units of TdT (New England Biolabs), 1 μg of BSA, and 200 pmol of dATP were then added to the cDNA in a 20-μL reaction volume, incubated for 1 h at 37°C, followed by the inactivation of the enzyme for 10 min at 70°C. The blocking step was performed by adding 100 pmol of biotin-11-ddATP (Perkin Elmer) and 5 units of TdT to the heat-denatured A-tailed reaction in a 10-μL volume (final volume being 30 μL), incubating for 1 h at 37°C, followed by the inactivation of the enzyme for 20 min at 70°C. The tailed and 3′-blocked cDNA was supplemented with 1 pmol of 3′ dideoxy-blocked oligonucleotide (TCACTATTGTTGAGAACGTTGGCCTATAGTGAGTCGTATTACGCGCGGT[ddC]) to minimize DNA loss during the sample capture to the sequencing flow cells. The final DNA was then loaded directly to the flow cells without additional cleaning steps. Purified cDNA from the 400 ng of RNA preparation was quantified using the OliGreen ssDNA assay kit (Invitrogen). Following the heat denaturation and snap-cooling of 60 ng of cDNA, 50 units of TdT (New England BioLabs), and 120 pmol of dATP were added to the cDNA in a 50-μL reaction volume, incubated for 1 h at 37°C and for 10 min at 70°C. The blocking step is performed by adding 50 pmol of biotin-11-ddATP (Perkin Elmer) and 40 units of TdT to the heat-denatured A-tailed reaction (final volume being 54 μL), incubating for 30 min at 37°C, followed by the enzyme inactivation for 20 min at 70°C. Template capture and sequencing was performed as described (Pushkarev et al. 2009) using the Helicos Genetic Analysis system, except that a modified hybridization buffer (available for purchase from Helicos BioSciences) was used to allow a higher fraction of templates to hybridize to flow cells. Furthermore, sample hybridization volume per channel was reduced from 100 μL to 15 μL by modifying the vacuum system of HeliScope Sample Loader to reduce the quantity of input material.
qRT-PCR validations
RNAs isolated and DNase I treated as described above were converted to cDNA using SuperScript III (Invitrogen) following manufacturer's instructions. A qRT-PCR assay for multiple genes was performed with the LightCycler (Roche Applied Science). Gapdh expression levels were used to normalize Ct values obtained for each gene. Primer sequences are provided in Supplemental Table S4.
Data analysis
Read filtering, S. cerevisiae reference sequences used, alignment (using the IndexDP algorithm), and transcript counting were done as described (Lipson et al. 2009). The mouse reference used was the MM9 assembly, and the human reference was the HG18 assembly downloaded from the UCSC Genome Browser (http://genome.ucsc.edu). The S. cerevisiae reference used was downloaded from Saccharomyces Genome Database (version 20070407) (http://www.yeastgenome.org). For the whole genome alignment of reads, the IndexDP Genomic alignment threshold used was 4.3 (Lipson et al. 2009). Using Bioconductor's edgeR (empirical analysis of DGE in R) package and RPKM (reads per kilobase of exon per million mapped reads) count values (Robinson et al. 2010), we identified 156 genes exhibiting significant gene expression changes (P-value cutoff 0.005) between ES versus B-iPS cells.
Acknowledgments
We thank Chris Hart, Kristen Kerouac, Daniel Jones, and Erik Hansen for technical assistance and discussions. A.G. is supported by an EMBO long-term postdoctoral fellowship. A.R. is an investigator of the Merkin Foundation for Stem Cell Research at the Broad Institute. B.E.B. is a Charles E. Culpeper Medical Scholar. A.R. and B.E.B. are Early Career Scientists of the Howard Hughes Medical Institute. This research was supported by funds from the Burroughs Wellcome Fund (B.E.B., A.R.), HHMI, and the National Human Genome Research Institute.
Footnotes
[Supplemental material is available online at http://www.genome.org. Sequence data from this study have been submitted to the NCBI Short Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession no. SRA009935.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.102129.109.
References
- Armour CD, Castle JC, Chen R, Babak T, Loerch P, Jackson S, Shah JK, Dey J, Rohl CA, Johnson JM, et al. Digital transcriptome profiling using selective hexamer priming for cDNA synthesis. Nat Methods. 2009;6:647–649. doi: 10.1038/nmeth.1360. [DOI] [PubMed] [Google Scholar]
- Che S, Ginsberg SD. Amplification of RNA transcripts using terminal continuation. Lab Invest. 2004;84:131–137. doi: 10.1038/labinvest.3700005. [DOI] [PubMed] [Google Scholar]
- Chin MH, Mason MJ, Xie W, Volinia S, Singer M, Peterson C, Ambartsumyan G, Aimiuwu O, Richter L, Zhang J, et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloonan N, Forrest AR, Kolle G, Gardiner BB, Faulkner GJ, Brown MK, Taylor DF, Steptoe AL, Wani S, Bethel G, et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods. 2008;5:613–619. doi: 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- Dean FB, Hosono S, Fang L, Wu X, Faruqi AF, Bray-Ward P, Sun Z, Zong Q, Du Y, Du J, et al. Comprehensive human genome amplification using multiple displacement amplification. Proc Natl Acad Sci. 2002;99:5261–5266. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberwine J, Yeh H, Miyashiro K, Cao Y, Nair S, Finnell R, Zettel M, Coleman P. Analysis of gene expression in single live neurons. Proc Natl Acad Sci. 1992;89:3010–3014. doi: 10.1073/pnas.89.7.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Cell Mol Biol. 2009;10:637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler U. Second-strand cDNA synthesis: mRNA fragments as primers. Methods Enzymol. 1987;152:330–335. doi: 10.1016/0076-6879(87)52038-9. [DOI] [PubMed] [Google Scholar]
- Hanna J, Markoulaki S, Schorderet P, Carey BW, Beard C, Wernig M, Creyghton MP, Steine EJ, Cassady JP, Foreman R, et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell. 2008;133:250–264. doi: 10.1016/j.cell.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TD, Buzby PR, Babcock H, Beer E, Bowers J, Braslavsky I, Causey M, Colonell J, Dimeo J, Efcavitch JW, et al. Single-molecule DNA sequencing of a viral genome. Science. 2008;320:106–109. doi: 10.1126/science.1150427. [DOI] [PubMed] [Google Scholar]
- He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. The antisense transcriptomes of human cells. Science. 2008;322:1855–1857. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto K, Bundo M, Ueda J, Nakano Y, Ukai W, Hashimoto E, Saito T, Kato T. Detection of chromosomal structural alterations in single cells by SNP arrays: A systematic survey of amplification bias and optimized workflow. PLoS One. 2007;2:e1306. doi: 10.1371/journal.pone.0001306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JM, Edwards S, Shoemaker D, Schadt EE. Dark matter in the genome: Evidence of widespread transcription detected by microarray tiling experiments. Trends Genet. 2005;21:93–102. doi: 10.1016/j.tig.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Linsen SE, de Wit E, Janssens G, Heater S, Chapman L, Parkin RK, Fritz B, Wyman SK, de Bruijn E, Voest EE, et al. Limitations and possibilities of small RNA digital gene expression profiling. Nat Methods. 2009;6:474–476. doi: 10.1038/nmeth0709-474. [DOI] [PubMed] [Google Scholar]
- Lipson D, Raz T, Kieu A, Jones DR, Giladi E, Thayer E, Thompson JF, Letovsky S, Milos P, Causey M. Quantification of the yeast transcriptome by single-molecule sequencing. Nat Biotechnol. 2009;27:652–658. doi: 10.1038/nbt.1551. [DOI] [PubMed] [Google Scholar]
- Marchetto MC, Yeo GW, Kainohana O, Marsala M, Gage FH, Muotri AR. Transcriptional signature and memory retention of human-induced pluripotent stem cells. PLoS One. 2009;4:e7076. doi: 10.1371/journal.pone.0007076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Murthy KK, Mahboubi VS, Santiago A, Barragan MT, Knoll R, Schultheiss HP, O'Connor DT, Schork NJ, Rana BK. Assessment of multiple displacement amplification for polymorphism discovery and haplotype determination at a highly polymorphic locus, MC1R. Hum Mutat. 2005;26:145–152. doi: 10.1002/humu.20199. [DOI] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F, Platt AR, Jones DR, Reifenberger JG, Sass LE, McInerney P, Thompson JF, Bowers J, Jarosz M, Milos PM. Direct RNA sequencing. Nature. 2009;431:814–818. doi: 10.1038/nature08390. [DOI] [PubMed] [Google Scholar]
- Perocchi F, Xu Z, Clauder-Munster S, Steinmetz LM. Antisense artifacts in transcriptome microarray experiments are resolved by actinomycin D. Nucleic Acids Res. 2007;35:e128. doi: 10.1093/nar/gkm683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer GP, Steigerwald SD, Mueller PR, Wold B, Riggs AD. Genomic sequencing and methylation analysis by ligation mediated PCR. Science. 1989;246:810–813. doi: 10.1126/science.2814502. [DOI] [PubMed] [Google Scholar]
- Pinard R, de Winter A, Sarkis GJ, Gerstein MB, Tartaro KR, Plant RN, Egholm M, Rothberg JM, Leamon JH. Assessment of whole genome amplification-induced bias through high-throughput, massively parallel whole genome sequencing. BMC Genomics. 2006;7:216. doi: 10.1186/1471-2164-7-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushkarev D, Neff NF, Quake SR. Single-molecule sequencing of an individual human genome. Nat Biotechnol. 2009;27:847–852. doi: 10.1038/nbt.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosok O, Sioud M. Systematic identification of sense–antisense transcripts in mammalian cells. Nat Biotechnol. 2004;22:104–108. doi: 10.1038/nbt925. [DOI] [PubMed] [Google Scholar]
- Spiegelman S, Burny A, Das MR, Keydar J, Schlom J, Travnicek M, Watson K. DNA-directed DNA polymerase activity in oncogenic RNA viruses. Nature. 1970;227:1029–1031. doi: 10.1038/2271029a0. [DOI] [PubMed] [Google Scholar]
- Subkhankulova T, Livesey FJ. Comparative evaluation of linear and exponential amplification techniques for expression profiling at the single-cell level. Genome Biol. 2006;7:R18. doi: 10.1186/gb-2006-7-3-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, Seifert M, Borodina T, Soldatov A, Parkhomchuk D, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- Talseth-Palmer BA, Bowden NA, Hill A, Meldrum C, Scott RJ. Whole genome amplification and its impact on CGH array profiles. BMC Res Notes. 2008;1:56. doi: 10.1186/1756-0500-1-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi K, Kajiyama T, Kambara H. Quantitative analysis of gene expression in a single cell by qPCR. Nat Methods. 2009;6:503–506. doi: 10.1038/nmeth.1338. [DOI] [PubMed] [Google Scholar]
- Telenius H, Carter NP, Bebb CE, Nordenskjold M, Ponder BA, Tunnacliffe A. Degenerate oligonucleotide-primed PCR: General amplification of target DNA by a single degenerate primer. Genomics. 1992;13:718–725. doi: 10.1016/0888-7543(92)90147-k. [DOI] [PubMed] [Google Scholar]
- Van Gelder RN, von Zastrow ME, Yool A, Dement WC, Barchas JD, Eberwine JH. Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc Natl Acad Sci. 1990;87:1663–1667. doi: 10.1073/pnas.87.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Cui X, Schmitt K, Hubert R, Navidi W, Arnheim N. Whole genome amplification from a single cell: Implications for genetic analysis. Proc Natl Acad Sci. 1992;89:5847–5851. doi: 10.1073/pnas.89.13.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]