Abstract
The standard, well-established sample preparation protocol to release N-linked glycans from glycoproteins for downstream analysis requires relatively long deglycosylation times (from several hours to overnight) and relatively high endoglycosidase concentration (1:250 – 1:500 enzyme:substrate molar ratio). In this paper, we significantly improve this standard protocol by the use of pressure cycling technology (PCT) to increase the speed and decrease the relative amount of PNGase F during the release of N-linked glycans from denatured glycoproteins. With the application of pressure cycling from atmospheric to as high as 30 kPsi, >95% release of the asparagine linked glycans from bovine ribonuclease B, human transferrin and polyclonal human immunoglobulin was rapidly achieved in a few minutes using as low as 1:2500 enzyme:substrate molar ratio. The deglycosylation rate was first examined by SDS-PAGE at the protein level. The released glycans were then quantitated by capillary electrophoresis with laser induced fluorescence detection (CE-LIF). This new sample preparation protocol readily supports large scale glycan analysis of biopharmaceuticals with rapid deglycosylation times.
INTRODUCTION
Glycosylation, one of the most important co- and post-translational modifications of proteins, plays a significant role in numerous biological processes including protein folding and stability, cellular adhesion, signaling and disease.1 Furthermore, glycan profiles of biotherapeutics provide essential information on efficacy and safety; 2 therefore, the biotech industry places great emphasis on batch to batch glycosylation profile analysis to assure consistent bioactivity 3 and to avoid immunogenicity. 4 Given its biological and therapeutic significance, rapid, robust and accurate glycan analysis methods are of high interest. The most frequently used carbohydrate separation and analysis techniques are liquid chromatography, capillary electrophoresis, mass spectrometry and NMR. 5–12
The three major types of glycosylation in living systems are the N-, O- and C-linked sugar structures, which in each case can be highly diverse. 13 In this paper, we focus on the release of N-linked glycans, which typically employs the endoglycosidase PNGase F (Peptide-N4-(acetyl-β-glucosaminyl)-asparagine amidase). 1 Although denaturation of glycoproteins prior to PNGase F digestion increases deglycosylation efficiency, the conventional digestion process is still time-consuming, usually requiring several hours to overnight and enzyme:substrate molar ratios of 1 to 250 – 1 to 500 14. Methods such as microwave assisted deglycosylation have been shown to lead to complete deglycosylation of monoclonal antibodies in 10 min; however, glycan release from other glycoproteins, such as RNase B, have required up to 1 hour of irradiation with the use of relatively high 1:50 enzyme:substrate molar ratio. 15 In addition, there have been recent reports on the use of immobilized PNGase F enzyme reactors in capillary columns, 16, 17 even on an integrated microfluidic chip for rapid deglycosylation. 8
A new method that has the potential to accelerate enzyme catalyzed digestion is pressure cycling technology (PCT). 18 The basis of this method is the application of alternating cycles of atmospheric and high pressure of up to tens of kpsi (1 kpsi = 6.895 MPa). 19 Recent studies have shown enhanced speed of digestion using trypsin, chymotrypsin and pepsin under pressure cycling. 20 It is believed that the high pressure changes protein conformation and forces the penetration of water molecules into the protein interior (especially into cavities); thereby, leading to unfolding. 21 Cyclization between high and atmospheric pressures on the other hand enhances the probability of the access of the digestion enzyme to the digestion site. 20 As an example, ultrafast in-solution tryptic digestion of complex protein mixtures was achieved in less than 60 sec. by applying pressure cycling technology. 22 It is important to note that PCT is usually conducted at mild temperatures (room temperature to 37°C), offering an additional advantage for the analysis of heat sensitive molecules, such as sialylated glycan structures. 23
In this paper, we demonstrate rapid (minutes) and efficient (>95%) deglycosylation of glycoproteins by means of pressure cycling up to the 30 kPsi level using 5–10 times less PNGase F than standard methods. In addition to the advantages of speed, PCT did not cause decomposition (e.g., desialylation) of the glycan structures under investigation. PCT assisted deglycosylation was followed by SDS-PAGE at the protein level, and by capillary electrophoresis 24 at the glycan level. The PCT approach can also be coupled with HPLC and mass spectrometry.
EXPERIMENTAL SECTION
Chemicals
Maltose, maltopentaose, citric acid, lithium hydroxide, ammonium hydroxide, acetic acid, 8-aminopyrene-1,3,6-trisulfonic-acid trisodium salt (APTS), sodium cyanoborohydrate (1 M solution in tetrahydrofuran), ribonuclease B (RNase B, from bovine pancrease), transferrin and asialotransferrin (both human), IgG (human, polyclonal), sodium dodecylsulfate (SDS), PNGase F (proteomics grade, from Elizabethkingia meningoseptica) and Triton X-100 were purchased from Sigma-Aldrich (St. Louis, MO). HPLC grade water was from Baker (Phillipsburg, NJ) and acetonitrile from Fisher Scientific (Fair Lawn, NJ).
Deglycosylation using pressure cycling technology (PCT)
PNGase F enzyme (50 U, Sigma-Aldrich) was dissolved in 100 μL HPLC grade water. For the glycan release experiments, stock solutions of 2 mg/mL RNase B, 20 mg/mL IgG and 2 mg/mL tranferrin (in 50 mM phosphate buffer, pH 7.5) were used. 5-μL aliquot of denaturing buffer (2 % SDS in 100 mM 2-mercapthethanol solution) was added to 50 μL of each protein stock solution, followed by incubation at 100°C for 10 minutes. After this denaturing step, the solution was cooled to room temperature, and 50 μL of 50 mM phosphate buffer (pH 7.5) was added. 5 μL of 10% Triton X-100, an enzyme friendly surfactant, was also added to the reaction mixture. The deglycosylation efficiency was determined by adding, in separate experiments, 0.25, 0.5, 1.0, 1.5 and 2.0 units of PNGase F to the RNase B solution (enzyme:substrate molar ratios of 1:10000, 1:5000, 1:2500, 1:1666 and 1:1250, respectively). After the addition of the enzyme, each reaction mixture was placed in Teflon reactor tubes (3 mm i.d.), carefully sealed and inserted into the cartridge of the pressure cycling device (Barocycler NEP 2320, Pressure Biosciences Inc., South Easton, MA). Each pressure cycle comprised a 50 sec. pressurized and a 10 sec. atmospheric period. Control experiments at atmospheric-only conditions, using the same reaction mixtures as in pressure cycle assisted deglycosylation, were also carried out. Both the pressure cycle assisted and control deglycosylation reactions were conducted at 37°C. For each data point, 3 μL aliquots from the PCT process and the control reaction were immediately diluted with 7 μL HPLC grade water and mixed with 2 μL of sample buffer and 2 μL of reducing agent (Invitrogen, Carlsbad, CA), followed by incubation at 100°C for 5 minutes to stop the reaction. The degree of protein deglycosylation was first assessed by SDS-PAGE (NuPAGE, Novex 4–12 % Bis-Tris Gel, 1.5 mm, Invitrogen), visualized by Coomassie Brilliant Blue staining. The band intensities in the SDS-PAGE images were quantified by ImageJ software (NIH, Bethesda, MD). All SDS-PAGE separations were done in triplicates, and the RSD of the quantitation was determined to be < 3%. After PNGase F treatment, the deglycosylated proteins were precipitated by the addition of 300 μL of ice cold ethanol and centrifuged at 11,000 g for 20 minutes. The supernatant was used for the glycan labeling and CE experiments.
Oligosaccharide labeling and purification of the labeled glycans
The released oligosaccharides and the solutions of sugar standards (maltose and maltopentaose) were evaporated to dryness in a centrifugal vacuum evaporator (Centrivap, Labconco Co., Kansas City, MO). APTS labeling of the glycans followed a previously published protocol. 25 The reactions were stopped by the addition of 200 μL of HPLC water, and a 40 μL aliquot was diluted with 360 μL of acetonitrile prior to the purification process of the labeled glycans. Normal phase polyamide resin filled pipette tips (5 μL bed volume, PhyNexus, San Jose, CA) were used to remove the residual (unconjugated) APTS from the reaction mixture. A semiautomated 12 channel pipettor was employed in the clean-up process, and the intake/expel cycles of the system were controlled by the PhyTip operating software (PhyNexus). The tips were washed four times with 200 μL of 95% acetonitrile by applying eight intake/expel cycles. The captured glycans were then eluted with 50 μL of 20% acetonitrile, evaporated to dryness in the centrifugal vacuum evaporator (Labconco) and dissolved in 10 μL of HPLC grade water.
CE-LIF analysis
All capillary electrophoretic experiments were carried out using a laboratory-built system, comprising a high voltage power supply (30 kV, Spellman High Voltage Electronics Corporation, Hauppauge, NY) and a ZETALIF laser induced fluorescent detector (Picometrics, Toulouse, France) equipped with a solid state 488 nm laser (Cyan, Picarro Inc., Sunnyvale, CA). The total length of the separation capillary (50 μm i.d. PVA coated capillary, Agilent Technologies, Santa Clara, CA) was 60 cm, with 40 cm effective length up to the detection point with a pre-fixed ellipsoid lens. Separations were carried out in 15 mM acetate buffer (pH 4.75) at ambient temperature by applying 250 V/cm electric field strength. Samples were injected hydrostatically (Δ 10 cm/20 sec). All capillary electrophoresis separations were done in triplicates and the RSD of quantitation was < 2.5%.
RESULTS AND DISCUSSION
Effect of pressure cycling on PNGase F digestion of glycoproteins
Pressure cycle assisted deglycosylation studies in a closed tube (100 μL) utilized automated cycling between high pressure (up to 30 kpsi) and atmospheric pressure in order to study the enzymatic reaction between the endoglycosidase PNGase F and glycoproteins. As mentioned in the Introduction, alternation between high and atmospheric pressure resulted in rapid unfolding and refolding of the target proteins, thus, increased the probability of the access of the digestion enzyme to the digestion site. 20, 21, 26
In our initial experiments, bovine pancreatic ribonuclease B (RNase B) was used as a model target glycoprotein in the PCT assisted PNGase F digestion reaction. RNase B, a globular protein with four disulfide bonds, has a single N-glycosylation site at Asn34, where five to nine mannose residues can be attached to the chitobiose core, i.e. Man5–9GlcNAc2.27 It has been reported that globular proteins with several disulfide bridges are resistant to the necessary unfolding required to providing full access to enzymes for digestion. 22 For this reason, RNase B was reduced by mercaptoethanol and heat denatured (boiling for 5 min) in the presence of SDS prior to PCT assisted PNGase F treatment. Considering the findings in earlier reports, 28 we evaluated the pressure cycling levels between 5 and 30 kPsi, for which access to the cleavage sites could be facilitated, without affecting enzyme activity.
The effect of PNGase F concentration on the digestion rate was initially studied by using, in separate experiments, 1:10,000; 1:5000; 1:2500; 1:1666 and 1:1250 enzyme:substrate molar ratios, respectively, with pressure cycling at 20 kPsi (50 sec. pressure/10 sec. atmospheric) for 20 minutes in the presence of a non-ionic detergent. Standard methods suggest the use of approximately 1:250 – 1:500 enzyme:substrate molar ratio. Deglycosylation was initially monitored at the protein level by following the migration time shift in SDS-PAGE using Coomassie Brilliant Blue staining as a measure of relative protein amount. With the use of 1:10000 and 1:5000 enzyme:substrate molar ratios, the reaction rate (expressed by the band intensity of the remaining glycosylated form divided by the sum of the band intensities of the glycosylated and deglycosylated forms) was too slow, i.e., even with pressure cycling assistance >60 % of RNase B still remained glycosylated after 20 minutes of PCT treatment, as seen in Table 1. On the other hand, using 1:2500 enzyme:substrate molar ratio and pressure cycling at the 20 kPsi level for 20 min at 37°C resulted in a high level of deglycosylation. In this instance, only 7% of RNase B remained glycosylated, while, under atmospheric conditions, 20% was still in the glycosylated form. Applying 1:1666 and 1:1250 enzyme:substrate molar ratios further accelerated the deglycosylation rate for both the atmospheric and pressure cycle assisted deglycosylation, as shown in Table 1.
Table 1.
Effect of the PNGase F concentration on deglycosylation efficiency under PCT vs. atmospheric reaction conditions.
| PNGase F: Substrate ratio | 1:10000 | 1:5000 | 1:2500 | 1:1666 | 1:1250 |
|---|---|---|---|---|---|
| PCT (% intact RNase B) | 83 | 71 | 7 | 7 | 7 |
| Atmospheric (% intact RNase B) | 84 | 69 | 20 | 7 | 8 |
Deglycosylation rate is presented as the percent of remaining intact RNase B calculated from the intensities of the Coomassie Brilliant Blue stained SDS-PAGE bands. Reaction conditions: pressure cycle level 20 kPsi, reaction time 20 min at 37°C.
The importance of a non-ionic detergent in the deglycosylation reaction mixture under pressure cycling conditions was also studied. Standard procedures suggest the use of 1:250 – 1:500 enzyme:substrate molar ratios and incubation from 2 hours to overnight at 37°C in order to deglycosylate several nanomoles of denatured glycoprotein in the presence of a non-ionic detergent. 14 The presence of a non-ionic detergent (0.45% Triton X-100) was examined under the same pressure cycling conditions (1:2500 enzyme:substrate molar ratio, 20 kPsi, 20 min), using SDS-PAGE. Under PCT conditions, 7% and 12% of the RNase B remained glycosylated with and without Triton X-100 in the reaction mixture, respectively. This result suggests that PCT assisted deglycosylation also benefitted from the addition of a non-ionic detergent to the reaction mixture to counteract the possible inhibitory effect of SDS on PNGase F activity, as in standard atmospheric deglycosylation. 29
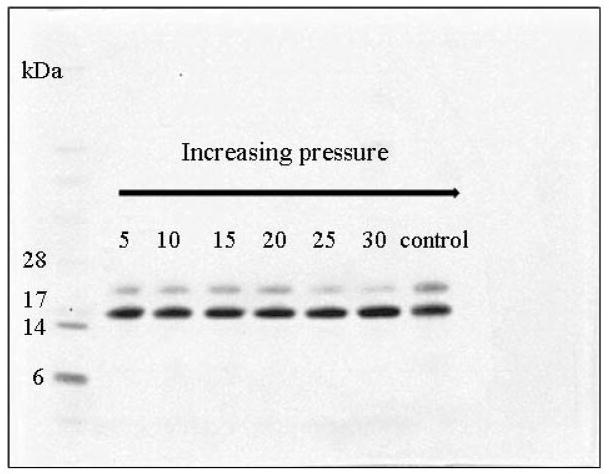
Figure 1 shows the effect of the maximum PCT pressure, in the range of 5 – 30 kPsi, (50 sec high pressure/10 sec atmospheric pressure) on the deglycosylation rate using 1:2500 enzyme:substrate molar ratio in the presence of Triton X-100, with only 5 minutes cycling time. The resulting deglycosylation efficiency was assessed by SDS-PAGE using Coomassie Brilliant Blue staining of the deglycosylated (lower band ~15 kDa) and glycosylated (upper band ~18 kDa) forms of RNase B. The increase of the cycle pressure resulted in a faster deglycosylation rate, as evidenced by the gradual diminishing of the 18 kDa band (upper band) compared to the control (atmospheric pressure) experiment. The intensity ratios of the deglycosylated and intact bands showed that with 30 kPsi, >95% deglycosylation of RNase B was obtained in 5 minutes.
Figure 1.
The effect of the maximum pressure level of pressure cycling on PNGase F mediated deglycosylation of the N-linked sugars from RNase B (Coomassie Brilliant Blue stained SDS-PAGE image). The bands at ~15 kDa and ~18 kDa represent the deglycosylated and intact forms of RNase B, respectively. Deglycosylation of denatured RNase B was carried out at 37°C for 5 minutes with 1:2500 enzyme:substrate molar ratio in the presence of Triton X-100. Control: 5 min deglycosylation at atmospheric pressure and 37°C. Pressure cycles: 50 s pressure/10 s atmospheric. Left lane: SDS-PAGE protein sizing standards: aprotinin (6 kDa), lysozyme 14 kDa), myoglobin (17 kDa) and carbonic anhydrase (28 kDa).
Based on these promising results, we next turned to evaluate the efficiency of pressure cycle assisted deglycosylation with respect to the quantitative amount of individual released glycans. Both released glycans and internal standards were first APTS labeled and then analyzed by CE-LIF.
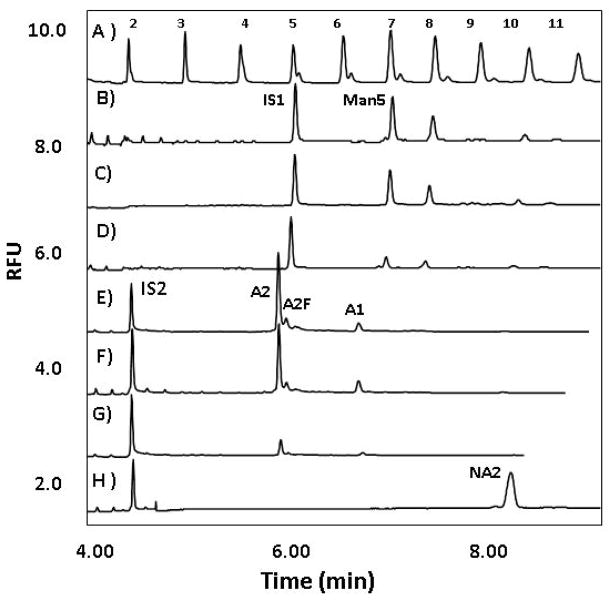
Figure 2 shows the electrophoretic profiles of the APTS labeled maltooligosaccharide ladder standard (trace A) as well as the APTS labeled glycans released from RNase B (traces B, C and D) and transferrin (traces E, F and G). Among all peaks in trace B, corresponding to Man 5 to Man 9 structures, 27 Man 5 (GlcNAc2Man5, see in Scheme 1) was selected for quantitative evaluation. APTS labeled maltopentaose was used as the internal standard (IS1) since it migrated close to the Man 5 structure. The peak height ratio of Man 5 to IS1 with pressure cycling assisted deglycosylation was 0.8 (1:2500 enzyme:substrate molar ratio, 30 kPsi level, 5 min, trace B). This ratio, under standard atmospheric deglycosylation conditions (37°C, 3 h, trace C), where full deglycosylation is expected, resulted in the same value (0.8), suggesting >95% deglycosylation with pressure cycling. Figure 2, trace D shows the result of a control deglycosylation experiment at atmospheric pressure for 5 minutes, clearly indicating significantly lower deglycosylation yield than with PCT (ratio of peak heights: 0.2). Comparison of the released glycan profiles and peak height ratios with PCT and atmospheric conditions (Figure 2, traces B and C, respectively) suggest that, as expected, the endoglycosidase activity of PNGase F was apparently not compromised at 30 kPsi at the reaction temperature of 37°C in the presence of Triton X-100.
Figure 2.
Comparative CE analysis of APTS labeled released glycans from RNase B and human transferrin by PCT and atmospheric deglycosylation using 1:2500 enzyme:substrate molar ratio. A) APTS labeled maltooligosaccharide ladder; B) RNase B glycans released at 30 kPsi pressure cycling level for 5 min (peaks in the order of their increasing migration times: Man 5 – Man 9); C) RNase B glycans released at atmospheric pressure for 3 hours; D) RNase B glycans released at atmospheric pressure for 5 minutes; E) transferrin glycans released at 30 kPsi pressure cycling level for 20 minutes; F) transferrin glycans released at atmospheric pressure for 3 hours; G) transferrin glycans released at atmospheric pressure for 20 minutes; H) asialotransferrin glycans released at atmospheric pressure for 3 hours. IS1: APTS derivatized maltopentaose, IS2: APTS derivatized maltose. Man 5, A2, A2F, A1 and NA2 structures are shown in Scheme 1. All separations were carried out in a PVA coated capillary (effective length: 40 cm, i.d.: 50 μm) at ambient temperature in 25 mM acetate (pH 4.75) buffer applying 250 V/cm field strength. Injection: hydrostatic, Δ 10 cm/10 sec.
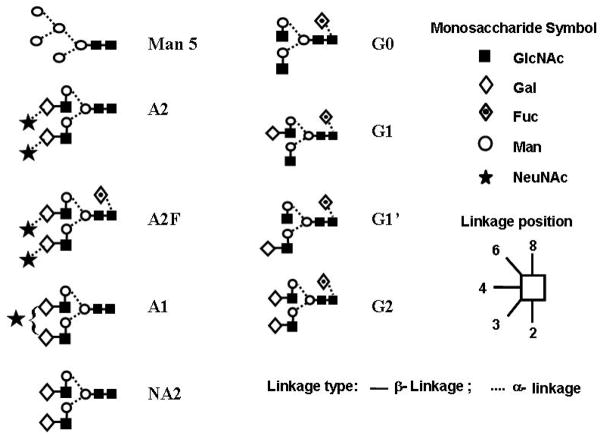
Scheme 1.
Schematic representation of the glycan structures used. Symbols adapted from, 34 linkages from right to left.
Pressure cycling assisted release of sialylated complex glycan structures
The application of high pressure during PCT assisted deglycosylation raised the concern of possible loss of terminal sialic acids. The stability of the sialic acid residues, a commonly considered issue in glycan analysis, 5 was studied in this work by analyzing the sugar structures after their PCT assisted release from transferrin, a highly sialylated glycoprotein. Human transferrin, a serum β-globulin, has a molecular mass of roughly 80 kDa and contains two N-linked glycosylation sites. 30 Because of its large size and multiple N-linked glycosylation sites, we increased the PCT time to 20 minutes, keeping the other parameters (30 kPsi, 37°C and enzyme:substrate molar ratio of 1:2500) constant. Trace E in Figure 2 depicts the results of PCT conditions, while traces F and G represent atmospheric release conditions for 3 hours (standard) and 20 minutes, respectively. Trace H in Figure 2 shows the electropherogram of the APTS labeled asialotransferrin glycans for comparative purpose. Because of the electrophoretically faster migrating sialylated structures of the transferrin glycans, a smaller size internal standard (APTS labeled maltose, IS2) was used. Structural assignment of the peaks in traces E-H was based on their glucose unit values (GU) calculated from previous data. 31, 32 Scheme 1 and Table 2 show the structures and the corresponding GU values for the significant peaks in Figure 2, marked as Man 5, A2, A2F, A1 and NA2. The peak height ratio of A2 to IS2 was 1.5 with PCT (20 min at 30 kPa, trace E), 1.4 for standard 3 hours atmospheric deglycosylation (trace F) where greater than 95% glycan release is expected, and only 0.2 for the control 20 min deglycosylation at atmospheric pressure (trace G). This result suggests full deglycosylation of transferrin in 20 min at 30 kPsi pressure cycling level. The fact that the NA2 peak (asialo-galactosylated biantennary glycan, trace H) did not appear in traces E, F and G further suggests no desialylation in any of these structures due to the high pressure. We anticipate that increase in the PNGase F molar ratio level in the reaction mixture would increase the reaction rate, therefore, leading to shorter deglycosylation time.
Table 2.
Measured and calculated glucose unit (GU) values of the Man 5 and transferrin glycan structures following the methods reported in ref 31, 32
| Glycan Structures | GU Units | |
|---|---|---|
| measured | calculated | |
| Man 5 | 6.92 | 6.82 |
| A2 | 4.80 | 4.60 |
| A2F | 5.01 | 4.94 |
| A1 | 6.48 | 6.52 |
| NA2 | 9.93 | 9.94 |
Structures are shown in Scheme 1.
Release of IgG glycans
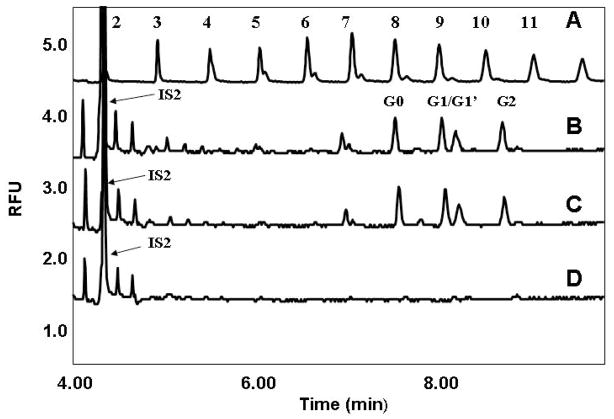
Immunglobulins (IgG) are a major class of biotherapeutics, and CE is one of the standard validated methods for their analysis due to its excellent resolving power and high separation speed. 12, 33 IgGs are large glycoproteins with a single conserved N-glycosylation site at Asn297 on both heavy chains in the CH2 domain of the Fc region. Analogous to the deglycosylation process of the other model glycoproteins, the IgG sample was thermally denatured in the presence of SDS, and the disulfide bonds were reduced prior to endoglycosidase digestion. The denaturation process resulted in some precipitation; however, this did not affect PNGase F mediated glycan release, which was carried out in the resulting suspension. We speculate that, under high pressure conditions, the precipitated aggregate partially disintegrated, resulting in more exposure and consequently improved access of the enzyme to the glycosylation sites. The samples were analyzed by CE-LIF using APTS labeled maltose as an internal standard (IS2). Figure 3 compares the electropherogram of the APTS labeled maltooligosaccharide ladder (trace A) with the APTS labeled IgG glycans released by PCT (30 kPa, 5 min, trace B), and by the standard 3 hours atmospheric deglycosylation (trace C), both at 37°C with 1:2500 enzyme:substrate molar ratio. The G0 (see Scheme 1) to IS2 (maltose) peak height ratios were 0.08 in both instances. Note that since the amount of internal standard was the same as in previous experiments, but the molar glycosylation ratio of IgG was significantly less (only ~2–3%), as expected, the peak height ratios were considerably lower. Trace D in Figure 3 depicts the control deglycosylation experiment at atmospheric pressure for 5 minutes showing essentially no glycan release. Figures 2 and 3 clearly demonstrate the accelerating effect of pressure cycling on PNGase F mediated digestion of glycoproteins. The rapid release of IgG glycans can be of particular interest in biopharmaceutical quality control/quality assurance analysis.
Figure 3.
Comparative CE analysis of APTS labeled released glycans from polyclonal human IgG using PCT and atmospheric deglycosylation with 1:2500 enzyme:substrate molar ratio. Traces: A) APTS labeled maltooligosaccharide ladder; B) IgG glycans released at 30 kPsi pressure cycling level for 5 min; C) IgG glycans released at atmospheric pressure for 3 hours; D) IgG glycans released at atmospheric pressure for 5 minutes. IS2: APTS derivatized maltose. All other conditions were the same as in Fig 2. Structures of G0, G1, G1′ and G2 are shown in Scheme 1.
CONCLUSION
In this paper, we have demonstrated that pressure cycling technology (PCT) can significantly improve carbohydrate analysis with respect to speed as well as endoglycosidase consumption during PNGase F – mediated release of N-linked glycans. We consider that the high pressure facilitated conformation changes of the target glycoprotein, which in conjunction with the alternation between high and atmospheric pressure probably resulted in rapid unfolding and folding, increasing the accessibility of the endoglycosidase to the cleavage sites. Pressure cycling technology with the use of 1:2500 enzyme:substrate molar ratio at 30 kPsi quantitatively released the asparagine linked glycans from bovine ribonuclease B and polyclonal human immunoglobulin (both having a single glycosylation site) in 5 minutes (compared to the industry standard of approximately 1:250 – 1:500 enzyme:substrate molar ratio in a 2 hours to overnight reaction). Large glycoproteins with multiple glycosylation sites, such as human transferrin, required longer (20 minutes) PCT treatment to obtain >95% glycan removal at the above enzyme to substrate level. A higher amount of enzyme could be applied to reduce the deglycosylation time. The target glycoproteins required reduction of the disulfide bridges and heat denaturation prior to PCT treatment. PCT assisted deglycosylation benefitted from the addition of a non-ionic detergent to the reaction mixture, which counteracted the possible inhibitory effect of SDS on the endoglycosidase activity. Importantly, pressure cycling did not appear to lead to any apparent loss of sialic acid residues. The microliter scale reaction volume and PCT assisted deglycosylation alleviated possible precipitation related issues that otherwise could represent a problem in integrated microcapillary systems. Importantly, PCT offers the possibility of simultaneous processing of 12 samples, thus high throughput glycan analysis can be envisioned by its combination with multicapillary electrophoresis systems 25. Although, we have only demonstrated the utilization of PCT assisted glycan release with electrophoresis, this method could obviously be used in conjunction with other well established carbohydrate characterization techniques of LC, MS and NMR.
Acknowledgments
This research was supported by NIH Grant GM 15847 (B.L.K.). The authors thank Dr Alex Lazarev of Pressure Bioscience for his assistance in the PCT measurements and Pressure Biosciences and PhyNexus for their gifts of equipment. Contribution No. 958 from the Barnett Institute.
References
- 1.Varki A, Cummings R, Esko J, Freeze H, Stanley P, Bertozzi CR, Hart GW, Etzler ME. Essentials of Glycobiology. 2. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2009. [PubMed] [Google Scholar]
- 2.Jefferis R. Expert Opin Biol Ther. 2007;7:1401–1413. doi: 10.1517/14712598.7.9.1401. [DOI] [PubMed] [Google Scholar]
- 3.Gennaro LA, Salas-Solano O. Anal Chem. 2008;80:3838–3845. doi: 10.1021/ac800152h. [DOI] [PubMed] [Google Scholar]
- 4.Li H, d’Anjou M. Curr Opin Biotechnol. 2009;20:678–684. doi: 10.1016/j.copbio.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 5.Townsend RR, Hotchkiss AT. Techniques in Glycobiology. Marcel Dekker; New York: 1997. [Google Scholar]
- 6.Takahashi N. J Chromatogr A. 1996;720:217–225. doi: 10.1016/0021-9673(95)00328-2. [DOI] [PubMed] [Google Scholar]
- 7.Ruhaak LR, Deelder AM, Wuhrer M. Anal Bioanal Chem. 2009;394:163–174. doi: 10.1007/s00216-009-2664-5. [DOI] [PubMed] [Google Scholar]
- 8.Bynum MA, Yin H, Felts K, Lee YM, Monell CR, Killeen K. Anal Chem. 2009;81:8818–8825. doi: 10.1021/ac901326u. [DOI] [PubMed] [Google Scholar]
- 9.Wuhrer M, de Boer AR, Deelder AM. Mass Spectrom Rev. 2009;28:192–206. doi: 10.1002/mas.20195. [DOI] [PubMed] [Google Scholar]
- 10.Rudd PM, Colominas C, Royle L, Murphy N, Hart E, Merry AH, Hebestreit HF, Dwek RA. Proteomics. 2001;1:285–294. doi: 10.1002/1615-9861(200102)1:2<285::AID-PROT285>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 11.Kang P, Mechref Y, Novotny MV. Rapid Commun Mass Spectrom. 2008;22:721–734. doi: 10.1002/rcm.3395. [DOI] [PubMed] [Google Scholar]
- 12.Ma S, Nashabeh W. Anal Chem. 1999;71:5185–5192. doi: 10.1021/ac990376z. [DOI] [PubMed] [Google Scholar]
- 13.Varki A, Freeze HH, Manzi AE. Overview of glycoconjugate analysis. 2009/08/19. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.www.prozyme.com; GK80115.
- 15.Sandoval VN, Arellano F, Arnott D, Raab H, Vandlen R, Lill JR. Int J Mass Spectrom. 2007;259:117–123. [Google Scholar]
- 16.Krenkova J, Lacher NA, Svec F. J Chromatogr A. 2009;1216:3252–3259. doi: 10.1016/j.chroma.2009.02.036. [DOI] [PubMed] [Google Scholar]
- 17.Palm AK, Novotny MV. Rapid Commun Mass Spectrom. 2005;19:1730–1738. doi: 10.1002/rcm.1979. [DOI] [PubMed] [Google Scholar]
- 18.Chicon R, Belloque J, Recio I, Lopez-Fandino R. J Dairy Res. 2006;73:121–128. doi: 10.1017/S0022029905001664. [DOI] [PubMed] [Google Scholar]
- 19.Ringham H, Bell RL, Smejkal GB, Behnke J, Witzmann FA. Electrophoresis. 2007;28:1022–1024. doi: 10.1002/elps.200600434. [DOI] [PubMed] [Google Scholar]
- 20.Belloque J, Chicon R, Lopez-Fandino R. J Agric Food Chem. 2007;55:5282–5288. doi: 10.1021/jf070170w. [DOI] [PubMed] [Google Scholar]
- 21.Meersman F, Dobson CM, Heremans K. Chem Soc Rev. 2006;35:908–917. doi: 10.1039/b517761h. [DOI] [PubMed] [Google Scholar]
- 22.Lopez-Ferrer D, Petritis K, Hixson KK, Heibeck TH, Moore RJ, Belov ME, Camp DG, 2nd, Smith RD. J Proteome Res. 2008;7:3276–3281. doi: 10.1021/pr7008077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guttman A, Chen FT, Evangelista RA, Cooke N. Anal Biochem. 1996;233:234–242. doi: 10.1006/abio.1996.0034. [DOI] [PubMed] [Google Scholar]
- 24.Guttman A. Nature (London) 1996;380:461–462. doi: 10.1038/380461a0. [DOI] [PubMed] [Google Scholar]
- 25.Laroy W, Contreras R, Callewaert N. Nat Protoc. 2006;1:397–405. doi: 10.1038/nprot.2006.60. [DOI] [PubMed] [Google Scholar]
- 26.Mozhaev VV, Lange R, Kudryashova EV, Balny C. Biotechnol Bioeng. 1996;52:320–331. doi: 10.1002/(SICI)1097-0290(19961020)52:2<320::AID-BIT12>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 27.Guttman A, Pritchett T. Electrophoresis. 1995;16:1906–1911. doi: 10.1002/elps.11501601314. [DOI] [PubMed] [Google Scholar]
- 28.Cano MP, Hernandez A, De Ancos B. J Food Sci. 1997;62:85–88. [Google Scholar]
- 29.Tanner MJ, Anstee DJ, Mallinson G, Ridgwell K, Martin PG, Avent ND, Parsons SF. Carbohydr Res. 1988;178:203–212. doi: 10.1016/0008-6215(88)80112-5. [DOI] [PubMed] [Google Scholar]
- 30.Spik G, Bayard B, Fournet B, Strecker G, Bouquelet S, Montreuil J. FEBS Lett. 1975;50:296–299. doi: 10.1016/0014-5793(75)80513-8. [DOI] [PubMed] [Google Scholar]
- 31.Guttman A, Herrick S. Anal Biochem. 1996;235:236–239. doi: 10.1006/abio.1996.0118. [DOI] [PubMed] [Google Scholar]
- 32.Guttman A, Ulfelder KW. J Chromatogr A. 1997;781:547–554. doi: 10.1016/s0021-9673(97)00724-3. [DOI] [PubMed] [Google Scholar]
- 33.Briggs JB, Keck RG, Ma S, Lau W, Jones AJ. Anal Biochem. 2009;389:40–51. doi: 10.1016/j.ab.2009.03.006. [DOI] [PubMed] [Google Scholar]