Abstract
Although the complex process of ribosome assembly in the nucleolus is beginning to be understood, little is known about how the ribosomal subunits move from the nucleolus to the nuclear membrane for transport to the cytoplasm. We show here that large ribosomal subunits move out from the nucleolus and into the nucleoplasm in all directions, with no evidence of concentrated movement along directed paths. Mobility was slowed compared with that expected in aqueous solution in a manner consistent with anomalous diffusion. Once nucleoplasmic, the subunits moved in the same random manner and also sometimes visited another nucleolus before leaving the nucleus.
INTRODUCTION
In eukaryotes, rRNA transcription and ribosome assembly take place in the nucleolus. Nascent ribosomes then exit the nucleolus and move into the cytoplasm. Multiple ribosomal proteins assemble with rRNA in the nucleolus and may facilitate proper processing of the pre-rRNA primary transcript (Fatica and Tollervey, 2002). Additionally, a significant number of nonribosomal proteins bind to these formative ribosomes in the nucleolus and also in the nucleoplasm after the nascent ribosomal subunits leave the nucleolus (Gadal et al., 2001; Kuersten et al., 2001; Nissan et al., 2002). The binding of particular proteins in a prescribed order is probably necessary for nucleocytoplasmic transport of the processed ribosomal subunits (e.g., Milkereit et al., 2001). The nuclear export of both the large and small ribosomal subunits has been shown to be dependent, at least indirectly, on the Ran-GTPase cycle and the exportin CRM1, and it is likely that CRM1-mediated export of 60S subunits requires the adaptor protein NMD3 (Moy and Silver, 1999; Ho et al., 2000; Gadal et al., 2001; Thomas and Kutay, 2003; Trotta et al., 2003). In situ hybridization experiments in fission yeast have indicated that rRNA may accumulate along short tracks when near the nuclear pores but nonetheless appears to exit from all the pores, not just those near the nucleolus (Léger-Silvestre et al., 1999). In situ hybridization studies in mammalian cells have primarily addressed the distribution of rRNA within the nucleolus (Huang, 2002) rather than the routes of extranucleolar rRNA traffic, because although high signal representing rRNA is routinely detected in both the nucleolus and the cytoplasm, nucleoplasmic signal which might represent ribosomes moving to the nuclear periphery is not easily detectable by in situ hybridization (Puvion-Dutilleul et al., 1991; Lazdins et al., 1997; J.C.R. Politz, unpublished observations). Therefore, very little is known about the spatial pattern or mechanism of rRNA movement from the nucleolus into the nucleoplasm and then to the nuclear pores for export (Cullen, 2000; Kuersten et al., 2001; Lei and Silver, 2002), although all three classes of eukaryotic RNAs, rRNA, mRNA, and pol III transcripts have been shown to exit from all the nuclear pores (Dworetzky and Feldherr, 1988; Pante et al., 1997; Mattaj and Englmeier, 1998).
In previous studies in live cells (Politz et al., 1998, 1999), we investigated the intranuclear movement of poly(A) RNA and found that a substantial fraction moves randomly throughout the interchromosomal space, even under conditions of ATP depletion. This suggested that these RNAs do not move along directed paths in the nucleoplasm. An electron microscopic study has demonstrated that a specific pol II transcript, the Balbiani ring 2 mRNA, also distributes randomly throughout the nucleoplasm after transcription (Singh et al., 1999; Daneholt, 1999). These findings opened the possibility that other classes of RNA may also move freely throughout the nucleus (Politz and Pederson, 2000) and led us to investigate the movement of rRNA out of the nucleolus and into the nucleoplasm. rRNA makes up >80% of total cell RNA and ∼104 ribosomal subunits are synthesized and transported per minute in growing mammalian cells (Lewis and Tollervey, 2000; Kuersten et al., 2001). Thus, its abundance makes rRNA an attractive target for tracking studies. Equally important, we reasoned that since the site of rRNA transcription, the nucleolus, can be readily identified microscopically, the movement of transcripts could be followed away from their known birth site.
Using our previously developed method to follow the movement of endogenous RNAs, which uses complementary oligodeoxynucleotides labeled with caged fluorochromes as hybridization tags (Politz et al., 1999, 2003; Politz, 1999), we have followed the movement of 28S rRNA out of the nucleolus and into the surrounding nucleoplasm in cultured rat myoblasts. To our knowledge, these are the first experiments in which the movement of a specific endogenous RNA has been directly observed in the nucleus. We found that the signal moved out from the nucleolus in all directions to fill the nucleoplasmic space, in a manner characteristic of diffusion. Once nucleoplasmic, the tagged ribosomal subunits still exhibited random movement, and sometimes revisited nucleoli.
MATERIALS AND METHODS
Cell Growth and Oligo Uptake
L6 rat myoblasts were grown in Dulbecco's modified Eagle's minimum essential medium (DMEM) with 10% fetal bovine serum on 25-mm round glass coverslips (placed in 35-mm dishes) to ∼60% confluency using standard tissue culture techniques. A mixture of the five fluorescently labeled oligos or caged (prefluorescent) oligos listed below were then introduced to cells using Lipofectamine 2000 (Invitrogen Corp., Carlsbad, CA) according to manufacturer's instructions (6 μl/35-mm dish and a final total oligo concentration of 0.2 μM in OptiMEM). After 2 h, the medium was replaced with fresh DMEM (with serum) and the cells were incubated for another 30 min to 1 h. Immediately before imaging, the medium was changed to Leibovitz's L15 medium (Life Technologies, Rockville, MD) with 10% serum.
Oligodeoxynucleotides complementary to 28S rRNA were as follows (see Gerbi, 1996 and DeRijk et al., 1999; for database and nomenclature information also see Politz et al., 2002): Oligo 1 in loop E11_1 (D7b): G*TACCGGCAC*GGACGCC*CGCGGCGCCCA*C; Oligo 2 in loop E9_1 (D-7a): C*GAGGGCAACGGAGGCCA*CGCCCG*CCCT*C; Oligo 3 in loop B13_1 (D1): G*ACGCCACAT*TCCCGCGCC*CGGCGCGCG*C; Oligo 4 in loop C1_1 (D2): C*CGCGCCGCCGGG*TCAATCC*CCGGGCGG*C; and Oligo 5 in loops H1_2, H1_3 (D12): A*GGCTC*CCGCACCGGACCCCGG*CCCGAC*C, where the asterisk indicates positions of aminohexyl-modified thymidine residues coupled during synthesis (Integrated DNA Technologies, Iowa City, IA).
HPLC-purified oligos were labeled with either fluorescein or caged-fluorescein (caged-fl; CMNB2AF in Mitchison et al., 1994) as described by Politz and Singer (1999).
In Situ Transcription and In Situ Hybridization
In situ reverse transcription to detect hybridization of rDNA oligos was performed as described (Politz and Singer, 1999) except for the following. In addition to digoxigenin-labeled dUTP, biotin-labeled dATP and dCTP were added to the reverse transcription mix to a final concentration of 50 μM each. Also, incubation with a mixture of antidigoxigenin and antibiotin antibodies (1:250 dilution of each) was at 4°C overnight in a humidified chamber, and 0.5% normal sheep serum and 0.5% normal goat serum instead of 1% BSA was used in the washes before and after antibody binding. In situ hybridization experiments were performed exactly as described (Politz et al., 2002).
Imaging Microscopy and Processing
A rapid wide-field epifluorescence imaging system previously described (Rizzuto et al., 1998; see also Politz et al., 2003) was used to photolytically uncage and follow the movement of the oligo-tagged 28S rRNA as previously described for poly(A) RNA tracking experiments (Politz et al., 1999). Briefly, the caged-fl oligos taken up by living cells were uncaged by a 65-msec exposure to an argon laser beam (λ = 360 nm) directed through a pinhole inserted into the epifluorescence optical path and focused to a 1–2-μm diameter spot in either the nucleolus or the nucleoplasm. The 360-nm power flux was ∼15 W/μm2. The uncaged fluorescein was then excited with 488 nm light from an argon/krypton laser, and either 2D time series (taken every 500 msec) or time series of 3D stacks for restoration (31 planes, 0.25-μm focus shift, repeated every 500 msec) were captured. Cells were not visibly changed or damaged by the photoactivation and imaging protocol. Image analysis, including diffusion coefficient calculations, determination of % uncaged signal remaining at the site, and constrained interative deconvolutions were all performed as previously described (Cardullo et al., 1991; Carrington et al., 1995; Politz et al., 1999).
RESULTS
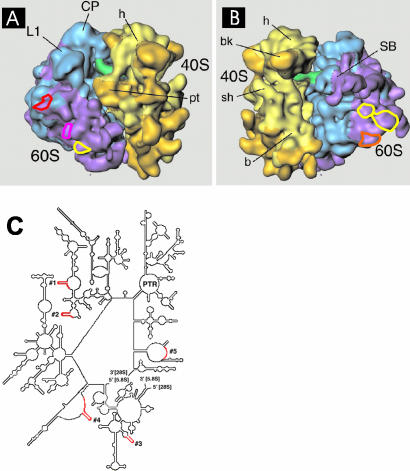
Oligodeoxynucleotides complementary to particular regions of rat 28S rRNA were used as hybridization probes to follow the movement of 28S rRNA within the nucleus of living cells. Target 28S rRNA sequences were selected based on several criteria. First, regions were chosen within expansion sequences, i.e., regions not present in prokaryotic 23S rRNA (Gerbi, 1996; Dube et al., 1998), to decrease the likelihood that the hybridized oligos would lie at functional sites of the 60S subunit. The majority of these expansion sequences lie at sites on the 60S subunit that are oriented away from the interface with the 40S subunit (see Beckman et al., 2001). Second, within these regions, the selected sequences were ones thought to be near the surface of the ribosome, i.e., ones containing nuclease sensitive sites or subject to chemical modification in whole ribosomes, and/or sequences near a binding site for a ribosomal protein known to localize to the ribosomal surface (Han et al., 1994; Holmberg and Nygård, 1997; Dube et al., 1998; Lieberman and Noller, 1998), but yet not near the face that contacts the small subunit (Holmberg et al., 1994b; Merryman et al., 1999). In this way we reasoned that the oligos might find their targets on both incompletely assembled ribosomal precursors as well as on nascent 60S subunits in the nucleolus. Oligos 1 and 2 are targeted to expansion sequences near the binding site for ribosomal protein L25, which is thought to be near the surface of the ribosome. No other proteins have been shown to bind to these particular regions and an antisense oligo targeted to a nearby region was shown to accumulate in nucleoli of mouse cells (Paillasson et al., 1997). Oligo 3 is targeted to an expansion region near the 5′ end of the 28S rRNA molecule where an insertion has been shown to be viable in yeast (Musters et al., 1989). Oligo 4 is targeted to a region of the large highly variable expansion sequence D2 (Gerbi, 1996), where insertion of a marker sequence in Tetrahymena is known to be viable (Sweeney et al., 1996). Oligo 5 is targeted to a region in the expansion sequence D12 that is known to be available for chemical modification in mouse ribosomes, indicating that this region may be uncovered near the surface of the ribosome (Holmberg et al., 1994a). Also, an insert at this site is viable in Tetrahymena (Sweeney et al., 1996). Figure 1, A and B, shows the approximate location of these expansion sequences on the surface of the ribosome. Figure 1C shows the five hybridization sites on the folded 28S rRNA molecule.
Figure 1.
Targeted hybridization sites on the 60S ribosomal subunit. (A and B) Colored circles show the locations of the expansion sequences for which complementary oligonucleotides probes were designed. (A) Yellow: oligo 1; pink: oligo 2; red: oligo 3. (B) Yellow: oligo 4; orange: oligo 5 (see MATERIALS AND METHODS for sequences of each oligo). The cryo-EM maps of the S. cerevisiae 80S ribosome are taken from Spahn et al. (2001) and reproduced with permission of CELL Press (Cambridge, MA). (C) 28S secondary structure map (yeast) showing regions targeted for oligo hybridization in red.
Oligodeoxynucleotides complementary to these five regions of 28S rRNA, each 33 nucleotides in length, were synthesized with four, approximately evenly spaced aminohexyl-modified thymidines (see MATERIALS AND METHODS), and these sites were labeled with fluorescein as described (Politz and Singer, 1999; Politz et al., 2002). RNA hybrids formed with oligos labeled in this manner are less susceptible to degradation in vivo, perhaps because the evenly spaced aminohexyl arms interfere with RNase H binding (Ueno et al., 1997; J.C.R. Politz, unpublished results).
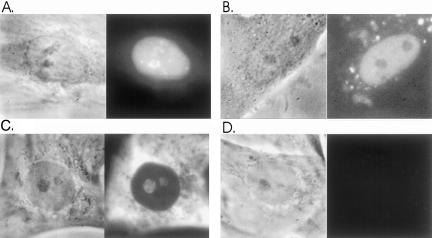
Rat L6 myoblasts were allowed to take up a Lipofectamine-bound mixture of all five oligos for 2 h. The medium was changed and after 1 h, cells were examined using digital imaging microscopy on a microscope stage maintained at 37°C (see MATERIALS AND METHODS). Fluorescent signal representing the rRNA oligos was found in the nucleus of transfected cells and additionally was often concentrated in the nucleolus (Figure 2A). Signal was also present in the cytoplasm at lower levels (unpublished data). In parallel experiments it was observed that oligo(dT) or oligo(dA), or oligos containing repeating CTG or CAG sequences did not concentrate in the nucleolus, and in fact, appeared to be excluded from the nucleolus (Figure 2B and our unpublished results). In standard in situ hybridization experiments with fixed cells, the 28S antisense oligos generated signal in the nucleolus and the cytoplasm as expected (Figure 2C; see also Politz et al., 2002), whereas only background levels of signal were detected with control oligos (Figure 2D).
Figure 2.
Live cell uptake and in situ hybridization of anti-28S rRNA oligos in L6 myoblasts. Cells were allowed to take up Lipofectamine-bound fluorescein-labeled oligos for 2 h as described in MATERIALS AND METHODS. Cells were incubated in fresh medium for 1 h and then examined using digital imaging microscopy. (A) Phase and fluorescent images of live cell nucleus after uptake of the five anti-28S rRNA oligos. Cytoplasmic signal is not visible because the image is scaled so that nuclear detail can be seen. (The bright nuclear spots are not SC35 rich speckles [our unpublished results] and are too plentiful to be Cajal bodies; similar bright spots are observed after oligo(dT) uptake.) (B) Phase and fluorescent images of a live cell nucleus that has taken up, as a control oligo, a 33mer repeat of CAG labeled in the same way as the oligos in A. (C) Phase image and in situ hybridization of anti-rRNA oligos to fixed cells. (D) Phase image and in situ hybridization of CAG control oligos (see B) to fixed cells.
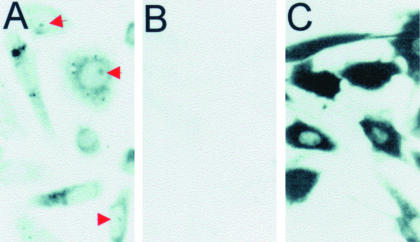
We next used an in situ reverse transcription assay (Eberwine et al., 1992; Politz et al., 1995) to confirm that the antisense oligos were hybridized to their target 28S rRNA regions in live cells. Cells were again allowed to take up oligo, the medium was changed (see MATERIALS AND METHODS) and the cells were fixed. Hybridized oligo was detected using an in situ reverse transcription reaction. This assay takes advantage of the fact that only hybridized oligo can act as a primer for incorporation of labeled dNTPs by reverse transcriptase; whereas unhybridized oligo cannot (Politz et al., 1995). Figure 3A shows that signal representing rDNA oligo hybridization was observed in the cytoplasm, and in many cases, also in the nucleolus (red arrows). Only background levels of signal were observed in cells that were not exposed to these oligos (Figure 3B). At higher magnification, the intranucleolar pattern of hybridization appeared generally similar to the fluorescence pattern we observe in live cells, with certain small lobules within the nucleolus showing the most intense signal (unpublished data).
Figure 3.
Detection of anti-rRNA oligo hybridization after cellular uptake. Cells were allowed to take up either anti-rRNA oligos (A) or oligo(dT) (C) and were then subjected to in situ reverse transcription to detect sites of oligo hybridization (Politz, 1999; Politz et al., 1999; see also MATERIALS AND METHODS). This assay exploits the fact that only hybridized oligo will prime reverse transcription and incorporation of labeled nucleotides; unhybridized oligo will not. Dark signal represents sites at which incorporated label, and thus hybridized oligo, is detected. Red arrows in A point to label present in nucleoli. (B) Results when the cells were not exposed to oligo.
The mixture of all five antisense oligos was next labeled with caged fluorescein (caged-fl; Mitchison et al., 1994) and introduced into cells as before. The caging groups are two o-nitrobenzyl moieties covalently linked to fluorescein via photolabile ether bonds. These groups chemically lock the fluorochrome in its nonfluorescent tautomer until photolysis releases the caging groups (Mitchison et al., 1994; Politz, 1999; Politz et al., 2003). The caged-fl rDNA oligos, hybridized to 28S rRNA in the cell, were uncaged in a small 1–2-μm diameter spot using a 360-nm wavelength laser line that was directed through a pinhole and then into the microscope objective. The movement of the resultant fluorescent rRNA was followed as it moved out from the uncaging spot and the 2D signal distribution was recorded every 500 msec using high-speed digital microscopy (Rizzuto et al., 1998; Politz et al., 1999, 2003). Unless otherwise noted, cells were kept at 37°C throughout the experiment.
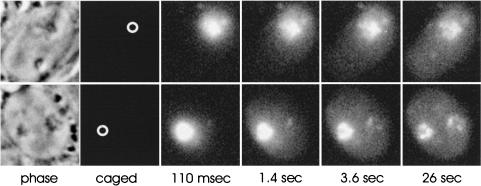
Before uncaging, only background levels of fluorescence were detected. When the uncaging beam was directed to nucleoli (which were visualized using phase contrast, Figure 4, top left, uncaging site circled), the resulting signal was observed to move out in all directions from the nucleolus, and a portion of the signal reached the nuclear periphery by 3.6 s. This pattern of movement, out in all directions from the nucleolar site of uncaging, was consistently observed in >100 cells examined (see also video supplement to Figure 4). No evidence of linear paths of signal moving toward a subset of nuclear pores was observed. However, in some cases, a progressive accumulation of signal at a second nucleolus (that was not uncaged) was observed (Figure 4, bottom panels). We ascertained that uncaged signal was distributed inside the nucleolus and throughout the nucleoplasm in three dimensions by optically sectioning cells after uncaging and subjecting the resulting image stacks to iterative deconvolution analysis (Carrington et al., 1995). Uncaged signal appeared in all midplanes at all time points, indicating that uncaged signal was distributed throughout the interior of the nucleolus as well as throughout the entire nucleoplasm (unpublished data).
Figure 4.
Movement of 60S subunits out from the nucleolus. Cells were allowed to take up a mixture of the five caged-fluorescein labeled anti-rRNA oligos as described in MATERIALS AND METHODS. After a 1-h incubation in fresh medium, oligo in the nucleolus was then uncaged and the movement of the signal was tracked over time using high-speed digital imaging techniques (see MATERIALS AND METHODS). The signal moved out in all directions from the nucleolus (top panel) and in some cases the uncaged nucleolar signal moved to the other nucleolus in the nucleus (bottom panel). See also supplemental video.
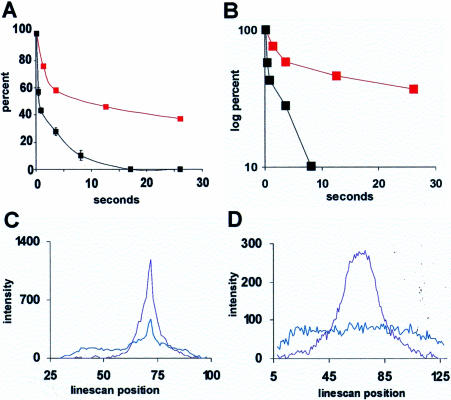
In a typical experiment, an average of 63% (range 35–72%) of the signal left the nucleolus (Figure 5A) within the 30 s observation period. In contrast, the unhybridized control oligo(dA) left the site much more rapidly; the vast majority was dispersed by 5 s (Figure 5A). The semilog plot in Figure 5B more clearly illustrates the different rates of departure of the control oligo(dA) and the considerably more slowly-moving hybridized rRNA oligos. To analyze the pattern of the rRNA signal movement from the site in more detail, pixel intensities were measured along lines drawn across the nucleus and the nucleolar uncaging site at the various time points (example in Figure 5C). Signal moved away from the site in a Gaussian distribution, indicative of random movement away from the nucleolus (broad shoulders on blue line in Figure 5C). A fraction of signal stayed at the nucleolar uncaging site for the duration of the assay period and was often represented by a peak in the center of the plot (Figure 5C, blue line).
Figure 5.
Measurement of signal intensity after uncaging. (A) The red curve shows the average percentage of uncaged anti-rRNA oligos remaining at a nucleolar uncaging site compared with the black curve showing the percentage of a nonhybridizing control oligo (oligo(dA); see Politz et al., 1999) remaining after uncaging in the nucleoplasm (control oligos do not localize to the nucleolus). The red curve here represents the average of 11 cells uncaged in one experiment, and similar results were obtained in four other experiments. (B) Same data as in A on a semilog plot. (C) Pixel intensities measured along a line drawn across the nucleus and the nucleolar uncaging site showing a Gaussian distribution of signal intensity at 110 msec (purple line) and 30 s (blue line) after uncaging. (D) Line intensity plot through center of an uncaged spot in nucleoplasm at 110 msec (purple) and 30 s (blue) after uncaging. The spatial distribution of signal after nucleoplasmic uncaging was similar at both 37 and 23°C.
We also measured the movement of 28S rRNA signal within the nucleoplasm (by uncaging away from nucleoli) in similar experiments and again found that the signal moved out from the uncaging site in all directions in a Gaussian profile to fill the nucleoplasm (Figure 5D), with no evidence for directed tracks of signal moving away from the site. We also sometimes observed that a portion of signal uncaged in the nucleoplasm subsequently became concentrated in nucleoli (unpublished data).
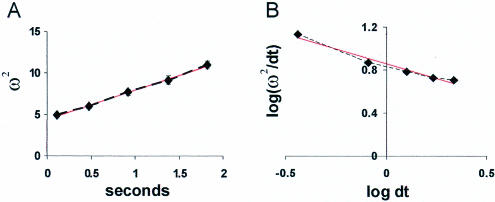
To measure the mobility of the 60S subunits as they moved from the nucleolus into the nucleoplasm, we calculated the mean square displacement of signal (as ω2, the mean square Gaussian width of the signal distribution; see Cardullo et al., 1991) at different times after uncaging at a nucleolus and plotted the results vs. time. As shown in Figure 6A, ω2 varied linearly with time, as expected for a diffusive process. The slopes of these plots predict that 60S ribosomal subunits move away from the nucleolus with an average apparent diffusion coefficient of 0.31 μm2/s (SD, ± 0.15 μm2/s). The nonhybridizing oligo(dA) reached the nuclear membrane too rapidly to allow measurement of a diffusion coefficient using this method; however, we earlier had estimated it to be ∼26 μm2/s using fluorescent recovery after photobleaching (Politz et al., 1998).
Figure 6.
Mobility and anomalous diffusion characteristics of 60S subunits. The pixel intensity along lines drawn through the center of a nucleolar uncaging site was plotted at different times after uncaging (i.e., Figure 5C). The mean square displacement (ω2) over time was then calculated as the average of the radius of the uncaged signal distribution measured at the points at which the intensity had fallen to e–2 of the maximum intensity within the uncaged spot (because the signal is distributed in a Gaussian). (A) Values from a typical experiment plotted vs. time. In this experiment, the apparent average diffusion coefficient estimated from the slope of the line (slope = 8D; see Cardullo et al., 1991) was 0.44 μm2/s. (B) Log-log plot of ω2/dt vs. dt shows anomalous diffusion. Values from the experiment shown in A fall on a straight line with slope of 2/dw – 1 (see Saxton, 1994), where the anomalous diffusion exponent dw equals 4.5. The red lines show a linear least-squares fit to each plot.
Biological processes that involve the consumption of metabolic energy typically display rate differences of 2.0–3.0-fold over a decade of temperature. When the same experiments and analyses as shown in Figures 4 and 5 were repeated at 23°C, rather than at 37°C, approximately the same fraction of signal left the nucleolus during the 30-s assay period, and a similar average apparent diffusion coefficient was observed (0.34 μm2/s, SD, ±0.35 μm2/s). This similar mobility at both 23 and 37°C suggests that the rate of 28S rRNA movement from the nucleolus is not metabolic energy-dependent.
Because the diffusion coefficient measured here was much slower than that predicted for a 60S subunit diffusing in aqueous solution (which we calculate to be ∼10 μm2/s), and because about one third of the uncaged signal did not leave the uncaging site during the assay period, we considered the possibility that the diffusion of the 60S subunits was slowed by collisions and/or retention within nuclear barriers or structures (e.g., chromatin) and therefore was more properly regarded as the phenomenon known as anomalous diffusion. When the log (ω2/dt) is plotted vs. log dt, the degree of anomalous diffusion can be determined, and information about the obstacle concentration is also obtainable in some cases (Saxton, 1994, 2001; Platani et al., 2002). We found that 60S subunit diffusion was indeed anomalous in the nucleoplasm; the log-log plots were linear with a very steep slope (Figure 6B), instead of the zero slope that would be seen with unconstrained diffusion. Anomalous diffusion exponents calculated from these curves were very large (range 4.5–20.7, whereas the exponent in normal diffusion is 2), which indicates that the concentration of diffusion obstacles in the nucleoplasm is very high.
DISCUSSION
We have analyzed the movement of a specific endogenous RNA in the nucleus of live cells. Using oligos labeled with caged fluorochromes as hybridization tags, we followed the movement of 28S rRNA (presumed to be in 60S ribosomal subunits) during transit from the nucleolus into the nucleoplasm. We found that signal moved away from the nucleolus and into the nucleoplasm in all directions in a random manner, with the characteristics of diffusion. It is noteworthy that the transport of rRNA within the nucleolus, i.e., before egress into the nucleoplasm, appears to be nonrandom when analyzed at the electron microscopic level of resolution (Thiry et al., 2000). This degree of intranucleolar spatial heterogeneity would not have been expected to be detectable in the present experiments, and our results are not at all incompatible with these previous findings.
It is important to emphasize that the targeted rRNA in these experiments was the endogenous RNA. It was not transcribed from a plasmid, modified with reporter sequences or microinjected. Although each of these methods offers useful opportunities to learn about the behavior of RNA in cells (Pederson, 2001a; Chartrand et al., 2001), the method used here allows the direct observation of the behavior of an endogenous RNA which has been transcribed and complexed with all required cellular proteins at the proper time, in the true native setting. Therefore, the movement and localization patterns of the RNA revealed using this technique are very likely to reflect the actual biological situation.
Using this method, it is important that the oligos used as tags hybridize to the target RNA but do not interfere with its normal behavior and that the hybridized signal can be distinguished from free oligo. As detailed in RESULTS, the oligos were chosen to hybridize to sequences within the eukaryotic expansion regions of the 28S rRNA molecule (sequences not present in prokaryotic ribosomes) and which additionally have been shown or predicted to be nonessential for eukaryotic ribosome function. It is improbable, therefore, that these oligos interfere with normal ribosome activity in live cells.
To ensure that the signal we were tracking represented the targeted rRNA molecules, we established that the oligo tags were actually hybridized to the RNA inside the live cell. After oligo uptake and a change of medium to wash out excess oligo (see MATERIALS AND METHODS), we used an in situ reverse transcription assay to reveal sites of hybridized oligo and found that oligo which had concentrated in the nucleolus was indeed hybridized to RNA. This assay does not allow the determination of the percent of total oligo hybridized, but earlier work (Politz et al., 1995) had determined that a 30-min incubation in fresh medium after oligo uptake allowed the majority of free oligo to become displaced from the cell surface. Additionally, any free oligo that might remain at the uncaging site does not interfere with measurements of RNA movement because it rapidly disperses to undetectable levels in a few seconds (Figure 5, A and B; Politz et al., 1999) so only hybridized (slower moving) oligos are tracked. Finally, it should be noted that there is no detectable population of naked 28S rRNA in the nucleus (e.g., Warner and Soeiro, 1967), making it very probable that the signal we are tracking represents bona fide 60S subunits.
In the experiments described here, the uncaged signal moved away from the nucleolus and filled the nucleus in <10 s. This moving population of 60S ribosomal subunits (∼60% of the nucleolar signal that was uncaged) moved away from the nucleolus in all directions, and the mean square displacement of signal was linearly proportional to time. This allows an estimation of a mean average diffusion coefficient of ∼0.3 μm2/s. The rate of movement away from the nucleolus did not change when the same uncaging and tracking experiments were carried out at 23°C. This observation is consistent with diffusive movement. If the movement out of the nucleolus were dependent on a metabolic energy source, the temperature drop (310–296°K) should have slowed the rate of movement by more than twofold (expected Q10 = 2–3). Therefore, our results suggest that a majority of 60S subunits move from the nucleolus into the nucleoplasm in a manner characteristic of diffusion. We cannot rule out, however, that the movement of a smaller fraction of 60S subunits may be dependent on metabolic energy.
The diffusion coefficient of 0.3 μm2/s estimated here can be compared with a predicted diffusion coefficient in aqueous solution of 10 μm2/s for 60S ribosomal subunits. The apparent mobility of the large subunit in the nucleus is thus substantially slowed compared with that of its diffusion in aqueous solution. This could partially reflect movement through a nuclear milieu that is more viscous than aqueous solution. However, the results of these experiments, along with the work of others, suggests a second interpretation, viz., that particle mobility is slowed by encounters with other nuclear structures or particles. This is usually called anomalous diffusion (Saxton, 1994, 2001; Feder et al., 1996). Diffusive-like movement has now been observed in the nucleus of live cells for several nuclear proteins, including nucleolar proteins (Pederson, 2000; Misteli, 2001; Pederson, 2001b) as well as poly(A) RNA (Politz et al., 1998, 1999). In fluorescent photobleaching experiments and in the uncaging experiments described here, the measured average mobility of the various nuclear particles is usually at least fivefold slower than that observed in solution. However, when more detailed analyses are carried out, it has been found in many cases that the entire population of molecules is not moving at this reduced rate, as one would expect for movement through a viscous solution, but instead, multiple populations of molecules with different mobilities are present (e.g., Politz et al., 1998; Wachsmuth et al., 2000; Platani et al., 2002; Pederson, 2002). This is consistent with anomalous subdiffusion, where mobility is constrained either by transient binding to and/or collisions with nuclear entities or by corralling within confinement zones, both of which phenomena impede free diffusion and give rise to multiple subpopulations of molecules moving with different mobilities (Feder et al., 1996; Saxton, 2001).
Our present results indicate that the behavior of nuclear 60S subunits is most consistent with anomalous diffusion. A log-log plot of ω2/dt vs. dt reveals anomalous diffusion as a line with negative slope (Saxton, 1994), and the more negative the slope, the more anomalous the diffusion (and the higher the obstacle concentration). The plots obtained here show highly negative slopes, approaching –1, which indicates the 60S subunits are diffusing through a high concentration of barriers, which slows the average mobility of the population. Furthermore, we do not see a recovery to a flat line with a slope of zero at longer time points, which would define a cross-over point and give information about obstacle concentration and size. This might be because the mobility of the 60S subunits is too rapid to allow detection of this cross-over point before signal reaches the nuclear membrane.
In the case of some nuclear particles, the movement of a fraction of the population appears to be metabolic-energy dependent, whereas another fraction appears not to be. Cajal bodies transiently bind chromatin in an energy-dependent process (Platani et al., 2002) and the movement of some PML bodies requires energy (Muratani et al., 2002). Although our results indicate that a majority of 60S ribosomal subunits leaving the nucleolus are undergoing free diffusion into the nucleoplasm, they do not preclude the possibility that a subset of these particles moves in a metabolic energy-dependent manner.
An unanticipated observation was that signal that had been uncaged in the nucleolus sometimes visited other nucleoli. Similarly, signal uncaged in the nucleoplasm sometimes visited nucleoli. This suggests that there is free exchange of nucleolar and nucleoplasmic ribosomal components at some level. This is not too surprising if one considers the nature of diffusion which mandates that, absent a boundary, some molecular movement will occur in both directions, even if ribosomal components are more concentrated in the nucleolus than in the nucleoplasm. Another major component of the nucleolus, fibrillarin, has also recently been found to freely exchange between nucleolar and nucleoplasmic sites (Phair and Misteli, 2000; Snaar et al., 2000; Chen and Huang, 2001). The results reported here strongly support the notion that even the movements of nascent ribosomes, which are synthesized, assembled, and transported to the cytoplasm at the rate of ∼4000/min in mammalian cells, are governed by the simple laws of diffusion.
Supplementary Material
Acknowledgments
We sincerely thank Tim Mitchison (Harvard Medical School) for the very generous gift of the caged-fluorescein used in these experiments and Christina Alavian from our laboratory for cheerful help with in situ reverse transcription experiments. Additionally, we thank Kevin Fogarty for expert help with image analysis and Joachim Frank (Wadsworth Center, State University of New York) for permission to use the ribosome structures shown in Figure 1. This work was supported by National Institutes of Health Grants GM-21595 and GM-60551, which require us to state that the content of our article is not the official position of the US Government.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–06–0395. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-06-0395.
Online version of this article contains video material for some figures. Online version is available at www.molbiolcell.org.
References
- Beckman, R., Spahn, C.M.T., Frank, J., and Blobel, G. (2001). The active 80S ribosome-Sec61 complex. Cold Spring Harbor Symp. Quant. Biol. 3, 543–554. [DOI] [PubMed] [Google Scholar]
- Cardullo, R.A., Mungovan, R.M., and Wolf, D.E. (1991). Imaging membrane organization and dynamics. In: Biophysical and Biochemical Aspects of Fluorescence Spectroscopy, ed. T.D. Dewey, New York: Plenum Publishing, 231–260.
- Carrington, W.A., Lynch, R.M., Moore, E.D., Isenberg, G., Fogarty, K.E., and Fay, F.S. (1995). Superresolution three-dimensional images of fluorescence in cells with minimal light exposure. Science 268, 1483–1487. [DOI] [PubMed] [Google Scholar]
- Chartrand, P., Singer, R.H., and Long, R.M. (2001). RNP localization and transport in yeast. Annu. Rev. Cell Dev. Biol. 17, 297–310. [DOI] [PubMed] [Google Scholar]
- Chen, D., and Huang, S. (2001). Nucleolar components involved in ribosome biogenesis cycle between the nucleolus and nucleoplasm in interphase cells. J. Cell Biol. 153, 169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen, B.R. (2000). Nuclear RNA export pathways. Mol. Cell Biol. 20, 4181–4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneholt, B. (1999). Pre-mRNP particles: from gene to nuclear pore. Curr. Biol. 9, R412–R415. [DOI] [PubMed] [Google Scholar]
- De Rijk, P., Robbrecht, E., De Hoog, S., Caers, A., Van de Peer, Y., and De Wachter, R. (1999). Database on the structure of the large subunit ribosomal RNA. Nucleic Acids Res. 27, 174–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube, P., Bacher, G., Stark, H., Mueller, F., Zemlin, F., van Heel, M., and Brimacombe, R. (1998). Correlation of the expansion segments in mammalian rRNA with the fine structure of the 80S ribosome; a cryoelectron microscopic reconstruction of the rabbit reticulocyte ribosome at 21 Å resolution. J. Mol. Biol. 279, 403–421. [DOI] [PubMed] [Google Scholar]
- Dworetzky, S.I., and Feldherr, C.M. (1988). Translocation of RNA-coated gold particles through the nuclear pores of oocytes. J. Cell Biol. 106, 575–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberwine, J., Spencer, C., Miyashiro, K., Mackler, S., and Finnell, R. (1992). Complementary DNA synthesis in situ: methods and applications. Methods Enzymol. 216, 80–100. [DOI] [PubMed] [Google Scholar]
- Fatica, A., and Tollervey, D. (2002). Making ribosomes. Curr. Opin. Cell Biol. 14, 313–318. [DOI] [PubMed] [Google Scholar]
- Feder, T.J., Brust-Mascher, I., Slattery, J.P., Baird, B., and Webb, W.W. (1996). Constrained diffusion or immobile fraction on cell surfaces: a new interpretation. Biophys. J. 70, 2767–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadal, O., Strauß, D., Braspenning, J., Hoepfner, D., Petfalski, E., Philippsen, P., Tollervey, D., and Hurt, E. (2001). A nuclear AAA-type ATPase (Rix7p) is required for biogenesis and nuclear export of 60S ribosomal subunits. EMBO J. 20, 3695–3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbi, S. (1996). Expansion segments: regions of variable size that interrupt the universal core secondary structure of ribosomal RNA. In: Ribosomal RNA: Structure, Evolution, Processing, and Function in Protein Biosynthesis, ed. R.A. Zimmermann and A.E. Dahlberg, New York: CRC Press. 71–87.
- Han, H., Schepartz, A., Pellegrini, M., and Dervan, P.B. (1994). Mapping RNA regions in eukaryotic ribosomes that are accessible to methidiumpropyl-EDTA-Fe(II) and EDTA-Fe(II). Biochemistry 33, 9831–9844. [DOI] [PubMed] [Google Scholar]
- Ho, J.H., Kallstrom, G., and Johnson, A.W. (2000). Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J. Cell Biol. 151, 1057–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg, L., and Nygård, O. (1997). Mapping of nuclease-sensitive sites in native reticulocyte ribosomes: an analysis of the accessibility of ribosomal RNA to enzymatic cleavage. Eur. J. Biochem. 247, 160–168. [DOI] [PubMed] [Google Scholar]
- Holmberg, L., Melander, Y., and Nygård, O. (1994b). Probing the conformational changes in 5.8S, 18S and 28S rRNA upon association of derived subunits into complete 80S ribosomes. Nucleic Acids Res. 22, 2776–2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg, L., Melander, Y., and Nygård, O. (1994a). Probing the structure of mouse Ehrlich ascites cell 5.8S, 18S and 28S ribosomal RNA in situ. Nucleic Acids Res. 22, 1374–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, S. (2002). Building an efficient factory: where is pre-rRNA synthesized in the nucleolus? J. Cell Biol. 157, 739–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuersten, S., Ohno, M., and Mattaj, I.W. (2001). Nucleocytoplasmic transport: Ran, Beta and beyond. Trends Cell Biol. 11, 497–503. [DOI] [PubMed] [Google Scholar]
- Lazdins, I.B., Delannoy, M., and Sollner-Webb, B. (1997). Analysis of nucleolar transcription and processing domains and pre-rRNA movements by in situ hybridization. Chromosoma 105, 481–495. [DOI] [PubMed] [Google Scholar]
- Léger-Silvestre, I., Trumtel, S., Noaillac-Depeyre, J., and Gas, N. (1999). Functional compartmentalization of the nucleus in the budding yeast Saccharomyces cerevisiae. Chromosoma 108, 103–113. [DOI] [PubMed] [Google Scholar]
- Lei, E.P., and Silver, P.A. (2002). Protein and RNA export from the nucleus. Dev. Cell 2, 261–272. [DOI] [PubMed] [Google Scholar]
- Lewis, J.D., and Tollervey, D. (2000). Like attracts like: getting RNA processing together in the nucleus. Science 288, 1385–1389. [DOI] [PubMed] [Google Scholar]
- Lieberman, K.R., and Noller, H.F. (1998). Ribosomal protein L15 as a probe of 50 S ribosomal subunit structure. J. Mol. Biol. 284, 1367–1378. [DOI] [PubMed] [Google Scholar]
- Mattaj, I.W., and Englmeier, L. (1998). Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem. 67, 265–306. [DOI] [PubMed] [Google Scholar]
- Merryman, C., Moazed, D., Daubresse, G., and Noller, H.F. (1999). Nucleotides in 23 S rRNA protected by the association of 30 S and 50 S ribosomal subunits. J. Mol. Biol. 285, 107–113. [DOI] [PubMed] [Google Scholar]
- Milkereit, P., Gadal, O., Podtelejnikov, A., Trumtel, S., Gas, N., Petfalski, E., Tollervey, D., Mann, M., Hurt, E., and Tschochner, H. (2001). Maturation and intranuclear transport of pre-ribosomes requires Noc proteins. Cell 105, 499–509. [DOI] [PubMed] [Google Scholar]
- Misteli, T. (2001). Protein dynamics: implications for nuclear architecture and gene expression. Science 291, 843–847. [DOI] [PubMed] [Google Scholar]
- Mitchison, T.J., Sawin, K.E., and Theriot, J.A. (1994). Caged fluorescent probes for monitoring cytoskeleton dynamics. In: Cell Biology: A Laboratory Handbook, Vol. 3, ed. J.E. Celis, New York: Academic Press, 65–74. [Google Scholar]
- Moy, T.I., and Silver, P.A. (1999). Requirements for the nuclear export of the small ribosomal subunit. J. Cell Sci. 115, 2985–2995. [DOI] [PubMed] [Google Scholar]
- Muratani, M., Gerlich, D., Janicki, S.M., Gebhard, M., Eils, R., and Spector, D.L. (2002). Metabolic-energy-dependent movement of PML bodies within the mammalian cell nucleus. Nat. Cell Biol. 4, 106–110. [DOI] [PubMed] [Google Scholar]
- Musters, W., Venema, J., van der Linden, G., van Heerikhuizen, H., Klootwijk, J., and Planta, R.J. (1989). A system for the analysis of yeast ribosomal DNA mutations. Mol. Cell. Biol. 9, 551–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissan, T.A., Baßler, J., Petfalski, E., Tollervey, D., and Hurt, E. (2002). 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 21, 5539–5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paillasson, S., Van De Corput, M., Dirks, R.W., Tanke, H.J., Robert-Nicoud, M., and Ronot, X. (1997). In situ hybridization in living cells: detection of RNA molecules. Exp. Cell Res. 231, 226–233. [DOI] [PubMed] [Google Scholar]
- Pante, N., Jarmolowski, A., Izaurralde, E., Sauder, U., Baschong, W., and Mattaj, I.W. (1997). Visualizing nuclear export of different classes of RNA by electron microscopy. RNA 3, 498–513. [PMC free article] [PubMed] [Google Scholar]
- Pederson, T. (2000). Diffusional protein transport within the nucleus: a message in the medium. Nat. Cell Biol. 2, E73–E74. [DOI] [PubMed] [Google Scholar]
- Pederson, T. (2001a). Fluorescent RNA cytochemistry: tracking gene transcripts in living cells. Nucleic Acids Res. 29, 1013–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson, T. (2001b). Protein mobility within the nucleus—what are the right moves? Cell 104, 635–638. [DOI] [PubMed] [Google Scholar]
- Pederson, T. (2002). Dynamics and genome-centricity of interchromatin domains. Nat. Cell Biol. 4, E287–E291. [DOI] [PubMed] [Google Scholar]
- Phair, R.D., and Misteli, T. (2000). High mobility proteins in the mammalian cell nucleus. Nature 404, 604–609. [DOI] [PubMed] [Google Scholar]
- Platani, M., Goldberg, I., Lamond, A.I., and Swedlow, J.R. (2002). Cajal bodies dynamics and association with chromatin are ATP-dependent. Nat. Cell Biol. 4, 502–508. [DOI] [PubMed] [Google Scholar]
- Politz, J.C., Taneja, K.L., and Singer, R.H. (1995). Characterization of hybridization between synthetic oligodeoxynucleotides and RNA in living cells. Nucleic Acids Res. 23, 4946–4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz, J.C., Browne, E.S., Wolf, D.E., and Pederson, T. (1998). Intranuclear diffusion and hybridization state of oligonucleotides measured by fluorescence correlation spectroscopy. Proc. Natl. Acad. Sci. USA 95, 6043–6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz, J.C., Tuft, R.A., Pederson, T., and Singer, R.H. (1999). Movement of nuclear poly(A) RNA throughout the interchromatin space in living cells. Curr. Biol. 9, 285–291. [DOI] [PubMed] [Google Scholar]
- Politz, J.C. (1999). Use of caged fluorochromes to track macromolecular movement in living cells. Trends Cell Biol. 9, 284–287. [DOI] [PubMed] [Google Scholar]
- Politz, J.C., and Singer, R.H. (1999). In situ reverse transcription for the detection of hybridization between oligonucleotides and their intracellular targets. Methods 18, 281–285. [DOI] [PubMed] [Google Scholar]
- Politz, J.C., and Pederson, T. (2000). Movement of mRNA from transcription site to nuclear pores. J. Struct. Biol. 129, 252–257. [DOI] [PubMed] [Google Scholar]
- Politz, J.C., Lewandowski, L.B., and Pederson, T. (2002). Signal recognition particle RNA within the nucleolus concentrates at sites different than those of ribosome synthesis. J. Cell Biol. 159, 411–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz, J.C.R., Tuft, R.A., and Pederson, T. (2003). Photoactivation-based labeling and in vivo tracking of RNA molecules in the nucleus. In: Live Cell Imaging: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Press (in press). [DOI] [PubMed]
- Puvion-Dutilleul, F., Bachellerie, J.-P., and Puvion, E. (1991). Nucleolar organization of HeLa cells as studied by in situ hybridization. Chromosoma 100, 395–409. [DOI] [PubMed] [Google Scholar]
- Rizzuto, R., Carrington, W., and Tuft, R.A. (1998). Digital imaging microscopy of living cells. Trends Cell Biol. 8, 288–92. [DOI] [PubMed] [Google Scholar]
- Saxton, M. (1994). Anomalous diffusion due to obstacles: a Monte Carlo study. Biophys. J. 66, 394–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton, M. (2001). Anomalous subdiffusion in fluorescence photobleaching recovery: a Monte Carlo study. Biophys. J. 81, 2226–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, O.P., Björkroth, B., Masich, S., Wieslander, L., and Daneholt, B. (1999). The intranuclear movement of Balbiani ring premessenger ribonucleoprotein particles. Exp. Cell Res. 251, 135–146. [DOI] [PubMed] [Google Scholar]
- Snaar, S., Wiesmeijer, K., Jochemsen, A.G., Tanke, H.J., and Dirks, R.W. (2000). Mutational analysis of fibrillarin and its mobility in living human cells. J. Cell Biol. 151, 653–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn, C.M.T., Beckmann, R., Eswar, N., Penczek, P.A., Sali, A., Blobel, G., and Frank, J. (2001). Structure of the 80S ribosome from Saccharomyces cerevisiae—tRNA-ribosome and subunit-subunit interactions. Cell 107, 373–386. [DOI] [PubMed] [Google Scholar]
- Sweeney, R., Fan, Q., and Yao, M.-C. (1996). Antisense ribosomes: rRNA as a vehicle for antisense RNAs. Proc. Natl. Acad. Sci. USA 93, 8518–8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiry, M., Cheutin, T., O'Donohue, M.-F., Kaplan, H., and Ploton, D. (2000). Dynamics and three dimensional localization of ribosomal RNA within the nucleolus. RNA 6, 1750–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, F., and Kutay, U. (2003). Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway. J. Cell Sci. 116, 2409–2419. [DOI] [PubMed] [Google Scholar]
- Trotta, C.R., Lund, E., Kahan, L., Johnson, A.W., and Dahlberg, J.E. (2003). Coordinated nuclear export of 60S ribosomal subunits and NMD3 in vertebrates. EMBO J. 22, 2841–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno, Y., Kumagai, I., Haginoya, N., and Matsuda, A. (1997). Effects of 5-(N-aminohexyl)carbamoyl-2′deoxyuridine on endonuclease stability and the ability of oligodeoxynucleotide to activate RNase H. Nucleic Acids Res. 25, 3777–3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachsmuth, M., Waldeck, W., and Langowski, J. (2000). Anomalous diffusion of fluorescent probes inside living cell nuclei investigated by spatially resolved fluorescence correlation spectroscopy. J. Mol. Biol. 298, 677–689. [DOI] [PubMed] [Google Scholar]
- Warner, J.R., and Soeiro, R. (1967). Nascent ribosomes from HeLa cells. Proc. Natl. Acad. Sci. USA 58, 1984–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.