Abstract
Allergic rhinitis (AR), chronic rhinosinusitis (CRS), and asthma are prevalent airway diseases that can have a substantial impact on a patient’s quality of life. Mass spectrometry analyses of biological fluids can effectively screen for proteins associated with disease processes, however, initial detection of diagnostic proteins is difficult due to protein complexity and dynamic range. To enhance the detection of lower abundance proteins, intact nasal lavage fluid (NLF) proteins from nonpolypoid AR and from asthmatic CRS patients were extensively fractionated prior to LC/MS/MS analysis. Pooled NLF samples were processed to remove low molecular weight molecules and high abundance plasma proteins. Anion exchange chromatography (AX) followed by RP-LC further separated the remaining intact NLF proteins. The resulting fractions were digested with trypsin and the peptides analyzed by LC/MS/MS. Spectra were searched with Mascot, Sequest, and X!Tandem to obtain peptide identifications and subsequently analyzed by Scaffold software to identify parent proteins with at least 99% confidence. The 197 identified proteins are compared to those previously cited in the literature and the workflow evaluated to determine the usefulness for detection of lower abundance proteins. This is the first extensive list of NLF proteins generated from CRS patients with coexisting asthma.
Keywords: Allergic rhinitis, Chronic rhinosinusitis, Nasal lavage, Protein fractionation, Proteomics
1 Introduction
Rhinitis and sinusitis comprise a large group of disorders that impact tens of millions of people in the U.S. and worldwide. Symptoms can range from mild rhinorrhea and sneezing in allergic rhinitis (AR) to debilitating mucus compaction and tissue remodeling in chronic rhinosinusitis (CRS). AR and CRS are separately associated with asthma, another prevalent and potentially debilitating airway disease [1]. Together, AR, CRS, and asthma, with their characteristic eosinophilic inflammation, account for tens of billions of annual costs [2, 3]. Current therapeutic options can control mild AR and asthma, but more severe disease requires continuous immunosuppressive therapy. For CRS, no standard FDA-approved therapy exists and endoscopic sinus surgery is a common last resort intervention.
Proteomic studies identify proteins using electrophoretic, chromatographic, and mass spectrometry techniques. Differential proteomics can be used to define biomarkers or molecular pathways associated with disease pathology. The serum proteome has been studied most frequently; however, other human proteomes, such as cerebrospinal fluid, saliva, urine, bronchoalveolar fluid, and nasal lavage fluid (NLF), are appearing in the literature [4, 5]. There are four main components to a proteomic study: (1) sample collection and protein extraction, (2) protein or peptide fractionation, (3) detection of peptides or proteins, and (4) interpretation and protein identification. Experimental design and implementation are difficult as each of these components require optimization for individual proteomes; often times with limited sample numbers and protein quantities.
The complexity of biological fluids, dynamic range of the proteins, and the presence of high abundance proteins are problematic in proteomic studies but can be addressed by various fractionation techniques. Numerous gel-based and gel-free protein fractionation and multidimensional approaches exist; all have distinctive advantages and drawbacks. 2-DE gel fractionation has a limited dynamic range of proteins, thus decreasing detection of low abundance proteins, very small or large proteins, as well as basic and hydrophobic proteins. Gel-free approaches overcome the dynamic range problem and can be directly coupled to MS and automated for high throughput analysis. Commonly, two or more fractionation techniques are combined, either off-line or on-line with MS, to obtain adequate protein or peptide separation for detection of lower abundance proteins. Previous proteomic studies reveal the necessity for complementary multidimensional fractionation for more complete protein identification profiles.
Proteomic studies of NLF can provide essential information to understand the pathophysiologic pathways, determine diagnostic biomarkers, and design effective therapeutic treatments for rhinosinusitis conditions. Numerous techniques have been used to identify NLF proteins and determine differences associated with certain disease states or chemical exposures. Lindahl et al. identified several proteins in NLF using 2-DE in combination with western blotting or N-terminal sequencing [6-9] and more recently using 2-DE with MS to compare NLF proteins from healthy controls and epoxy workers exposed to dimethylbenzylamine [10, 11], and from nonsmokers and smokers [12]. This group and that of Bryborn et al. [13] also used 2-DE with MS to compare NLF from healthy controls and AR patients [14]. Johannesson et al. used 2-DE and LC/MS/MS to identify NLF proteins that form adducts with hexahydrophthalic anhydride, a highly allergenic chemical [15], while Casado et al. used LC/MS/MS to compare protein profiles of sinusitis patients before and after antibiotic treatment [16]. In a follow-up study, protein profiles before and after nasal provocation identified several new proteins in NLF not previously observed by the 2-DE studies [17]. A recent review discusses the importance of separation techniques prior to MS analysis of NLF proteins [18], and Tewfik et al. used prefractionation and peptide labeling techniques to compare the nasal mucus proteins from CRS and control subjects [19]. Thus, these studies utilized a variety of proteomic techniques, and although the combined efforts identified numerous NLF proteins, no single workflow appeared to provide optimal protein identifications.
In this study, a combination of affinity, anion exchange (AX), and reverse phase separation techniques were used to extensively fractionate intact NLF proteins from AR or asthmatic CRS patients prior to digestion and LC/MS/MS analysis. Our main goal was to identify new, lower abundance proteins. Protein lists are presented along with discussion of the advantages and disadvantages of the experimental workflow.
2 Materials and Methods
2.1 Chemicals and reagents
Ammonium bicarbonate (NH4HCO3), DTT, trifluoroethanol (TFE), glacial acetic acid (HAc), formic acid, iodoacetamide (IA), and NaCl were purchased from Sigma-Alrich, (Milwaukee, WI), urea from Bio-Rad Laboratories (Hercules, CA), Zwittergent 316 (Z316) detergent from Calbiochem (San Diego, Ca), and sequencing grade modified trypsin from Promega Corporation (Madison, WI). Burdick and Jackson HPLC grade water and ACN were ordered from VWR International, Inc. (North Mankato, MN).
2.2 Sample collection
NLF samples were obtained from 9 and 6 patients diagnosed with nonpolypoid AR and CRS with coexisting asthma, respectively, after obtaining their informed consent and via an Institutional Review Board-approved protocol. Exclusion criteria included pregnancy or lactation or a history of smoking, immunodeficiency or cystic fibrosis, airway bacterial or viral infection, antibiotic therapy in the past week, systemic glucocorticoids in the past three months, or allergy immunotherapy in the past year. Intranasal medications, non-prescribed medications, or systemic medications to treat AR or CRS were stopped at least four days before and inhaled medications at least one day before NLF collection. Without breathing or swallowing by the subject and with their neck tilted 30 degrees back from the vertical, 5 mL of sterile isotonic saline was instilled into one nostril, a 10 second wait was observed, and the lavage solution was expelled into a collection beaker. The lavage solution was immediately transferred to a sterile conical tube and placed on ice. This was repeated for the other nostril. The ice-cold nasal lavage was gently rocked at room temperature for 15 minutes and then centrifuged at 800 x g for 10 min at 4° C. Nasal lavage supernatant, excluding pelleted cellular material and any buoyant mucus, was collected. Protein concentrations were determined on individual lavage supernatant samples by the Bradford assay (BSA reference standard; Bio-Rad Laboratories, Hercules, CA). Pooled samples for each group were prepared by combining 100 to 350 μg of protein from individual patients to provide a total protein content of 2 mg in 14.3 ml and 1.7 mg in 10.7 ml for AR and CRS NLF, respectively. Pooled samples were frozen at −80° C until fractionation.
2.3 Abundant plasma protein depletion
The pooled lavage samples were thawed, desalted, and concentrated using an iCON Concentrator (20 mL maximum volume and 9 kDa molecular weight cut-off (MWCO); Pierce, Rockford, IL) to ~200 μl. The retentate was diluted with Agilent Buffer A (Agilent Technologies; Wilmington, DE), filtered through an UltraFree-MC spin column (Millipore Corporation; Bedford, MA), and applied directly to a preconditioned Multiple Affinity Removal Column (MARS; 4.6×100 mm; HU6; Agilent Technologies) according to manufacture protocol. Samples were kept on ice or cooled in refrigerated centrifuges at all times. The collected flow-through depleted fraction was desalted, washed with 20 mM Tris, pH 8.2, concentrated to ~200 μl using an iCON Concentrator (7 mL; 9 kDa MWCO), and injected directly onto the AX column.
2.4 Anion exchange fractionation (AX)
AX was performed using a BioSuite Q-PEEK column (4.6 × 50 mm; 10μ; Waters Corporation; Milford, MA) on a 10ADVP HPLC system (Shimadzu Scientific Instruments; Columbia, MD). The entire desalted MARS depleted protein sample was loop injected directly onto the column. Mobile phase buffer composition was 20 mM Tris, pH 8.2 for buffer A and 20 mM Tris, pH 8.2 with 0.5 M NaCl for buffer B. Separation was achieved with a gradient of 0-50% B over 30 minutes; 50-100% B over 15 minutes; 100% B for 10 minutes; followed by 15 minutes of re-equilibration at 0% B. The separation was monitored at 214 nm. One minute fractions were collected over 70 minutes (400 μl/min) and frozen at −80° C prior to reverse-phase separation.
2.5 Reverse-Phase fractionation (RP)
AX fractions were thawed and 190 μg of preweighed dry urea powder and 4 μl HAc added to each tube and incubated for 30 min at room temperature. RP separation was achieved on a Macroporous Reverse-Phase High Recovery Protein Column (mRP-C18; 2.1 × 75mm; 5μ; 80° C; Agilent Technologies; Foster City, CA) with mobile phases of 0.1% TFA in H2O (pump A) and 0.08% TFA in ACN (pump B). The denatured AX fractions were loaded onto the column and washed for 6 minutes with mobile phase A prior to a separation gradient of 5-30% B over 17 minutes; 30-55% B over 20 minutes; 55-100% B over 10 minutes; 100% B over 10 minutes; followed by re-equilibration at 5% B for 13 minutes. Separation was achieved with a flow rate of 200 μl/min. One-minute fractions were collected and up to 5 fractions combined according to the 220 nm UV intensity (1357 AR fractions; 690 CRS fractions). All fractions were frozen, dried on a SpeedVac, and stored at −20° C.
2.6 Tryptic Digestion
RP fractions were reconstituted with 5 μl TFE and 25 μl 40 mM NH4HCO3. Proteins were reduced with 20 mM DTT and alkylated with 40 mM IA at room temperature for 1 hour. The fractions were diluted with NH4HCO3 buffer prior to addition of trypsin (0.5μg) and incubated overnight at 37° C. Digested fractions were dried and stored at −20° C until LC/MS/MS analysis.
2.7 LC/MS/MS analysis of peptides
Automated LC-MS/MS analyses were performed on a linear ion trap mass spectrometer (LTQ; ThermoFinnigan San Jose, CA ) interfaced to a Paradigm MS4 autosampler and liquid chromatograph (Michrom BioResources Inc, Auburn,CA) using a 75μm × 10 cm ProteoPepII C18 PicoFrit nanoflow column (New Objective Inc, Woburn, MA). The 2047 digested fractions were reconstituted into 18 μl of sample buffer (98:2 H2O:ACN; 0.1% formic acid; 0.0005% Z316) and 15 μl loaded onto a 250 nL OPTI-PAK trap (Optimize Technologies, Oregon City, OR) custom packed with Michrom Magic C8 solid phase particles. (Michrom Bioresources, Auburn, CA). Mobile phase A consisted of water/formic acid (99.9/0.1 by volume) and mobile phase B of ACN/formic acid (99.9/0.1 by volume). The LC method employed a gradient of 5 to 60% B over 30 minutes, 60 to 80% B over 3 minutes, followed by re-eqilibrium at 5% B for 10 minutes, with a column flow of 0.300 ul/min. The ion trap experiment was set for data dependent triple play consisting of a full scan for ions in mass range of 400-1400 m/z, triggering to 10 amu profile mode zoom scan then MS/MS mode on the full scan ions with intensities exceeding a preset threshold of 2000 counts. The MS/MS spectra were aquired with a 2.5 mass unit isolation width, target ion population of 2 ×104 ions, two microscans, maximum ionization fill time of 200 ms, normalized collision energy of 35%, activation Q of 0.25, and activation time of 30 ms. Once ions were selected for MS/MS, they were subsequently excluded for 120 seconds allowing 2 repeats. The exclusion window was 1.5 mass units below and above the exclusion mass. The maximum scan time was ~0.9 seconds depending on the ion injection time automatically determined by the instrument. The MS/MS spectra obtained from the 1357 AR and 690 CRS fractions were combined into one AR and one CRS MS/MS data file and converted to DTA files using Bioworks software (version 3.2; ThermoFinnigan).
2.8 Data analysis and protein identification
The Ar and CRS DTA files were analyzed using Mascot (Matrix Science, London, UK; version 2.1.03), Sequest (ThermoFinnigan, San Jose, CA; version 27, rev. 12v) and X!Tandem (www.thegpm.org; version 2006.09.15.3). All three programs searched the Sprot_20070320_human_r.fasta.hdr database; variable modifications included methionine oxidation and N-acetylation of terminus and cysteines. Tolerances of 2.0 Da (precursor) and 1.0 Da (fragment) were utilized while full tryptic cleavage was required and up to two missed cleavages allowed. Scaffold (version Scaffold-01_05_14, Proteome Software Inc., Portland, OR) was used to compile and assign probability scores. Peptide identifications were accepted if they could be established at greater than 95.0% probability as specified by the Peptide Prophet algorithm [20]. Protein identifications were accepted if they could be established at greater than 99.0% probability and contained at least 2 identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm [21].
3 Results and discussion
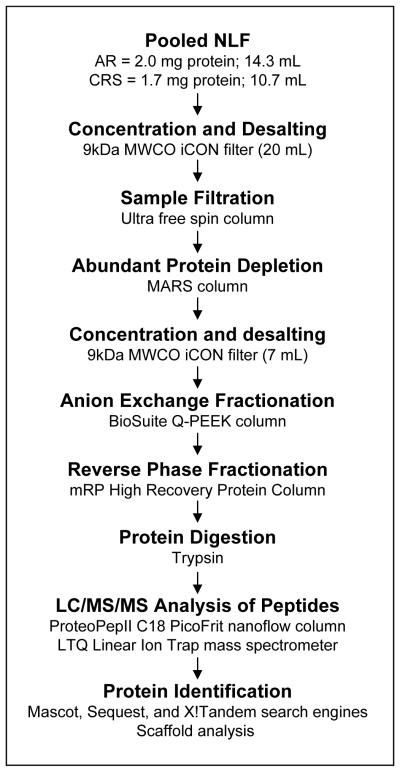
The aim of this study was to obtain an extensive list of lower abundance proteins in NLF from nonpolypoid AR and from asthmatic CRS patients (Table 1). Identifying the proteins specific to NLF can be complicated by contaminating plasma proteins from lesions or capillary breakthrough into the nasal passages. The plasma proteins increase the complexity of NLF and make it difficult to detect lower abundance proteins. Our approach was to maximize the separation of intact NLF proteins prior to digestion to increase the number of peptides per protein detected and obtain more protein identifications. The individual techniques used in our protocol have been standardized by the manufacturers and other groups. To this end, we used a combination of affinity, AX, and RP chromatography prior to LC/MS/MS analysis of the digested peptides to identify proteins found in NLF from patients diagnosed with AR or CRS with coexisting asthma (Figure 1).
Table 1. Demographics of Nasal Lavage Fluid Donors.
NLF samples were obtained from 9 and 6 patients diagnosed with nonpolypoid AR and CRS with coexisting asthma, respectively. All subjects had limited confounding health issues and were off of AR- or CRS-related treatments before donating their NLF sample.
| Sample ID |
Age (yrs) |
Sex | Race | Asthma dx* |
Serum IgE** | Blood eosinophils (% / # x 109/L) |
Aspirin sensitive |
Nasal Polyps |
|---|---|---|---|---|---|---|---|---|
| AR | ||||||||
| 1 | 37 | F | unknown | No | +cat | 1.41 / 0.12 | No | No |
| 2 | 24 | F | Hispanic | No | ++++HDM | 1.68 / 0.10 | No | No |
| 3 | 25 | F | unknown | No | +/−cat;+SRW;++Alt | 2.16 / 0.11 | No | No |
| 4 | 22 | F | White | No | +cat;++TG;++HDM | 5.27 / 0.43 | No | No |
| 5 | 28 | F | White | No | +HDM;++SRW | 3.00 / 0.30 | No | No |
| 6 | 32 | F | White | No | +SRW;++cat;++TG | 3.27 / 0.28 | No | No |
| 7 | 38 | F | White | No | − (patient reported oak) | 5.46 / 0.46 | unknown | No |
| 8 | 34 | F | White | No | +/−TG;+HDM;++SRW | unknown | No | No |
| 9 | 20 | M | White | No | +/−cat;+TG;++SRW;++HDM | 2.90 / 0.19 | No | No |
| CRS | ||||||||
| 1 | 37 | M | White | Yes | +cat;+SRW;+TG;+HDM;+++Alt | 9.65 / 0.44 | No | Yes |
| 2 | 48 | M | White | Yes | +/−TG;+Alt | 1.66 / 0.12 | No | Yes |
| 3 | 33 | M | White | Yes | − | 6.66 / 0.50 | No | No |
| 4 | 45 | F | White | Yes | − (patient reported horse hair) | 4.89 / 0.40 | No | Yes |
| 5 | 33 | M | White | Yes* | +/−cat;+SRW;+HDM | 4.59 / 0.41 | No | No |
| 6 | 41 | F | White | Yes | − | 0.23 / 0.04 | No | Yes |
asthma diagnosis based primarily on a positive bronchial methacholine challenge test
RAST tests for IgE specific for Cat Epithelium, Short Ragweed, Timothy Grass, House dust mite (DF), and Alternaria;
− = neg., <0.35; +/− = Equivocal, 0.35-0.70; + = 0.71-3.50, ++ = 3.51-17.5, +++ = 17.6-50, ++++ = 50.1-100 kU/L
Figure 1. Workflow schematic.
Pooled NLF sample proteins were concentrated, desalted, and extensively fractionated by affinity (MARS Column), anion exchange (BioSuite Q-PEEK Column), and RP (Macroporous RP High Recovery Protein Column) chromatography prior to LC/MS/MS analysis and protein identification from the tryptic peptides.
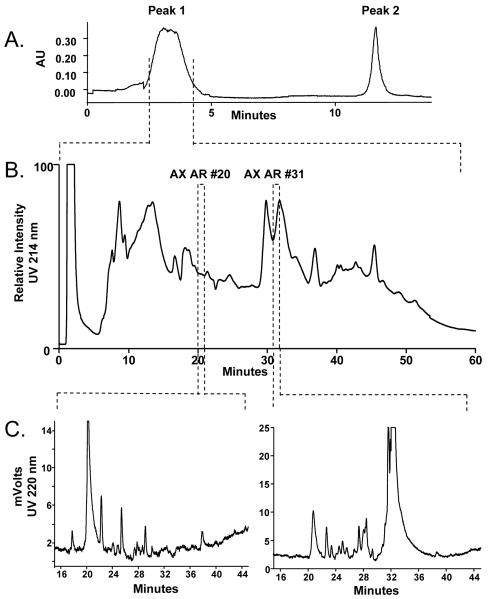
Because individual NLF samples had low protein concentrations compared to other biological fluids and variable protein concentrations and volumes (52 to 407 μg/mL and 2.5 to 8.0 mL among the 15 NLF samples), individual sample volumes providing 100 to 350 μg of protein were pooled to obtain a 2 mg (14.3 mL AR) or 1.7 mg (10.7 mL CRS) total protein preparation. Initial sample preparation included the use of iCON spin filters to reduce the volume and desalt the pooled NLF samples. The manufacturer claims >90% recovery of proteins using their suggested protocol. The iCON retentates were filtered through an Ultra-free spin column to clarify and remove particulates before injection onto the MARS column. The removal of the top six abundant plasma proteins (albumin, transferrin, IgG, IgA, anti-trypsin, haptoglobin; 85% total plasma protein; 99% efficiency) by the MARS column was important to offset the variability in the concentration of contaminating plasma proteins found in each individual NFL sample[22]. Figure 2A shows the elution profile of the AR NLF proteins off the MARS column. Peak 1 area shows that the majority of the proteins in the AR NFL sample were not retained on the column, whereas, Peak 2 area shows the contaminating plasma proteins that were retained and then eluted off the column. The volume from the depleted protein fraction (Peak 1; 2.5 mL) was reduced and desalted using a smaller volume iCON filter (7 mL). The initial large NLF volumes (14.3 and 10.7 ml) and the post MARS column volumes (2.5 ml) required spin filters with large surface areas, and even with extensive washes of the filters, protein losses were significant. Bradford analysis of the concentrated, desalted protein fractions prior to AX fractionation showed substantial reduction of protein with only 73% and 78% protein recovered for AR and CRS, respectively. Protease inhibitors were not used in order to minimize the complexity of the NLF, however, the formation of truncated and degradation products could have also contributed to a reduced recovery of intact proteins.
Figure 2. Chromatographic profiles of the pooled AR NLF sample.
A.) Elution profile from the MARS column. Peak 1 is the fraction depleted of the six abundant plasma proteins. Peak 2 contains the bound plasma proteins. B.) The UV profile of separated Peak 1 proteins on the AX column using a salt gradient. One minute fractions were collected. C.) The UV profiles of AX fractions 20 and 31 on the mRP-C18 column. One to 5 minute fractions were collected for tryptic digestion.
AX was chosen as the first chromatographic fractionation following MARS depletion due to its efficient protein separation and the minimal sample handling needed to prepare the fractions for the subsequent RP separation. The proteins were separated by charge using a salt gradient as described. Figure 2B shows the AX chromatogram obtained of the AR NLF proteins present in Peak 1 (Figure 2A) off the MARS column. Although some of the larger peaks eluted over several minutes, 1 minute fractions were collected in order to maintain maximum separation of the low abundance proteins. The comparison of AX fractions AR20 and AR 31 fractions, highlighted in Figure 2B, was used as an example to demonstrate the overlap of proteins identified in distinct AX fractions.
The AX fractions were prepared for RP separation according to manufacturer protocol. Typically, recovery of proteins on RP columns is low due to the irreversible absorption onto the stationary phase (30-80% efficiency). The stationary phase of the mRP-C18 column was designed to minimize absorption resulting in better peak resolution and protein recovery (>98%) [23]. Figure 2C shows the differences in the UV absorbance profiles from the RP separations of the AX AR20 and AX AR31 fractions from Figure 2B. The UV intensities from each RP analysis determined whether adjacent fractions (up to five) were pooled together prior to trypsin digestion. This resulted in 1357 fractions for the AR sample and 690 fractions for the CRS sample. According to the UV absorbencies shown in Figure 2C, 10 and 17 RP fractions were collected from the example AX AR20 and AX AR31 fractions, respectively. Since a single protein may be found in adjacent RP fractions, the individual MS/MS spectra obtained from each RP fraction (n=10 and 17) were combined prior to protein identification to increase the number of peptide hits for each protein. The top 12 non-keratin identified proteins from these two AX fractions are listed in Table 2. Lactotransferrin and lipocalin were the most abundant proteins found in AX AR20 and AX AR31 fractions, respectively. The two AX fractions contained common proteins, (e.g. desmoplakin and zinc-α-2-glycoprotein), although the elution times differed by 11 minutes. The identified proteins also displayed a wide range of pIs not consistent with AX separation properties. These discrepancies may be explained by peak broadening effects from decreased resolution on the AX column or the presence of truncated or modified protein products which differ in size and pI from the native state protein. Covalent or noncovalent interactions between an AX bound protein and other proteins could also contribute to the unexpected range of pIs. Due to the overlap of proteins observed in both the AX and RP fractions, the MS/MS spectra obtained from the 1357 AR and 690 CRS fractions were combined into one AR and one CRS data file prior to protein identification.
Table 2. Proteins identified from AR AX fractions 20 and 31.
The top 12 non-keratin proteins from each fraction are ranked according to the number of unique peptides used for identification. Overlap of proteins and diversity in pIs may be due to protein degradation or noncovalent protein interactions.
| Protein accession number |
SwissProt accession number |
Protein name | Protein molecular weight (Da) |
pI | Number of Unique Peptides |
|
|---|---|---|---|---|---|---|
|
Allergic Rhinitus Anion Exchange Fraction #20
| ||||||
| 1 | TRFL_HUMAN | P02788 | Lactotransferrin precursor | 78164 | 8.47 | 22 |
| 2 | SPB4_HUMAN | P48594 | Serpin B4 (Squamous cell carcinoma antigen 2) | 44837 | 5.86 | 9 |
| 3 | DESP_HUMAN | P15924 | Desmoplakin | 331765 | 6.44 | 7 |
| 4 | B2MG_HUMAN | P61769 | Beta-2-microglobulin precursor | 13696 | 6.07 | 5 |
| 5 | S10A9_HUMAN | P06702 | Protein S100-A9 (Calgranulin-B) | 13224 | 5.71 | 5 |
| 6 | ZA2G_HUMAN | P25311 | Zinc-alpha-2-glycoprotein precursor | 33854 | 5.58 | 5 |
| 7 | SMR3A_HUMAN | Q99954 | Submaxillary gland androgen-regulated protein | 14100 | 10.35 | 4 |
| 8 | SPR2B_HUMAN | P35325 | Small proline-rich protein 2B | 7956 | 8.81 | 4 |
| 9 | SPB3_HUMAN | P29508 | Serpin B3 (Squamous cell carcinoma antigen 1) | 44547 | 6.35 | 4 |
| 10 | ACTB_HUMAN | P60709 | Actin, cytoplasmic 1 | 41719 | 5.29 | 4 |
| 11 | S10A8_HUMAN | P05109 | Protein S100-A8 (Calgranulin-A) | 10816 | 6.51 | 4 |
| 12 | TFF3_HUMAN | Q07654 | Trefoil factor 3 precursor | 8622 | 5.13 | 3 |
|
| ||||||
|
Allergic Rhinitus Anion Exchange Fraction #31
| ||||||
| 1 | LCN1_HUMAN | P31025 | Lipocalin-1 precursor (Von Ebner gland protein) | 19232 | 5.26 | 28 |
| 2 | ZA2G_HUMAN | P25311 | Zinc-alpha-2-glycoprotein precursor | 33854 | 5.58 | 11 |
| 3 | PROL4_HUMAN | Q16378 | Proline-rich protein 4 precursor | 15078 | 7.17 | 6 |
| 4 | ACTC_HUMAN | P68032 | Actin, alpha cardiac | 42002 | 5.23 | 6 |
| 5 | TFF3_HUMAN | Q07654 | Trefoil factor 3 precursor | 8622 | 5.13 | 5 |
| 6 | TRFL_HUMAN | P02788 | Lactotransferrin precursor | 78164 | 8.47 | 5 |
| 7 | LACRT_HUMAN | Q9GZZ8 | Extracellular glycoprotein lacritin precursor | 14228 | 5.17 | 3 |
| 8 | WFDC2_HUMAN | Q14508 | WAP four-disulfide core domain protein | 12974 | 4.5 | 3 |
| 9 | SMR3A_HUMAN | Q99954 | Submaxillary gland androgen-regulated protein | 14100 | 10.35 | 3 |
| 10 | TCO1_HUMAN | P20061 | Transcobalamin-1 precursor | 48177 | 4.85 | 3 |
| 11 | DESP_HUMAN | P15924 | Desmoplakin | 331765 | 6.44 | 3 |
| 12 | FABPE_HUMAN | Q01469 | Fatty acid-binding protein, epidermal | 15015 | 6.82 | 3 |
The AX and RP fractionation methods were fast and provided extensive separation of the NLF proteins; however, the trypsin digestion of >2000 fractions was time consuming and proved to be the major drawback of the workflow. Two digestion issues were recognized. First, to maintain stability, the RP fractions were lyophilized and stored at −20° C, and while TFE was used to enhance solubility prior to digestion, protein recoveries may have varied, especially in tubes containing small amounts of protein. Second, the digestion protocol was manually prepared in batches (~200 fractions/day), and although the protocol was consistently applied, digestion efficiencies may have fluctuated.
LTQ-MS instrumentation was used for LC/MS/MS analysis because of its speed and sensitivity. In order to detect the maximum number of peptides, especially those found at low levels, a longer gradient was used. The automation of the instrument was advantageous in handling the large number of digest fractions (25 samples/day); however, the weeks needed to complete the analyses required the replacement of cartridges, columns, and silica tubing introducing more variables into the study.
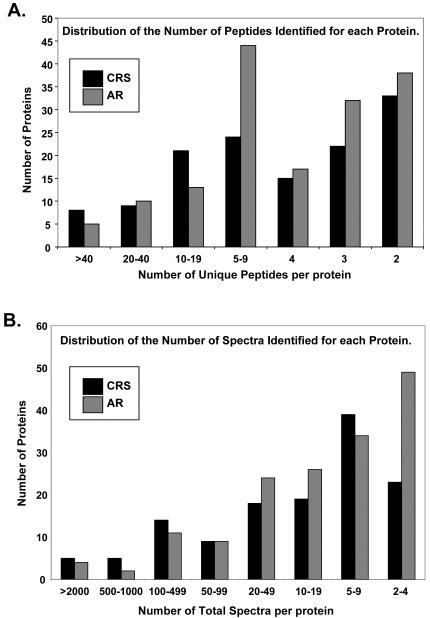
As observed in Table 2, peptides from the same proteins were found in several RP fractions. As a result of protein overlap in both the AX and RP fractions, all of the individual MS/MS spectra were combined prior to peptide identification in order to enhance the number of peptide hits per protein and increase the confidence level of protein identifications. The implementation of the three search engines Mascot, Seaquest, and X!Tandem, provided a more comprehensive list; however, this required more time and computing power (~2.5 million MS/MS spectra). The initial protein lists generated by Scaffold included 202 and 163 protein hits for AR and CRS NLF, respectively. Proteins identified by the reverse database were removed. Upon manual inspection of the remaining spectral data, it was noted that a number of the identified peptides with higher charge states (3+ or 4+) and molecular mass (>2500 Da) had poor quality or questionable MS/MS spectra and were also removed. The revised list included 156 and 129 proteins identified for AR and CRS NLF, respectively. Of the total proteins observed, 88 proteins were identified from both AR and CRS NLF. Figure 3 illustrates the number of unique peptides (two required per protein) and the number of total spectra used to identify the AR and CRS NLF proteins. The expectation from the extensive protein fractionation was an increase in the number of peptides detected for each protein; however, Figure 3A shows that over 50% of the AR and CRS proteins were identified from two to four unique peptides. The majority of proteins identified from 20 or more unique peptides were keratins or plasma proteins. Figure 3B shows the distribution of total spectra collected per identified protein. Each unique peptide could produce more than one spectrum due to peptide concentration and/or more than one peptide charge state. A majority of the proteins were identified using nine or fewer spectra; again, the keratins and plasma proteins constituted the higher numbers of total spectra.
Figure 3. Distribution of the number of unique peptides and total spectra per protein for the CRS and AR proteins identified.
A.) Over 50% of the proteins were identified from 2 to 4 unique peptides. Most proteins identified from 20 or more peptides were keratins or plasma proteins. B.) The distribution of total spectra ranged from 2 to >2000; the majority of proteins were identified from 9 or less spectra. Keratins and plasma proteins dominated the proteins with high spectral counts.
Keratins are observed in proteomic studies, often as a result of sample preparation contamination, but can be present at high levels in NLF due to the airway mucosa environment. Table 3 lists the identified keratins. Of note is the large number of total spectra detected from the keratins which reduced the detection of lower level peptides. A similar problem was observed due to the presence of plasma proteins that were not depleted by the MARS column. Table 4 illustrates the higher level of these proteins in CRS NLF compared to AR NLF. This could be the result of a protocol problem, such as a poorly equilibrated MARS column, or more likely, denaturation or degradation of the plasma proteins in one or more of the individual CRS NLF samples. The increased levels of the MARS targeted proteins in the final depleted CRS NLF sample may have contributed to a decrease in the number of proteins identified compared to AR NLF.
Table 3. Keratins identified from CRA and AR NLF.
The number of unique peptides and total spectra used to identify keratins are noted for CRS and AR, respectively. The high numbers of total spectra from certain keratins interfere with the detection of lower abundance peptides.
| Protein accession number |
Protein name | Number of unique peptides CRS, AR |
Number of total spectra CRS, AR |
|
|---|---|---|---|---|
| 1 | K1C10_HUMAN | Keratin, type I cytoskeletal 10 | 50, 57 | 3195, 5280 |
| 2 | K1C12_HUMAN | Keratin, type I cytoskeletal 12 | 0, 2 | 0, 2 |
| 3 | K1C13_HUMAN | Keratin, type I cytoskeletal 13 | 12, 11 | 47, 20 |
| 4 | K1C14_HUMAN | Keratin, type I cytoskeletal 14 | 23, 27 | 299, 330 |
| 5 | K1C16_HUMAN | Keratin, type I cytoskeletal 16 | 30, 33 | 618, 427 |
| 6 | K1C17_HUMAN | Keratin, type I cytoskeletal 17 | 18, 15 | 50, 64 |
| 7 | K1C19_HUMAN | Keratin, type I cytoskeletal 19 | 0, 4 | 0, 5 |
| 8 | K1C9_HUMAN | Keratin, type I cytoskeletal 9 | 59, 52 | 3130, 2271 |
| 9 | K1H1_HUMAN | Keratin, type I cuticular Ha1 | 15, 16 | 142, 95 |
| 10 | K1H2_HUMAN | Keratin, type I cuticular Ha2 | 4, 0 | 4, 0 |
| 11 | K1H4_HUMAN | Keratin, type I cuticular Ha4 | 26, 19 | 148, 72 |
| 12 | K1H5_HUMAN | Keratin, type I cuticular Ha5 | 11, 8 | 47, 22 |
| 13 | K1H6_HUMAN | Keratin, type I cuticular Ha6 | 10, 5 | 39, 14 |
| 14 | K1H7_HUMAN | Keratin, type I cuticular Ha7 | 3, 0 | 11, 0 |
| 15 | K1H8_HUMAN | Keratin, type I cuticular Ha8 | 2, 4 | 2, 6 |
| 16 | K1HB_HUMAN | Keratin, type I cuticular Ha3-II | 0, 4 | 0, 16 |
| 17 | K22E_HUMAN | Keratin, type II cytoskeletal 2 epi | 70, 70 | 2566, 3057 |
| 18 | K22O_HUMAN | Keratin, type II cytoskeletal 2 oral | 3, 0 | 4, 0 |
| 19 | K2C1_HUMAN | Keratin, type II cytoskeletal 1 | 77, 59 | 7115, 5954 |
| 20 | K2C1B_HUMAN | Keratin, type II cytoskeletal 1b | 2, 3 | 7, 10 |
| 21 | K2C3_HUMAN | Keratin, type II cytoskeletal 3 | 7, 0 | 9, 0 |
| 22 | K2C4_HUMAN | Keratin, type II cytoskeletal 4 | 6, 12 | 19, 19 |
| 23 | K2C5_HUMAN | Keratin, type II cytoskeletal 5 | 33, 22 | 350, 230 |
| 24 | K2C6A_HUMAN | Keratin, type II cytoskeletal 6A | 44, 38 | 565, 504 |
| 25 | K2C6B_HUMAN | Keratin, type II cytoskeletal 6B | 9, 6 | 64, 26 |
| 26 | K2C6D_HUMAN | Keratin, type II cytoskeletal 6D | 2, 2 | 3, 4 |
| 27 | K2C79_HUMAN | Keratin, type II cytoskeletal 79 | 0, 3 | 0, 4 |
| 28 | KPRP_HUMAN | Keratinocyte proline-rich protein | 15, 9 | 62, 21 |
| 29 | KR101_HUMAN | Keratin-associated protein 10-1 | 0, 2 | 0, 3 |
| 30 | KR103_HUMAN | Keratin-associated protein 10-3 | 2, 2 | 3, 4 |
| 31 | KR132_HUMAN | Keratin-associated protein 13-2 | 3, 0 | 4, 0 |
| 32 | KRA24_HUMAN | Keratin-associated protein 2-4 | 4, 3 | 30, 14 |
| 33 | KRA43_HUMAN | Keratin-associated protein 4-3 | 0, 2 | 0, 2 |
| 34 | KRA47_HUMAN | Keratin-associated protein 4-7 | 0, 2 | 0, 3 |
| 35 | KRA49_HUMAN | Keratin-associated protein 4-9 | 2, 0 | 2, 0 |
| 36 | KRA98_HUMAN | Keratin-associated protein 9-8 | 3, 0 | 4, 0 |
| 37 | KRT33A_HUMAN | Keratin, Type I hair 3A | 9, 8 | 26, 13 |
| 38 | KRT33B_HUMAN | Keratin, Type I hair 3B | 6, 0 | 19, 0 |
| 39 | KRT81_HUMAN | Keratin type II cuticular Hb1 | 25, 24 | 154, 103 |
| 40 | KRT82_HUMAN | Keratin type II cuticular Hb2 | 10, 4 | 57, 15 |
| 41 | KRT83_HUMAN | Keratin type II cuticular Hb3 | 2, 4 | 9, 9 |
| 42 | KRT84_HUMAN | Keratin type II cuticular Hb4 | 24, 7 | 69, 11 |
| 43 | KRT85_HUMAN | Keratin type II cuticular Hb5 | 43, 25 | 229, 100 |
| 44 | KRT86_HUMAN | Keratin type II cuticular Hb6 | 4, 3 | 13, 13 |
Table 4. Plasma proteins identified from CRS and AR NLF.
The number of unique peptides and total spectra used to identify plasma proteins not retained on the MARS column are noted for CRS and AR, respectively. Denatured or modified proteins found in one or more individual NLF samples may have led to ineffective binding to the MARS column. The presence of plasma proteins is more evident in the CRS NLF compared to the AR NLF.
| Protein accession numbers |
SwissProt accession number |
Protein name | Number of unique peptides CRS,AR |
Number of total spectr CRS,AR |
|
|---|---|---|---|---|---|
| 1 | A1AT_HUMAN | P01009 | Alpha-1-antitrypsin precursor | 22, 4 | 211, 6 |
| 2 | ALBU_HUMAN | P02768 | Serum albumin precursor | 80, 34 | 3094, 118 |
| 3 | HPT_HUMAN | P00738 | Haptoglobin precursor | 18, 0 | 188, 0 |
| 4 | HPTR_HUMAN | P00739 | Haptoglobin-related protein precursor | 3, 0 | 12, 0 |
| 5 | HV303_HUMAN | P01764 | Ig heavy chain V-III region VH26 precursor | 2, 0 | 4, 0 |
| 6 | IGHA1_HUMAN | P01876 | Ig alpha-1 chain C region | 14, 6 | 334, 21 |
| 7 | IGHA2_HUMAN | P01877 | Ig alpha-2 | 3, 0 | 12, 0 |
| 8 | IGHG1_HUMAN | P01857 | Ig gamma-1 chain C region | 14, 6 | 440, 17 |
| 9 | IGHG2_HUMAN | P01859 | Ig gamma-2 chain C region | 4, 0 | 81, 0 |
| 10 | IGHG3_HUMAN | P01860 | Ig gamma-3 chain C region | 2, 0 | 5, 0 |
| 11 | IGHG4_HUMAN | P01861 | Ig gamma-4 chain C region | 2, 0 | 6, 0 |
| 12 | IGJ_HUMAN | P01591 | Immunoglobulin J chain | 5, 0 | 23, 0 |
| 13 | KAC_HUMAN | P01834 | Ig kappa chain C region | 7, 7 | 213, 13 |
| 14 | LAC_HUMAN | P01842 | Ig lambda chain C regions | 4, 2 | 166, 4 |
| 15 | TRFE_HUMAN | P02787 | Serotransferrin precursor | 58, 7 | 618, 10 |
Table 5 lists the remaining 138 AR and CRS NLF proteins identified in our study of which 41 have been identified in previous proteomic NLF studies. Of these 138 proteins, 52 were detected in both CRS and AR NLF, 25 in only CRS NLF, and 61 in only AR NLF. The number of unique peptides used for protein identification is also given in Table 5 for CRS and AR NLF, respectively. The extensive fractionation was expected to enhance the number of unique peptides detected per protein; however, 81 of the 138 proteins in CRS and/or AR NLF were identified with only 2 or 3 unique peptides. Although the effectiveness of our workflow can not be unequivocally determined, many of the peptides for these 81 proteins may have gone undetected without the fractionation prior to digestion.
Table 5. Protiens identified from CRS and AR NLF (minus the keratins and plasma proteins).
The number of unique peptides used for protein identification is given for CRS and AR NLF, respectively (>99% protein probability score with ≥2 peptides per protein). Of the 138 proteins listed, 81 were identified in CRS and/or AR NLF with only 2 or 3 unique peptides; 52 proteins were detected in both CRS and AR NLF, 25 in only CRS NLF, and 61 in only AR NLF. The proteins identified in previous studies are referenced.
| Protein Accession Number |
SwissProt Accession Number |
Protein Name | Molecular Weight |
Number of Unique Peptides CRS,AR |
Reference | |
|---|---|---|---|---|---|---|
| 1 | 1433B_HUMAN | P31946 | 14-3-3 protein beta/alpha | 28065 | 0, 3 | |
| 2 | 1433S_HUMAN | P31947 | 14-3-3 protein sigma | 27757 | 4, 4 | |
| 3 | 1433Z_HUMAN | P63104 | 14-3-3 protein zeta/delta | 27728 | 3, 0 | |
| 4 | A1AG1_HUMAN | P02763 | Alpha-1-acid glycoprotein 1 precursor |
23494 | 2, 0 | [6,7,10,19] |
| 5 | A2MG_HUMAN | P01023 | Alpha-2-macroglobulin precursor |
163258 | 0, 16 | [7,10,11,17] |
| 6 | AACT_HUMAN | P01011 | Alpha-1-antichymotrypsin precursor |
47635 | 5, 0 | [6,16] |
| 7 | ACTA_HUMAN | P62736 | Actin, aortic smooth muscle | 41992 | 0, 19 | [17] |
| 8 | ACTB_HUMAN | P60709 | Actin, cytoplasmic 1 | 41720 | 7, 8 | [13,16,17] |
| 9 | ACTN3_HUMAN | Q08043 | Alpha-actinin-3 | 103279 | 0, 3 | |
| 10 | ACTS_HUMAN | P68133 | Actin, alpha skeletal muscle | 42034 | 0, 2 | |
| 11 | AL3A1_HUMAN | P30838 | Aldehyde dehydrogenase, dimeric NADP-preferring |
50361 | 0, 2 | [17] |
| 12 | ALDOA_HUMAN | P04075 | Fructose-bisphosphate aldolase A |
39403 | 2, 6 | |
| 13 | AMYC_HUMAN | P19961 | Alpha-amylase 2B precursor | 57692 | 2, 0 | [19] |
| 14 | AMYS_HUMAN | P04745 | Salivary alpha-amylase precursor |
57750 | 0, 3 | |
| 15 | ANXA1_HUMAN | P04083 | Annexin A1 | 38697 | 4, 5 | [7,9,10,12,16] |
| 16 | ANXA2_HUMAN | P07355 | Annexin A2 | 38588 | 9, 7 | [17] |
| 17 | APOA1_HUMAN | P02647 | Apolipoprotein A-I precursor | 30760 | 0, 4 | [7,10,12- 14,16,17,24] |
| 18 | APRV1_HUMAN | Q53RT3 | Retroviral-like aspartic protease 1 precursor |
36973 | 3, 0 | |
| 19 | ARGI1_HUMAN | P05089 | Arginase-1 | 34718 | 0, 3 | |
| 20 | AT2A1_HUMAN | O14983 | Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 |
110236 | 0, 4 | |
| 21 | ATPA_HUMAN | P25705 | ATP synthase subunit alpha, mitochondrial pre |
59734 | 0, 5 | |
| 22 | ATPB_HUMAN | P06576 | ATP synthase subunit beta, mitochondrial precursor |
56542 | 0, 7 | |
| 23 | B2MG_HUMAN | P61769 | Beta-2-microglobulin precursor |
13697 | 2, 5 | [12,14,17] |
| 24 | BLMH_HUMAN | Q13867 | Bleomycin hydrolase | 52544 | 0, 2 | |
| 25 | CALL5_HUMAN | Q9NZT1 | Calmodulin-like protein 5 | 15903 | 10, 8 | |
| 26 | CALM_HUMAN | P62158 | Calmodulin (CaM) | 16820 | 2, 0 | |
| 27 | CASPE_HUMAN | P31944 | Caspase-14 precursor | 27662 | 3, 3 | |
| 28 | CF058_HUMAN | Q6P5S2 | Uncharacterized protein C6orf58 precursor |
37909 | 0, 2 | |
| 29 | CO1A1_HUMAN | P02452 | Collagen alpha-1(I) chain precursor |
138893 | 7, 3 | |
| 30 | CO1A2_HUMAN | P08123 | Collagen alpha-2(I) chain precursor |
129271 | 5, 0 | |
| 31 | CO3_HUMAN | P01024 | Complement C3 precursor | 187131 | 0, 7 | [7,10,16,17] |
| 32 | COF1_HUMAN | P23528 | Cofilin-1 | 18485 | 0, 4 | |
| 33 | CP343_HUMAN | Q9HB55 | Cytochrome P450 3A43 | 57655 | 2, 0 | |
| 34 | CYC_HUMAN | P99999 | Cytochrome c | 11731 | 0, 2 | |
| 35 | CYTA_HUMAN | P01040 | Cystatin-A | 10989 | 4, 6 | |
| 36 | CYTB_HUMAN | P04080 | Cystatin-B | 11121 | 2, 0 | |
| 37 | CYTM_HUMAN | Q15828 | Cystatin-M precursor | 16493 | 0, 2 | |
| 38 | CYTN_HUMAN | P01037 | Cystatin-SN precursor | 16343 | 0, 2 | [10,12,13] |
| 39 | CYTS_HUMAN | P01036 | Cystatin-S precursor | 16197 | 4, 0 | [8,10,12- 14,19,24] |
| 40 | DCD_HUMAN | P81605 | Dermcidin precursor | 11266 | 5, 5 | |
| 41 | DEF1_HUMAN | P59665 | Neutrophil defensin 1 precursor |
10183 | 3, 3 | |
| 42 | DESP_HUMAN | P15924 | Desmoplakin | 331763 | 26, 26 | |
| 43 | DMBT1_HUMAN | Q9UGM3 | Deleted in malignant brain tumors 1 protein precursor |
260707 | 0, 5 | [17] |
| 44 | DSC1_HUMAN | Q08554 | Desmocollin-1 precursor | 100028 | 0, 2 | |
| 45 | DSG1_HUMAN | Q02413 | Desmoglein-1 precursor | 113699 | 8, 12 | |
| 46 | EF1A1_HUMAN | P68104 | Elongation factor 1-alpha 1 | 50123 | 5, 4 | |
| 47 | EF2_HUMAN | P13639 | Elongation factor 2 | 95322 | 0, 2 | |
| 48 | ENOA_HUMAN | P06733 | Alpha-enolase | 47152 | 6, 6 | [13] |
| 49 | EPIPL_HUMAN | P58107 | Epiplakin | 553070 | 0, 3 | |
| 50 | FABPE_HUMAN | Q01469 | Fatty acid-binding protein, epidermal |
15146 | 9, 5 | |
| 51 | FETUA_HUMAN | P02765 | Alpha-2-HS-glycoprotein precursor |
39305 | 3, 0 | [6,7,10] |
| 52 | FILA_HUMAN | P20930 | Filaggrin | 435127 | 18, 6 | |
| 53 | G3P_HUMAN | P04406 | Glyceraldehyde-3- phosphate dehydrogenase |
36035 | 10, 5 | |
| 54 | G3PT_HUMAN | O14556 | Glyceraldehyde-3- phosphate dehydrogenase |
44482 | 0, 3 | |
| 55 | GRP78_HUMAN | P11021 | 78 kDa glucose-regulated protein precursor |
72316 | 0, 2 | |
| 56 | GSTP1_HUMAN | P09211 | Glutathione S-transferase P | 23339 | 3, 2 | |
| 57 | H2A2A_HUMAN | Q6FI13 | Histone H2A type 2-A | 14078 | 0, 2 | [16] |
| 58 | H4_HUMAN | P62805 | Histone H4 | 11349 | 3, 5 | [16,17] |
| 59 | HBA_HUMAN | P69905 | Hemoglobin subunit alpha | 15240 | 8, 0 | [17,19] |
| 60 | HBB_HUMAN | P68871 | Hemoglobin subunit beta | 15980 | 6, 3 | [13,17,19] |
| 61 | HEMO_HUMAN | P02790 | Hemopexin precursor | 51658 | 0, 6 | [6,7,10,13] |
| 62 | HORN_HUMAN | Q86YZ3 | Hornerin | 282354 | 24, 8 | |
| 63 | HPTR_HUMAN | P00739 | Haptoglobin-related protein precursor |
38990 | 3, 0 | |
| 64 | HS90A_HUMAN | P07900 | Heat shock protein HSP 90- alpha |
84645 | 0, 2 | |
| 65 | HSPB1_HUMAN | P04792 | Heat-shock protein beta-1 | 22765 | 4, 5 | |
| 66 | INVO_HUMAN | P07476 | Involucrin | 68447 | 2, 0 | |
| 67 | KAD2_HUMAN | P54819 | Adenylate kinase isoenzyme 2, mitochondrial |
26461 | 0, 2 | |
| 68 | KCRM_HUMAN | P06732 | Creatine kinase M-type | 43083 | 0, 3 | |
| 69 | KPYM_HUMAN | P14618 | Pyruvate kinase isozymes | 57920 | 4, 3 | |
| 70 | LACRT_HUMAN | Q9GZZ8 | Extracellular glycoprotein lacritin precursor |
14228 | 2, 5 | [16] |
| 71 | LCN1_HUMAN | P31025 | Lipocalin-1 precursor | 19232 | 3, 29 | [8,10,12- 17,19,24] |
| 72 | LDHA_HUMAN | P00338 | L-lactate dehydrogenase A chain |
36671 | 3, 0 | |
| 73 | LEG7_HUMAN | P47929 | Galectin-7 | 15057 | 6, 10 | |
| 74 | LMNA_HUMAN | P02545 | Lamin-A/C | 74123 | 2, 2 | |
| 75 | LPLC1_HUMAN | Q8TDL5 | Long palate, lung and nasal epithelium carcinoma- associated protein 1 pre |
52442 | 0, 13 | [17] |
| 76 | LYSC_HUMAN | P61626 | Lysozyme C precursor | 16518 | 4, 7 | [7,10,12,14,17, 19] |
| 77 | MDHM_HUMAN | P40926 | Malate dehydrogenase, mitochondrial precursor |
35513 | 0, 3 | |
| 78 | MUC_HUMAN | P01871 | Ig mu chain C region | 49538 | 2, 0 | |
| 79 | MUC5B_HUMAN | Q9HC84 | Mucin-5B precursor | 590465 | 0, 5 | [17,19] |
| 80 | MYG_HUMAN | P02144 | Myoglobin | 17166 | 2, 3 | |
| 81 | MYH1_HUMAN | P12882 | Myosin-1 | 223103 | 0, 12 | |
| 82 | MYH13_HUMAN | Q9UKX3 | Myosin-13 | 223667 | 2, 0 | |
| 83 | MYH2_HUMAN | Q9UKX2 | Myosin-2 | 223032 | 0, 4 | |
| 84 | MYH4_HUMAN | Q9Y623 | Myosin-4 | 223000 | 0, 25 | |
| 85 | MYH6_HUMAN | P13533 | Myosin-6 | 223677 | 0, 2 | |
| 86 | MYH9_HUMAN | P35579 | Myosin-9 | 226520 | 2, 3 | |
| 87 | NGAL_HUMAN | P80188 | Neutrophil gelatinase- associated lipocalin pre |
22570 | 0, 2 | [17] |
| 88 | PDLI5_HUMAN | Q96HC4 | PDZ and LIM domain protein 5 |
63953 | 0, 3 | |
| 89 | PGAM1_HUMAN | P18669 | Phosphoglycerate mutase 1 | 28786 | 0, 2 | |
| 90 | PGK1_HUMAN | P00558 | Phosphoglycerate kinase 1 | 44597 | 0, 3 | |
| 91 | PIGR_HUMAN | P01833 | Polymeric-immunoglobulin receptor precursor |
83295 | 19, 9 | [16,17,19] |
| 92 | PIP_HUMAN | P12273 | Prolactin-inducible protein precursor |
16555 | 10, 4 | [10,12- 15,17,24] |
| 93 | PKP1_HUMAN | Q13835 | Plakophilin-1 | 82845 | 3, 3 | |
| 94 | PLAK_HUMAN | P14923 | Junction plakoglobin | 81613 | 16, 13 | |
| 95 | PLUNC_HUMAN | Q9NP55 | Protein Plunc precursor | 26696 | 2, 7 | [10,12,14,17, 24] |
| 96 | PRB2_HUMAN | P02812 | Basic salivary proline-rich protein 2 |
37259 | 3, 3 | |
| 97 | PRB3_HUMAN | Q04118 | Basic salivary proline-rich protein 3 precursor |
30917 | 3, 6 | |
| 98 | PRB4S_HUMAN | P10163 | Basic salivary proline-rich protein 4 allele S precursor |
25089 | 0, 2 | |
| 99 | PRDX1_HUMAN | Q06830 | Peroxiredoxin-1 | 22092 | 0, 2 | |
| 100 | PRDX2_HUMAN | P32119 | Peroxiredoxin-2 | 21874 | 2, 0 | |
| 101 | PROL4_HUMAN | Q16378 | Proline-rich protein 4 precursor |
15106 | 0, 6 | [16,17] |
| 102 | PRPC_HUMAN | P02810 | Salivary acidic proline-rich phosphoprotein 1/2 pre |
16998 | 2, 3 | |
| 103 | S10A6_HUMAN | P06703 | Protein S100-A6 | 10162 | 0, 2 | |
| 104 | S10A7_HUMAN | P31151 | Protein S100-A7 | 11440 | 10, 7 | [13] |
| 105 | S10A8_HUMAN | P05109 | Protein S100-A8 | 10817 | 6, 3 | [14,19] |
| 106 | S10A9_HUMAN | P06702 | Protein S100-A9 | 13224 | 7, 4 | [10-14, 16,19,24] |
| 107 | S10AB_HUMAN | P31949 | Protein S100-A11 | 11723 | 2, 0 | |
| 108 | SEMG1_HUMAN | P04279 | Semenogelin-1 precursor | 52112 | 15, 0 | |
| 109 | SEMG2_HUMAN | Q02383 | Semenogelin-2 precursor | 65425 | 6, 0 | |
| 110 | SH3L3_HUMAN | Q9H299 | SH3 domain-binding glutamic acid-rich-like pro |
10419 | 0, 2 | |
| 111 | SMR3A_HUMAN | Q99954 | Submaxillary gland androgen-regulated protein 3 homolog A precursor |
14100 | 0, 4 | |
| 112 | SPB12_HUMAN | Q96P63 | Serpin B12 | 46260 | 2, 0 | |
| 113 | SPB3_HUMAN | P29508 | Serpin B3 | 44548 | 15, 9 | [17] |
| 114 | SPB4_HUMAN | P48594 | Serpin B4 | 44837 | 7, 5 | |
| 115 | SPR1A_HUMAN | P35321 | Cornifin-A | 9864 | 3, 2 | |
| 116 | SPR1B_HUMAN | P22528 | Cornifin-B | 9869 | 2, 0 | |
| 117 | SPR2B_HUMAN | P35325 | Small proline-rich protein 2B | 7957 | 4, 2 | |
| 118 | SPR2G_HUMAN | Q9BYE4 | Small proline-rich protein 2G | 8139 | 0, 3 | |
| 119 | STAT_HUMAN | P02808 | Statherin precursor | 7286 | 0, 2 | [10,11,14,24] |
| 120 | TBA1_HUMAN | P68366 | Tubulin alpha-1 chain | 49906 | 0, 4 | |
| 121 | TBA3_HUMAN | Q71U36 | Tubulin alpha-3 chain | 49877 | 3, 0 | |
| 122 | TBB2A_HUMAN | Q13885 | Tubulin beta-2A chain | 49889 | 0, 4 | |
| 123 | TCO1_HUMAN | P20061 | Transcobalamin-1 precursor | 48177 | 0, 3 | [17,19] |
| 124 | TFF3_HUMAN | Q07654 | Trefoil factor 3 precursor | 8622 | 0, 7 | |
| 125 | TGM3_HUMAN | Q08188 | Protein-glutamine gamma- glutamyltransferase E pre |
76615 | 3, 2 | |
| 126 | THIO_HUMAN | P10599 | Thioredoxin | 11719 | 0, 2 | |
| 127 | TNNT3_HUMAN | P45378 | Troponin T, fast skeletal muscle |
31807 | 0, 2 | |
| 128 | TPIS_HUMAN | P60174 | Triosephosphate isomerase | 26651 | 4, 3 | |
| 129 | TPM1_HUMAN | P09493 | Tropomyosin-1 alpha chain | 32692 | 0, 7 | |
| 130 | TRFL_HUMAN | P02788 | Lactotransferrin precursor | 78164 | 11, 47 | [7,10,12,14- 17,19] |
| 131 | TYB4_HUMAN | P62328 | Thymosin beta-4 | 5034 | 0, 3 | |
| 132 | TYPH_HUMAN | P19971 | Thymidine phosphorylase precursor |
49937 | 0, 3 | |
| 133 | U773_HUMAN | Q96DA0 | Protein UNQ773/PRO1567 precursor |
19582 | 0, 6 | |
| 134 | UBIQ_HUMAN | P62988 | Ubiquitin | 8547 | 4, 2 | |
| 135 | UTER_HUMAN | P11684 | Uteroglobin precursor | 9976 | 0, 3 | [7,9,10,11,19, 24] |
| 136 | VIME_HUMAN | P08670 | Vimentin | 53635 | 2, 0 | |
| 137 | WFDC2_HUMAN | Q14508 | WAP four-disulfide core domain protein 2 precursor |
12974 | 0, 6 | [15] |
| 138 | ZA2G_HUMAN | P25311 | Zinc-alpha-2-glycoprotein precursor |
33854 | 10, 13 | [10,12,13,16, 17,19] |
While the validity of directly comparing the two protein lists obtained from AR and CRS NLF is suspect, there are some obvious differences. Five proteins, α-2-macroglobulin, actin, long palate, lung and nasal epithelium carcinoma-associated protein, myosin 1, and myosin 4, were identified from AR NLF with 16, 19, 13, 12, and 25 unique peptides, respectively, but were not observed in the asthmatic CRS NLF. Similarly, lipocalin and lactotransferrin were identified from AR NLF with 29 and 47 peptides, respectively, compared to the CRS NLF at 3 and 11 unique peptides, respectively. In contrast, semenogelin 1 and 2 were identified in asthmatic CRS NLF with 15 and 6 unique peptides, respectively, but were not observed in AR NLF. Future studies are needed to determine the significance of these differences and how these newly identified proteins may be associated with rhinosinusitis diseases.
4 Concluding Remarks
In this study, we contribute lists of 197 NLF proteins identified from patients with nonpolypoid AR or CRS with coexisting asthma. A portion of these proteins have been identified previously; however, the newly identified proteins can be further studied to determine their association with rhinosinusitis disease and/or their potential as disease biomarkers. Although difficulties, mainly protein loss and manual digestion of numerous fractions, may limit the practicality of this exact work flow for future analyses of biological fluids, in general, extensive protein prefractionation appears to have increased our detection of lower abundance NFL proteins.
Acknowledgements
The authors wish to thank Melinda Miller for MARS depletion sample preparation; Kay Bachman, RN and Sharone Rustad, RN for assistance with patient recruitment and nasal lavage fluid collection; and Mrs. Diana Ayerhart for manuscript preparation. Funding provided by NIH R21 AI058208 and Mayo Foundation.
Abbreviations
- AR
allergic rhinitis
- CRS
chronic rhinosinusitis
- NLF
nasal lavage fluid
- AX
anion exchange
- TFE
trifluoroethanol
- HAc
glacial acetic acid
- IA
iodoacetamide
- MWCO
molecular weight cut off
- MARS
multiple affinity removal column
- mRP-C18
macroporous reverse-phase high recovery protein column
- LTQ
linear ion trap
5 References
- [1].Smart BA. Clin. Rev. Allergy Immuno.l. 2006;30:153–164. doi: 10.1385/CRIAI:30:3:153. [DOI] [PubMed] [Google Scholar]
- [2].Bukstein D, Kraft M, Liu AH, Peters SP. J. Allergy Clin. Immunol. 2006;118:S1–15. doi: 10.1016/j.jaci.2006.08.002. [DOI] [PubMed] [Google Scholar]
- [3].Sasama J, Sherris DA, Shin SH, Kephart GM, Kern EB, Ponikau JU. Curr. Opin. Otolaryngol. Head Neck Surg. 2005;13:2–8. doi: 10.1097/00020840-200502000-00003. [DOI] [PubMed] [Google Scholar]
- [4].Hu S, Loo JA, Wong DT. Proteomics. 2006;6:6326–6353. doi: 10.1002/pmic.200600284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Thongboonkerd VE. Proteomics of Human Body Fluids Principals, Methods, and Applications. Springer; 2007. [Google Scholar]
- [6].Lindahl M, Stahlbom B, Tagesson C. Electrophoresis. 1995;16:1199–1204. doi: 10.1002/elps.11501601200. [DOI] [PubMed] [Google Scholar]
- [7].Lindahl M, Stahlbom B, Svartz J, Tagesson C. Electrophoresis. 1998;19:3222–3229. doi: 10.1002/elps.1150191828. [DOI] [PubMed] [Google Scholar]
- [8].Lindahl M, Stahlbom B, Tagesson C. Electrophoresis. 1999;20:3670–3676. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3670::AID-ELPS3670>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- [9].Lindahl M, Svartz J, Tagesson C. Electrophoresis. 1999;20:881–890. doi: 10.1002/(SICI)1522-2683(19990101)20:4/5<881::AID-ELPS881>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- [10].Lindahl M, Stahlbom B, Tagesson C. Electrophoresis. 2001;22:1795–1800. doi: 10.1002/1522-2683(200105)22:9<1795::AID-ELPS1795>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- [11].Lindahl M, Irander K, Tagesson C, Stahlbom B. Biomarkers. 2004;9:56–70. doi: 10.1080/13547500410001662005. [DOI] [PubMed] [Google Scholar]
- [12].Ghafouri B, Stahlbom B, Tagesson C, Lindahl M. Proteomics. 2002;2:112–120. [PubMed] [Google Scholar]
- [13].Bryborn M, Adner M, Cardell LO. Respir. Res. 2005;6:118. doi: 10.1186/1465-9921-6-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ghafouri B, Irander K, Lindbom J, Tagesson C, Lindahl M. J. Proteome Res. 2006;5:330–338. doi: 10.1021/pr050341h. [DOI] [PubMed] [Google Scholar]
- [15].Johannesson G, Lindh C, Nielsen J, Bjork B, Rosquist S, Jonsson B.Al. Toxicol. Appl. Pharmacol. 2004;194:69–78. doi: 10.1016/j.taap.2003.09.001. [DOI] [PubMed] [Google Scholar]
- [16].Casado B, Pannell LK, Viglio S, Iadarola P, Baraniuk JN. Electrophoresis. 2004;25:1386–1393. doi: 10.1002/elps.200305862. [DOI] [PubMed] [Google Scholar]
- [17].Casado B, Pannell LK, Iadarola P, Baraniuk JN. Proteomics. 2005;5:2949–2959. doi: 10.1002/pmic.200401172. [DOI] [PubMed] [Google Scholar]
- [18].Casado B. Curr. Allergy Asthma Rep. 2004;4:224–229. doi: 10.1007/s11882-004-0030-4. [DOI] [PubMed] [Google Scholar]
- [19].Tewfik MA, Latterich M, DiFalco MR, Samaha M. Am. J. Rhinol. 2007;21:680–685. doi: 10.2500/ajr.2007.21.3103. [DOI] [PubMed] [Google Scholar]
- [20].Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Anal. Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- [21].Nesvizhskii AI, Keller A, Kolker E, Aebersold R. Anal. Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- [22].Mrozinski P, Haiying C, Zolotarjova N, Szafranski C, Barrett W, Bailey J, Boyes B, Agilent Technologies, Inc. 2005. 5989-2881EN. [Google Scholar]
- [23].Barrett W, Martosella J, Zolotarjova N, Yang L, Szafranski C, Nicol G, Agilent Technologies, Inc. 2005. [Google Scholar]
- [24].Ghafouri B, Kihlstrom E, Stahlbom B, Tagesson C, Lindahl M. Biochem. Soc. Trans. 2003;31:810–814. doi: 10.1042/bst0310810. [DOI] [PubMed] [Google Scholar]