Abstract
To determine the expression of components in Toll-like receptors (TLRs)/)Nod-like receptors (NLRs)/inflammasome/caspase-1/interleukin pathway, we examined the expression profiles of those genes by analyzing the data from expression sequence tag cDNA cloning and sequencing. We made several important findings: (1) Among 11 tissues examined, vascular tissues and heart express fewer types of TLRs and NLRs than immune and defense tissues including blood, lymph nodes, thymus and trachea; (2) Brain, lymph nodes and thymus do not express proinflammatory cytokines IL-1β and IL-18 constitutively, suggesting that these two cytokines need to be upregulated in the tissues; and (3) Based on the expression data of three characterized inflammasomes (NALP1, NALP3 and IPAF inflammasome), the examined tissues can be classified into three tiers: the first tier tissues including brain, placenta, blood and thymus express inflammasome(s) in constitutive status; the second tier tissues have inflammasome(s) in nearly-ready expression status (with the requirement of upregulation of one component); the third tier tissues, like heart and bone marrow, require upregulation of at least two components in order to assemble functional inflammasomes. Our original model of three-tier expression of inflammasomes would suggest a new concept of tissues’ inflammation privilege, and provides an insight to the differences among tissues in initiating acute inflammation in response to stimuli.
Keywords: vascular cells, inflammation, apoptosis, innate immune responses, atherosclerosis
INTRODUCTION
Chronic systemic vascular inflammation is an essential requirement for the progression of atherosclerotic pathogenesis in patients(1). However, detailed mechanisms underlying initiation of atherogenic inflammation remain poorly defined. “Traditional” risk factors of atherosclerosis include hyperlipidemia, high density lipoprotein, cigarette smoking, diabetes, hypertension and obesity(2). Recent reports from Wang's laboratory among others’ teams confirmed that hyperhomocysteinemia also acts as an independent factor in accelerating atherosclerosis, etc(3-7). In addition, our laboratory showed that similar to the suppression of autoimmune diseases and anti-tumor immune response by CD4+CD25highFoxp3+ regulatory T cells (Tregs)(8, 9), innate immune response-dominant vascular inflammation is also suppressed by Tregs(10, 11). Epidemiological studies suggest that infected bacteria-derived endotoxinemia at levels as low as 50 pg/ml constitutes a strong risk factor for the development of atherosclerosis(12). However, an essential question remains to be addressed is how vascular cells sense infection and metabolic stress and initiate vascular cell inflammation(11, 13). Continuous improvement of our understanding on atherogenic vascular inflammation will lead to the development of novel therapeutics for this disease and other inflammatory diseases.
Toll-like receptors (TLRs) belong to the pathogen-associated molecular patterns’ (PAMPs) receptor families (PAMP-Rs) and are initiators of inflammation driven by exogenous PAMPs and endogenous sterile tissue insults. TLR-mediated responses cause pathology. For example, proatherogenic TLR2 responses to unknown endogenous or unknown endemic exogenous agonists are mediated by non-bone marrow derived cells including vascular cells(13). The depletion of TLR2 in low density lipoprotein receptor deficient (LDLR−/−) mice, an atherosclerotic mouse model, led to a significant reduction (50%) of lesion size in both the aortic sinus and the aorta of mice fed with hypercholesterolemic diet(14). Similarly, the depletion of TLR4 in apolipoprotein E deficient (ApoE−/−) mice, another atherosclerosis mouse model, results in significant reduction in atherosclerotic lesion size and macrophages in lesion(15). These results suggest that TLRs may initiate inflammatory signals by recognizing atherogenic metabolic risk factors presented in these atherogenic models. In addition to binding to TLRs, some PAMPs are also recognized by a family of cytosolic nucleotide binding and oligomerization domain (NOD)-like receptors (NLRs)(16), another groups of PAMP-Rs. Some NLRs are involved in the recognition of microbial molecules and/or endogenous factors released from tissue destruction. This recognition can lead to activation of caspase-1 (a proinflammatory caspase), and subsequent proteolytic conversion of potent proinflammatory cytokines interleukin-1β (IL-1β) and IL-18 from their precursors pro-IL-1β and pro-IL-18, respectively. The proteolytic conversion of IL-1β and IL-18 is mediated by a cytosolic caspase 1-activating protein complex, termed as inflammasome(17).
Due to the functional significance of PAMP-Rs in bridging pro-atherogenic risk factors to the initiation of vascular inflammation, the tissue expressions and their regulatory mechanisms of PAMP-Rs become very important. As an aspect of the regulation of TLR function, the expression of TLRs is upregulated in response to peptidoglycan and lipopolysaccharide (LPS) priming, suggesting that the expression of TLRs is under regulation of their responses to stimuli via poorly defined mechanisms(18). However, the expression profiles of these newly identified TLRs, NLRs, inflammasome components, caspases, and IL-1β in vascular tissues have not been examined thoroughly. In this study, we examined the hypothesis that these PAMP-Rs and inflammation signal sensing molecules have differential expression patterns in vascular and other tissues. Our results indicate the functional differences of PAMP-Rs in sensing PAMPs in the tissues and the differences in assembling inflammasomes.
MATERIALS AND METHODS
1. Tissue expression profiles of genes encoding PAMP-Rs, inflammasome components, proinflammatory caspases and proinflammatory cytokines
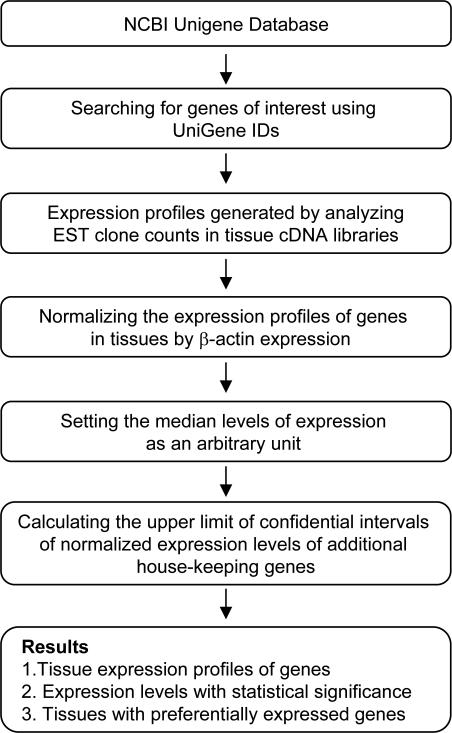
An experimental data mining strategy was adopted to analyze the expression profiles of mRNA transcripts of TLRs, NLRs, inflammasome components, inflammatory caspases, proinflammatory cytokines IL-1β and IL-18 in vasculature-related tissues by mining human expression sequence tag (EST) database (Fig. 1)(19), which resulted from cDNA cloning from various tissue cDNA libraries and DNA sequencing followed by sequence analysese and deposited in the National Institutes of Health (NIH)/National Center of Biotechnology Information (NCBI) Unigene (http://www.ncbi.nlm.nih.gov/sites/entrez?db=unigene)(20). The arbitrary units of the gene expression were calculated by normalizing the transcripts per million of gene of interest with that of house-keeping gene β-actin in any given tissue. The confidential intervals of the expression variation of house-keeping genes were generated by calculating the mean and 2 times the standard deviation of the arbitrary units of three randomly selected house-keeping genes normalized by β-actin in given tissues (legend of Fig. 22). If the expression variation of a given gene in various tissues was larger than the upper limit of the confidential intervals (the mean plus 2 times the standard deviations) of the house-keeping genes, the high expression levels of genes in the tissues were statistically significant. Any given gene transcript, if lower than one per million, was technically presented as no expression.
Fig. 1.
The flow chart of database mining analysis of gene expression profiles using NCBI/UniGene database.
2. Expression profile of genes in response to stimulus with proinflammatory cytokine
To examine gene expression data set in response to stimulation with inflammatory cytokine, we analyzed the data set in the NCBI/Gene expression omnibus (GEO) data sets [(GDS) accession numbers: GDS1542 and GDS1543]. The data were collected in analyzing human umbilical vein endothelial cells (HUVEC) stimulated with tumor necrosis factor-alpha (TNF-α) for five hours(21).
RESULTS
1. PAMP-Rs are differentially expressed in tissues
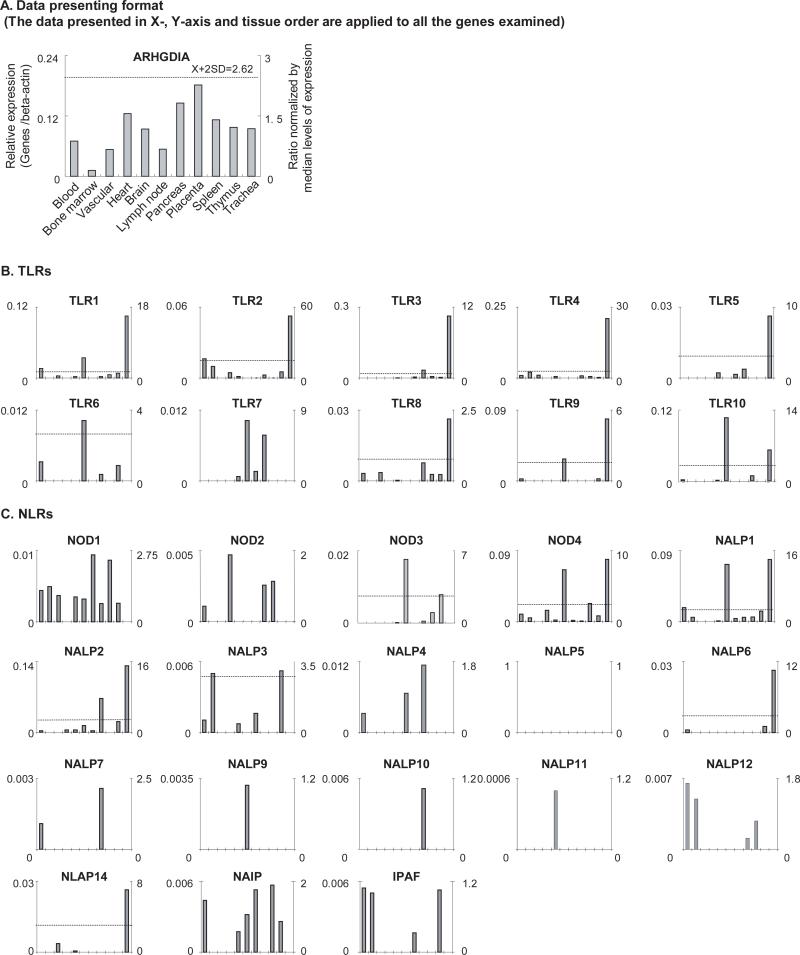
To determine the expression of PAMP-Rs’ gene transcripts in human tissues, we adopted a database mining method. The copy number per million transcripts was calculated based on the experimental data of expression sequence tag (EST) cDNA cloning and sequencing that were deposited in the NCBI UniGene database. In addition, since the gene expression data were normalized by the same β-actin expression data, the arbitrary units of gene expression (Fig. 2A, left Y-axis) were comparable among genes.
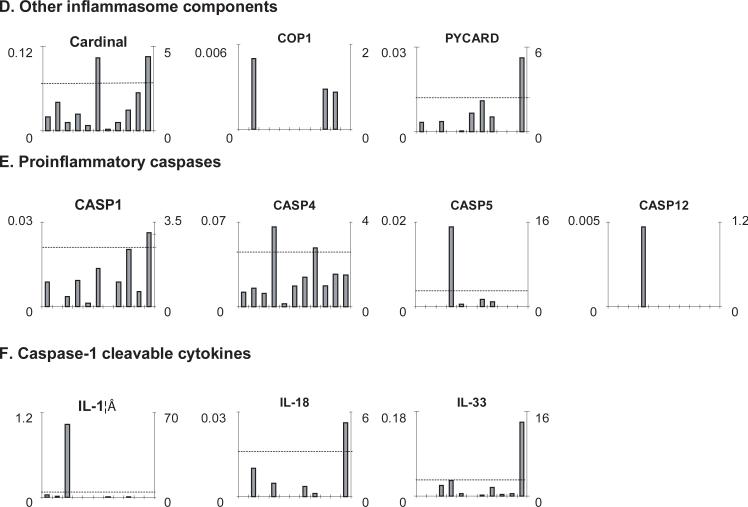
Fig. 2. The gene expression profiles of TLRs, NLRs, other inflammasome components, proinflammatory caspases and caspase-1 cleaved cytokines.
A. Data presenting format. As an example, the gene expression profiles of house-keeping gene Rho GDP dissociation inhibitor (GDI) alpha in the eleven tissues including vascular tissue, blood, heart, trachea and immune system tissues are presented, with the tissue names shown in the bottom of the figure. The gene expression data were normalized by the β-actin expression data from the same tissue, which are presented in the left Y-axis. The expression ratios among tissues were generated by normalizing the arbitrary units of the gene in the tissue with the median levels of the arbitrary units of the gene in all the tissues, which are presented in the right Y-axis. In order to define confidential intervals for statistically higher expression levels of given genes, we calculated the confidential intervals of tissue expression [the mean X + 2 × standard deviations (SD) = 2.62] for three common house keeping genes including Rho GDP dissociation inhibitor alpha (ARHGDIA, Hs.159161), glyceraldehyde-3-phosphate dehydrogenase (GAPDH, Hs.544577) and ribosomal protein S27a (RPS27A, Hs.311640). The expression variations of given genes in tissues, when they were larger than 2.62 folds, were defined as the high expression levels with statistical significance (the right Y-axis). B. The expression profiles of TLRs. The X-axis indicates the eleven tissues including vascular tissue, blood, heart, trachea and immune system tissues with the order the same as that shown in Fig. 2A. C. The expression profiles of NLRs. The X-axis indicates the eleven tissues including vascular tissue, blood, heart, trachea and immune system tissues with the order the same as that shown in Fig. 2A. D. The expression profiles of other inflammasome components. The X-axis indicates the eleven tissues including vascular tissue, blood, heart, trachea and immune system tissues with the order the same as that shown in Fig. 2A. E. The expression profiles of proinflammatory caspases. The X-axis indicates the eleven tissues including vascular tissue, blood, heart, trachea and immune system tissues with the order the same as that shown in Fig. 2A. The follows are the human gene ID numbers in the NCBI/UniGene database: β-actin, Hs.520640; TLR1, Hs.654532; TLR2, Hs.519033; TLR3, Hs.657724; TLR4, Hs.174312; TLR5, Hs.604542; TLR6, Hs.662185; TLR7, Hs.659215; TLR8, Hs.660543; TLR9, Hs.87968; TLR10, Hs.120551; NOD1, Hs.405153; NOD2, Hs.592072; NOD3 (NLRC3), Hs.592091; NOD4 (NLRC5), Hs.528836; NALP1 (NLRP1), Hs. 652273; NALP2 (NLRP2), Hs.369279; NALP3 (NLRP3), Hs.159483; NALP4 (NLRP4), Hs.631533; NALP5 (NLRP5), Hs.356872 (no expression data available in the tissues examined); NALP6 (NLRP6), Hs.352611; NALP7 (NLRP7), Hs.351118; NALP8 (NLRP8), Hs.446925 (no tissue expression data available); NALP9 (NLRP9), Hs.661568; NALP10 (NLRP10), HS.449636; NALP11 (NLRP11), Hs.375039; NALP12 (NLRP12), Hs.631573; NALP13 (NLRP13), Hs.446924 (no tissue expression data available); NALP14 (NLRP14), Hs.449637; NAIP, Hs.654500; IPAF (NLRC4), Hs.574741; Cardinal (CARD8), Hs.446146; COP1 (CARD16), Hs.348365; PYCARD (ASC), Hs.499094; Caspase-1 (CAS1), Hs.2490; Caspase-4 (CAS4), Hs.138378; Caspase-5 (CAS5), Hs.213327; Caspase-12 (CAS12), Hs.476989; IL-1β, Hs.126256; IL-18, Hs.83077; and IL-33, Hs.348390.
We first examined the expression of 10 human TLR gene transcripts in 11 tissues, including blood, bone marrow, vascular, heart, brain, lymph nodes, pancreas, placenta, spleen, thymus and trachea (this order of tissues was applied in the X-axis of all the gene expression sub-figures in Fig. 2). In Fig. 2B, the expression levels of TLR1, TLR3, TLR4, TLR9, and TLR10 (> 0.09 arbitrary units, the left axis) were higher than those of other TLRs in the tissues examined. The expression levels of TLRs in tissues were different (p<0.05), in the scale as much as 25 folds (TLR4 in thymus). As shown in Table 1, tissues had different TLR expression. Lymphoid system, lymph nodes, spleen and thymus, peripheral blood, trachea, placenta, and brain expressed more types of TLRs than that of internal tissues including vascular system, bone marrow and heart, suggesting the sensing function of TLRs in these defense tissues for exogenous and endogenous PAMPs. These results correlate with previous report that endothelial cells of normal artery constitutively express low levels of at least nine TLRs(22). The correlation of the results obtained by experimental data mining here with previous report suggests that the results from this data mining analysis are informative of the gene expression.
Table 1.
TLRs are differentially expressed in human tissues
| Tissue | Protein |
|---|---|
| 1. Blood | TLR1, TLR2, TLR4, TLR6, TLR8, TLR9, TLR10 |
| 2. Bone marrow | TLR2, TLR4 |
| 3. Vascular | TLR1, TLR4, TLR8 |
| 4. Heart | TLR2 |
| 5. Brain | TLR1, TLR2, TLR3, TLR4, TLR5, TLR7, TLR8, TLR10 |
| 6. Lymph nodes | TLR1, TLR6, TLR7, TLR9, TLR10 |
| 7. Pancreas | TLR3, TLR5, TLR7 |
| 8. Placenta | TLR1, TLR2, TLR3, TLR4, TLR5, TLR6, TLR7, TLR8 |
| 9. Spleen | TLR1, TLR3, TLR4, TLR8, TLR10 |
| 10. Thymus | TLR1, TLR2, TLR3, TLR4, TLR6, TLR8, TLR9 |
| 11. Trachea | TLR1, TLR2, TLR3, TLR4, TLR5, TLR8, TLR9, TLR10 |
We also determined the expression of 18 NLR gene transcripts in the tissues. In Fig. 2C, the expression levels of NOD4, NALP1, NALP2 (>0.09 arbitrary units), were higher than those of other NLRs in the tissues examined. The expression levels of NLRs in tissues were different (p<0.05) in the scale as much as 15 folds (NOD3, NALP1 and NALP2). Lymph nodes and trachea were found to have the highest expression levels of NALP1 , which correlate well with Kummer et al report(23). Bone marrow and thymus had the highest expression levels of NALP3, which suggests the potential roles of NALP3 in triggering innate immune responses in guarding important adaptive immunity developing organs if other components of NALP3 inflammasome are available. As shown in Table 2, tissues had different NLR expression. Similar to the expression of TLRs, lymphoid system, lymph nodes, spleen and thymus, peripheral blood, trachea, placenta, and brain expressed more NLRs than internal tissues including vascular system, bone marrow and heart, suggesting the sensing function of NLRs in these defense tissues for exogenous and endogenous PAMPs. In addition, many more types of NLR than TLR were expressed in bone marrow, suggesting the functional importance of NLRs in this tissue. Furthermore, vascular tissue and heart disproportionally expressed fewer types TLRs and NLRs than other tissues, suggesting a relative “inflammation privilege” of these tissues against potential destruction mediated by inflammation and innate immune responses.
Table 2.
NLRs are differentially expressed in human tissues
| Tissue | Protein |
|---|---|
| 1. Blood | NOD1, NOD2, NOD4, NALP1, NALP2, NALP3, NALP4, NALP6, NALP7, NALP12, NAIP, IPAF |
| 2. Bone marrow | NOD1, NOD4, NALP1, NALP3, NALP12, IPAF, NOD1, NALP14 |
| 3. Vascular | |
| 4. Heart | NOD2, NOD4, NALP2 |
| 5. Brain | NOD1, NOD3, NOD4, NALP1, NALP2, NALP3, NALP11, NALP14 , NAIP |
| 6. Lymph nodes | NOD1, NOD3, NOD4, NALP1, NALP2, NALP4, NALP9, NAIP |
| 7. Pancreas | NOD1, NOD4, NALP1, NALP2, NALP3 , IPAF , NAIP |
| 8. Placenta | NOD1, NOD2, NOD3, NOD4, NALP1, NALP2, NALP4, NALP7, NALP10, NALP12, NAIP |
| 9. Spleen | NOD1, NOD2, NOD3,NOD4, NALP1, NALP12, NAIP |
| 10. Thymus | NOD1, NOD3, NOD4, NALP1, NALP2, NALP3, NALP6, IPAF,NAIP |
| 11. Trachea | NOD4, NALP1, NALP2, NALP6, NALP14 |
We then determined the expression of four inflammatory caspases (caspase-1, caspase-4, caspase-5, and caspase-12) (Fig. 2E)(24), three caspase-1 cleavable inflammatory cytokines (IL-1β, IL-18, and IL-33)(Fig. 2F), inflammasome components (PYCARD (ASC) and cardinal, and an inflammasome regulator COP1) (Fig. 2D). In Fig. 2E, the expression levels of caspase-1, caspase-4 and caspase-5 (>0.02 arbitrary units) were higher than that of caspase-12 in the tissues examined. As shown in Table 3, caspase-1 and caspase-4 were expressed in most of the tissues examined. The expression levels of caspase-4, caspase-5 and probably caspase-12 in heart were significantly higher than those in other tissues (p<0.05). Of note, the highest expression of IL-1β was found in vascular whereas highest expressions of IL-18 and IL-33 were found in trachea (Fig. 2F). The cardinal gene was expressed in every tissue examined; with the highest expression in lymph nodes and trachea. The wide expression of PYCARD (ASC) was found in blood, vascular, brain, lymph nodes, pancreas, placenta, and trachea, while an inflammasome regulator COP1 was limitedly expressed in bone marrow, spleen and thymus.
Table 3.
Inflammatory caspases, IL-1β, IL-18, IL-33, PYCARD and COP1 are differentially expressed in human tissues
| Tissue | Protein |
|---|---|
| 1. Blood | CAS1, CAS4, PYCARD, IL-1β, Cardinal |
| 2. Bone marrow | CAS4, IL-1β, IL-18, COP1, Cardinal |
| 3. Vascular | CAS1, CAS4, PYCARD, IL-1β, IL-33, Cardinal |
| 4. Heart | CAS1, CAS4, CAS5, CAS12, IL-18, IL-33, Cardinal |
| 5. Brain | CAS1, CAS4, CAS5, PYCARD, IL-33, Cardinal |
| 6. Lymph nodes | CAS1, CAS4, PYCARD, Cardinal |
| 7. Pancreas | CAS4, CAS5, PYCARD, IL-1β, IL-18, IL-33, Cardinal |
| 8. Placenta | CAS1, CAS4, CAS5, PYCARD, IL-18, IL-33, Cardinal |
| 9. Spleen | CAS1, CAS4, IL-1β, IL-33, COP1,Cardinal |
| 10. Thymus | CAS1, CAS4, IL-33, COP1,Cardinal |
| 11. Trachea | CAS1, CAS4, PYCARD, IL-18, IL-33, Cardinal |
2. Less tissues constitutively express inflammasomes than those tissues in which the expression of inflammasomes can be induced
A previous report showed that in diseased vessels, the pattern of TLR expression is characterized by markedly upregulated expression of TLR1, TLR2, TLR4, and TLR6 by endothelial cells and infiltrated macrophages(25), suggesting PAMP-Rs could be unregulated in ECs. We examined a hypothesis that inflammasome expression profiles can be used in categorizing the examined tissues into three tiers. To test the hypothesis, we focused on the three well characterized inflammasomes(24): NALP1 inflammasome, NALP3 inflammasome and IPAF inflammasome. The rationale to focus on three inflammasomes IPAF, NALP1 and NALP3 was that the putative function of the majority of the 21 human NLRs and NLR-related genes in activating caspase-1 has not been confirmed except for those three(26). NALP1 inflammasome consists of four components, NALP1, PYCARD (ASC)(27), caspase-1 and caspase-5, and functions as primary mediator of susceptibility to anthrax lethal toxin(28); NALP3 inflammasome consists of three components, NALP3, PYCARD and caspase-1, and can sense various stimuli including anti-viral compounds R837 and R848, bacterial mRNA, gout-associated crystals, bacterial toxins derived from Listeria M., staphylococcus A. and shigella F, etc(29); IPAF inflammsome consists of three components, IPAF(30), NAIP and caspase-1 and functions to sense flagellin derived from Legionella P., Salmonella T., pseudomonas A. and shigella F(29). Based on the tissue expression profiles of these components (Table 4), three tiers of tissues can be categorized for these three inflammasomes. The first tier of tissues with constitutively expressed (“stand-by”) inflammasomes included brain, blood, placenta and thymus. For the significance, blood, placenta and thymus, are functionally involved in innate and adaptive immune responses (Fig. 2C). The second tier of tissues with potentially inducible expression of one inflammasome component included brain, blood, pancreas, vascular, lymph nodes, placenta, thymus, trachea and spleen. The third tier of tissues with presumably inducible expression of at least two inflammasome components. These results suggest that more tissues adopt inducible expression status for inflammasomes. Of note, some tissues, for example, placenta, constitutively expresses NALP1 inflammasome but may express inflammasomes NALP3 and IPAF in an inducible manner, which places this tissue into the classifications. The results may also suggest that different types of inflammasomes may play various roles in tissues.
Table 4.
The three tier expression status of inflammasome types in tissues
| Inflammasome type | Tissue |
|---|---|
| First tier (“ready to go” expression status with all components) | |
| NALP1 inflammasome (NALP1, PYCARD, caspase-1 and caspase-5) | Brain, placenta |
| NALP3 inflammasome (NALP3, PYCARD and caspase-1, Cardinal ) | Blood, brain |
| IPAF inflammasome (IPAF, NAIP, caspase-1) | Blood, Thymus |
| Second tier (nearly-ready expression status that requires one component missing) | |
| NALP1 inflammasome | Blood, pancreas, placenta, trachea |
| NALP3 inflammasome | Vascular, lymph nodes, pancreas, placenta, thymus, trachea |
| IPAF inflammasome | Brain, lymph node, pancreas, placenta, spleen |
| Third tier (expression status that requires upregulation of more than one components) | |
| NALP1 inflammasome | Bone marrow, vascular, heart, spleen, thymus |
| NALP3 inflammasome | Bone marrow, heart, spleen, trachea |
| IPAF inflammasome | Bone marrow, vascular, heart, placenta, trachea |
3. The expressions of inflammatory cytokines IL-1β and IL-18 are upregulated by inflammatory cytokine TNF-α
We then examined a hypothesis that the expressions of some TLRs, NLRs, caspases, inflammasome components and inflammatory cytokines IL-1β and IL-18 in vascular endothelial cells are inducible by inflammation stimuli. To test this hypothesis, we analyzed a gene microarray data set banked in the NCBI-Gene Expression Omnibus (GEO) Repository, in which the expression levels of these genes in human umbilical vein endothelial cells (HUVEC) in the absence or presence of TNF-α for 5 hours can be compared. In Table 5, we calculated the gene expression change index using the expression levels of genes in TNF-α stimulated HUVEC over the expression levels of genes in unstimulated HUVEC control. As controls, the expression of house-keeping gene β-actin was not changed with a ration of 1.1, suggesting that the experiments of RNA sampling and microarray were well performed. Furthermore, to confirm the capability of TNF-α in activating endothelial cells(11) in this microarray, the change index of EC activation markers vascular cell adhesion molecule-1 (VCAM1) and intercellular cell adhesion moleculeam-1 (ICAM1) were examined, in Table 5, and they were upregulated 38.7 folds and 19.4 folds, respectively. The results suggest that the HUVEC activation by TNF-α was well performed, indicating that the data sets were qualified for our mining analysis.
Table 5.
The expressions of TLRs, NLRs, caspases, IL-1β, IL-18 in HUVECs are modulated by inflammatory cytokine TNF-α
| Gene | NCBI No. | Change folds |
|---|---|---|
| TLR1 | AL050262 | 1.27 |
| TLR2 | NM_003264 | 0.93 |
| TLR3 | NM_003265 | 1.34 |
| TLR5 | AF051151 | 1.11 |
| TLR6 | NM_006068 | 1.43 |
| TLR7 | NM_016562 | 3.81 |
| NALP1 | NM_021730 | 1.24 |
| NALP2 | AF298547 | 0.57 |
| NALP3 | NM_004895 | 0.89 |
| Caspase-1 | U13698 | 1.02 |
| Caspase-4 | U25804 | 1.14 |
| Caspase-5 | NM_004347 | 0.57 |
| IL-1β | NM_000576 | 1.83 |
| IL-18 | NM_001562 | 2.37 |
| House-keeping gene control | ||
| β-actin | NM_001101 | 1.10 |
| EC activation control | ||
| VCAM1 | NM_001078 | 38.71 |
| ICAM1 | NM_000201 | 19.44 |
# Change folds= the expression levels of gene in TNF-α-stimulated HUVEC over the expression levels of gene in unstimulated HUVEC control
The expressions of TLR1, TLR3, TLR6 and TLR7 in HUVEC were upregulated by TNF-α stimulation by 1.3-, 1.3-, 1.4- and 3.8-folds, respectively, whereas the expressions of TLR2 and TLR5 were not changed (<1.1 of the β-actin change index). In addition, the expression of NALP1 was slightly upregulated by 1.24-folds whereas the expressions of NALP2 and NALP3 were not upregulated (Table 5). Moreover, the expressions of caspase-1, caspase-4 and caspase-5 were not upregulated. Interestingly, the expressions of IL-1β and IL-18 were significantly upregulated by 1.8- and 2.4-folds, respectively, suggesting that the upregulation of inflammatory cytokine transcripts is an early event in endothelial cells in response to TNF-α stimulation. Since this data set includes gene expression data before TNF-α stimulation and 5 hours after stimulation, the expression of other genes may possibly be (1) modulated by TNF-α in the longer stimulation time; (2) mediated by signals derived from other signal pathways; and (3) cell-type specific signals. Future work is needed to determine the potential induction of inflammasome components with (a) more cell types of vascular cells including aortic endothelial cells, microvascular endothelial cells and vascular smooth muscle cells, (b) more inflammatory stimuli and (c) longer stimulation time.
DISCUSSION
Recent progress in characterization of PAMP-Rs and inflammasomes has further emphasized the importance of proinflammatory cytokine IL-1β signaling in bridging atherogenic risk factors to initiate inflammation(31). However, constitutive expression levels and expression readiness of PAMP-Rs, inflammasome components and proinflammtory caspases in tissues remained poorly defined. By analyzing cDNA cloning and DNA sequencing data from tissue cDNA libraries, we studied expression profiles of TLRs, NLRs, inflammasome components, inflammatory caspases, and caspase-1 cleavable inflammatory cytokines. Since this data is collected from cDNA cloning and DNA sequencing experiments, rather than theoretical data derived from computer modeling, the data require no further experimental verification. Since EST databases have been established based on precise DNA sequencing data, the data obtained by EST database mining are more precise in providing the tissue expression profiles of genes than traditional hybridization- and primer annealing-based approaches like Northern blots and RT-PCRs.
Upregulation of some inflammasome components and TLRs in HUVECs stimulated by TNF-α is just one example to show that, in principle, the upregulation of inflammasome related gene expression is modulated potentially by inflammatory signals. Previous reports supported this finding and showed that several pathological conditions and signaling pathways modulate expression of TLRs/NLRs/inflammasome/caspase-1/IL- pathway. First, systemic levels of TLR2 and TLR4 are higher in acute myocardial infarction patients than in stable angina patients and controls showing normal coronary angiogram(32). However, our results showed that TLR2 and TLR4 expressions were lower in vascular tissues and heart than in other tissues. Taken together, our and other results suggest that (1) acute myocardial infarction-induced stress may upregulate TLR2 and TLR4 expressions in vascular tissues and heart; (2) stimulation via upregulated TLR2 and TLR4 are associated with progress of myocardial infarction; Second, murine endothelial cells express membrane TLR2 and respond to TLR2 ligands, but human endothelial cells normally will not respond unless they are first primed with inflammatory stimulation, which triggers translocation of TLR2 to the cell surface(33). In addition to the posttranslational regulatory manner, our results showed that TLR2 expression in vascular tissues is low, suggesting potential upregulation of TLR2 via a transcription manner in the tissues; Third, PYCARD (ASC) is upregulated by various inflammatory cytokines including IL-1β, interferon-γ (IFN-γ), TNF-α, lipopolysaccharide (LPS) and Fas ligand (34). The upregulated expression of the NALP3 inflammasome, due to gene polymorphisms, is associated with interleukin-1β production and severe inflammation(35). Of note, upregulation of TLRs, NLRs, caspases and inflammasome components may not share the same pathways. Endothelial cells after exposure to carbon monoxide, a ubiquitous environmental pollutant, undergo cell death with the characteristics of activation of caspase-1 but not caspase-3 (36) probably via endogenous p53 pathway(37). Unlike caspase-1, the expression of caspase-11 is LPS-inducible, and thus, it is reasonable to postulate that other members of the family are regulated at the transcriptional or translational level by extracellular stimuli (24).
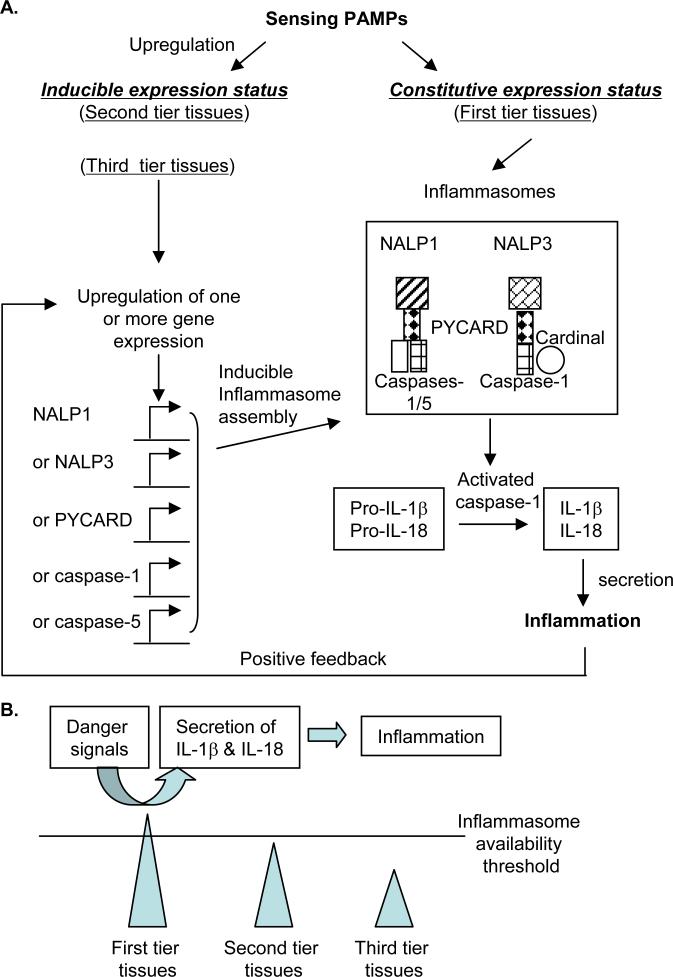
After analyzing the data from EST cDNA cloning and sequencing we have made several important findings: (1) Among 11 tissues examined, vascular tissues and heart express fewer types of TLRs and NLRs than immune system tissues including blood, lymph nodes, thymus and trachea; (2) Brain, lymph nodes and thymus do not express proinflammatory cytokines IL-1β and IL-18 constitutively, suggesting that these two cytokines need to be upregulated in response to inflammatory stimuli in the tissues; and (3) based on the expression data of three characterized inflammasomes (NALP1, NALP3 and IPAF inflammasomes), the examined tissues can be classified into three tiers: the first tier tissues including brain, placenta, blood and thymus express inflammasome(s) in constitutive status; the second tier tissues have inflammasome(s) in nearly-ready expression status (with the requirement of upregulation of one component); the third tier tissues like heart and bone marrow, require upregulation of at least two components in order to assemble functional inflammasomes. Based on the expression readiness of inflammasomes in tissues, we propose a new working model of three-tier responsive expression of inflammasomes in tissues and suggest a new concept of third tier tissues’ inflammation privilege, which provides an insight on the differences of tissues in initiating acute inflammations (Fig. 3). This model suggests that (a) the first-tier tissues with constitutively expressed inflammasomes initiate inflammation quicker than the second-tier and third-tier tissues; and (b) the second tier tissues (requiring one component upregulation) including vascular tissue and the third tier tissues including heart (requiring more than one component upregulation) are in an inducible expression status of inflammasomes. The inducible expressions of inflammasomes are presumably mediated through various signal pathways and the interplay between the signal pathways, which take longer time and overcome a higher threshold than the first tier tissue in initiating inflammation. Traditional concept of immune privilege suggests a protective mechanism from autoimmune destruction based on no expression of antigen-presenting self-major compatibility complex (MHC) molecules in tissues(38). No expression of self-MHCs in immune privilege tissues including testis results in failing in presenting self-antigens from these tissues to stimulate host immune system, thereby protecting immune privilege tissues from autoimmune destruction. Similarly, we propose a new concept of tissues’ inflammation privilege that emphasizes a protective mechanism against tissue destruction mediated by inflammasome/IL-1β-based innate immune responses. In this new concept of tissues’ inflammation privilege, vascular tissue and heart disproportionally express fewer types of TLRs and NLRs and may only inducibly express inflammasomes, which can be explained by this “inflammation privilege” of the tissue against uncontrolled inflammation destruction mediated by inflammasome-based innate immune responses(39). Our new concept and model may also explain the potential differences between cardiovascular tissues and other tissues in initiation of acute inflammation. The first-tier tissues may have higher probability to have acute inflammation than the second-tier and third-tier tissues. Future work is required to elucidate the specific roles of different tiers of tissue inflammasomes in vascular inflammation and atherogenesis and the regulatory signal pathways.
Fig. 3. The schematic representation of a working model of three-tier expression of inflammasomes in response to danger signals in tissues.
The two inflammasomes NALP1 and NALP3 are presented as examples. The NALP3 inflammasome component Cardinal is constitutively expressed in the tissues examined so Cardinal is not included in the list of potentially upregulated genes.
ACKNOWLEDGEMENTS
I am very grateful to Michael Jan and Xinyu Xiong for critical reading.
REFERENCES
- 1.Hansson GK. Inflammation, atherosclerosis, and coronary artery disease. N Engl J Med. 2005;352:1685–1695. doi: 10.1056/NEJMra043430. [DOI] [PubMed] [Google Scholar]
- 2.Ross R. Atherosclerosis. In: Wyngaarden J, Smith LH, Bennett JC, editors. Cecil Textbook of Medicine. W.B. Saunders Company; Philadelphia, London, Toronto, Montreal, Sydney, Tokyo: 1992. pp. 293–298. [Google Scholar]
- 3.Wang H, Jiang X, Yang F, et al. Hyperhomocysteinemia accelerates atherosclerosis in cystathionine beta-synthase and apolipoprotein E double knock-out mice with and without dietary perturbation. Blood. 2003;101:3901–3907. doi: 10.1182/blood-2002-08-2606. [DOI] [PubMed] [Google Scholar]
- 4.Jiang X, Yang F, Tan H, et al. Hyperhomocystinemia impairs endothelial function and eNOS activity via PKC activation. Arterioscler Thromb Vasc Biol. 2005;25:2515–2521. doi: 10.1161/01.ATV.0000189559.87328.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tan H, Jiang X, Yang F, et al. Hyperhomocysteinemia inhibits post-injury reendothelialization in mice. Cardiovasc Res. 2006;69:253–262. doi: 10.1016/j.cardiores.2005.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liao D, Tan H, Hui R, et al. Hyperhomocysteinemia decreases circulating high-density lipoprotein by inhibiting apolipoprotein A-I Protein synthesis and enhancing HDL cholesterol clearance. Circ Res. 2006;99:598–606. doi: 10.1161/01.RES.0000242559.42077.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jamaluddin MD, Chen I, Yang F, et al. Homocysteine inhibits endothelial cell growth via DNA hypomethylation of the cyclin Agene. Blood. 2007;110:3648–3655. doi: 10.1182/blood-2007-06-096701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan Y, Chen Y, Yang F, et al. HLA-A2.1-restricted T cells react to SEREX-defined tumor antigen CML66L and are suppressed by CD4+CD25+ regulatory T cells. Int J Immunopathol Pharmacol. 2007;20:75–89. doi: 10.1177/039463200702000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang XF. Factors regulating apoptosis and homeostasis of CD4+CD25highFOXP3+ regulatory T cells are new therapeutic targets. Front Biosci. 2008;13:1472–1499. doi: 10.2741/2775. [DOI] [PubMed] [Google Scholar]
- 10.Xiong Z, Song J, Yan Y, et al. Higher expression of Bax in regulatory T cells increases vascular inflammation. Front Biosci. 2008;13:7143–7155. doi: 10.2741/3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiong Z, Yan Y, Song J, et al. Expression of TCTP antisense in CD25(high) regulatory T cells aggravates cuff-injured vascular inflammation. Atherosclerosis. 2009;203:401–408. doi: 10.1016/j.atherosclerosis.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stoll LL, Denning GM, Weintraub NL. Potential role of endotoxin as a proinflammatory mediator of atherosclerosis. Arterioscler Thromb Vasc Biol. 2004;24:2227–2236. doi: 10.1161/01.ATV.0000147534.69062.dc. [DOI] [PubMed] [Google Scholar]
- 13.Tobias PS, Curtiss LK. Toll-like receptors in atherosclerosis. Biochem Soc Trans. 2007;35:1453–1455. doi: 10.1042/BST0351453. [DOI] [PubMed] [Google Scholar]
- 14.Mullick AE, Tobias PS, Curtiss LK. Modulation of atherosclerosis in mice by Toll-like receptor 2. J Clin Invest. 2005;115:3149–3156. doi: 10.1172/JCI25482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michelsen KS, Wong MH, Shah PK, et al. Lack of Toll-like receptor 4 or myeloid differentiation factor 88 reduces atherosclerosis and alters plaque phenotype in mice deficient in apolipoprotein E. Proc Natl Acad Sci U S A. 2004;101:10679–10684. doi: 10.1073/pnas.0403249101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tschopp J, Martinon F, Burns K. NALPs: a novel protein family involved in inflammation. Nat Rev Mol Cell Biol. 2003;4:95–104. doi: 10.1038/nrm1019. [DOI] [PubMed] [Google Scholar]
- 17.Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 18.Matsuda N, Yamazaki H, Takano K, et al. Priming by lipopolysaccharide exaggerates acute lung injury and mortality in responses to peptidoglycan through up-regulation of Toll-like receptor-2 expression in mice. Biochem Pharmacol. 2008;75:1065–1075. doi: 10.1016/j.bcp.2007.10.036. [DOI] [PubMed] [Google Scholar]
- 19.Ng B, Yang F, Huston DP, et al. Increased noncanonical splicing of autoantigen transcripts provides the structural basis for expression of untolerized epitopes. J Allergy Clin Immunol. 2004;114:1463–1470. doi: 10.1016/j.jaci.2004.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pires Martins R, Leach RE, Krawetz SA. Whole-body gene expression by data mining. Genomics. 2001;72:34–42. doi: 10.1006/geno.2000.6437. [DOI] [PubMed] [Google Scholar]
- 21.Viemann D, Goebeler M, Schmid S, et al. TNF induces distinct gene expression programs in microvascular and macrovascular human endothelial cells. J Leukoc Biol. 2006;80:174–185. doi: 10.1189/jlb.0905530. [DOI] [PubMed] [Google Scholar]
- 22.Edfeldt K, Swedenborg J, Hansson GK, et al. Expression of toll-like receptors in human atherosclerotic lesions: a possible pathway for plaque activation. Circulation. 2002;105:1158–1161. [PubMed] [Google Scholar]
- 23.Kummer JA, Broekhuizen R, Everett H, et al. Inflammasome components NALP 1 and 3 show distinct but separate expression profiles in human tissues suggesting a site-specific role in the inflammatory response. J Histochem Cytochem. 2007;55:443–452. doi: 10.1369/jhc.6A7101.2006. [DOI] [PubMed] [Google Scholar]
- 24.Martinon F, Tschopp J. Inflammatory caspases and inflammasomes: master switches of inflammation. Cell Death Differ. 2007;14:10–22. doi: 10.1038/sj.cdd.4402038. [DOI] [PubMed] [Google Scholar]
- 25.Yan ZQ, Hansson GK. Innate immunity, macrophage activation, and atherosclerosis. Immunol Rev. 2007;219:187–203. doi: 10.1111/j.1600-065X.2007.00554.x. [DOI] [PubMed] [Google Scholar]
- 26.Opitz B, Hippenstiel S, Eitel J, et al. Extra- and intracellular innate immune recognition in endothelial cells. Thromb Haemost. 2007;98:319–326. [PubMed] [Google Scholar]
- 27.Masumoto J, Taniguchi S, Ayukawa K, et al. ASC, a novel 22-kDa protein, aggregates during apoptosis of human promyelocytic leukemia HL-60 cells. J Biol Chem. 1999;274:33835–33838. doi: 10.1074/jbc.274.48.33835. [DOI] [PubMed] [Google Scholar]
- 28.Boyden ED, Dietrich WF. Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat Genet. 2006;38:240–244. doi: 10.1038/ng1724. [DOI] [PubMed] [Google Scholar]
- 29.Ye Z, Ting JP. NLR, the nucleotide-binding domain leucine-rich repeat containing gene family. Curr Opin Immunol. 2008;20:3–9. doi: 10.1016/j.coi.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 30.Poyet JL, Srinivasula SM, Tnani M, et al. Identification of Ipaf, a human caspase-1-activating protein related to Apaf-1. J Biol Chem. 2001;276:28309–28313. doi: 10.1074/jbc.C100250200. [DOI] [PubMed] [Google Scholar]
- 31.Yang X-F, Ying Yin, Hong Wang. Vascular inflammation and atherosclerosis are activated via receptors for PAMPs and suppressed by regulatory T cells. Drug Discovery Today Therapeutic Strategies. 2008;5:125–142. doi: 10.1016/j.ddstr.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ishikawa Y, Satoh M, Itoh T, et al. Local expression of Toll-like receptor 4 at the site of ruptured plaques in patients with acute myocardial infarction. Clin Sci (Lond) 2008;115:133–140. doi: 10.1042/CS20070379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shuang C, Wong MH, Schulte DJ, et al. Differential expression of Toll-like receptor 2 (TLR2) and responses to TLR2 ligands between human and murine vascular endothelial cells. Journal of endotoxin research. 2007;13:281–296. doi: 10.1177/0968051907085096. [DOI] [PubMed] [Google Scholar]
- 34.Shiohara M, Taniguchi S, Masumoto J, et al. ASC, which is composed of a PYD and a CARD, is up-regulated by inflammation and apoptosis in human neutrophils. Biochem Biophys Res Commun. 2002;293:1314–1318. doi: 10.1016/S0006-291X(02)00384-4. [DOI] [PubMed] [Google Scholar]
- 35.Verma D, Lerm M, Blomgran Julinder R, et al. Gene polymorphisms in the NALP3 inflammasome are associated with interleukin-1 production and severe inflammation: relation to common inflammatory diseases? Arthritis Rheum. 2008;58:888–894. doi: 10.1002/art.23286. [DOI] [PubMed] [Google Scholar]
- 36.Thom SR, Fisher D, Xu YA, et al. Adaptive responses and apoptosis in endothelial cells exposed to carbon monoxide. Proc Natl Acad Sci U S A. 2000;97:1305–1310. doi: 10.1073/pnas.97.3.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gupta S, Radha V, Furukawa Y, et al. Direct transcriptional activation of human caspase-1 by tumor suppressor p53. J Biol Chem. 2001;276:10585–10588. doi: 10.1074/jbc.C100025200. [DOI] [PubMed] [Google Scholar]
- 38.Yang F, Yang XF. New concepts in tumor antigens: their significance in future immunotherapies for tumors. Cell Mol Immunol. 2005;2:331–341. [PubMed] [Google Scholar]
- 39.Streilein JW, Stein-Streilein J. Does innate immune privilege exist? J Leukoc Biol. 2000;67:479–487. doi: 10.1002/jlb.67.4.479. [DOI] [PubMed] [Google Scholar]