Abstract
We report the identification of a recurrent 520-kbp 16p12.1 microdeletion significantly associated with childhood developmental delay. The microdeletion was detected in 20/11,873 cases vs. 2/8,540 controls (p=0.0009, OR=7.2) and replicated in a second series of 22/9,254 cases vs. 6/6,299 controls (p=0.028, OR=2.5). Most deletions were inherited with carrier parents likely to manifest neuropsychiatric phenotypes (p=0.037, OR=6). Probands were more likely to carry an additional large CNV when compared to matched controls (10/42 cases, p=5.7×10-5, OR=6.65). Clinical features of cases with two mutations were distinct from and/or more severe than clinical features of patients carrying only the co-occurring mutation. Our data suggest a two-hit model in which the 16p12.1 microdeletion both predisposes to neuropsychiatric phenotypes as a single event and exacerbates neurodevelopmental phenotypes in association with other large deletions or duplications. Analysis of other microdeletions with variable expressivity suggests that this two-hit model may be more generally applicable to neuropsychiatric disease.
Introduction
The majority of known recurrent genomic disorders result from non-allelic homologous recombination (NAHR) or unequal crossing-over between large and highly identical segmental duplications (>10 kbp) 1. Specific human chromosomes (e.g. 7, 15, 16, 17, and 22) are enriched for interspersed segmental duplications 2, and, as a result, multiple genomic disorders have already been assigned to these “hotspot” regions of the genome 3,4. The short arm of human chromosome 16 is particularly enriched for large segmental duplications that have arisen specifically during human-great ape evolution 5-8. In the last three years, at least three microdeletion/microduplication syndromes have been characterized whose breakpoints map within chromosome 16 segmental duplications. These include a 500-kbp microdeletion/duplication of 16p11.2 associated with autism spectrum disorders and intellectual disability 9,10, a large microdeletion encompassing 16p11.2-p12.2 in patients with a wide spectrum of phenotypic features 11, and distal 16p13.11 rearrangements in patients with autism, intellectual disability, and other neurodevelopmental phenotypes 12-14.
We recently performed a genome-wide meta-analysis comparing the frequency of large deletion/duplication events in individuals with neurocognitive/psychiatric disabilities to that in the control population 15. This meta-analysis suggested a potentially fourth example of a duplication-mediated microdeletion of ∼600 kbp on chromosome 16p12.1 that was found in 5/6,860 individuals affected by schizophrenia or autism compared to 0/5,674 controls 15. Therefore, we sought to systematically characterize this particular microdeletion in two large cohorts (discovery set and replication set) of children with intellectual disability (n=11,873 and 9,254, respectively) in comparison to controls (n=8,540 and 6,299, respectively). Our data suggest that the 16p12.1 microdeletion is a risk factor for neurodevelopmental disease that also acts in concert with other large copy-number variants (CNVs) to modify neuropsychiatric phenotypes, thereby supporting a “two-hit” model for the generation of severe cognitive deficits involving this region. Analysis of other microdeletions suggests that this model may help to explain the variability in expressivity of recurrent CNVs associated with neuropsychiatric phenotypes.
Results
Pathogenic association of 16p12.1 microdeletion
From our initial discovery cohort, we identified 20 cases with a 520.8-kbp 16p12.1 microdeletion (build36, chr16:21854025-22374785) among 11,873 individuals with indications of intellectual disability/developmental delay (ID/DD) and congenital malformation (Fig. 1, see Methods). In contrast, CNV studies on 8,540 controls identified only two individuals, both from the GAIN cohort, with the 16p12.1 microdeletion (Table 1). Thus, 16p12.1 microdeletions are significantly enriched in the panel of children with developmental delay studied here (Fisher's exact test, p=0.0009, OR=7.2). To replicate this association, we evaluated CNV data from an independent set of 9,254 individuals with ID/DD and 6,299 controls (see Methods). The microdeletion was identified in 22/9,254 cases and 6/6,299 controls, confirming a significant enrichment of 16p12.1 microdeletion in the affected individuals (Fisher's exact test, p=0.028, OR=2.5) (Table 1, Supplementary Fig. 1). For the combined set (21,127 cases and 15,199 controls), the pathogenic association of 16p12.1 microdeletion was highly significant (Fisher's exact test, p=8.6 ×10-5, OR=3.78) (Table 1).
Fig. 1. High-resolution array-based CGH characterization of 16p12.1 microdeletion.
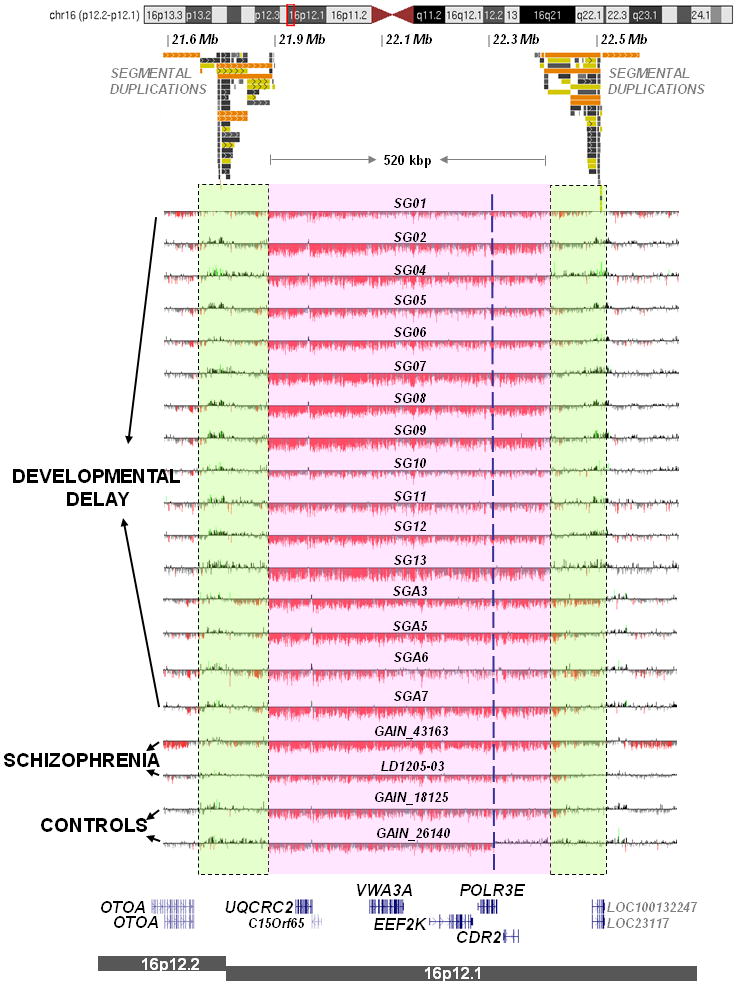
Validation of 16p12.1 microdeletions, in a representative set of cases, using high-resolution tiling-path custom array-based CGH is shown. Probes with log2 ratios above or below a threshold of 1.5 standard deviations from the normalized mean log2 ratio are colored green (duplication) or red (deletion), respectively. Dotted lines represent breakpoint regions. SG01-13 and SGA3-SGA7 are cases with indications of developmental delay or cognitive disability, sample 43163 is from GAIN schizophrenia study, and LD1205-03 has schizophrenia and intellectual disability (from family LD1205). Note, samples 26140 and 18125 were analyzed as part of the GAIN control cohort for schizophrenia. It is noteworthy that one control (26140) was retrospectively diagnosed with a major depressive disorder. Six RefSeq genes map within the 16p12.1 microdeletion. Four cases (SG04, SG07, SG11, affected with hypoplastic left heart syndrome, and LD1205-03 diagnosed with schizophrenia) were sequenced for CDR2, EEF2K, and UQCRC2; no recessive mutations were identified.
Table 1. Frequency of 16p12.1 microdeletion in cases and controls.
| Cases | Controls | Significance | ||||
|---|---|---|---|---|---|---|
| ID/DD cohort | del 16p12.1 | total | del 16p12.1 | total | p-value | OR |
| Discovery set1 | 20 | 11,873 | 2 | 8,540 | 0.0009 | 7.2 |
| Replication set2 | 22 | 9,254 | 6 | 6,299 | 0.028 | 2.5 |
| Combined3 | 42 | 21,127 | 8 | 15,199 | 0.000086 | 3.78 |
| Schizophrenia cohort | 3 | 3,061 | 8 | 15,199 | 0.27 | 1.86 |
The 16p12.1 microdeletion was originally identified based on a meta-analysis that identified 5 cases from 6,860 individuals with autism, schizophrenia and developmental delay compared to 0 observations in a control group of 5,674 (p=0.049). None of these cases were used in the discovery set although the original control group was expanded in the discovery set.
For the replication set all controls and cases were independent.
Combined set composed of discovery set + replication set.
We also examined 3,061 individuals diagnosed with schizophrenia and identified three 16p12.1 microdeletions, all of which were sporadic schizophrenia cases (Table 1). Significant enrichment for the 16p12.1 event was not, however, observed specifically in the schizophrenia cases compared to 15,199 total controls (Fisher's exact test, p=0.27, OR=1.86), although we note that this is perhaps indicative of a lack of statistical power in the schizophrenia panel rather than a true lack of disease association (only 3,061 individuals contrasted with the 21,127 individuals with developmental delay). It is interesting, in this regard, that in one family where both schizophrenia and mental retardation phenotypes were segregating, the 16p12.1 deletion was only observed among patients diagnosed with both psychosis and severe intellectual disability (see Supplementary Note, family LD1205).
Using high density and targeted array-based comparative genomic hybridization (CGH) experiments (see Methods), we mapped the 16p12.1 microdeletion breakpoints in 37 individuals to two large blocks of segmental duplications (Fig. 1, Supplementary Fig. 1). Within these blocks we identified a 68-kbp duplicon in direct orientation (with 99.5% identity) in relation to its paralog in the distal breakpoint region (Supplementary Table 1). Since misalignment of directly oriented duplicons during meiosis predisposes to non-allelic homologous recombination (NAHR) events resulting in microdeletions 1, the 68-kbp duplicon likely mediates the recurrent 16p12.1 rearrangements observed in patients. Numerous studies of the region using FISH (Fig. 2), single-nucleotide polymorphism (SNP) microarrays 16, and sequencing-based approaches 17 reveal that the 68-kbp duplicon varies in copy number and that the entire region may be inverted in some individuals, similar to that observed with the 17q21.31 microdeletion 18. Preliminary analyses suggest that particular structural configurations may be more predisposed to 16p12.1 rearrangement (data not shown). Due to the complexity at this locus and high copy-number variation of the duplications, we could not refine the boundary of the breakpoints below 100 kbp by array-based CGH. Interestingly, the microdeletion in one control sample (GAIN_26140) was 102.9 kbp shorter in length and did not span the CDR2 (cerebellar degeneration related 2) gene (Fig. 1).
Fig. 2. Genomic structure of 16p12.1 region.
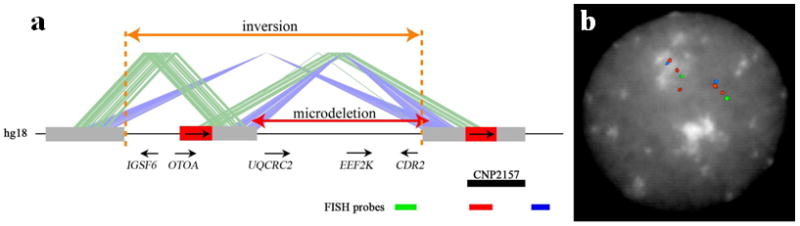
(a) A schematic of the 16p12.1 region shows the location of the microdeletion flanked by directly oriented 68-kbp segmental duplication blocks (red boxes). The segmental duplication blocks (red and gray boxes) are connected by the green and blue lines to indicate direct or inverted orientation, respectively. Also shown are representative genes in the region with the transcriptional direction. CNP indicates the copy-number polymorphism annotated by SNP genotyping 16 for this region (CNP2157). (b) FISH analysis was performed on lymphoblast cell line from GM18956 utilizing fosmid probes mapping to the 68-kbp duplicon (WIBR2-2031K01, shown in red) and the flanking unique regions (WIBR2-3632J22 in green and WIBR2-1829F15 in blue). Results show that the 68-kbp duplicon is polymorphic, i.e. it has a variable number of copies and that the orientation of the region is inverted in this HapMap sample compared to the human genome reference assembly. High copy numbers of the segmental duplications have complicated mapping of the inversion breakpoint for this region.
Phenotypic evaluation and parental analysis
Evaluation of available medical records from the ID/DD cohort showed that multiple phenotypic features are associated with the 16p12.1 microdeletion (Table 2, Supplementary Tables 2 and 3). While not all clinical features were completely recognizable in very young patients, we observed developmental delay and learning disability in most cases. All 15 individuals older than 12 months had speech delay. Craniofacial and skeletal abnormalities were observed in 22/23 cases (Fig. 3). Growth retardation was documented in 9/22 cases, while 7/20 individuals also had microcephaly (Table 2). (Note: the denominator varies because not all patients could be ascertained for all features). Furthermore, congenital cardiac disease was observed in 7/21 cases, of which four cases were specifically diagnosed with a hypoplastic left heart syndrome (Table 2, Supplementary Table 3). Seizure disorders, manifesting in various forms, including West syndrome, febrile seizures, or seizure-like episodes, were observed in 8/22 cases, and hypotonia was present in 10/21 cases (Supplementary Note). Psychiatric and behavioral abnormalities were also documented in 9/16 affected children. Non-typical facial gestalt and variable clinical presentations suggest that this microdeletion is non-syndromic.
Table 2. Frequency of phenotypic features in individuals with 16p12.1 deletions.
| 01 | 02 | 03 | 04 | 05 | 06 | 07 | 08 | 09 | 10 | 11 | 12 | 13 | A1 | A2 | A3 | A5 | A6 | A7 | A8 | A9 | A28 | 25514 | Frequency | % | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Developmental delay | + | + | + | N | + | + | + | + | + | + | N | + | + | + | + | + | + | N | N | N | + | + | + | 18/18 | 100% |
| Speech delay* | + | + | + | N | + | + | + | + | + | + | N | N | + | N | + | + | + | N | N | N | + | N | + | 15/15 | 100% |
| Craniofacial, Skeletal features | + | + | + | + | + | + | + | - | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 22/23 | 96% |
| Growth retardation | - | + | - | - | - | + | - | + | + | + | + | N | - | + | - | - | - | - | - | - | + | + | - | 9/22 | 41% |
| Microcephaly | + | + | - | - | - | + | + | + | + | - | - | N | - | N | - | - | - | - | - | - | N | + | - | 7/20 | 35% |
| Congenital cardiac defect | - | - | - | + | - | - | + | - | + | - | + | N | - | N | - | - | - | # | + | + | - | - | - | 7/21 | 33% |
| Hypoplastic heart | - | - | - | + | - | - | - | - | - | - | + | N | - | N | - | - | - | - | + | + | - | - | - | 4/21 | 19% |
| Seizures | + | + | - | - | + | @ | - | + | - | - | - | + | - | N | - | + | - | † | - | - | - | - | - | 8/22 | 36% |
| Psychiatric/behavioral features | - | + | - | N | - | + | - | + | + | - | N | N | + | N | + | - | - | N | N | N | + | + | + | 9/16 | 56% |
| Hearing loss | - | - | - | N | N | - | - | - | - | N | - | N | - | N | - | - | + | + | - | N | - | + | - | 3/17 | 18% |
| Hypotonia | + | + | + | - | + | - | - | + | + | - | - | N | - | N | + | + | - | - | - | - | + | - | + | 10/21 | 48% |
| Sacral dimple or tethered cord | - | - | - | - | - | - | - | - | + | + | + | N | - | N | - | - | - | - | + | - | - | - | - | 4/21 | 19% |
phenotype present,
phenotype absent,
N = phenotype not assessed.
myoclonus,
speech delay was not evaluated in very young cases,
patent ductus arteriosus and patent foramen ovale resolved on a follow-up examination,
slowing on EEG. Gray shaded columns represent patients who carry “two hits.”
Fig. 3. Representative photographs of individuals with 16p12.1 microdeletion.
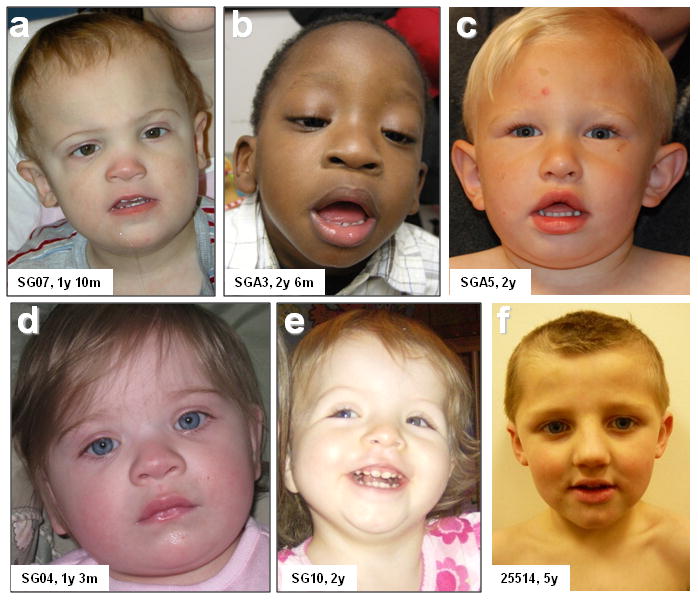
Facial features of patient SG07 at 22 months (a), patient SGA3 at 2.5 y (b), patient SGA5 at 2 y (c), patient SG04 at 15 months (d), patient SG10 at 2 y (e), and patient 25514 at 5 y (f) are shown. Specific consents were obtained to publish these patient photographs.
We documented that 6/20 individuals with 16p12.1 microdeletion from the discovery set had an additional chromosomal abnormality or large (>500 kbp) CNV (Fig. 4, Supplemental Table 4, Table 3). The frequency (30%) of such double-hit CNVs was increased 7.5-fold in the 16p12.1 microdeletion cases (Fisher's exact test, p=0.0005, OR=9.7) when compared to controls conditioned for a large CNV (deletion or duplication) first hit (9/217 or 4.1%) (Table 3). In the replication set, the double-hit frequency was also enriched in the cases (4/22, 18.2%) compared to controls (12/254, 4.7%) (Fisher's exact test, p=0.029, OR=4.48) (Supplementary Table 2, Supplementary Fig. 2). Overall in the combined set, there was a significant (Fisher's exact test, p=5.7×10-5, OR=6.65) enrichment of double hits among 16p12.1 deletion carriers (10/42, 24%) compared to controls (21/471, 4.4%) (Table 3). Significant enrichment for double hits in 16p12.1 cases was also observed when the controls were conditioned to carry only a large deletion event as the first hit (Supplementary Table 5).
Fig. 4. Family pedigrees of probands with 16p12.1 microdeletion.
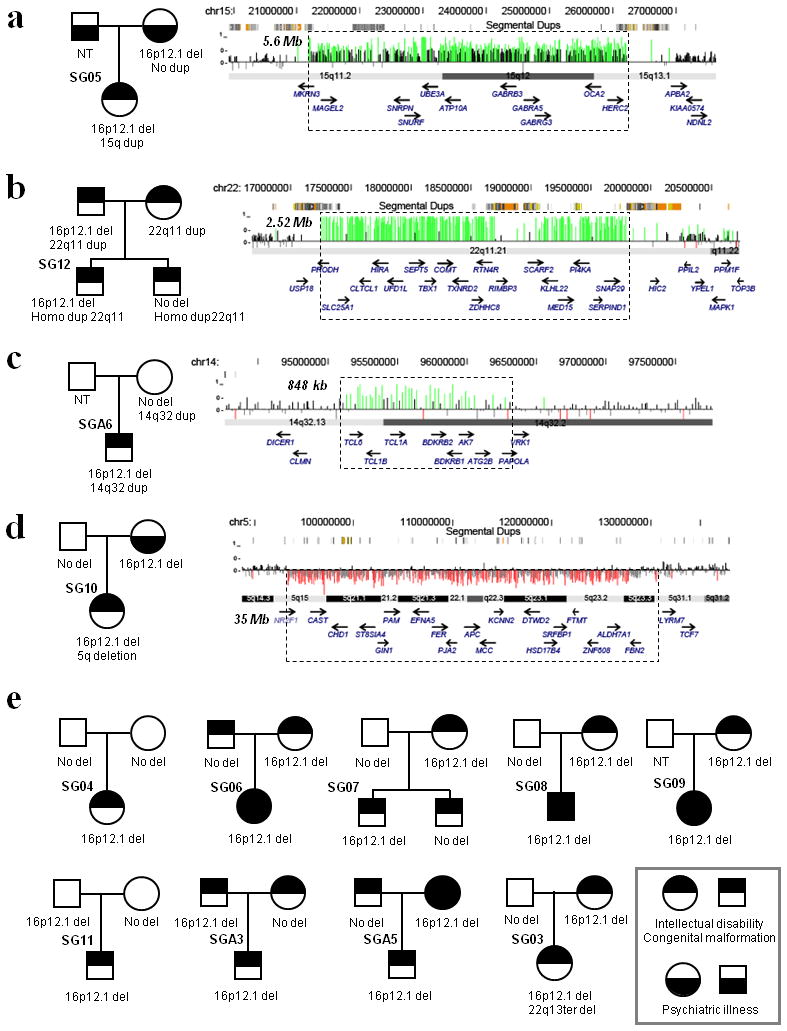
Large CNVs, outside of 16p12.1 region, in a representative set of individuals with 16p12.1 microdeletions are shown (a-d). The CNV regions are indicated by dotted lines and the cytogenetic extent and size are labeled. We utilized a 135K NimbleGen array to identify these CNVs (with average probe density of 2.5 kbp in regions flanked by segmental duplications and a genomic backbone of 35 kbp). CNV calls were made using a Hidden Markov Model CNV-calling algorithm described previously 15. Also shown are the pedigrees of individuals with 16p12.1 microdeletion with known available parental information (e). Circles indicate females and squares indicate males. Intellectual disability and congenital malformation category also includes congenital heart defects and seizures. Psychiatric illness includes depression or bipolar disorder, attention deficit hyperactive disorder, and abnormal behaviors. Note that there is an excess of transmitting parents with the microdeletion who also manifested with a phenotype. NT = not tested.
Table 3. Enrichment for ‘second-hit’ CNVs among 16p12.1 microdeletion carriers.
| Two large CNVs | Total | Percent | Significance | OR | |
|---|---|---|---|---|---|
| Discovery set cases | 6 | 20 | 30% | p=0.0005 | 9.7 |
| Discovery controls* | 9 | 217 | 4.1% | ||
| All discovery set controls | 9 | 2493 | 0.4% | ||
| Replication set cases | 4 | 22 | 18.2% | ||
| Replication controls* | 12 | 254 | 4.7% | p=0.029 | 4.48 |
| All replication set controls | 12 | 2792 | 0.42% | ||
| Combined cases | 10 | 42 | 23.8% | ||
| Combined controls* | 21 | 471 | 4.4% | p=0.000057 | 6.65 |
| All controls | 12 | 5285 | 0.39% |
Conditioned on presence of one large CNV (>500 kbp) and finding of a second hit
In the cases where the second-hit CNV is associated with a described syndrome (DECIPHER 19, ECARUCA 20, or published literature), two-hit 16p12.1 carriers manifested more severe or distinct phenotypes than the typical features of the syndrome (Supplementary Note). For example, patient SG03 carries a 16p12.1 microdeletion in addition to a 22q13 terminal deletion. The single del22q13ter event has been reported in Phelan-McDermid syndrome 21 and is associated with autism spectrum disorders 22, developmental delay, hypotonia, normal to accelerated growth, and dysmorphic features including macrocephaly/dolichocephaly and micrognathia. However, SG03 manifests with learning disability, microcephaly, exotropia, and periventricular abnormalities, with no autistic features yet apparent (age 2 years 3 months). SG10 carries a 35-Mbp deletion on chromosome 5q15q23.2 along with the 16p12.1 microdeletion. Previously, Lindgren and colleagues reported on two individuals carrying a del5q15q23.2 with craniofacial features and benign adenomatous polyps in the intestine 23. SG10 has developmental delay, significant growth retardation, overt craniofacial features (ptosis, hypertelorism, bifid uvula, exotropia, ectopic pupils, wide nasal bridge, and smooth philtrum), hypoactivity, and seizures—a more severe phenotype than that reported from a single second hit. Patient SGA2 carries a 1.3-Mbp duplication on chromosome 2p13p12 in addition to the 16p12.1 microdeletion and has overt clinical features, including impaired intellectual function, behavior problems, growth anomalies, craniofacial features, and café-au-lait spots. While dup2p13p12 has been reported to be associated with primary cutaneous lymphoma 24, there have been no reports of developmental defect or craniofacial anomalies. SGA6 carries an 848-kbp duplication on chromosome 14q32.1 along with the 16p12.1 microdeletion. Additionally, he has had a mutation identified in the BRAF gene (F468S), consistent with a diagnosis of cardiofaciocutaneous syndrome (CFCS). The patient presents with a diverse range of severe clinical features including craniofacial anomalies, complete agenesis of corpus callosum, renal and cardiac defects as well as Hirschsprung disease (Supplementary Table 3). These features are more severe than has been described for CFCS 25 or a de novo dup14q32.1 case reported in association with schizophrenia 26.
To test if the patients inherited the 16p12.1 microdeletion from their parents, we were able to obtain DNA from 23 sets of parents. The 16p12.1 microdeletion was inherited in 22/23 cases (17 maternal, 5 paternal) with one case confirmed as being de novo (Fig. 4, Supplementary Fig. 4). Of the seven double-hit cases where inheritance could be assessed, 6/7 large CNV second hits were inherited and one large CNV arose de novo. In most cases (5/7), the large CNV second hit was inherited from the other parent who did not carry a 16p12.1 microdeletion or it arose de novo. In one case, SG12, a transmission of a heterozygous dup22q11.21 from both the parents to the homozygote proband was documented (Fig. 4). SGA30 inherited both the events from the mother (Supplementary Table 6).
Parental phenotypic information was obtained during the initial visit prior to CNV testing from 11/13 inherited cases. In two cases clinical inquiries were made only after the child was found to carry the microdeletion (Supplementary Table 7). Carrier parents with the 16p12.1 microdeletion were more likely (p=0.037, OR=6, Fisher's exact test) to present with learning disability, depression or bipolar disorders, or seizures than the non-carrier parents (Table 2, Fig. 4, Supplementary Table 8). We found only three cases, SG11, SGA9 and 25514, where the microdeletion was inherited from an apparently normal parent (Supplementary Table 9). Compared to the carrier parents, the index cases manifested with more severe, clinically recognizable manifestations, including severe speech and motor disability, recognizable facial dysmorphology, and systemic organ abnormalities (Table 2, Fig. 3, Fig. 4, Supplementary Note). Finally, we assessed the SNP genotypes for chromosome 16p in 17 probands and four available parents to investigate if there is one specific sequence haplotype that predisposes to this microdeletion. Results showed that the microdeletion occurred on different haplotype backgrounds consistent with the recurrent nature of this rearrangement as opposed to inheritance from a common ancestor (Supplementary Note).
Discussion
Previously, we undertook a large meta-analysis study of copy-number variants in individuals diagnosed with intellectual disability, autism, and schizophrenia, identifying 16p12.1 region as a potential pathogenic locus (five cases and no controls) 15. However, this study lacked the power to definitively identify a disease association and/or delineate the phenotypic consequences of the microdeletion. Here we utilized the CNV data from one of the largest collections of individuals with intellectual disability and developmental delay. We identified 42 index cases with developmental delay, craniofacial dysmorphology, and congenital heart defects and three sporadic and one familial case of overt schizophrenia carrying a similar-sized microdeletion of 16p12.1; we found only eight control individuals carrying a 16p12.1 deletion. While all cases appear to share the segmental duplication-mediated 520-kbp minimal deletion region, it is noteworthy that one deletion-carrying control was retrospectively diagnosed with a major depressive disorder and also had an atypical, smaller deletion (417 kbp). Removal of this control sample from our analysis strengthens the association between 16p12.1 microdeletion and disease (Fisher's exact test, p=1.6×10-5, OR=4.3). Comparison to other genomic disorders in our cohort suggests that the incidence of 16p12.1 microdeletion is ∼1/15,000 live births, similar to that of Smith-Magenis syndrome (Table 4). However, the subtle clinical features and variable phenotypes associated with the single 16p12.1 event likely lead to under-ascertainment or misdiagnoses.
Table 4. Percentage of second hits and de novo rates of microdeletions.
| Disorder | Cases (n) | Total cases | Incidence (%) | de novo | de novo (%) | Second hits | Second hit (%) | p-value |
|---|---|---|---|---|---|---|---|---|
| Smith-Magenis syndrome | 25 | 20,647 | 0.12 | 3/3 | 100% | 0 | 0% | - |
| 17q21.31 deletion | 29 | 20,647 | 0.14 | 7/7 | 100% | 0 | 0% | - |
| Williams syndrome | 60 | 20,647 | 0.29 | 5/5 | 100% | 3 | 5% | 0.52 |
| DiGeorge syndrome | 113 | 20,647 | 0.54 | 13/17 | 76.4% | 9 | 8% | 0.10 |
| 15q13.3 deletion | 66 | 20,647 | 0.32 | 6/25 | 24% | 6 | 9.1% | 0.1 |
| 16p11.2 deletion | 91 | 20,647 | 0.44 | 26/35 | 74.3% | 9 | 9.9% | 0.038 |
| 1q21.1 deletion | 98 | 25,866 | 0.37 | 15/45 | 33% | 11 | 11.2% | 0.012 |
| 22q11.2 duplication | 61 | 20,647 | 0.30 | 2/21 | 9.5% | 9 | 14.8% | 0.003 |
| 16p12.1 deletion | 42 | 21,127 | 0.20 | 1/23 | 4.3% | 10 | 23.8% | 5.7×10-5 |
| Controls1 | 471 | 5285 | 21 | 4.4% | ||||
| Controls (unconditioned)2 | 5285 | 21 | 0.39% |
For comparison, controls were conditioned to have at least one large CNV (>500 kbp) and then the number of second hits in these cases were counted.
controls were not conditioned for first hit. This gives an estimate of two hits in the general population compared to affected individuals.
Most of our pediatric cases had indications of developmental delay/learning disability and congenital abnormalities. However, variable phenotypes associated with the 16p12.1 microdeletion include congenital heart defects, seizures, and severe growth abnormalities (Supplementary Note). We also identified five adult individuals with a diagnosis of schizophrenia and found that 23% of the probands inherited the microdeletion from a carrier parent with manifestations of psychiatric disease. Carrier parents were significantly more likely (p=0.037) to manifest other neurologic/neuropsychiatric features (learning disability, depression, bipolar disorders, and seizures) than non-carriers. While the fact that these parents were recruited as a result of their child's referral constitutes an ascertainment bias, we note that any such bias should apply to both parents equally, and yet only 5/14 non-carrier parents had any clinical symptoms (Supplementary Table 9). A detailed clinical examination is therefore warranted in parents for disorders such as 16p12.1 microdeletion to understand the relationship between the various neurodevelopmental and cognitive phenotypes. Within probands that have the microdeletion, we observed a 6-fold excess of double CNV hits (10/42, 24%) compared to normal controls when conditioned for at least one large CNV (21/471, 4%) and more than a 60-fold excess as compared to the general population. Compared to the classical phenotypes associated with the known pathogenic locus, each of these children exhibited additional or more severe phenotypes (Supplementary Note). Collectively, these data suggest that phenotypes associated with the 16p12.1 microdeletion show variable expressivity dependent upon the genetic background.
We conclude that the deletion of 16p12.1 is a significant, independent risk factor for intellectual disability and developmental delay that also acts in concert with other factors to modify neurological phenotypes. We propose a “two-hit” model, wherein a secondary insult is necessary during development to result in a more severe clinical manifestation as pediatric disease. The second hit could potentially be another CNV, a disruptive single basepair mutation in a phenotypically-related gene, or an environmental event influencing the phenotype. It has been previously noted in a number of studies that genetic factors originally identified to associate with a specific neurological disease are often subsequently identified in patients with distinct illness (for example, del15q13.3 associated with epilepsy, schizophrenia, and intellectual disability) 27-29.
The prevalence of schizophrenia in individuals with a learning disability is reported to be three times that of the general population and several studies suggest that the co-morbidity of schizophrenia in learning-disabled patients is mainly due to a greater tendency of schizophrenic patients to develop cognitive delay 30-32. The observation, as seen in family LD1205, that cognitive impairment is seen only among siblings with the 16p12.1 deletion is also consistent with the hypothesis that the 16p12.1 deletion exacerbates the phenotypic consequences of other heritable neurological disease risk factors within this multiplex family (Supplementary Note). Notably, about 70% of individuals with autism also present with learning disability 33. The “two-hit” model we propose may also help to explain the significant co-morbidity that exists among cognitive impairment, schizophrenia and autism 34 or the underlying phenotypic variability reported for several recurrent microdeletions 29,35.
To test this, we investigated the occurrence of two hits more broadly among other genomic disorders. We first reanalyzed the recently reported 1q21.1 microdeletion for the presence of a second large CNV. We found a 40-fold enrichment for two hits (4/25, 16%) among cases with 1q21.1 microdeletion when compared to controls. The phenotypes of these four cases were variable, ranging from severe neurological deficit and craniofacial abnormalities to severe schizophrenia without cognitive impairment (Supplementary Note). Based on this finding, we expanded our analysis considering both syndromic and non-syndromic genomic disorders for the frequency of double hits. Our analysis for two hits in nine genomic disorders shows that 16p12.1 microdeletion ranks as the top recurrent CNV enriched for two hits (Table 4). Notably, the proportion of cases with a second hit is generally much higher (9-24% of the cases) for recurrent microdeletions or microduplications where variable expressivity has been reported, including del15q13.3, del16p11.2, dup22q11.2, and del16p12.1. We find an inverse correlation between the proportion of cases that are de novo versus the prevalence of the second hit. For example, only one de novo case (∼4%) of the 16p12.1 microdeletion has been reported and this microdeletion shows the greatest fraction of double hits. In contrast, we observed either a low level or no double hits among canonical syndromes such as Williams and Smith-Magenis syndromes (Table 4) where almost all cases arise de novo. We note, however, that we did not find a significant enrichment for two hits among cases with 22q11.2 deletion—similar to that observed by Bassett and colleagues 36. In general, these findings provide additional support for the two-hit hypothesis, with elevated double-hit rates among pathogenic CNVs with clearly variable penetrance and expressivity. We propose that phenotypic variability of microdeletions such as 16p12.1 and 1q21.1 are subject to substantial modification (or are themselves modifiers).
Materials and Methods
Cases and controls
We obtained CNV data for 16p12.1 microdeletion analysis on 14,454 cases comprising two phenotypically distinct cohorts from four independent sources: (1) DNA samples (n=11,393) with indications primarily of intellectual disability/developmental delay and congenital malformation (ID/DD cohort) submitted to Signature Genomic Laboratories during the period of 2007-2008 for CNV analysis (Supplementary Note); (2) CNV data from individuals diagnosed with schizophrenia (n=416) analyzed on the NimbleGen HD2 array-based comparative genomic hybridization (CGH) platform at Cold Spring Harbor Laboratories; (3) 2,645 DNA samples from individuals with schizophrenia analyzed on the Affymetrix 6.0 platform by the Genetic Association Information Network (GAIN) project for the study of schizophrenia (phs000021.v2.p1); and (4) 480 individuals from Italy and Australia diagnosed with neurodevelopmental anomalies for CNV analysis using a custom targeted “hotspot” NimbleGen array (Supplementary Note). (5) For the replication study, we analyzed CNV data from DNA from 9,254 individuals with ID/DD submitted to Signature Genomic Laboratories in 2009. In addition, we included 96 individuals affected with neurocognitive features and schizophrenia from 26 multiplex families. These families were interviewed, diagnosed, and sampled as previously described 37,38. Informed consent was also obtained from a subset of individuals with 16p12.1 microdeletions to perform sequencing analysis of candidate genes within the 16p12.1 microdeletion region. Phenotypic information about probands' parents was obtained based on family history information gathered during the initial clinic visit prior to genetic testing (Supplementary Note).
Copy-number variation data on controls (n=8,540) consisted of six sets: (1) 671 individuals of European descent with no family history or first-degree relative with amyotrophic lateral sclerosis, ataxia, autism, brain aneurysm, dystonia, Parkinson's disease, or schizophrenia; (2) 936 middle-aged (40–70 years) individuals of European descent living in the United States tested for statin response and cholesterol levels; (3) 886 individuals from the Human Genome Diversity Panel (HGDP) 15; (4) control individuals (n=3,181) used for a large study of schizophrenia 35; (5) 446 schizophrenia control samples; and (6) data obtained from 2,420 GAIN controls utilized for a genome-wide association study of schizophrenia. The GAIN cohort was collected to represent unrelated cases or controls. Their health and ethnicity were evaluated through a web-based questionnaire and ascertained by source of sample, geographic representativeness, comparability of cases and controls, and comprehensiveness of trait and phenotypic definitions 39. For the replication study, we utilized published CNV data on 2,792 individuals (2,792/2,998 passed QC) from Welcome Trust Case Control Consortium (WTCCC) controls 40,41 and CNV data from 2,026 individuals excluded for neurological disorders 42. The University of Washington Committee on Research Involving Human Subjects and the Institutional Review Board approved this study.
Despite the platform heterogeneity in CNV detection, we and others have shown that these platforms have comparable sensitivity and specificity for large CNV events (>500 Kbp) (Itsara et al., 2009). We compared the frequency of probands with large two large CNVs to that observed in the 2,493 controls 15 (Supplementary Note). We utilized 2,493 control samples from set 1 (n=671), set 2 (n=936), and set 3 (n=886), described previously, from the discovery set of controls for this purpose. Less than 1% (9/2,493) of our controls had two or more events in excess of 500 kbp. Since selecting 16p12.1 deletion probands requires to be at least one hit already present, we only considered those control individuals that harbored at least one large (>500 kbp) CNV as a comparison set. Among the 2,493 controls, 217 individuals have at least one event >500 kbp and of these 217 individuals only 9 have double hits for a general population frequency of 4.1% (Table 3). We assessed the frequency of two hits in the replication set of 22 cases with the 16p12.1 microdeletions and compared to the replication set controls (n=2,792) where individual sample identifiers were available for controls 41. In the replication cohort, we identified 7 patients with a second CNV. Four (18%) of these patients carried a second hit greater than 500 kbp in length. We also analyzed the WTCCC data for the second hit 41. No >500-kbp second hit was identified in the three WTCCC controls with the 16p12.1 microdeletion.
Copy-number variation detection
Microarray-based comparative genomic hybridization was performed with a whole-genome bacterial artificial chromosome (BAC) microarray chip (SignatureChipWG®), an oligo-based (SignatureChipOS®) chip (Agilent Technologies) or NimbleGen ‘hotspot’ array, and validated by fluorescent in situ hybridization (FISH) (Supplementary Note) 43. To refine the breakpoints of the 16p12.1 deletions identified by whole-genome BAC/oligo arrays, a custom, high-density oligonucleotide array (NimbleGen) was used (Supplementary Note). All high-density microarray hybridization experiments were performed as described previously 44 using a single, unaffected male (GM15724 [Coriell]) as reference. For the schizophrenia cohort, DNA samples were evaluated for large CNVs (>100 kb) with whole-genome NimbleGen HD2 arrays, Affymetrix 6.0, or Representational Oligonucleotide Microarray Analysis (ROMA) 45. The replication set controls were analyzed using Illumina Human Hap550 chip 42, Illumina Quad61, or using Affymetrix GeneChip 500K 41.
16p12 genome structure analysis
Metaphase spreads were obtained from a HapMap lymphoblast cell line (Coriell Cell Repository). FISH experiments were performed using fosmid clones directly labeled by nick-translation with Cy3-dUTP (Perkin-Elmer), Cy5-dUTP (Perkin-Elmer), and fluorescein-dUTP (Enzo). Briefly, 300 ng of labeled probe were used for the FISH experiments; hybridization was performed at 37°C in 2×SSC, 50% (v/v) formamide, 10% (w/v) dextran sulphate, and 3 μg sonicated salmon sperm DNA in a volume of 10 μL. Post-hybridization washing was at 60°C in 0.1×SSC (three times, high stringency). Nuclei were simultaneously DAPI stained. Digital images were obtained using a Leica DMRXA2 epifluorescence microscope equipped with a cooled CCD camera (Princeton Instruments). DAPI, Cy3, Cy5 and fluorescein fluorescence signals, detected with specific filters, were recorded separately as grayscale images. Pseudo-coloring and merging of images were performed using Adobe Photoshop software. A minimum of 50 interphase cells were scored for each experiment.
Supplementary Material
Acknowledgments
We thank the subjects and their families for participating in this study. We thank Dr. Michael Whyte for providing cardiological evaluation (for case SG07). We thank Andrew Singleton, Luigi Ferrucci and Ronald Krauss for sharing the control CNV data generated with support from the Intramural Research Program of the National Institute on Aging, National Heart, Lung, and Blood Institute (PARC project), National Institutes of Health, Department of Health and Human Services. For the schizophrenia study, genotyping of samples was also provided through the genetic association information network (GAIN). The datasets used for the analyses described in this manuscript were obtained from the database of genotype and phenotype (dbGaP) found at http://www.ncbi.nlm.nih.gov/gap through dbGaP accession number phs000021.v2.p1. Samples and associated phenotype data for the genome-wide association of schizophrenia study were provided by the Molecular Genetics of Schizophrenia Collaboration (PI: P.V. Gejman, Evanston Northwestern Healthcare (ENH) and Northwestern University, Evanston, Illinois, USA). We also thank Joshua Smith for SNP genotyping; Tin Louie for bioinformatics support; Catarina Campbell, Luis Perez-Jurado, George Kirov, M. Katherine Rudd, Christa Lese-Martin for useful discussions and Tonia Brown, Peter Sudmant, and Jacob Kitzman for critical review of the manuscript. This work was supported by NIH grant HD065285 (EEE), a grant from the Simons Foundation (SFARI 137578 to EEE) and a NARSAD Award (MCK). GMC is supported by a Merck, Jane Coffin Childs Memorial Fund post-doctoral fellowship. EEE is an investigator of the Howard Hughes Medical Institute.
Footnotes
Accession code: dbGaP (genotype data): GAIN study of schizophrenia (phs000021.v2.p1)
Author Contributions: This study was designed by S.G., L.G.S., and E.E.E. J.A.R., B.C.B., and L.G.S. supervised array-CGH experiments at Signature Genomics. J.A.R. coordinated clinical data collection. T.W., S.E.M., D.E.D., D.L.L., J.S., L.E.deL., and M.-C.K. contributed to schizophrenia data collection and analysis. S.G., L.V. and C.B. performed high-density array-CGH experiments. G.M.C. and A.I. analyzed control CNV data. S.G., F.A. and J.M.K. performed genome structure analysis. F.A. performed FISH experiments. S.G. and P.S. sequenced and analyzed candidate genes. S.R.B. and B.L.B. performed haplotype analysis. K.P., D.M. F., G.C.G., J.J.W., A.A., D.D.W., P.R.M., J.D., B.P.G., S.A.E., R.S., V.C.B., W.S., M.T.M., J.J.H., B.N.F., C.H., J.P.J., J.R.O., J.B.M., U.S., L.F.E., D.E-K., J.L.G., J.K., B.S., Y.L., A.B., D.M.M., E.H.Z., M.A.D., T.H.S., E.H., K.L.F., M.F., C.R., and J.G., provided clinical information. H.C.M. provided 1q21.1 data. S.G., G.M.C., M.-C.K. and E.E.E. contributed to data interpretation. S.G. and E.E.E. wrote the manuscript.
Competing Financial Interest: Evan E. Eichler is a Pacific Biosciences SAB member. Jill A. Rosenfeld and Blake C. Ballif are employees of Signature Genomic Laboratories, LLC. Lisa G. Shaffer is an employee of, owns shares in, and sits on the Members' Board of Signature Genomic Laboratories, LLC.
References
- 1.Lupski JR. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:417–22. doi: 10.1016/s0168-9525(98)01555-8. [DOI] [PubMed] [Google Scholar]
- 2.Marques-Bonet T, Girirajan S, Eichler EE. The origins and impact of primate segmental duplications. Trends Genet. 2009 doi: 10.1016/j.tig.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mefford HC, Eichler EE. Duplication hotspots, rare genomic disorders, and common disease. Curr Opin Genet Dev. 2009;19:196–204. doi: 10.1016/j.gde.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet. 2009;10:451–81. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eichler EE, et al. Divergent origins and concerted expansion of two segmental duplications on chromosome 16. J Hered. 2001;92:462–8. doi: 10.1093/jhered/92.6.462. [DOI] [PubMed] [Google Scholar]
- 6.Johnson ME, et al. Recurrent duplication-driven transposition of DNA during hominoid evolution. Proc Natl Acad Sci U S A. 2006;103:17626–31. doi: 10.1073/pnas.0605426103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson ME, et al. Positive selection of a gene family during the emergence of humans and African apes. Nature. 2001;413:514–9. doi: 10.1038/35097067. [DOI] [PubMed] [Google Scholar]
- 8.Loftus BJ, et al. Genome duplications and other features in 12 Mb of DNA sequence from human chromosome 16p and 16q. Genomics. 1999;60:295–308. doi: 10.1006/geno.1999.5927. [DOI] [PubMed] [Google Scholar]
- 9.Kumar RA, et al. Recurrent 16p11.2 microdeletions in autism. Hum Mol Genet. 2008;17:628–38. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- 10.Weiss LA, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–75. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 11.Ballif BC, et al. Discovery of a previously unrecognized microdeletion syndrome of 16p11.2-p12.2. Nat Genet. 2007;39:1071–3. doi: 10.1038/ng2107. [DOI] [PubMed] [Google Scholar]
- 12.Hannes FD, et al. Recurrent reciprocal deletions and duplications of 16p13.11: the deletion is a risk factor for MR/MCA while the duplication may be a rare benign variant. J Med Genet. 2009;46:223–32. doi: 10.1136/jmg.2007.055202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mefford HC, et al. A method for rapid, targeted CNV genotyping identifies rare variants associated with neurocognitive disease. Genome Res. 2009 doi: 10.1101/gr.094987.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ullmann R, et al. Array CGH identifies reciprocal 16p13.1 duplications and deletions that predispose to autism and/or mental retardation. Hum Mutat. 2007;28:674–82. doi: 10.1002/humu.20546. [DOI] [PubMed] [Google Scholar]
- 15.Itsara A, et al. Population analysis of large copy number variants and hotspots of human genetic disease. Am J Hum Genet. 2009;84:148–61. doi: 10.1016/j.ajhg.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCarroll SA, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet. 2008;40:1166–74. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 17.Kidd JM, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zody MC, et al. Evolutionary toggling of the MAPT 17q21.31 inversion region. Nat Genet. 2008;40:1076–83. doi: 10.1038/ng.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Firth HV, et al. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am J Hum Genet. 2009;84:524–33. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feenstra I, et al. European Cytogeneticists Association Register of Unbalanced Chromosome Aberrations (ECARUCA); an online database for rare chromosome abnormalities. Eur J Med Genet. 2006;49:279–91. doi: 10.1016/j.ejmg.2005.10.131. [DOI] [PubMed] [Google Scholar]
- 21.Phelan MC, et al. 22q13 deletion syndrome. Am J Med Genet. 2001;101:91–9. doi: 10.1002/1096-8628(20010615)101:2<91::aid-ajmg1340>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 22.Durand CM, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39:25–7. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindgren V, Bryke CR, Ozcelik T, Yang-Feng TL, Francke U. Phenotypic, cytogenetic, and molecular studies of three patients with constitutional deletions of chromosome 5 in the region of the gene for familial adenomatous polyposis. Am J Hum Genet. 1992;50:988–97. [PMC free article] [PubMed] [Google Scholar]
- 24.Mao X, et al. Genetic alterations in primary cutaneous CD30+ anaplastic large cell lymphoma. Genes Chromosomes Cancer. 2003;37:176–85. doi: 10.1002/gcc.10184. [DOI] [PubMed] [Google Scholar]
- 25.Roberts A, et al. The cardiofaciocutaneous syndrome. J Med Genet. 2006;43:833–42. doi: 10.1136/jmg.2006.042796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu B, et al. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet. 2008;40:880–5. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- 27.Helbig I, et al. 15q13.3 microdeletions increase risk of idiopathic generalized epilepsy. Nat Genet. 2009;41:160–2. doi: 10.1038/ng.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharp AJ, et al. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat Genet. 2008;40:322–8. doi: 10.1038/ng.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stefansson H, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–6. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turner TH. Schizophrenia and mental handicap: an historical review, with implications for further research. Psychol Med. 1989;19:301–14. doi: 10.1017/s0033291700012344. [DOI] [PubMed] [Google Scholar]
- 31.Moorhead TW, et al. Voxel-based morphometry of comorbid schizophrenia and learning disability: analyses in normalized and native spaces using parametric and nonparametric statistical methods. Neuroimage. 2004;22:188–202. doi: 10.1016/j.neuroimage.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 32.Sanderson TL, Best JJ, Doody GA, Owens DG, Johnstone EC. Neuroanatomy of comorbid schizophrenia and learning disability: a controlled study. Lancet. 1999;354:1867–71. doi: 10.1016/s0140-6736(99)01049-1. [DOI] [PubMed] [Google Scholar]
- 33.Fombonne E. Epidemiological trends in rates of autism. Mol Psychiatry. 2002;7(2):S4–6. doi: 10.1038/sj.mp.4001162. [DOI] [PubMed] [Google Scholar]
- 34.Woodberry KA, Giuliano AJ, Seidman LJ. Premorbid IQ in schizophrenia: a meta-analytic review. Am J Psychiatry. 2008;165:579–87. doi: 10.1176/appi.ajp.2008.07081242. [DOI] [PubMed] [Google Scholar]
- 35.Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–41. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bassett AS, Marshall CR, Lionel AC, Chow EW, Scherer SW. Copy number variations and risk for schizophrenia in 22q11.2 deletion syndrome. Hum Mol Genet. 2008;17:4045–53. doi: 10.1093/hmg/ddn307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DeLisi LE, et al. Clinical characteristics of schizophrenia in multiply affected Spanish origin families from Costa Rica. Psychiatr Genet. 2001;11:145–52. doi: 10.1097/00041444-200109000-00006. [DOI] [PubMed] [Google Scholar]
- 38.DeLisi LE, et al. Genome-wide scan for linkage to schizophrenia in a Spanish-origin cohort from Costa Rica. Am J Med Genet. 2002;114:497–508. doi: 10.1002/ajmg.10538. [DOI] [PubMed] [Google Scholar]
- 39.Manolio TA, et al. New models of collaboration in genome-wide association studies: the Genetic Association Information Network. Nat Genet. 2007;39:1045–51. doi: 10.1038/ng2127. [DOI] [PubMed] [Google Scholar]
- 40.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kirov G, et al. Support for the involvement of large copy number variants in the pathogenesis of schizophrenia. Hum Mol Genet. 2009;18:1497–503. doi: 10.1093/hmg/ddp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shaikh TH, et al. High-resolution mapping and analysis of copy number variations in the human genome: a data resource for clinical and research applications. Genome Res. 2009;19:1682–90. doi: 10.1101/gr.083501.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bejjani BA, et al. Use of targeted array-based CGH for the clinical diagnosis of chromosomal imbalance: is less more? Am J Med Genet A. 2005;134:259–67. doi: 10.1002/ajmg.a.30621. [DOI] [PubMed] [Google Scholar]
- 44.Selzer RR, et al. Analysis of chromosome breakpoints in neuroblastoma at sub-kilobase resolution using fine-tiling oligonucleotide array CGH. Genes Chromosomes Cancer. 2005;44:305–19. doi: 10.1002/gcc.20243. [DOI] [PubMed] [Google Scholar]
- 45.Sebat J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–9. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


