Abstract
Photoreceptor cells achieve high sensitivity, reliably detecting single photons, while limiting the spontaneous activation events responsible for dark noise. We used proteomic, genetic, and electrophysiological approaches to characterize Retinophilin (RTP) (CG10233) in Drosophila photoreceptors and establish its involvement in dark-noise suppression. RTP possesses membrane occupation and recognition nexus (MORN) motifs, a structure shared with mammalian junctophilins and other membrane-associated proteins found within excitable cells. We show the MORN repeats, and both the N- and C-terminal domains, are required for RTP localization in the microvillar light-gathering organelle, the rhabdomere. RTP exists in multiple phosphorylated isoforms under dark conditions and is dephosphorylated by light exposure. An RTP deletion mutant exhibits a high rate of spontaneous membrane depolarization events in dark conditions but retains the normal kinetics of the light response. Photoreceptors lacking neither inactivation nor afterpotential C (NINAC) myosin III, a motor protein/kinase, also display a similar dark-noise phenotype as the RTP deletion. We show that NINAC mutants are depleted for RTP. These results suggest the increase in dark noise in NINAC mutants is attributable to lack of RTP and, furthermore, defines a novel role for NINAC in the rhabdomere. We propose that RTP is a light-regulated phosphoprotein that organizes rhabdomeric components to suppress random activation of the phototransduction cascade and thus increases the signaling fidelity of dark-adapted photoreceptors.
Introduction
The Drosophila photoresponse is a very fast and highly controlled G-protein-coupled cascade and has proven to be a preeminent model system to investigate prevalent and conserved neuronal molecular signaling mechanisms (Wang and Montell, 2007; Hardie and Postma, 2008; Katz and Minke, 2009). These processes include receptor–G-protein–arrestin interactions, phosphoinositide signaling, calcium-mediated regulation, activation of transient receptor potential (TRP) channels, and the organization of multiple molecular components within signaling complexes. In the Drosophila photoreceptor, microvilli project from the cell body of photoreceptor cells in structures called rhabdomeres. The rhabdomeres contain key phototransduction components, including Rhodopsin, G-protein, phospholipase C, and the Ca2+-permeable channels TRP and TRPL (TRP-like). Some of the signaling molecules are organized by an additional rhabdomeric specific scaffolding protein, inactivation no afterpotential D (INAD) (Shieh and Niemeyer, 1995; Tsunoda et al., 1997; Montell, 1998). This spatial organization likely contributes to the exceptionally fast kinetic response of Drosophila phototransduction (Zuker, 1996).
Protein phosphorylation/dephosphorylation cycles play key roles in photoreceptor signaling and cell biology. Rhodopsin (Steele et al., 1992; Byk et al., 1993), arrestin 1 and arrestin 2 (Matsumoto et al., 1994), and INAD (Matsumoto et al., 1999) are among the proteins phosphorylated in light conditions. Protein kinases expressed in the photoreceptor include neither inactivation nor afterpotential C (NINAC), a hybrid protein containing both a kinase and myosin III motor domain. NINAC is required for maintenance of the microvillar ultrastructure and the normal kinetics of the phototransduction response (Porter et al., 1992; Porter and Montell, 1993; Liu et al., 2008).
Many molecular components involved in Drosophila phototransduction were identified by isolating mutations altering the visual response (Pak, 1995). A complementary approach has been the characterization of gene products expressed predominantly, if not exclusively, within retinal tissues (Yamada et al., 1990; Zuker, 1996; Xu et al., 2004; Takemori et al., 2007). A Drosophila gene named retinophilin (rtp) (CG10233) has been repeatedly identified as an eye-enriched gene (Hyde et al., 1990; Arbeitman et al., 2002; Xu et al., 2004; Yang et al., 2005; Takemori et al., 2007). An rtp homolog has been identified in mammals and is expressed in the retina and CNS (Mecklenburg, 2007). The predicted RTP proteins contain four membrane occupation and recognition nexus (MORN) domains. These motifs were first identified in junctophilins, proteins that act to bring the plasma membrane into close contact with internal cellular membranes in excitable cells (Takeshima et al., 2000).
Here, we describe the role of RTP in Drosophila photoreceptors. We show that RTP is a rhabdomeric, light-regulated phosphoprotein. Unlike most other light-regulated phosphoproteins of Drosophila, light favors RTP dephosphorylation. To understand RTP function in the photoreceptor, we generated and analyzed a null rtp mutant. These mutants showed a striking increase in dark noise reminiscent of one of the phenotypes previously reported in the ninaC-null mutant (Hofstee et al., 1996). Furthermore, RTP protein was absent in the ninaC mutant, thus accounting for the dark-noise phenotype of the ninaC mutant. We propose RTP organizes rhabdomeric components to suppress random activation of the phototransduction cascade and thus increases the signaling fidelity of dark-adapted photoreceptors.
Materials and Methods
Fly strains.
The deficiency stocks Df(3R)5156, Df(3R)5142, Df(3R)5147, and Df(3R)3-4 were obtained from the Bloomington Stock Center, the piggyBac insertion strains PBacFMms19e00210 and PBac{RB}CG12163e00209 were obtained from the Exelixis Collection at the Harvard Medical School, and phototransduction mutant stocks were from collections maintained in our laboratories or obtained from the laboratories of Craig Montell (The Johns Hopkins University, Baltimore, MD) and William Pak (Purdue University, West Lafayette, IN). The Df(3R)rtp1 deletion chromosome was generated as described in Results using the methodology developed by Parks et al. (2004) and the oligonucleotide primers shown in supplemental Table S1 (available at www.jneurosci.org as supplemental material). The host for creation of all transgenic strains was w1118.
Antibody generation.
Primers described in supplemental Table S1 (available at www.jneurosci.org as supplemental material) were used to PCR amplify rtp sequence from a full-length cDNA clone (Mecklenburg, 2007). Treatment with EcoR1 and HindIII allowed ligation into the pET32(a+) expression vector (Novagen). After sequence verification, fusion protein was produced from this vector in Bl-21 Escherichia coli hosts and purified over His-Bind columns as described by the manufacturer (Novagen) and used to raise antibodies in mice.
Protein blots.
Drosophila heads were homogenized in solubilization buffer (60 mm Tris-HCl, pH 6.8, 20% SDS, 0.0004% bromophenol blue, 10% β-mercaptoethanol, 20% glycerol) and incubated for 1 h at 37°C. Homogenates were loaded at a concentration of one head per lane on NuPage 4–12% Bis-Tris gels (Invitrogen). After electrophoresis, proteins were transferred to Immobilon-P polyvinylidene difluoride membranes (Millipore) for 1 h using a Mini Trans-Blot electrophoretic transfer cell (Bio-Rad). Membranes were blocked for 1 h, washed, and probed with primary antibody (1:1000 dilution) overnight. Blots were washed and then probed with appropriate secondary antibodies, either anti-rabbit or anti-mouse IgG horseradish peroxidase (GE Healthcare) for 1 h. All washes were for four times for 5 min each time with TBST, pH 7.4. Detection was with ECL Western Blotting Detection Reagents per the manufacturer's protocol (GE Healthcare).
Generation of transgenic flies.
rtp (CG10233) constructs were generated by PCR amplifying w1118 genomic DNA or a previously described cDNA (Mecklenburg, 2007) using primers described in supplemental Table S1 (available at www.jneurosci.org as supplemental material). The native rtp promoter-RTP construct was made by PCR amplifying genomic DNA with the primers listed in supplemental Table S1 (available at www.jneurosci.org as supplemental material) and cloning into the pENTR/D-TOPO entry vector (Invitrogen). The rtp cDNA product was also cloned into pENTR/D-TOPO. The deletion chimeras were synthesized using the rtp pENTR/D-TOPO plasmid and primers 5′ to the pENTR/D-TOPO attL1 site and 3′ to the attL2 site, to generate chimera fusions as described in supplemental Table S1 (available at www.jneurosci.org as supplemental material). The entry clones and deletion chimera products were recombined into Terence Murphy Gateway-compatible Drosophila transformation vectors, obtained from the Drosophila Genomics Resource Center (Indiana University, Bloomington, IN). We used pTGW for GFP-RTP and pTWR for RTP-RFP and the deletion constructs. To construct the rtp genomic rescue P element vector, the BAC clone BACR01D10 was obtained from the Berkeley Drosophila Genome Project and used as template with primers listed in supplemental Table S1 (available at www.jneurosci.org as supplemental material) to PCR amplify a EcoR1–rtp–NotI genomic fragment possessing 1965 bp upstream of the ATG start site and 780 bp after the TGA stop site. The PCR product was placed in pCaSpeR4 (Thummel et al., 1988). The Mms19 rescue plasmid was made in a similar manner, using an Xho1 site found within a PCR product to create an XhoI–Mms19–NotI genomic fragment possessing 1350 bp upstream of the ATG start site and 891 bp beyond the TAG stop site. The coding capacity of all constructs was sequence verified and the plasmids were introduced into w1118 flies using standard P element transformation techniques.
Histology.
For visualization of green fluorescent protein (GFP) and red fluorescent protein (RFP) in whole-mount ommatidia, retinas were dissected and individual ommatidia dissociated into PBS by raking retina tissue with a tungsten needle. Samples were viewed on a Zeiss fluorescent microscope with a Plan-Apochromomat 63×, numerical aperture (NA) 1.40 objective. For visualization of GFP and RFP within the pseudopupil structure, live flies were viewed on a Zeiss Stemi SV 11 stereozoom microscope equipped with fluorescent optics. For visualization of GFP and RFP in retinal sections, Drosophila heads were removed and fixed in 4% paraformaldehyde/5% sucrose overnight at 4°C. The samples were then washed three times for 10 min in 5% sucrose/PBS, pH 7.4, and incubated overnight at 4°C. Heads were transferred to 30% sucrose/PBS overnight, and placed in 30% sucrose/PBS: Tissue-Tek OCT compound (1:1) for 4 h at room temperature. The samples were embedded in Tissue-Tek OCT compound sectioned at 10 μm and examined on a Bio-Rad MRC 1024 confocal microscope equipped with a Nikon Diaphot 200 and PlanApochromomat 60×, NA 1.40 objective. For electron microscopy, animals reared on a 12 h light/dark cycle and at <1 or 7 d after eclosion were fixed for 4 h in 2% paraformaldehyde/2% glutaraldehyde in 0.75 m Na cacodylate at room temperature, postfixed overnight in 2% tannic acid, dehydrated, and embedded as described by Ahmad et al. (2007). Thin sections were cut and stained in uranyl acetate and lead citrate and viewed on a Hitachi 600 transmission electron microscope.
Two-dimensional gel electrophoresis.
The Canton-S wild-type Drosophila melanogaster strain was dark adapted for 12 h before sample preparation. Light-adapted flies were reared under identical conditions before illumination under white fluorescent room light for 2 min. Flies were rapidly frozen in liquid nitrogen. Fly heads were separated from bodies by vortexing and immersed in −20°C acetone and held at −20°C for >72 h. Retinal tissue was dissected as described previously (Matsumoto et al., 1982). A dissected tissue sample of 150 eyes was homogenized in 100 μl of a lysis solution containing 8 m urea, 4% CHAPS (3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate), 0.2% (w/v) Bio-Lyte 3/10 (Bio-Rad), and 5% (v/v) β-mercaptoethanol. All reagents for two-dimensional electrophoresis were purchased from GE Healthcare, and other chemicals were purchased from Sigma-Aldrich unless specified. After centrifugation at 15,000 × g for 7 min, the supernatants were loaded onto Immobiline Dry Strips (pH 3–10, 13 cm in length; GE Healthcare). Isoelectric focusing (IEF) was performed using a IPGphor (GE Healthcare) with a cup loading strip holder. IEF was performed at 500 V for 5 min, 4000 V for 1.5 h, and 8000 V up to a total of 20,000 V · h. The strips were incubated for 30 min in SDS equilibration buffer [50 mm Tris-HCl, pH 8.8, 6 m urea, 30% (v/v) glycerol, 2% (w/v) SDS, and 2% (w/v) DTT] and transferred onto 11% SDS-polyacrylamide gels. After electrophoresis, gels were subjected to fluorescence staining with Pro-Q Diamond phosphoprotein dye (Invitrogen) initially and then stained with CBB R-250. Fluorescence images were captured by Molecular Imager FX (Bio-Rad).
Mass spectrometry.
Protein spots were manually excised from the gels and subjected to in-gel digestion with sequence-grade modified trypsin (Promega) as previously described (Takemori et al., 2007). Peptide mass fingerprinting (PMF) analysis was performed matrix-assisted laser desorption/ionization (MALDI)-time of flight (TOF) mass spectrometer (Voyager; Applied Biosystems). PMF data were submitted to the MASCOT program (Matrix Science). Database searches were performed against the National Center for Biotechnology Information nonredundant database using the following parameters: (1) the Drosophila protein database, (2) unlimited protein molecular weight and pI ranges, (3) the presence of protein modifications including acrylamide modification of cysteine, methionine oxidation, protein N terminus acetylation, and pyro-glutamic acid, and (4) peptide mass tolerance of ±0.25 Da. Tandem mass (MS/MS) analysis was performed using MALDI-quadrupole ion trap (QIT)-TOF mass spectrometer (AXIMA QIT; Shimadzu/Kratos). The theoretical fragment ions resulting from collision induced dissociation fragmentation of the phosphopeptides were calculated by MS-Product program (http://prospector.ucsf.edu/). For on-target alkaline phosphatase treatment, the peptide samples were dissolved in 0.5 μl of 50 mm ammonium bicarbonate, pH 8.9, containing 0.01 U of calf intestinal alkaline phosphatase (Sigma-Aldrich) on the MALDI target plate. Samples were then incubated for 10 min at room temperature. Dephosphorylation reaction was stopped by the addition of 0.5 μl matrix solution [2% (w/v) 2,5-dihydroxybenzoic acid in 50% (v/v) acetonitrile/0.1% (v/v) trifluoroacetic acid].
Electrophysiology.
Electroretinographic recordings were made from white-eyed flies maintained in 12 h light/dark cycle using standard protocols (Larrivee et al., 1981). For whole-cell photoreceptor recordings, dissociated ommatidia were prepared as previously described (Hardie et al., 2002) from newly eclosed adult flies and transferred to the bottom of a recording chamber on an inverted Nikon Diaphot microscope. The bath contained the following (in mm): 120 NaCl, 5 KCl, 10 N-Tris-(hydroxymethyl)-methyl-2-amino-ethanesulfonic acid, 4 MgCl2, 1.5 CaCl2, 25 proline, and 5 alanine, pH 7.15. The intracellular pipette solution was as follows (in mm): 140 K-gluconate, 10 N-Tris-(hydroxymethyl)-methyl-2-amino-ethanesulphonic acid, 4 magnesium-ATP, 2 MgCl2, 1 NAD, and 0.4 sodium-GTP, pH 7.15. All chemicals were obtained from Sigma-Aldrich. Whole-cell voltage-clamp recordings were made at room temperature (20 ± 1°C) at −70 mV (including correction for −10 mV junction potential) using electrodes of resistance ∼10–15 MΩ. Series resistance values were generally <30 MΩ and were routinely compensated to >80%. Data were collected and analyzed using Axopatch 200 or 2D amplifiers and pCLAMP8, 9, or 10 software (Molecular Devices). Quantum bumps and spontaneous dark events were analyzed using the Minianalysis program (Jaejin Software), using a threshold criterion of 1.5 pA. Multipeaked overlapping events were included in estimation of event rate, but excluded from analysis of event amplitudes. Photoreceptors were stimulated via a green light-emitting diode; intensities were calibrated in terms of effectively absorbed photons by counting quantum bumps at low intensities.
Image analysis.
Digital images of all fluorescent micrographs were obtained directly from a digital camera mounted on the microscopes. Other digital images (stained gels, electron photomicrographs, and detection of proteins after electrophoresis using antibodies) were obtained from photographic film using a digital scanner. All digital images were manipulated using Adobe Photoshop CS3 software (Adobe Systems) for the purpose of scaling and uniform adjustments of contrast and brightness.
Results
RTP undergoes light-regulated phosphorylation in the compound eye
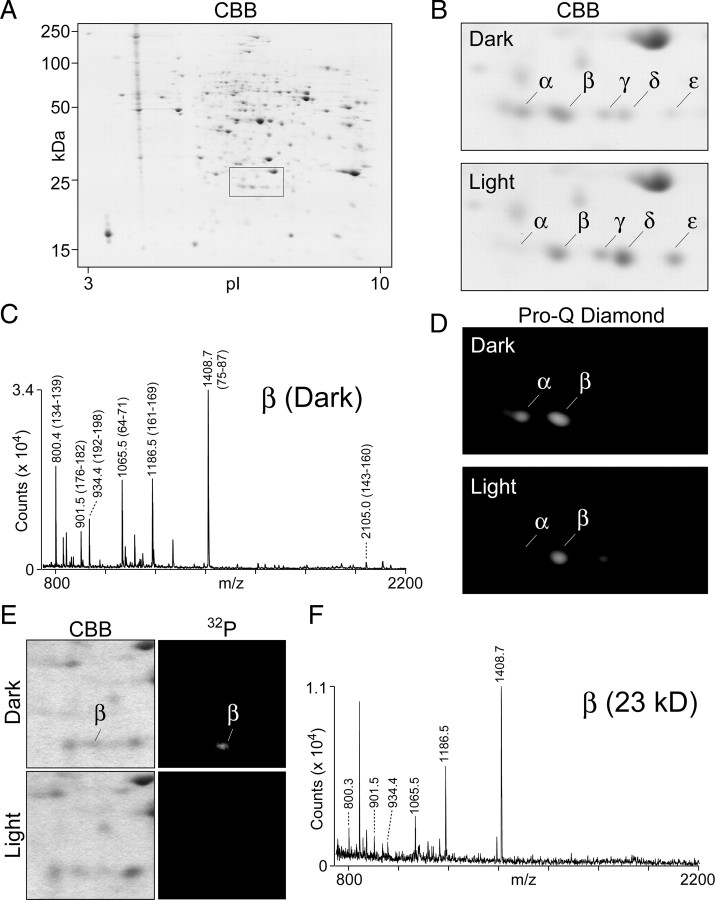
rtp transcripts were previously identified as highly enriched in retinal tissues (Hyde et al., 1990; Xu et al., 2004). RTP protein expression in the retina was confirmed by antibody reactivity (Mecklenburg, 2007) and proteomics analysis (Takemori et al., 2007). To extend the characterization of RTP, we examined the two-dimensional gel profile of RTP in dark- and light-adapted eyes. A representative two-dimensional gel image of the extracted eye proteins is shown in Figure 1A, and an enlarged view of the boxed region is shown in Figure 1B. Within this boxed region, individual protein spots are labeled as α, β, γ, δ, and ε. A comparison of the two-dimensional gel protein profile from dark- and light-adapted conditions revealed that the dark-adapted sample contained a marked increase in the α spot and corresponding reduction of the δ and ε spots (Fig. 1B). PMF analyses were performed to characterize each of these spots. The PMF trace of the dark-adapted β spot is shown in Figure 1C; the PMF traces of the other spots are shown in supplemental Figure S1A–F (available at www.jneurosci.org as supplemental material), and the results are summarized in Table 1. This analysis established that the RTP protein is the major component of the α, β, γ, δ, and ε protein spots.
Figure 1.
Identification and characterization of RTP proteins. A, The profile of retinal proteins in dark-adapted flies as analyzed by two-dimensional gel electrophoresis is shown. The boxed area identifies the location of RTP isoforms. B, The RTP region from two-dimensional profiles of retinal proteins prepared from dark- and light-adapted flies. The spots labeled α, β, γ, δ, and ε all contain the RTP protein (see Table 1). The abundance of the α spot was increased, and the δ and ε spots were decreased in the dark-adapted sample relative to the light-adapted sample. CBB, Coomassie brilliant blue. C, PMF analyses of the β spot excised from two-dimensional gel of the dark-adapted flies. The tryptic fragments labeled on the trace corresponded to peptide sequence of RTP and established that the major protein present in the β spot is RTP. The analysis was also carried on selected α, β, γ, δ, and ε spots and is summarized in Table 1. The PMF traces are shown in supplemental Figure S1 (available at www.jneurosci.org as supplemental material). D, In-gel phosphorylation analysis of the two-dimensional gel region containing RTP. The fluorescent phosphorylation sensor dye Pro-Q Diamond identified both the α and β spots in the dark-adapted profile as positive for phosphor staining. Weaker phosphor staining was seen in the light-adapted sample, and the phosphorylated α spot is greatly reduced. E, The RTP region of dark-adapted and light-adapted flies in CBB-stained two-dimensional gel (left panels) and 32P incorporation (right panels) from the analysis performed by Matsumoto and Pak (1984). The marked β spot in the dark-adapted sample showed the highest level of 32P labeling. Although the 25-year-old experiment showed more limited resolution of the spots than possible with current technology, both results show that RTP phosphorylation is reduced in the light-treated samples. F, PMF analysis of the dark-adapted β spot from the archived two-dimensional gel shown in E. The labeled tryptic fragments were derived from RTP (as identified in C), demonstrating that the major protein present in the 32P-labeled β spot was RTP.
Table 1.
Identification of RTP in two-dimensional gel spots by PMF analysis
| Spota | Protein | MOWSE(PMF)b | Matchedpeptide | Matched peptidecoverage (%) | Sequencecoverage (%) |
|---|---|---|---|---|---|
| α (DA) | Retinophilin | 141 | 8 | 73 | 51 |
| β (DA) | Retinophilin | 146 | 8 | 80 | 51 |
| β (LA) | Retinophilin | 141 | 8 | 73 | 51 |
| γ (DA) | Retinophilin | 111 | 7 | 64 | 41 |
| γ (LA) | Retinophilin | 111 | 7 | 64 | 41 |
| δ (LA) | Retinophilin | 161 | 9 | 75 | 60 |
| ϵ (LA) | Retinophilin | 146 | 8 | 80 | 51 |
| β (23 kDa) | Retinophilin | 127 | 7 | 88 | 41 |
aSpot IDs corresponding to those shown in Figure 1.
bMASCOT PMF score >60 corresponding to p < 0.05.
To investigate the basis of the distinct isoelectric properties of the multiple RTP forms, we conducted an in-gel phosphorylation assay with the fluorescent phosphorylation sensor dye, Pro-Q Diamond. The phosphor staining was prominent for the α and β spots in the dark-adapted extracts (Fig. 1D). Previously, Matsumoto and Pak (1984) identified a 23 kDa protein highly labeled in vivo by 32P when flies were dark adapted but not when flies were light adapted (Fig. 1E). We used mass spectrometry to analyze the 23 kDa phosphorylated spot from this archived two-dimensional gel according to the method of Matsumoto and Komori (1999). The analysis established that RTP was the 23 kDa phosphoprotein identified in the previous experiment (Fig. 1F, Table 1).
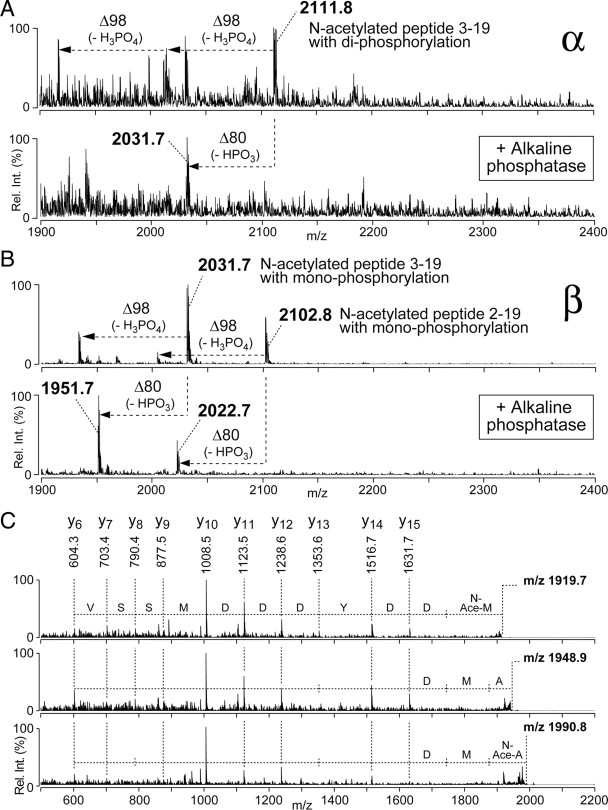
To further characterize RTP phosphorylation states, the α and β spots from dark-adapted flies shown in Figure 1B were subjected to MALDI-QIT-TOF MS analysis. The analysis of phosphorylated peptides enriched from the in-gel tryptic digests of the α spot (Fig. 2A, top trace) revealed a peptide ion peak at m/z 2111.8 with marker fragmentation loss of 98 and 196 Da, indicative of loss of a single H3PO4 (Qin and Chait, 1997) and two H3PO4 units, respectively. In-gel tryptic digests of the β spot (Fig. 2B, top trace) revealed two peptide ion peaks, at m/z 2031.7 and 2102.8, each showing marker fragmentation loss indicative of loss of a single H3PO4. In additional support of the finding that the α and β spots are phosphopeptides, alkaline phosphatase treatment caused an −80 Da mass shift (Fig. 2A,B, bottom traces), indicating phosphor removal. These peptides contain amino acids 2 through 19 or 3 through 19 of RTP, establishing that phosphorylation occurs within the N-terminal region. These data showed that RTP within the α spot possessed two phosphorylated amino acids, whereas RTP in the β spot possessed a single phosphorylated amino acid. Thus, the β spot present in both light- and dark-adapted eyes is a monophosphorylated RTP species intermediate between the nonphosphorylated (γ, δ, ε) and doubly phosphorylated α spot.
Figure 2.
Different phosphorylation states and N-terminal modifications are found in RTP isoforms. A, Enriched phosphopeptides derived from the α spot in Figure 1B were subjected to on-target alkaline phosphatase treatment using MALDI-QIT-TOF MS. The α spot contained a peptide ion at m/z 2111.8 corresponding to peptide 3–19 of RTP (top trace). Two peaks, corresponding to single and double loss of 98 Da, characteristic of the loss of one or two neutral H3PO4, were also present. After on-target alkaline phosphatase treatment (bottom trace), a dephosphorylated peptide was generated by the loss of a phosphate group (HPO3; −80 Da). B, Enriched phosphopeptides derived from the β spot in Figure 1B were subjected to on-target alkaline phosphatase treatment using MALDI-QIT-TOF MS. The β spot contained two peptide ions at m/z 2102.8 and m/z 2031.7, corresponding to RTP peptides 2–19 and 3–19, respectively (top trace), each showing single loss of 98 Da caused by loss of H3PO4. After on-target alkaline phosphatase treatment (bottom trace), a dephosphorylated peptide was generated by the loss of a phosphate group (HPO3; −80 Da). C, MS/MS analyses on the RTP N-terminal peptide ions m/z 1919.7, 1948.9, and 1990.8. The N-terminal amino acid in the m/z 1919.8 was acetylated Met3, the N-terminal amino acid in the m/z 1948.9 was unmodified Ala2, and N-terminal amino acid in m/z 1990.8 ion was acetylated Ala2.
MS/MS analysis of peptide ions derived from the N-terminal fragment of RTP revealed additional sources of variation. Figure 2C displays three selected ion peaks, the m/z 1919.7 and 1990.8 ions from the δ spot (supplemental Fig. S1E, available at www.jneurosci.org as supplemental material) and the m/z 1948.9 ion from the ε spot (supplemental Fig. S1F, available at www.jneurosci.org as supplemental material). These data showed that either Ala2 (1990.8 and 1948.9 peaks) or Met3 (1919.7 peak) was the N-terminal amino acid in RTP isoforms. RTP was found both with and without N-terminal acetylation, providing a second source of variation. The 1990.8 ion peak contained acetylated Ala2, whereas the 1948.9 peak was the same Ala2 fragment lacking acetylation.
RTP is a rhabdomeric protein
To extend previous observations of RTP localization within the retina, we examined native RTP promoter expression by constructing a carboxy-GFP-tagged version of the RTP gene. As shown in Figure 1A, GFP was localized to the rhabdomere, and no GFP fluorescence was detected within the central brain. Expression was detected in the rhabdomeres of the outer photoreceptor cells R1–R6, as well as in the rhabdomere of the central cell (Fig. 3B). To control RTP expression in photoreceptors and other cell types, we created UAS constructs placing rtp in frame with GFP and RFP coding sequences and under GAL4 transcriptional control. Using the pRh1-GAL4 driver specifying expression in the R1–R6 photoreceptor cells (Tabuchi et al., 2000) and the RTP-RFP construct, we found that RTP was efficiently trafficked to the rhabdomere (Fig. 3C). Localization therefore was not influenced by artificial expression with a UAS driver. The same flies also expressed the RH1 rhodopsin tagged with GFP (RH1-GFP). The companion image detecting RH1-GFP (Fig. 3D) and the merged RTP-RFP/RH1-GFP image (Fig. 3E) are also shown. The pseudopupil observed for RTP-RFP, RH1-GFP, and the merged image for this genotype are shown in Figure 1F–H. Together, these images demonstrated the colocalization of RTP-RFP with RH1 along the entire length of the rhabdomeric microvilli. RTP was not detected in other photoreceptor cell compartments.
Figure 3.
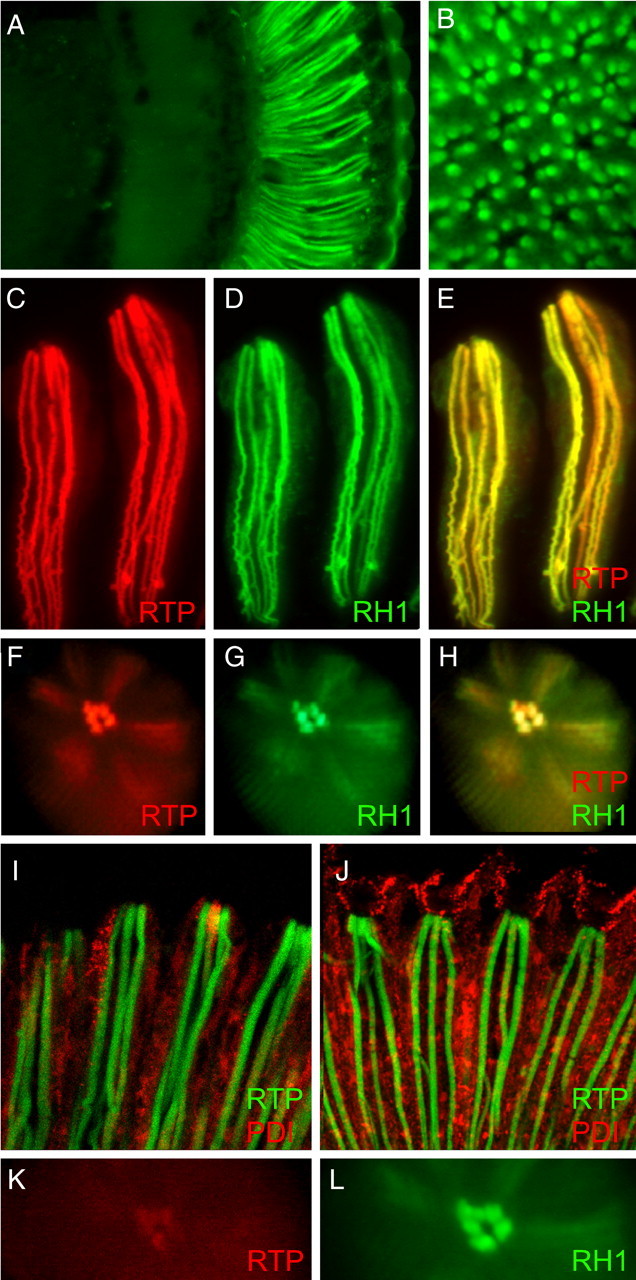
RTP protein is exclusively found in the rhabdomeres of photoreceptors. A, A frontal section through the adult head shows RTP-GFP is only detected within the retina when the rtp promoter controls expression. B, A cross-section through the retina demonstrates that RTP-GFP is expressed in the rhabdomeres of all photoreceptor cells. C–H, The coexpression of RTP-RFP and RH1-GFP under pRh1-Gal4 control in photoreceptor cells R1–R6 shows RTP-RFP colocalized with RH1-GFP within the photoreceptor rhabdomere. C–E show two isolated ommatidial bundles of photoreceptors, and F–H show deep pseudopupil images taken from living flies. RFP, GFP, and merged channel images are shown. I, Photoreceptors coexpressing both GFP-RTP and PDI-RFP under pRh1-GAL4 in photoreceptor cells R1–R6 showed GFP-RTP also localized to rhabdomere and showed no overlap with cytoplasmic PDI-RFP. J, Photoreceptors coexpressing both GFP-RTP and PDI-RFP under GMR-GAL4 in all retinal cells showed only photoreceptor rhabdomeres retain RTP while PDI expression was expanded to other cell types of retina. K, pRh1-Gal4-driven expression of a ΔN-terminal RTP-RFP protein showed that RTP protein lacking the N-terminal domain was not stable in photoreceptor rhabdomere. The signal was very weak at young ages and lost by 1 d of age. L, RH1-GFP expression in the same eye displayed in K. The Rh1 rhodopsin localizes within the rhabdomere.
A second UAS-driven rtp transgene was constructed that placed GFP at the N terminus (GFP-RTP). The localization of the GFP-RTP fusion protein within photoreceptors was again examined using the pRh1-GAL4 driver. A longitudinal retinal section of this genotype (Fig. 3I) revealed the GFP-RTP protein also was localized to the rhabdomere. Thus, neither the type of fluorescent protein tag, nor attachment to the N or C terminus of RTP, influenced the rhabdomeric localization.
The genotype displayed in Figure 3I also contained a UAS controlled proline diisomerase (PDI) PDI-RFP transgene designed to mark the endoplasmic reticulum (ER). The PDI-RFP transgene was assembled from three domains: the coding information for the amino signal sequence of the ER-resident protein PDI, the RFP ORF, and the coding information for the C-terminal KDEL ER retention domain of PDI. When pRh1-Gal4 was used to express PDI-RFP in R1–R6 photoreceptors, PDI-RFP was found widely distributed within cytoplasmic compartments consistent with localization within ER membranes. GFP-RTP showed no overlap with PDI-RFP, establishing that the tagged versions of RTP were detected only within the rhabdomeric membranes and was not associated with the ER or other cytoplasmic membranes.
To assay the stability of RTP in other retinal cell types, we expressed GFP-RTP and RTP-RFP constructs with the GMR-GAL4 (Freeman, 1996) driver. Glass multimer receptor (GMR) allows expression of UAS controlled genes in all classes of retinal cells of the compound eye and ocelli, the simple eyes dorsally located on the head. Under GMR control, the GFP-RTP and RTP-RFP proteins were detected in the rhabdomeres of all the photoreceptors of the compound eye and within the ocelli. However, we did not detect GFP-RTP and RTP-RFP within any other cell type. Figure 3J displays a section through a retina with GMR-controlled expression of GFP-RTP and PDI-RFP. The expanded PDI-RFP expression, relative to pRh1-restricted expression in photoreceptors (Fig. 3I), was most evident in pigment cells. These cells extend further distal than photoreceptors, surrounding the noncellular pseudocone located on top of the rhabdomere (Cagan and Ready, 1989). The apparent lack of RTP in nonphotoreceptor cells suggests that the stability of the RTP protein was dependent on its association with other components of the photoreceptor rhabdomeres.
To define the role of specific RTP sequences in rhabdomeric localization, we constructed a series of RTP-RFP deletions. Three deletions individually removed the N-terminal domain, the four MORN repeats, and the carboxy domain. Two smaller deletions removed either MORN repeats 1 and 2 or MORN repeats 3 and 4. As done previously with the full-length RTP, each deletion construct was simultaneously expressed with RH1-GFP under pRh1-GAL4 control. None of the deletions was stably maintained in the rhabdomere. At young ages, low levels of the deletion lacking the amino-domain was detected in the rhabdomere (Fig. 3K), but this signal was lost by 24 h after eclosion. Weaker signal was also detected in young flies expressing the deletion lacking all four MORN repeats. No signal could be detected from the other deletions, while RH1-GFP remained localized to the rhabdomere in all cases (Fig. 3L). These experiments indicated that all three domains of the RTP protein are needed for rhabdomeric targeting or stabilization.
Construction of rtp1, an rtp (CG10233)-null mutant
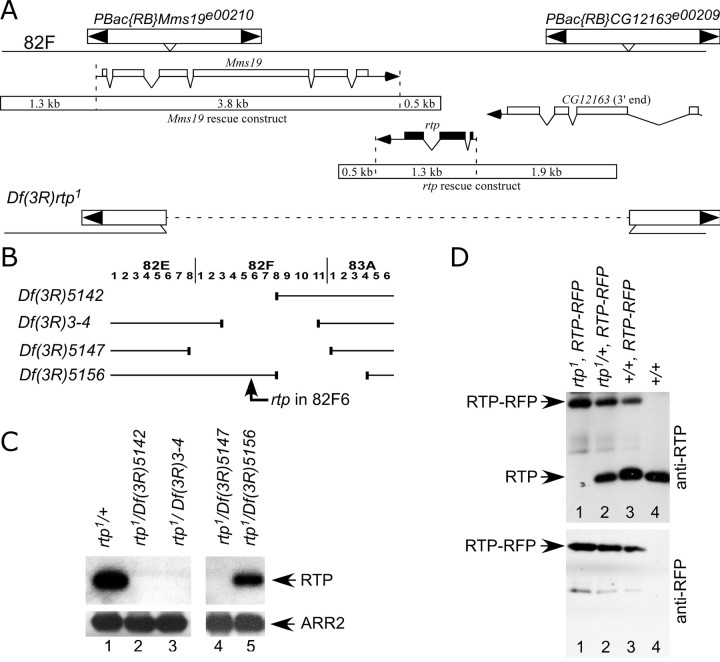
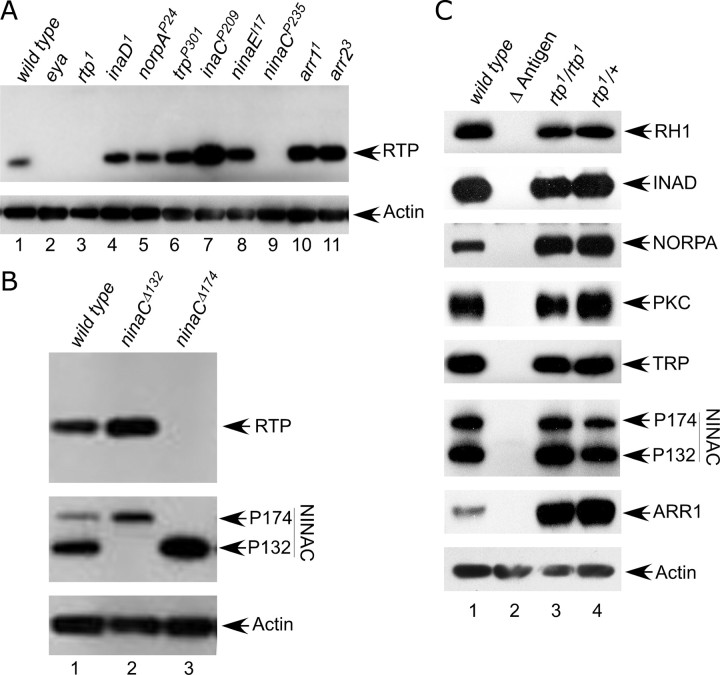
We used the Flp recombinase target (FRT)-based deletion approach developed by Parks et al. (2004) to generate a mutant strain lacking the RTP protein. FLP directed recombination between the FRT-bearing piggyBac insertions, PBac{RB}Mms19e00210 and PBac{RB}CG12163e00209, produced a deletion chromosome, Df(3R)rtp1, that disrupted the genes Mms19 and CG12163 and deleted rtp (Fig. 4A). Two-sided PCR and other PCR-based diagnostics were used to verify construction of Df(3R)rtp1. The homozygous lethality of the Df(3R)rtp1 chromosome could not be rescued by chromosome deficiencies uncovering these three genes, establishing that at least one of the three genes was essential for viability. Mms19 was considered most likely to be lethal based on genetic analysis, including the observation that the PBac{RB}Mms19e00210 chromosome was homozygous lethal. Therefore, to attempt rescue of the lethality associated with Df(3R)rtp1, we isolated a 5.6 kb genomic DNA fragment containing the Mms19+ gene (Fig. 4A) and used this DNA to create transgenic Mms19+ flies. The Mms19+ transgene rescued the lethality associated with PBac{RB}Mms19e00210. A series of genetic crosses showed that two different Mms19+ transgenes, one on the second and one on the third chromosome, were able to rescue the lethality associated with the Df(3R)rtp1 deletion chromosome. This allowed us to create an rtp-null homozygous strain, either by placing Df(3R)rtp1 and the Mms19+ transgene on the same third chromosome or by inclusion of Mms19+ on the second chromosome and Df(3R)rtp1 on the third chromosome. The rtp-null mutation synthesized in this way will be referred to as rtp1 in this report.
Figure 4.
Creation and analysis of the rtp1 mutant. A, The genomic region that contains CG10233 (rtp) is flanked by the PBac insertions PBac{RB}Mms19e00210 and PBac{RB}CG12163e00209. FLP-directed recombination between these insertions produced a deletion chromosome, Df(3R)rtp1, that deleted CG10233 and disrupted the Mms19 and CG12163 genes. The 5.6 kb genomic DNA fragment containing the Mms19+ gene (Mms19 rescue construct) that rescued the zygotic lethality of Df(3R)rtp1 and the 3.7 kb genomic DNA used in the rtp rescue construct are also shown. B, Diagram showing the four deficiency chromosomes used in the analysis of the rtp gene. The location of the rtp gene in 82F6 is indicated. The rtp gene was missing in all deletions except Df(3R)5156. C, Top panels, Protein blots show that rtp/+ and rtp1/Df 5156 retain RTP protein, but RTP protein was absent in all rtp1/Df genotypes expected to lack the rtp gene. Bottom panels, Reprobing the protein blot with ARR2 antibodies demonstrated that similar levels of retinal proteins were loaded in all lanes. D, Protein blot analysis of the RTP-RFP fusion protein. The RTP-RFP 50 kDa protein was identified by both anti-RTP and anti-RFP antibodies (lanes 1–3). The levels of RTP-RFP protein was greatest when no native RTP is expressed (lane 1) but diminished in a dose-responsive manner when one copy (lane 2) or two copies (lane 3) of the native RTP were also expressed.
RTP protein is absent in rtp1 mutants
To confirm that rtp1 deleted the rtp gene, we used protein blots to assess the presence of the RTP protein in Drosophila strains with different combinations of rtp1, larger deficiencies in the 82F region, and the RTP-RFP construct. Figure 4B illustrates the location of rtp and the breakpoints of the chromosomal deficiencies used in the analysis. Figure 4C, lane 1, shows that RTP protein was detected in rtp1/+ heterozygotes. RTP protein was also detected in rtp1/Df(3R)5156 flies (lane 5) because of the retention of the 82F6 region and the rtp+ gene on the Df(3R)5156 chromosome. However, RTP protein was absent in all three strains of rtp1/Df lacking the 82F region and therefore deleted for the rtp gene (lanes 2–4).
The anti-RTP serum used in these protein blots was generated against the carboxy domain of RTP and therefore expected to detect the RTP-RFP tagged construct described above. We tested flies homozygous for rtp1 but expressing instead RTP-RFP. Figure 4D, lane 1, shows, as expected, the 23 kDa RTP protein was missing, and instead a 50 kDa protein, corresponding to the predicted size of the RTP-RFP fusion protein, was present. To confirm the identity of the 50 kDa protein, anti-RFP antibodies were used to detect the same 50 kDa protein specifically in the strains expressing RTP-RFP (Fig. 4D, bottom panel). We extended this analysis in an effort to estimate the levels of expression of the RTP-RFP fusion protein. In Figure 4D, top panel, a comparison of lanes 1 and 4 suggested the abundance of the RTP-RFP protein was similar to the 23 kDa protein when only one form was expressed. In lanes 2 and 3, the RTP-RFP was simultaneously expressed with one and two copies, respectively, of the rtp+ wild-type gene. The abundance of the RTP-RFP protein was reduced with increasing levels of the 23 kDa protein, suggesting that the overall levels of RTP were maintained at a steady-state level within the photoreceptor cell. The observation that RTP-RFP protein competed with native RTP was consistent with the expectation that RTP-RFP was properly localized in the photoreceptor cell.
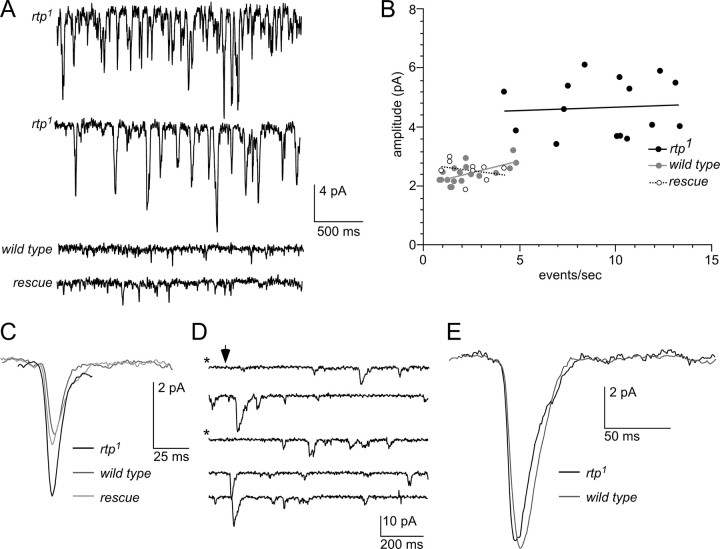
rtp mutants show high levels of spontaneous dark noise
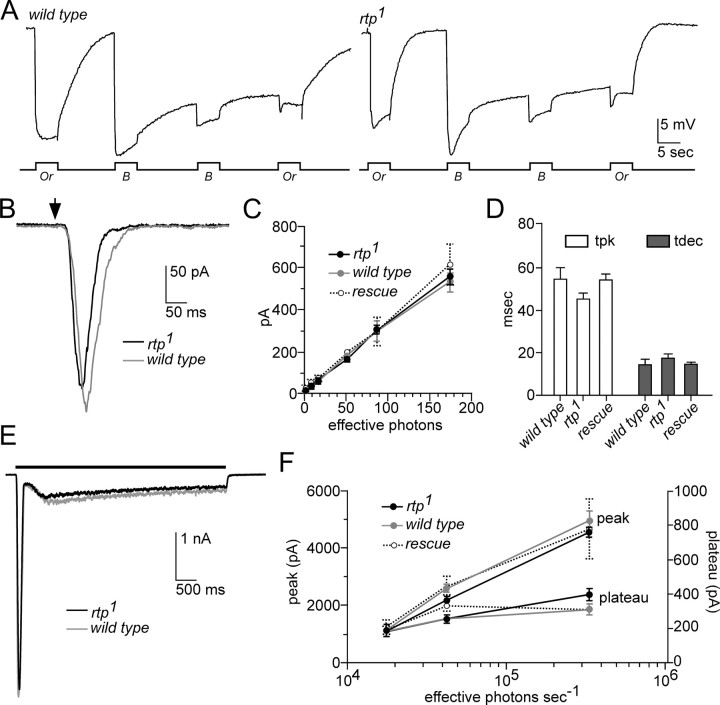
Electroretinograms revealed no detectable alterations in the sustained light response or the prolonged depolarizing afterpotential of rtp1 mutants (Fig. 5A). To further evaluate the role of RTP in light-regulated photoreceptor physiology, we performed whole-cell voltage-clamp recordings from dissociated ommatidia. In these experiments, we examined rtp-null flies of the genotype w; rtp1/Df(3R)5142 (abbreviated to rtp1), comparing them with both wild-type (w) and rtp1 flies carrying the genomic rtp+ rescue construct (Figs. 5, 6, labeled “rescue”) as an additional control. Responses of the null rtp1 flies to brief dim light flashes of varying intensity were similar to controls (Fig. 5B–D). Although there was a tendency for slightly faster time to peak (tpk) in rtp1 mutants (45 ms compared with 55 ms), this did not reach statistical significance (Fig. 5C,D). The waveforms of sustained responses to a bright (up to ∼3 × 105 effective photons s−1) 5 s light stimulus were also similar to wild type (Fig. 5E), with both peak and plateau responses showing no statistically significant differences (Fig. 5F).
Figure 5.
The rtp1 light response is similar to wild type. A, The electroretinogram response of white-eyed wild type (left) and the rtp1 mutant (right) were similar (n > 6). The protocol consisted of 5 s orange (Or) or blue (B) light stimuli administered in the indicated order. B, The responses of rtp1 and wild-type photoreceptors to a 1 ms test flash (arrow) containing ∼60 effective photons were similar in whole-cell recordings. C, The amplitude of the light response to increasing intensities of light stimuli was similar in rtp1 (n = 6) and wild-type photoreceptors (n = 4). The analysis of an additional control, the rtp1 mutant genotype containing an rtp+ transgene, is labeled as “rescue” (n = 4). Mean ± SEM is shown. D, Time to peak (tpk) and time to 50% decay (tdec) in rtp1 (n = 16) and the control strains (wild type, n = 5; rescue, n = 11) were similar. Mean ± SEM is shown. There was a tendency for slightly faster tpk values in rtp1, but this was not statistically significant (two-tailed t test). E, The rtp1 waveform of sustained responses to a bright (∼3 × 105 effective photons s−1) 5 s light stimulus was similar to wild type. F, Statistical analysis of the amplitude of the sustained response waveform in rtp1 (n = 6), wild type (n = 5), and the rescue genotype (n = 5, except at highest intensity, n = 2). Mean ± SEM is shown. rtp1 and the control strains showed similar plateau and peak response values. All data in B–F were obtained from whole-cell recordings of photoreceptors from dissociated ommatidia, voltage clamped at −70 mV.
Figure 6.
High spontaneous bump rates in the rtp1 mutant. A, A high rate of spontaneous dark events was present in the rtp1 mutant (top 2 traces from different cells) relative to both wild type (lower middle trace), and the rtp rescue genotype (bottom trace). B, Scatter plot of event rate against mean event amplitude for rtp1 cells (n = 15); wild type (n = 18) and rescue controls (n = 9). All rtp1 mutant photoreceptors are distinct from controls; regression lines show no correlation between event rate and amplitude. C, The spontaneous dark events (average waveform of >50 events in a representative cell of each genotype) in the rtp1 mutant, wild type, and the rescue genotype. Overall kinetics was similar, but event amplitude in rtp1 was approximately twice that of controls. D, Representative traces of light-induced bumps elicited by brief dim flashes containing on average approximately one effective photon (arrow) in the rtp1 mutant. The starred traces were judged to be “failures.” E, The light-induced bumps (average waveform of 30 bumps; aligned by rising phase) in rtp1 mutant and a control cell were indistinguishable. Shown are data from one cell (for additional traces and analysis, see text and supplemental Fig. S2, available at www.jneurosci.org as supplemental material).
Although the main features of the light response appeared normal, whole-cell recordings from rtp1 mutant photoreceptors invariably showed a conspicuously high level of spontaneous dark noise consisting of small quantum bump-like events (Fig. 6A, top two traces). As previously reported (Hardie et al., 2002), spontaneous dark events were also present in wild-type photoreceptors and rtp rescue controls (Fig. 6A, bottom two traces) but occurred at a much lower rate and were smaller in amplitude. An event detection algorithm (Minanalysis) was used to quantify the event rates and amplitudes, yielding amplitudes in rtp1 mutants approximately two times larger than in wild type (mean, 4.7 ± 0.9 pA; n = 15 cells) and rates approximately five times faster (mean, 9.4 ± 2.8 events s−1). Both parameters were highly significantly different, with both rate and amplitude having p < 10−10 t test values. Although this analysis may overestimate amplitude and underestimate rate because of the inability to distinguish coincident overlapping events, such errors are likely to be small (<10% of events) because of the short duration (t1/2 ∼ 15 ms) of the events. Furthermore, a scatterplot of event rate versus amplitude for all data clearly showed that rtp1 cells lie in a different parameter space from controls, even for cells at the end of the ranges in which event rates were similar (Fig. 6B). The average waveform of the events was also reconstructed by manually selecting “clean” single events from mutant cells with relatively low rates of spontaneous events. After aligning and averaging, the averaged waveform was again clearly larger than spontaneous dark events in controls (Fig. 6C). The pronounced increase in dark noise was quantitatively rescued in the <rtp+>; rtp1 rescue genotype, in which both amplitude and rate of dark events were not statistically different from seen in wild type (Figs. 6B), with wild type versus rescue t tests values of p = 0.99 (rate) and p = 0.4 (amplitude). These results established that deletion of the rtp gene was solely responsible for the dark-noise phenotype.
The macroscopic response to brief flashes containing up to several hundred effective photons in Drosophila photoreceptors is the strict linear summation of the underlying quantum bumps, with the overall waveform representing the convolution of the quantum bump and its latency distribution (Henderson et al., 2000). Hence, the fact that macroscopic flash responses in rtp1 mutants were similar to wild type in both amplitude and kinetics, strongly suggests that the underlying quantum bumps were also similar to wild type. A rigorous analysis of light-evoked quantum bumps in rtp1 was not possible because of the high rate of spontaneous events. Nevertheless, in one cell with a relatively low rate of spontaneous dark events, individual quantum bumps in response to dim flashes could be reliably resolved with a quantum efficiency similar to wild-type controls (Fig. 6D). Quantum bumps in this cell were indistinguishable from controls in amplitude (mean ± SD, 9.3 ± 3.3 pA; n = 30; compared with 9.4 pA in control) and waveform (Fig. 6E) and larger than the ongoing spontaneous events in this cell (mean ± SD, 3.9 ± 1.6 pA, n = 197, at 4.8 events s−1). In other cells, the spontaneous noise precluded detailed analysis, but averaging responses to flashes containing on average approximately only a single effective photon (as calibrated in controls) indicated that the single photon sensitivity was almost as large as controls (mean ± SEM, 65 ± 19%; n = 4) (supplemental Fig. S2, available at www.jneurosci.org as supplemental material). Such recordings also directly demonstrate that, although light-induced bumps can still be seen rising above the background noise, the signal-to-noise ratio in rtp1 mutants around threshold must be severely reduced.
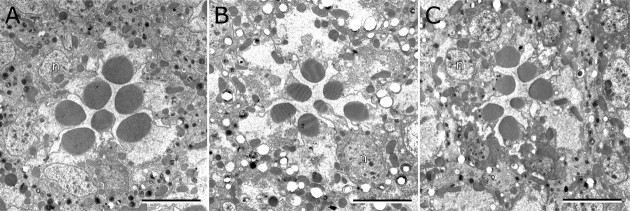
While performing these whole-cell recordings, we found that rtp1 cells exhibited an average capacitance of 50 pF, significantly less than the wild-type value of 63 pF (t test, p = 0.003). This result suggested a corresponding reduction in the membrane surface area of the microvillar rhabdomeres, which account for the majority of the whole-cell capacitance. To investigate this further, we used electron microscopy to examine rtp photoreceptor morphology. Indeed, cross-sections of rtp1 photoreceptors at the young ages used in the electrophysiological preparations showed a reduced rhabdomere surface area relative to the wild type (Fig. 7A,B). No additional structural defects were found in rtp1 photoreceptors at 7 d of age (Fig. 7C) when reared under a 12 h light/dark cycle.
Figure 7.
Morphology of the rtp1 mutant retina. Cross-section of wild type (A), rtp1 mutant (B) photoreceptors at <1 d after eclosion and rtp1 mutant photoreceptors at 7 d (C) are shown. The sections are at comparable depths, indicated by the presence of R1–R6 photoreceptor nuclei (n) in all images. The rtp+ genotype was rtp1/+; the rtp mutant genotype was rtp1/Df(3R)5142. Scale bars, 5 μm.
RTP protein stability is dependent on NINAC
The electrophysiological results showing rtp1 mutants alter photoreceptor membrane excitability suggested that RTP may interact with rhabdomeric proteins responsible for phototransduction. To further investigate this possibility, we examined RTP protein content in mutant photoreceptors lacking proteins involved in the phototransduction process. In all cases, we examined young flies carrying null or very severe alleles that result in no detectable protein expression. These results, summarized in Figure 8A, show that the NINAC protein was necessary for the retention of RTP in photoreceptors. NINAC is reported to bind to the INAD scaffolding protein; however, the core components in the INAD signaling complex, INAD, inactivation no afterpotential C (INAC), TRP, and no receptor potential A (NORPA), responsible for membrane depolarization events did not have a major effect on RTP expression. Also, RTP expression was not dependent on the presence of the RH1 rhodopsin, ARR1, or ARR2.
Figure 8.
RTP expression in mutants lacking other rhabdomeric components and rhabdomeric protein expression in the rtp1 mutant. A, The RTP protein expression was absent in ninaCP235, but was expressed in other phototransduction proteins found in the rhabdomere. Top panel, The first three lanes at left: wild type, eyes absent (eya1). eya1 and rtp1 are controls for antibody specificity. All mutants were homozygotes. The apparent increased expression of RTP in inaCP209 was not confirmed in subsequent experiments. Bottom panel, The blot was reprobed with anti-actin antibody as a protein loading control. B, RTP expression was dependent on activity of the NINAC P174 rhabdomeric protein. Top panel, Flies lacking only the NINAC P132 cytoplasmic form (lane 2) expressed RTP, but flies lacking only the NINA P174 rhabdomeric form (lane 3) did not express RTP. Middle panel, The blot was reprobed with NINAC antibodies to confirm identity of the ninaC Δ132 and ninaCΔ174 strains. Bottom panel, The blot was reprobed with anti-actin antibody as a protein loading control. C, RTP protein was not necessary for stable expression of other phototransduction proteins. The mutant alleles described in A were used as antigen-null (Δ Antigen) controls.
Two isoforms of the NINAC protein are present in photoreceptor cells. NINAC P132 is found in the cell body, and P174 localizes to the rhabdomeres (Porter et al., 1992). To further investigate the process by which NINAC controlled RTP expression, we examined RTP expression in ninaC genotypes that specifically delete either of these two isoforms. Figure 8B, top panel, shows that RTP remained at high levels in ninaCΔ132, missing only the cell body-specific isoform P132. In contrast, RTP was absent in the ninaCΔ174 allele missing the rhabdomeric isoform P174 but not the cell body P132 protein. We then created ninaCP235; rtp1 flies that expressed RTP-RFP under control of the Rh1 rhodopsin promoter. Whereas control ninaCP235/+ flies showed high levels of RTP-RFP expression, no expression was seen in homozygous ninaCP235 flies. The finding that RTP-RFP also required NINAC for stable expression supported the previous conclusion that RTP-RFP retained the normal expression properties of the native RTP protein.
Conversely, we also tested whether the expression and stability of other rhabdomeric phototransduction proteins were dependent on RTP. Figure 8C shows the rtp1 deletion mutant retained high protein levels of all core members of the signalplex including INAD, NORPA, PKC, and TRP, and also NINAC, ARR1, ARR2, and the RH1 rhodopsin. Thus, RTP was not required for the expression or stability of these rhabdomeric proteins.
Discussion
rtp (CG10233) has been repeatedly identified as a gene expressed predominantly in the Drosophila eye (Hyde et al., 1990; Arbeitman et al., 2002; Xu et al., 2004; Yang et al., 2005). The analysis of such retinal specific genes has greatly advanced our understanding of the molecular mechanisms underlying the invertebrate phototransduction process (Pak, 1995; Zuker, 1996; Wang and Montell, 2007). For this reason, we sought to characterize this gene and identify the role of the RTP protein in photoreceptor biology. The rtp gene has been recently studied with respect to a role in phagocytosis under the gene name undertaker (Cuttell et al., 2008). We retained the original rtp (retinophilin) gene name in this report. This is justified by the exceptionally high level of retinal enrichment of RTP, the MORN domain homologies with junctophilin, and the retinal phenotype reported here.
Several key Drosophila photoreceptor proteins show light-dependent cycling between phosphorylated and dephosphorylated states. These include Rhodopsin, TRP, INAD, ARR1, and ARR2 (for review, see Wang and Montell, 2007; Hardie and Postma, 2008). Whereas these proteins are phosphorylated in response to light, RTP is the first phosphoprotein characterized that shows higher phosphorylation levels when the fly is maintained in dark conditions. Matsumoto and Pak (1984) first identified a 32P-labeled 23 kDa retinal phosphoprotein in flies kept in the dark for 1 h, but not from flies maintained in the light. Using analytical mass spectrometry, we show this protein, retrieved from the protein analysis contained in the 1984 report, is RTP. Improvements in two-dimensional gel electrophoresis technologies allowed us to resolve multiple RTP forms. These results indicate that the most acidic form of RTP, the α spot, is phosphorylated at two sites within the N-terminal domain. The β spot is phosphorylated at a single site within the N-terminal domain, and the δ and ε spots contain nonphosphorylated RTP. Thus, the presence of RTP carrying different number of phosphates is a major reason that distinct forms of RTP are observed in the two-dimensional gel analysis.
N-terminal modifications are a second mechanism for generating multiple forms of RTP. One RTP species is acetylated at Ala2, presumably by proteolytic removal of Met1 (start) as commonly seen for N-terminal acetylation events (Polevoda and Sherman, 2000). A second species contains N-terminal acetylation at Met3. This second subgroup may represent the uncommon removal of two amino acids before the acetylation, or translation initiation at Met3 and direct acetylation. Additional species of RTP were identified that lacked acetylation with Ala2 present at the N terminus. Acetylation of the N-terminal sites is not critical to RTP stability or localization because our GFP-RTP fusion protein, in which neither Met is available for N-terminal modification, correctly traffics to the rhabdomere.
N-terminal and C-terminal RTP fusion constructs confirm the previous observation (Mecklenburg, 2007) that RTP is a rhabdomeric protein, hence positioned to interact with other components of the phototransduction process. Analysis of ninaC mutants shows the rhabdomeric isoform of NINAC, P174, is absolutely required for expression of RTP in photoreceptors. It is unlikely that this interaction is being modulated at the transcriptional level, because expression of RTP from an ectopic promoter (pRh1-GAL4→UAS-RTP) is still dependent on NINAC. Rather, NINAC P174 is found within the rhabdomere and therefore likely acts in the rhabdomere to stabilize RTP. The requirement for NINAC P174 in stabilization of RTP likely accounts for the failure to detect RTP protein when expressing the RTP gene in other retinal cell types.
To determine the role of RTP, we created rtp mutant flies and examined photoreceptor cell electrophysiology. Both the electroretinogram and whole-cell voltage-clamped macroscopic flash responses were very similar to wild type, indicating no obvious defects in excitation or response inactivation. However, a major defect in rtp mutant cells was a high level of dark noise caused by a high rate of spontaneous, small quantum bump-like events. In wild-type photoreceptors, similar, but smaller and less frequent dark events are believed to result from the spontaneous activation of the Gq protein releasing active Gq α subunits, triggering the generation of miniature quantum bump-like events (Minke and Stephenson, 1985; Hardie et al., 2002; Elia et al., 2005). Compared with wild type, the rtp mutants show an approximately fivefold increase in dark event rate and an approximately twofold increase in amplitude. The mechanistic basis for the increase in dark noise requires additional study, but assuming the events are also generated by spontaneous G-protein activation, at least two, not mutually exclusive possibilities should be considered. First, RTP might be required to suppress the rate of spontaneous G-protein activations, and second, RTP might reduce the probability that spontaneously activated G-protein α subunits can cause sufficient downstream activation to overcome threshold for bump generation.
Significantly, a very similar increase in spontaneous dark noise was reported in ninaC mutants (Hofstee et al., 1996), although the molecular basis was unknown. Our finding that NINAC P174 is required for RTP stabilization strongly suggests that the increased rate of spontaneous bump-like events observed in the ninaC mutant is attributable to the reduction in RTP. Also, loss of RTP may contribute to the reduced rhabdomere found in the ninaC mutants (Matsumoto et al., 1987) as rtp mutants also show a reduction in rhabdomere volume. Interestingly, other reported features of the ninaC phenotype, which include light-dependent retinal degeneration, and response inactivation defects (Porter et al., 1992; Porter and Montell, 1993; Liu et al., 2008), were not reproduced in rtp mutants, highlighting the multifunctional nature of the NINAC protein.
A prominent feature of the RTP protein is the presence of four MORN repeats. Originally identified in junctophilin, proteins containing MORN sequences have now been found in both plant and animal systems. Common themes emerging from these studies are that MORN sequences are necessary for stable interactions with plasma membranes (Takeshima et al., 2000; Gubbels et al., 2006) and that MORN sequences bind to phospholipids (Ma et al., 2006; Im et al., 2007). Structural data from the MORN-containing protein histone methyltransferase SET7/9 (Wilson et al., 2002) suggest that an individual MORN repeat consists of short β-pleated sheet regions folded back on themselves to create a relatively flat surface area. MORN repeats positioned in tandem further expand the protein surface area. The extended flat topology of the MORN repeats might allow RTP to organize and stabilize phospholipids and other components at the membrane surface. Our deletion construct analysis showed that RTP lacking all four MORN repeats was weakly detectable only in young flies; RTP proteins lacking only two of the four MORN repeats were not detectable. These results are consistent with another study showing no clear relationship between MORN repeat number and protein stability or localization (Takeshima et al., 2000). In addition to four MORN repeats, RTP contains an N-terminal sequence of 63 aa and a C-terminal sequence of 42 aa. Deletion constructs lacking each of these regions were poorly maintained in the photoreceptor, showing that motifs in all regions are required for RTP stability.
RTP shares MORN repeat elements with junctophilins, which stabilize the junctional complex anchoring the plasma membrane to the sarcoplasmic reticulum in excitable cells (Takeshima et al., 2000). Drosophila possesses a junctophilin homolog in addition to RTP, and the microvillar distribution of RTP suggests a different role than described for junctophilin. It is noteworthy that both proteins are localized to structures mediating membrane depolarization because of Ca2+ release or influx. The presence of RTP in the rhabdomere suggests a different role in photoreceptors than in macrophages in which RTP has been localized to the ER, phagosomes, and the plasma membrane (Cuttell et al., 2008).
Light-dependent dephosphorylation is not unique to RTP in visual systems. Drosophila dMoesin, localized at the interface of the rhabdomere and cytoplasm, is regulated by light-driven dephosphorylation and controls the movement of rhabdomeric components (Chorna-Ornan et al., 2005). Two types of mammalian photoreceptor proteins also exhibit dark-adapted phosphorylation. The first is the centrin family, found at the connective cilium. Phosphorylation of centrin decreases the affinity for Gβγ subunits allowing different distribution of Gβγ under light and dark conditions (Trojan et al., 2008). The second dark-phosphorylated protein found in vertebrate photoreceptors is phosducin. In a phosphorylated state, phosducin also shows reduced binding of Gβγ protein subunits (Lee et al., 2004; Song et al., 2007). Although the phosphorylation of both types of proteins alters the binding affinity for G-protein subunits and may provide a means of adapting the photoreceptor to different light conditions, recent data suggest that phosducin also directly affects G-protein levels (Krispel et al., 2007). Elia et al. (2005) reported that an imbalance of Gq subunits increased the rate of spontaneous dark bumps in Drosophila, and translocation of Gqα and other photoreceptor components during the light response has been documented (Kosloff et al., 2003; Cronin et al., 2004; Frechter et al., 2007). The possibility that rtp mutants alter the cellular distribution or assemblage of phototransduction components needs to be investigated.
Our analysis establishes a role for RTP in suppressing spontaneous dark noise. Light perception requires an adequate signal-to-noise ratio; hence under very dim light conditions the suppression of spontaneous events is essential to increased sensitivity. A testable hypothesis is that RTP phosphorylation is responsible for the suppression of the dark bumps, perhaps by altering the ability of RTP to bind with or affect the position of other transduction components. Our analysis demonstrates a novel role for the NINAC-encoded myosin III in retinal protein stabilization. This interaction will be examined by identifying RTP binding partners and ectopic expression of NINAC and RTP. Furthermore, our experiments demonstrate the function of a protein containing MORN repeats in Drosophila photoreceptor cells providing an additional attractive experimental system to study the role of this conserved protein motif.
Footnotes
This work was supported by Biotechnology and Biological Sciences Research Council Grant BB/D007585/1 (R.C.H.), National Institutes of Health (NIH) Grants EY06595, EY13877, EY12190, and RR17703 (H.M.), and NIH Grant EY06808 (J.E.O.). We thank Craig Montell, Armin Huber, and Alex Kiselev for supplying antibodies used in this study, Benjamin Currie, Karen Hibbard, Kathleen Mitchell, and Yeona Chun for assistance in analyzing the rtp1 chromosomal region, Lidiya Orlichenko for construction of PDI-RFP, and Sheila Adams for electron microscopic analysis.
References
- Ahmad ST, Natochin M, Artemyev NO, O'Tousa JE. The Drosophila rhodopsin cytoplasmic tail domain is required for maintenance of rhabdomere structure. FASEB J. 2007;21:449–455. doi: 10.1096/fj.06-6530com. [DOI] [PubMed] [Google Scholar]
- Arbeitman MN, Furlong EE, Imam F, Johnson E, Null BH, Baker BS, Krasnow MA, Scott MP, Davis RW, White KP. Gene expression during the life cycle of Drosophila melanogaster. Science. 2002;297:2270–2275. doi: 10.1126/science.1072152. [DOI] [PubMed] [Google Scholar]
- Byk T, Bar-Yaacov M, Doza YN, Minke B, Selinger Z. Regulatory arrestin cycle secures the fidelity and maintenance of the fly photoreceptor cell. Proc Natl Acad Sci U S A. 1993;90:1907–1911. doi: 10.1073/pnas.90.5.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagan RL, Ready DF. The emergence of order in the Drosophila pupal retina. Dev Biol. 1989;136:346–362. doi: 10.1016/0012-1606(89)90261-3. [DOI] [PubMed] [Google Scholar]
- Chorna-Ornan I, Tzarfaty V, Ankri-Eliahoo G, Joel-Almagor T, Meyer NE, Huber A, Payre F, Minke B. Light-regulated interaction of Dmoesin with TRP and TRPL channels is required for maintenance of photoreceptors. J Cell Biol. 2005;171:143–152. doi: 10.1083/jcb.200503014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin MA, Diao F, Tsunoda S. Light-dependent subcellular translocation of Gqalpha in Drosophila photoreceptors is facilitated by the photoreceptor-specific myosin III NINAC. J Cell Sci. 2004;117:4797–4806. doi: 10.1242/jcs.01371. [DOI] [PubMed] [Google Scholar]
- Cuttell L, Vaughan A, Silva E, Escaron CJ, Lavine M, Van Goethem E, Eid JP, Quirin M, Franc NC. Undertaker, a Drosophila Junctophilin, links Draper-mediated phagocytosis and calcium homeostasis. Cell. 2008;135:524–534. doi: 10.1016/j.cell.2008.08.033. [DOI] [PubMed] [Google Scholar]
- Elia N, Frechter S, Gedi Y, Minke B, Selinger Z. Excess of Gβe over Gqαe in vivo prevents dark, spontaneous activity of Drosophila photoreceptors. J Cell Biol. 2005;171:517–526. doi: 10.1083/jcb.200506082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frechter S, Elia N, Tzarfaty V, Selinger Z, Minke B. Translocation of Gqα mediates long-term adaptation in Drosophila photoreceptors. J Neurosci. 2007;27:5571–5583. doi: 10.1523/JNEUROSCI.0310-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman M. Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell. 1996;87:651–660. doi: 10.1016/s0092-8674(00)81385-9. [DOI] [PubMed] [Google Scholar]
- Gubbels MJ, Vaishnava S, Boot N, Dubremetz JF, Striepen B. A MORN-repeat protein is a dynamic component of the Toxoplasma gondii cell division apparatus. J Cell Sci. 2006;119:2236–2245. doi: 10.1242/jcs.02949. [DOI] [PubMed] [Google Scholar]
- Hardie RC, Postma M. Phototransduction in microvillar photoreceptors of Drosophila and other invertebrates. In: Masland R, Albright T, editors. The senses—a comprehensive reference. Vision. Vol 1. San Diego: Academic; 2008. pp. 77–130. [Google Scholar]
- Hardie RC, Martin F, Cochrane GW, Juusola M, Georgiev P, Raghu P. Molecular basis of amplification in Drosophila phototransduction: roles for G protein, phospholipase C, and diacylglycerol kinase. Neuron. 2002;36:689–701. doi: 10.1016/s0896-6273(02)01048-6. [DOI] [PubMed] [Google Scholar]
- Henderson SR, Reuss H, Hardie RC. Single photon responses in Drosophila photoreceptors and their regulation by Ca2+. J Physiol. 2000;524:179–194. doi: 10.1111/j.1469-7793.2000.00179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofstee CA, Henderson S, Hardie RC, Stavenga DG. Differential effects of ninaC proteins (p132 and p174) on light-activated currents and pupil mechanism in Drosophila photoreceptors. Vis Neurosci. 1996;13:897–906. doi: 10.1017/s0952523800009147. [DOI] [PubMed] [Google Scholar]
- Hyde DR, Mecklenburg KL, Pollock JA, Vihtelic TS, Benzer S. Twenty Drosophila visual system cDNA clones: one is a homolog of human arrestin. Proc Natl Acad Sci U S A. 1990;87:1008–1012. doi: 10.1073/pnas.87.3.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im YJ, Davis AJ, Perera IY, Johannes E, Allen NS, Boss WF. The N-terminal membrane occupation and recognition nexus domain of Arabidopsis phosphatidylinositol phosphate kinase 1 regulates enzyme activity. J Biol Chem. 2007;282:5443–5452. doi: 10.1074/jbc.M611342200. [DOI] [PubMed] [Google Scholar]
- Katz B, Minke B. Drosophila photoreceptors and signaling mechanisms. Front Cell Neurosci. 2009;3:2. doi: 10.3389/neuro.03.002.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosloff M, Elia N, Joel-Almagor T, Timberg R, Zars TD, Hyde DR, Minke B, Selinger Z. Regulation of light-dependent Gqα translocation and morphological changes in fly photoreceptors. EMBO J. 2003;22:459–468. doi: 10.1093/emboj/cdg054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krispel CM, Sokolov M, Chen YM, Song H, Herrmann R, Arshavsky VY, Burns ME. Phosducin regulates the expression of transducin βγ subunits in rod photoreceptors and does not contribute to phototransduction adaptation. J Gen Physiol. 2007;130:303–312. doi: 10.1085/jgp.200709812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrivee DC, Conrad SK, Stephenson RS, Pak WL. Mutation that selectively affects rhodopsin concentration in the peripheral photoreceptors of Drosophila melanogaster. J Gen Physiol. 1981;78:521–545. doi: 10.1085/jgp.78.5.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BY, Thulin CD, Willardson BM. Site-specific phosphorylation of phosducin in intact retina. Dynamics of phosphorylation and effects on G protein βγ dimer binding. J Biol Chem. 2004;279:54008–54017. doi: 10.1074/jbc.M405669200. [DOI] [PubMed] [Google Scholar]
- Liu CH, Satoh AK, Postma M, Huang J, Ready DF, Hardie RC. Ca2+-dependent metarhodopsin inactivation mediated by calmodulin and NINAC myosin III. Neuron. 2008;59:778–789. doi: 10.1016/j.neuron.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Lou Y, Lin WH, Xue HW. MORN motifs in plant PIPKs are involved in the regulation of subcellular localization and phospholipid binding. Cell Res. 2006;16:466–478. doi: 10.1038/sj.cr.7310058. [DOI] [PubMed] [Google Scholar]
- Mann M, Ong SE, Grønborg M, Steen H, Jensen ON, Pandey A. Analysis of protein phosphorylation using mass spectrometry: deciphering the phosphoproteome. Trends Biotechnol. 2002;20:261–268. doi: 10.1016/s0167-7799(02)01944-3. [DOI] [PubMed] [Google Scholar]
- Matsumoto H, Komori N. Protein identification on two-dimensional gels archived nearly two decades ago by in-gel digestion and matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Anal Biochem. 1999;270:176–179. doi: 10.1006/abio.1999.4054. [DOI] [PubMed] [Google Scholar]
- Matsumoto H, Pak WL. Light-induced phosphorylation of retina-specific polypeptides of Drosophila in vivo. Science. 1984;223:184–186. doi: 10.1126/science.6419348. [DOI] [PubMed] [Google Scholar]
- Matsumoto H, O'Tousa JE, Pak WL. Light-induced modification of Drosophila retinal polypeptides in vivo. Science. 1982;217:839–841. doi: 10.1126/science.7100927. [DOI] [PubMed] [Google Scholar]
- Matsumoto H, Isono K, Pye Q, Pak WL. Gene encoding cytoskeletal proteins in Drosophila rhabdomeres. Proc Natl Acad Sci U S A. 1987;84:985–989. doi: 10.1073/pnas.84.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto H, Kurien BT, Takagi Y, Kahn ES, Kinumi T, Komori N, Yamada T, Hayashi F, Isono K, Pak WL, Jackson KW, Tobin SL. Phosrestin I undergoes the earliest light-induced phosphorylation by a calcium/calmodulin-dependent protein kinase in Drosophila photoreceptors. Neuron. 1994;12:997–1010. doi: 10.1016/0896-6273(94)90309-3. [DOI] [PubMed] [Google Scholar]
- Matsumoto H, Kahn ES, Komori N. The emerging role of mass spectrometry in molecular biosciences: studies of protein phosphorylation in fly eyes as an example. Novartis Found Symp. 1999;224:225–244. doi: 10.1002/9780470515693.ch13. discussion 244–228. [DOI] [PubMed] [Google Scholar]
- Mecklenburg KL. Drosophila retinophilin contains MORN repeats and is conserved in humans. Mol Genet Genomics. 2007;277:481–489. doi: 10.1007/s00438-007-0211-7. [DOI] [PubMed] [Google Scholar]
- Minke B, Stephenson RS. The characteristics of chemically induced noise in Musca photoreceptors. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 1985;156:339–356. [Google Scholar]
- Montell C. TRP trapped in fly signaling web. Curr Opin Neurobiol. 1998;8:389–397. doi: 10.1016/s0959-4388(98)80066-4. [DOI] [PubMed] [Google Scholar]
- Pak WL. Drosophila in vision research. The Friedenwald Lecture. Invest Ophthalmol Vis Sci. 1995;36:2340–2357. [PubMed] [Google Scholar]
- Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, Huppert K, Tan LR, Winter CG, Bogart KP, Deal JE, Deal-Herr ME, Grant D, Marcinko M, Miyazaki WY, Robertson S, Shaw KJ, Tabios M, Vysotskaia V, Zhao L, Andrade RS, et al. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat Genet. 2004;36:288–292. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]
- Polevoda B, Sherman F. Nα-terminal acetylation of eukaryotic proteins. J Biol Chem. 2000;275:36479–36482. doi: 10.1074/jbc.R000023200. [DOI] [PubMed] [Google Scholar]
- Porter JA, Montell C. Distinct roles of the Drosophila ninaC kinase and myosin domains revealed by systematic mutagenesis. J Cell Biol. 1993;122:601–612. doi: 10.1083/jcb.122.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter JA, Hicks JL, Williams DS, Montell C. Differential localizations of and requirements for the two Drosophila ninaC kinase/myosins in photoreceptor cells. J Cell Biol. 1992;116:683–693. doi: 10.1083/jcb.116.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Chait BT. Identification and characterization of posttranslational modifications of proteins by MALDI ion trap mass spectrometry. Anal Chem. 1997;69:4002–4009. doi: 10.1021/ac970489n. [DOI] [PubMed] [Google Scholar]
- Shieh BH, Niemeyer B. A novel protein encoded by the InaD gene regulates recovery of visual transduction in Drosophila. Neuron. 1995;14:201–210. doi: 10.1016/0896-6273(95)90255-4. [DOI] [PubMed] [Google Scholar]
- Song H, Belcastro M, Young EJ, Sokolov M. Compartment-specific phosphorylation of phosducin in rods underlies adaptation to various levels of illumination. J Biol Chem. 2007;282:23613–23621. doi: 10.1074/jbc.M701974200. [DOI] [PubMed] [Google Scholar]
- Steele FR, Washburn T, Rieger R, O'Tousa JE. Drosophila retinal degeneration C (rdgC) encodes a novel serine/threonine protein phosphatase. Cell. 1992;69:669–676. doi: 10.1016/0092-8674(92)90230-a. [DOI] [PubMed] [Google Scholar]
- Tabuchi K, Sawamoto K, Suzuki E, Ozaki K, Sone M, Hama C, Tanifuji-Morimoto T, Yuasa Y, Yoshihara Y, Nose A, Okano H. GAL4/UAS-WGA system as a powerful tool for tracing Drosophila transsynaptic neural pathways. J Neurosci Res. 2000;59:94–99. [PubMed] [Google Scholar]
- Takemori N, Komori N, Thompson JN, Jr, Yamamoto MT, Matsumoto H. Novel eye-specific calmodulin methylation characterized by protein mapping in Drosophila melanogaster. Proteomics. 2007;7:2651–2658. doi: 10.1002/pmic.200700343. [DOI] [PubMed] [Google Scholar]
- Takeshima H, Komazaki S, Nishi M, Iino M, Kangawa K. Junctophilins: a novel family of junctional membrane complex proteins. Mol Cell. 2000;6:11–22. doi: 10.1016/s1097-2765(00)00003-4. [DOI] [PubMed] [Google Scholar]
- Thummel CS, Boulet AM, Lipshitz HD. Vectors for Drosophila P-element-mediated transformation and tissue culture transfection. Gene. 1988;74:445–456. doi: 10.1016/0378-1119(88)90177-1. [DOI] [PubMed] [Google Scholar]
- Trojan P, Rausch S, Giessl A, Klemm C, Krause E, Pulvermüller A, Wolfrum U. Light-dependent CK2-mediated phosphorylation of centrins regulates complex formation with visual G-protein. Biochim Biophys Acta. 2008;1783:1248–1260. doi: 10.1016/j.bbamcr.2008.01.006. [DOI] [PubMed] [Google Scholar]
- Tsunoda S, Sierralta J, Sun Y, Bodner R, Suzuki E, Becker A, Socolich M, Zuker CS. A multivalent PDZ-domain protein assembles signalling complexes in a G-protein-coupled cascade. Nature. 1997;388:243–249. doi: 10.1038/40805. [DOI] [PubMed] [Google Scholar]
- Wang T, Montell C. Phototransduction and retinal degeneration in Drosophila. Pflugers Arch. 2007;454:821–847. doi: 10.1007/s00424-007-0251-1. [DOI] [PubMed] [Google Scholar]
- Wilson JR, Jing C, Walker PA, Martin SR, Howell SA, Blackburn GM, Gamblin SJ, Xiao B. Crystal structure and functional analysis of the histone methyltransferase SET7/9. Cell. 2002;111:105–115. doi: 10.1016/s0092-8674(02)00964-9. [DOI] [PubMed] [Google Scholar]
- Xu H, Lee SJ, Suzuki E, Dugan KD, Stoddard A, Li HS, Chodosh LA, Montell C. A lysosomal tetraspanin associated with retinal degeneration identified via a genome-wide screen. EMBO J. 2004;23:811–822. doi: 10.1038/sj.emboj.7600112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Takeuchi Y, Komori N, Kobayashi H, Sakai Y, Hotta Y, Matsumoto H. A 49-kilodalton phosphoprotein in the Drosophila photoreceptor is an arrestin homolog. Science. 1990;248:483–486. doi: 10.1126/science.2158671. [DOI] [PubMed] [Google Scholar]
- Yang Z, Edenberg HJ, Davis RL. Isolation of mRNA from specific tissues of Drosophila by mRNA tagging. Nucleic Acids Res. 2005;33:e148. doi: 10.1093/nar/gni149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker CS. The biology of vision of Drosophila. Proc Natl Acad Sci U S A. 1996;93:571–576. doi: 10.1073/pnas.93.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]