Abstract
A decade of structural genomics, the large-scale determination of protein structures, has generated a wealth of data and many important lessons for structural biology and for future large-scale projects. These lessons include a confirmation that it is possible to construct large-scale facilities that can determine the structures of a hundred or more proteins per year, that these structures can be of high quality, and that these structures can have an important impact. Technology development has played a critical role in structural genomics, the difficulties at each step of determining a structure of a particular protein can be quantified, and validation of technologies is nearly as important as the technologies themselves. Finally, rapid deposition of data in public databases has increased the impact and usefulness of the data and international cooperation has advanced the field and improved data sharing.
Keywords: international cooperation, protein structure, X-ray crystallography, nuclear magnetic resonance
INTRODUCTION
What is Structural Genomics?
Structural genomics is the large-scale determination of protein structures. In the late 1990s many structural biologists realized that major breakthroughs in technologies for structure determination, combined with the success of genome projects, laid a foundation for a systematic worldwide effort to determine the structures of proteins (12, 22, 42, 63, 66). Public and private funding agencies in the United States, Europe, and Japan sponsored workshops on what this new field might accomplish and how international cooperation could help (50, 53). This led to support for major structural genomics efforts in the United States [the NIH Protein Structure Initiative (PSI), http://www.nigms.nih.gov/Initiatives/PSI/], Europe (The Protein Structure Factory, http://www.proteinstrukturfabrik.de/, and SPINE, http://www.spineurope.org/), and Japan [The National Project on Protein Structural and Functional Analyses (Protein 3000) and RIKEN Structural Genomics/Proteomics Initiative; http://protein.gsc.riken.jp/]. Over the next decade many additional structural genomics efforts were started around the world, two of the largest of which are the Canadian-U.K.-Swedish Structural Genomics Consortium (http://www.sgc.utoronto.ca/) and the Japanese Targeted Proteins Research Program (http://www.tanpaku.org/). The TargetDB database at http://sg.pdb.org/target_centers.html has a current list of structural genomics efforts and their status, and a volume and a review devoted to structural genomics have recently been published (19, 59).
Technologies Forming a Foundation for Structural Genomics
Structural genomics is a technology-driven field. Key foundations for structural genomics included developments in macromolecular X-ray crystallography and NMR along with general systems for cloning and expression of proteins. In X-ray crystallography the increasing use of tunable synchrotron radiation in collecting anomalous diffraction data on protein crystals, the use of selenomethionine in crystallographic phase determination (68), automation of nanodroplet crystallization (11, 52), cryo-cooling techniques that reduced radiation damage to those crystals (30, 49), and automation of the structure determination process for macromolecular crystallography (47, 62) suggested that determining a protein structure once crystals were available would be increasingly straightforward. In the NMR field, the availability of cryo-probe high-field NMR devices (32), the development of multidimensional techniques for data acquisition (60), and the introduction of automated procedures for data analysis (8, 28, 43) similarly suggested that NMR structure determination was headed toward ever-higher throughput. Generalized systems for cloning and expression of proteins, particularly the addition of histidine tags at the N or C termini of recombinant proteins (48), were becoming increasingly used and allowed the use of generic techniques such as nickel-affinity chromatography, suggesting that standardized protocols might be developed that could be used for protein production in a factory-like setting.
Genomic Sequences and Structural Genomics
In parallel with these major technical developments, the availability of genomic sequences from dozens of organisms and the promise of sequences of many more over the following years were major stimulants for the idea of structural genomics. It was widely recognized that these genomic sequences could be used to identify sets of protein structures that would be highly informative, such as all the structures from a particular organism, representatives of all unique protein folds, representatives of all unique protein families, or all members of a biochemical pathway (12, 22, 35, 42, 63, 69) (http://www.thermus.org/e_index.htm). Combined with the technological advances, this led to a great deal of excitement about the idea of large-scale protein structure determination, or “structural genomics” or “structural proteomics” as it has variously come to be called.
Challenges in Structural Genomics
It was recognized early on that genomic sequencing was fundamentally different from structure determination because the diversity among proteins and their solubility and physical properties is much greater than that of fragments of DNA, so that the challenge in determining all protein structures from an organism was vastly more difficult than that of determining the genome sequence for that organism. Nevertheless, the idea of completeness and of structural coverage of all proteins was an important part of much of the thinking in structural genomics (50, 53).
The biggest challenges in structural genomics have largely been clear from the early days of the field: Production of proteins in a soluble form and crystallization (for X-ray crystallography) or suitability for production of high-quality spectra (for NMR) were bottlenecks in all structural biology laboratories. Moreover, only a few structures of membrane proteins had been determined by that time.
What was not known at the start of structural genomics was whether it would be possible to overcome bottlenecks in protein expression and structure determination for various classes of proteins, and whether large-scale centers could build pipelines that could routinely determine the structures of targeted proteins. The cost of structure determination by conventional means was generally estimated to be in the range of $100,000–$300,000 per protein structure, and it was anticipated that cost savings could be obtained by large-scale structure determination, but the extent of possible savings was unclear (20). Similarly, the importance of structures of proteins of unknown function was not known. The role of rapid deposition of data was not known but was generally expected to be a key one. It was generally expected that technology development would be a critical part of structural genomics (58), but it was not clear at the start of structural genomics how much development in this area would actually occur. Finally, at the start of structural genomics the importance of international collaboration was unclear, although experience with the genome projects suggested that cooperation would be highly beneficial.
WHAT CAN BE ACCOMPLISHED BY STRUCTURAL GENOMICS?
Table 1 lists key lessons from a decade of structural genomics. One of the most important of these is that it is indeed possible to construct large-scale facilities that can determine the structures of a hundred or more proteins per year. This has been accomplished in efforts around the world, including each of the current four U.S. PSI Large-Scale Centers [Midwest Center for Structural Genomics, http://www.mcsg.anl.gov/; Joint Center for Structural Genomics (JCSG), http://www.jcsg.org/; Northeast Structural Genomics Consortium, http://www.nesg.org; and the New York SGX Research Center for Structural Genomics, http://www.nysgrc.org/nysgrc], the Japanese RIKEN structural genomics effort and the Canadian-UK-Swedish Structural Genomics Consortium.
Table 1.
Lessons from structural genomics
| Lessons |
|---|
| 1. It is possible to construct large-scale facilities that can determine the structures of a hundred or more proteins per year. |
| 2. The difficulties at each step of determining a structure of a particular protein can be quantified. |
| 3. Structures from structural genomics can have an important impact on scientific research. |
| 4. Rapid deposition of data in public databases increases the impact and usefulness of the data. |
| 5. Technology development has played a critical role in structural genomics. |
| 6. Validation of technologies is nearly as important as the technologies themselves. |
| 7. Structures from structural genomics are of high quality. |
| 8. International cooperation advances the field and improves data sharing. |
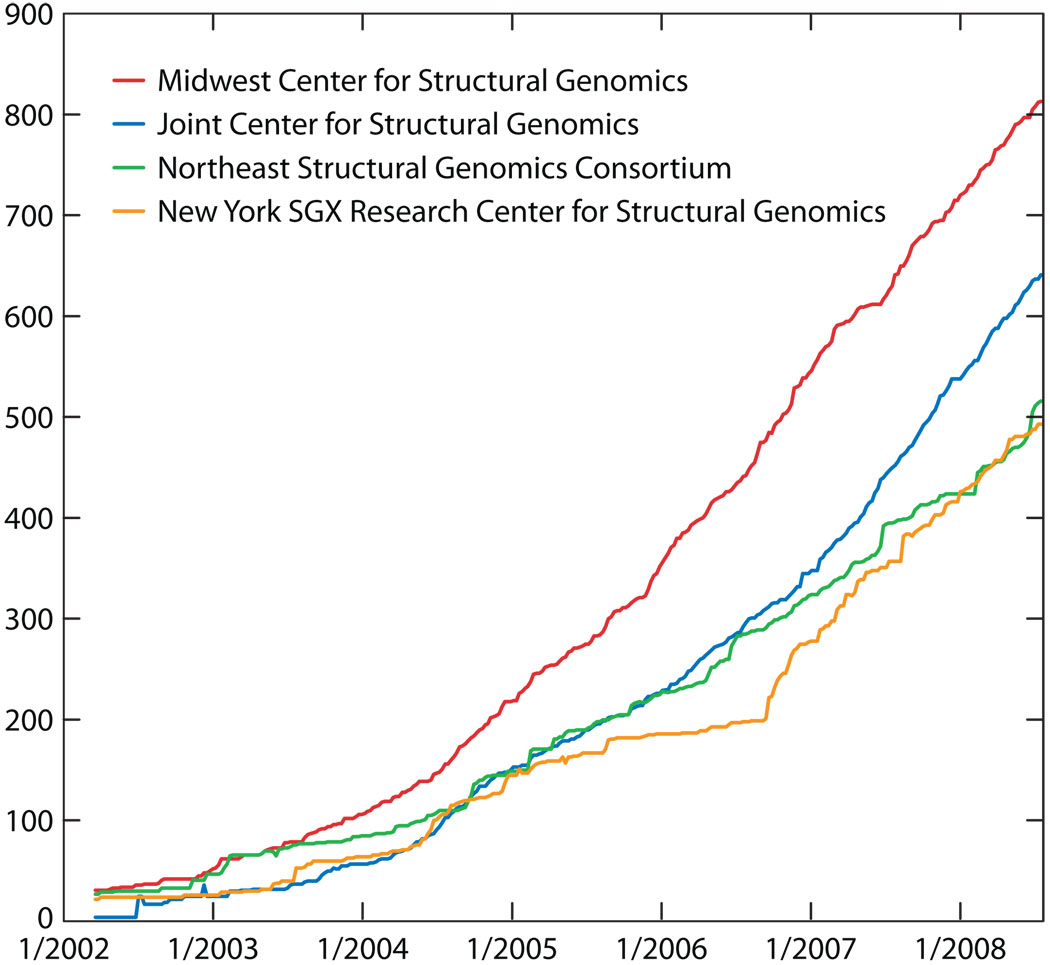
The U.S. PSI-2 Large-Scale Centers provide a good set of examples of what is possible when a concerted effort is made to produce a large number of structures from prokaryotic organisms (13). Each of the four Centers has been operating since 2001. The rates of depositing new protein structures into the Protein Data Bank (PDB) (4, 5) for these Centers have increased dramatically during this period, from an average of about 25 structures per year each in 2002 to an average of 155 each per year in 2007 (Figure 1). The corresponding cost per structure has decreased from about $500,000 to about $70,000, as calculated by dividing the total cost of the projects by the number of structures produced (13, 16).
Figure 1.
PDB depositions by NIH Protein Structure Initiative Large-Scale Centers (see http://www.mcsg.anl.gov for a current plot).
Similar rates of structure determination and costs have been achieved by RIKEN and by the Structural Genomics Consortium (SGC). During the fiscal years 2002–2006 RIKEN deposited 2675 structures into the PDB, with an average cost per structure of approximately $55,000. These structures included many small protein structures determined by NMR, as well as many eukaryotic proteins. Focusing on eukaryotic proteins, the SGC has deposited 750 structures into the PDB since 2003 (http://www.sgc.utoronto.ca/structures/target_progress.php), a particularly notable accomplishment as more than 600 of these structures were from human cells and included two membrane proteins. The cost per structure was approximately $135,000. Together RIKEN and the SGC account for over 50% of newly determined structures of human proteins.
Together, these structural genomics efforts have deposited (as of August 2008) some 6048 structures into the PDB (see http://targetdb.pdb.org/). These 6048 structures represent about 11.5% of all structures in the PDB. The successes of these large structural genomics projects show that it is indeed possible to create a pipeline for large-scale protein structure determination.
SUCCESS RATES: WHICH STRUCTURES WILL BE EASY TO DETERMINE AND WHICH WILL BE DIFFICULT?
A second major lesson from structural genomics is that the difficulties at each step of determining a structure of a particular protein can be quantified. There are two aspects of this type of analysis. One aspect is that the probability of overall success for a particular step or for obtaining a structure can be estimated. The second and more important aspect is that the relative probabilities of success for trying various approaches at a particular stage can be estimated. The development of ways to estimate the effectiveness of various approaches toward successful structure determination is a major step forward for structural biology because it allows a rational approach to choosing what methods to try for any particular protein.
Success Rates for Individual Steps in Structure Determination
One of the earliest impacts of structural genomics on the structural biology field was the ability to identify overall success rates for the major steps in structure determination. Early on, structural genomics efforts around the world agreed to post the status of their targeted structures on the TargetDB Web site maintained by the PDB (http://sg.pdb.org/). This allowed anyone to count, for example, the number of targets that were cloned, purified, crystallized, had NMR data collected, or were deposited into the PDB at any time. For the first time a quantitative measure of success rates for each step in structure determination was available. Table 2 shows the success rates for the major steps in structure determination as of July 2008. Each of these steps is (on average) successful from about one-third to two-thirds of the time, with the lowest success rate at the steps between expressing a protein and purifying it (which include obtaining the protein in a soluble form and successfully isolating the pure protein) and between purified protein and obtaining useful crystals or NMR spectra. This type of analysis has been important because it shows which steps need to be improved most (obtaining soluble protein, crystallization, and obtaining samples suitable for high-quality NMR spectra) and which are relatively successful (protein expression and structure determination).
Table 2.
Success rates for major steps in structure determination
| Status | Total number of targets | % Success (step) | % Success (overall) |
|---|---|---|---|
| Cloned | 125,316 | 100 | 100 |
| Expressed | 83,115 | 66.3 | 66.3 |
| Purified | 29,409 | 35.4 | 23.5 |
| Diffraction-quality crystals or NMR spectrum | 8,690 | 29.5 | 6.9 |
| In PDB | 5,811 | 66.9 | 4.6 |
A more sophisticated approach has been developed as well in which success rates, both overall and for individual steps, are analyzed as functions of the physical properties of proteins (where these physical properties are inferred from their amino acid sequences). Success rates are highly dependent on the isoelectric point, hydrophobicity, and propensity for disordered structure of a protein (17, 24, 55). Owing to this, protein sequences have been classified as optimal, suboptimal, average, difficult, and very difficult to get to the stage of crystallization (55). Such a classification can be used to estimate how difficult a particular structure would be to determine, but more importantly it can be used to decide the relative allocation effort of various proteins. For example, if an investigator is not concerned about the amino acid differences between a pair of proteins, then work can be focused on the one with the higher probability of success. Alternatively, a large-scale effort can continually reprioritize entire targeted sets of proteins on the basis of their current relative probabilities of success.
Identifying the Best Approach to Take at Each Step in Structure Determination
Prior to structural genomics efforts, every structural biology laboratory had accumulated in-house a set of techniques that could be applied to obtain soluble protein, crystals and X-ray data, or NMR data. The order in which these techniques might be applied, and the point at which a project might be abandoned, would largely depend on the personal experience of the investigator and anecdotal evidence gathered from the experiences of other investigators. Success in structure determination depended strongly on the ability of the investigator to synthesize this evidence and identify the best approaches to apply to the problem at hand. This paradigm is now changing because structural genomics provides the opportunity for a systematic evaluation of success rates for various approaches to overcoming bottlenecks.
One of the earliest applications of this approach was the identification of conditions likely to lead to protein crystallization. The thousands of proteins produced by structural genomics efforts allowed the evaluation of relative effectiveness of many precipitants and additives, leading to new standardized sets of screens for protein crystallization (14, 45, 46). Similar crystallization data, combined with information on the physical properties of proteins obtained from their amino acid sequences, also led to the ability to predict the overall probability of crystallization success depending on the isoelectric point or the hydrophobicity of a protein (14, 56).
An important use of systematic information about relative success rates for different approaches is in the choice of which approach to apply next after failure at some step in structure determination (36). For example, if soluble protein is not obtained upon expression of a cloned gene, possible choices might include changing the expression vector or host, expressing the protein in the presence of chaperones or cofactors, expressing domains, or engineering the protein sequence to increase solubility. A good way to decide which of these to try would be to balance the probability of success of each possible approach with the cost in time or effort of applying that approach, weighting the benefits and cost according to how important they are to the circumstance at hand. This type of approach is made possible by detailed analyses of the chances of success for a wide variety of approaches at each step of structure determination.
Synthesis of Experience from Structural Genomics Laboratories Around the World
The systematic analysis of chances of success at each step in structure determination has had qualitative practical outcomes in addition to the quantitative ones described above. The combined experience of a large group of structural genomics efforts has led to many protocols for carrying out these steps, including general protocols with recommendations for each step in structure determination (2, 21, 27).
VALUE OF STRUCTURES FROM STRUCTURAL GENOMICS
The next major lesson from structural genomics is that the structures that result from such a worldwide effort can have an important impact. From the start of structural genomics efforts it has been recognized that the choice of which protein structures to determine is a critical one. Many approaches have been suggested for choosing which proteins to target, and the structural genomics efforts around the world have had a range of emphases. The most fundamental choice has been whether to target proteins with identified biochemical or biological importance to provide structural information directly applicable to their functions, or to target proteins representative of large families of related proteins to provide a coarse level of structural information for the entire families.
The U.S. PSI-1 and PSI-2 have been largely targeted at determining structures of representative proteins from protein families with many members (http://www.nigms.nih.gov/Initiatives/PSI). Centers supported by PSI-1 also targeted proteins from Mycobacterium tuberculosis (the TB Structural Genomics Consortium, http://www.doe-mbi.ucla.edu/TB/) and from human parasites (the Structural Genomics of Pathogenic Protozoa Consortium; http://www.sgpp.org/) as well as complete coverage of the Thermotoga maritima genome (JCSG, http://www.jcsg.org). Two of the PSI-2 Specialized Centers are targeting membrane proteins (the Center for Structures of Membrane Proteins, http://csmp.ucsf.edu; and the New York Consortium on Membrane Protein Structure, http://www.nycomps.org) and one is focused on structures of eukaryotic proteins (the Center for Eukaryotic Structural Genomics, http://www.uwstructuralgenomics.org).
The SPINE-1 and SPINE-2 efforts targeted eukaryotic macromolecular structures and complexes relevant to human disease (http://www.spineurope.org; http://www.spine2.eu/SPINE2/), and the Protein Structure Factory effort targeted eukaryotic proteins. The RIKEN structural genomics efforts targeted coverage of the complete genome of Thermus thermophilus as well as structure determination of eukaryotic proteins with identified function. The SGC has targeted human proteins of therapeutic interest, and the Japanese Targeted Proteins Research Program has targeted protein structures for understanding fundamental biology, medical importance, food, and the environment.
New large-scale efforts in the United States are targeting proteins from a variety of human pathogens (the Center for Structural Genomics of Infectious Diseases, http://www.csgid.org; and the Seattle Structural Genomics Center for Infectious Diseases, http://ssgcid.org). Other efforts around the world have targeted proteins to identify their functions (the Structure 2 Function project, http://s2f.umbi.umd.edu/, and the Yeast Structural Genomics pilot project, http://genomics.eu.org/spip/).
Some of the structures determined by structural genomics projects were targeted on the basis of their importance and have had obvious individual impacts. These include, for example, structures of a protein secretion apparatus from Pseudomonas aeruginosa (44), structures of an aquaporin (64), and structures of hormone receptor-ligand interactions (9). Proteins from M. tuberculosis, targeted by the TB Structural Genomics Consortium, are of clear interest because of their potential as targets for antituberculosis therapeutics. This consortium has determined structures of 116 different proteins from M. tuberculosis (3), including at least 9 that are active targets for therapeutics in pharmaceutical companies (J. Sacchettini, personal communication).
Other proteins have been targeted in structural genomics efforts simply because they were proteins of unknown function and the investigators hoped that determination of their structures would hint at their biochemical and cellular functions (23, 54). In some cases, particularly those in which a ligand or cofactor has been discovered bound to a protein in crystals, this has been successful (38). In other cases the structures were similar to those of characterized proteins of known function, allowing information from the characterized proteins to be transferred (69) to the targeted proteins (26, 61). In still other cases, a combination of methods had to be applied to identify the function of a protein (51).
Efforts to determine the structures of all proteins from a microorganism, while not yielding completeness, have nevertheless given a picture of the range of structures involved in supporting a living organism (13, 31, 34, 35, 54) (http://www.thermus.org/e_index.htm). More importantly, they form a foundation for future efforts to develop detailed models of each step in metabolism, regulation, and other cellular functions. The availability of structural information is a prerequisite for atomic-level simulations of these processes and makes it possible to think about developing such comprehensive models.
Another important avenue through which structures from structural genomics efforts will have increasing impacts is by adding to the database of structures in the PDB. A useful way to look at this is that these structures are being determined now so that they will be available when they are needed. An example is the structure of TM0936 from T. maritima, determined by the JCSG, which provided key information for identification of the function of this protein by docking potential intermediates some five years later (29). A different type of example is that structures from structural genomics are being used as molecular replacement models for the determination of new proteins of identified interest. The contribution of structural genomics to the novel structures in the PDB has been increasing, with structural genomics efforts now determining about 50% of the structures that represent new protein families (15, 37, 40).
IMPORTANCE OF RAPID DEPOSITION OF DATA
The experience from structural genomics strengthens another important lesson made clear earlier by the genomic sequencing efforts. Rapid deposition of data in public databases vastly increases the impact and usefulness the data. In the case of structural genomics, the international community agreed at the outset (see http://www.isgo.org) to deposit the identities of structures that were targeted on the TargetDB Web site (http://sg.pdb.org/) and to continually update this target list with the status of each target. As discussed above, this has made analyses of success rates possible. Additionally, this target information has facilitated efforts to avoid duplication of effort, as a number of structural genomics groups have had a policy of scanning TargetDB on a regular basis and discontinuing work on structures that are solved or nearly solved (16).
The structural genomics community also agreed at the outset to deposit structures and raw data (e.g., structure factors for crystallographic data) into the PDB (http://www.pdb.org) promptly upon completion of the structures (with up to six months delay in exceptional cases). This rapid deposition of structural information has made the results of structural genomics efforts rapidly accessible to the broader community.
Beyond the simple listing of target status and deposition of the final structures and data, structural genomics efforts have made a systematic effort to deposit and make publicly accessible the methods used to produce proteins and determine their structures as well as to make materials such as expression clones generally available. For example, the U.S. PSI has developed a KnowledgeBase portal (http://kb.psi-structuralgenomics.org/KB/) that is intended to allow general access to data, flow charts of structure determination, methods, intermediate data files, structures, and interpretations of structures. Additionally, the JCSG and SGC (http://www.thesgconline.org/) have extensive annotations of structures available online. For SGC targets that have been deprioritized, the SGC Web site contains the methods used to determine structures and the availability of clones.
IMPORTANCE OF TECHNOLOGY DEVELOPMENT
It has been clear from the beginning of structural genomics efforts that technology development made structural genomics possible, that continued development would be necessary for the success of high-throughput structure determination, and that developments in structural genomics would be applicable to other areas of structural and general biology. One lesson from structural genomics is that technology development is indeed important. Another lesson is that the systematic validation of the utilities of new technologies is almost as important as the technologies themselves.
The high-throughput needs of structural genomics have spurred efforts to develop diverse sets of technologies. These include the development of high-throughput cell-free systems for protein expression (33, 65), methods for improving solubilities of proteins (67), highly parallel, small-volume screening systems for protein crystallization (6, 52), automation of X-ray data collection (18, 25, 57), and automated macromolecular structure determination procedures for X-ray crystallography (1, 41) and NMR (39). These developments have had an impact on all of structural biology. Because of the influence of structural genomics, a user of most beamlines for macromolecular X-ray crystal structure determination can now expect to have available a highly automated system that allows robotic mounting of crystals on the X-ray beam and automated screening of a set of crystals to find those that show the best diffraction. This has completely changed the strategy of data collection and has vastly increased the potential throughput of structure determination at X-ray beamlines.
QUALITY OF STRUCTURES FROM STRUCTURAL GENOMICS
At the start of structural genomics there was some concern that the quality of structures obtained from high-throughput efforts might be lower than that for structures determined in individual researchers’ laboratories. A powerful lesson from structural genomics is that in general the structures determined in structural genomics pipelines are similar in quality compared with those determined by nonstructural genomics research (10). For X-ray structures the quality is typically higher for structural genomics, whereas for NMR structures the quality is somewhat lower for structural genomics (7). In retrospect the high quality of structures from structural genomics is not surprising, as structural genomics projects can devote the necessary effort to develop standardized procedures for structure determination and quality control.
ROLE OF INTERNATIONAL COOPERATION
A final lesson from structural genomics is the importance of international cooperation, particularly during the early stages of the field. During the first several years of structural genomics efforts, a series of workshops were held, first on the feasibility and potential impact of structural genomics and later on data sharing and collaboration. The Argonne workshop in 1998 was a defining workshop for the field (53). It was attended by structural biologists, bioinformaticians, and representatives of funding agencies, and it helped identify the possible approaches to targeting proteins for structure determination along with the tools that were available and those that would be needed.
A set of meetings sponsored by the Wellcome Trust, the U.S. NIH, and the Japanese Ministry of Science was critical in defining the environment in which structural genomics was to be carried out. At these meetings, held in 2000 and 2001, a charter for the International Structural Genomics Organization (ISGO; http://www.isgo.org) was drafted. The participants agreed on the guidelines and principles of ISGO and that structural genomics efforts around the world would follow them. These guidelines included the rapid deposition of structural information with the raw data supporting them. The international nature of these agreements was critical in this process. Structural biologists in individual countries used it to convince their respective governments that as the rest of the world was going to agree to rapid deposition, so should they (31). The resulting agreements have been a key motivating force for target status reporting and rapid deposition for structural genomics efforts worldwide.
The atmosphere of collaboration fostered by the structural genomics community and the ISGO has led to international meetings on structural genomics (ICSG 2000 in Yokohama, Japan; ICSG 2002 in Berlin; ICSG 2004 in Washington, DC; ICSG 2006 in Beijing; ICSG 2008 in Oxford, U.K.; see http://www.isgo.org) and to workshops designed to share technologies such as cell-free expression and new methods in NMR structure determination.
PERSPECTIVES
Structural genomics is now a mature field, with highly successful large-scale centers around the world and thousands of structures determined. The lessons learned from structural genomics are important not only for this field but also for other fields. The fact that high-throughput centers have been developed that can successfully carry out the complicated and difficult task of macromolecular structure determination has implications for future efforts in other challenging fields, from proteomics to cell biology. The developments in methods for the identification of which structures will be feasible, the recognition of the importance of systematic validation of methods, and approaches for estimating the relative efficacy of different procedures are applicable to almost any field. The important roles of international cooperation, data and method sharing, and rapid data deposition are key lessons as well for all disciplines generating large amounts of data. Efforts worldwide have now shown that structural genomics is possible and practical and how it can be carried out. The future for structural genomics is to continue to apply this powerful approach to determine structures that are of both current and long-term interest, providing a foundation for understanding macromolecules whose biological roles are known now and for those whose roles will be identified in the future.
ACKNOWLEDGMENTS
The authors are grateful to the many colleagues around the world who initiated and developed the field of structural genomics. Los Alamos National Laboratory requests that the publisher identify this article as work performed under the auspices of the U.S. Department of Energy. Los Alamos National Laboratory strongly supports academic freedom and a researcher’s right to publish; as an institution, however, the Laboratory does not endorse the viewpoint of a publication or guarantee its technical correctness.
Glossary
- Structural genomics
large-scale determination of protein structures, typically using robotics in many steps of the process, often carried out by consortia of research efforts; also known as structural proteomics
- Structure determination
identifying the shape of a protein, normally represented by the coordinates of the nonhydrogen atoms in a model of the protein
- U.S. National Institutes of Health Protein Structure Initiative (U.S. NIH PSI)
PSI-1 was the first five-year program, devoted to pilot-scale initiatives, and PSI-2 included large-scale structural genomics efforts
- SPINE and SPINE-2
Structural Proteomics in Europe
- Crystallization
proteins can be crystallized by adding salts or other compounds to a solution containing the purified protein
- Production of proteins in a soluble form
to crystallize a protein or to obtain an NMR spectrum of a protein, usually the protein must first be purified, separating it from other proteins, and remain soluble, retaining its 3D shape and not aggregating
- Protein Data Bank (PDB)
an open repository of structural information on proteins
- NMR spectra
used to determine structures of proteins by identifying pairs of atoms that are close together in the structure
- Structural genomics pipeline
an integrated procedure for determining the structures of proteins
- ISGO
International Structural Genomics Organization
Footnotes
The U.S. Government has the right to retain a nonexclusive, royalty-free license in and to any copyright covering this paper.
DISCLOSURE STATEMENT
The authors are not aware of any biases that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Adams PD, Grosse-Kunstleve RW, Hung L-W, Ioerger TR, McCoy AJ, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 2.Alzari PM, Berglund H, Berrow NS, Blagova E, Busso D, et al. Implementation of semiautomated cloning and prokaryotic expression screening: the impact of SPINE. Acta Crystallogr. D. 2006;62:1103–1113. doi: 10.1107/S0907444906029775. [DOI] [PubMed] [Google Scholar]
- 3.Baker EN. Structural genomics as an approach towards understanding the biology of tuberculosis. J. Struct. Funct. Genomics. 2007;8:57–65. doi: 10.1007/s10969-007-9020-9. [DOI] [PubMed] [Google Scholar]
- 4.Berman HM, Battistuz T, Bhat TN, Bluhm WF, Bourne PE, et al. The Protein Data Bank. Acta Crystallogr. D. 2002;58:899–907. doi: 10.1107/s0907444902003451. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein FC, Koetzle TF, Williams GJB, Meyer EF, Brice MD, et al. Protein Data Bank—computer-based archival file for macromolecular structures. J. Mol. Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 6.Berry IM, Dym O, Esnouf RM, Harlos K, Meged R, et al. SPINE high-throughput crystallization, crystal imaging and recognition techniques: current state, performance analysis, new technologies and future aspects. Acta Crystallogr. D. 2006;62:1137–1149. doi: 10.1107/S090744490602943X. [DOI] [PubMed] [Google Scholar]
- 7.Bhattacharya A, Tejero R, Montelione G. Evaluating protein structures determined by structural genomics consortia. Proteins. 2007;66:778–795. doi: 10.1002/prot.21165. [DOI] [PubMed] [Google Scholar]
- 8.Bhavesh N, Panchal S, Hosur R. An efficient high-throughput resonance assignment procedure for structural genomics and protein folding research by NMR. Biochemistry. 2001;40:14727–14735. doi: 10.1021/bi015683p. [DOI] [PubMed] [Google Scholar]
- 9.Billas IML, Iwema T, Garnier JM, Mitschler A, Rochel N, Moras D. Structural adaptability in the ligand-binding pocket of the ecdysone hormone receptor. Nature. 2003;426:91–96. doi: 10.1038/nature02112. [DOI] [PubMed] [Google Scholar]
- 10.Brown EN, Ramaswamy S. Quality of protein crystal structures. Acta Crystallogr. D. 2007;63:941–950. doi: 10.1107/S0907444907033847. [DOI] [PubMed] [Google Scholar]
- 11.Brown J, Walter TS, Carter L, Abrescia NGA, Aricescu AR, et al. A procedure for setting up high-throughput nanolitre crystallization experiments. II. Crystallization results. J. Appl. Crystallogr. 2003;36:315–318. [Google Scholar]
- 12.Burley SK, Almo SC, Bonanno JB, Capel M, Chance MR, et al. Structural genomics: beyond the Human Genome Project. Nat. Genet. 1999;23:151–157. doi: 10.1038/13783. [DOI] [PubMed] [Google Scholar]
- 13.Burley SK, Joachimiak A, Montelione GT, Wilson IA. Contributions to the NIH-NIGMS Protein Structure Initiative from the PSI Production Centers. Structure. 2008;16:5–11. doi: 10.1016/j.str.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Canaves JM, Page R, Wilson IA, Stevens RC. Protein biophysical properties that correlate with crystallization success in Thermotoga maritima: maximum clustering strategy for structural genomics. J. Mol. Biol. 2004;344:977–991. doi: 10.1016/j.jmb.2004.09.076. [DOI] [PubMed] [Google Scholar]
- 15.Chandonia JM, Brenner SE. The impact of structural genomics: expectations and outcomes. Science. 2006;311:347–351. doi: 10.1126/science.1121018. [DOI] [PubMed] [Google Scholar]
- 16.Chandonia JM, Kim S-H, Brenner SE. Target selection and deselection at the Berkeley Structural Genomics Center. Proteins Struct. Funct. Bioinform. 2006;62:356–370. doi: 10.1002/prot.20674. [DOI] [PubMed] [Google Scholar]
- 17.Christendat D, Yee A, Dharamsi A, Kluger Y, Savchenko A, et al. Structural proteomics of an archaeon. Nat. Struct. Mol. Biol. 2000;7:903–909. doi: 10.1038/82823. [DOI] [PubMed] [Google Scholar]
- 18.Cohen AE, Ellis PJ, Miller MD, Deacon AM, Phizackerley RP. An automated system to mount cryo-cooled protein crystals on a synchrotron beamline, using compact sample cassettes and a small-scale robot. J. Appl. Crystallogr. 2002;35:720–726. doi: 10.1107/s0021889802016709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edwards A. Large-scale structural biology of the human proteome. Annu. Rev. Biochem. 2009;78 doi: 10.1146/annurev.biochem.78.070907.103305. In press. [DOI] [PubMed] [Google Scholar]
- 20.Fletcher L. Efforts to commercialize structural genomics may be limited. Nat. Biotechnol. 2000;18 doi: 10.1038/80215. 1036-36. [DOI] [PubMed] [Google Scholar]
- 21.Fox BG, Goulding C, Malkowski MG, Stewart L, Deacon A. Structural genomics: from genes to structures with valuable materials and many questions in between. Nat. Methods. 2008;5:129–132. doi: 10.1038/nmeth0208-129. [DOI] [PubMed] [Google Scholar]
- 22.Gaasterland T. Structural genomics: bioinformatics in the driver’s seat. Nat. Biotechnol. 1998;16:625–627. doi: 10.1038/nbt0798-625. [DOI] [PubMed] [Google Scholar]
- 23.Gilliland GL, Teplyakov A, Obmolova G, Tordova M, Thanki N, et al. Assisting functional assignment for hypothetical Haemophilus influenzae gene products through structural genomics. Curr. Drug Targets Infect. Disord. 2002;2:339–353. doi: 10.2174/1568005023342281. [DOI] [PubMed] [Google Scholar]
- 24.Goh C-S, Lan N, Douglas SM, Wu B, Echols N, et al. Mining the structural genomics pipeline: identification of protein properties that affect high-throughput experimental analysis. J. Mol. Biol. 2004;336:115–130. doi: 10.1016/j.jmb.2003.11.053. [DOI] [PubMed] [Google Scholar]
- 25.Gonzalez A, Moorhead P, McPhillips SE, Song J, Sharp K, et al. Web-Ice: integrated data collection and analysis for macromolecular crystallography. J. Appl. Crystallogr. 2008;41:176–184. [Google Scholar]
- 26.Graille M, Quevillon Cheruel S, Leulliot N, Zhou C, de La Sierra Gallay I, et al. Crystal structure of the YDR533c S. cerevisiae protein, a class II member of the Hsp31 family. Structure. 2004;12:839–847. doi: 10.1016/j.str.2004.02.030. [DOI] [PubMed] [Google Scholar]
- 27.Graslund S, Nordlund P, Weigelt J, Bray J, Gileadi O, et al. Protein production and purification. Nat. Methods. 2008;5:135–146. doi: 10.1038/nmeth.f.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guntert P. Automated NMR protein structure calculation. Prog. NMR Spectrosc. 2003;43:105–125. [Google Scholar]
- 29.Hermann JC, Marti-Arbona R, Fedorov AA, Fedorov E, Almo SC, et al. Structure-based activity prediction for an enzyme of unknown function. Nature. 2007;448:775–779. doi: 10.1038/nature05981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hope H. Cryocrystallography of biological macromolecules—a generally applicable method. Acta Crystallogr. B. 1988;44:22–26. doi: 10.1107/s0108768187008632. [DOI] [PubMed] [Google Scholar]
- 31.Iino H, Naitow H, Nakamura Y, Nakagawa N, Agari Y, et al. Crystallization screening test for the whole-cell project on Thermus thermophilus HB8. Acta Crystallogr. F. 2008;64:487–491. doi: 10.1107/S1744309108013572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kennedy MA, Montelione GT, Arrowsmith CH, Markley JL. Role for NMR in structural genomics. J. Struct. Funct. Genomics. 2002;2:155–169. doi: 10.1023/a:1021261026670. [DOI] [PubMed] [Google Scholar]
- 33.Kigawa T, Yabuki T, Matsuda N, Matsuda T, Nakajima R, et al. Preparation of Escherichia coli cell extract for highly productive cell-free protein expression. J. Struct. Funct. Genomics. 2004;5:63–68. doi: 10.1023/B:JSFG.0000029204.57846.7d. [DOI] [PubMed] [Google Scholar]
- 34.Kim S-H, Shin D-H, Kim R, Adams P, Chandonia J-M. Structural genomics of minimal organisms: pipeline and results. Methods Mol. Biol. 2008;426:475–496. doi: 10.1007/978-1-60327-058-8_32. [DOI] [PubMed] [Google Scholar]
- 35.Kuramitsu S, Kawaguchi S, Hiramatsu Y. Database of heat-stable proteins from Thermus thermophilus HB8. Protein Eng. 1995;8:964. doi: 10.1002/elps.11501601176. [DOI] [PubMed] [Google Scholar]
- 36.Lesley SA, Wilson IA. Protein production and crystallization at the Joint Center for Structural Genomics. J. Struct. Funct. Genomics. 2005;6:71–79. doi: 10.1007/s10969-005-2897-2. [DOI] [PubMed] [Google Scholar]
- 37.Levitt M. Growth of novel protein structural data. Proc. Natl. Acad. Sci. USA. 2007;104:3183–3188. doi: 10.1073/pnas.0611678104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liger D, Graille M, Zhou C-Z, Leulliot N, Quevillon-Cheruel S, et al. Crystal structure and functional characterization of yeast YLR011wp, an enzyme with NAD(P)H-FMN and ferric iron reductase activities. J. Biol. Chem. 2004;279:34890–34897. doi: 10.1074/jbc.M405404200. [DOI] [PubMed] [Google Scholar]
- 39.Liu GH, Shen Y, Atreya HS, Parish D, Shao Y, et al. NMR data collection and analysis protocol for high-throughput protein structure determination. Proc. Natl. Acad. Sci. USA. 2005;102:10487–10492. doi: 10.1073/pnas.0504338102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu J, Montelione GT, Rost B. Novel leverage of structural genomics. Nat. Biotechnol. 2007;25:849–851. doi: 10.1038/nbt0807-849. [DOI] [PubMed] [Google Scholar]
- 41.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution—from diffraction images to an initial model in minutes. Acta Crystallogr. D. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 42.Montelione GT, Anderson S. Structural genomics: keystone for a human proteome project. Nat. Struct. Biol. 1999;6:11–12. doi: 10.1038/4878. [DOI] [PubMed] [Google Scholar]
- 43.Moseley HN, Monleon D, Montelione GT. Automatic determination of protein backbone resonance assignments from triple resonance nuclear magnetic resonance data. Methods Enzymol. 2001;339:91–108. doi: 10.1016/s0076-6879(01)39311-4. [DOI] [PubMed] [Google Scholar]
- 44.Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, et al. Avirulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 2006;312:1526–1530. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Newman J, Egan D, Walter TS, Meged R, Berry I, et al. Towards rationalization of crystallization screening for small- to medium-sized academic laboratories: the PACT/JCSG plus strategy. Acta Crystallogr. D. 2005;61:1426–1431. doi: 10.1107/S0907444905024984. [DOI] [PubMed] [Google Scholar]
- 46.Page R, Grzechnik SK, Canaves JM, Spraggon G, Kreusch A, et al. Shotgun crystallization strategy for structural genomics: an optimized two-tiered crystallization screen against the Thermotoga maritima proteome. Acta Crystallogr. D. 2003;59:1028–1037. doi: 10.1107/s0907444903007790. [DOI] [PubMed] [Google Scholar]
- 47.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 48.Porath J, Carlsson J, Olsson I, Belfrage G. Metal chelate affinity chromatography, a new approach to protein fractionation. Nature. 1975;258:598–599. doi: 10.1038/258598a0. [DOI] [PubMed] [Google Scholar]
- 49.Rodgers DW. Cryocrystallography. Structure. 1994;2:1135–1140. doi: 10.1016/s0969-2126(94)00116-2. [DOI] [PubMed] [Google Scholar]
- 50.Sali A. Meeting on 100,000 protein structures for the biologist (Avalon, New Jersey, USA) Nat. Struct. Biol. 1998;5:1029–1032. doi: 10.1038/4136. [DOI] [PubMed] [Google Scholar]
- 51.Sanishvili R, Yakunin AF, Laskowski RA, Skarina T, Evdokimova E, et al. Integrating structure, bioinformatics, and enzymology to discover function: BioH, a new carboxylesterase from Escherichia coli. J. Biol. Chem. 2003;278:26039–26045. doi: 10.1074/jbc.M303867200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Santarsiero BD, Yegian DT, Lee CC, Spraggon G, Gu J, et al. An approach to rapid protein crystallization using nanodroplets. J. Appl. Crystallogr. 2002;35:278–281. [Google Scholar]
- 53.Shapiro L, Lima CD. The Argonne Structural Genomics Workshop: Lamaze class for the birth of a new science. Structure. 1998;6:265–267. doi: 10.1016/s0969-2126(98)00030-6. [DOI] [PubMed] [Google Scholar]
- 54.Shin DH, Hou J, Chandonia J-M, Das D, Choi I-G, et al. Structure-based inference of molecular functions of proteins of unknown function from Berkeley Structural Genomics Center. J. Struct. Funct. Genomics. 2007;8:99–105. doi: 10.1007/s10969-007-9025-4. [DOI] [PubMed] [Google Scholar]
- 55.Slabinski L, Jaroszewski L, Rodrigues APC, Rychlewski L, Wilson IA, et al. The challenge of protein structure determination–lessons from structural genomics. Protein Sci. 2007;16:2472–2482. doi: 10.1110/ps.073037907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smialowski P, Schmidt T, Cox J, Kirschner A, Frishman D. Will my protein crystallize? A sequence-based predictor. Proteins. 2006;62:343–355. doi: 10.1002/prot.20789. [DOI] [PubMed] [Google Scholar]
- 57.Snell G, Cork C, Nordmeyer R, Cornell E, Meigs G, et al. Automated sample mounting and alignment system for biological crystallography at a synchrotron source. Structure. 2004;12:537–545. doi: 10.1016/j.str.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 58.Stevens RC, Yokoyama S, Wilson IA. Global efforts in structural genomics. Science. 2001;294:89–92. doi: 10.1126/science.1066011. [DOI] [PubMed] [Google Scholar]
- 59.Sussman JL, Silman I. Structural Proteomics and Its Impact on the Life Sciences. Hackensack, NJ: World Sci.; 2008. [Google Scholar]
- 60.Szyperski T, Braun D, Banecki B, Wuthrich K. Useful information from axial peak magnetization in projected NMR experiments. J. Am. Chem. Soc. 1996;118:8146–8147. [Google Scholar]
- 61.Teplyakov A, Pullalarevu S, Obmolova G, Doseeva V, Galkin A, et al. Crystal structure of the YffB protein from Pseudomonas aeruginosa suggests a glutathione-dependent thiol reductase function. BMC Struct. Biol. 2004;4:5. doi: 10.1186/1472-6807-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr. D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Terwilliger TC, Waldo G, Peat TS, Newman JM, Chu K, Berendzen J. Class-directed structure determination: foundation for a Protein Structure Initiative. Protein Sci. 1998;7:1851–1856. doi: 10.1002/pro.5560070901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tornroth-Horsefield S, Wang Y, Hedfalk K, Johanson U, Karlsson M, et al. Structural mechanism of plant aquaporin gating. Nature. 2006;439:688–694. doi: 10.1038/nature04316. [DOI] [PubMed] [Google Scholar]
- 65.Vinarov DA, Lytle BL, Peterson FC, Tyler EM, Volkman BF, Markley JL. Cell-free protein production and labeling protocol for NMR-based structural proteomics. Nat. Methods. 2004;1:149–153. doi: 10.1038/nmeth716. [DOI] [PubMed] [Google Scholar]
- 66.Vitkup D, Melamud E, Moult J, Sander C. Completeness in structural genomics. Nat. Struct. Biol. 2001;8:559–566. doi: 10.1038/88640. [DOI] [PubMed] [Google Scholar]
- 67.Waldo GS, Standish BM, Berendzen J, Terwilliger TC. Rapid protein-folding assay using green fluorescent protein. Nat. Biotechnol. 1999;17:691–695. doi: 10.1038/10904. [DOI] [PubMed] [Google Scholar]
- 68.Yang W, Hendrickson WA, Crouch RJ, Satow Y. Structure of RNase H phased at 2 Å resolution by MAD analysis of the selenomethionyl protein. Science. 1990;249:1398–1405. doi: 10.1126/science.2169648. [DOI] [PubMed] [Google Scholar]
- 69.Yokoyama S, Hirota H, Kigawa T, Yabuki T, Shirouzu M, et al. Structural genomics in Japan. Nat. Struct. Biol. 2000;7 Suppl.:943–945. doi: 10.1038/80712. [DOI] [PubMed] [Google Scholar]