Abstract
Because of its eukaryotic nature, simple fermentation requirements, and pliable genetics, there have been many attempts at improving recombinant protein production in S. cerevisiae. These strategies typically involve altering the expression of a native protein thought to be involved in heterologous protein trafficking. Usually, these approaches yield three to ten-fold improvements over wild-type strains and are almost always specific to one type of protein. In this study, a library of mutant alpha mating factor 1 leader peptides (MFα1pp) is screened for the enhanced secretion of a single-chain antibody. One of the isolated mutants is shown to enhance the secretion of the scFv up to sixteen-fold over wild-type. These leaders also confer a secretory improvement to two other scFvs as well as two additional, structurally unrelated proteins. Moreover, the improved leader sequences, combined with strain engineering, allow for a one-hundred eighty fold improvement over previous reports in the secretion of full length, functional, glycosylated human IgG1. The production of full-length IgG1 at milligram per liter titers in a simple, laboratory-scale system will significantly expedite drug discovery and reagent synthesis while reducing antibody cloning, production, and characterization costs.
Keywords: heterologous protein secretion, S. cerevisiae, directed evolution, secretory leader, IgG
Introduction
Commonly used as a secretory peptide (Brake 1984; Kjaerulff and Jensen 2005; Lee et al. 2002; Oka et al. 1999), the MFα1pp secretory leader is made of two distinct pieces: the pre region and the pro region. The first nineteen residues make up the pre region that serves to direct the nascent polypeptide to the endoplasmic reticulum. Although the exact amino acid sequence varies widely, pre regions from different proteins do often share some common motifs. Pre regions typically consist of a positively charged N-terminus, followed by a hydrophobic middle sequence, and then a polar C-terminus (Brodsky 1998). It is no surprise, then, that mutations to these residues can severely compromise ER targeting and secretion (Rapoport et al. 1996). Although yeast can translocate polypeptides into the endoplasmic reticulum co-translationally or post-translationally, it is believed that proteins under the direction of the MFα1pp are typically translocated post-translationally (Lodish 2000). Upon extrusion into the ER, the pre sequence is cleaved by signal peptidase leaving a smaller molecular weight proprotein. At this point, N-linked glycosylation is added to three asparagine residues on the pro sequence. Although not absolutely essential to secretion, these glycosylations do appear to play a role in facilitating ER to Golgi transport (Caplan et al. 1991; Ellgaard and Helenius 2003). In addition to inter-organelle transport, the pro region may play a role in ER translocation as insulin-like growth factor is inefficiently translocated when expressed with the pre region alone (Chaudhuri et al. 1992). In addition to these roles, the pro region is thought to facilitate aspects of late secretory processing such as COPII vesicle sorting and vacuolar targeting. In fact the vacuole is a common, although undesirable, fate for secretion-directed recombinant protein. As the proprotein migrates through the Golgi, it may encounter a member of a family of vacuolar sorting proteins (VPS) that actively direct the proprotein from the Golgi to the vacuole. These proteins are normally involved in the trafficking of native vacuole-localized proteins but may engage and consequently “mis-sort” heterologous proteins into the vacuole as well. (Caplan et al. 1991; Chaudhuri et al. 1992; Otte and Barlowe 2004; Zhang et al. 2001). Ultimately, the pro region is cleaved by the late Golgi protease Kex2 before the mature protein is trafficked to the surface.
The MFα1pp is commonly used to direct the secretion of a number of different heterologous proteins in a number of hosts including S. cerevisiae, S. pombe, P. pastoris, and mammalian cells, and its success as a secretory leader depends a great deal on the protein being directed (Kjaerulff and Jensen 2005; Lee et al. 2002; Oka et al. 1999). Secretory leaders have previously been engineered by iterative processes of rational design and empirical optimization, specifically for the secretion of insulin precursor. These synthetic leaders imparted as much as a 2.5-fold improvement in insulin precursor secretion over the wild-type alpha mating factor 1 prepro (Kjeldsen et al. 1997). Other synthetic prepros have been developed for the secretion of hEGF which direct secretion at levels similar to those obtained using the native yeast invertase prepro (Clements 1991). However, to this point, it has not been demonstrated whether a native secretory leader can be engineered through directed evolution to enhance the secretion of a heterologous protein. In this study, an evolutionary approach is taken to dramatically improve the secretion of a single chain antibody fragment (scFv) as well as a full-length IgG, a molecule that has previously been secreted from S. cerevisiae in titers of only 50 μg/L (Horwitz et al. 1988). It is the hope that this approach can be used to augment the more traditional methodologies of expression maturation as well as create a production platform for high-value proteins such as IgG.
Materials and Methods
Alpha Factor Prepro Library Construction
The alpha mating factor leader was PCR-amplified from genomic DNA isolated from a prep (Zymo Research, Orange, CA) of the yeast strain BJ5464α (matα ura3-52 leu2Δ1 his3Δ200 pep4∷HIS3 prb1Δ1.6Rcan1 GAL) using the primers 5′-atatgcatgcatgagatttccttcaatttttact and 5′-atatgctagctcttttatccagctgtaccccttc, which introduce SphI and NheI (New England Biolabs, Beverly, MA) restriction sites respectively. The leader was then re-amplified with the primers 5′-aagtgatagtggtgtttgagtcataacgacgtcgctagccttgtcatcgtcgtcctgtagtcaaagagtctggg and 5′-ttgttaatatacctctatactttaacgtcaaggagaaaaaacccgcatgc, which introduced a FLAG tag 3′ to the leader sequence. This leader was then subcloned 3′ to the GAL10 promoter and 5′ to the 4m5.3 scFv ORF (Midelfort et al. 2004) to create the tryptophan auxotrophic, low-copy, CEN shuttle vector WTappF4m5.3. Error prone PCR mutagenesis was performed on the SphI/NheI excised leader sequence using the second set of primers described above. Ten micrograms of SphI/NheI cut WTappF4m5.3 acceptor vector was combined with fifty micrograms of the gel purified PCR product and transformed into the yeast BJ5464α (matα ura3-52, trp1 leu2Δ1 his3Δ200 pep4∷HIS3 prb1Δ1.6R can1 GAL) over ten electroporations to yield a library of approximately 1×108 clones (Colby et al. 2004).
Library Screening
The library was passaged into five milliliters of SD-CAA (2% glucose, 0.67% yeast nitrogen base, 0.54% Na2HPO4, 0.86% NaH2PO4·H2O, and 0.5% casein amino acids) and grown to an OD600 of 12 before being induced in YPG/BSA (2% galactose, 2% peptone, 1% yeast extract, and 0.025% bovine serum albumin) for twelve hours at 30°C. The cells were then labeled with fluorescein and analyzed by the cell surface secretion assay (CeSSA) (Rakestraw 2006). Briefly, this assay uses target covalently linked to the yeast cell wall to capture and display the associated binding protein secreted from the cell. For the application described here, labeling the cell with fluorescein allows for the capture of the femtomolar affinity anti-fluorescein scFv 4m5.3. It has been shown previously that this assay is an effective tool for selections for improved heterologous protein secretion. After elution from the assay, the cells were labeled with 50μl 10μg/ml M2 anti-FLAG antibody (Sigma, St. Louis, MO) for twenty minutes on ice. After washing once in 1 ml PBS/BSA (1g/L bovine serum albumin), the cells were labeled in 50 μl 20 μg/mL Alexa-Fluor610-R-phycoerythrin goat anti-mouse IgG (Invitrogen, Chicago, IL). The library was sorted on a Cytomation MoFlo Cell Sorter (Cytomation Inc., Fort Collins, CO) or a BD FACSaria (Becton Dickinson, San Diego, CA), and the cells collected in SD-CAA media supplemented with 50 U/mL penicillin and 50 μg/mL streptomycin (Gibco/Invitrogen). After outgrowth of the culture, the cells were induced again and the process repeated for a total of six rounds of selection.
Mutant Characterization
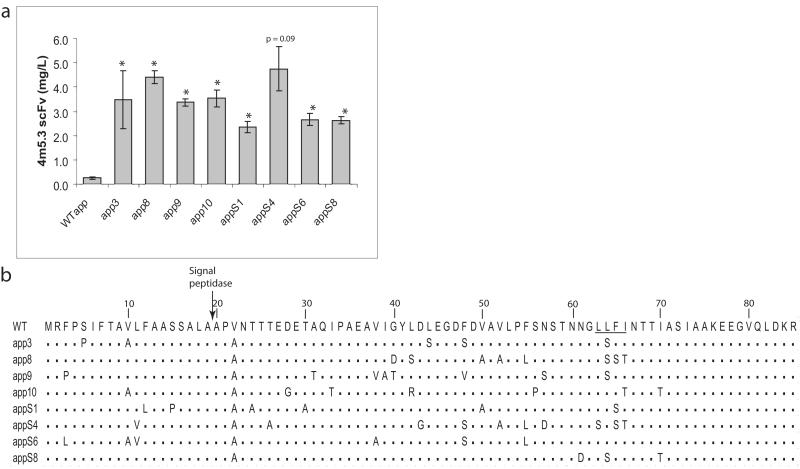
Isolated clones were grown in SD-CAA overnight and then induced at saturation overnight in YPG/BSA at 30°C. 500μl of the supernatant was used to quantify 4m5.3 scFv concentration with the fluorescein quench titration assay (Midelfort et al. 2004). Plasmids from particularly productive clones were isolated using the Zymoprep Yeast Plasmid Miniprep Kit and amplified in XL-1 Blue chemically competent E. coli (Stratagene, Carlsbad, CA). The mutant leaders were sequenced, and the isolated plasmid transformed into a fresh strain of BJ5464α using the EZ Yeast Transformation Kit (Zymo Research) and assayed again. The data presented in Figure 1a are from this second characterization.
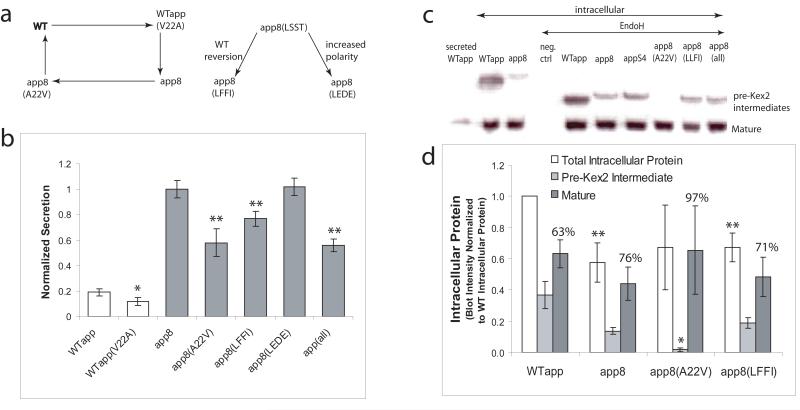
Figure 1.
(a) Secreted 4m5.3 scFv from individual clones isolated from the mutagenized alpha factor leader library are compared to the wild-type leader (WTαpp); error bars indicate one standard deviation of three trials (two trials for the S4 leader). Student′s t-tests comparing average 4m5.3 secretion between each mutant leader and WTαpp returning p<0.05 are indicated by an asterisk unless otherwise indicated. (b) Sequences of eight of the most productive clones; the signal peptidase site and LLFI motif are indicated.
Site-Directed Mutagenesis
Site-directed mutagenesis was performed by PCR amplification of the entire mutant plasmid using complementary primers containing the desired point mutations in a 30μl reaction. The amplification was digested with one microliter of DpnI (New England Biolabs), and two microliters were transformed into XL-1 Blue. The transformants were then sequenced to confirm the desired mutations. Leader sequences with the necessary mutations were then subcloned into a new expression vector to ensure the fidelity of the unsequenced plasmid. The plasmids were then transformed back into fresh BJ5464α.
Intracellular Western Blots
Intracellular protein was isolated using a TCA protein precipitation protocol. After a fourteen-hour induction in YPG/BSA, 2×107 cells were resuspended in 100μl 20% TCA extraction buffer (20 mM TrisCl, pH 7.9; 50 mM ammonium acetate, 2 mM EDTA) containing Halt Protease Inhibitor (Pierce, Rockford, IL). The cells were added to 1.5 mL vials containing ~600 μl of glass beads (Sigma) and 100 μl TCA and beaten with 2×50 second bursts on a bead beater with one minute incubations on ice between the bead treatments. After beating, the lysate was withdrawn and centrifuged for five minutes at 12,000×g. The supernatant was discarded and the pellet containing the precipitated protein was resuspended in 100 μl TCA resuspension buffer (3% SDS, 100 mM Tris base, pH 11, 3 mM DTT).
Seventeen microliters of resuspension was boiled in 2 μl of denaturation buffer (New England Biolabs) for ten minutes. Following denaturation, 2 μl of Buffer G5 was added followed by 3 μl of EndoHf (New England Biolabs) followed by a 1.5 hour incubation at 37°C. After the addition of loading buffer, the protein was run on a 4-12% Bis-Tris gradient gel (Invitrogen), transferred to a nitrocellulose membrane (BioRad, Hercules, CA), and then incubated in 5% milk/TBST with 1:4000 anti-FLAG HRP (Sigma). A 1:500 dilution of anti-yeast phosphoglycerate kinase antibody (Invitrogen) was also added as an internal loading control. After a one-hour incubation at room temperature and subsequent washings in TBST, a 1:1000 dilution of goat anti-mouse HRP antibody (Sigma) was added and the blot incubated for twenty minutes before development using the SuperSignal West Femto Substrate (Pierce) and imaging on a FluorS phosphorescence imager (BioRad).
VPS and DER1 Chromosomal Deletions
Yeast chromosomal deletions were made using an antibiotic cassette disruption technique based on previously described protocols (Johnston et al. 2002; Rakestraw and Wittrup 2006; Wach et al. 1994). In summary the KanMX gene conferring G418 resistance to yeast was amplified by PCR with primers that contain approximately fifty bases of homology to the 5′ ends of both strands of the target gene. The PCR product was then transformed into yeast by electroporation where the cassette inserted itself into the target site on the chromosome by homologous recombination. Transformants were then selected by growth on plates containing G418. The location of the integration was confirmed by colony PCR using one primer with homology to sequence just outside of the insertion site and one primer with homology to the KanMX insertion cassette.
Gene Construction and Expression of Additional Heterologous Proteins
Genes from IL-2 2-4(Q126R) (Rao 2005), D1.3, horseradish peroxidase (HRP), and disulfide-stabilized sm3E (Graff 2004) were amplified by PCR introducing NheI and XhoI sub-cloning restriction sites on the ends. The amplifications were cloned into the mutant prepro expression vectors and transformed into BJ5464α. After overnight growth in SD-CAA, the cells were induced for 24 hours in YPG/BSA at 30°C after which 17 μl of supernatant for the D1.3 and IL-2 2-4 (Q126R) cultures was withdrawn and analyzed by Western blotting (protein expressions done in triplicate). FLAG-BAP fusions (Sigma) were used for quantification. HRP secretion was quantified by enzymatic assay (Lipovsek 2007).
The human IgG1 constant heavy chain was PCR amplified from vector 6-23 IgG (Yeung 2007), a derivative of the human IgG1 kappa expression vector pPNL501 (generous gift of Michael Feldhaus) introducing MluI and XhoI sites on the 5′ and 3′ ends respectively. The heavy chain was then subcloned into the vector WTαppD1.3 using the MluI and XhoI subcloning sites. The 4m5.3 variable region was PCR amplified from WTαppF4m5.3 with primers introducing MluI and NheI sites with which it was subcloned into the new heavy chain vector. The light chain was PCR amplified from 6-23 introducing 5′ and 3′ NheI and XhoI restriction sites respectively, which were used for subcloning into the WTαpp4m5.3 vector to make the 6-23 light chain yeast vector. The 4m5.3 light chain variable region was amplified from WTαppF4m5.3 using primers introducing NheI and BsiWI sites used to subclone the VL behind the WT prepro. Mutant prepro leaders were inserted by amplifying the leaders introducing SphI and NheI sites and cloning them in front of the IgG heavy or light chain ORF.
IgG vectors were transformed into the yeast BJ5464α which were grown overnight in five milliliter cultures in SD-CAA at 30°C. The saturated culture was then induced in phosphate buffered YPG/BSA (YPG, 0.54% Na2HPO4, 0.86% NaH2PO4·H2O) for three days at 20°C. For integration the heavy and light chains were subcloned into pRS304 and pRS306 respectively with KpnI and SacI digests. Five micrograms of vector were linearized by restriction digest in the nutritional marker and transformed into BJ5464α or YVH10 (BJ5464α containing an integrated, GAPDH promoted, yeast PDI gene) (Robinson et al. 1994) using electroporation.
Expression and purification of 4m5.3 hIgG1
Wild-type 4m5.3 or 4m5.3 N297Q heavy and light chain vectors were transformed into YVH10 and selected for growth on SD-CAA plates. A single colony was inoculated into 5 ml SD-CAA and grown overnight at 30°C until saturation (OD600 ~ 5), and then used to inoculate 1 L of SD-CAA in a shake flask at 30 °C for ~ 24 hrs. Upon saturation (OD600 ~ 5), cells were pelleted and resuspended in 1 L of phosphate buffered YPG and allowed to secrete for 72 hrs at 20°C. Cell culture supernatant was clarified by centrifugation followed by vacuum filtration, then concentrated and exchanged into PBS, pH 7.4. IgG was purified from concentrated supernatant by Protein A (Pierce) followed by anti-FLAG (Sigma) affinity chromatography.
For characterization of IgG glycosylation, 50 μg of purified IgG was treated with EndoH (New England Biolabs) and/or jack bean mannosidase (Prozyme, San Leando, CA), then re-purified with Protein A agarose (Pierce).
Cloning of 225 and sm3E as hIgG1 chimeras
The 225 variable regions were PCR-amplified from the yeast surface display vector pCT-225, expressing 225 as a scFv (225 scFv DNA graciously provided by Winfried Wels). The oligos 5-agtcacacgcgtcaggtacaactgaagcagtcagg and 5′-tcatacgctagcagcggaaacggtgaccagggtcccttgg were used to amplify the 225 VH with 5′ and 3′ MluI and NheI sites respectively for ligation into the hIgG1 heavy chain backbone; the oligos 5′-caacgtgctagcgacatcctgctgacccagtctccag and 5′-atgtaccgtacgtttgagctccagcttggtcccagc were used to amplify the 225 VL with 5′ and 3′ NheI and BsiWI sites respectively for ligation into the light chain backbone. Similarly, the sm3E variable regions were PCR-amplified from the yeast secretion vector sm3E-His.
Flow cytometric analysis of IgG labeling
1 × 107 YVH10 cells were washed three times with 500 μl carbonate buffer (4.2% NaHCO3 and 0.034% NaCO3, pH 8.4), incubated for 30 min with either 4 μg/μl NHSPEG-fluorescein or NHS-PEG-biotin (Nektar), and then washed three times with 1000 μl PBS/BSA. Fluorescein-labeled yeast were then incubated with 10 μg/ml 4m5.3 hIgG1 or a hIgG1 kappa polyclonal control antibody (Sigma) for 60 min at room temperature, washed, and then labeled with a 1:100 dilution of goat anti-human phycoerythrin conjugate (Rockland, Gilbertsville, PA) for 30 min on ice. For competition experiments, 10 μg/ml 4m5.3 hIgG1 was pre-incubated with 10 μM fluorescein (Pierce) for 60 min at room temperature and the mixture then incubated with fluorescein-labeled yeast.
A431NS cells (ATCC; kind gift of Jennifer Cochran), a derivative of the EGFR-overexpressing epidermoid carcinoma cell line A431 (Girard 1973), were maintained in DMEM plus 10% FBS. At approximately 80% confluency, cells were trypsinized, washed three times with ice cold PBS/BSA, and counted on a hemocytometer. 2 × 105 cells were incubated with 5 μg/ml yeast-produced, purified 225 hIgG1 or hIgG1 kappa control antibody for 60 min on ice, washed, and then labeled with a 1:100 dilution of goat anti-human PE for 30 min on ice. For competition experiments, 5 μg/ml yeast-produced 225 hIgG1 was pre-incubated with 1 μM EGFR ectodomain (404SG) (Kim 2006) for 60 min on ice before the mixture was incubated with A431NS cells. Alternatively, A431NS cells were pre-incubated with 50 μg/ml commercial murine 225 IgG (Lab Vision, Fremont, CA) for 60 min on ice before yeast-produced, purified 225 IgG was added to a final concentration of 5 μg/ml (resulting in a final murine 225 concentration of 25 μg/ml). The yeast 225 IgG was then labeled with a PE conjugated goat anti-human antibody as described above.
LS174T cells (ATCC), a CEA-overexpressing colorectal adenocarcinoma cell line, were maintained in MEM supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. At approximately 80% confluency, cells were trypsinized, washed three times with ice cold PBS/BSA, and counted on a hemocytometer. 2 × 105 cells were incubated with 10 nM yeast-produced sm3E hIgG1 or hIgG1 kappa control antibody for 60 min on ice, washed, and then labeled with a 1:100 dilution of goat anti-human PE for 30 min on ice. For competition experiments, LS174T cells were sm3E IgG was added to a final concentration of 10 nM resulting in a final competitor scFv concentration of 100 nM.
Student′s t-Tests
All studies were done in triplicate unless otherwise stated. When comparing between two populations, the Student′s t-test was performed using the null hypothesis that the sample means of both populations are the same. That is, the test returned the probability (denoted in the text and figures as “p”) that the means of the two populations are equivalent. The two data set means that are being compared and the relevant p-values are given in the figure captions or text.
Results
For the mutant leader selections, the fluorescein-binding single chain antibody 4m5.3 (Boder et al. 2000) was used as a model heterologous protein. The 4m5.3 scFv was cloned under the direction of a library of S. cerevisiae alpha mating factor 1 prepro signal sequence peptides preceding a Kex2p recognition site for leader peptide cleavage. The library was created through error prone PCR mutagenesis of the native prepro sequence followed by transformation into yeast via homologous recombination with the 4m5.3 ORF to yield a library of approximately 108 variants. The selection for improved secretion was carried out using the Cell Surface Secretion Assay (CeSSA) (Manz et al. 1995; Rakestraw 2006). The CeSSA technique uses cell surface-bound antigen to capture the secreted scFv against the antigen (in this case using a fluorescein antigen covalently bound to the library of cells secreting the anti-fluorescein scFv 4m5.3). This assay has been shown to be an effective way to select for the improved secretion of the scFv. After the last round of sorting, plasmid DNA was isolated and retransformed into fresh cells that were then tested for 4m5.3 scFv secretion. All of the isolated and characterized mutant leaders imparted improvement in 4m5.3 secretion over the wild-type leader (WTαpp) with the most productive mutant leaders yielding up to a sixteen-fold improvement (the eight best clones are shown in Figure 1a). The isolated leaders were sequenced and analyzed for similarities (sequences of the eight best leader sequences are shown in Figure 1b). The sequence data show definite trends in leader mutations. The 22nd valine residue in the WTαpp is universally changed to an alanine residue. Moreover, the hydrophobic LLFI motif spanning the 63rd to the 66th residue is mutated to residues with more polar side chains. The impact of these two types of mutations will be addressed later.
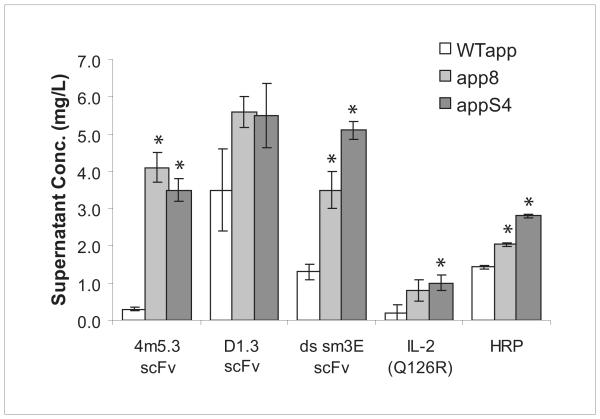
One of the greatest difficulties in generating hyperproductive strains for secretion is the inability to develop strains with enhanced secretion independent of the protein being expressed. To test whether the αpp8 and αppS4 leaders isolated from the selection could be used to improve the secretion of proteins other than 4m5.3 scFv, the two leaders were used to direct the secretion of the lysozyme binding scFv D1.3 and a disulfide-stabilized version of the carcinoembryonic antigen (CEA) binding scFv sm3E (Graff 2004). The data show that the αpp8 and αppS4 leaders enhance the secretion of both types of single-chains (Figure 2). Additionally, the two leaders were used to enhance the secretion of horseradish peroxidase as well as a poorly expressing mutant of interleukin-2, IL-2 (Q126R). These results demonstrate that leader-imparted improvements in heterologous protein secretion can be generalized to other scFvs as well as structurally unrelated proteins.
Figure 2.
Secretion of the scFvs 4m5.3, D1.3, and sm3E as well as the IL-2 mutant 2-4(Q126R) and HRP was assayed under the direction of the WT, αpp8, and αppS4 leaders. Student′s t-tests comparing average secretion titers between the WTαpp and the mutant leaders for each protein returning p<0.05 are indicated with an asterisk. Error bars indicate one standard deviation.
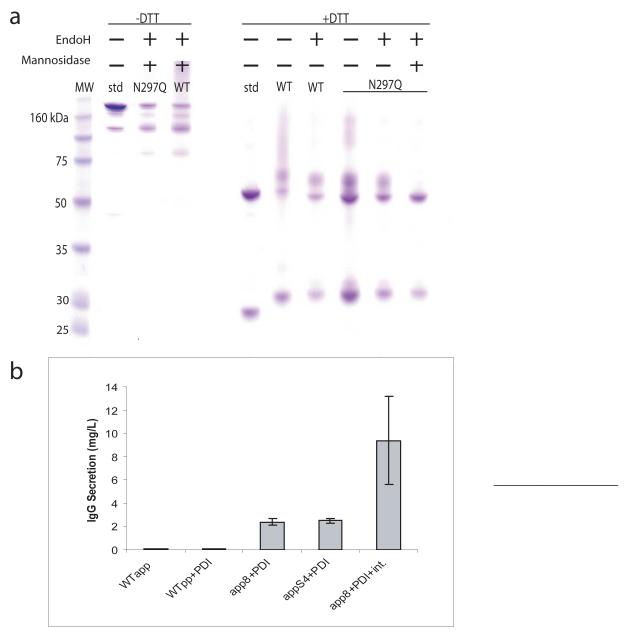
Immunoglobulin fragments such as scFvs and Fabs have been successfully expressed in yeast (Gasser 2007). However, with previously reported yields as low as 50 μg/L, S. cerevisiae has not been shown to secrete the full-length IgG in titers sufficient for use as an expression host (Horwitz et al. 1988). In hopes to remedy this shortcoming, the αpp8 and αppS4 leaders were used to direct the secretion of the full-length heavy chain and the N-terminally FLAG-epitope-tagged light chain of a mouse/human chimeric 4m5.3 IgG1. These chains were expressed from two different auxotrophically marked low copy plasmids. Protein gels of protein A and FLAG-purified supernatant show a band consistent in size with a full-length human IgG1 (Figure 3a) that is reactive with both anti-human and anti-FLAG antibodies when probed by Western blot (data not shown). Additionally, smaller molecular weight bands consisting of partially assembled IgG are purified. When reduced with DTT, the IgG bands collapse into a 30 kDa light chain band (containing the FLAG epitope tag), two Fc-containing bands around 50 kDa, as well as a good deal of higher molecular weight protein. Treating the IgG with the N-glycosidase EndoH results in the two major forms of the heavy chain exhibiting a slightly increased mobility. The new mobility of these two bands is consistent with that of the DTT-reduced, non-N-glycosylated 4m5.3 N297Q mutant, which removes the lone N-linked glycosylation site in hIgG1, demonstrating that the fully assembled IgG contains N-linked Fc glycosylation. The Fc site is the only N-linked glycosylation site in the final, fully processed form of the molecule. Higher molecular weight, EndoH sensitive forms of the N297Q mutant (as well as some of the higher molecular weight WT IgG) are likely non-Kex2 cleaved protein still retaining the N-glycosylated pro region. Further treatment of the IgG with mannosidase causes the upper heavy chain band to collapse into the lower band, suggesting that a subpopulation of the secreted heavy chain has some O-linked glycosylation in addition to the typical N-linked Fc glycosylation.
Figure 3.
(a) SDS-PAGE of Protein A and FLAG purified 4m5.3 IgG1 generated with the αpp8 leader. Abbreviations: std (control human IgG1 standard), WT (wild-type 4m5.3 IgG with app8 leader), N297Q (4m5.3 N297Q non N-glycosylated mutant with app8 leader). (b) The secretion of a 4m5.3 IgG1 mouse/human chimera was improved by strain manipulation. The heavy and light chain genes were expressed separately on two CEN plasmids and transformed into BJ5464α cells or cells carrying an additional, integrated copy of PDI. The genes were also integrated (Int.) into the chromosomes of PDI-overexpressing cells in place of CEN plasmid expression. Error bars indicate one standard deviation.
4m5.3 IgG secretion was further enhanced through manipulation of the host strain. IgG secretion directed by the αpp8 and αppS4 leaders was compared to the wild-type leader with and without the overexpression of protein disulfide isomerase (PDI), a chaperone previously shown to be beneficial to scFv expression (Figure 3b) (Robinson et al. 1994; Shusta et al. 1998). Both improved leaders, in combination with PDI, enhanced IgG secretion 40-fold over the wild-type leader alone with about 25-fold of that improvement due to the mutant leaders. In previous studies, it has been shown that heterologous protein expression can put significant negative selection pressure on low-copy plasmid retention resulting in as much as 50% of cells to be plasmid negative (see (Boder et al. 2000) and (Rakestraw and Wittrup 2006) for comparison of a yeast surface displayed protein utilizing genes expressed from low-copy plasmids versus integrations). To compensate for expression instability caused by the dual plasmid system, chromosomal integrations of the two genes were made resulting in an additional four-fold improvement in secretion. The total yield of 4m5.3 IgG secreted from the enhanced expression strain in five mL culture was determined to be approximately 9 mg/L by a fluorescein quench assay and quantitative Western blotting. The total yield of a one-liter shake flask culture after protein A and FLAG purification was determined to be approximately 1.5 mg. Combined, these strategies impart a 180-fold improvement in IgG secretion over previously reported yields making S. cerevisiae a productive host for the expression of full-length IgG.
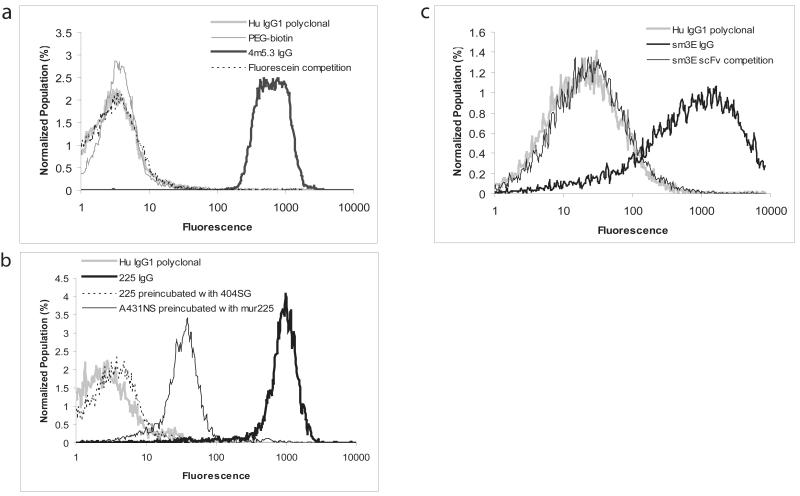
To test the functionality and specificity of the yeast-produced IgG, purified 4m5.3 IgG was used to bind fluorescein-labeled cells. Specificity was measured by labeling the cells with biotin instead of fluorescein. Additionally, the 4m5.3 IgG was preincubated with 10μM fluorescein before being added to the cells (Figure 4a). A human, type-matched, polyclonal IgG was also used as a negative control in the labeling experiments. To test the flexibility of the yeast host for the generation of IgGs with different variable domains, the variable heavy and light chains of the anti-EGFR 225 IgG and the anti-CEA scFv sm3.E were cloned into the yeast IgG vector to express as hIgG1 chimeras. Antibody titers for 225 and sm3E in 1 L cultures were similar to or better than the yields derived for 4m5.3 expression. The yeast-produced 225 IgG binds to the EGFR-expressing A431NS cell line. Competition experiments show that the IgG does not show strong binding when cells are pre-incubated with murine 225 or when the IgG is pre-incubated with soluble 404SG, a yeast produced version of the extracellular domain of EGFR (Figure 4b). Additionally, the sm3E IgG labels the CEA-expressing LS174T cell line but is blocked by pre-incubating the cells with sm3E scFv (Figure 4c). Functional expression of these three antibodies, derived from different mouse variable genes and further altered by chimerization, suggests that this system can robustly express full-length antibodies regardless of their family or origin.
Figure 4.
(a) Labeling of fluorescein-conjugated yeast with purified 4m5.3 IgG followed by an anti-human PE-conjugated antibody and analysis by flow cytometry. A type-matched, non-specific, human IgG1 kappa polyclonal antibody (Hu IgG1 polyclonal) and cells labeled with biotin (PEG-biotin) instead of fluorescein were used as negative controls. Pre-incubation of the 4m5.3 IgG with 10 μM fluorescein was also done to demonstrate specificity. (b) Labeling of the EGFR expressing cell line A431NS with yeast-produced 225 IgG. Specificity was determined through competition assays: yeast 225 IgG was pre-incubated with yeast-produced EGFR ectodomain (404SG), or the A431NS cells were pre-labeled with murine 225 IgG. (c) Labeling of the CEA presenting cell line LS174T with sm3E IgG. Specificity was demonstrated by pre-incubating the cells with the sm3E scFv.
The αpp8 and αppS4 leaders have been shown to improve the secretion of a range of proteins. The isolated leaders (listed in Figure 1b) also exhibit some common sequence motifs. The two most common types of mutations in the improved leaders are the A22V mutation in the early pro region and a tendency toward more polar residues in the WTαpp LLFI motif found in the 63rd to 66th residues of the prepro. To examine the effects of the V22A mutation, a mutagenic cycle was performed on the leader at this position (Figure 5a). In this cycle, the V22A mutation that is ubiquitous in the improved leaders was also mutated in the WTαpp. Additionally, the alanine at the 22nd position of the mutant leader αpp8 was reverted back to the wild-type valine. Looking at the secretory output of the mutants in the V22A mutagenesis cycle (Figure 5b), it is apparent how important the V22A mutation is to the αpp8 leader, yet this mutation does not benefit the WTαpp leader at all. Although the V22A mutation is essential to the enhanced activity of the αpp8 leader, the improvement is only fully realized within the context of the other mutations. This dependence is an important realization as it implies that the mutations found in the αpp8 leader work in concert with one another. Therefore, it may be supposed that toggling the 22nd residue back and forth between valine and alanine could have different effects on protein trafficking depending on whether it is done in the context of the WTαpp or αpp8 leader. Perhaps the V22A mutation overcomes a secretory bottleneck that is only encountered when the other αpp8 mutations are present. Alternatively, the V22A mutation might interact directly with another mutated residue on the leader in order to be effective. Because of this synergy between the various mutations, it is more relevant to look at individual mutations in the αpp8 leader in hopes of ascertaining the contribution of each mutation to the secretory improvement imparted by the leader as a whole. In following this strategy, the polar LSST motif in the αpp8 leader was reverted back to the wild-type LFFI or mutated to the more polar but bulkier residues LEDE. Together, these data show that there is a functional preference for more polar residues in place of the native LLFI motif in the mutant leaders.
Figure 5.
(a) Mutational analysis of improved leaders. The WT Val22 was mutated to alanine, and the Ala22 in αpp8 was mutated back to the WT valine. The effect of the hydrophobicity of the 63rd-66th residues on secretion was also examined by mutating αpp8(LSST) to the wild-type LFFI or the more polar LEDE. (b) Secretory competencies of the valine and hydrophobicity mutants in (a), as well as that of the combination of all the αpp8 mutations (i.e. A22V, LSST to LLFI, and L55F, denoted “all”) were normalized to the secretory output of αpp8. Average secretion titers using mutant leaders compared to WTαpp (white bars) with p<0.05 are indicated by one asterisk. Average secretory output with mutant leaders compared to αpp8 (gray bars) with p<0.05 are indicated by two asterisks. Error bars indicate one standard deviation. (c) Secreted 4m5.3 scFv from the WTαpp (WT secreted) as well as intracellularly accumulated 4m5.3 (lanes 2-10) from cell lysate with and without EndoH treatment. (d) Total intracellular protein and its distribution into the pre-Kex2 cleaved and mature forms as determined by Western blot band intensity for the WT, αpp8, αpp8(V22A), and αpp8(LFFI) mutant leaders. The data have been normalized to WTαpp total intracellular protein, and one standard deviation of two trials is shown. Student′s t-tests comparing average total intracellular protein between WTαpp and αpp8 with p<0.15 are indicated by two asterisks. T-tests comparing the average percentage of pre-Kex2 cleaved protein between the αpp8 mutants and αpp8 returning p<0.05 are indicated with one asterisk.
Although the secreted forms from both wild-type and mutant leaders are consistent with mature 4m5.3 scFv in size, companion intracellular Western blots show that there are two forms of intracellular scFv: a glycosylated, higher molecular weight, pre-Kex2 proprotein form in addition to a full-length, mature form (Figure 5c). Intracellular immunofluorescent microscopy suggests that a considerable amount of intracellular protein is localized to a large organelle consistent with the vacuole in size and structure (data not shown). The vacuole has previously been shown to be a common destination for heterologous protein that has been mis-sorted from the Golgi (Coughlan et al. 2004; Rakestraw and Wittrup 2006; Steube et al. 1991; Zhang et al. 2001). A survey of intracellular 4m5.3 scFv and its distribution between the pre-Kex2 form and the mature form was made by analyzing the band intensity of Western blots for two separate sets of experiments (Figure 5d). The amount of protein found in the pre-Kex2 and mature forms as well as the total intracellular protein (pre-Kex2 plus mature) for the WTαpp, αpp8, and two of the αpp8 mutagenic cycle leaders are shown. For easier comparison, all of the data have been normalized to the total intracellular protein found from the WTαpp leader. The percent of total intracellular protein made of the mature form for each leader is also shown. This analysis of intracellular 4m5.3 considered with the effects of the leaders on secretion in Figure 5b show that on the whole, the αpp8 leader redistributes total protein from the intracellular forms to the supernatant when compared to WTαpp. Furthermore, changing the alanine at the 22nd position in the αpp8 leader back to the wild-type valine causes a drastic reduction in pre-Kex2 protein accumulation compared to αpp8. This result suggests that this alanine in the αpp8 leader may reduce Kex2 cleavage efficiency somehow resulting in preferential trafficking to the surface when the pro region is finally cleaved (no proprotein was found in the supernatant). Alternatively, an alanine at the 22nd position may give the αpp8-led protein more secretory pathway residence time, which is a characteristic previously correlated with improved secretion (Kjeldsen et al. 1997; Luo 2002). In any case, the αpp8(A22V) mutation reveals a secretory bottleneck characterized, in part, by relatively less pre-Kex2 intermediate than the unmutated αpp8 leader. The alanine in the 22nd position of the αpp8 leader is a key part of overcoming this bottleneck. The αpp8 LLFI reversion has little effect on total intracellular protein accumulation or intracellular protein distribution compared to αpp8.
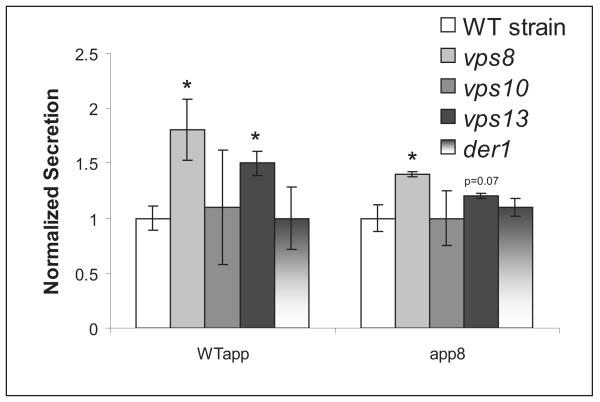
Because vacuolar mis-targeting seems to be a common fate for heterologous protein including the WTαpp 4m5.3 scFv here, it may be that the wild-type leader itself contains sequence elements that make it more likely to engage one of the vacuolar sorting proteins. This engagement would cause the protein to be trafficked to the vacuole instead of the surface. We hypothesized that the αpp8 leader removes the sequence elements responsible for this vacuolar mistrafficking causing more protein to be secreted. To test this hypothesis, chromosomal deletions of three vacuolar sorting proteins often implicated in heterologous protein trafficking, VPS8, VSP10, and VPS13 (Bowers and Stevens 2005; Cowles et al. 1997; Harsay and Bretscher 1995; Harsay and Schekman 2002; Zhang et al. 2001) were made in the expression strain. The VPS8 and VPS13 mutations do improve the secretion of wild-type leader scFv almost two-fold, suggesting that vacuolar sorting could play some role in the retention of protein directed by the WTαpp (Figure 6). However, the VPS mutations do not account for all of the secretory improvement seen in the αpp8 leader. Deletions of the ER associated degradation (ERAD) protein DER1 (Knop 1996) or the vacuolar sorting protein VPS10 had no effect on secretion.
Figure 6.
Three vacuolar sorting genes, VPS8, VPS10, VPS13, as well as an ERAD associated gene, DER1, were deleted, and the resulting 4m5.3 secretory competency under the WT and αpp8 leaders was tested. The data are normalized to the secretion in the WT strain for each leader. Student′s t-tests comparing average secretion between the mutant deletion strains and the WT strain for each leader yielding p<0.05 are indicated by an asterisk. Error bars indicate one standard deviation.
Conclusions
The secretory leaders described here have shown enhanced heterologous protein secretion in S. cerevisiae for a variety of proteins at levels that are as good as or better than previously reported results. These improvements come from an ensemble of mutations that affect the distribution of protein between non-Kex2 processed and mature forms within the cell and ultimately the distribution between total intracellular protein and the supernatant. The results also suggest leader engineering may be a more general approach than genomic mutations which typically enhance productivity for only one particular protein. This methodology seems to yield more productive strains and to be applicable to a wider range of proteins. A further approach would be to combine this method with traditional genomic approaches to strain improvement, which, in this example, yielded a 180-fold improvement over previously reported results for the secretion of full-length IgG1 when combined with PDI overexpression. The amount of IgG secreted using the improved leader makes full-length antibody production accessible for those wanting to use small scale, easily fermentable microorganisms to produce usable, purifiable amounts of IgG in time spans of a week or less. This ability stands in stark contrast to the weeks to months necessary for hybridoma generation and mammalian cell fermentation yet produces comparable yields to E. coli-based methods without the need for lysis or extensive purification (Mazor 2007; Simmons 2002). Furthermore, these yeast-produced antibodies retain their binding characteristics and specificities even when expressed with an array of variable domains. The techniques presented here illustrate a new strategy and method for the selection of clones and strains with improved heterologous secretion. Moreover, the success of IgG secretion in yeast facilitates candidate therapeutic characterization, reagent generation, and drug target validation.
Acknowledgements
The authors would like to thank Pacific Northwest National Laboratories, MIT Flow Cytometry Core, MIT Biopolymers Lab, Ben Hackel, Annie Gai, Mike Schmidt, Anne Robinson, Andy Yeung, Jennifer Cochran, NIH CA96504 and CA101830.
Contributor Information
J. Andy Rakestraw, MIT Department of Biological Engineering.
Stephen L. Sazinsky, MIT Department of Biological Engineering.
Andrea Piatesi, MIT Department of Biological Engineering.
Eugene Antipov, MIT Department of Biological Engineering.
Works Cited
- Boder ET, Midelfort KS, Wittrup KD. Directed evolution of antibody fragments with monovalent femtomolar antigen-binding affinity. Proc Natl Acad Sci U S A. 2000;97(20):10701–5. doi: 10.1073/pnas.170297297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers K, Stevens TH. Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 2005;1744(3):438–54. doi: 10.1016/j.bbamcr.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Brake AJ, et al. Alpha-factor-directed synthesis and secretion of mature foreign proteins in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1984;81:4642–6. doi: 10.1073/pnas.81.15.4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky JL. Targeting to and Translocation across the Endoplasmic Reticulum Membrane. In: Phoenix DA, editor. Protein Targeting and Translocation. Princeton University Press; Princeton, NJ: 1998. pp. 169–191. [Google Scholar]
- Caplan S, Green R, Rocco J, Kurjan J. Glycosylation and structure of the yeast MF alpha 1 alpha-factor precursor is important for efficient transport through the secretory pathway. J Bacteriol. 1991;173(2):627–35. doi: 10.1128/jb.173.2.627-635.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri B, Steube K, Stephan C. The pro-region of the yeast prepro-alpha-factor is essential for membrane translocation of human insulin-like growth factor 1 in vivo. Eur J Biochem. 1992;206(3):793–800. doi: 10.1111/j.1432-1033.1992.tb16986.x. [DOI] [PubMed] [Google Scholar]
- Clements JM, Catlin JH, Price MJ, Edwards RM. Secretion of human epidermal growth factor from Saccharomyces cerevisiae using synthetic leader sequences. Gene. 1991;106(2):267–71. doi: 10.1016/0378-1119(91)90209-t. [DOI] [PubMed] [Google Scholar]
- Colby DW, Kellogg BA, Graff CP, Yeung YA, Swers JS, Wittrup KD. Engineering antibody affinity by yeast surface display. Methods Enzymol. 2004;388:348–58. doi: 10.1016/S0076-6879(04)88027-3. [DOI] [PubMed] [Google Scholar]
- Coughlan CM, Walker JL, Cochran JC, Wittrup KD, Brodsky JL. Degradation of mutated bovine pancreatic trypsin inhibitor in the yeast vacuole suggests post-endoplasmic reticulum protein quality control. J Biol Chem. 2004;279(15):15289–97. doi: 10.1074/jbc.M309673200. [DOI] [PubMed] [Google Scholar]
- Cowles CR, Snyder WB, Burd CG, Emr SD. Novel Golgi to vacuole delivery pathway in yeast: identification of a sorting determinant and required transport component. Embo J. 1997;16(10):2769–82. doi: 10.1093/emboj/16.10.2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellgaard L, Helenius A. Quality control in the endoplasmic reticulum. Nat Rev Mol Cell Biol. 2003;4(3):181–91. doi: 10.1038/nrm1052. [DOI] [PubMed] [Google Scholar]
- Gasser B, Mattanovich D. Antibody production with yeasts and fungi: on the road to large scale? Biotechnol Letters. 2007;29:201–12. doi: 10.1007/s10529-006-9237-x. [DOI] [PubMed] [Google Scholar]
- Girard DJ, Aaronson SA, Todaro GJ, Amstein P, Kersey JH, Dosik H, Parks WP. In vitro cultivation of human tumors: establishment of cell lines derived from a series of solid tumors. J. Natl Cancer Inst. 1973;51(5):1417–1423. doi: 10.1093/jnci/51.5.1417. [DOI] [PubMed] [Google Scholar]
- Graff CP, Chester K, Begent R, Wittrup KD. Directed evolution of an anti-carcinoembryonic antigen with a 4-day monovalent dissociation half-time at 37 degrees C. Protein Eng. Des. Sel. 2004;17(4):293–304. doi: 10.1093/protein/gzh038. [DOI] [PubMed] [Google Scholar]
- Harsay E, Bretscher A. Parallel secretory pathways to the cell surface in yeast. J Cell Biol. 1995;131(2):297–310. doi: 10.1083/jcb.131.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harsay E, Schekman R. A subset of yeast vacuolar protein sorting mutants is blocked in one branch of the exocytic pathway. J Cell Biol. 2002;156(2):271–85. doi: 10.1083/jcb.200109077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz AH, Chang CP, Better M, Hellstrom KE, Robinson RR. Secretion of functional antibody and Fab fragment from yeast cells. Proc Natl Acad Sci U S A. 1988;85(22):8678–82. doi: 10.1073/pnas.85.22.8678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M, Riles L, Hegemann JH. Gene disruption. Methods Enzymol. 2002;350:290–315. doi: 10.1016/s0076-6879(02)50970-8. [DOI] [PubMed] [Google Scholar]
- Kim YS, Bhandari R, Cochran JR, Kuriyan J, Wittrup KD. Directed evolution of the epidermal growth factor receptor extracellular domain for expression in yeast. Proteins. 2006;62(4):1026–1035. doi: 10.1002/prot.20618. [DOI] [PubMed] [Google Scholar]
- Kjaerulff S, Jensen MR. Comparison of different signal peptides for secretion of heterologous proteins in fission yeast. Biochem Biophys Res Commun. 2005;336(3):974–82. doi: 10.1016/j.bbrc.2005.08.195. [DOI] [PubMed] [Google Scholar]
- Kjeldsen T, Pettersson AF, Hach M, Diers I, Havelund S, Hansen PH, Andersen AS. Synthetic leaders with potential BiP binding mediate high-yield secretion of correctly folded insulin precursors from Saccharomyces cerevisiae. Protein Expr Purif. 1997;9(3):331–6. doi: 10.1006/prep.1996.0695. [DOI] [PubMed] [Google Scholar]
- Knop M, Finger A, Braun T, Hellmuth Kl, Wolf DH. Der1, a Novel Protein Specifically Required for Endoplasmic Reticulum Degradation in Yeast. EMBO J. 1996;15(4):753–63. [PMC free article] [PubMed] [Google Scholar]
- Lee MA, Cheong KH, Shields D, Park SD, Hong SH. Intracellular trafficking and metabolic turnover of yeast prepro-alpha-factor-SRIF precursors in GH3 cells. Exp Mol Med. 2002;34(4):285–93. doi: 10.1038/emm.2002.40. [DOI] [PubMed] [Google Scholar]
- Lipovsek DL, Antipov E, Armstrong KA, Olsen MJ, Klibanov AM, Tidor B, Wittrup KD. Selection of Horseradish Peroxidase Variants with Enhanced Enantioselectivity by Yeast Surface Display. Chemistry and Biology. 2007;14(10):1176–1185. doi: 10.1016/j.chembiol.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Lodish H, Berk A, Zipursky SL, Matsudaira P, Baltimore D, Darnell . In: Molecular Cell Biology. Tenney S, editor. W.H. Freeman and Co.; New York, NY: 2000. [Google Scholar]
- Luo W-j, Xiao-hua Gong, Amy Chang. An ER Membrane Protein, Sop4, Facilitates ER Export of the Yeast Plasma Membrane. Traffic. 2002;3:730–739. doi: 10.1034/j.1600-0854.2002.31005.x. [DOI] [PubMed] [Google Scholar]
- Manz R, Assenmacher M, Pfluger E, Miltenyi S, Radbruch A. Analysis and sorting of live cells according to secreted molecules, relocated to a cell-surface affinity matrix. Proc Natl Acad Sci U S A. 1995;92(6):1921–5. doi: 10.1073/pnas.92.6.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazor Y, Van Blarcom T, Mabry R, Iverson BL, Georgiou G. Isolation of Engineered, Full-Length Antibodies from Libraries Expressed in Escherichia coli. Nature Biotechnology. 2007 doi: 10.1038/nbt1296. online publication. [DOI] [PubMed] [Google Scholar]
- Midelfort KS, Hernandez HH, Lippow SM, Tidor B, Drennan CL, Wittrup KD. Substantial energetic improvement with minimal structural perturbation in a high affinity mutant antibody. J Mol Biol. 2004;343(3):685–701. doi: 10.1016/j.jmb.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Oka C, Tanaka M, Muraki M, Harata K, Suzuki K, Jigami Y. Human lysozyme secretion increased by alpha-factor pro-sequence in Pichia pastoris. Biosci Biotechnol Biochem. 1999;63(11):1977–83. doi: 10.1271/bbb.63.1977. [DOI] [PubMed] [Google Scholar]
- Otte S, Barlowe C. Sorting signals can direct receptor-mediated export of soluble proteins into COPII vesicles. Nat Cell Biol. 2004;6(12):1189–94. doi: 10.1038/ncb1195. [DOI] [PubMed] [Google Scholar]
- Rakestraw A, Wittrup KD. Contrasting secretory processing of simultaneously expressed heterologous proteins in Saccharomyces cerevisiae. Biotechnol Bioeng. 2006;93(5):896–905. doi: 10.1002/bit.20780. [DOI] [PubMed] [Google Scholar]
- Rakestraw JA, Baskaran AR, Wittrup KD. A Flow Cytometric Assay for Screening Improved Heterologous Protein Secretion in Yeast. Biotechnology Progress. 2006;22(4):1200–1208. doi: 10.1021/bp0600233. [DOI] [PubMed] [Google Scholar]
- Rao BM, Driver I, Lauffenburger DA, Wittrup KD. High-affinity CD25-binding IL-2 mutants potently stimulate T-cell growth. Biochemistry. 2005;44(31):10696–10701. doi: 10.1021/bi050436x. [DOI] [PubMed] [Google Scholar]
- Rapoport TA, Jungnickel B, Kutay U. Protein transport across the eukaryotic endoplasmic reticulum and bacterial inner membranes. Annu Rev Biochem. 1996;65:271–303. doi: 10.1146/annurev.bi.65.070196.001415. [DOI] [PubMed] [Google Scholar]
- Robinson AS, Hines V, Wittrup KD. Protein disulfide isomerase overexpression increases secretion of foreign proteins in Saccharomyces cerevisiae. Biotechnology (N Y) 1994;12(4):381–4. doi: 10.1038/nbt0494-381. [DOI] [PubMed] [Google Scholar]
- Shusta EV, Raines RT, Pluckthun A, Wittrup KD. Increasing the secretory capacity of Saccharomyces cerevisiae for production of single-chain antibody fragments. Nat Biotechnol. 1998;16(8):773–7. doi: 10.1038/nbt0898-773. [DOI] [PubMed] [Google Scholar]
- Simmons LCea. Expression of full-length immunoglobulins in Escherichia coli: rapid and efficient production of aglycosylated antibodies. J Immunol Methods. 2002;263:133–47. doi: 10.1016/s0022-1759(02)00036-4. [DOI] [PubMed] [Google Scholar]
- Steube K, Chaudhuri B, Marki W, Merryweather JP, Heim J. Alpha-factor-leader-directed secretion of recombinant human-insulin-like growth factor I from Saccharomyces cerevisiae. Precursor formation and processing in the yeast secretory pathway. Eur J Biochem. 1991;198(3):651–7. doi: 10.1111/j.1432-1033.1991.tb16063.x. [DOI] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10(13):1793–808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Yeung YA, et al. Isolation and characterization of human antibodies targeting human aspartyl (asparaginyl) beta-hydroxylase. Hum Antibodies. 2007;16:163–76. [PubMed] [Google Scholar]
- Zhang B, Chang A, Kjeldsen TB, Arvan P. Intracellular retention of newly synthesized insulin in yeast is caused by endoproteolytic processing in the Golgi complex. J Cell Biol. 2001;153(6):1187–98. doi: 10.1083/jcb.153.6.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]