Abstract
Stem cell fate is influenced by a number of factors and interactions that require robust control for safe and effective regeneration of functional tissue. Coordinated interactions with soluble factors, other cells, and extracellular matrices define a local biochemical and mechanical niche with complex and dynamic regulation that stem cells sense. Decellularized tissue matrices and synthetic polymer niches are being used in the clinic, and they are also beginning to clarify fundamental aspects of how stem cells contribute to homeostasis and repair, for example, at sites of fibrosis. Multi-faceted technologies are increasingly required to produce and interrogate cells ex vivo, to build predictive models, and ultimately to enhance stem cell integration in vivo for therapeutic benefit.
Control over stem cell trafficking, survival, proliferation, and differentiation within a complex in vivo milieu is extremely challenging. In studies of animal models and humans where stem cell engraftment has been quantified after injection, only a few percent of cells remain after several days or weeks [eg.(1)]. Many clinical trials are nonetheless underway, particularly with adult bone marrow derived mesenchymal stem cells (MSC) which are being investigated as treatments for diseases of non-hematopoietic tissues – primarily myocardial infarction and peripheral ischemia (2). Although FDA approval for human testing of cells differentiated from embryonic stem cells (ESC) is a recent landmark for the field (3), two widely reported clinical cases highlight some of the technical opportunities and challenges with stem cells in soft tissue repair. One patient in Spain was successfully transplanted with a re-engineered trachea in 2008: donor trachea was first decellularized using a detergent (without denaturing the collagenous matrix), and then this scaffold was re-cellularized in a rotating bioreactor using MSC-derived cartilage-like cells (4). Long term safety and efficacy will be important to monitor and understand. Indeed, in a second case, the cerebellum of a boy with ataxia telangiectasia (AT) was injected with human fetal neural stem cells (NSC), and four years later, a glio-neuronal brain tumor of stem cell origin was found (5). Upon implantation, stem cells and their derived lineages encounter a multitude of cues that can influence cell fate. Efforts to parse the molecular mechanisms for translation from bench to clinic will increasingly benefit from a wide range of new and established technologies. Here we briefly review salient features of microenvironments, mechanics, and material systems that are being pursued to control stem cells for both basic insight and application.
Niche interactions and in vitro designs
The niche is the in vivo microenvironment that regulates stem cell survival, self-renewal, and differentiation. Key niche components and interactions include growth factors, cell-cell contacts, and cell-matrix adhesions (Fig. 1A). The interplay of these niche factors is particularly important to comprehend if any desired stem cell response is to be made robust for therapy – i.e. resistant to the many types of perturbations encountered by cells delivered in vivo.
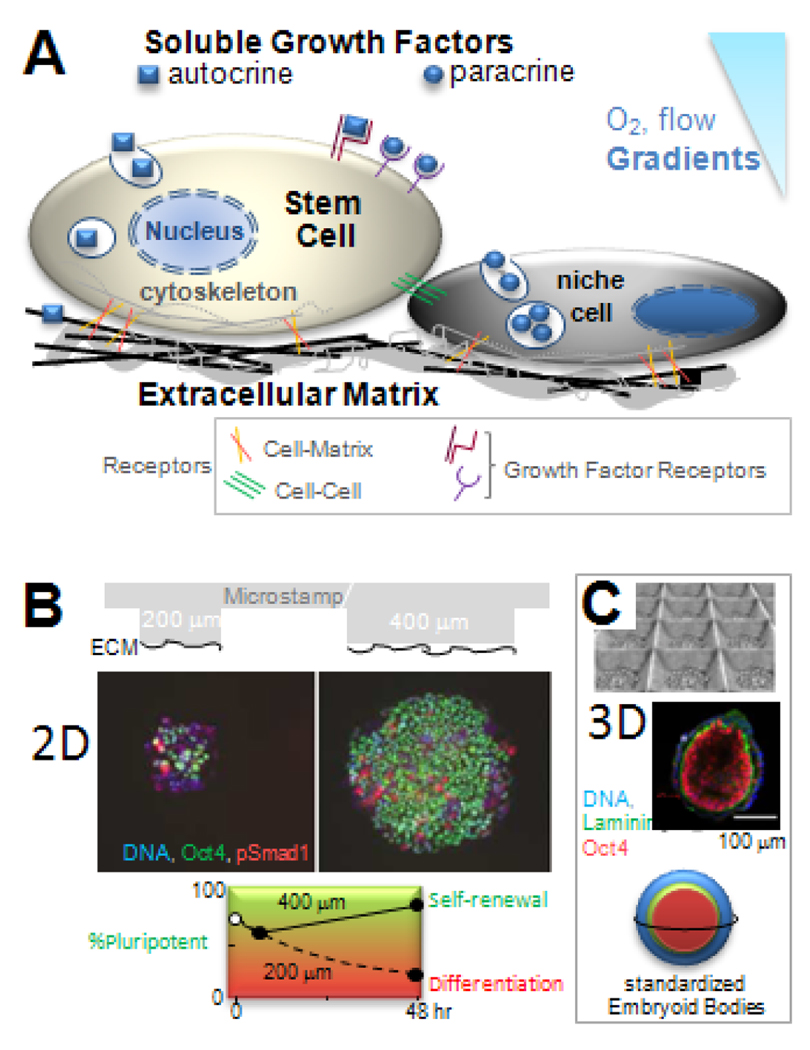
Fig 1. Stem cell niche and designs.
(A) Soluble and matrix bound factors combine with cell-cell contact, cell-matrix adhesion, and gradients (such as in O2) to direct cell fate. (B) Substrates with 2D micro-patterns of Extracellular Matrix, ECM, control the size of ESC colonies and pluripotency of ESC based on immunofluorescence for the transcription factor Oct4 (15). Plot: large islands increase the pluripotent population that undergoes self-renewal, while small islands inhibit lead to differentiation with a time constant of 24 hr. (C) Substrates with micro-wells help to standardize the diameter of quasi-spherical, 3D embryoid bodies (21). Cells and ECM (laminin) exhibit spherically symmetric pattern, with pluripotent cells in the inside.
Growth factors added to culture or secreted by stem cells and nearby niche cells are often potent in their effects on cell fate, and so in embryonic development, growth factors are tightly regulated in space and time (6). In culture, one means of controlling niche interactions in 2D is with micropatterns of extracellular matrix (ECM) islands, which limit diffusion of secreted growth factors within and between islands and also limit the modulating effects of matrix and cell contacts. With human-ESC, for example, islands of ECM made by microstamping onto a substrate (Fig. 1B) have demonstrated a minimal island size for maintenance and expansion of the pluripotent state; the mechanism is based in part on an antagonism between two different members of the Transforming Growth Factor-β (TGF-β) superfamily (7). Such factors can affect the secreting cell (autocrine) or other cells (paracrine), and so for spatiotemporal control of concentrations, microfluidic devices have been made which continuously wash away secreted factors while perfusing known concentrations of active factors. With human-NSC (hNSC), for example, microfluidic control of growth factor within a single culture chamber showed very clearly that the amount of proliferation and the fraction of differentiated cells follow a strict inverse relationship (8). Indeed, most terminally differentiated cells do not proliferate, whereas stem cells and progenitor cells do, but this distinction can be difficult to sort out in standard culture, where most cells crawl around semi-randomly, dynamically changing both exposure to any gradients in growth factor and contact with other cells. In order to directly assess any modulating role of cell-cell contacts in soluble factor signaling, micromechanical devices have been made to reversibly move cells into contact (7). The results suggest that different cell types might need to come into contact before a given cell type will respond to locally secreted factors.
Extracellular matrix not only mediates cell attachment and presents key cues to cells (see below), but it often also binds growth factors, limiting their diffusion. This can be mimicked by synthetically tethering a growth factor to a substrate (9), which has been used to enhance survival of MSC (10) and to regulate select transcriptional networks in mouse-ESC (mESC) (11).
Adhesion of MSC and ESC to matrix or other cells is essential for viability – individual cells do not survive in suspension, but adhesive signals might be controlled just as well or better with synthetic mimics. Such materials could conceivably replace nonhuman niche cells or animal-derived matrix products (eg. Matrigel™) in common use with human stem cells. In an early combinatorial study with human-ESC (hESC) and muscle-derived stem cells, rigid spots of 576 different combinations of 25 different acrylate-based polymers were arrayed and found to combine with soluble factors in exerting wide-ranging effects on cell attachment, proliferation, and lineage induction (12). For 3-D cultures, cross-linked hyaluronic acid (HA) hydrogels proved unique in supporting hESC growth in undifferentiated masses (i.e. embryoid bodies), possibly because HA is a prominent ECM polymer in embryonic development (13). Embryoid bodies can also be sculpted to well-controlled diameters with polymer microwells and other methods (Fig. 1C) (14, 15). Size control is important to minimize gradients in oxygen and other physical or chemical factors that regulate stem cell fate (16). Nonetheless, cell segregation and differentiation within embryoid bodies (and embryos too) still need to be understood more deeply, with mechanisms likely rooted in the multiple pathways that a cell uses to sense its microenvironment (17).
Forces, matrix elasticity, and fibrosis
Whether in vitro or in vivo, cells generate force and are often exposed to force – and both can influence stem cell fates. The very first stages of cell differentiation in embryogenesis are indeed blocked upon knockout of ubiquitous force-generating myosins (18). Flowing fluids also generate forces on any object in the flow (you feel such forces when you hold your hand out of a car window), and fluid forces typical of blood flow have been found to initiate an endothelial program in isolated ESC (19). Mechanical deformations or strains are also common in solid tissues (eg. beating heart, dilating arteries), and imposing substrate strains of just 5% can induce MSC differentiation toward smooth muscle (20). Mechanistically, a force f will strain any physically linked protein and affect the kinetic rate k of a protein-protein interaction or conformation change (21) as
Stem cells may well possess more than the typical ensemble of force-coupled signaling pathways as a means to sensitize themselves to microenvironments that range – physically – from flowing fluids and strained tissues to solid tissues of varied elasticity (Fig. 2A). Indeed, when MSC are grown on firm gels that mimic the elasticity of muscle and that are coated with collagen-I, myogenic markers were upregulated, whereas when MSC are grown on rigid gels that mimic pre-calcified bone, the cells appear osteogenic (21). Added induction factors augment or oppose this programming by matrix. With NSC likewise, neuron differentiation is favored on soft matrices that mimic normal brain, whereas differentiation into glial is promoted on harder matrices that typify glial scars (22). These latter examples – and topographical patterns as well (23) – suggest that carefully made materials can prime the expansion of specific progenitors.
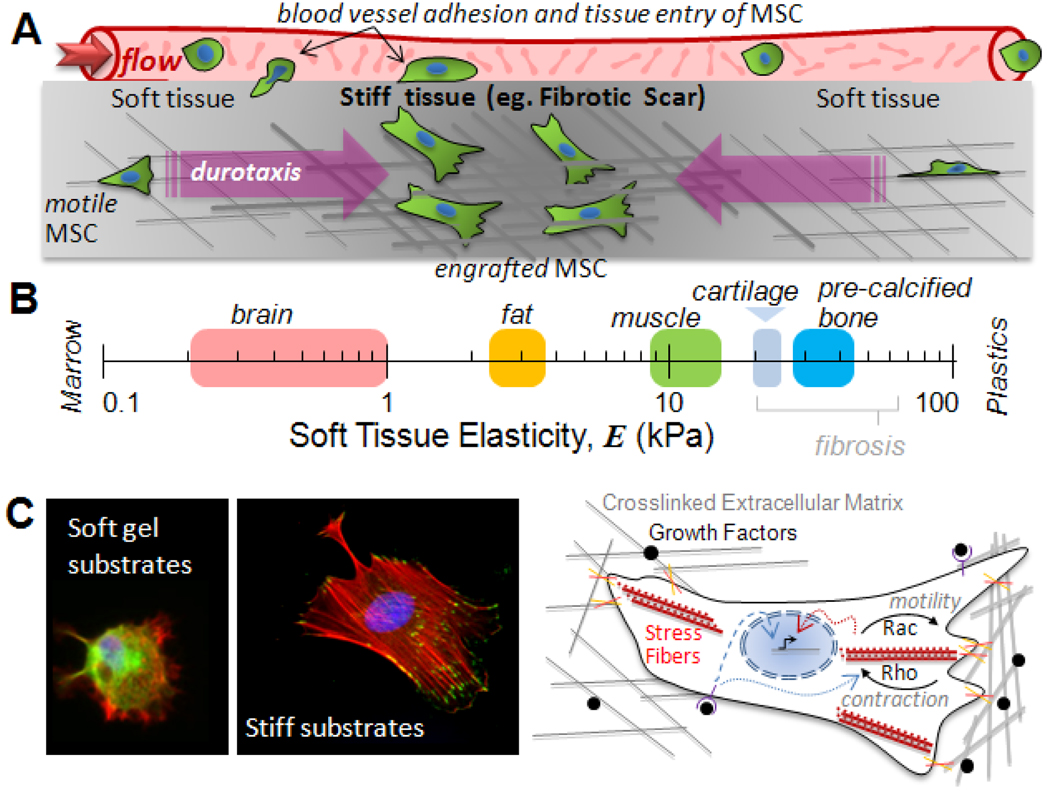
Fig 2. Forces in stem cell trafficking.
(A) To extravasate from the circulation and invade a tissue, stem cells must adhere strongly to the vessel wall and withstand high flow forces. Within a tissue, additional physical factors can direct motile cells, including durotaxis into stiff, fibrotic regions of tissue where cells engraft. (B) Soft tissue elasticity scale ranging from soft brain (72), fat (73), and striated muscle (25), to stiff cartilage (E ~ 20–30 kPa at the scale of adhesions (74)) and pre-calcified bone (25). (C) In vitro substrates that mimic soft and stiff tissue microenvironments (left) show that cells anchor more strongly to stiff substrates, building focal adhesions and actin-myosin stress fibers. Schematic (right) shows matrix adhesion and growth factors influence both cell physiology and lineage. Signals from growth factor receptors not only propagate into the nucleus (dashed blue arrow) and direct transcription (black arrow), but also affect Rho-GTPase activity (dotted blue arrow). Rac drives motility forward, and Rho regulates contraction of stress fibers (red), and both can also influence gene expression (dotted red arrow).
In vivo, when stem cells egress from their niche into the circulation (24) or when stem cells are injected intravenously as part of a therapeutic regimen, fluid forces push and drag the cells (Fig. 2A), which opposes adhesion to the vessel wall, while tissue entry requires stem cell motility and might benefit from the high deformability of stem cell nuclei (25). The processes are similar to those widely studied with leukocytes and metastatic cancer cells [eg. (26)]. The latter comparison is especially intriguing in that metastatic ‘capture’ depends strongly on at least one matrix fibrosis factor (lysyl oxidase which catalyzes crosslinking of collagen) and fibrotic scarring is a challenge common to many regenerative goals with stem cells.
Recent mechanical measurements of fibrotic scars that develop after an acute myocardial infarction (27) or with more chronic stimuli (28) show that the fibrotic tissue is locally rigidified by at least several-fold compared to normal tissue. Atomic Force Microscopy probing gives cell-scale elasticities of E ~ 20–60 kPa for fibrotic wounds in soft tissues (27, 29), which exceeds E for soft tissues and overlaps with cartilage and pre-calcified bone (Fig. 2B).
Rigid fibrotic tissue can in and of itself contribute a homing signal. Based on the use of various gel matrices with well-controlled elasticity and matrix ligand and density, most if not all cells are found to adhere, spread, assemble their cytoskeleton (Fig. 2C), and anchor more strongly to stiff substrates compared to soft substrates (30). In a gradient of elasticity, cells therefore accumulate on stiffer substrates in a process called ‘durotaxis’ (31), which might constitute a biophysical basis for why MSC home to sites of injury and fibrosis (1). Matrix can also be a more potent differentiation cue for MSC than standard induction cocktails (32, 33). Whereas MSC in an infarct scar in mice have been reported to generate bone in the heart (34), MSC are also often found to attenuate scar formation (1, 27). Recent models of a ‘scar in a dish’ have shown that the well-studied differentiation of fibroblasts to myofibroblasts requires both a stiff matrix (E > 20 kPa) and TGF-β, with growth factor release from the ECM dependent on cell contraction driven unfolding of the ECM complex that sequesters the TGF-β (29). Growth factor regulation by matrix and cell force has yet to be reported for stem cells, but developmentally critical cell-cell interactions appear to mediate forced unfolding of Notch receptor at the membrane. Force opens a cleavage site that ultimately liberates a Notch fragment which diffuses into the nucleus to affect transcription (35).
Materials pervade stem cell biology, often unintentionally. Isolation and growth of stem cells on rigid tissue culture plastic tend to promote spreading of cells rich in actin-myosin stress fibers. With similarly rigid surfaces, if the area of MSC contact is controlled with adhesive patterns (36), it is found that mixed induction cocktails that induce both fat and bone lead to adipogenesis on small islands (which minimize matrix contact) and osteogenesis on large islands (which maximize contractile anchorage). Mechanisms for sensing of matrix as well as for motility continue to be clarified, but growth factor and integrin coupled roles for Rac and Rho in stem cell motility, contractility, and anchorage are well-documented. In hematopoietic stem cells (HSC), Rac isoforms are key regulators of engraftment and marrow retention, with Rac activation occurring by β1-integrin adhesion to matrix and also via stimulation by factors including stromal derived factor (SDF-1) (37). A similar, prototypical coupling of anchorage signaling pathways is also revealed in the phosphoproteome of MSC induced by growth factors (38).
Consistent with the strict requirement for nonmuscle myosin in differentiation within embryos (18), pharmacological inhibition of myosin in MSC blocks all lineages on both rigid and compliant substrates (21, 36) with measurable effects on folding and assembly of the proteome (39). Inhibition of the Rho kinase effector, ROCK, resulting in deactivation of myosin, also blocks all lineages on rigid substrates (36), though not on compliant substrates (21). Inhibition of ROCK in dissociated hESC (40) also dramatically enhances survival; no effects on ESC differentiation have been reported. Such pharmacological perturbations of the cytoskeleton have been known for years to affect mesenchymal lineages such as chondrocytes (41), but effects can couple to matrix. The recent success in trachea reconstruction (4) likely benefited from suitable matrix signals for chondrogenesis in the detergent-decellularized donor trachea, and while detergents might or might not preserve tissue mechanics, not all tissue regeneration can use such methods.
Physical perspectives on normal and diseased tissues help to generate new hypotheses. As a leading cause of death, heart disease is a principal target of stem cell therapies (2), but cardiogenesis involves a complex interplay of mechanochemical factors. Embryo-derived cardiomyocytes maintain their spontaneous beating on substrates with elasticity less than or equal to that of normal heart tissue, but the cells twitch and stop beating on stiff matrices that mechanically mimic a fibrotic scar (42). Studies of neonatal cardiomyocytes further show that ROCK inhibition selectively blocks cell dysfunction on stiff substrates (43). Therefore, future experiments with cardiomyocytes derived from ESC or induced pluripotent stem cells require close attention to the matrix microenvironment.
Synthetic niches in vivo
An original assumption in the stem cell field was that transplanted cells would directly participate in the building of tissues; however, it is now clear that paracrine effects are also important (44). Advanced clinical applications of MSC are seen with immune/inflammatory conditions including Crohn's Disease of the bowel and Graft versus Host Disease (GvHD) that can result from bone marrow transplants (1). The MSC treatments do not exploit tissue building abilities, but instead appear to exert immunomodulatory functions that extend to wound-healing and reduced scar formation. These applications serve to highlight again the importance of tight control over stem cell purity and differentiation, a need for strategies to enhance survival and engraftment of transplanted cells, and the possibility that distinct strategies may be useful for transplantation, depending on the specific functional outcome(s) sought.
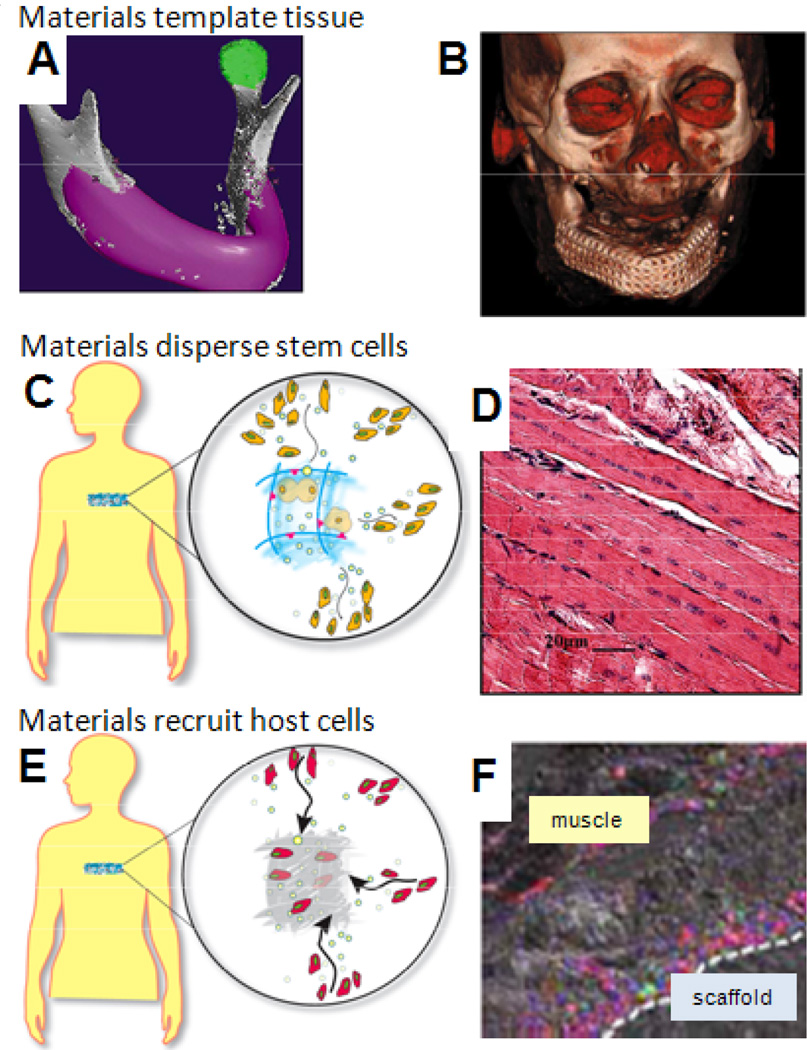
An appealing approach to address some of the challenges involved in stem cell transplantation is the development and use of materials systems that create specialized niches for the cells. Limitations of conventional cell infusions or injections include poor delivery and retention of cells at the intended site or cell death due to loss of anchorage (anoikis). A variety of naturally-derived materials and synthetic polymers are currently in development as vehicles for stem cell transplantation due to their ability to provide adhesion for interacting cells; control over the presentation of adhesion sites (e.g., density of peptides to bind integrins) by these materials improves transplanted cell survival and participation in tissue regeneration (45, 46). Material architecture can be used to further template the structure of tissues (Fig. 3A,B) formed from interacting stem cells (47), an approach that has been used to form tissue patches to enhance cardiac function in animal models (48) and to format a new mandible in a human patient (49). There has not yet been a demonstration that substrate mechanical properties regulate stem cell fate in vivo, although the demonstration that gel mechanical properties regulate capillary formation in vivo (50) supports this concept. One complication is that the mechanical properties and degradation rates of synthetic niches are typically coupled, and the degradation rate has been demonstrated to enhance bone regeneration by transplanted stem cells (51).
Fig 3. Synthetic niches in vivo.
(A) Materials can fill a specific anatomic defect (pink) to localize transplanted cells and serve as a scaffolding for formation of new tissue. (B) A mandible formed in a patient used a metal and polymer structure that was seeded with MSC and cytokines (49). (C) A designed material niche maintains stem cell viability and proliferation, while promoting outward migration at an appropriate stage of differentiation. (D) Dispersion of stem cells from a niche into regenerating skeletal muscle (60). (E) Recruitment of host stem cells for subsequent homing to sites of tissue injury. (F) Mice with GFP bone marrow-derived cells (green) show regenerating muscle infiltrated with cells that are dual-labeled for endothelial cell marker CD31 (red), indicating neo-vascularization (75).
Growth factors or cytokines can be provided in a localized manner either with controlled release particle systems (eg. (52)) or from the material scaffolds used as synthetic stem cell niches. Cardiomyocyte function has been dramatically improved by coordinated release of Insulin-like Growth Factor (IGF) from the transplantation vehicle (46), as has bone formation by mesenchymal stems with release of TGF-β family proteins (53). However, full regeneration will only result from mechanical, vascular, and neural integration of the regenerating and surrounding tissues. Materials presenting angiogenic factors can enhance local vascularization (54), and increase the survival of transplanted stem cells and subsequent regeneration (55). Materials may also be used as carriers of ESC-derived endothelial cell progenitors that form new vascular networks (56). Nervous system integration can also be enhanced by materials that provide gradients of neurotrophic factors (57) and by implantation adjacent to a transected nerve (58).
Instead of anchoring transplanted cells to a specific location with a material, stem cell niches might be mimicked to regulate the proliferation, differentiation, and dispersal of daughter cells into the surrounding tissue to participate in regeneration or provide trophic factors over a large volume (Fig. 3C,D). Because the stage of cell differentiation, exposure to morphogens and cytokines, and implant site conditions all regulate stem cell function after transplantation (59, 60), this niche approach seems broadly useful. The ability of satellite cell-derived myoblasts to promote skeletal muscle regeneration following injury was recently enhanced with a material that activated resident cells, but prevented terminal differentiation until the cells exited the material (61). This approach may be most useful in the wide dispersion of stem cell populations that secrete trophic factors which influence host cells. Material-directed endothelial progenitors will even orchestrate regional revascularization and salvage of necrotic limbs (62).
Potentially useful cell populations already exist in the body and attracting these cells to a desired anatomic site (Fig. 3E, F) has the potential to provide new therapeutic options. Cost and complexity should be reduced compared to approaches with ex vivo bioreactors. With bone repair for example, implantation of simple biomaterials without growth factors has shown how potent the in vivo milieu can be in generating native-like bone (63) – in spite of risks of implantation injury. Mobilization of endothelial progenitor cells from bone marrow with either G-CSF or with drugs such as Statins is also being tested in ischemic conditions (1).
Systems biology and translation
Fate decisions in vivo with HSC and MSC appear tightly regulated by physiological demand. In addition to all of the extracellular cues highlighted earlier, stimulus-response can be complicated by tissue-specific patterns of ligand and receptor expression (64) as well as by sequential autocrine and paracrine inductive loops (65) that arise as cell populations develop and adapt (66). Genome-wide studies (67) and mathematical models (68) have begun to generate relevant network models, but only a few attempts have been made to unravel inter-cellular signaling (69, 70). Cells (as opposed to proteins or genes) could be considered as nodes in networks that are linked by cell contact, by shared matrix, and by secreted factors. Integrating immune cells, particularly natural killer cells and macrophages, into such networks will be important to understand how implanted cells either engraft and contribute to tissue or else are cleared (71).
Stem cell studies beyond those cited offer important perspectives on the opportunities and additional challenges that lie ahead. Soluble factor strategies combined with attention to cell adhesion and mechanics seem likely to synergize with both synthetic material niches and nonetheless, tissue-derived matrices to play essential roles in making stem cell therapies safe, efficacious, and routine.
Acknowledgments
Grants from the NIH and NSF are very gratefully acknowledged. Images of MSC in Fig.2C courtesy of A. Zajac (Univ. Penn.). Assistance of E. Silva (Harvard) with Fig. 3 is acknowledged.
References and Notes
- 1.Pittenger M, Martin B. Circ. Res. 2004;95:9. doi: 10.1161/01.RES.0000135902.99383.6f. [DOI] [PubMed] [Google Scholar]
- 2.NIH. 2009 http://clinicaltrials.gov.
- 3.Alper J. Nat. Biotechnol. 2009;27:213. doi: 10.1038/nbt0309-213a. [DOI] [PubMed] [Google Scholar]
- 4.Macchiarini P, et al. Lancet. 2008;372:2023. [Google Scholar]
- 5.Amariglio N, et al. PLoS Med. 2009;6:221. [Google Scholar]
- 6.Murry C, Keller G. Cell. 2008;132:661. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 7.Peerani R, et al. EMBO J. 2007;26:4744. doi: 10.1038/sj.emboj.7601896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung B, et al. Lab On A Chip. 2005;5:401. doi: 10.1039/b417651k. [DOI] [PubMed] [Google Scholar]
- 9.Hui E, Bhatia S. Proc. Natl. Acad. Sci. U. S. A. 2007;104:5722. doi: 10.1073/pnas.0608660104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan V, et al. Stem Cells. 2007;25:1241. doi: 10.1634/stemcells.2006-0320. [DOI] [PubMed] [Google Scholar]
- 11.Alberti K, et al. Nat. Methods. 2008;5:645. doi: 10.1038/nmeth.1222. [DOI] [PubMed] [Google Scholar]
- 12.Anderson D, Levenberg S, Langer R. Nat. Biotechnol. 2004;22:863. doi: 10.1038/nbt981. [DOI] [PubMed] [Google Scholar]
- 13.Gerecht S, et al. Proc. Natl. Acad. Sci. U. S. A. 2007;104:11298. doi: 10.1073/pnas.0703723104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McDevitt T, Palecek S. Curr. Opin. Biotechnol. 2008;19:527. doi: 10.1016/j.copbio.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ungrin MD, Joshi C, Nica A, Bauwens C, Zandstra PW. PLoS ONE. 2008;3:e1565. doi: 10.1371/journal.pone.0001565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parmar K, Mauch P, Vergilio JA, Sackstein R, Down JD. Proc. Natl. Acad. Sci. U. S. A. 2007;104:5431. doi: 10.1073/pnas.0701152104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engler AJ, Humbert PO, Wehrle-Haller B, Weaver VM. Science. 2009;324:208. doi: 10.1126/science.1170107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conti MA, Even-Ram S, Liu C, Yamada KM, Adelstein RS. J. Biol. Chem. 2004;279:41263. doi: 10.1074/jbc.C400352200. [DOI] [PubMed] [Google Scholar]
- 19.Yamamoto K, et al. Am. J. Physiol. Heart Circ. Physiol. 2005;288:H1915. doi: 10.1152/ajpheart.00956.2004. [DOI] [PubMed] [Google Scholar]
- 20.Kurpinski K, Chu J, Hashi C, Li S. Proc. Natl. Acad. Sci. U. S. A. 2006;103:16095. doi: 10.1073/pnas.0604182103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006;126:677. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 22.Saha K, et al. Biophys. J. 2008;95:4426. doi: 10.1529/biophysj.108.132217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dalby MJ, et al. Nature Materials. 2007;6:997. doi: 10.1038/nmat2013. [DOI] [PubMed] [Google Scholar]
- 24.Pitchford S, Furze R, Jones C, Wengner A, Rankin S. Cell Stem Cell. 2009;4:62. doi: 10.1016/j.stem.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 25.Pajerowski JD, Dahl KN, Zhong FL, Sammak PJ, Discher DE. Proc. Natl. Acad. Sci. U. S. A. 2007;104:15619. doi: 10.1073/pnas.0702576104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Erler J, et al. Nature. 2006;440:1222. doi: 10.1038/nature04695. [DOI] [PubMed] [Google Scholar]
- 27.Berry MF, et al. Am J Physiol Heart Circ Physiol. 2006;290:H2196. doi: 10.1152/ajpheart.01017.2005. [DOI] [PubMed] [Google Scholar]
- 28.Georges P, et al. Am. J. Physiol. Gastro. 2007;293:G1147. doi: 10.1152/ajpgi.00032.2007. [DOI] [PubMed] [Google Scholar]
- 29.Wipff P, Rifkin D, Meister J, Hinz B. J. Cell Biol. 2007;179:1311. doi: 10.1083/jcb.200704042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Discher DE, Janmey P, Wang YL. Science. 2005;310:1139. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 31.Lo C, Wang H, Dembo M, Wang Y. Biophys. J. 2000;79:144. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bennett K, et al. BMC Genomics. 2007;8:380. doi: 10.1186/1471-2164-8-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benoit DSW, Schwartz MP, Durney AR, Anseth KS. Nature Materials. 2008;7:816. doi: 10.1038/nmat2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Breitbach M, et al. Blood. 2007;110:1362. doi: 10.1182/blood-2006-12-063412. [DOI] [PubMed] [Google Scholar]
- 35.Kopan R, Ilagan MXG. Cell. 2009;137:216. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Dev Cell. 2004;6:483. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 37.Williams DA, Zheng Y, Cancelas JA. Small Gtpases in Disease, Pt B. vol. 439. 2008. p. 365. [DOI] [PubMed] [Google Scholar]
- 38.Kratchmarova I, Blagoev B, Haack-Sorensen M, Kassem M, Mann M. Science. 2005;308:1472. doi: 10.1126/science.1107627. [DOI] [PubMed] [Google Scholar]
- 39.Johnson CP, Tang HY, Carag C, Speicher DW, Discher DE. Science. 2007;317:663. doi: 10.1126/science.1139857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watanabe K, et al. Nat. Biotechnol. 2007;25:681. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- 41.Zanetti NC, Solursh M. J. Cell Biol. 1984;99:115. doi: 10.1083/jcb.99.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Engler AJ, et al. J. Cell Sci. 2008;121:3794. doi: 10.1242/jcs.029678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jacot JG, McCulloch AD, Omens JH. Biophys. J. 2008;95:3479. doi: 10.1529/biophysj.107.124545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gnecchi M, Zhang Z, Ni A, Dzau V. Circ. Res. 2008;103:1204. doi: 10.1161/CIRCRESAHA.108.176826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alsberg E, et al. J. Dent. Res. 2003;82:903. doi: 10.1177/154405910308201111. [DOI] [PubMed] [Google Scholar]
- 46.Davis M, et al. Circulation. 2005;111:442. doi: 10.1161/01.CIR.0000153847.47301.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Langer R, Vacanti JP. Science. 1993;260:920. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- 48.Zimmermann W, et al. Nat. Med. 2006;12:452. doi: 10.1038/nm1394. [DOI] [PubMed] [Google Scholar]
- 49.Warnke P, et al. Lancet. 2004;364:766. doi: 10.1016/S0140-6736(04)16935-3. [DOI] [PubMed] [Google Scholar]
- 50.Mammoto A, et al. Nature. 2009;457:1103. doi: 10.1038/nature07765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simmons C, Alsberg E, Hsiong S, Kim W, Mooney D. Bone. 2004;35:562. doi: 10.1016/j.bone.2004.02.027. [DOI] [PubMed] [Google Scholar]
- 52.Takehara N, et al. J. Am. Coll. Cardiol. 2008;52:1858. doi: 10.1016/j.jacc.2008.06.052. [DOI] [PubMed] [Google Scholar]
- 53.Park IH, et al. Nature. 2008;451:141. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 54.Trentin D, Hall H, Wechsler S, Hubbell J. Proc. Natl. Acad. Sci. U. S. A. 2006;103:2506. doi: 10.1073/pnas.0505964102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang Y, Kaigler D, Rice K, Krebsbach P, Mooney D. J. Bone Miner. Res. 2005;20:848. doi: 10.1359/JBMR.041226. [DOI] [PubMed] [Google Scholar]
- 56.Levenberg S, et al. Nat. Biotechnol. 2005;23:879. doi: 10.1038/nbt1109. [DOI] [PubMed] [Google Scholar]
- 57.Kapur T, Shoichet M. J. Biomed. Mater. Res. 2004;68A:235. doi: 10.1002/jbm.a.10168. [DOI] [PubMed] [Google Scholar]
- 58.Dhawan V, Lytle I, Dow D, Huang Y, Brown D. Tissue Eng. 2007;13:2813. doi: 10.1089/ten.2007.0003. [DOI] [PubMed] [Google Scholar]
- 59.Collins C, et al. Cell. 2005;122:289. doi: 10.1016/j.cell.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 60.Laflamme MA, et al. Nat. Biotechnol. 2007;25:1015. doi: 10.1038/nbt1327. [DOI] [PubMed] [Google Scholar]
- 61.Hill E, Boontheekul T, Mooney D. Proc. Natl. Acad. Sci. U. S. A. 2006;103:2494. doi: 10.1073/pnas.0506004103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Silva E, Kim E, Kong H, Mooney D. Proc. Natl. Acad. Sci. U. S. A. 2008;105:14347. doi: 10.1073/pnas.0803873105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stevens M, et al. PNAS. 2005;102:11450. doi: 10.1073/pnas.0504705102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kluger Y, et al. Proc. Natl. Acad. Sci. U. S. A. 2004;101:6508. doi: 10.1073/pnas.0401136101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Janes K, Lauffenburger D. Curr. Opin. Chem. Biol. 2006;10:73. doi: 10.1016/j.cbpa.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 66.Kirouac D, Zandstra P. Curr. Opin. Biotechnol. 2006;17:538. doi: 10.1016/j.copbio.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 67.Muller F, et al. Nature. 2008;455:401. doi: 10.1038/nature07213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chickarmane V, Enver T, Peterson C. PLoS Comp. Biol. 2009;5:e1000268. doi: 10.1371/journal.pcbi.1000268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Frankenstein Z, Alon U, Cohen I. Biol. Direct. 2006;1:32. doi: 10.1186/1745-6150-1-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rendl M, Lewis L, Fuchs E. PLoS Biol. 2005;3:1910. doi: 10.1371/journal.pbio.0030331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Takenaka K, et al. Nat. Immunol. 2007;8:1313. doi: 10.1038/ni1527. [DOI] [PubMed] [Google Scholar]
Additional References in Figure Legends
- 72.Flanagan LA, Ju YE, Marg B, Osterfield M, Janmey PA. Neuroreport. 2002;13:2411. doi: 10.1097/01.wnr.0000048003.96487.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Patel PN, Smith CK, Patrick CW. J Biomed Mater Res A. 2005;73:313. doi: 10.1002/jbm.a.30291. [DOI] [PubMed] [Google Scholar]
- 74.Stolz M, et al. Nat Nanotechnol. 2009;4:186. doi: 10.1038/nnano.2008.410. [DOI] [PubMed] [Google Scholar]
- 75.Fischbach C, et al. Proc Natl Acad Sci U S A. 2009;106:399. doi: 10.1073/pnas.0808932106. [DOI] [PMC free article] [PubMed] [Google Scholar]