Abstract
Fluorescent quantum dots coated with zinc(II)-dipicolylamine coordination complexes can selectively stain a rough Escherichia coli mutant that lacks an O-antigen element and permit optical detection in a living mouse leg infection model.
Recently, we discovered that small, fluorescent probes with 10 zinc(II) dipicolylamine (Zn-DPA) units as targeting ligands act as universal stains for most, if not all, strains of bacteria.1,2 The Zn-DPA ligands have a strong affinity for the anionic phospholipids and related phosphorylated amphiphiles that are ubiquitous on the bacterial cell surface.3 As part of a program 15 to create extremely bright fluorescent probes for detection of bacterial contaminantion in the environment and in vivo imaging of living animals, we are currently evaluating nanoparticle scaffolds. The fluorescnt CdSe/ZnS core/shell nanoparticles known as Quantum Dots (QDs) are promising 20 imaging agents with several attractive features such as broad absorption, narrow emission bands, extreme brightness, and high photostability.4 The commercial availability of streptavidin-coated QDs (up to one hundred times brighter than streptavidin–FITC)5 and the technical simplicity of 25 mixing these nanoparticles with biotinylated targeting ligands is an attractive way to generate imaging probes.6 It appears to be a straightforward strategy with large targeting ligands, such as biotinylated-antibodies (MW ~ 150 kD), whose highly specific recognition abilities hardly change upon immobilization to the QD surface. Indeed, antibody-QD probes have recently been shown to selectively target the surfaces of Escherichia coli,7 Salmonella typhimurium,8 Mycobacterium bovis,9 and oral bacteria.10 However, the cell binding outcome when small biotinylated targeting ligands are 35 used is not likely to be so predictable. The ligand recognition properties may change substantially depending on structural variables, e.g., the steric size and polarity of the polymeric material that coats the QD and supports the immobilized streptavidin, the streptavidin loading level, accessibility of the 40 cell surface target, multivalent complementarity between target and ligand, biotin-ligand linker length, etc.
Here, we report that a relatively small Zn-DPA targeting ligand exhibits altered bacterial cell surface recognition properties when it is attached to a QD. Specifically, we have 45 treated the biotinylated Zn-DPA probe, 1 (MW 0.7 kD),11 with separate samples of streptavidin-coated QDs and created a suite of extremely bright fluorescent imaging probes (Figure 1) whose bacterial affinity is determined by the cell surface topology. The probes can distinguish different mutants of the 50 same bacterial species. This toplogical information is complementary to that gained from smaller molecular probes, like the Gram stain and labeled antibiotics, which target different binding locations on the bacterial cell.12
Figure 1.
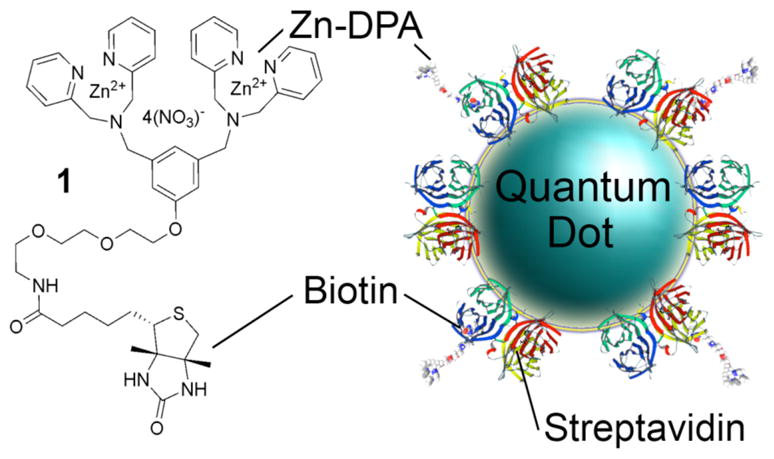
Association of Zn-DPA-biotin conjugate 1 with a streptavidin–coated QD.
A two step procedure was followed to achieve bacterial 55 staining. First, the Zn-DPA biotin conjugate 1 (4 μM) and streptavidin-coated red quantum dots (RQD, em. 655 nm, 1 μM) were mixed to give the 1-RQD nanoparticle complex. Next, the complex was added to separate samples of three E. coli strains; JM83, UTI89, and AO16. After washing with 60 buffer, the cells were examined using fluorescence microscopy. Intense surface staining was observed with the E. coli JM83 (Figure 2) which is a rough strain (derivative of E. coli K12) that lacks the branched, O-antigen polysaccharide component extending from the lipopolysaccharide (LPS) in 65 the exterior monolayer of the cell’s outer membrane.13 No cell staining was obtained with the smooth E. coli UTI89 and AO16 strains that have wild type LPS and extended O-antigen polysaccharides composed of ~ 200 sugar units.14 Repeating these staining experiments with streptavidin-coated green 70 quantum dots (i.e., 1-GQD, em: 565 nm) produced the same micoscopy results (Figure 2).¶ It appears that the UTI89 and AO16 cell surfaces are protected by a “lawn” of O-antigen polysaccharides that prevent access of the relatively large nanoparticles to the phopsphorylated “lipid A” portion of LPS 75 buried in the outer membrane. Thus, for Gram negative E. coli, the nanoscale probes, 1-RGD and 1-GQD, are staining indicators of O-antigen length on the cell surface.
Figure 2.
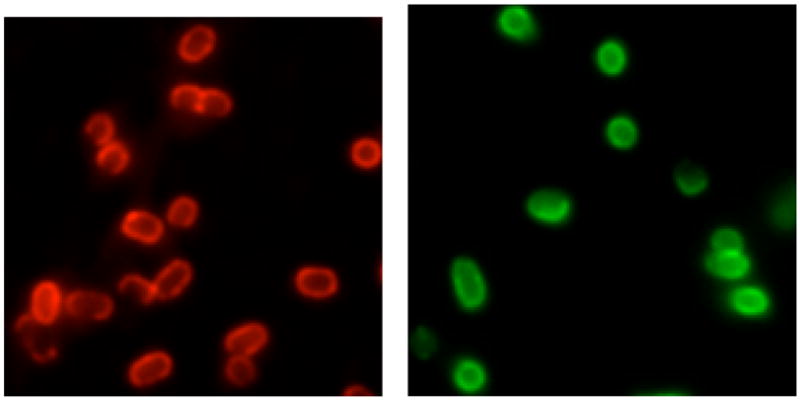
Fluorescence micrographs of rough E. coli JM83 cells stained with 1-RQD (left) and 1-GQD (right).
It is worth noting that cell staining does not occur if the order of reagent addition is reversed. That is, no bacterial 80 staining is observed if the E. coli strains are treated first with the Zn-DPA-biotin conjugate 1 and then the streptavidin-coated RQD. It appears that the streptavidin-coated RQD cannot reach the biotin group on 1 after it binds to the bacteria. It is possible that the bacteria remove the biotinylated 1 from the surface via promicuous biotin transport systems,15 which would explain why there is no staining even with the E. coli JM83 cells that lack sterically protecting O-antigen polysacchrides.
The preformed 1-RQD and 1-GQD complexes were also tested for staining of Gram-positive Staphylococcus aureus NRS11 and Enterococcus facaelis cells. No cell binding was observed which contrasts with the intense staining obtained previously using small fluorescent Zn-DPA probes.1 These results suggest that anionic phospholipids in the Gram-positive bacterial membrane are crucial binding targets. The membrane is protected by a thick, surrounding cell wall which contains pores that are too small (maximum diameter of around 10 nm)16 to allow passage of the functionalized quantum dots (hydrodynamic diameter 15–20 nm).17
The bacterial staining results indicate that tethering Zn-DPA affinity ligands to relatively large QDs produces fluorescent probes that can detect differences in cell surface topology. It should be possible to employ these extremely bright probes in highly sensitive multicolored staining schemes for rapid identification of bacterial species and mutant strains in contaminated samples. An added feature with streptavidin-coated QDs is the ability to quickly create a suite of multiplexed QD probes with different biotinylated ligands. These probes can be incorporated into staining arrays and analysed by pattern recognition methods.18 This staining array technology is much faster than classical plating and culturing methods, and is well suited for “point-of-care” medical applications.
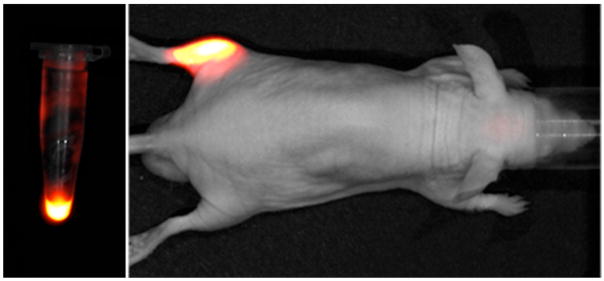
A concurrent goal of the study was to determine the feasibility of Zn-DPA coated QD probes for in vivo imaging of bacterial infection in living mice. Optical imaging of bacteria is emerging as an effective method to study pre-clinical models of infection.19 Bacteria may be genetically encoded with luminescent proteins that signal their presence, but when these reporters are unavailable, the bacteria must be labeled with a synthetic molecular probe. To achieve maximum tissue penetration, the excitation and emission light should be in the window of 650–900 nm.20 While a streptavidin-coated quantum dot tuned to emit at 800 nm (NIRQD) would seem to be ideal, its optimal excitation wavelength is below 500 nm,6 which is problematic for in vivo imaging because penetration depth is diminished and there is substantial autofluorescence. Moving to longer excitation wavelengths substantially decreases the fluorescence brightness. So although Near-IR QDs have been reported by others as fluorescent probes for imaging of lymph nodes21 and tumors,22 it was not clear to us that in vivo brightness would be greater than that observed previously with a Near-IR Cy-7 fluorophore.2a Therefore, we conducted the following bacterial labeling and in vivo imaging experiment. E. coli JM83 cells (~108 cells) were treated with the preformed probe 1-NIRQD and the sample centrifuged and washed twice. The labeled bacterial cells were imaged using an IVIS Lumina imaging station and a Near-IR imaging filter set (Ex: 635±20 nm, Em: 840±30 nm, Low Binning, Fstop 1, acquisition time 10 s). The left panel in Figure 3 shows a false colored fluorescent image of the labeled JM83 cells in an eppendorf tube (as expected, 1-NIRQD does not stain UTI89 and AO16 strains). Further analysis of these cells by fluorescence microscopy showed that the fluorescence was localized on the bacterial cell surface (essentially the same staining micrographs as Figure 2). These labeled bacterial cells were injected into the rear left leg of a living nude mouse and the entire animal was imaged after five minutes. The right panel in Figure 3 shows a photographic image of the mouse with the Near-IR fluorescence overlayed. Region of interest (ROI) analysis of the mouse fluorescence image indicated that the Near-IR signal from the site of bacterial infection is approximately 10-fold greater than the background autofluorescence from the mouse’s back. While this level of contrast is potentially very useful, it is only 1.5 times higher than that obtained when the bacteria are labeled with a Zn-DPA probe containing an Near-IR Cy-7 fluorophore.2a Compared to organic dyes, the brightness advantage of visible wavelength QD probes is undisputed,5 however, the photophysical advantages of NIRQD for in vivo imaging are less apparent.‡ In any case, non-covalent labeling of bacteria with Near-IR Zn-DPA probes is technically straightfoward and produces very bright in vivo images that should enable studies of bacteria motility and antibiotic efficacy in a mouse leg infection model.19,23
Figure 3.
Left: Fluorescence image of an eppendorf tube containing E. coli JM83 cells labeled with 1-NIRQD. Right: Nude mouse injected in the left rear calf muscle with the 1-NIRQD labeled E. coli sample from the left panel.
Acknowledgments
This work was supported by the NIH (GM059078) and Sandia National Laboratories. Sandia is a multiprogram laboratory operated by Sandia Corporation, a Lockheed Martin Company, for the United States Department of Energy’s National Nuclear Security Administration under contract DE-AC04-94AL8500. We thank D. R. Dixon (US Army Dental and Trauma Research Detachment) for JM83 and AO16 cell lines, and S. J. Hultgren at Washington University in St Louis for UTI89 cells.
Footnotes
Control experiments demonstrated that the streptavidin-coated QDs 10 alone do not stain any of the E. coli strains, but all all three strains were intensely stained with small probes composed of Zn-DPA ligands attached to organic fluorophores.
As probes for in vivo imaging QDs may have advantages such as 15 multivalency but they also have potential drawbacks, such as toxicity and slow rates of renal clearance. For further discussion, see reference 17.
References
- 1.Leevy WM, Johnson JR, Lakshmi C, Morris J, Marquez M, Smith BD. Chem Commun. 2006:1595. doi: 10.1039/b517519d. [DOI] [PubMed] [Google Scholar]
- 2.(a) Leevy WM, Gammon ST, Jiang H, Johnson JR, Maxwell DJ, Jackson EN, Marquez M, Piwnica-Worms D, Smith BD. J Am Chem Soc. 2007;128:16476. doi: 10.1021/ja0665592. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) DiVittorio KM, Leevy WM, O’Neil EJ, Johnson JR, Vakulenko S, Morris JD, Rosek KD, Serazin N, Hilkert S, Hurley S, Marquez M, Smith BD. Chem Bio Chem. 2008;9:286. doi: 10.1002/cbic.200700489. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Leevy WM, Gammon ST, Johnson JR, Lampkins AJ, Jiang H, Marquez M, Piwnica-Worms D, Suckow MA, Smith BD. Bioconj Chem. 2008 doi: 10.1021/bc700376v. ASAP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ratledge C, Wilkinson SG, editors. Microbial Lipids. Vol. 1. Academic Press; London: 1988. [Google Scholar]
- 4.(a) Du W, Wang Y, Luo Q, Liu BF. Anal Bioanal Chem. 2006;386:444. doi: 10.1007/s00216-006-0541-z. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S. Science. 2005;307:538. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hahn MA, Tabb JS, Krauss TD. Anal Chem. 2005;77:4861. doi: 10.1021/ac050641i. [DOI] [PubMed] [Google Scholar]
- 6.The streptavidin-coated QDs used in this study were purchased from Invitrogen Corporation, and the photophysical properties are summarized in the supporting imformation.
- 7.Su XL, Li Y. Anal Chem. 2004;76:4806. doi: 10.1021/ac049442+. [DOI] [PubMed] [Google Scholar]
- 8.Yang L, Li Y. Analyst. 2006;131:394. doi: 10.1039/b510888h. [DOI] [PubMed] [Google Scholar]
- 9.Otsuka Y, Hanaki KI, Zhao J, Ohtsuki R, Toyooka K, Yoshikura H, Kuratsuii T, Yamamoto K, Kirikae T. Jpn J Infect Dis. 2004;57:183. [PubMed] [Google Scholar]
- 10.Chalmers NI, Palmer RJ, Jr, Du-Thumm L, Sullivan R, Shi W, Kolenbrander PE. Appl Environ Microbiol. 2007;73:630. doi: 10.1128/AEM.02164-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanshaw RG, Lakshmi C, Lambert TN, Smith BD. ChemBiochem. 2005;12:2214. doi: 10.1002/cbic.200500149. [DOI] [PubMed] [Google Scholar]
- 12.(a) Beveridge TJ. Biotechnic Histochem. 2001;76:111. [PubMed] [Google Scholar]; (b) Tiyanont K, Doan T, Lazarus MB, Fang X, Rudner DZ, Walker S. Proc Natl Acad Sci USA. 2006;103:11033. doi: 10.1073/pnas.0600829103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.(a) Kuhnert P, Nicolet J, Frey J. Appl Environ Microbiol. 1995;61:4135. doi: 10.1128/aem.61.11.4135-4139.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ishiwa A, Komano T. J Bacteriol. 2003;185:5192. doi: 10.1128/JB.185.17.5192-5199.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.(a) Bainbridge BW, Page RC, Darveau RP. Infect Immun. 1997;65:4801. doi: 10.1128/iai.65.11.4801-4805.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Chen SL, Hung CS, Xu J, Reigstad CS, Magrini V, Sabo A, Blasiar D, Bieri T, Meyer RR, Ozersky P, Armstrong JR, Fulton RS, Latreille JP, Spieth J, Hooton TM, Mardis ER, Hultgren SJ, Gordon JI. Proc Nat Acad Sci USA. 2006;103:5977. doi: 10.1073/pnas.0600938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walker JR, Altman E. Appl Environ Microbiol. 2005;71:1850. doi: 10.1128/AEM.71.4.1850-1855.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meroueh SO, Bencze DH, Lee M, Fisher JF, Stemmler TL, Mobashery S. Proc Nat Acad Sci. 2006;103:4404. doi: 10.1073/pnas.0510182103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi HS, Liu W, Misra W, Tanaka E, Zimmer EJP, Ipe BI, Bawendi MG, Frangioni JV. Nature Biotchnol. 2007;25:1165. doi: 10.1038/nbt1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phillips RL, Miranda OR, You C-C, Rotello VM, Bunz UH. Angew Chem Int Ed. 2008;47 doi: 10.1002/anie.200703369. [DOI] [PubMed] [Google Scholar]
- 19.Leevy WM, Serazin N, Smith BD. Drug Disc Today: Disease Mod. 2008 doi: 10.1016/j.ddmod.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bashkatov AN, Genina EA, Kochubey VI, Tuchin VV. J Phys D: Appl Phys. 2005;38:2543. [Google Scholar]
- 21.Parungo CP, Colson YL, Kim SW, Kim S, Cohn LH, Bawendi MG, Frangioni JV. Chest. 2005;127:1799. doi: 10.1378/chest.127.5.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cai W, Dong-Woon S, Chen K, Gheysens O, Cao Q, Wang SX, Gambhir SS, Chen X. Nano Lett. 2006;6:669. doi: 10.1021/nl052405t. [DOI] [PubMed] [Google Scholar]
- 23.Kadurugamuwa JL, Sin LV, Yu J, Francis KP, Kimura R, Purchio T, Contag PR. Antimicrob Agents Chemother. 2003;47:3130. doi: 10.1128/AAC.47.10.3130-3137.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]