Abstract
Mus81 is a highly conserved substrate specific endonuclease. Human Mus81 cleaves Holliday junctions, replication forks, and 3′ flap substrates in vitro, suggesting a number of possible in vivo functions. We show here that the abundance of human Mus81 peaks in S-phase and remains high in cells that have completed DNA replication and that Mus81 is a predominantly nuclear protein, with super accumulation in nucleoli. Two RecQ related DNA helicases BLM and WRN that are required for recombination repair in human cells colocalize with Mus81 in nucleoli. However, the nucleolar retention of Mus81 is not dependent on the presence of BLM or WRN, or on ongoing transcription. Mus81 is recruited to localized regions of UV damage in S-phase cells, but not in cells that are blocked from replicating DNA or that have completed replication. The retention of human Mus81 at regions of UV-induced damage specifically in S-phase cells suggest that the enzyme is recruited to the sites at which replication forks encounter damaged DNA. The nucleolar concentration of Mus81 suggests that it is required to repair problems that arise most frequently in the highly repetitive nucleolar DNA. Together these data support a role for Mus81 in recombination repair in higher eukaryotes.
INTRODUCTION
DNA is constantly damaged by endogenous reactive metabolites, exogenous chemicals, and radiation. A network of cell cycle checkpoints and DNA repair mechanisms maintain integrity of the genome and prevent the deleterious consequences of genetic degeneration (Zhou and Elledge, 2000). Several distinct repair pathways have evolved to repair the many different forms of damage that DNA can suffer (reviewed in Hoeijmakers, 2001). Damage that affects only one strand of the duplex can be repaired using the information present on the remaining intact strand, accordingly, helix distorting injuries, such as UV light–induced pyrimidine dimers and bulky chemical adducts, are predominantly repaired by nucleotide excision repair (NER). Excision of the damaged strand involves ∼20 polypeptides including several proteins encoded by the xeroderma pigmentosum XP disease-causing genes. Two endonuclease activities are required: XPG is needed to make a first incision 3′ to the damaged site and XPF-ERCC1 is required to make a second, 5′ incision. A region of 25–30 nucleotides surrounding the damaged site is removed, and the undamaged strand is used to fill in the gap (Hoeijmakers, 2001). In addition to its role in NER, XPF-ERCC1 is implicated in interstrand cross-link removal, in removing nonhomologous termini from recombination intermediates and in gene replacement (Weeda et al., 1997; Adair et al., 2000; Kuraoka et al., 2000; Sargent et al., 2000; Niedernhofer et al., 2001). Other forms of damage that affect both strands cannot be repaired by copying an intact strand of the duplex. Double-strand breaks (DSB) are caused by ionizing radiation, free radical damage, and by many chemical agents. Two distinct mechanisms exist for the repair of DSBs. Before replication, when only one copy of the genome is available, nonhomologous end-joining (NHEJ) can be used to repair DSBs. NHEJ involves the ligation of two DNA ends with little or no homology. Despite the potential this mechanism has for the loss of information, it is clear that NHEJ is a significant DSB repair pathway in human cells (Critchlow and Jackson, 1998). An alternative to using the end-joining strategy is to repair DSBs using another DNA duplex of identical, or near identical, sequence as a template for extension of broken ends. After replication, when two copies of the genome are available, homologous recombination is favored (Karran, 2000; Jasin, 2002). Homologous recombination involves resection of one strand of the damaged duplex, invasion of highly homologous or identical duplex DNA and extension of the broken strand using one strand of the intact duplex as a template. The process of recombination repair is frequently used to repair or recapture replication forks in prokaryotes and evidence that is also used by eukaryotes in accumulating (McGlynn and Lloyd, 2002).
Phenotypic analysis of two different yeasts suggests that Mus81 is primarily involved in recombination repair (Boddy et al., 2000; Interthal and Heyer, 2000). Mutants of Mus81 are synthetically lethal with homologues of the RecQ helicase, Sgs1 and Rqh1 in budding yeast and fission yeast, respectively (Boddy et al., 2000; Mullen et al., 2001), and Mus81 is required to tolerate replication problems, including those caused by defects in replicative DNA polymerases and depletion of nucleotide pools (Boddy et al., 2000; Interthal and Heyer, 2000). A role for Mus81 in the resolution of Holliday junctions in fission yeast is supported by the observation that disruptants of Mus81 or of its binding partner Eme1 fail to complete meiosis and that the meiotic defect can be suppressed by expression of the bacterial protein, RusA (Boddy et al., 2001), a Holliday junction resolvase (Bolt and Lloyd, 2002). By contrast, meiotic defects in budding yeast Mus81-Mms4 mutants are severe in some strains, but not in others (Interthal and Heyer, 2000; de Los Santos et al., 2001; Kaliraman et al., 2001), deletion of Mus81-Mms4 has a modest effect on meiotic recombination rates (de Los Santos et al., 2001), and recombinant Mus81-Mms4, purified from bacteria, has weak activity on X-structures in vitro (Kaliraman et al., 2001; Whitby et al., 2003). The exact substrates that Mus81-associated endonucleases act on in vivo remain controversial (Boddy et al., 2001; Chen et al., 2001; Haber and Heyer, 2001; Kaliraman et al., 2001; Constantinou et al., 2002; Doe et al., 2002; Whitby et al., 2003).
Biochemical analysis of the endonuclease activity of human Mus81 has shown that it acts on X-shaped DNA duplex oligonucleotides that are used to represent Holliday junctions, Y-shaped duplexes that are used to represent replication forks and 3′ flaps that represent the structures generated during single-strand annealing and during NER (Chen et al., 2001; Constantinou et al., 2002). Mus81 is one of two activities that can process Holliday junctions into linear duplexes in human cell extracts (Constantinou et al., 2002). These two resolvases have distinct biochemical and enzymatic properties, suggesting that they have distinct functions in vivo (Constantinou et al., 2002).
To gain insight into the role Mus81 plays in DNA repair in human cells, we have examined the expression profile and subcellular distribution of human Mus81. We report that the abundance of Mus81 increases as cell progress through the cell cycle, with Mus81 being most abundant in replicative and postreplicative cells. We show that Mus81 is a nuclear protein and that it is enriched in nucleoli. Two human RecQ helicases, BLM and WRN, which are implicated in recombination repair, are also enriched in nucleoli. However, the nucleolar accumulation of Mus81 is not dependent on the presence of either BLM or WRN. In addition, we find that Mus81 is retained at regions of UV damage when cells are in S-phase at the time of UV irradiation. Mus81 is not recruited to regions of DNA damage in cells that are UV-irradiated before or on completion of DNA replication. The retention of Mus81 in nucleoli, the increased abundance in replicating and postreplicative cells, and the observation that it localizes to regions of UV damage specifically in human cells that are undergoing DNA replication suggest that the Mus81-endonuclease is required to resolve problems that arise during DNA replication.
MATERIALS AND METHODS
Cell Lines and Culture
HeLa cells, SV40 transformed normal fibroblasts (GM678G), BS (GM08505 and GM01492), WS (AG11395), and XPA (GM04429F) from Coriell Institute for Medical Research (Camden, NJ) and normal human fibroblasts (WI38) from ATCC (Manassas, VA) were grown in DMEM or MEM supplemented with 10% fetal bovine serum (FBS), 100 μg/ml penicillin, and streptomycin. HeLa cells were synchronized at the G1/S boundary by double-thymidine block: cells were grown in the presence of 2 mM thymidine for 16 h; after 8 h growth in normal medium, 2 mM thymidine was added to the culture and cells were again grown for 16 h. At the time of release, cells were washed twice in 37°C DMEM and returned to growth for the indicated times. WI38 cells were synchronized by growth in MEM supplemented with 0.1% FBS for 3 d and then splitting cells at low density and culturing in MEM with 10% FBS. Where indicated, 2 mM hydroxyurea (Sigma, St. Louis, MO) or 10 nM camptothecin (Sigma) was added to complete culture medium 8 h before harvesting. Cells were UV irradiated using a UV-Stratalinker 18000 (Stratagene, Carlsbad, CA) set to deliver 60 J m–2 of 254-nm light. For local UV irradiation, an isopore polycarbonate membrane filter (Millipore, Billerica, MA) containing 10-μm-diameter pores was placed on top of the cells. After exposure to UV light, the filter was removed, and cells were cultured for 15 min before harvesting.
Immune-fluorescence Microscopy
For immune-fluorescence studies, cells were grown on coverslips for 24 h. Before fixation with 4% formaldehyde, cells were washed in phosphate-buffered saline (PBS) to remove medium, or the in situ fractionation procedure was performed as described in Mirzoeva and Petrini (2001). Coverslips were washed PBS, incubated in cytoskeleton buffer (10 mM piperazine-N,N′-bis(2-ethanesulfonic acid), PIPES, pH 6.8; 100 mM NaCl; 300 mM sucrose; 3 mM MgCl2; 1 mM EGTA; 0.5% Triton X-100) for 5 min on ice, followed by incubation in cytoskeleton stripping buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 1% Tween 40, 0.5% sodium deoxycholate) for 5 min on ice. After several washes with ice-cold PBS, the cells were fixed in 4% formaldehyde for 10 min and permeabilized in 0.5% Triton X-100 solution for 15 min at room temperature as previously described. Cells were blocked with 10% FBS in PBS for 1 h and incubated with primary antibody overnight in 4°C and with secondary antibody for1hat room temperature. All antibodies were diluted in 1% bovine serum albumin. Cells were then washed, counterstained with 4′,6′-diamidino-2-phenylindole (DAPI), and mounted in 70% glycerol. Primary antibody dilutions were as follows: Mus81 1:50, Ha 1:1000, BLM 1:50, WRN 1:100, nucleophosmin/B23 1:100, Lamin 1:100, TDM-2 1:50. Fluorescein isothiocyanate–conjugated anti-mouse or anti-rabbit IgG, or Alexa-fluor 546–conjugated IgG (Molecular Probes, Eugene, OR) were used at 1:500 and 1:1000, respectively. For DNase I treatment, cells were permeabilized with 0.5% Triton X-100 in PBS and incubated with 5 U RNAse-free DNAse I (Roche, Indianapolis, IN) in 0.1 M sodium acetate, 5 mM MgSO4, at 37°C for 10 min before fixing. Images were captured using a charge-couples device camera (Photometrics, Tucson, AZ), gray scale images were processed using Adobe Photoshop 7.0 (Adobe, San Jose, CA).
Where indicated HeLa cells were transfected with 2 μg of pCDNA-3Ha-Mus81 plasmid (Chen et al., 2001) using Effectene (Qiagen, Valencia, CA).
Antibodies and Western Blotting
Antibodies to Mus81 were generated by immunizing a rabbit with GST-Mus81 purified from bacteria. The resulting sera were affinity purified over GST-Mus81 that had been cross-liked to glutathione-Sepharose using dimethylpimelimidate (Harlow and Lane, 1988), as previously described (Chen et al., 2001). Antibodies to BLM (7790), tubulin (5546) and cyclin B (752) were from Santa Cruz Biotechnology (Santa Cruz, CA), WRN (100–161) from Novus Biologicals (Littleton, CO), nucleophosmin/B23 (18-7288) from Zymed (South San Francisco, CA). UV-induced cyclobutane pyrimidine dimers were detected using the mouse monoclonal TDM-2 antibody (Mizuno et al., 1991) kindly provided by Dr. Matsunaga (Kanazawa University). Antibodies to lamins A and C was kindly provided by Dr. Gerace (The Scripps Research Institute). For Western analysis cells were lysed in 20 mM HEPES, pH 7.4, 150 mM NaCl, 5% glycerol, 0.1% NP-40, 0.1% β-mercaptoethanol, 0.5 mM phenylmethylsulphonyl fluoride, and 5 μg/ml leupeptin, pepstatin, and aprotinin. Lysates were cleared by centrifugation at 10,000 × g for 10 min. Protein concentration of the supernatants was determined using Bradford reagent (Bio-Rad, Hercules, CA). Cell lysate, 50 μg, was resolved on 10% acrylamide-SDS gels. Immune-blots were incubated in Mus81 (1:500), cyclin B (1:1000), BLM (1:1000), or tubulin (1:1000) followed by horseradish peroxidase–conjugated anti-rabbit or anti-mouse (Promega, Madison WI). Chemoluminescence (Pierce, Rockford, IL) was used to detect the respective proteins.
RESULTS
Mus81 Abundance Increases in S-phase
To investigate the function of Mus81 in human cells, we first studied its expression and subcellular location. A polyclonal antibody was raised against the full-length protein expressed in bacteria (Chen et al., 2001). Affinity-purified antibodies recognize a band of the expected molecular weight. The abundance of Mus81 was examined in HeLa cells that had been synchronized at the G1/S-boundary by double-thymidine synchronization. Mus81 abundance was low in thymidine-arrested cells and increased as cells progressed through S-phase and into G2 (Figure 1A). Samples from the same experiment were probed with antibody to cyclin B. As expected, cyclin B showed a gradual increase in abundance as cells progressed into S-phase and G2. A considerable reduction in the abundance of cyclin B was seen between 9 and 10 h when the majority of cells passed through mitosis. Likewise, the abundance of Mus81 decreased before the majority of cells passed through mitosis. The top portion of the same gel was probed for the presence of BLM. BLM, the protein defective in Bloom's syndrome (BS), is a RecQ-like helicase that acts in homologous recombination and suppresses illegitimate recombination (Enomoto, 2001; Hickson, 2003). As shown previously, the BLM protein accumulates as cells progress through the cell cycle, with the protein being most abundant late in the cell cycle (Sanz et al., 2000; Bischof et al., 2001). The coincident increased abundance of Mus81 and BLM in replicative and postreplicative cells, suggests a role for both proteins in cells that contain two copies of the genome. The possibility that the increased abundance of Mus81 seen here was a consequence of damage that might occur when cells are cultured in the presence of thymidine was considered. To settle this, the abundance of Mus81 was examined in normal human fibroblasts (WI38) that were synchronized by serum starvation and release. As shown in Figure 1B, the abundance of Mus81 was low in G0-arrested cells, and did not increase until 16–20 h after serum stimulation. The abundance of Mus81 continued to increase until 32 h at which time the majority of cells had entered or had completed S-phase. As seen in HeLa cells, the increase in abundance of Mus81 in WI38 cells closely paralleled that of cyclin B, although the relative change is less for Mus81. In view of the fact that the cells in this experiment were synchronized without exogenous DNA damage it is likely that the increased abundance of Mus81 reflects a requirement for the protein as cells progress through an unperturbed cell cycle.
Figure 1.
The abundance of Mus81 is increased in replicative and postreplicative cells. (A) The abundance of Mus81 was monitored in extracts from HeLa cells after release from a double-thymidine block. Samples were harvested at the indicated time points, and the abundance of Mus81 was monitored by Western analysis. The top portion of the same gel was probed for the presence of BLM, the protein altered in Bloom's syndrome. The timing of mitosis was determined by monitoring the disappearance of cyclin B. (B) The abundance of Mus81 was low in serum-starved WI38 cells and increased gradually as cells progress through S-phase. Contact inhibited serum-starved human fibroblasts were stimulated to reenter the cell cycle by seeding cells at low density in medium containing 10% serum. The abundance of cyclin B, a highly periodic protein that is expressed in late S phase and G2 is shown for comparison (middle panel). Anti-tubulin immune-blots confirmed that equivalent amounts of cell extract were present in all samples (bottom panel).
Mus81 Is Retained in Nucleoli
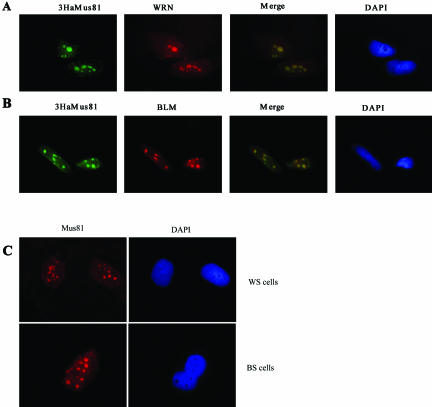
The subcellular distribution of endogenous Mus81 was investigated using affinity-purified antibodies. Indirect immune-fluorescence of fibroblasts (see Figures 3 and 5) or epithelial cells (see Figure 2) revealed a predominantly nuclear staining pattern. Several regions of increased intensity were seen within the nucleus; data shown later indicate that these regions correspond to nucleoli (see Figure 4). Retention of repair enzymes to specific regions of damaged DNA has previously been reported and the site or circumstance of retention has provided insight into the function of several repair protein. Specifically, a number of NER enzymes are recruited to regions of UV irradiation (Katsumi et al., 2001; Mone et al., 2001; Volker et al., 2001), and several recombination repair enzymes are recruited to distinct irradiation induced foci (Maser et al., 1997; Scully et al., 1997; Paull et al., 2000). In some cases the retention of a subpool of protein has been more easily seen after in situ fractionation (Mirzoeva and Petrini, 2001). Therefore we tested the idea that Mus81 might be preferentially retained in nucleoli by procedures that wash away the majority of nuclear Mus81. As shown in Figure 2B, a fraction of Mus81 was preferentially retained in nucleoli after in situ extraction. Prior treatment with agents that damage DNA, specifically UV irradiation, hydroxyurea (HU), and camptothecin (CPT), increased the proportion of cells in the population with strong nucleolar staining (our unpublished results) but did not affect the subnuclear distribution of Mus81 (Figure 2B). As shown below, a similar pattern of nucleolar retention was seen using antibody to the Ha-epitope and cells that had been transfected with plasmid encoding 3HaMus81 (Figure 3, A and B). The pattern of Mus81 retention in subnuclear bodies was reminiscent of that previously reported for the RecQ helicases BLM and WRN (Marciniak et al., 1998; Sanz et al., 2000; Yankiwski et al., 2000). The genes encoding BLM and WRN are mutated in the cancer-prone disorder BS and the cancer prone, progeroid disorder Werner's syndrome (WS), respectively. These syndromes are characterized by a high degree of genomic instability, and cells derived from BS and WS patients show defects in DNA replication. BLM and WRN helicases are both thought to function in replication restart and have been shown to branch migrate Holliday junctions in vitro (Constantinou et al., 2000; Karow et al., 2000a).
Figure 3.
Nucleolar retention of Mus81 is independent of the BLM and WRN helicases. Cells that had grown on coverslips for 24 h were subjected to in situ extraction before fixation as above. (A). 3HaMus81 and WRN colocalize in several subnuclear spots in pCDNA-3HaMus81–transfected HeLa cells. (B) 3HaMus81-colocalizes BLM in several subnuclear spots in pCDNA-3HaMus81–transfected HeLa cells. (C) Nucleolar retention of endogenous Mus81 is not abrogated in BS or WS cells.
Figure 5.
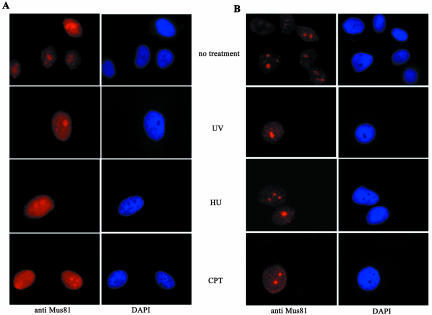
Mus81 is retained at localized regions of UV damage during S-phase. XPA cells were grown on coverslips and synchronized at the G1/S boundary by double-thymidine block. At the indicated times a UV-opaque filter containing pores of ∼10-μm-diameter was placed over cells that were exposed to 60 mJ m–2 254-nm light and then cultured for 15 min before in situ fraction and fixation. Cells were stained for Mus81 and with TDM-2 to detect the regions of UV-induced CPD. (A) A typical panel of cells that were in thymidine at the time of irradiation and at 5, 8, and 11 h after release from the thymidine block. (B) Quantification of the coincident localization of Mus81 and TDM-2 staining. Thirty to 35 cells were counted for each time point from two independent synchronizations.
Figure 2.
Mus81 is a nuclear protein that is retained in nucleoli after in situ extraction. Indirect immune fluorescence analysis of HeLa cells using affinity purified antibody to Mus81 revealed a predominantly nuclear pattern of staining with increased intensity in subnuclear bodies. (A) Samples on the left-hand side were fixed after a brief wash in PBS to remove extracellular proteins only. (B) Samples on the righthand side were extracted with detergent containing buffers to remove soluble proteins before fixation (Mirzoeva and Petrini, 2001). Samples were costained with DAPI to reveal DNA. Treatment of cells with UV-light, HU, or CTP before fixation had little effect on the localization of Mus81.
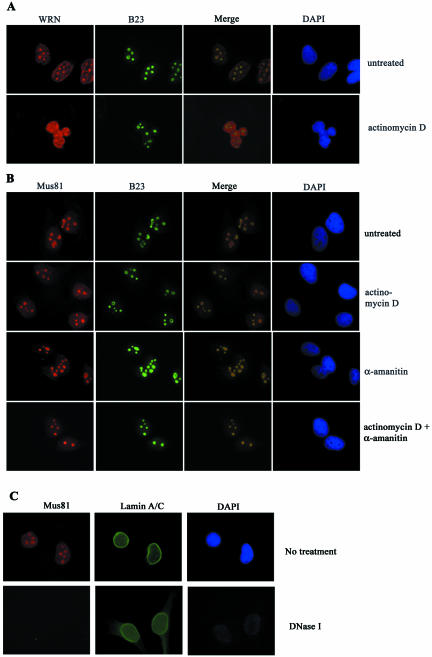
Figure 4.
Mus81 is retained in the nucleoli of actinomycin D– and α-amanitin–treated cells, but not in DNAse I–treated cells. HeLa cells were incubated with 50 ng/ml actinomycin D and/or 0.5 μg/ml α-amanitin for 1 h before harvest. After in situ extraction and fixation, samples were costained for WRN or Mus81, the nucleolar protein, nucleophosmin/B23, and for DNA. (A) Nucleolar WRN became predominately nuclearplasmic after treatment with actinomycin D. (B) The nucleolar staining of Mus81 was not affected by actinomycin D or α-amanitin treatment. (C) Digestion of DNA results in the loss of nucleolar Mus81 staining.
If Mus81 is acting on the same structures as BLM and WRN, it might be expected to colocalize with these proteins, and if Mus81 associates with the helicases its accumulation in the nucleolus might be dependent on their presence. We therefore examined the pattern of Mus81 in cells that had been costained with antibody to WRN or BLM. The antibodies against BLM AND WRN were raised in rabbits; therefore the location of 3Ha-Mus81 was determined using mouse monoclonal anti-Ha. As previously reported WRN was found to localize predominantly in 3–5 subnuclear regions (Figure 3A; Gray et al., 1998; Marciniak et al., 1998). The identity of these spots as nucleoli was confirmed by staining with antibody to human nucleolar protein nucleophosmin/B23 (Figure 4A). Merged images of Mus81 and nuclophosmin/B23 staining reveal the two proteins within nucleoli and suggest that they are concentrated in distinct subnucleolar domains (Figure 4B). Costaining of cells with Mus81 and WRN antibodies revealed coincident localization of the two proteins (Figure 3A). Costaining of Mus81 with BLM also showed significant colocalization of the two proteins (Figure 3B). Antibody to BLM also stained smaller, more punctate regions of the nucleus that costain with antibodies to the PML protein (Johnson et al., 2000; Sanz et al., 2000; Bischof et al., 2001; and our unpublished results). PML bodies were found to stain weakly or not at all with Mus81 antibody. The nucleolar distribution of Mus81 was examined in cells from patients with BS and with WS that lack detectable BLM (Neff et al., 1999) and WRN (Moser et al., 2000) protein, respectively. As shown in Figure 3C, the nucleolar staining of Mus81 was unaltered in these cell-lines, indicating that the recruitment of Mus81 to nucleoli is not dependent on the presence of either of these helicases.
Nucleoli are enriched in regions of the genome that encode rRNA genes. The highly repetitive nature of the rDNA loci is susceptible to recombination. In budding yeast, mutants in the Sgs1 helicase, which is a sequence homologue of BLM and WRN, result in increased genetic instability, particularly at the rDNA locus (Sinclair and Guarente, 1997; Sinclair et al., 1997). Thus, one explanation for the abundance of WRN in nucleoli is that it is required to suppress recombination at highly repetitive sequences in the nucleolus (Marciniak et al., 1998). Nucleoli are also regions of high transcriptional activity and the presence of WRN in nucleoli correlates with on-going transcription (Shiratori et al., 2002). Furthermore, WS cells have reduced rates of transcription that can be complimented by introduction of the wild-type protein (Balajee et al., 1999). Therefore the abundance of WRN in nucleoli might reflect a requirement for WRN in efficient transcription. To determine if the nucleolar accumulation of Mus81 required on-going transcription, HeLa cells were treated with actinomycin D and α-amanitin and the location of Mus81, WRN, and the nucleolar marker nucleophosmin/B23 was determined (Figure 4, A and B). As previously reported, treatment with 50 ng/ml actinomycin D caused the majority of WRN to relocate to the nucleoplasm with less retention of the protein in the nucleoli (Shiratori et al., 2002; Figure 4A). In contrast to WRN, the nucleolar staining of Mus81 was unaffected by actinomycin D. Exposure to the RNA polymerase II inhibitor, α-amanitin, and simultaneous treatment with actinomycin D and α-amanitin did not affect the nucleolar localization of Mus81, (Figure 4B). These data confirmed that nucleolar retention of Mus81 is independent of WRN and suggest that its presence in the nucleolus does not require on-going transcription. Because the nucleolar staining of Mus81 was resistant to detergent extraction, to transcriptional inhibitors, and to the absence of BLN or WRN, we sought evidence that nucleolar retention of Mus81 was dependent on the presence of DNA. To do this, cells were exposed to mild detergent and digested with DNAse I before fixation. DAPI staining revealed that the majority of DNA was released from nuclei, whereas staining with antibody to lamins A and C showed that proteins of the nuclear envelope were not disrupted by this treatment (Dwyer and Blobel, 1976). As shown in Figure 4C, the majority of Mus81 was released by DNAse I treatment. Thus, the presence of Mus81 in nucleoli is directly or indirectly dependent on the presence of DNA.
Mus81 Is Recruited to Regions of UV Damage
The retention of Mus81 within discrete subnuclear bodies suggested that the protein is partially compartmentalized in the absence of DNA damage. A similar pattern was seen after DNA damage (Figure 2); however, the agents used above damage DNA throughout the nucleus and retention to the whole nucleus can be difficult to monitor. Polycarbonate filters containing pores of defined size have been used to restrict UV damage to confined areas of individual nuclei, and several proteins are transiently recruited to specific sites of damage (Katsumi et al., 2001; Mone et al., 2001; Volker et al., 2001). To determine if Mus81 can be recruited to regions of damage, cells were irradiated through polycarbonate filters containing 10-μm-diameter pores. SV40-transformed cells with a defect in XPA were used for this analysis because they are deficient in NER (Mone et al., 2001). UV-induced damage was visualized by immune-fluorescence labeling using the cyclobutane pyrimidine dimer (CPD) specific mAb TDM2 (Mizuno et al., 1991; Mori et al., 1991). Preliminary analysis showed that Mus81 was retained at regions of UV damage in 20–30% of the cells in an asynchronous population (our unpublished results), suggesting that retention might be related to cell cycle position.
To determine if Mus81 is recruited to UV-damaged regions in a cell cycle–dependent manner, cells were synchronized at the G1/S boundary by double-thymidine block. Arrested cells, or cells that had been released for 5, 8, or 11 h, were UV irradiated, and 15 min later samples were fixed and stained for regions of damage and for Mus81. When thymidine-arrested cells were UV irradiated, regions of damage were clearly seen using the TDM-2 antibody. Mus81 was found only in nucleoli. Costaining of Mus81 and the damage specific antibody was only seen in cells in which the UV-damaged region happened to contain nucleoli (Figure 5A, top panel). By contrast, 5 h after thymidine release, Mus81 was found to colocalize with the regions of damage in >90% of cells. Mus81 staining within the nucleoli was still clearly visible in these cells, suggesting that recruitment of Mus81 to the UV-damaged regions does not significantly deplete the pool of nucleolar Mus81. Eight hours after release from thymidine 42% of cells retained Mus81 at regions of UV irradiation. In cells that had been released from thymidine for 11 h, Mus81 was found in regions of UV-induced damage in <10% of cells. Quantification of the coincidence of regions of damage and Mus81 staining is shown in Figure 5B. The recruitment of Mus81 to damaged areas of the nucleus in S-phase cells suggests that Mus81 is recruited to regions of damage when replication forks encounter a region of UV-damaged template. The observation that it is rarely recruited to regions of damage in thymidine-arrested cells suggests it is not recruited to regions of UV damage per se.
DISCUSSION
Mus81 is a subunit of a highly conserved, structure specific endonuclease that is thought to function in a homologous recombination repair pathway. In support of this idea, we find that human Mus81 is expressed primarily in S-phase and G2, when homologous recombination is operational. We have previously reported that the abundance of Mus81 increases after DNA damage and that its abundance is greatest in cells treated with agents that interfere with progression through S-phase (Chen et al., 2001). Here, we report that the abundance of Mus81 increases in the S-phase of cells that have not been exposed to exogenous DNA damaging agents, suggesting that Mus81 participates in normal S-phase functions. The abundance of Mus81 drops quite abruptly at the time cells enter mitosis. The drop in Mus81 protein slightly precedes the disappearance of cyclin B, which is degraded late in mitosis (Pines and Hunter, 1989). A similar pattern of expression was seen for the BLM helicase, a protein that is also implicated in homologous recombination in human cells (Bischof et al., 2001). The abundance of Mus81 is highest in cells that have partially or completely replicated the genome, supporting the idea that Mus81 abundance increases in cells that are competent for homologous recombination repair. We found that Mus81 is preferentially retained both in the nucleoli of damaged and undamaged cells. A similar pattern of subnuclear localization was seen in a number of cell lines. The significance of the nucleolar location is not entirely clear; it could simply suggest nucleolar storage of Mus81. However, the observation that Mus81 is found in the same place as WRN and BLM support the idea that Mus81 and the helicases function in related processes. BS cells show marked genomic instability; in particular, they have elevated rates of recombination between sister chromatids and homologous chromosomes (reviewed in Enomoto, 2001; Hickson, 2003). This instability is thought to result from defective processing of DNA replication intermediates. The most obvious phenotype associated with WS is premature ageing, but these patients are also highly susceptible to early onset cancer and cells from WS patients have defects in S-phase progression and mitotic recombination (Prince et al., 2001). Both BLM and WRN have been shown to promote the ATP-dependent translocation of Holliday junctions in vitro (Constantinou et al., 2000; Karow et al., 2000a). Thus the phenotype of BS and WS cells could arise from a failure to branch migrate Holliday junctions and thus from a failure to restore replication forks. This hypothesis is supported by the observation that the WRN, BLM, and Mus81 accumulate in regions of highly repetitive recombinogenic DNA (Sinclair and Guarente, 1997; Sinclair et al., 1997; Marciniak et al., 1998; Sanz et al., 2000). The BLM helicase is found in PML bodies and in nucleoli of somatic cells (Sanz et al., 2000; Bischof et al., 2001) and is associated with synapsed chromosomes in meiotic cells (Walpita et al., 1999; Moens et al., 2000).
In this analysis, we have found that Mus81 and BLM colocalize in nucleoli. Consistent with other reports (Johnson et al., 2000; Bischof et al., 2001), we also found BLM in smaller more punctate regions that costain with antibody to PML (our unpublished results) in cells that were not extracted before fixation. We did not find any evidence that Mus81 specifically accumulates in PML bodies. However, it is worth noting that before in situ extraction, Mus81 is present throughout the nucleus, and an accumulation in or association with PML bodies could have been missed in this analysis. Mus81 is retained in the nucleoli of WS and BS cells that lack detectable expression of these proteins (Neff et al., 1999; Moser et al., 2000), and thus retention of Mus81 in nucleoli is not dependent on WRN or BLM. This could be explained if Mus81 and WRN act in alternative pathways to process a common replication intermediate that frequently arises in nucleoli. A model for the function of BLM and WRN in suppressing recombination is based on the observation that these helicase bind and migrate Holliday junctions in vitro. If Mus81 resolves Holliday junctions that cannot be pushed back into productive replication forks, Mus81 would be needed in the same circumstances as the helicases, but function independently of them (Boddy et al., 2001). Support for this model comes from the observation that simultaneous disruption of Mus81 and Sgs1 or Rqh1 is lethal in budding and fission yeast, respectively (Boddy et al., 2000; Mullen et al., 2001). Unlike WRN, Mus81 was not released from nucleoli on treatment with actinomycin D or α-amanitin, conditions that are commonly used to suppress transcription (Casse et al., 1999). Even simultaneous treatment with actinomycin D and α-amanitin did not detectably affect the nucleolar retention of Mus81. By contrast, Mus81 was released from nucleoli after treatment with DNAse I; thus it is likely that, Mus81 is retained in nucleoli by binding to DNA. Mus81 has helix-hairpin-helix motifs found in many DNA-binding proteins (Aravind et al., 1999; Boddy et al., 2000; Interthal and Heyer, 2000; Chen et al., 2001), and Mus81 itself may bind the nucleolar DNA directly; however, the possibility that Mus81 is retained on nucleolar DNA through association with other proteins cannot been discounted.
UV light predominantly induces pyrimidine dimers and other bulky adducts that are efficiently removed by NER. If these lesions are not repaired and a replication fork runs into the damaged site, a double-strand end will be generated. (Michel et al., 1997; Limoli et al., 2000; Oakley et al., 2001). We did not find evidence that Mus81 is recruited to regions of UV-induced damage in cells that are held in thymidine; however, in cells that have been released from the block and allowed to progress through S-phase, Mus81 was frequently retained at damaged regions of the nucleus. The simplest explanation for this is that Mus81 is not required to repair UV-induced damage as such, but that it is required at damaged regions when replication is on-going. If progression of a replication fork is blocked by damage on the template, the leading and lagging strands may anneal forming a chicken foot or Holliday junction structure (Higgins et al., 1976). The formation of a chicken foot structure allows extension of the leading strand; thus by using the lagging strand as template, the damaged site can be bypassed. After this, RecQ helicases are hypothesized to migrate the Holliday junction back into a replication fork (Karow et al., 2000b), and replication can resume beyond the damaged site. In these circumstances, the damaged site is not immediately repaired, but replication is not blocked and no potentially mutagenic repair has occurred. Cells that lack the RecQ helicase have elevated rates of recombination; thus it is assumed that the helicase limits recombination by favoring the restoration of the fork. One model for Mus81 function is that it acts on Holliday junctions that are formed when replication forks encounter damage. The data presented here support such a model, but do not eliminate models in which Mus81 acts on other structures that might arise in UV-irradiated cells (Kaliraman et al., 2001; Doe et al., 2002; Whitby et al., 2003).
DOI: 10.1091/mbc.E03-05-0276.
References
- Adair, G.M., Rolig, R.L., Moore-Faver, D., Zabelshansky, M., Wilson, J.H., and Nairn, R.S. (2000). Role of ERCC1 in removal of long non-homologous tails during targeted homologous recombination. EMBO J. 19, 5552–5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind, L., Walker, D.R., and Koonin, E.V. (1999). Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 27, 1223–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balajee, A.S. et al. (1999). The Werner syndrome protein is involved in RNA polymerase II transcription. Mol. Biol. Cell 10, 2655–2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof, O., Kim, S.H., Irving, J., Beresten, S., Ellis, N.A., and Campisi, J. (2001). Regulation and localization of the bloom syndrome protein in response to DNA damage. J. Cell Biol. 153, 367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy, M.N., Gaillard, P.H., McDonald, W.H., Shanahan, P., Yates, J.R., and Russell, P. (2001). Mus81-eme1 are essential components of a holliday junction resolvase. Cell 107, 537–548. [DOI] [PubMed] [Google Scholar]
- Boddy, M.N., Lopez-Girona, A., Shanahan, P., Interthal, H., Heyer, W.D., and Russell, P. (2000). Damage tolerance protein mus81 associates with the FHA1 domain of checkpoint kinase cds1. Mol. Cell. Biol. 20, 8758–8766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolt, E.L., and Lloyd, R.G. (2002). Substrate specificity of RusA resolvase reveals the DNA structures targeted by RuvAB and RecG in vivo. Mol. Cell 10, 187–198. [DOI] [PubMed] [Google Scholar]
- Casse, C., Giannoni, F., Nguyen, V.T., Dubois, M.F., and Bensaude, O. (1999). The transcriptional inhibitors, actinomycin D and alpha-amanitin, activate the HIV-1 promoter and favor phosphorylation of the RNA polymerase II C-terminal domain. J. Biol. Chem. 274, 16097–16106. [DOI] [PubMed] [Google Scholar]
- Chen, X.B. et al. (2001). Human Mus81-associated endonuclease cleaves Holliday junctions in vitro. Mol. Cell. 8, 1117–1127. [DOI] [PubMed] [Google Scholar]
- Constantinou, A., Chen, X.B., McGowan, C.H., and West, S.C. (2002). Holliday junction resolution in human cells: two junction endonucleases with distinct substrate specificities. EMBO J. 21, 5577–5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinou, A., Tarsounas, M., Karow, J.K., Brosh, R.M., Bohr, V.A., Hickson, I.D., and West, S.C. (2000). Werner's syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep. 1, 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Critchlow, S.E., and Jackson, S.P. (1998). DNA end-joining: from yeast to man. Trends Biochem. Sci. 23, 394–398. [DOI] [PubMed] [Google Scholar]
- de Los Santos, T., Loidl, J., Larkin, B., and Hollingsworth, N.M. (2001). A role for MMS4 in the processing of recombination intermediates during meiosis in Saccharomyces cerevisiae. Genetics 159, 1511–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe, C.L., Ahn, J.S., Dixon, J., and Whitby, M.C. (2002). Mus81-Eme1 and Rqh1 involvement in processing stalled and collapsed replication forks. J. Biol. Chem. 277, 32753–32759. [DOI] [PubMed] [Google Scholar]
- Dwyer, N., and Blobel, G. (1976). A modified procedure for the isolation of a pore complex-lamina fraction from rat liver nuclei. J. Cell Biol. 70, 581–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto, T. (2001). Functions of RecQ family helicases: possible involvement of Bloom's and Werner's syndrome gene products in guarding genome integrity during DNA replication. J. Biochem. (Tokyo) 129, 501–507. [DOI] [PubMed] [Google Scholar]
- Gray, M.D., Wang, L., Youssoufian, H., Martin, G.M., and Oshima, J. (1998). Werner helicase is localized to transcriptionally active nucleoli of cycling cells. Exp. Cell Res. 242, 487–494. [DOI] [PubMed] [Google Scholar]
- Haber, J.E., and Heyer, W.D. (2001). The fuss about Mus81. Cell 107, 551–554. [DOI] [PubMed] [Google Scholar]
- Harlow, E., and Lane, D. (1988). Antibodies. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Hickson, I.D. (2003). RecQ helicases: caretakers of the genome. Nat. Rev. Cancer 3, 169–178. [DOI] [PubMed] [Google Scholar]
- Higgins, N.P., Kato, K., and Strauss, B. (1976). A model for replication repair in mammalian cells. J. Mol. Biol. 101, 417–425. [DOI] [PubMed] [Google Scholar]
- Hoeijmakers, J.H. (2001). Genome maintenance mechanisms for preventing cancer. Nature 411, 366–374. [DOI] [PubMed] [Google Scholar]
- Interthal, H., and Heyer, W.D. (2000). MUS81 encodes a novel helix-hairpin-helix protein involved in the response to UV- and methylation-induced DNA damage in Saccharomyces cerevisiae. Mol. Gen. Genet. 263, 812–827. [DOI] [PubMed] [Google Scholar]
- Jasin, M. (2002). Homologous repair of DNA damage and tumorigenesis: the BRCA connection. Oncogene 21, 8981–8993. [DOI] [PubMed] [Google Scholar]
- Johnson, F.B. et al. (2000). Association of the Bloom syndrome protein with topoisomerase IIIalpha in somatic and meiotic cells. Cancer Res. 60, 1162–1167. [PubMed] [Google Scholar]
- Kaliraman, V., Mullen, J.R., Fricke, W.M., Bastin-Shanower, S.A., and Brill, S.J. (2001). Functional overlap between Sgs1-Top3 and the Mms4-Mus81 endonuclease. Genes Dev. 15, 2730–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karow, J.K., Constantinou, A., Li, J.L., West, S.C., and Hickson, I.D. (2000a). The Bloom's syndrome gene product promotes branch migration of holliday junctions. Proc. Natl. Acad. Sci. USA 97, 6504–6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karow, J.K., Wu, L., and Hickson, I.D. (2000b). RecQ family helicases: roles in cancer and aging. Curr. Opin. Genet. Dev. 10, 32–38. [DOI] [PubMed] [Google Scholar]
- Karran, P. (2000). DNA double-strand break repair in mammalian cells. Curr. Opin. Genet. Dev. 10, 144–150. [DOI] [PubMed] [Google Scholar]
- Katsumi, S. et al. (2001). In situ visualization of ultraviolet-light-induced DNA damage repair in locally irradiated human fibroblasts. J. Invest. Dermatol. 117, 1156–1161. [DOI] [PubMed] [Google Scholar]
- Kuraoka, I., Kobertz, W.R., Ariza, R.R., Biggerstaff, M., Essigmann, J.M., and Wood, R.D. (2000). Repair of an interstrand DNA cross-link initiated by ERCC1-XPF repair/recombination nuclease. J. Biol. Chem. 275, 26632–26636. [DOI] [PubMed] [Google Scholar]
- Limoli, C.L., Giedzinski, E., Morgan, W.F., and Cleaver, J.E. (2000). Inaugural article: polymerase eta deficiency in the xeroderma pigmentosum variant uncovers an overlap between the S phase checkpoint and double-strand break repair. Proc. Natl. Acad. Sci. USA 97, 7939–7946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak, R.A., Lombard, D.B., Johnson, F.B., and Guarente, L. (1998). Nucleolar localization of the Werner syndrome protein in human cells. Proc. Natl. Acad. Sci. USA 95, 6887–6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser, R.S., Monsen, K.J., Nelms, B.E., and Petrini, J.H. (1997). hMre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks. Mol. Cell. Biol. 17, 6087–6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlynn, P., and Lloyd, R.G. (2002). Recombinational repair and restart of damaged replication forks. Nat. Rev. Mol. Cell. Biol. 3, 859–870. [DOI] [PubMed] [Google Scholar]
- Michel, B., Ehrlich, S.D., and Uzest, M. (1997). DNA double-strand breaks caused by replication arrest. EMBO J. 16, 430–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzoeva, O.K., and Petrini, J.H. (2001). DNA damage-dependent nuclear dynamics of the Mre11 complex. Mol. Cell. Biol. 21, 281–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno, T., Matsunaga, T., Ihara, M., and Nikaido, O. (1991). Establishment of a monoclonal antibody recognizing cyclobutane-type thymine dimers in DNA: a comparative study with 64M-1 antibody specific for (6–4) photoproducts. Mutat. Res. 254, 175–184. [DOI] [PubMed] [Google Scholar]
- Moens, P.B., Freire, R., Tarsounas, M., Spyropoulos, B., and Jackson, S.P. (2000). Expression and nuclear localization of BLM, a chromosome stability protein mutated in Bloom's syndrome, suggest a role in recombination during meiotic prophase. J. Cell Sci. 113, 663–672. [DOI] [PubMed] [Google Scholar]
- Mone, M.J., Volker, M., Nikaido, O., Mullenders, L.H., van Zeeland, A.A., Verschure, P.J., Manders, E.M., and van Driel, R. (2001). Local UV-induced DNA damage in cell nuclei results in local transcription inhibition. EMBO Rep. 2, 1013–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori, T., Nakane, M., Hattori, T., Matsunaga, T., Ihara, M., and Nikaido, O. (1991). Simultaneous establishment of monoclonal antibodies specific for either cyclobutane pyrimidine dimer or (6–4)photoproduct from the same mouse immunized with ultraviolet-irradiated DNA. Photochem. Photobiol. 54, 225–232. [DOI] [PubMed] [Google Scholar]
- Moser, M.J., Kamath-Loeb, A.S., Jacob, J.E., Bennett, S.E., Oshima, J., and Monnat, R.J., Jr. (2000). WRN helicase expression in Werner syndrome cell lines. Nucleic. Acids Res. 28, 648–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen, J.R., Kaliraman, V., Ibrahim, S.S., and Brill, S.J. (2001). Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics 157, 103–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff, N.F., Ellis, N.A., Ye, T.Z., Noonan, J., Huang, K., Sanz, M., and Proytcheva, M. (1999). The DNA helicase activity of BLM is necessary for the correction of the genomic instability of bloom syndrome cells. Mol. Biol. Cell 10, 665–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedernhofer, L.J. et al. (2001). The structure-specific endonuclease Ercc1-Xpf is required for targeted gene replacement in embryonic stem cells. EMBO J. 20, 6540–6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley, G.G. et al. (2001). UV-induced hyperphosphorylation of replication protein a depends on dna replication and expression of atm protein. Mol. Biol. Cell 12, 1199–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull, T.T., Rogakou, E.P., Yamazaki, V., Kirchgessner, C.U., Gellert, M., and Bonner, W.M. (2000). A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 10, 886–895. [DOI] [PubMed] [Google Scholar]
- Pines, J., and Hunter, T. (1989). Isolation of a human cyclin cDNA: evidence for cyclin mRNA and protein regulation in the cell cycle and for interaction with p34cdc2. Cell 58, 833–846. [DOI] [PubMed] [Google Scholar]
- Prince, P.R., Emond, M.J., and Monnat, R.J., Jr. (2001). Loss of Werner syndrome protein function promotes aberrant mitotic recombination. Genes Dev. 15, 933–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz, M.M., Proytcheva, M., Ellis, N.A., Holloman, W.K., and German, J. (2000). BLM, the Bloom's syndrome protein, varies during the cell cycle in its amount, distribution, and co-localization with other nuclear proteins. Cytogenet Cell Genet. 91, 217–223. [DOI] [PubMed] [Google Scholar]
- Sargent, R.G., Meservy, J.L., Perkins, B.D., Kilburn, A.E., Intody, Z., Adair, G.M., Nairn, R.S., and Wilson, J.H. (2000). Role of the nucleotide excision repair gene ERCC1 in formation of recombination-dependent rearrangements in mammalian cells. Nucleic Acids Res. 28, 3771–3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully, R., Chen, J., Ochs, R.L., Keegan, K., Hoekstra, M., Feunteun, J., and Livingston, D.M. (1997). Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell 90, 425–435. [DOI] [PubMed] [Google Scholar]
- Shiratori, M., Suzuki, T., Itoh, C., Goto, M., Furuichi, Y., and Matsumoto, T. (2002). WRN helicase accelerates the transcription of ribosomal RNA as a component of an RNA polymerase I-associated complex. Oncogene 21, 2447–2454. [DOI] [PubMed] [Google Scholar]
- Sinclair, D.A., and Guarente, L. (1997). Extrachromosomal rDNA circles–a cause of aging in yeast. Cell 91, 1033–1042. [DOI] [PubMed] [Google Scholar]
- Sinclair, D.A., Mills, K., and Guarente, L. (1997). Accelerated aging and nucleolar fragmentation in yeast sgs1 mutants. Science 277, 1313–1316. [DOI] [PubMed] [Google Scholar]
- Volker, M. et al. (2001). Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell 8, 213–224. [DOI] [PubMed] [Google Scholar]
- Walpita, D., Plug, A.W., Neff, N.F., German, J., and Ashley, T. (1999). Bloom's syndrome protein, BLM, colocalizes with replication protein A in meiotic prophase nuclei of mammalian spermatocytes. Proc. Natl. Acad. Sci. USA 96, 5622–5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeda, G. et al. (1997). Disruption of mouse ERCC1 results in a novel repair syndrome with growth failure, nuclear abnormalities and senescence. Curr. Biol. 7, 427–439. [DOI] [PubMed] [Google Scholar]
- Whitby, M.C., Osman, F., and Dixon, J. (2003). Cleavage of model replication forks by fission yeast mus81-eme1 and budding yeast mus81-mms4. J. Biol. Chem. 278, 6928–6935. [DOI] [PubMed] [Google Scholar]
- Yankiwski, V., Marciniak, R.A., Guarente, L., and Neff, N.F. (2000). Nuclear structure in normal and Bloom syndrome cells. Proc. Natl. Acad. Sci. USA 97, 5214–5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, B.B., and Elledge, S.J. (2000). The DNA damage response: putting checkpoints in perspective. Nature 408, 433–439. [DOI] [PubMed] [Google Scholar]