Abstract
Pro-apoptotic Bax is essential for RGC (retinal ganglion cell) death. Gene dosage experiments in mice, yielding a single wild-type Bax allele, indicated that genetic background was able to influence the cell death phenotype. DBA/2JBax+/− mice exhibited complete resistance to nerve damage after 2 weeks (similar to Bax−/− mice), but 129B6Bax+/− mice exhibited significant cell loss (similar to wild-type mice). The different cell death phenotype was associated with the level of Bax expression, where 129B6 neurons had twice the level of endogenous Bax mRNA and protein as DBA/2J neurons. Sequence analysis of the Bax promoters between these strains revealed a single nucleotide polymorphism (T129B6 to CDBA/2J) at position −515. A 1.5- to 2.5-fold increase in transcriptional activity was observed from the 129B6 promoter in transient transfection assays in a variety of cell types, including RGC5 cells derived from rat RGCs. Since this polymorphism occurred in a p53 half-site, we investigated the requirement of p53 for the differential transcriptional activity. Differential transcriptional activity from either 129B6 or DBA/2J Bax promoters were unaffected in p53−/− cells, and addition of exogenous p53 had no further effect on this difference, thus a role for p53 was excluded. Competitive electrophoretic mobility-shift assays identified two DNA–protein complexes that interacted with the polymorphic region. Those forming Complex 1 bound with higher affinity to the 129B6 polymorphic site, suggesting that these proteins probably comprised a transcriptional activator complex. These studies implicated quantitative expression of the Bax gene as playing a possible role in neuronal susceptibility to damaging stimuli.
Keywords: Bax, gene expression, glaucoma, neuronal apoptosis, retinal ganglion cells, susceptibility allele
Abbreviations: C/EBP, CCAAT/enhancer-binding protein; DMEM, Dulbecco’s modified Eagle’s medium; EMSA, electrophoretic mobility-shift assay; FBS, fetal bovine serum; FOXI1, Forkhead box I1; βgal, β-galactosidase; Luc, luciferase; ONC, optic nerve crush; RGC, retinal ganglion cell
INTRODUCTION
Neuronal apoptosis is widespread during the development of the nervous system and in chronic neurodegenerative diseases such as Huntington’s disease, amyotrophic lateral sclerosis, Parkinson’s disease and Alzheimer’s disease (Akhtar et al., 2004; Kermer et al., 2004; Lindholm et al., 2004; Conforti et al., 2007). RGC (retinal ganglion cell) death, and the degeneration of their axons in the optic nerve, is the principal characteristic of blinding optic neuropathies such as glaucoma (Nickells, 2007). Previously we, and others, have documented an essential role for Bax expression and the intrinsic apoptotic pathway in the process of RGC death in development, after acute nerve trauma and in a mouse model of chronic glaucoma (Isenmann et al., 1997; Mosinger Ogilvie et al., 1998; Isenmann et al., 1999; Li et al., 2000; Libby et al., 2005a).
The intrinsic apoptotic pathway involves mitochondrial dysfunction and is regulated by members of the Bcl2 gene family (Adams and Cory, 2007). In this pathway, the molecular events leading to cell death cause the release of cytochrome c through permeabilization of the mitochondria by the pro-apoptotic proteins BAX and BAK. The release of cytochrome c subsequently activates the caspase cascade via its association with pro-caspase 9 and the apoptosis protease activating factor-1 (Danial and Korsmeyer, 2004; Adams and Cory, 2007). In most cell types, the functions of BAX and BAK are similar, and therefore redundant with the exception of some cancer cells (Zhang et al., 2000) and many neurons (Knudson et al., 1995; Deckwerth et al., 1996; White et al., 1998), in which BAX is the central mediator of apoptosis. In neurons, the exclusive role for BAX may result from alternative splicing of Bak transcripts, which result in the translation of a truncated form of BAK (N-BAK) containing only the BH3 domain (Uo et al., 2005).
Mouse genetic studies manipulating the Bax gene showed that Bax knockout mice on a mixed 129/Sv and C57BL/6 background (129B6) (Knudson et al., 1995) exhibited reduced developmental neuronal death throughout the central and peripheral nervous system compared with wild-type mice (Deckwerth et al., 1996; Mosinger Ogilvie et al., 1998; White et al., 1998). Similarly, Bax deficiency prevented sympathetic neuron death in the absence of nerve growth factor in culture. In vivo, Bax deficiency promoted motor neuron survival following both facial and sciatic nerve axotomy, and RGC survival following ONC (optic nerve crush) (Deckwerth et al., 1996; Li et al., 2000; Sun and Oppenheim, 2003). In Bax+/− mice, however, developmental apoptosis of sympathetic and facial motor neurons, and RGCs, was comparable with wild-type mice. In addition, motor neuron death after facial nerve axotomy was comparable between Bax+/− and wild-type mice (Deckwerth et al., 1996; Mosinger Ogilvie et al., 1998). Collectively, these studies suggested that expression from a single copy of Bax was sufficient to execute apoptosis in mice with the 129B6 mixed genetic background.
Similar to 129B6 mice, complete Bax deficiency in the DBA/2J mouse line prevented RGC soma death during glaucoma and after ONC (Libby et al., 2005a). DBA/2J mice develop chronic secondary glaucoma at 10–12 months of age characterized by elevated intraocular pressure, optic nerve degeneration, and the progressive loss of RGCs (Chang et al., 1999; Libby et al., 2005b; Schlamp et al., 2006). In contrast with 129B6 mice, however, DBA/2J mice heterozygous for the Bax allele did not exhibit significant RGC death shortly after ONC and in glaucoma, whereas wild-type animals did (Libby et al., 2005a). Thus reduced Bax expression, at least on some genetic backgrounds, was able to significantly affect RGC soma susceptibility to optic nerve damage.
In the present study, we examine the underlying cause for the differential cell death phenotype between 129B6 and DBA/2J mice heterozygous for the mutant Bax allele. This difference is associated with higher levels of latent Bax mRNA and protein in the neurons of 129B6 mice. Promoter analysis also identified a single nucleotide polymorphism in the Bax promoter of each strain, which can significantly alter the level of expression of this gene, and affect the binding affinity of nuclear proteins.
MATERIALS AND METHODS
Animals
Mice used in the present study were maintained and handled in accordance with the guidelines established by the Association for Research in Vision and Ophthalmology Statement on Animals in Research and overseen by the Animal Care and Use Committee at the University of Wisconsin. A colony of mice deficient for Bax (Knudson et al., 1995) was established from breeders obtained from the late Dr Stanley Korsmeyer and maintained as an inbred population. In this line, the null allele was initially generated in 129/Sv-derived embryonic stem cells, which were introduced into C57BL/6 embryos. We have designated these mice as 129B6 because of their mixed genetic background. DBA/2J mice congenic for the Bax null allele (Libby et al., 2005a) were generously provided by Dr Simon John (Bar Harbor, ME, U.S.A.). FVB mice were a gift from Dr Paul Lambert (School of Medicine and Public Health, University of Wisconsin, Madison, WI, U.S.A.).
ONC and ganglion cell counting
ONC surgery was performed as described previously (Li et al., 1999). ONC causes synchronous RGC death in the ganglion cell layer, which accounts for 50–60% of the neurons in this layer. Only the left eye of each mouse underwent surgery, leaving the other eye as a control. The loss of cells in the retinal ganglion cell layer was quantified as described previously (Li et al., 2007).
Cell culture
All cells were maintained in a humidified incubator at 37°C with 5% CO2 and media replaced every 2–3 days. NIH 3T3 cells, an immortalized murine fibroblast cell line, were a gift from Dr Donna Peters (Department of Pathology and Laboratory Medicine, University of Wisconsin, Madison, WI, U.S.A.). The cells were grown in DMEM (Dulbecco’s modified Eagle’s medium) containing 4.5 g/l glucose with l-glutamine (Cambrex) supplemented with 10% FBS (fetal bovine serum) (Atlanta Biologicals) and with 1% penicillin/streptomycin (Cambrex). NIH 3T3 cells were passaged at 70–80% confluency. RGC-5 cells (Krishnamoorthy et al., 2001), an immortalized rat retinal ganglion cell line, were a gift from Dr Neeraj Agarwal (Vision Research Program, National Eye Institute, Rockville, MD, U.S.A.). RGC-5 cells were cultured in DMEM containing 10% FBS, 1 g/l glucose and 1% penicillin/streptomycin. Primary lung fibroblasts from 2-month-old FVB mice (p53+/+ and p53−/− genotypes) and 3-month-old DBA/2JBax+/+ mice were isolated as described previously (Konigsberg et al., 2004). Briefly, lungs from each mouse were aseptically dissected and minced into ∼1 mm pieces. The tissue was washed three times with PBS containing 2% penicillin/streptomycin and treated with 0.25% trypsin for 10 min. The tissue was washed again with PBS and treated for 5 h with 169 units/mg of 0.3% collagenase type I (Worthington). The dissociated cells and tissue were split into three 10-cm-diameter plates in 4.5 g/l glucose-containing DMEM with l-glutamine, 15% FBS, and 1% penicillin/streptomycin. The resulting fibroblasts were allowed to become confluent before being passed. Cells were cryogenically frozen after 10 days in culture until use. All experiments using primary fibroblasts were performed at the third passage.
Clones and plasmids
A 3091 bp genomic fragment containing the DBA/2J or 129B6 Bax promoter region, including exon 1 and half of intron 1, was PCR-amplified using LA Taq (Takara). The primer sequences used were: 5′-GATTAGGTTGGCTTGTTTGTGG (forward) and 5′-CTAGTAGTGACAAGTAGCATGG (reverse). Nhe1 and Bgl II sites were engineered into the promoter regions by nested PCR to isolate 1370 bp directly upstream of the start codon using the primers 5′-GTGATCTTACGCTAGCTTCCTGCGTTCTGAGG (forward containing Nhe1) and 5′-CGAACTGTCTAGATCTCACTGCCGCTGCCTCTCG (reverse containing BglII). The nucleotide directly upstream of the start codon was considered −1 and the numbering of all other nucleotides is relative to this position. This fragment was directionally cloned into pGL3-Basic (Promega), a Luc (luciferase) expression vector, to create the DBA/2J–Luc and 129B6–Luc reporter constructs. The expression plasmid containing human p53 under the control of the CMV promoter (pC53-SN3) was a gift from Dr Mary Ellen Perry (National Cancer Institute, Rockville, MD, U.S.A.) and the plasmid containing 13 p53 response elements (Kern et al., 1992) with a Luc reporter (pG13-Luc) was a gift from Dr Bert Vogelstein (Sidney Kimmel Comprehensive Cancer Center, Johns Hopkins University, Baltimore, MD, U.S.A.). All clones were sequenced at the University of Wisconsin Biotechnology Center. The pGL3-Basic plasmid was used as a negative control for Luc experiments, pGL3-Control (Promega) was used as a positive control, and pSV-βgal (βgal, β-galactosidase) (Promega) was used to determine transfection efficiency.
RNA isolation and quantitative PCR
Total RNA was isolated from whole retina tissue and cultured cells using Tri-reagent (Molecular Research Center, Cincinnati, OH, U.S.A.). First-strand cDNA was synthesized using oligo-dT as a primer, and quantitative PCR was performed using the ABI (Applied Biosystems) 7300 real-time PCR system and ABI SYBR Green PCR Master Mix as described previously (Pelzel et al., 2006). Standard curves were generated for each product using cloned cDNAs for Bax, Bcl-X and the S16 ribosomal protein to quantify the abundance of cDNA in each sample. The Bax coding region (627 bp fragment) was isolated from DBA/2J and 129B6 retinal cDNA with the following primers: 5′-ACCCGCCGAGAGGCAGCG (forward) and 5′-CACAGTCCCAGGCAGTGGG (reverse) and blunt-end cloned the SmaI site of pBK-CMV (Stratagene). BclX was blunt-end cloned into pBK-CMV using the primers 5′-CATCTCACCTACCAGTCA (forward) and 5′-GTCAGAGTGGATGGTCAGT (reverse). A cDNA for S16 (199 bp fragment) was also blunt-end cloned using 129B6 cDNA and primers 5′-CACTGCAAACGGGGAAATGG (forward) and 5′-TGAGATGGACTGTCGGATGG (reverse).
For standard curves, a dilution series of cloned Bax, Bcl-X and S16 templates ranging from 102 to 109 copies were used. The quantitative PCR cycling parameters were: 1 cycle of 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Data collection was taken at the 60°C annealing/extension phase. In order to ensure the presence of a single product, a dissociation curve was performed after each run and products were visualized on ethidium bromide-stained agarose gels. Data were collected from threshold values using the automatic function of the 7300 System Sequence Detection Software program. The primers used to quantify Bax cDNA yielded a 220 bp fragment that spanned intron 3: 5′-TTCATCCAGGATCGAGCAGG (forward) and 5′-CATCAGCAAACATGTCAGC (reverse). The primers used to quantify BclX cDNA yielded a 275 bp fragment: 5′-GCATCGTGGCCTTTTTCTCC (forward) and 5′-CGACTGAAGAGTGAGCCCAG (reverse). All cDNA levels were normalized to murine S16 ribosomal protein cDNA, which was quantified using the same primers that were used to clone S16 above. All primers crossed at least one intron/exon boundary. All PCR products generated with these primers were also sequenced to verify identity.
Transfections
NIH 3T3 cells were plated into 60 cm plates at a density of 1×105 cells/plate. RGC-5 cells and primary fibroblasts were plated into six-well plates with a density of 3×104 cells/well. All cells were transfected 24 h after initial plating. Transfections were performed using the Tfx-50 transfection reagent (Promega) with a 2:1 transfection reagent/DNA ratio for primary fibroblasts and RGC-5 cells and a 3:1 ratio for NIH 3T3 cells. Plasmid DNAs used for transfection were 2 μg of either DBA/2J–Luc or 129B6–Luc along with 1 μg of pSV-βgal to control for transfection efficiency. Empty pGL3-Basic was used as a negative control in all experiments and pGL3-Control was used as a positive control. For transfections into p53−/− fibroblasts, Luc reporter plasmids were co-transfected with pSV-βgal and 1 μg of either pGL3-Basic or pC53-SN3 plasmids. For primary fibroblasts and RGC-5 cells, the transfection medium was replaced after 5 h with complete medium in order to increase viability. In NIH 3T3 cells, however, complete medium was added directly to the transfection medium after 1 h. Reporter gene activity was typically measured 48 h after transfection, except where noted. Luc activity was measured with Luc assay reagent (Promega) using a Turner TD-20e Luminometer (Sunnyvale, CA) and β-gal activity was measured using β-gal assay reagent (Promega) and reading the absorbance at 420 nm in a spectrophotometer.
EMSA (electrophoretic mobility-shift assay)
EMSA and unlabelled competition EMSA were performed using double-stranded 30 bp oligonucleotides encompassing the polymorphic site from −499 to −528 of the 129B6 or DBA/2J Bax promoters. When making probes for EMSA, complementary strands of the 30 bp 129B6 or DBA/2J Bax promoter regions (400 pmol) were annealed in 100 μl of annealing buffer (10 mM Tris/HCl, pH 7.5, and 20 mM NaCl). The double-stranded oligonucleotides (16 pmol) were end-labelled using [γ-32P]ATP (6000 Ci/mmol; Amersham) and T4 Polynucleotide Kinase (Promega). Free radioactivity was removed using illustraTM Microspin G-25 Columns (Amersham). Probes were counted in a Packard TRI CARB 2100TR liquid scintillation analyser (Canberra, Meriden, CT, U.S.A.). NIH 3T3 nuclear extract was purchased from Santa Cruz Biotechnology. Whole retinal nuclear protein isolations were performed as described previously (Andrews and Faller, 1991). Nuclear extracts (5 μg) were incubated for 10 min at room temperature (22°C) with binding buffer [5 mM MgCl2, 2.5 mM EDTA, 2.5 mM DTT (dithiothreitol), 250 mM NaCl and 50 mM Tris/HCl, pH 7.5] and 3 μg of dI-dC (Amersham) per lane. A 32P-labelled probe (600 000 c.p.m./lane; ∼0.12 pmol) was then added in the presence of excess (6.25- to 150-fold where noted) or absence of unlabelled competitor oligonucleotides and incubated for 20 min at room temperature. The reactions were run on a 5% polyacrylamide gel in 0.5× Tris-borate EDTA buffer at 250 V for 2 h. Gels were dried and exposed to a PhosphorImager screen (Molecular Dynamics), scanned on a Storm 860 scanner (Amersham), and band density was quantified using ImageQuant software v5.2 (Amersham).
Immunoblotting
Immunoblots were performed as described previously (Azarian et al., 1993) with a few modifications. Whole brain lysates (60 μg/lane) from 129B6 and DBA/2J mice were used in the assay. Protein concentration was quantified using a BCA (bicinchoninic acid) protein assay (Pierce). The following antibodies were utilized to detect specific antigens. Polyclonal antibodies developed in rabbit, anti-BAX antibody (B3428) and anti-actin antibody (A2066) were purchased from Sigma and used at 1:2000 and 1:100 dilutions respectively. The secondary antibody was alkaline-phosphatase-conjugated and the blot was visualized utilizing ECF Substrate (Amersham Biosciences). Blots were scanned on a Storm 860 scanner (Amersham), and band density quantified using ImageQuant software v5.2 (Amersham).
Quantitative analysis of BCL-X and BAX protein levels were examined using blot strips. Equal protein amounts of retinal lysates were each run in single oversized lanes on 12% polyacrylamide gels. After transblotting to Immobilon-P membranes, each lane was divided into three equal-sized strips, and each strip was individually probed with polyclonal antibodies for actin, BAX or BCL-X (sc-634, Santa Cruz Biotechnology, Santa Cruz, CA, U.S.A.; 1:1000 dilution). After probing with secondary antibody, the blots were developed using colorimetric staining for alkaline phosphatase activity in Nitro-Blue Tetrazolium and BCIP (5-bromo-4-chloroindol-3-yl phosphate). Staining for BCL-X and BAX was allowed to continue until one of the comparative strips had reached saturation.
In silico analysis
The 10 bp core sequence from the 129B6 Bax promoter, AGGTTTGCCT, was entered as a search term in a number of different transcription factor databases all available online: JASPAR (http://mordor.cgb.ki.se/cgi-bin/jaspar2005/jaspar_db.pl), TESS (http://www.cbil.upenn.edu/cgi-bin/tess/tess), TFSearch (http://www.cbrc.jp/research/db/TFSEARCH.html) and MatInspector from Genomatix (http://www.genomatix.de). For the initial screen of transcription factors putatively binding to the core sequence, any sequence not possessing at least 60% homology with the core was eliminated. Other parameters taken into account were transcription-factor-binding sequences that matched a portion of the core sequence, including the polymorphic site, with high sequence similarity and in the correct succession of base pairs. The likelihood of that particular transcription factor being expressed in mouse neurons was also considered.
Statistical analyses
For data presented as means±S.E.M., assessment for significant differences between groups was performed by Student’s t test. The data from quantified EMSA gels were plotted, best-fit curves generated, and IC50 values were calculated using Kaleidagraph 4 software (Synergy Software).
RESULTS
Genetically distinct mice, heterozygous for mutant Bax, display susceptible or resistant RGC death phenotypes in response to ONC
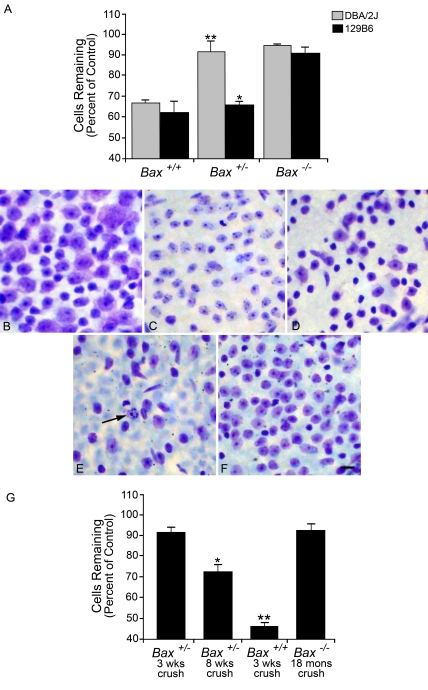
ONC in mice causes the selective death of RGCs that generally progresses to maximal cell loss by 3 weeks after surgery in wild-type animals (Li et al., 1999; Li et al., 2007). We initially investigated the effects of Bax gene dosage on RGC survival in 129B6 mice and DBA/2J mice at an intermediate term period (2 weeks) after ONC (Figure 1A). DBA/2JBax+/+ mice had 66% cells remaining, which represents a loss of 34.0±2.45% of the cells in the ganglion cell layer relative to the control retina (P<0.001). Likewise, 129B6Bax+/+ mice had 62.3% of their cells remaining, or a loss of 37.7±5.3% of the cells in the ganglion cell layer relative to control retinas (P<0.005). DBA/2JBax−/− and 129B6Bax−/− were completely resistant to the damaging effects of ONC (P>0.25 for DBA/2JBax−/−, P>0.10 for 129B6Bax−/− relative to control). Importantly, animals heterozygous for Bax exhibited distinctly different phenotypes. 129B6Bax+/− mice displayed significant cell death after ONC (P<0.005), similar to wild-type animals. DBA/2JBax+/− displayed no significant cell death (P>0.25), similar to Bax−/− animals. The results observed for 129B6Bax+/− and DBA/2JBax+/− were consistent with previously published reports of cell death after acute nerve trauma in these two lines (Deckwerth et al., 1996; Libby et al., 2005a). In order to examine if there was long-term protection of RGCs in Bax+/− mice, experiments examining RGC loss in resistant DBA/2JBax+/− mice at both 3 and 8 weeks post-ONC were performed (Figures 1B–1G). Mice at 3 weeks after ONC still showed dramatic preservation of cells in the ganglion cell layer (∼90% of control retinas), but a significant loss of cells by 8 weeks (72.3±3.4% cells remaining, P = 2.85×10−6). This loss of cells was still well below that for Bax+/+ mice at 3 weeks, which exhibited only 45.8±1.7% of the cells present in control retinas, and probably accounts for complete elimination of the RGC population in this retinal layer. Conversely, Bax−/− RGC somas survive up to 18 months after optic nerve damage, exhibiting more than 92% cells remaining at that point.
Figure 1. Retinal ganglion cell death after ONC in 129B6 and DBA/2J mice.
(A) Histograms of the mean±S.E.M. cells remaining in the retinal ganglion cell layer 2 weeks after ONC in DBA/2J and 129B6 mice. Both 129B6Bax−/− and DBA/2JBax−/− mice showed no significant loss of cells relative to control eyes (P>0.10 and P>0.25 respectively), whereas wild-type mice showed significant loss (P<0.005). DBA/2JBax+/− mice displayed a knockout phenotype, showing no significant loss (**P>0.25), whereas 129B6Bax+/− exhibited significant cell loss similar to wild-type animals (*P<0.001). After a prolonged period after ONC, however, resistant heterozygous mice also exhibit cell loss. (B–F) Nissl-stained cells in the ganglion cell layer of whole mounted mouse retinas. A region of approx. 104 μm2 of the superior mid-peripheral retina of each eye is shown. (B) Control retina from a Bax+/− mouse. (C) Retina from a Bax+/− mouse, 3 weeks after ONC. Minimal cell loss is observed, compared with control. (D) Retina from a Bax+/− mouse, 8 weeks after ONC. These mice exhibit moderate cell loss and partial atrophy, typified by cell soma shrinkage of the remaining ganglion cell population, which is common in Bax-deficient neurons after damage (Li et al., 2000; Sun and Oppenheim, 2003). (E) Retina from a Bax+/+ mouse, 3 weeks after ONC. These mice typically exhibit a maximum loss of ganglion cells by this stage. A cell undergoing apoptosis is indicated by the arrow. (F) Retina from a Bax−/− mouse, 18 months after ONC. Even at this extended time after a lesion to the nerve, there is minimal loss of cells in the ganglion cell layer. Note that Bax−/− animals have a higher density of ganglion cells than either wild-type or heterozygous mice because of the lack of Bax-dependent programmed cell death, which normally prunes approx. 50% of the ganglion cell population during the first few weeks of life (Mosinger Ogilvie et al., 1998; Pequignot et al., 2003). Scale bar, 15 μm. (G) Quantitative analysis of the change in cell numbers in experimental retinas 3 weeks, 8 weeks and 18 months after ONC. Results are presented as a percentage of cells (means±S.E.M.) counted in the control fellow eyes of all mice examined. Bax heterozygous animals were on the DBA/2J background, whereas Bax−/− mice were on the 129B6 background for this experiment. A total of five to seven mice were analysed for each condition, except for Bax−/− mice at 18 months, in which two mice were examined. (*P = 2.85×10−6 and **P = 7.78×10−18, both compared with control fellow eyes of mice used in each group).
DBA/2JBax+/− and 129B6Bax+/− mice express different neuronal levels of Bax
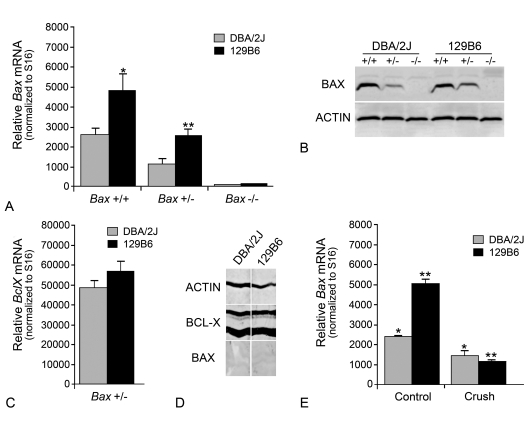
We then examined if the variation in susceptibilities between heterozygous DBA/2J and 129B6 mice was either due to a difference in BAX protein function caused by an amino acid substitution or the level of expression. Sequence analysis of cDNAs isolated from the DBA/2J and 129B6 strains yielded no difference in predicted amino acid structure (data not shown). This ruled out the possibility that a difference in BAX protein function as a consequence of an amino acid sequence difference was the underlying cause of the phenotype. We then investigated whether there was a difference in neuronal Bax expression. Quantitative PCR was used to quantify Bax mRNA levels in the retinas of 129B6 and DBA/2J mice (Figure 2A). As the number of RGCs that populate the ganglion cell layer in DBA/2J and 129B6 retinas are comparable, and Bax mRNA has been shown to be principally localized to the RGC layer of adult rats (Shin et al., 1999), we used whole retinal cDNA for analysis. In each strain, neuronal Bax mRNA levels were reduced by approx. 50% in heterozygous mice relative to wild-type mice. Bax−/− mice of either strain expressed no detectable Bax mRNA. Retinas from 129B6 mice contained twice the level of Bax mRNA compared with DBA/2J mice, such that 129B6Bax+/− and DBA/2JBax+/+ animals had similar levels of Bax transcripts (P = 0.46) (Figure 2A). Consistent with the mRNA levels, 129B6Bax+/+ neurons expressed 1.2- to 1.8-fold (range for three separate experiments) the BAX protein level as DBA/2JBax+/+ neurons (Figure 2B). Similarly, BAX protein measurements indicated that 129B6Bax+/− neurons had between 1.4- and 2.0-fold (range for three separate experiments) the amount of BAX protein as DBA/2JBax+/− neurons. Bax−/− mice expressed no detectable BAX protein (Figure 2B). These observations correlated the higher susceptibility to ONC observed in 129B6+/− mice (Figure 1A) with latent levels of Bax mRNA and protein.
Figure 2. DBA/2J and 129B6 mice differentially express Bax.
(A) Quantitative PCR analysis of latent Bax transcript levels (means±SEM) in 129B6 and DBA/2J mice. Values shown are the number of Bax mRNA molecules [per 2.5 pg of poly(A) RNA input] normalized to the number of S16 molecules in each sample. Reducing Bax gene dosage to one gene caused a 50% reduction of Bax transcripts (for 129B6, P = 0.004, for DBA/2J, P = 0.002, control retinas of Bax+/− mice compared with control retinas of Bax+/+ mice) and no transcripts were detected in knock-out mice. Similarly, 129B6 mice had approximately twice the amount of Bax mRNA in the retina (*129B6Bax+/+ compared with DBA/2JBax+/+ mice, P = 0.008; **129B6Bax+/− compared with DBA/2JBax+/− mice, P = 0.002). (B) Representative immunoblot showing relative Bax protein levels in DBA/2J and 129B6 neurons. Bax protein levels were 1.2- to 1.8-fold higher in 129B6Bax+/+ mice compared with DBA/2JBax+/+ mice (when normalized to actin in each lane). Similarly, 129B6Bax+/− mice expressed 1.4- to 2.0-fold as much Bax as DBA/2JBax+/− mice. No Bax protein was detected in Bax−/− mice. (C) Histogram of BclX transcript levels in the retinas of Bax heterozygous mice from each strain. Unlike Bax mRNA, no difference in BclX mRNA was detected between strains (P = 0.26), or between wild-type and knockout mice (data not shown). (D) Blot strips taken from the same lanes of retina homogenates of wild-type DBA/2J or 129B6 mice. To accurately assess the relative levels of BAX and BCL-X, the strips were developed equally, until BCL-X staining began to saturate the colorimetric reaction. BCL-X shows up as two prominent bands, probably reflecting modified and deamidated polypeptides (Johnstone, 2002). BAX levels, under these conditions, are just detectable. Chemiluminescence detection of BAX staining clearly shows, however, that this antibody specifically interacted with BAX protein in similar samples (see B). Actin was also evaluated as a loading control. (E) Histogram showing changes in Bax transcript abundance (means±S.E.M.), 4 days after ONC. Retinal transcript levels were examined at 4 days after injury when others have reported moderate increases in Bax mRNA after optic nerve axotomy (Näpänkangas et al., 2003). ONC caused a 42.0±13.2% decrease in Bax mRNA in DBA/2J mice relative to control eyes (*P = 0.003) and a 79.0±3.8% decrease in 129B6 mice relative to control eyes (**P = 4.6×10−5). The control retinas of 129B6Bax+/+ mice contained 2.2-fold the amount of Bax mRNA compared with control DBA/2JBax+/+ retinas (P = 2.0×10−4). Thus Bax expression was not differentially increased between strains, and instead showed a relative decrease after injury.
Even though the lower levels of latent Bax mRNA and protein in DBA/2J mice were associated with resistance to ONC in heterozygous animals, we investigated if other factors may also be affecting this phenotype. Since cell death is influenced in a stoichiometric fashion by the levels of anti-apoptotic and BH3-only members of the Bcl2 gene family, we also examined mouse retinas for the expression of antagonistic anti-apoptotic genes. Previously, we had described BclX as the dominant anti-apoptotic Bcl2 family member expressed in the retina (Levin et al., 1997). No significant difference in BclX transcript level was observed between Bax+/+ and Bax−/− of each strain (data not shown). Importantly, there was no significant difference in BclX transcript levels between 129B6Bax+/− and DBA/2JBax+/− (P = 0.26, Figure 2C). Furthermore, in direct quantitative analyses of the same tissues, BclX transcripts were also 5–10-fold more abundant than Bax transcripts (compare Figure 2A with Figure 2C). To verify that there was also in increased level of BCL-X protein, predicted by the mRNA levels, we examined relative BCL-X and BAX protein concentrations in retina homogenates of wild-type 129B6 and DBA/2J mice. Strips cut from the same lanes of sample run on SDS/12% polyacrylamide gels were individually probed with antibodies against BCL-X, BAX and actin (to control for loading differences). Strips developed for equal periods of time for BCL-X and BAX showed a strong reaction for BCL-X, but BAX levels were only just detectable (Figure 2D). Thus, BCL-X protein levels appeared to be in excess over BAX in both samples, consistent with the quantitative PCR data.
Some studies have suggested that Bax expression is increased in RGCs after optic nerve damage (Isenmann et al., 1997; Näpänkangas et al., 2003). Although this increase in expression has been refuted by others (McKinnon et al., 2002), we nevertheless explored the possibility that Bax was differentially up-regulated after crush in the two mouse lines. Quantitative analysis of Bax transcript levels in mouse retinas 4 days after ONC indicated that Bax mRNA levels were not induced in the retina (Figure 2E). Instead, each strain exhibited a significant decrease in transcript abundance (DBA/2J Control compared with 129B6 Control, P = 2×10−4; DBA/2J Control compared with DBA/2J Crush, P = 0.003; 129B6 Control compared with 129B6 Crush, P = 1×10−4).
A single-nucleotide polymorphism in the Bax gene promoter is associated with a decrease in transcriptional activity
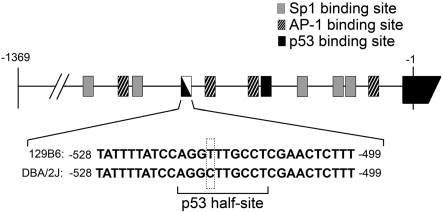
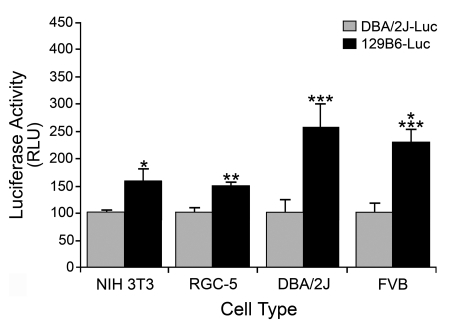
To investigate the mechanism underlying differential Bax expression, we examined the promoter regions for differences in cis-element sequences. For this, we cloned 1370 bp of the Bax promoter regions directly upstream of the translation start site from the DBA/2J and 129B6 mouse lines and compared them by sequence analysis. Both promoters were identical with the exception of a single T (in 129B6) to a C (in DBA/2J) polymorphism at position −515 upstream from the start of translation (Figure 3). This polymorphism was verified in six independent mice of each strain. Analysis of potential binding sites in this region showed that the polymorphism existed within a potential p53 half-site in the DBA/2J line. We tested the ability of this polymorphism to affect transcription in vitro by fusing 1370 bp of the promoter region of each strain to a Luc reporter gene. Transient transfection experiments into both non-neuronal NIH 3T3 cells and RGC-derived RGC-5 cells showed that 129B6–Luc consistently exhibited 1.5-fold the expression level of DBA/2J–Luc in immortalized cells (P = 3×10−4 and P = 1.71×10−12 respectively, Figure 4). Transient transfections into primary fibroblasts isolated from DBA/2J and FVB mice showed 129B6–Luc expression to be 2.5- and 2.3-fold higher than DBA/2J–Luc expression (P = 0.002 and P = 2.82×10−6 respectively), which was similar to the difference in Bax transcript levels between 129B6 and DBA/2J mice. Thus the higher expression observed in vitro from 129B6–Luc suggested that this polymorphism was, at least in part, responsible for the difference in Bax expression observed in vivo.
Figure 3. Diagram of 1370 bp of the murine Bax promoter region.
The Bax promoter region from the DBA/2J and 129B6 mouse lines was isolated, sequenced, and compared. The nucleotide directly upstream of the translation start site is indicated as −1 in the first exon and numbering of the murine Bax promoter is relative to this position. Depicted are the sequences of 30 bp of the 129B6 and DBA/2J promoter regions at positions −499 to −528. A single nucleotide polymorphism (outlined with a dashed box) was found at position −515. The core p53 half-site recognition sequence is Pu Pu Pu C A/T A/T G Py Py Py (where Pu, purine; Py, pyramidine) (El-Deiry et al., 1992). The DBA/2J promoter has a perfect consensus sequence (underlined). Other putative transcription factor binding sites are also shown as previously described (Igata et al., 1999).
Figure 4. Expression from 129B6–Luc is higher than DBA/2J–Luc in both immortalized non-neuronal and neuronal cell lines and primary fibroblasts.
DBA/2J–Luc or 129B6–Luc were transiently co-transfected with pSV-βgal into NIH 3T3 murine fibroblasts, RGC-5 cells, or primary fibroblasts derived from either DBA/2J mice or FVB mice. The histogram represents the mean Luc levels (in RLU, relative light units) normalized to βgal activity (±S.E.M.) of three to five experiments, with three to six replicates for each condition in each experiment. 129B6–Luc exhibited 1.5-fold the expression of DBA/2J–Luc in NIH 3T3 cells and RGC-5 cells (*P = 3×10−4 and **P = 1.71×10−12 respectively). In transient transfections of primary fibroblasts from DBA/2J and FVB mice, 129B6–Luc exhibited 2.5- and 2.3-fold the expression, respectively, of DBA/2J–Luc (***P = 0.002 and ****P = 2.82×10−6 respectively). These results are consistent with higher levels of Bax mRNA in neurons of 129B6 mice. Cells from 129B6 mice were excluded from this analysis because they exhibited poor growth kinetics in primary cultures.
The difference in the level of expression between DBA/2J–Luc and 129B6–Luc is not regulated by p53
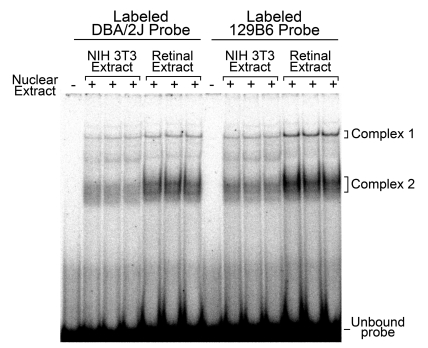
We first determined if nuclear proteins interacted with the polymorphic site. EMSAs were performed using radiolabelled 129B6 or DBA/2J probes spanning 30 bp, including the polymorphic site (see Figure 3), and nuclear extracts isolated from NIH 3T3 cells or retinal tissue. Retinal nuclear extracts were derived from either 129B6 or DBA/2J mice, both yielding the same result. Only data from the 129B6 extract is shown. NIH 3T3 and retinal nuclear extract produced two predominant shifted complexes, Complex 1 and Complex 2, compared with lanes with no nuclear extract (Figure 5). Although Complex 2 consisted of multiple bands, these bands behaved similarly in competition assays (data not shown) and were treated as a single complex during analyses. The polymorphism in the DBA/2J promoter was contained within a consensus p53 half-site. Two of these p53 half-sites, 0–13 bp apart, are typically required for effective p53 binding (El-Deiry et al., 1992). Others have shown, however, that p53 half-sites can mediate expression, which involved p53 binding, by remotely interacting with other cis-elements (Menendez et al., 2007). The change of a T(129B6) to a C(DBA/2J) at the −515 position would putatively increase p53 interactions with the DBA/2J half-site.
Figure 5. Both NIH 3T3 and retinal nuclear extracts produced two shifted complexes in EMSA.
A representative PhosphorImager-generated autoradiograph of an EMSA performed with 32P-labelled 129B6 or DBA/2J double-stranded probe (−499 to −528 sequence in Figure 3) is shown. No shifted complexes were formed in the absence of nuclear extract. Nuclear extracts from both fibroblasts and 129B6 retinal tissue (5 μg of total protein each) formed two shifted complexes denoted as Complex 1 and Complex 2. Three separate extracts, from each tissue source, are shown.
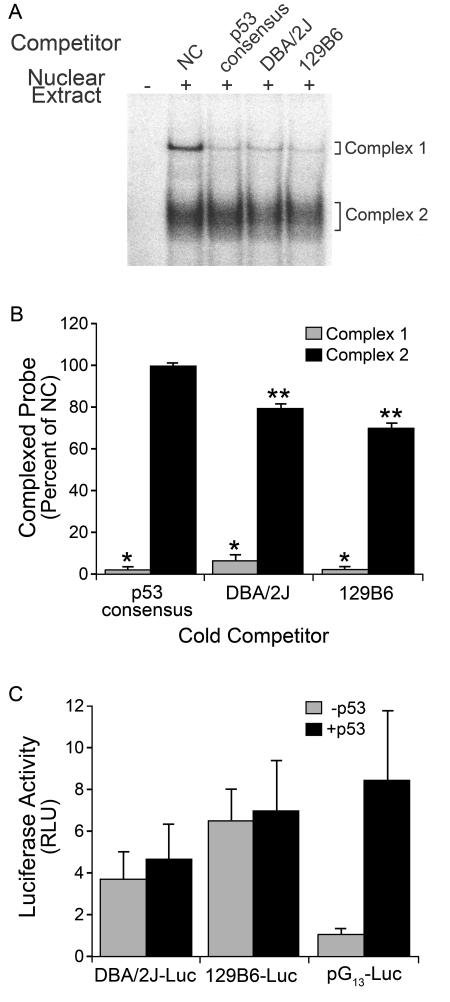
We initially tested the hypothesis that binding of p53, principally to the DBA/2J polymorphism, may be acting to repress transcription. To examine p53 involvement, unlabelled competition EMSA was first performed with a 50-fold molar excess of a concatemer of four p53 half-sites (Figures 6A and 6B). The unlabelled p53 half-site concatemer competed specifically with Complex 1 (P = 4.23×10−10). This result was consistent with possible involvement of p53, but importantly indicated that DNA–protein interactions of Complex 1 included the polymorphic region. To test the effects of p53 directly, we transiently transfected DBA/2J–Luc and 129B6–Luc into primary fibroblasts isolated from p53−/− FVB mice, to determine if the differential expression observed between the two promoters was lost in the absence of p53. In p53−/− fibroblasts, 129B6–Luc expression was approx. 1.5-fold higher than DBA/2J–Luc (P = 0.03), and in p53+/+ cells 129B6–Luc expression was twice as high as DBA/2J–Luc expression (P = 0.003, see FVB cells in Figure 4). As a control, we also co-transfected the cells with a plasmid expressing exogenous p53 (pC53-SN3). The addition of exogenous p53 did not significantly transactivate or repress either DBA/2J–Luc (P = 0.16, compared with no p53) or 129B6–Luc (P = 0.30) expression (Figure 6C). Conversely, a reporter construct with 13 p53 consensus sites (pG13–Luc) exhibited 8-fold more Luc expression after the addition of exogenous p53 compared with cells without p53 (P = 0.0001, Figure 6C). Importantly, differential expression between DBA/2J–Luc and 129B6–Luc was not affected by the presence or absence of p53. In order to confirm these data, we examined if the EMSA-binding pattern was altered in nuclear extracts isolated from p53−/− cells. Extracts from both p53+/+ and p53−/− fibroblasts (with an otherwise identical genetic background) yielded identical EMSA banding patterns, including Complex 1 (data not shown). Lastly, super-shifting either complex with a p53 antibody was unsuccessful (data not shown).
Figure 6. p53 does not regulate differential expression of the Bax promoter.
(A) PhosphorImager-generated autoradiograph of a competition EMSA using the 129B6 probe and 129B6 retina extract. In order to test possible p53 involvement, unlabelled competition EMSA was performed with a 50-fold molar excess of a concatemer of four p53 half-sites and compared with the DBA/2J and 129B6 unlabelled competitors at a 50-fold molar excess (consisting of the 30 bp highlighted in Figure 3). The unlabelled p53 half-site concatemer competed specifically with Complex 1, whereas DBA/2J and 129B6 oligos competed with both Complexes. NC, lane containing no competitor. (B) Histogram showing quantification of percentage inhibition of Complex 1 and 2 formation compared with NC (means±SEM). The amount of complexed probe in the presence of no competitor DNA was assigned a value of 100% and subsequent competitor lanes are expressed as a percentage of this. The p53 consensus sequence, DBA/2J sequence and 129B6 sequence all significantly competed Complex 1 (*P<0.01; P = 4.23×10−10, P = 5.17×10−8 and P = 2.13×10−10 respectively compared with NC. The p53 consensus sequence did not significantly compete Complex 2 (P = 0.45), whereas the DBA/2J and 129B6 sequences did (**P<0.01; P = 6.29×10−5 and P = 1.10×10−5 respectively). (C) Histogram of Luc activity in transient transfections of p53−/− fibroblasts (FVB mice). Bax–Luc constructs were co-transfected with pSV-βgal and either pGL-Basic (-p53) or pC53-SN3 (+p53). The 129B6–Luc construct had higher expression than the DBA/2J–Luc construct with or without addition of exogenous p53 (P = 0.03 and P = 0.006, respectively). Although there was a trend toward an increase in expression from DBA/2J–Luc and 129B6–Luc when co-transfected with human p53, compared with p53−/− cells transfected with a non-expressing control vector, the increase was not statistically significant (DBA/2J, P = 0.16; 129B6, P = 0.30). These results are consistent with other reports showing that p53 does not transactivate the murine Bax promoter (Igata et al., 1999; Schmidt et al., 1999; Thornborrow et al., 2002). The p53-responsive vector, pG13–Luc, exhibited a significant increase in expression with co-transfection of human p53 (P = 0.0001), indicating that the exogenous p53 was biologically active in these cells.
The single nucleotide polymorphism affects the binding affinity of nuclear proteins
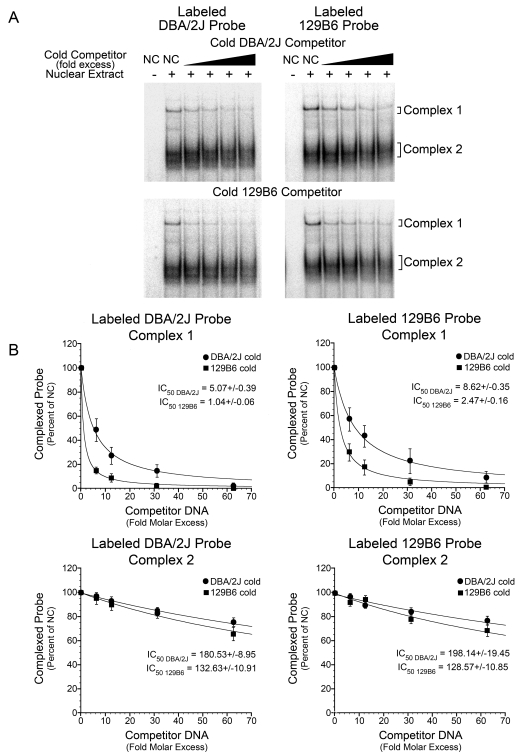
Because p53 was ruled out as a transcription factor that affected the difference in expression, we determined if the polymorphism affected the binding affinity of other nuclear proteins. Competition EMSA was performed to determine relative binding affinities for the polymorphic site. Complexes formed on radiolabelled DBA/2J or 129B6 probes were competed with increasing fold molar excess of either unlabelled DBA/2J DNA or unlabelled 129B6 DNA. The inhibition constants at half the labelled complex formation (IC50) were calculated using Kaleidagraph software from the values of band density in each lane. The values were plotted and a best-fit curve generated (Figure 7A). For Complex 1, the IC50 value for the DBA/2J radiolabelled probe competed with unlabelled DBA/2J DNA was 5.07±0.39 (fold molar excess) and with unlabelled 129B6 DNA was 1.04±0.06. The IC50 for the 129B6 radiolabelled probe compared with unlabelled DBA/2J DNA was 8.62±0.35 and compared with unlabelled 129B6 DNA was 2.47±0.16 (Figure 7B). Thus, 4–5-fold more unlabelled DBA/2J DNA, relative to unlabelled 129B6 DNA, was required to inhibit half of the radiolabelled complex from each probe. These results suggested that the protein(s) forming Complex 1 had a higher affinity for the 129B6 site. For Complex 2, the IC50 value of the DBA/2J radiolabelled probe co-competed with DBA/2J unlabelled DNA was 180.53±8.95 and with 129B6 unlabelled DNA was 132.63±10.91 (Figure 7B). The IC50 for Complex 2 of the 129B6 radiolabelled probe compared with the unlabelled DBA/2J DNA was 198.14±19.45 and compared with unlabelled 129B6 DNA was 128.57±10.85 (Figure 7B). The IC50 ratios of Complex 2 indicated that the protein(s) forming this complex exhibited similar binding affinities for both the DBA/2J and 129B6 DNAs. These data indicated Complex 1 was formed via DNA–protein interactions with the polymorphic site, whereas Complex 2 was probably formed with DNA–protein interactions elsewhere on the 30 bp sequence. To confirm this, we also performed unlabelled competition EMSA with separate regions of the 30 bp sequence (Figure 8). Complex 2 formation was inhibited most successfully with a unlabelled 10 bp DNA fragment upstream of the polymorphic site, suggesting that this region was principally involved in Complex 2 formation.
Figure 7. Nuclear proteins forming Complex 1 bind with higher affinity to the 129B6 polymorphic site.
(A) Representative PhosphorImager-generated autoradiographs of unlabelled co-competition experiments. Top two panels: 600000 c.p.m./lane (0.12 pmol) of radiolabelled DBA/2J probe (left panel) or radiolabelled 129B6 probe (right panel) were reacted with either no or 5 μg of 129B6 retinal nuclear extract. Complexes were also incubated with no competitor DNA (NC) or increasing molar excess of unlabelled DBA/2J competitor DNA. Increasing molar excess caused inhibition of the formation of radiolabelled Complex 1 and Complex 2 compared with lanes with no competitor DNA. Bottom two panels: same probes as the top two panels with increasing amounts of unlabelled 129B6 competitor DNA. (B) Graphs plotting fold molar excess of DNA compared with band density of complexed probe obtained from analysing PhosphorImager-generated autoradiographs using ImageQuant v5.2 software. Kaleidograph software was then used to generate graphs and calculate IC50 values. The mean (±S.E.M.) complex formed in three separate experiments are shown. The amount of complexed probe in the presence of no competitor DNA was assigned a value of 100% and subsequent competitor lanes are expressed as a percentage of this. As the fold molar excess competitor DNA increases, there is increased inhibition of radiolabelled complex formation. Top panels: radiolabelled DBA/2J probe (left) or radiolabelled 129B6 probe (right) compared with increasing fold molar excess of DBA/2J or 129B6 unlabelled competitor DNA for Complex 1. For Complex 1, the IC50 value of DBA/2J radiolabelled probe (left) co-competed with unlabelled DBA/2J DNA was 5.07±0.39 and with unlabelled 129B6 DNA was 1.04±0.06. The IC50 for 129B6 radiolabelled probe (right) compared with unlabelled DBA/2J DNA was 8.62±0.35 and compared with unlabelled 129B6 DNA was 2.47±0.16. Approx. 4–5-fold more DBA/2J unlabelled DNA relative to unlabelled 129B6 DNA was required to inhibit half of the radiolabelled complex. These data suggest that the protein(s) forming Complex 1 have a higher affinity for the 129B6 site. Bottom panels: radiolabelled DBA/2J probe (left) or radiolabelled 129B6 probe (right) versus increasing fold molar excess of DBA/2J or 129B6 unlabelled competitor DNA for Complex 2. The IC50 value of the DBA/2J radiolabelled probe (left) co-competed with DBA/2J unlabelled DNA was 180.53±8.95 and with 129B6 unlabelled DNA was 132.63±10.91. The IC50 for Complex 2 of the 129B6 radiolabelled probe (right) compared with unlabelled DBA/2J DNA was 198.14±19.45 and compared with unlabelled 129B6 DNA was 128.57±10.85. The IC50 ratios of Complex 2 indicate that the protein(s) forming this complex exhibit similar binding affinities for both the DBA/2J and 129B6 DNAs.
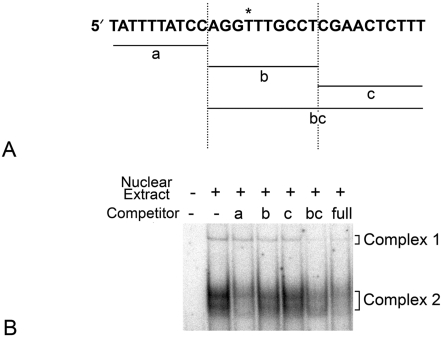
Figure 8. Complex 2 is formed by DNA–protein interactions involving nucleotides upstream of the polymorphic site.
(A) Shown is the 30 bp sequence delineating unlabelled competitor DNAs for the 129B6 polymorphic region. The polymorphic site is denoted by an asterisk. Fragments a, b and c span the individual regions shown. These fragments were used for competition as concatemers of three copies of each 10 bp region. (B) Representative PhosphorImager-generated autoradiograph of unlabelled co-competition experiments with different competitor DNAs reacting with 129B6 retinal extract. The radiolabelled 129B6 full-length probe was incubated in the presence or absence of unlabelled competitor DNAs. Each competitor DNA was used at a 150-fold molar excess of potential binding sites. Competitor a specifically competed Complex 2, while b competed Complex 1, but minimally competed Complex 2. Alternatively, oligo bc efficiently competed Complex 2. These results suggested Complex 2 was formed with interactions upstream of the polymorphic site, but may also involve interactions further downstream of this site.
DISCUSSION
Several neuronal populations require Bax for cell death during development. Bax deficiency resulted in an increase in neuronal numbers in regions such as the cerebellum, hippocampus and retina (Mosinger Ogilvie et al., 1998; White et al., 1998). Importantly, Bax gene dosage was shown to be critical for susceptibility to RGC death in the DBA/2J mouse glaucoma model and after ONC (Libby et al., 2005a).
Similarly, we demonstrated that retinal ganglion cell somas of DBA/2J mice heterozygous for the mutant Bax allele exhibited resistance to cell death in response to an acute crush lesion of the optic nerve, when assayed at 2 weeks after ONC. Unlike completely Bax-deficient cells, which exhibit essentially permanent resistance to optic nerve lesion, Bax+/− cells, which have short-term resistance to ONC (at 2 weeks), eventually die over a much longer time frame. A distinct cell death phenotype was observed for Bax+/− mice on the 129B6 background, however, in that these ganglion cells died with similar kinetics to wild-type cells.
The variation in cell death phenotype associated with Bax genotype appeared to be related to the relative level of latent Bax mRNA and protein in affected cells. Quantitative analysis of Bax transcript and protein levels showed that 129B6 neuronal populations (including retina and brain) expressed approximately twice the level of gene product than did equivalent cells of DBA/2J mice. Importantly, 129B6Bax+/− mice expressed approximately the same amount of Bax mRNA and protein as wild-type DBA/2J animals, thus providing one explanation for why animals of both these genotypes exhibited essentially normal levels of ganglion cell death. Taking the level of Bax expression in these mice as a starting point, two important observations are made with respect to the level of Bax and the ability of these neurons to execute apoptosis. Lowering Bax expression levels by half from this point, as was observed in DBA/2JBax+/− mice, was a sufficient reduction in BAX to make ganglion cells substantially resistant to ONC, at least in the short term. Conversely, doubling Bax expression levels from the same point, as was observed in 129B6Bax+/+ mice, had no increased effect on the rate of cell death. This latter observation is consistent with earlier reports that transgenic mice overexpressing wild-type Bax showed no increase in normal developmental programmed cell death (Bernard et al., 1998), and underscores a hypothetical concept that cells require a set threshold of Bax expression, which allows them to completely commit to the apoptotic program once it has been activated. This threshold may also be distinct from the levels of other members of the Bcl2 gene family. Both lines of mice, regardless of Bax genotype, expressed equivalent levels of BclX mRNA and protein, which, at the mRNA level, was also in approx. 5–10-fold molar excess. Similarly, preliminary experiments to quantify Bim transcripts, which encode a principal BH3-only protein involved in ganglion cell death (Näpänkangas et al., 2003; McKernan and Cotter, 2007) also showed no difference between strains (data not shown). Thus the correlation between cell death and Bax genotype appears to be mediated primarily by the level of Bax expression and not in conjunction with altered levels of other members of the Bcl2 gene family.
A caveat to the conclusion that lower Bax expression levels influence retinal ganglion cell in DBA/2J mice is that there are probably multiple interacting genes that influence the cell death response. These genes could also vary between different genetic backgrounds. For example, previously we reported that DBA/2J mice were more resistant to ONC than BALB/cByJ mice (Li et al., 2007). The difference in phenotype between the strains accounted for approx. 12% of the variance found in the loss of RGCs among mice after this procedure. Examination of the levels of Bax expression between strains showed that Bax was not the affecting allele, which was later attributed to a quantitative trait locus that mapped to chromosome 5 (Dietz et al., 2008). Interestingly, wild-type C57BL/6J and 129×1/SvJ, which both contribute to the genetic make-up of129B6 mice, exhibited a cell death phenotype statistically similar to DBA/2J mice. This suggests that influences attributable to different genetic background are not dramatically influencing the relationship between Bax expression and cell death we observed in the present study.
Experiments described in the present study establish a link between a single nucleotide polymorphism in the Bax promoter with the quantitative difference in Bax expression observed between the DBA/2J and 129B6 mouse lines. In transient transfection experiments, the 129B6 polymorphism roughly doubled the transcriptional activity of a Luc reporter gene. It is likely, however, that this cis-element is not the only factor affecting Bax gene expression quantitatively. Recently, for example, Whitney et al. (2009) described a quantitative and hereditary difference in amacrine cell number in the retinas of C57BL/6J and A/J mice. This quantitative trait was mapped to a locus on chromosome 7, near Bax, and was associated with a higher level of Bax transcripts in A/J mice. Surprisingly, A/J mice carry the −515(C) polymorphism, similar to DBA/2J mice, whereas C57BL/6J mice carry the −515(T) polymorphism, which is the opposite association of what we would predict from our data presented here. Although no in vitro transcription studies were conducted by this group, they hypothesized that differential transcription of the Bax allele may be due to enhanced p53 binding to the A/J allele. Since our studies have excluded p53 binding to this region, we predict that differences in genetic background can influence Bax transcription both at the level of the cis-element polymorphism and the transcriptional machinery interacting with it. Overall, our studies, combined with those of Whitney et al. (2009), underscore the importance of characterizing the transcription factors interacting with this polymorphic site.
Candidates for transcription factors that may interact with this site were examined. Although the polymorphism in the DBA/2J Bax promoter creates a perfect consensus p53 half-site (El-Deiry et al., 1992), our experiments clearly exclude p53 binding as a critical factor for the differential regulation of Bax. We then investigated if the affinity of other nuclear proteins was affected by the polymorphism. Our data suggested that the polymorphism in the Bax promoter was sufficient to cause a difference in binding affinity of nuclear proteins. Specifically, nuclear proteins were found to be binding with higher affinity to the 129B6 polymorphic site. Therefore, we predicted that an activator or an activation complex was interacting more efficiently with the 129B6 promoter, resulting in an increase in transcription. The identities of the binding proteins in this region are currently being investigated. Interestingly, artificial transcription factors have been designed to bind to a p53 half-site contained in the human BAX promoter with the intention of selectively up-regulating BAX and not other p53 targets (Falke et al., 2003). An artificial transcription factor containing five zinc-finger-binding motifs, 5ZFAV, was able to elicit significant transactivation from a 40 bp section of promoter containing the consensus p53 half-site. A caveat to this study, however, was that this portion of the promoter also contained three other imperfect p53 half-sites, which could have affected binding and therefore may not be analogous to the mouse polymorphic region.
Alternatively, a search of several transcription-factor-binding-site databases, using the consensus 10 bp surrounding the 129B6 polymorphism, identified other potential transcription factor-binding sites (Table 1). None of the sites was a perfect match. Nevertheless, the putative transcription factors identified were activators of transcription, consistent with our prediction that Complex 1 is a positive regulator of transcription. These potential transcription factors included C/EBP (CCAAT/enhancer-binding protein), FOXI1 (Forkhead box I1), and E2F1. E2F1 may be the most promising candidate because of the role it plays in apoptosis. Mice deficient in E2F1, for example, have excess T-cells owing to defects in thymocyte apoptosis, and E2F1 has been shown to activate the expression of Apaf-1. Importantly, increased E2F1 activity also resulted in the release of cytochrome c (Ginsberg, 2002). In microarrays, E2Fs have also been implicated in the up-regulation of members of the Bcl-2 gene family, including Bad and Bak (Ma et al., 2002).
Table 1. In silico analysis of transcription-factor-binding sites with similarity to the polymorphic region.
A search of transcription factors that can potentially bind the polymorphic region was conducted using the 10 bp core sequence from the 129B6 Bax promoter (AGGTTTGCCT). The databases searched included MatInspector, TESS, TFSEARCH and JASPAR. The sequences for putative transcription-factor-binding sites shown in this Table were chosen based on having the highest sequence similarity. Nucleotides that do not align with the polymorphic site are denoted in lower case, whereas exact matches are in upper case. The letter symbols used for nucleotide sequence are as follows: R = G or A, W = T or A, Y = C or T, S = G or C. EBP-45, enhancer-binding protein 45.
| Binding site similarity to AGGTTTGCCT | Factor | Notes |
|---|---|---|
| - t GTTTGC - - | EBP-45 or C/EBP | C/EBP found to bind to site, no evidence EBP-45 expressed in neurons (Petropoulos et al., 1991; McCauslin et al., 2006) |
| RRRcWWGYYY | p53 | Must have two of these half-sites within 13 bp of each other for p53 binding (El-Deiry et al., 1992) |
| - t RTTTRY - - | FOXI1 | Forkhead transcription factor (Overdier et al., 1997; Hulander et al., 1998) |
| - - - TTTSSCgc | E2F1 | Regulation of cell cycle and apoptosis induction (Helin et al., 1993; Ginsberg, 2002; Rabinovich et al., 2008) |
Although experimentally we have demonstrated that the level of Bax expression can profoundly affect cell death, we have only been able to detect this phenotype in mice that have been genetically manipulated to have one functional Bax gene. Interestingly, however, a similar phenotype has been described in humans. Saxena et al. (2002) noted that patients with chronic lymphocytic leukaemia had a high BCL2/BAX protein ratio. Further examination revealed that a single G to an A polymorphism 248 bp upstream from the translational start site was present in one copy of the BAX gene of 69% of the patients. This promoter polymorphism associated with late-stage disease and resistance to treatment. These investigators then demonstrated that this polymorphism was responsible for a 2.6-fold reduction in BAX expression (Moshynska et al., 2005). Although the G(−248)A polymorphism in the human BAX promoter lies outside of the polymorphism we have detected in the mouse Bax promoter, these observations validate the concept that mutations affecting BAX expression can influence cell death in disease. Based on our observations in mice, the same or similar polymorphisms in the human BAX gene may also contribute to susceptibility of cells in genetically complex neurodegenerative diseases.
ACKNOWLEDGEMENTS
We thank Dr Cassandra L. Schlamp for helpful discussions and design of Figures, Dr Simon John for DBA/2JBax+/− mice, Dr Leonard Levin (Department of Ophthalmology and Visual Sciences, University of Wisconsin, Madison, WI, U.S.A.) for RGC-5 cells, Dr Donna Peters for NIH 3T3 cells, Anny Shai and Dr Paul Lambert for FVBp53−/− mice and to Christopher Berndsen for help calculating IC50 values.
Footnotes
This work was supported by a grant from The National Eye Institute [grant number R01 EY012223], a Vision Research CORE grant [grant number P30 EY016665] to the Department of Ophthalmology and Visual Sciences, and an unrestricted grant from Research to Prevent Blindness, Inc.
REFERENCES
- Adams JM, Cory S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene. 2007;26:1324–1337. doi: 10.1038/sj.onc.1210220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar RS, Ness JM, Roth KA. Bcl-2 family regulation of neuronal development and neurodegeneration. Biochim Biophys Acta. 2004;1644:189–203. doi: 10.1016/j.bbamcr.2003.10.013. [DOI] [PubMed] [Google Scholar]
- Andrews NC, Faller DV. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of cells. Nuc Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azarian SM, Schlamp CL, Williams DS. Characterization of calpain II in the retina and photoreceptor outer segments. J Cell Sci. 1993;105:787–798. doi: 10.1242/jcs.105.3.787. [DOI] [PubMed] [Google Scholar]
- Bernard R, Dieni S, Rees S, Bernard O. Physiological and induced neuronal death are not affected in NSE-bax transgenic mice. J Neurosci Res. 1998;52:247–259. doi: 10.1002/(SICI)1097-4547(19980501)52:3<247::AID-JNR1>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Chang B, Smith RS, Hawes NL, Anderson MG, Zabaleta A, Savinova O, Roderick TH, Heckenlively JR, Davisson MT, John SWM. Interacting loci cause severe iris atrophy and glaucoma in DBA/2J mice. Nat Genet. 1999;21:405–409. doi: 10.1038/7741. [DOI] [PubMed] [Google Scholar]
- Conforti L, Adalbert R, Coleman MP. Neuronal death: where does the end begin? Trends Neurosci. 2007;30:159–166. doi: 10.1016/j.tins.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- Deckwerth TL, Elliot JL, Knudson CM, Johnson EM, Jr, Snider WD, Korsmeyer SJ. BAX is required for neuronal death after trophic factor deprivation and during development. Neuron. 1996;17:401–411. doi: 10.1016/s0896-6273(00)80173-7. [DOI] [PubMed] [Google Scholar]
- Dietz JA, Li Y, Chung LM, Yandell BS, Schlamp CL, Nickells RW. Rgcs1, a dominant QTL that affects retinal ganglion cell death after optic nerve crush in mice. BMC Neurosci. 2008;9:74. doi: 10.1186/1471-2202-9-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- Falke D, Fisher MR, Ye D, Juliano RL. Design of artificial transcription factors to selectively regulate the pro-apoptotic bax gene. Nucleic Acids Res. 2003;31:e10. doi: 10.1093/nar/gng010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg D. E2F1 pathways to apoptosis. FEBS Lett. 2002;529:122–125. doi: 10.1016/s0014-5793(02)03270-2. [DOI] [PubMed] [Google Scholar]
- Helin K, Wu CL, Fattaey AR, Lees JA, Dynlacht BD, Ngwu C, Harlow E. Heterodimerization of the transcription factors E2F-1 and DP-1 leads to cooperative trans-activation. Genes Dev. 1993;7:1850–1861. doi: 10.1101/gad.7.10.1850. [DOI] [PubMed] [Google Scholar]
- Hulander M, Wurst W, Carlsson P, Enerback S. The winged helix transcription factor Fkh10 is required for normal development of the inner ear. Nat Genet. 1998;20:374–376. doi: 10.1038/3850. [DOI] [PubMed] [Google Scholar]
- Igata E, Inoue T, Ohtani-Fujita N, Sowa Y, Tsujimoto Y, Sakai T. Molecular cloning and functional analysis of the murine bax gene promoter. Gene. 1999;238:407–415. doi: 10.1016/s0378-1119(99)00348-0. [DOI] [PubMed] [Google Scholar]
- Isenmann S, Wahl C, Krajewski S, Reed JC, Bähr M. Up-regulation of Bax protein in degenerating retinal ganglion cells precedes apoptotic cell death after optic nerve lesion in the rat. Eur J Neurosci. 1997;9:1763–1772. doi: 10.1111/j.1460-9568.1997.tb01534.x. [DOI] [PubMed] [Google Scholar]
- Isenmann S, Engel S, Gillardon F, Bähr M. Bax antisense oligonucleotides reduce axotomy-induced retinal ganglion cell death in vivo by reduction of Bax protein expression. Cell Death Differ. 1999;6:673–682. doi: 10.1038/sj.cdd.4400538. [DOI] [PubMed] [Google Scholar]
- Johnstone RW. Deamidation of Bcl-XL: a new twist in a genotoxic murder mystery. Mol Cell. 2002;10:695–697. doi: 10.1016/s1097-2765(02)00693-7. [DOI] [PubMed] [Google Scholar]
- Kermer P, Liman J, Weishaupt JH, Bahr M. Neuronal apoptosis in neurodegenerative diseases: from basic research to clinical application. Neurodegen Dis. 2004;1:9–19. doi: 10.1159/000076665. [DOI] [PubMed] [Google Scholar]
- Kern SE, Pietenpol JA, Thiagalingam S, Seymour A, Kinzler KW, Vogelstein B. Oncogenic forms of p53 inhibit p53-regulated gene expression. Science. 1992;256:827–830. doi: 10.1126/science.1589764. [DOI] [PubMed] [Google Scholar]
- Knudson CM, Tung KSK, Toutellotte WG, Brown GAJ, Korsmeyer SJ. Bax-deficient mice with lymphoid hyperplasia and male germ cell death. Science. 1995;270:96–99. doi: 10.1126/science.270.5233.96. [DOI] [PubMed] [Google Scholar]
- Konigsberg M, Lopez-Diazguerrero NE, Aguilar MC, Ventura JL, Gutierrez-Ruiz MC, Zentella A. Senescent phenotype achieved in vitro is indistinguishable, with the exception of Bcl-2 content, from that attained during the aging process. Cell Biol Int. 2004;28:641–651. doi: 10.1016/j.cellbi.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Krishnamoorthy RR, Agarwal P, Prasanna G, Vopat K, Lambert W, Sheedlo HJ, Pang I-H, Shade D, Wordinger RJ, Yorio T, Clark AF, Agarwal N. Characterization of a transformed rat retinal ganglion cell line. Mol Brain Res. 2001;86:1–12. doi: 10.1016/s0169-328x(00)00224-2. [DOI] [PubMed] [Google Scholar]
- Levin LA, Schlamp CL, Spieldoch RL, Geszvain KM, Nickells RW. Identification of bcl-2 family genes in the rat retina. Invest Ophthalmol Vis Sci. 1997;38:2545–2553. [PubMed] [Google Scholar]
- Li Y, Schlamp CL, Nickells RW. Experimental induction of retinal ganglion cell death in adult mice. Invest Ophthalmol Vis Sci. 1999;40:1004–1008. [PubMed] [Google Scholar]
- Li Y, Schlamp CL, Poulsen KP, Nickells RW. Bax-dependent and independent pathways of retinal ganglion cell death induced by different damaging stimuli. Exp Eye Res. 2000;71:209–213. doi: 10.1006/exer.2000.0873. [DOI] [PubMed] [Google Scholar]
- Li Y, Semaan SJ, Schlamp CL, Nickells RW. Dominant inheritance of retinal ganglion cell resistance to optic nerve crush in mice. BMC Neurosci. 2007;8:19. doi: 10.1186/1471-2202-8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libby RT, Li Y, Savinova OV, Barter J, Smith RS, Nickells RW, John SWM. Susceptibility to neurodegeneration in glaucoma is modified by Bax gene dosage. PLoS Genet. 2005a;1:17–26. doi: 10.1371/journal.pgen.0010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libby RT, Anderson MG, Pang I-H, Robinson ZH, Savinova OV, Cosma IM, Snow A, Wilson LA, Smith RS, Clark AF, John SWM. Inherited glaucoma in DBA/2J mice: pertinent disease features for studying the neurodegeneration. Vis Neurosci. 2005b;22:637–648. doi: 10.1017/S0952523805225130. [DOI] [PubMed] [Google Scholar]
- Lindholm D, Eriksson O, Korhonen L. Mitochondrial proteins in neuronal degeneration. Biochem Biophys Res Commun. 2004;321:753–758. doi: 10.1016/j.bbrc.2004.06.138. [DOI] [PubMed] [Google Scholar]
- Ma Y, Croxton R, Moorer RL, Jr, Cress WD. Identification of novel E2F1-regulated genes by microarray. Arch Biochem Biophys. 2002;399:212–224. doi: 10.1006/abbi.2002.2761. [DOI] [PubMed] [Google Scholar]
- McCauslin CS, Heath V, Colangelo AM, Malik R, Lee S, Mallei A, Mocchetti I, Johnson PF. CAAT/enhancer-binding protein δ and cAMP-response element-binding protein mediate inducible expression of the nerve growth factor gene in the central nervous system. J Biol Chem. 2006;281:17681–17688. doi: 10.1074/jbc.M600207200. [DOI] [PubMed] [Google Scholar]
- McKernan DP, Cotter TG. A critical role for Bim in retinal ganglion cell death. J Neurochem. 2007;102:922–930. doi: 10.1111/j.1471-4159.2007.04573.x. [DOI] [PubMed] [Google Scholar]
- McKinnon SJ, Lenhman DM, Kerrigan-Baumrind LA, Merges CA, Pease ME, Kerrigan DF, Ransom NL, Tahzib NG, Reitsamer HA, Levkovitch-Verbin H, Quigley HA, Zack DJ. Caspase activation and amyloid precursor protein cleavage in rat ocular hypertension. Invest Ophthalmol Vis Sci. 2002;43:1077–1087. [PubMed] [Google Scholar]
- Menendez D, Inga A, Snipe J, Krysiak O, Schonfelder G, Resnick MA. A single-nucleotide polymorphism in a half-binding site creates p53 and estrogen receptor control of vascular endothelial growth factor receptor 1. Mol Cell Biol. 2007;27:2590–2600. doi: 10.1128/MCB.01742-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshynska O, Moshynskyy I, Misra V, Saxena A. G125A single-nucleotide polymorphism in the human BAX promoter affects gene expression. Oncogene. 2005;24:2042–2049. doi: 10.1038/sj.onc.1208377. [DOI] [PubMed] [Google Scholar]
- Mosinger Ogilvie J, Deckwerth TL, Knudson CM, Korsmeyer SJ. Suppression of developmental retinal cell death but not photoreceptor degeneration in Bax-deficient mice. Invest Ophthalmol Vis Sci. 1998;39:1713–1720. [PubMed] [Google Scholar]
- Näpänkangas U, Lindqvist N, Lindholm D, Hallböök F. Rat retinal ganglion cells upregulate the pro-apoptotic BH3-only protein Bim after optic nerve transection. Mol Brain Res. 2003;120:30–37. doi: 10.1016/j.molbrainres.2003.09.016. [DOI] [PubMed] [Google Scholar]
- Nickells RW. From ocular hypertension to ganglion cell death: a theoretical sequence of events leading to glaucoma. Can J Ophthalmol. 2007;42:278–287. [PubMed] [Google Scholar]
- Overdier DG, Ye H, Peterson RS, Clevidence DE, Costa RH. The winged helix transcriptional activator HFH-3 is expressed in the distal tubules of embryonic and adult mouse kidney. J Biol Chem. 1997;272:13725–13730. doi: 10.1074/jbc.272.21.13725. [DOI] [PubMed] [Google Scholar]
- Pelzel HR, Schlamp CL, Poulsen GL, Ver Hoeve JN, Nork TM, Nickells RW. Decrease of cone opsin mRNA in experimetal ocular hypertension. Mol Vis. 2006;12:1272–1282. [PubMed] [Google Scholar]
- Pequignot MO, Provost AC, Salle S, Taupin P, Sainton KM, Marchant D, Martinou JC, Ameisen JC, Jais JP, Abitbol M. Major role of BAX in apoptosis during retinal development and in establishment of a functional postnatal retina. Dev Dynamics. 2003;228:231–238. doi: 10.1002/dvdy.10376. [DOI] [PubMed] [Google Scholar]
- Petropoulos I, Auge-Gouillou C, Zakin MM. Characterization of the active part of the human transferrin gene enhancer and purification of two liver nuclear factors interacting with the TGTTTGC motif present in this region. J Biol Chem. 1991;266:24220–24225. [PubMed] [Google Scholar]
- Rabinovich A, Jin VX, Rabinovich R, Xu X, Farnham PJ. E2F in vivo binding specificity: comparison of consensus versus nonconsensus binding sites. Genome Res. 2008;18:1763–1777. doi: 10.1101/gr.080622.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena A, Moshynska O, Sankaran K, Viswanathan S, Sheridan DP. Association of a novel single nucleotide polymorphism, G(−248)A, in the 5′ UTR of the BAX gene in chronic lymphocytic leukemia with disease progression and treatment resistance. Cancer Lett. 2002;187:199–205. doi: 10.1016/s0304-3835(02)00378-6. [DOI] [PubMed] [Google Scholar]
- Schlamp CL, Li Y, Dietz JA, Janssen KT, Nickells RW. Progressive ganglion cell loss and optic nerve degeneration in DBA/2J mice is variable and asymmetric. BMC Neurosci. 2006;7:66. doi: 10.1186/1471-2202-7-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T, Korner K, Karsunky H, Korsmeyer SJ, Muller R, Moroy T. The activity of the murine Bax promoter is regulated by Sp1/3 and E-box binding proteins, but not p53. Cell Death Differ. 1999;6:873–882. doi: 10.1038/sj.cdd.4400562. [DOI] [PubMed] [Google Scholar]
- Shin DH, Lee HY, Lee HW, Kim HJ, Lee E, Cho SS, Baik SH, Lee KH. In situ localization of p53, bcl-2 and bax mRNAs in rat ocular tissue. Neuroreport. 1999;10:2165–2167. doi: 10.1097/00001756-199907130-00030. [DOI] [PubMed] [Google Scholar]
- Sun W, Oppenheim RW. Response of motoneurons to neonatal sciatic nerve axotomy in Bax-knockout mice. Mol Cell Neurosci. 2003;24:875–886. doi: 10.1016/s1044-7431(03)00219-7. [DOI] [PubMed] [Google Scholar]
- Thornborrow EC, Patel S, Mastropietro AE, Schwartzfarb EM, Manfredi JJ. A conserved intronic response element mediates direct p53-dependent transcriptional activation of both the human and murine Bax genes. Oncogene. 2002;21:990–999. doi: 10.1038/sj.onc.1205069. [DOI] [PubMed] [Google Scholar]
- Uo T, Kinoshita Y, Morrison RS. Neurons exclusively express N-Bak, a BH3 domain-only Bak isoform that promotes neuronal apoptosis. J Biol Chem. 2005;280:9065–9073. doi: 10.1074/jbc.M413030200. [DOI] [PubMed] [Google Scholar]
- White FA, Keller-Peck CR, Knudson CM, Korsmeyer SJ, Snider WD. Widespread elimination of naturally occurring neuronal death in bax-deficient mice. J Neurosci. 1998;18:1428–1439. doi: 10.1523/JNEUROSCI.18-04-01428.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney IE, Raven MA, Ciobanu DC, Williams RW, Reese BE. Multiple genes on chromosome 7 regulate dopaminergic amacrine cell number in the mouse retina. Invest Ophthalmol Vis Sci. 2009;50:1996–2003. doi: 10.1167/iovs.08-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Yu J, Park BH, Kinzler KW, Vogelstein B. Role of BAX in the apoptotic response to anticancer agents. Science. 2000;290:989–995. doi: 10.1126/science.290.5493.989. [DOI] [PubMed] [Google Scholar]