Abstract
Next-generation sequencing technologies have made it possible to sequence targeted regions of the human genome in hundreds of individuals. Deep sequencing represents a powerful approach for the discovery of the complete spectrum of DNA sequence variants in functionally important genomic intervals. Current methods for single nucleotide polymorphism (SNP) detection are designed to detect SNPs from single individual sequence data sets. Here, we describe a novel method SNIP-Seq (single nucleotide polymorphism identification from population sequence data) that leverages sequence data from a population of individuals to detect SNPs and assign genotypes to individuals. To evaluate our method, we utilized sequence data from a 200-kilobase (kb) region on chromosome 9p21 of the human genome. This region was sequenced in 48 individuals (five sequenced in duplicate) using the Illumina GA platform. Using this data set, we demonstrate that our method is highly accurate for detecting variants and can filter out false SNPs that are attributable to sequencing errors. The concordance of sequencing-based genotype assignments between duplicate samples was 98.8%. The 200-kb region was independently sequenced to a high depth of coverage using two sequence pools containing the 48 individuals. Many of the novel SNPs identified by SNIP-Seq from the individual sequencing were validated by the pooled sequencing data and were subsequently confirmed by Sanger sequencing. We estimate that SNIP-Seq achieves a low false-positive rate of ∼2%, improving upon the higher false-positive rate for existing methods that do not utilize population sequence data. Collectively, these results suggest that analysis of population sequencing data is a powerful approach for the accurate detection of SNPs and the assignment of genotypes to individual samples.
With the availability of several next-generation sequencing platforms, the cost of DNA sequencing has dropped dramatically over the past few years and improvements in technology are expected to decrease the cost further (Shendure and Ji 2008). Next-generation sequencers, such as the Illumina Genome Analyzer (GA), can generate gigabases of nucleotides per day and have enabled the sequencing of complete individual human genomes (Bentley et al. 2008; Ley et al. 2008; Wang et al. 2008; Wheeler et al. 2008; McKernan et al. 2009). While the resequencing of complete human genomes still remains quite expensive, the targeted sequencing of specific genomic intervals in a large population of individuals is now feasible in an individual laboratory. Resequencing of coding sequences of genes in large populations has previously been shown to be useful for identifying multiple rare variants affecting quantitative traits (Cohen et al. 2004, 2006; Ji et al. 2008). Resequencing of genomic regions identified by genome-wide association studies in healthy and diseased populations represents a powerful strategy for assessing the contribution of rare variants to disease etiology. Nejentsev et al. (2009) have used this approach to identify four rare variants protective for type 1 diabetes.
For harnessing the capacity of next-generation sequencers for deep population resequencing, the first challenge is to selectively capture DNA from the region of interest. Recently, Craig et al. (2008) used long-range PCR and DNA barcodes to sequence specific regions of the human regions in multiple samples simultaneously using the Illumina GA. Harismendy et al. (2009) also used long-range PCR to sequence targeted regions of the human genome using multiple sequencing platforms to evaluate the feasibility of targeted population sequencing and the concordancy of variant calling between the different platforms. However, traditional sequence capture methods, such as long-range polymerase chain reaction (LR-PCR), are not adequate for capturing thousands of noncontiguous regions of the genome, e.g., all exons, in a large number of samples. Several high-throughput target capture methods have been developed (Hodges et al. 2007; Okou et al. 2007; Porreca et al. 2007; Turner et al. 2009).
After millions of reads have been generated by the sequencer, the next challenge is to identify genetic variants by mapping the reads to a reference sequence. A variety of tools have been developed that can efficiently align hundreds of millions of short reads to a reference sequence even in the presence of multiple errors in the reads (Li et al. 2008; Langmead et al. 2009; Li and Durbin 2009; Li et al. 2009b). Each base mismatch in an aligned read represents either a sequencing error or a single nucleotide variant in the diploid individual. To compensate for the high sequencing error rates of next-generation sequencing platforms, one requires the presence of multiple overlapping reads, each with a base different from the reference base for single nucleotide polymorphism (SNP) calling. Base quality values—probability estimates of the correctness of a base call—are particularly useful for distinguishing sequencing errors from SNPs. The Illumina sequencing system generates a phred-like quality score for each base call using various predictors of the sequencing errors. SNP calling methods for Illumina sequence data utilize these base quality values to compute the likelihood of different genotypes at each position using Bayesian or statistical models (Li et al. 2008, 2009a). Positions for which the most likely genotype is different from the reference genotype and which satisfy additional filters on neighborhood sequence quality, read alignment quality, etc. are reported as SNPs. However, sequencing errors for the Illumina GA are not completely random and are dependent on the local sequence context of the base being read, the position of the base in the read, etc. (Dohm et al. 2008; Erlich et al. 2008). Therefore, assuming independence between multiple base calls, each with a non-reference base, results in overcalling of SNPs, i.e., increased number of false-positives. To reduce the number of false variant calls, the MAQ SNP caller (Li et al. 2008) uses a dependency model to estimate an average error rate using all base quality scores.
MAQ and other SNP calling methods have enabled fairly accurate detection of SNPs from resequencing of individual human genomes (Bentley et al. 2008; Wang et al. 2008). However, there is potential for developing more accurate SNP detection methods, in particular, by taking advantage of sequence information from a population of sequenced individuals. Comparison of sequenced reads for a potential variant site across multiple individuals has the potential to differentiate systematic sequencing errors from real SNPs. Patterns of mismatched bases (bases not matching the reference base) resulting from systematic sequencing errors are likely to be shared across individuals. On the other hand, the profiles of mismatched bases between individuals with and without a SNP are likely to be distinct. Comparison of read alignments across multiple individuals also has the potential to filter out SNPs that are an artifact of inaccurate read alignments. We present a probabilistic model that leverages sequence data from a population of individuals, each sequenced separately, for detecting single nucleotide variants and also assigning genotypes to each individual in the population.3 Our method recalibrates each base quality value by adding a population error correction to the Illumina base error probability. This correction is computed using the distribution of mismatched bases across all sequenced individuals. The recalibrated base quality values are then used to compute genotype probabilities for each individual under a simple Bayesian model that assumes independence between base calls. Finally, positions in the sequence with one or more individuals showing evidence for harboring a non-reference allele are identified as SNPs. Craig et al. (2008) described a similar approach for SNP detection using sequence data from multiple individuals where they used Bayes factors to compare the fraction of reads with the alternate allele across multiple individuals. Sites at which one or more individuals have a fraction of reads with the alternate allele sufficiently greater than the average were identified as SNPs. Our model is much more general and can take advantage of the complete information about each base call, i.e., base quality value, position in the read containing the base, and the strand to which the read aligns to.
To evaluate our population SNP detection method, we analyzed sequence data from a 200-kilobase (kb)-long region on chromosome 9p21 that was sequenced to a median depth of 45× in 48 individuals using the Illumina Genome Analyzer (O Harismendy, V Bansal, N Rahim, X Wang, N Heintzman, B Ren, EJ Topol, and KA Frazer, in prep.). We demonstrate that our method can accurately detect SNPs with a low false-positive rate (∼2%) and a low false-negative rate in comparison to SNP detection from individual sequence data using MAQ. By comparing genotype calls between replicate samples, we show a 98.8% accuracy for sequence-based genotyping using our method.
Results
For SNP detection using sequence data from one individual, there is no distinction between common or rare variants. In contrast, SNPs with a high minor allele frequency should be easier to detect by sequencing a population of individuals. The more challenging task is to distinguish rare SNPs from sequencing errors. Systematic sequencing errors can result in a few individuals being called as heterozygous for a particular position using individual sequence reads. Other individuals in the population also carry a small number of reads with the alternate allele, but may be below the threshold for being classified as heterozygous. For real SNPs, individuals homozygous for the reference allele are unlikely to demonstrate reads with the alternate allele. Using population sequence information, one can potentially distinguish between false SNPs due to sequencing errors and real SNPs.
Before presenting the formal description, we use an example to illustrate the intuition behind our approach. Consider a potential SNP site in a population of individuals with reference allele A and alternate allele B. Each individual in a population has one of three possible genotypes: AA, AB, and BB, assuming the locus is biallelic. Due to sequencing errors, even individuals with the genotype AA can have reads with the B allele. Consider the case where one individual has seven reads with the A allele and three reads with the B allele. If none of the other sequenced individuals in a population carries a read with the B allele, it is very likely that the position represents a real variant. If many individuals carry a small number of reads with the B allele, then the position could represent a common SNP or all reads with the B allele could be attributed to systematic sequencing error in reading the base A as B. However, if the three B alleles are in the same position in the reads, then the SNP is unlikely to be a real one. In general, comparison of the distribution (strand to which read is aligned, position in read, and base quality score) of mismatched bases across multiple individuals can reveal whether the reads with an alternate allele in a single individual represent a SNP or are an artifact of sequencing errors.
Framework for population SNP detection
For each potential SNP position, the reads from all individuals that cover this position are partitioned into bins based on the position of the variant site in the read, the base quality score and the strand of the reference sequence that the reads align to. Non-reference base calls for individuals who are homozygous for the reference base at the SNP position represent sequencing errors. For each bin, our method utilizes the base counts from such individuals to recalibrate the Illumina base quality values by adding a population error correction to the base error probability. The rationale behind this approach is that systematic base-call errors in the sequencing (if any) are likely to be clustered in one or more bins, since such errors are dependent upon the local sequence context (the nucleotides flanking the base being read), the base being read, position of the base within the read, etc. (Dohm et al. 2008; Erlich et al. 2008). In contrast, non-reference base calls that represent a variant allele are expected to be more or less randomly distributed across the bins. Therefore, the correction is expected to lower the base quality values of non-reference base calls that represent systematic sequencing errors and not affect base calls that represent alternate alleles. Lowering the base quality values of erroneous base calls is likely to decrease the probability of calling systematic sequencing errors as SNPs.
For computing the population error correction, we partition the reads from all sequenced individuals covering a potential SNP site into 36 × 2 × 3 bins. Each bin corresponds to one of the 36 sequencing cycles, one of the two strands to which the read aligns (forward/reverse), and one of the three intervals for the base quality value (0–9, 10–19, and 20–30). For each bin, we used the base calls from individuals who are homozygous for the reference base R to compute an empirical error probability Pb of each base call being incorrect (see Methods). For each base, let Pq denote the probability of the base call being incorrect using the Illumina base quality value. We define the combined error probability as
Here, (1 − Pq) · Pb · wb is the population error correction and wb is the weight of the contribution of the empirical error probability and is proportional to the square root of the number of reads in the bin containing the base call.
Algorithm for SNP detection and genotyping (SNIP-Seq)
The algorithm starts by assigning a genotype to each individual in the population using the posterior probability distribution of the genotypes, which is computed using the Illumina base quality values under a simple Bayesian model (see Methods). For a potential SNP with reference allele A and alternate allele B, each individual is assigned one of the three genotypes: AA, AB, or BB. Using the base calls for the individuals with the AA genotype, the algorithm computes the population-derived correction for each bin using the formula described previously and recalibrates the base quality values for each base call. A genotype is sampled for each individual using the new base quality values under the Bayesian model. This procedure is repeated iteratively until the genotype assignments do not change between consecutive iterations. The complete algorithm for iteratively sampling the genotypes, re-estimating the base quality values, and detecting SNPs is as follows:
-
For each potential variant site in the sequenced region:
Set the base quality value for each base call to the Illumina quality value
-
For k = 1, 2,…
a. Sample a genotype for each individual from the posterior distribution using a heterozygote prior of 0.001.
b. Recalibrate the quality score for each base call using genotypes for all individuals.
If the genotype of any individual is different from the reference, identify position as a SNP.
Sample a genotype for each individual from the posterior distribution computed using a heterozygote prior of 0.2.
Accuracy of SNP detection
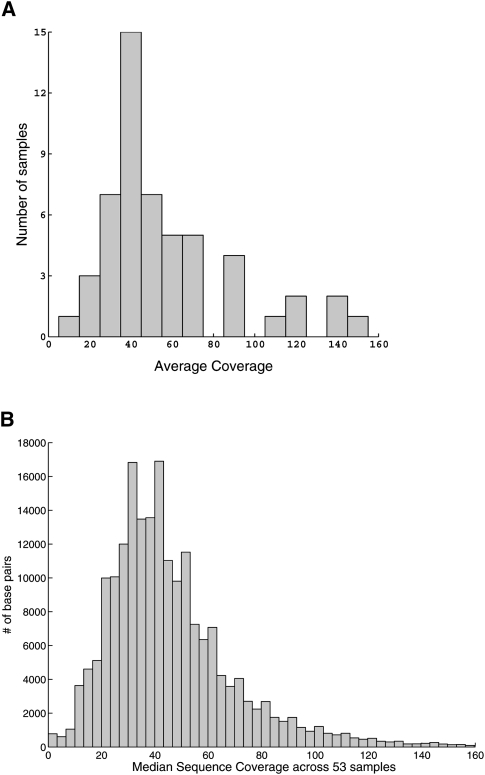
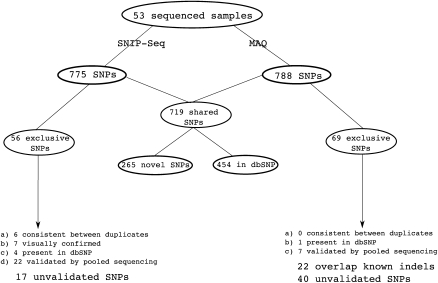
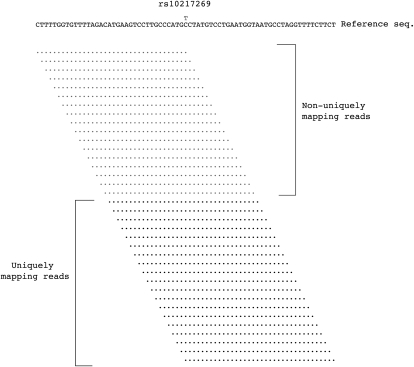
We applied our method to sequence data on 48 individuals from a 200-kb region on chromosome 9p21. The individuals were sequenced on two runs of the Illumina GA using barcoded adapters at a median coverage of ∼45× (for the distribution of average sequence coverage for each of the sequenced samples and the distribution of the median sequence coverage at each position across the targeted region, see Fig. 1). Five of the 48 individuals were sequenced twice resulting in a total of 53 sequenced samples (for details of sequencing, see Methods). We used the MAQ aligner to align the reads to the reference sequence and filtered read alignments for uniquely mapping reads (see Methods). Our SNP detection method, SNIP-Seq (single nucleotide polymorphism identification from population sequence data), identified 775 single nucleotide variants across the 53 samples (Fig. 2). We compared the identified SNPs to variants in dbSNP (build 129), 458 variants (59%) matched previously identified SNPs with the identical alternate allele. A large fraction (80%) of the 317 novel variants was observed in a small number of samples (170 in one sample and 84 in two samples). For comparison, we also applied the default MAQ SNP calling method (threshold of Q20) to detect SNPs in each of the 53 sequenced samples. This identified 788 SNPs, of which 455 (58%) overlapped known SNPs in dbSNP and 719 SNPs were shared with SNIP-Seq (including 265 novel variants not present in dbSNP). Of the 56 SNPs exclusive to SNIP-Seq, six were concordant between duplicate samples suggesting that they are real, seven were singletons (detected in only one sample) that showed clear evidence for the presence of an alternate allele (four or more unique start-site reads), and four were present in dbSNP (Fig. 2). Three of the four SNPs present in dbSNP were located close to the boundary of a repetitive sequence (for an illustration, see Fig. 3). SNPs near repetitive sequences are typically filtered out by individual SNP calling methods to avoid false SNPs.
Figure 1.
(A) Distribution of the average sequence coverage for the 53 sequenced samples. (B) Distribution of the median sequence coverage for each position across the 53 samples.
Figure 2.
Comparison of the set of SNPs identified by SNIP-Seq and MAQ from the 53 samples.
Figure 3.
SNP calling at the edge of a repetitive sequence. SNP rs10217269 is at the boundary of a repetitive sequence with 17 of the 36 36-mers covering the position aligning to multiple locations in the reference sequence. However, 19 36-mers align uniquely to this position (allowing for 0–1 mismatches) and hence can be used by SNIP-Seq to detect a SNP at this site.
Considering the 69 SNPs exclusive to MAQ, only one was present in dbSNP. This SNP had low coverage (10×) and was below the threshold to call SNPs using SNIP-Seq. Further analysis of these 69 SNPs revealed that 22 overlapped previously known indels (dbSNP 129) and had read alignment patterns consistent with indels, clearly indicating that these represent indel variants that are classified as SNPs, due to inaccurate alignments. For 33 (71.7%) of the 46 SNPs exclusive to MAQ (excluding those that overlap known indels and the one SNP present in dbSNP), the genotype consensus score (maximum over all samples classified as heterozygous) was below Q30. In comparison, only 19 (7.5%) of the 265 novel SNP common to MAQ and SNIP-Seq had a best genotype consensus score less than Q30. This suggests enrichment for false-positives in the set of SNPs with a MAQ consensus score below Q30. Increasing the threshold from Q20 to Q30 for MAQ would reduce the number of false-positives, but also filter out some real SNPs.
To estimate SNIP-Seq false-negative rates, one of the 48 individuals was sequenced to a very high depth of coverage (300×) using one lane of the Illumina GA. At such a high sequence depth, all SNPs should be easily identifiable. We identified 260 SNPs in this individual using a simple SNP calling method (see Methods). Of these, 253 were detected in the same individual sequenced as part of the indexed population sequencing. Each of the seven missed SNPs either had low sequence coverage in the indexed sample (4/7) or was present in low complexity sequence (5/7). Comparison to dbSNP, the high overlap between the SNP calls from MAQ and SNIP-Seq and results from the deep sequencing of one individual indicate that our method has a low false-negative rate of ∼2%–3%.
Validation of novel SNPs using pooled sequencing data and Sanger sequencing
To validate novel SNP calls exclusively identified by MAQ and our algorithm SNIP-Seq, we utilized pooled sequencing data from the 200-kb region for 50 individuals. These 50 individuals (the 48 individuals sequenced individually and two additional individuals) were sequenced in two pools of 25 individuals each using one run of the Illumina GA. Both pools had an average sequence coverage greater than ∼2000×, resulting in each individual having a very high depth of coverage. We identified potential SNPs from this data by comparing allele counts between the two pools for every position in the reference sequence using the Fisher's exact test (see Methods). Using this approach, we detected 700 SNPs of which 394 represent previously known SNPs (dbSNP v129) and 645 match SNPs identified by MAQ or SNIP-Seq using the population sequencing. A large fraction of the SNPs that were not identified using this approach either represented SNPs present in equal numbers of individuals in the two pools or singletons (data not shown). The substantial overlap with the individual SNP calls suggested that this simple approach could accurately identify SNPs from pooled sequencing data and could therefore be used to validate novel SNP calls.
Twenty-four SNPs exclusive to SNIP-Seq were shared with pooled SNP calls. For each of these SNPs, the alternate alleles between the individual sequencing data and the pooled sequencing were identical. Excluding two of these SNPs that were also present in dbSNP, pooled sequencing supported an additional 22 novel SNPs called by SNIP-Seq. In comparison, only seven SNPs exclusively identified by MAQ showed evidence for the presence of a SNP from the pooled sequencing data. The pooled sequencing was done independently of the indexed sequencing and we used a completely different method to detect SNPs from the pooled sequence data. Hence, the pooled SNP calls are unlikely to be biased in favor of either SNIP-Seq or MAQ. Nevertheless, the subset of SNP calls supported by the pooled sequencing data was further validated by PCR amplification and Sanger sequencing. Twenty-six of the 29 SNPs (20/22 for SNIP-Seq and 6/7 for MAQ) were successfully amplified and Sanger sequenced (Supplemental Table 1). For SNIP-Seq, all 20 SNPs were confirmed to be heterozygous by Sanger sequencing with the identical alternate allele. For MAQ, two of the six SNPs were heterozygous, three were homozygous for the reference allele, and one SNP turned out be a 5-bp homozygote indel.
Thus, of the 56 exclusive SNIP-Seq SNPs, 39 (69.6%) have independent evidence that they are likely real (see Fig. 2). Whereas for the 69 exclusive MAQ SNPs, only three (4.3%) have independent evidence supporting they are real. Overall, the greater fraction of SNPs exclusive to SNIP-Seq that are validated point to a lower false-positive rate for SNIP-Seq. Although it is difficult to obtain precise estimates for the false-positive rate for SNP calling, we estimate that our method has a low false-positive rate4 ∼2% (17 of 775 SNPs).
Accuracy of sequence-based genotype calls
An important application of population resequencing is to determine the genotype at SNPs for each individual in a population. Accurate genotyping of SNPs is crucial for sequencing-based association studies to be feasible. We assessed the accuracy of the sequencing derived genotype calls using two different strategies. First, we evaluated the sequence-based genotypes at 21 SNPs (present in dbSNP) that were also genotyped in the 48 sequenced individuals using the Sequenom MassARRAY platform (see Methods). Of 1035 genotype calls common between the Sequenom genotypes and SNIP-Seq genotype calls, 34 were discordant. These discordancies could be further categorized as 28 heterozygote undercalls (heterozygote call by one method called as reference homozygote by other) and six heterozygote overcalls (heterozygote called as reference homozygote by other). Analysis of the sequence coverage at these 34 genotypes in the sequenced samples showed that 15 had coverage below 10×, 26 had sequence coverage below 20×, and none had coverage greater than 30×. Therefore, low sequence coverage could explain virtually all discordancies and 98.1% of SNIP-Seq genotype calls with sequence coverage ≥10× were concordant with the array-based genotype calls.
Although known SNPs can be genotyped using genotyping arrays, certain SNPs can fail genotyping due to the presence of hidden variants (Bentley et al. 2008). Sequencing represents the most direct method for the simultaneous discovery and genotyping of SNPs and has been shown to be robust to the presence of hidden variants. In order to evaluate the accuracy of genotype calling between duplicate samples at all identified SNPs, we took advantage of sequence data from five individuals that were sequenced twice on the Illumina GA. For each pair of duplicate samples, we compared the genotype calls at each position in the reference sequence. We restricted the comparison to sites for which both samples had at least 10× coverage and those which were called as heterozygote or alternate homozygote in at least one of the two samples. In total, we had 1060 pairs of comparisons for the five pairs of duplicate samples using the SNIP-Seq algorithm. Only 13 of these pair of genotype calls were discordant, which corresponds to an error rate of 1.2%. The 13 discordancies represented nine heterozygote undercalls and four heterozygote overcalls. In contrast, 31 of 1035 (3.1%) pairs of genotype calls were discordant for the MAQ variant calls. These represented 24 heterozygote undercalls, six heterozygote overcalls, and one genotype classified as reference homozygote in one duplicate and as alternate homozygote in the other. The different number of comparisons is due to the different sets of SNPs reported by the two methods. These results demonstrate that the population-based approach has a significantly lower discordancy rate for genotype assignments between duplicate samples and an absolute accuracy of 98.8%. This greater accuracy is due to improved genotype calling and in part due to the greater accuracy in detecting SNPs, since false SNPs are more likely to be discordant between duplicates.
SNP detection using base counts
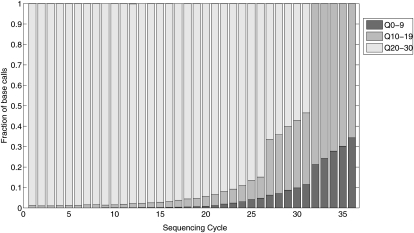
Our method uses the base calls in each bin for computing the population error correction that is used to recalibrate each base quality value in the bin. For computing an accurate estimate of the population error value, each bin should contain a sufficient number of base calls. In the most general model described earlier, each sequencing cycle corresponds to a separate bin. For 53 samples, each sequenced to an average coverage of 45×, the average number of base calls in each bin5 was ∼33. When the number of sequenced samples is not very large, the number of base calls in each bin can be increased by reducing the granularity of the bins. For example, sequencing cycles with similar error rates can be grouped together. To illustrate the flexibility of our population SNP calling framework, we considered a binning approach where the base calls for each position were partitioned into two bins, one for each strand. This approach ignored position-specific error rates. Therefore, we only considered bases at positions 2–26 in reads, since the average sequencing error rate increased significantly after cycle 26 (see Fig. 4). In addition, we removed base calls with quality scores below 10. Using this model, our method identified 742 SNPs in the 53 samples. 739 of these were identical to the SNPs detected in the 53 samples using SNIP-Seq under the more general model. The reduced number of detected SNPs can be explained by the reduction in the effective coverage. The small number of novel SNPs likely represents false variants since the general model is more effective in filtering out false-positives.
Figure 4.
Distribution of base quality values for each sequencing cycle for one of the 53 sequenced samples.
Application of SNIP-Seq to additional population sequencing data sets
To further demonstrate the ability of SNIP-Seq to accurately identify SNPs while achieving a low false-positive rate, we applied it to population sequence data obtained from the targeted sequencing of ∼190 kb of the human genome in 42 individuals using 36-bp single end Illumina reads (Scripps Genomic Medicine, unpub.). The targeted regions were amplified using LR-PCR and sequenced using barcoded adapters in a similar manner as for the chromosome 9p21 population data set. The average sequence coverage was 80 ± 32 across the 42 individuals. We applied SNIP-Seq (with the same parameters as used for the 9p21 data set) to identify SNPs from the sequenced population of individuals. SNIP-Seq identified 673 SNPs in the sequenced population of which 378 (56.2%) were present in dbSNP and 634 (94.2%) were also identified by MAQ (at a Q20 threshold). Thirty-two of the 39 SNPs exclusive to SNIP-Seq were singleton heterozygotes. Further analysis of these 39 SNPs showed that five were present in dbSNP with the identical alternate allele, seven were singletons identified by MAQ as SNPs but had a consensus score below 20, and four singletons had the MAQ consensus genotype as heterozgote, but were filtered out. An additional 10 singleton SNPs had strong evidence for the presence of the alternate allele with 90%–96% of the reads with the alternate allele across the population concentrated in only one individual. Of the remaining 13 SNPs, one was classified as heterozygote in all samples (suggesting some type of systematic error) and eight SNPs minimally satisfied the thresholds used for identifying SNPs using SNIP-Seq. These results indicate that the false-positive rate of SNIP-Seq on this data set is not more than 2%–3% and similar to that for the 9p21 data set.
The 1000 Genomes project (http://1000genomes.org/) has sequenced the coding regions of 1000 genes in several hundred individuals (Pilot Project 3). We applied SNIP-Seq to identify SNPs from this population sequence data. SNIP-Seq identified 626 SNPs in 120 individuals across ∼150 kb of targeted regions on chromosome 1 (for details, see Supplemental material). Of these, 304 (48.6%) represented previously known SNPs (dbSNP 129). Further analysis showed that 46% of the known cSNPs were nonsynonymous. In comparison, 55% of the novel cSNPs were nonsynonymous. For coding SNPs, the ratio of transitions to transversions was 2.99, consistent with previous observations (Freudenberg-Hua et al. 2003). While it is difficult to estimate the accuracy of SNIP-Seq on this data without validation of the novel SNP calls, it demonstrated the general applicability of our method.
Discussion
Deep sequencing of genomic regions represents a powerful approach to identify the complete spectrum of DNA sequence variants. With the availability of ultrahigh-throughput sequencing platforms and efficient target capture methods, sequence-based association scans are likely to become common in the near future. Accurate detection and genotyping of SNPs is crucial for using population sequencing to detect rare, as well as common, variants that increase susceptibility to common diseases. The short read lengths and the high error rates of reads generated by the Illumina GA pose new computational challenges for the accurate detection of SNPs. Many methods have been developed for aligning short reads with multiple errors to a reference sequence and SNP calling for the Illumina GA (Li et al. 2008, 2009a,b; Malhis et al. 2009). In particular, MAQ represents an efficient, easy-to-use and popular tool for read alignment and SNP calling. However, MAQ and other SNP detection methods have been designed for calling SNPs using individual sequence data and do not take advantage of sequence data from a population of individuals.
We have proposed a novel approach to SNP detection that leverages sequence data from multiple individuals at the same locus. For each potential SNP, SNIP-Seq utilizes the set of base calls across all samples to recalibrate base quality values, identifies SNPs in each sample individually using the recalibrated base quality values and subsequently assigns genotypes to each sample at each SNP site. For sampling genotypes in each sample, we use a simple Bayesian model that assumes independence between multiple base calls. Systematic sequencing errors can result in inflated base quality values, which in turn, can result in a high false-positive rate for SNP calling under the simple Bayesian model. However, the recalibration is designed to lower the base-quality values of nonrandom sequencing errors, which in turn is expected to lower the probability of calling such sequencing errors as SNPs using the simple Bayesian model. Using sequence data from a 200-kb region of the human genome sequenced in 48 individuals, we have demonstrated the accuracy of our method for both SNP detection and genotype assignment. Comparison to previously reported variants in dbSNP and validation of novel variants using pooled sequencing data strongly suggests that our method has a lower false-positive rate, as well as a lower false-negative rate, in comparison to methods that call SNPs individually for each sample. We estimate that our method has a low false SNP detection rate of less than 2%. It is important to note that this represents an estimate of the population false-positive rate rather than the individual false-positive rate. Almost all false variant calls correspond to singleton variants (called in one sample), while a large fraction of the real SNPs are common in the population. Therefore, the individual false SNP positive rate is much lower than the population false-positive rate. Similarly, the greater the population frequency of the non-reference allele for a SNP, the more likely it is to be detected from population sequencing. Therefore, our false-negative rate of 2%–3% implies a higher population false-negative rate.
There are several advantages to SNP detection using a population of sequenced samples. Population-based SNP detection can automatically filter out SNPs that represent artifacts of systematic sequencing errors. Additionally, once a SNP has been identified in an individual, relaxed criteria can be used to assign genotypes (detect SNPs at low coverage) in other individuals. Population sequence data can be used for SNP detection even in the absence of accurate base quality values if a large number of individuals are sequenced. We have demonstrated the power of population SNP calling using sequencing data from the Illumina GA. However, our method represents a generic framework for population SNP calling, which could be adapted for sequencing data from other platforms. We note that existing methods for individual SNP calling could also be modified to leverage sequence data from a population of individuals.
Finally, our method is designed to detect SNPs in a population of individuals where each individual has been sequenced to a moderate depth of coverage (10–20×) on the same sequencing platform. It does not attempt to combine evidence for the presence of a SNP from multiple samples and is therefore not suited for SNP detection from very low coverage population sequence data. It also does not utilize information from multiple SNPs simultaneously to identify SNPs or call genotypes. Statistical methods have been developed to impute genotypes at untyped SNPs or call missing genotypes using the correlations between alleles at neighboring SNPs in a population of individuals (Marchini et al. 2007; Servin and Stephens 2007). Li et al. (2009c) have used simulations to show that imputation methods can also be used for polymorphism detection and genotype calling from the sequencing of a large number of individuals at very low coverage, e.g., 400 individuals at 2–4× coverage per individual. However, in order to detect variants and impute genotypes, such methods require the sequencing of a population of individuals with similar ancestry and also require the variant allele to be observed in the population a sufficient number of times. In comparison, SNIP-Seq aims to accurately identify all rare variants (with low false-positive and low false-negative rates), including those present in a single individual in the population, and accurately assign genotypes. A large fraction of false-positives and false-negatives correspond to singletons and for such variants, imputation-based methods are unlikely to provide additional information.
Methods
Population sequencing data
We used SNIP-Seq to analyze sequence data from 48 individuals in a 196-kb interval on chromosome 9p21. This region contains SNPs associated with Coronary Artery Disease (CAD) and Type 2 Diabetes (T2D) in genome-wide association studies. Fifty individuals were sequenced as part of a study to identify and characterize the sequence variants in this region (O Harismendy, V Bansal, N Rahim, X Wang, N Heintzman, B Ren, EJ Topol, and KA Frazer, in prep.). The targeted region (NCBI36 chr 9: 21,996,845–22,193,741) was amplified using 42 LR-PCR amplicons in each of the 50 individuals. The amplicons from each sample were pooled in equimolar amounts and the pool subjected to DNA library preparation as previously described (Harismendy et al. 2009) with the following modification: we used a 4-bp DNA barcode indexing adapter in the library preparation, similar to indexes used by Craig et al. (2008). We pooled five to six sequencing libraries indexed with distinct barcodes in one lane of Illumina GA I and sequenced for 40 cycles. Forty-eight of the 50 individuals were sequenced with barcodes and were used for evaluating our SNP calling method SNIP-Seq. Five of the 48 individuals were sequenced twice resulting in a total of 53 sequenced samples. The median coverage of the sequencing for the 53 samples was 45× (Fig. 1).
Read mapping and SNP calling using MAQ
We used MAQ version 0.6.8 (http://maq.sourceforge.net/) to align the 36-bp reads (after removing the 4-bp barcode) for each sample to the reference sequence of the targeted region using default parameters. MAQ calculates the genotype probabilities for each position in an individual using the sequenced reads and positions for which the most likely genotype is different from the reference homozygote genotype are called as potential variant sites. We also used the default MAQ SNP calling filters to identify SNPs and the corresponding genotypes for each individual. The default MAQ SNP caller imposes a Q20 threshold for calling a site as a variant site in an individual. For each position that was reported by MAQ to be a SNP in one or more individuals, we used the most likely genotype from the MAQ consensus calls to assign genotypes to each individual at that position.
SNP detection using SNIP-Seq
For SNP calling using SNIP-Seq, we utilized the subset of reads with three or fewer mismatches to the reference sequence. Reads with a MAQ mapping quality of zero were filtered out for SNP calling. Such reads have two or more equally good alignments to the reference sequence. For each potential SNP, reads with the reference allele were allowed to have at most two mismatches, while reads that carried the alternate allele were allowed to have a maximum of three mismatches. This was done to eliminate potential bias in favor of reads with the reference allele, since reads with the reference allele have one fewer mismatch to the reference sequence than reads with the alternate allele. Additionally, for a potential SNP site, we also filtered out reads with the reference allele if the corresponding read with the non-reference allele did not have a unique alignment to the reference sequence. The motivation for doing this was to remove bias in favor of reads with the reference allele at any potential SNP site. Additionally, within each aligned read, we utilized base calls at positions 2–31, since the error rate in the last five bases increased dramatically (see Fig. 4). Base calls at the first position were discarded since the quality values of non-reference base calls were significantly lower than quality values for reference base calls. Base calls with an Illumina phred-scaled quality value below 10 were not used for SNP calling. The average error rate across the 53 sequenced samples (restricted to the filtered set of base calls) was 0.007 ± 0.0018. Figure 4 shows the cycle-to-cycle distribution of quality values for one of the 53 sequenced samples.
For each potential variant site, SNIP-Seq was initialized with the list of filtered base calls for each of the sequenced samples. The algorithm iteratively sampled genotypes for each sample using the quality values and recalibrated base quality values using the genotype assignments for all samples. The algorithm was allowed to run for a maximum of five iterations. In practice, however, the algorithm typically converged in a few iterations. SNPs called by the algorithm SNIP-Seq were filtered further to remove SNPs with low read counts and false SNPs likely to be artifacts of inaccurate alignments. We required a minimum of reads to call a SNP (5× or greater), a minimum number of unique start site reads with the alternate allele (one from each strand or at least three from one strand), and at least one read with the alternate allele present outside the ends of the read (first and last 5 bp of the read). If any one of the sequenced samples passed these filters, the site was identified as a SNP.
Recalibration of base quality values
For each bin, we used the base calls from individuals who were called as being homozygous for the reference base R to compute the four probabilities Pr(R → R′) (R′ = {A, C, T, G}), where Pr(R → R′) is the probability of reading the base R as R′. We define Pr(R → R′) = N(R′)/N, where N equals the total number of base calls in the bin and N(R′) is the number of R′ base calls. For non-reference base calls, Pb, i.e., the empirical error probability equals Pr(R → R′), while for reference base calls, Pb equals 1 − Pr(R → R′). Pq, the probability of the base call being incorrect using the Illumina quality value was computed as 10−0.1×Q, where Q is the phred-scaled base quality value. For recalibrating base quality values using Equation 1, we used 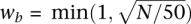 , which corresponds to a maximum weight of 1 for bins with 50 or more base calls. Alternate ways of combining the two error probabilities could be used, such as by sampling the error probability from a β distribution derived from the base counts with a prior based on the quality value.
, which corresponds to a maximum weight of 1 for bins with 50 or more base calls. Alternate ways of combining the two error probabilities could be used, such as by sampling the error probability from a β distribution derived from the base counts with a prior based on the quality value.
Individual genotype likelihoods considering all aligned reads
Let 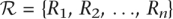 represent the set of aligned reads covering a position p in a sample. Let A and B be the two most common bases at this position (determined using the allele counts for all sequenced samples). For a diploid individual, consider the three possible genotypes corresponding to the two alleles A and B: AA, AB, and BB. We want to find the most likely genotype given the set of reads
represent the set of aligned reads covering a position p in a sample. Let A and B be the two most common bases at this position (determined using the allele counts for all sequenced samples). For a diploid individual, consider the three possible genotypes corresponding to the two alleles A and B: AA, AB, and BB. We want to find the most likely genotype given the set of reads 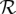 . Assuming independence between sequencing errors from multiple reads, we can define:
. Assuming independence between sequencing errors from multiple reads, we can define:
and
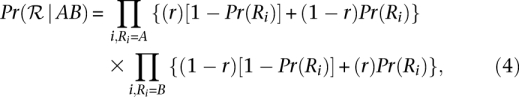 |
where Pr(Ri) is the probability that the base in read Ri is incorrect and r is the probability of sampling the chromosome with the “A” allele. Pr(Ri) corresponds to the sequencing error probability and can be derived from the base quality value Q as 10−0.1×Q. Assuming equal likelihood of sampling the two chromosomes, the righthand side of Equation 4 reduces to 0.5n, where n is the number of reads with the nucleotide A or B at position p. The posterior probability of each of the three genotypes {AA, AB, BB}, conditional on the observed read data, can be computed using Bayes rule:
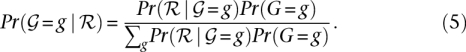 |
We note that similar equations for an independent SNP calling model have been described earlier (Supplemental material, Sec. 4.1; Li et al. 2008). To actually compute the likelihoods, we need to specify the prior probability of observing a heterozygote genotype. Similar to previous methods (Li et al. 2009a), the prior probability of observing a heterozygote, i.e., Pr(AB) was set to 0.001 and the homozoygote priors Pr(AA) and Pr(BB) equal to [1 − Pr(AB)]/2 for each sample. For SNPs that have been identified in one or more individuals in the population, we increase the prior heterozygote probability to 0.2 to assign genotypes to each sample. More sophisticated prior probabilities can be chosen using additional information about the frequency of different classes of nucleotide substitutions or knowledge of SNPs from previous studies (Li et al. 2009a).
SNP calling in a single individual sequenced to high depth
One individual was sequenced to an average sequence coverage of ∼300×, using one lane of the Illumina GA. For each position, we used the simple Bayesian model (described in the previous section) with a heterozygote prior of 0.001 for computing the posterior likelihoods of the three genotypes corresponding to the two most common bases. We did not consider base calls with a quality score below 10, filtered out reads with more than three mismatches to the reference sequence, and only considered bases at positions 2–31 in the reads.
Pooled sequencing and SNP detection
The targeted region was amplified separately in the 50 individuals (including the 48 individuals sequenced using indexing) using the 42 LR-PCR amplicons. The amplicons were then pooled in equimolar amounts to form two pools of 25 individuals each. A DNA sequencing library was then prepared from each pool as described by O Harismendy, V Bansal, N Rahim, X Wang, N Heintzman, B Ren, EJ Topol, and KA Frazer (in prep.). We sequenced each pool on four lanes of the Illumina GA1 sequencer for 36 cycles using version one recipes. After pooling the reads from the four lanes and aligning them to the targeted reference sequence, we obtained an average coverage of 2000× per pool, resulting in an average depth of 80× per sample.
We used the MAQ alignment program to align the reads to the reference sequence. We only considered reads with three or less mismatches and base calls with quality scores of 10 or greater for computing allele counts. For each position s in the reference sequence, we computed the two allele counts ai and bi (i = 1, 2) for each of the two pools. The allele counts were computed separetely for the two strands. We used the Fisher's exact test to compute a P-value for the significance of the difference in allele counts between the two pools. Positions with a P-value below 0.01 were reported as potential SNPs. We required significant evidence of difference in allele counts between the two pools from both strands (each strand P-value ≤ 0.2). This approach identified 700 single nucleotide variants of which 390 were present in dbSNP and 645 overlapped SNPs identified by MAQ or our method SNIP-Seq.
Validation of SNPs by Sanger sequencing
We attempted to validate each of the 29 novel SNPs that were supported by the pooled sequencing data. Amplicons were designed using Primer3 (http://frodo.wi.mit.edu/primer3/) with default parameters for 26 SNPs (20 for SNIP-Seq and six for MAQ). For each SNP, PCR amplification was performed in one sample with the strongest evidence for the presence of the variant allele. Amplification was carried out in 50-μL reactions with 1× Phusion HF buffer, 200 μM dNTPs, 0.5 μM forward and reverse primers, 100 ng of DNA, and 0.02 U/μL Phusion polymerase (Finnzyme). The reaction was then cycled with the following conditions: initial denaturation at 98°C for 30 sec; seven cycles at 98°C for 5 sec, 70°C for 15 sec, and 72°C for 30 sec; 30 cycles at 98°C for 5 sec, 65°C for 15 sec, and 72°C for 30 sec; final extension at 72°C for 5 min. PCR products was purified using AMPure SPRI magnetic beads (Beckman). Samples were sequenced by Sanger chemistry by Eton Biosciences and all variants were manually called by visual inspection.
Genotyping of 21 SNPs
Twenty-one SNPs (selected from dbSNP to represent a range of minor allele frequencies) from the 200-kb region on chromosome 9p21 were genotyped using the Sequenom MassARRAY platform in 48 individuals. PCR assays and extension primers for these SNPs were designed using the MassARRAY Assay Design software, version 3.1 (Sequenom). SNPs were genotyped using the iPLEX Gold assay, based on multiplex PCR followed by a single base primer extension reaction. The mass of the primer extension products, correlating to genotype, as determined using matrix assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry. Final genotypes were called using the MassArray Type, version 4.0.
SNIP-Seq implementation and download
The SNIP-Seq method has been implemented in python and is available for download from the Supplemental material and also from the website http://polymorphism.scripps.edu/∼vbansal/software/SNIP-Seq/. It accepts read alignments in the generic SAM format from which it creates pileup files for each sequenced sample. The input files to SNIP-Seq consist of a set of pileup files and it outputs the list of detected SNPs and a genotype for each sample.
Acknowledgments
This work was supported by Scripps Genomic Medicine and the Scripps Translational Science Institute under the Clinical Translational Science Award (NIH U54RR02504-01).
Footnotes
We use the term population sequencing for the sequencing of multiple individuals rather than the sequencing of a population of individuals with a similar genetic background.
False-positive rate is defined as the fraction of SNPs called across all samples in the population that are not real.
Assuming 72 bins corresponding to 36 bp reads and the two strands.
[Supplemental material is available online at http://www.genome.org. The SNIP-Seq method is freely available at http://polymorphism.scripps.edu/∼vbansal/software/SNIP-Seq/.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.100040.109.
References
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- Cohen JC, Pertsemlidis A, Fahmi S, Esmail S, Vega GL, Grundy SM, Hobbs HH. Multiple rare variants in NPC1L1 associated with reduced sterol absorption and plasma low-density lipoprotein levels. Proc Natl Acad Sci. 2006;103:1810–1815. doi: 10.1073/pnas.0508483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig DW, Pearson JV, Szelinger S, Sekar A, Redman M, Corneveaux JJ, Pawlowski TL, Laub T, Nunn G, Stephan DA, et al. Identification of genetic variants using bar-coded multiplexed sequencing. Nat Methods. 2008;5:887–893. doi: 10.1038/nmeth.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohm JC, Lottaz C, Borodina T, Himmelbauer H. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Res. 2008;36:e105. doi: 10.1093/nar/gkn425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlich Y, Mitra PP, delaBastide M, McCombie WR, Hannon GJ. Alta-Cyclic: A self-optimizing base caller for next-generation sequencing. Nat Methods. 2008;5:679–682. doi: 10.1038/nmeth.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freudenberg-Hua Y, Freudenberg J, Kluck N, Cichon S, Propping P, Nöthen MM. Single nucleotide variation analysis in 65 candidate genes for CNS disorders in a representative sample of the European population. Genome Res. 2003;13:2271–2276. doi: 10.1101/gr.1299703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harismendy O, Ng PC, Strausberg RL, Wang X, Stockwell TB, Beeson KY, Schork NJ, Murray SS, Topol EJ, Levy S, et al. Evaluation of next generation sequencing platforms for population targeted sequencing studies. Genome Biol. 2009;10:R32. doi: 10.1186/gb-2009-10-3-r32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges E, Xuan Z, Balija V, Kramer M, Molla MN, Smith SW, Middle CM, Rodesch MJ, Albert TJ, Hannon GJ, et al. Genome-wide in situ exon capture for selective resequencing. Nat Genet. 2007;39:1522–1527. doi: 10.1038/ng.2007.42. [DOI] [PubMed] [Google Scholar]
- Ji W, Foo JN, O'Roak BJ, Zhao H, Larson MG, Simon DB, Newton-Cheh C, State MW, Levy D, Lifton RP, et al. Rare independent mutations in renal salt handling genes contribute to blood pressure variation. Nat Genet. 2008;40:592–599. doi: 10.1038/ng.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley TJ, Mardis ER, Ding L, Fulton B, McLellan MD, Chen K, Dooling D, Dunford-Shore BH, McGrath S, Hickenbotham M, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Li Y, Fang X, Yang H, Wang J, Kristiansen K, Wang J. SNP detection for massively parallel whole-genome resequencing. Genome Res. 2009a;19:1124–1132. doi: 10.1101/gr.088013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics. 2009b;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009c;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhis N, Butterfield YS, Ester M, Jones SJ. Slider–maximum use of probability information for alignment of short sequence reads and snp detection. Bioinformatics. 2009;25:6–13. doi: 10.1093/bioinformatics/btn565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- McKernan KJ, Peckham HE, Costa G, McLaughlin S, Tsung E, Fu Y, Clouser C, Dunkan C, Ichikawa J, Lee C, et al. Sequence and structural variation in a human genome uncovered by short-read, massively parallel ligation sequencing using two-base encoding. Genome Res. 2009;19:1527–1541. doi: 10.1101/gr.091868.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nejentsev S, Walker N, Riches D, Egholm M, Todd JA. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009;324:387–389. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okou DT, Steinberg KM, Middle C, Cutler DJ, Albert TJ, Zwick ME. Microarray-based genomic selection for high-throughput resequencing. Nat Methods. 2007;4:907–909. doi: 10.1038/nmeth1109. [DOI] [PubMed] [Google Scholar]
- Porreca GJ, Zhang K, Li JB, Xie B, Austin D, Vassallo SL, LeProust EM, Peck BJ, Emig CJ, Dahl F, et al. Multiplex amplification of large sets of human exons. Nat Methods. 2007;4:931–936. doi: 10.1038/nmeth1110. [DOI] [PubMed] [Google Scholar]
- Servin B, Stephens M. Imputation-based analysis of association studies: Candidate regions and quantitative traits. PLoS Genet. 2007;3:e114. doi: 10.1371/journal.pgen.0030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- Turner EH, Ng SB, Nickerson DA, Shendure J. Methods for genomic partitioning. Annu Rev Genomics Hum Genet. 2009;10:263–284. doi: 10.1146/annurev-genom-082908-150112. [DOI] [PubMed] [Google Scholar]
- Wang J, Wang W, Li R, Li Y, Tian G, Goodman L, Fan W, Zhang J, Li J, Zhang J, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, He W, Chen YJ, Makhijani V, Roth GT, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]