Abstract
Prk1p is a serine/threonine kinase involved in the regulation of the actin cytoskeleton organization in the yeast Saccharomyces cerevisiae. Previously, we have identified LxxQxTG as the phosphorylation site of Prk1p. In this report, the recognition sequence for Prk1p is investigated more thoroughly. It is found that the presence of a hydrophobic residue at the position of P-5 is necessary for Prk1p phosphorylation and L, I, V, and M are all able to confer the phosphorylation at various efficiencies. The residue flexibility at P-2 has also been identified to include Q, N, T, and S. A homology-based three-dimensional model of the kinase domain of Prk1p provided some structural interpretations for these substrate specificities. The characterization of the [L/I/V/M]xx[Q/N/T/S]xTG motif led to the identification of a spectrum of potential targets for Prk1p from yeast genome. One of them, Scd5p, which contains three LxxTxTG motifs and is previously known to be important for endocytosis and actin organization, has been chosen to demonstrate its relationship with Prk1p. Phosphorylation of Scd5p by Prk1p at the three LxxTxTG motifs could be detected in vitro and in vivo, and deletion of PRK1 suppressed the defects in actin cytoskeleton and endocytosis in one of the scd5 mutants. These results allowed us to conclude that Scd5p is likely another regulatory target of Prk1p.
INTRODUCTION
The actin cytoskeleton is required for a number of fundamental processes in eukaryotic cells such as cell motility and intracellular trafficking of proteins and membrane-bound vesicles. In the yeast Saccharomyces cerevisiae, the actin cytoskeleton is essential for cell growth (Pruyne and Bretscher, 2000; Chang and Peter, 2003). The actin structures that are involved in the polarized growth in yeast are the cortical actin patches, which serve to define the site of cell growth, and the cytoplasmic actin cables, which act as “tracks” for myosin-dependent vesicle movement (Pruyne and Bretscher, 2000). These structures are highly dynamic and undergo frequent redistribution throughout the cell cycle (Adams and Pringle, 1984; Kilmartin and Adams, 1984; Lew and Reed, 1993).
It is also well documented that the actin cytoskeleton in yeast is intimately associated with endocytosis, a retrograde membrane-trafficking process during which portions of the plasma membrane along with extracellular materials are internalized into cytoplasm and delivered to endosomal compartments. A group of proteins whose cellular localization pattern coincides with that of cortical actin patches have been shown to be required for both the normal morphology of actin patches and the process of endocytosis (Geli and Riezman, 1998). Of particular importance among them is Pan1p, the yeast homolog of a mammalian protein named Eps15. The role of Pan1p in the actin cytoskeleton organization and endocytosis seems to be multifold, acting as an adaptor to coordinate multivalent interactions of several cortical actin patch-localized proteins on one hand, and as an activator of the Arp2/3 complex to promote the actin polymerization on the other (Tang and Cai, 1996; Wendland et al., 1996, 1999; Tang et al., 1997, 2000; Wendland and Emr, 1998; Duncan et al., 2001).
The rapid and orderly rearrangement of the actin cytoskeleton during the cell cycle suggests that there exist efficient mechanisms of regulation in the actin organization in yeast. Although these mechanisms remain largely unknown, the recent identification of a protein kinase, Prk1p, as an important regulator of the actin cytoskeleton organization and endocytosis has begun to shed some light on them (Cope et al., 1999; Zeng and Cai, 1999; Smythe and Ayscough, 2003). Based on genetic and biochemical analyses, Prk1p has been considered to be a negative regulator of some of the proteins that are required for proper structure and function of the actin cytoskeleton. These proteins include Pan1p, End3p, Sla1p, Ent1p, and Ent2p (Zeng and Cai, 1999; Watson et al., 2001; Zeng et al., 2001), and likely others (see below). Loss-of-function mutations of PRK1 suppressed the pan1 and end3 mutations and caused a delay in the actin polarization and bud formation at the early stage of the cell cycle (Zeng and Cai, 1999). Overexpression of PRK1 led to cell death accompanied by gross actin abnormalities (Zeng and Cai, 1999). Prk1p is able to phosphorylate Pan1p and Sla1p in vitro in a sequence-specific manner (Zeng and Cai, 1999; Zeng et al., 2001). The recognition motif of Prk1p has been identified as LxxQxTG (Zeng and Cai, 1999). Both Q and G are essential for recognition and phosphorylation, whereas the importance of the residue L has not been investigated (Zeng and Cai, 1999).
Prk1p belongs to a new family of the serine/threonine kinase, which also include Ark1p and Akl1p from yeast, and a few homologous kinases from higher eukaryotic organisms (Smythe and Ayscough, 2003). One of the members of this family, AAK1, is important for the process of endocytosis in mammalian cells and phosphorylates the AP2 complexes with similar sequence specificity as Prk1p (Conner and Schmid, 2002; Ricotta et al., 2002). It is therefore possible that the mechanism of regulation by Prk1p-like kinases is highly conserved from yeast to mammalian cell systems.
In this study, we have defined the motif sequence required for Prk1p recognition and phosphorylation more accurately through systematic mutagenesis. The new motif, [L/I/V/M]xx[Q/N/T/S]xTG, widened the spectrum of potential Prk1p targets present in the yeast genome. Scd5p, which contains three clustered copies of LxxTxTG and is previously known to be required for actin cytoskeleton organization and endocytosis, has been investigated for its relationship with Prk1p. Evidence is presented to suggest that Scd5p may be another regulatory target of Prk1p.
MATERIALS AND METHODS
Strains, Media, and General Methods
The yeast strains used in this study are listed in Table 1. Yeast cells were grown in standard yeast extract-peptone-dextrose (YEPD) or dropout medium supplemented with appropriate amino acids required for plasmid maintenance (Rose et al., 1990). In experiments requiring the expression of genes under the GAL1 promoter, raffinose instead of dextrose was used as the carbon source and galactose was later added for GAL1 induction. Bacterial strains were grown on standard media supplemented with 100 μg/ml ampicillin to maintain plasmids. Genetic and recombinant DNA manipulations were performed according to standard recombinant techniques (Sambrook et al., 1989; Rose et al., 1990).
Table 1.
Yeast strains used in this study
| Strain | Genotype |
|---|---|
| W303-1B | MATα ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3-1 |
| YMC427 | MATa ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3-1 prk1Δ::URA3 |
| YMC446 | MATα ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3-1 scd5-1 (scd5::SCD5PBM2Δ-LEU2) |
| YMC447 | MATα ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3-1 prk1Δ::URA3 scd5-1 |
| YMC448 | MATα ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3-1 scd5::SCD5-HA-LEU2 |
| YMC449 | MATα ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3-1 scd5::SCD5AAA-HA-LEU2 |
Plasmid and Strain Construction
The plasmid constructs used in this study are shown in Table 2. Mutations in pSCD5PBM2Δc, pSCD5AAAc-HA, and variants of GST-R15, GST-YAP1801c, GST-ENT1c, and GST-SCD5m were generated either by one-step polymerase chain reaction (PCR) with mutagenic primers or by sequential PCR mutagenesis as described previously (Cormack, 1997).
Table 2.
Plasmid constructs used in this study
| Construct | Description |
|---|---|
| pGAL-PRK1 | PRK1 under GAL1 promoter control in pRS316 (Zeng et al., 2001). |
| pGAL-PRK1D158Y | PRK1D158Y under GAL1 promoter control in pRS316 (Zeng et al., 2001). |
| pGAL-HA-PRK1 | The HA-Prk1p expressing construct (Zeng et al., 2001). |
| pGAL-HA-PRK1D158Y | The HA-Prk1D158Yp expressing construct (Zeng et al., 2001). |
| pGAL-STE3-EGFP | STE3 coding region was generated by PCR, cloned in frame with a C-terminal EGFP epitope followed by the ADH1 terminator, and placed under GAL1 promoter control in pRS314. |
| pGEX-R15-WT | GST-R15; The DNA coding region for Pan1p (564-846 aa) containing the 15th Pan1p repeat was generated by PCR and cloned in frame into pGEX-4T-1 (Zeng and Cal, 1999). |
| pGEX-R15-T/S | GST-R15 variant with T to S mutation at P0 position. |
| PGEX-R15-L/A | GST-R15 variant with L to A mutation at P-5 position. |
| pGEX-R15-LP-6 | GST-R15 variant with PP-6 to L and LP-5 to A mutations. |
| pGEX-R15-LP-4 | GST-R15 variant with TP-4 to L and LP-5 to A mutations. |
| pGEX-R15-LP-3 | GST-R15 variant with AP-3 to L and LP-5 to A mutations. |
| pGEX-R15-L/I | GST-R15 variant with L to I mutation at P-5 position. |
| pGEX-R15-L/M | GST-R15 variant with L to M mutation at P-5 position. |
| pGEX-R15-L/P | GST-R15 variant with L to P mutation at P-5 position. |
| pGEX-R15-L/V | GST-R15 variant with L to V mutation at P-5 position. |
| pGEX-R15-L/T | GST-R15 variant with L to T mutation at P-5 position. |
| pGEX-R15-L/S | GST-R15 variant with L to S mutation at P-5 position. |
| pGEX-R15-L/N | GST-R15 variant with L to N mutation at P-5 position. |
| pGEX-R15-L/F | GST-R15 variant with L to F mutation at P-5 position. |
| pGEX-R15-L/W | GST-R15 variant with L to W mutation at P-5 position. |
| pGEX-R15-LN | GST-R15 variant with Q to N mutation at P-2 position. |
| pGEX-R15-IN | GST-R15 variant with LP-5 to I and QP-2 to N mutations. |
| pGEX-R15-VN | GST-R15 variant with LP-5 to V and QP-2 to N mutations. |
| pGEX-R15-MN | GST-R15 variant with LP-5 to M and QP-2 to N mutations. |
| pGEX-R15-NA | GST-R15 variant with QP-2 to N and TP0 to A mutations. |
| pGEX-R15-LT | GST-R15 variant with Q to T mutation at P-2 position. |
| pGEX-R15-IT | GST-R15 variant with LP-5 to I and QP-2 to T mutations. |
| pGEX-R15-VT | GST-R15 variant with LP-5 to V and QP-2 to T mutations. |
| pGEX-R15-MT | GST-R15 variant with LP-5 to M and QP-2 to T mutations. |
| pGEX-R15-TA | GST-R15 variant with QP-2 to T and TP0 to A mutations. |
| pGEX-R15-LS | GST-R15 variant with Q to S mutation at P-2 position. |
| pGEX-R15-IS | GST-R15 variant with LP-5 to I and QP-2 to S mutations. |
| pGEX-R15-VS | GST-R15 variant with LP-5 to V and QP-2 to S mutations. |
| pGEX-R15-MS | GST-R15 variant with LP-5 to M and QP-2 to S mutations. |
| pGEX-R15-SA | GST-R15 variant with QP-2 to S and TP0 to A mutations. |
| pGEX-YAP1801WT | GST-YAP1801c; The DNA coding region for Yap1801p (407-637 aa) was generated by PCR and cloned in frame into pGEX-4T-1. |
| pGEX-YAP1801T/A | GST-YAP1801c variant with T413A mutation. |
| pGEX-YAP1801AAA | GST-YAP1801c variant with T413A, T427A, and T453A mutations. |
| pGEX-YAP1801TAA | GST-YAP1801c variant with T427A and T453A mutations. |
| pGEX-YAP1801ATA | GST-YAP1801c variant with T413A and T453A mutations. |
| pGEX-YAP1801AAT | GST-YAP1801c variant with T413A and T427A mutations. |
| pGEX-ENT1WT | GST-ENT1c; The DNA coding region for Ent1p (272-454 aa) was generated by PCR and cloned in frame into pGEX-4T-1. |
| pGEX-ENT1AT | GST-ENT1c variant with T395A mutation. |
| pGEX-ENT1TA | GST-ENT1c variant with T415A mutation. |
| pGEX-ENT1AA | GST-ENT1c variant with T395A and T415A mutations. |
| pGEX-ENT1AAAAA | GST-ENT1c variant with T346A, T366A, T395A, T415A, and T427A mutations. |
| pGEX-ENT1TAAAA | GST-ENT1c variant with T366A, T395A, T415A, and T427A mutations. |
| pGEX-ENT1ATAAA | GST-ENT1c variant with T346A, T395A, T415A, and T427A mutations. |
| pGEX-ENT1AATAA | GST-ENT1c variant with T346A, T366A, T415A, and T427A mutations. |
| pGEX-ENT1AAATA | GST-ENT1c variant with T346A, T366A, T395A, and T427A mutations. |
| pGEX-ENT1AAAAT | GST-ENT1c variant with T346A, T366A, T395A, and T415A mutations. |
| pGEX-SCD5WT | GST-SCD5m; The DNA coding region for Scd5p (409-585 aa) was generated by PCR and cloned in frame into pGEX-4T-1. |
| pGEX-SCD5AAA | GST-SCD5m variant with T416A, T450A, and T490A mutations. |
| pGEX-SCD5TAA | GST-SCD5m variant with T450A and T490A mutations. |
| pGEX-SCD5ATA | GST-SCD5m variant with T416A and T490A mutations. |
| pGEX-SCD5AAT | GST-SCD5m variant with T416A and T450A mutations. |
| pSCD5PBM2Δc | The DNA coding region for Scd5p (79-872 aa) containing mutations on its 2nd PP1-binding motif (KKVRF to AKAAA) was generated by PCR and cloned into pRS305. |
| pSCD5c-HA | The DNA coding region for Scd5p (79-872 aa) was generated by PCR and cloned in frame with a C-terminal HA epitope followed by the ADH1 terminator in pRS305. |
| pSCD5AAAc-HA | The DNA coding region for Scd5p (79-872 aa) with T416A, T450A, and T490A mutations was generated by PCR and cloned in frame with a C-terminal HA epitope followed by the ADH1 terminator in pRS305. |
The strain YMC427 (prk1Δ) was generated as described previously (Zeng and Cai, 1999). To obtain YMC446 (scd5-1), YMC448, and YMC449, plasmids pSCD5PBM2Δc, pSCD5c-HA, and pSCD5AAAc-HA were linearized within the SCD5 gene by BclI digestion and integrated into W303-1B, respectively. The integration was confirmed by PCR (Huxley et al., 1990) and sequencing analysis. YMC447 (prk1Δ scd5-1) was a progeny of a diploid made of a cross between YMC427 and YMC446.
Preparation of Glutathione S-Transferase (GST) Fusion Proteins, Immunoprecipitation, In Vitro Kinase Assay, and Protein Extraction by Trichloroacetic Acid (TCA) Precipitation
To make GST-fusion proteins, the DNA coding regions were obtained by PCR and fused in-frame to a bacterial GST expression vector pGEX-4T-1 (Amersham Biosciences, Singapore). The plasmids were transformed into Escherichia coli strain DH5α. Expression and purification of the fusion proteins were performed according to Zeng and Cai (1999). Preparation of yeast extracts, immunoprecipitation, immunoblotting of the hemagglutinin (HA)-tagged proteins, and in vitro kinase assay with GST-fusion proteins as substrates were performed as described previously (Zeng and Cai, 1999; Zeng et al., 2001). To measure the relative phosphorylation intensity, the images of substrate protein bands were taken with Nikon COOLPIX995 digital camera and imported into the Multi-Analysis software (Bio-Rad, Hercules, CA) for normalization. The bands were then excised from the dried gel and transferred to scintillation counting vials. Five milliliters of Ecoscint-scintillation liquid (National Diagnostics, Yorkshire, United Kingdom) was dispensed into each vial and counted with LKB 1219 RackBeta automatic liquid scintillation counter (PerkinElmer Wallac, Turku, Finland).
To analyze the gel mobility change of Scd5p upon PRK1 overexpression, yeast cells (YMC448 and YMC449) containing various plasmids (pRS316, pGAL-PRK1, and pGAL-PRK1D158Y) were grown at 30°C to log phase followed by addition of galactose to 2%. After incubation for 5 h, 4 OD600 of cells were collected and washed once with ice-cold water. The cells were suspended in 400 μl of lysis buffer (925 mM NaOH, 3.75% β-mercaptoethanol) and incubated on ice for 20 min. Cell lysates were mixed with equal volume of 50% TCA and incubated on ice for 10 min, followed by centrifugation at high speed for 5 min. The pellets were washed once with ice-cold phosphate-buffered saline (PBS) and dissolved in 100 μl of protein loading buffer. Ten microliters of 1 M Tris-HCl (pH 8.0) was finally added to each sample for pH adjustment.
Structural Modeling
A three-dimensional model of the kinase domain (residues 23–298) of Prk1p (National Center for Biotechnology Information accession number NP_012171) was generated from the SWISSMODEL (Guex and Peitsch, 1997) automated protein structure homology-modeling server (http://www.expasy.org/swissmod/SWISS-MODEL.html). Eighty-nine templates from different serine/threonine and tyrosine kinases with sequence identity ranging from 25 to 40% were used. Subsequent modeling used Sybyl 6.8 (Tripos, St. Louis, MO) and all energy calculations invoked the Kollman force field. Holding the backbone constant, the model of Prk1p was minimized, followed by an alignment with the crystal structure of a serine/threonine kinase (2PHK) possessing a bound peptide. The bound peptide was extracted into our model and substituted to PLTAQKTGF, the LxxQxTG motif of Pan1p present in the in vitro substrate R15. Essential interactions necessary for phosphotransfer were modeled according to reported observations (Madhusudan et al., 1994). The peptide's P0 (Thr) residue was placed in proximity to the catalytic residue ASP158 and the peptide-anchoring residue LYS160. Peptide residues P-2 (Gln) to P0 (Thr) together with kinase residues within 8-Å proximity were energetically refined. The carboxyl group of ASP158 was measured to be ∼1.7Å from the hydroxyl hydrogen of P0 (Thr). GLN207 was found to establish a critical H-bond (∼1.8Å) with P-2 (Gln). Twelve pockets were identified on the surface of the kinase by SiteID in Sybyl. The closest pocket to P-2 (Gln) is hydrophobic, being lined by residues ILE117, MET120, and ILE238. P-5 (Leu) was docked into this pocket using DOCK from sybyl6.8. Thereafter, the link between P-5 (Leu) and P-2 (Gln) was constructed and further minimization was performed. A conformational search was performed to determine the optimal side chain conformation of P-5 in the pocket. Finally, a MOLCAD surface was created to display the docked conformation with the use of Sybyl6.8.
Endocytosis Assays
The lucifer yellow (LY) uptake assay was performed as described previously (Dulic et al., 1991) with minor modifications. Cells were grown at 25°C in YEPD to early log phase, and cultures were kept at 25°C or preshifted to 37°C for 15 min before addition of lucifer yellow CH dilithium (Sigma-Aldrich) to 5 mg/ml. After incubation for 2 h at 25°C or 37°C, cells were collected and washed five times with PBS containing 10 mM sodium azide and 50 mM sodium fluoride, followed by suspension in Vectashield mounting medium (Vector Laboratories, Burlingame, CA) and observation by fluorescein isothiocyanate and Nomarski optics with a Leica DMAXA microscope equipped with a Hamamatsu C4742–98 digital camera.
To observe the internalization of Ste3p, cells containing pGAL-STE3-EGFP were grown at 25°C in dropout medium supplemented with raffinose to early log phase followed by addition of galactose to 2%. After incubation at 25°C for 45 min, cultures were kept at 25°C or preshifted to 37°C for 15 min before addition of glucose to 3%. Samples were taken at 15-min intervals at 25°C or 37°C immediately upon addition of glucose, washed three times with PBS containing 10 mM sodium azide and 50 mM sodium fluoride, suspended in Vectashield mounting medium, and visualized with the Leica DMAXA microscope.
Actin Staining
Staining of actin filaments with rhodamine-phalloidin (Molecular Probes, Eugene, OR) was performed as described previously (Adams and Pringle, 1991) with minor modifications. Cells were grown at 25°C in YEPD to early log phase and then kept at 25°C or shifted to 37°C. After incubation for 4 h, cells were collected and suspended in fixation solution (3.7% formaldehyde, 100 mM KH2PO4, 100 mM K2HPO4) for 15 min. Cells were then washed two times with PBS and resuspended in PBS containing 0.1% Triton X-100 for 15 min. After washing again with PBS for two times, cells were incubated with PBS containing rhodamine-phalloidin (1:100) at 25°C for 30 min. Cells were finally washed with PBS for four times and suspended in Vectashield mounting medium before visualization with the Leica DMAXA microscope.
RESULTS
Prk1p Could Not Phosphorylate Its Target Sequence with Threonine Replaced by Serine
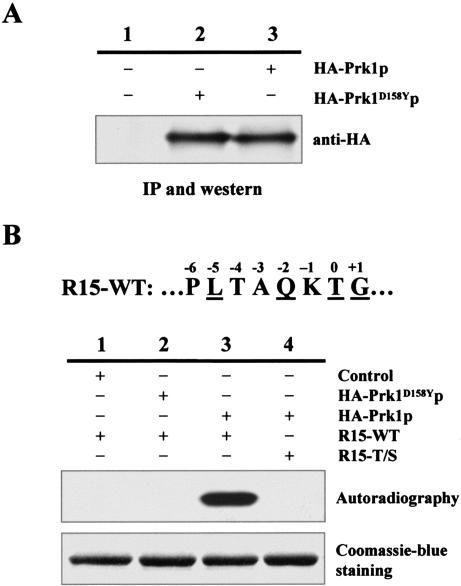
Our previous studies have characterized the consensus phosphorylation site of Prk1p as LxxQxTG (Zeng and Cai, 1999). It is conspicuous that Pan1p contains 15 copies of this motif but not a single copy of LxxQxSG, suggesting that Prk1p may only be able to phosphorylate threonine but not serine in this context. To test this possibility, we carried out in vitro kinase assays using a mutated substrate of Prk1p that had the threonine replaced by a serine in this motif. This substrate was derived from R15, a GST-fused Pan1p fragment (amino acids 564–846) that contained a single LxxQxTG motif and had been used as a standard substrate for Prk1p in the previous in vitro kinase assays (Zeng and Cai, 1999). For convenience of description, the threonine in the LxxQxTG motif was given a position number P0, and the residues upstream or downstream of P0 were designated as P–n or P+n, respectively (Figure 1B). In the control experiments, the immunoprecipitated HA-Prk1p (Figure 1A, lane 3) was able to phosphorylate the GST-fusion protein R15-WT (Figure 1B, lane 3), whereas the anti-HA immunoprecipitates from cells containing either untagged Prk1p (Figure 1A, lane 1), or HA-tagged kinase-dead mutant of Prk1p (HA-Prk1D158Yp; Figure 1A, lane 2), failed to phosphorylate the same protein (Figure 1B, lanes 1 and 2). Prk1p, on the other hand, was unable to phosphorylate R15-T/S, an R15 variant with a T-to-S mutation at P0 (Figure 1B, lane 4). This result confirmed our speculation that the Prk1p phosphorylation motif, as had been identified, was threonine-specific.
Figure 1.
Inability of Prk1p to phosphorylate serine in its recognition motif. (A) Western blotting of Prk1p and its kinase-dead mutant Prk1D158Yp (Zeng and Cai, 1999). The HA-tagged proteins were immunoprecipitated from equal amounts of cell extracts with the rabbit anti-HA antibody and analyzed on immunoblots. (B) Prk1p was unable to phosphorylate LxxQxSG. The immunoprecipitated proteins and substrates used in each reaction are as indicated. Phosphorylation results were shown as autoradiography, and the input substrates were visualized by Coomassie Blue staining. The amino acid sequence of the 15th LxxQxTG motif of Pan1p (R15-WT) is shown on the top with each residue labeled according to their position relative to the phosphorylation target T.
Hydrophobic Residues at P-5 Are Required for Prk1p Recognition
So far, we have determined the importance of Q, T, and G of the LxxQxTG motif in Prk1p phosphorylation. To examine the stringency of L for Prk1p recognition, the L residue of R15-WT at P-5 was mutated into A and subjected to in vitro kinase assay. As shown in Figure 2A, the resulting protein R15-L/A could not be phosphorylated by Prk1p (Figure 2A, lane 1). In addition, introduction of an L into other positions (P-6, P-4, and P-3) of R15-L/A also failed to confer Prk1p phosphorylation (Figure 2A, lanes 2, 4, and 5). These results indicate that there is a stringent requirement for leucine at P-5, but not other positions, for the motif to be recognized by Prk1p.
Figure 2.
Analysis of sequence requirement at P-5 for Prk1p recognition. (A) The requirement for leucine at P-5 for Prk1p recognition. GST-fusion proteins of R15-WT and its mutants were prepared and subjected to in vitro kinase assay. R15-L/A (lane 1) carried an L to A mutation at P-5, whereas mutants in lanes 2, 4, and 5 had an additional mutation that introduced L into positions of P-6, P-4, and P-3, respectively. (B) Phosphorylation of the [L/I/V/M]xxQxTG motifs by Prk1p. GST-fusion proteins of R15-WT and its mutants bearing various substitutions at P-5 as indicated were prepared and subjected to in vitro kinase assay. The immunoprecipitated HA-Prk1p was used as the kinase in all reactions of A and B. Phosphorylation results were shown as autoradiography and the input substrates were visualized by the Coomassie Blue staining. (C) Comparison of phosphorylation efficiency of the [L/I/V/M/F/P/W/A]xxQxTG motifs. The bands of the substrates were excised from the gel for scintillation counting after autoradiography and the results were normalized against the amounts of the substrates. Phosphorylation efficiencies of each motif were calculated as the percentage of reading relative to that of the LxxQxTG motif.
The Prk1p recognition motif LxxQxTG occurs numerous times in Pan1p and Sla1p, clustered within certain domains that are involved in protein–protein interactions (Zeng and Cai, 1999; Zeng et al., 2001). Interestingly, multiple additional copies of QxTG, in which the P-5 position is other residues than L, are also present within these domains. These multiple QxTG motifs in Pan1p and Sla1p may represent additional patterns of Prk1p recognition sequence. To investigate this possibility, the L residue of R15-WT at P-5 was mutated into each of the every P-5 variation found in Pan1p and Sla1p, namely, I, M, P, V, T, A, S, and N. The in vitro kinase assays showed that three of these residues, I, M, and V, were able to confer Prk1p phosphorylation (Figure 2B, lanes 3, 4, and 6). As these residues are all hydrophobic, two additional hydrophobic residues, F and W, were also tested for their ability to support Prk1p phosphorylation. As shown in Figure 2B, neither R15-L/F nor R15-L/W could serve as an efficient substrate for Prk1p (Figure 2B, lanes 10 and 11). Furthermore, introduction of other residues to P-5 did not result in identification of additional Prk1p recognition patterns (our unpublished data).
Although all the P-5 residues suitable for Prk1p phosphorylation were hydrophobic, they differed in phosphorylation efficiency (Figure 2C). Among them, L is most optimal. This is consistent with the fact that LxxQxTG is present most frequently in the known Prk1p substrates Pan1p and Sla1p (Zeng and Cai, 1999; Zeng et al., 2001). After L, in a descending order, are I, V, and M, with recognition efficiencies of 36, 15, and 13%, respectively, of that of L.
Identification of N, T, and S as Additional P-2 Residues
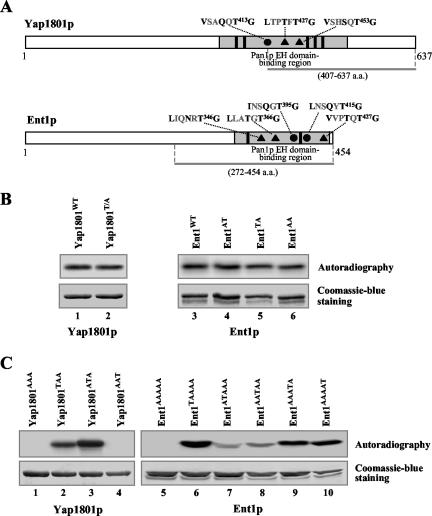
Using R15 variants as substrates, we identified [I/V/M]xxQxTG motifs as the new phosphorylation site of Prk1p. To verify this finding, we chose Yap1801p and Ent1p, two yeast proteins that contain some of these newly identified motifs, to test their ability to act as substrates of Prk1p in vitro. Yap1801p was originally identified as one of the yeast homologs of the mammalian clathrin assembly protein AP180 from a yeast two-hybrid screen by using the Eps15-homology (EH) domains of Pan1p as a bait (Wendland and Emr, 1998). The same screen also identified a pair of yeast epsins, Ent1p and Ent2p (Wendland et al., 1999). Yap1801p contains one (VxxQxT413G), whereas Ent1p has two (IxxQxT395G and LxxQxT415G) Prk1p recognition motifs in their Pan1p EH domain-binding regions (Figure 3A). Recently, Ent1p has been shown to exhibit Prk1p-dependent phosphorylation on its [L/I]xxQxTG motifs in vivo (Watson et al., 2001). Nevertheless, it is of interest to test whether it can be directly phosphorylated by Prk1p in vitro.
Figure 3.
Identification of N, T, and S as additional P-2 residues for Prk1p recognition. (A) Schematic diagrams of Yap1801p and Ent1p showing the distribution of Prk1p phosphorylation sites. The QxTG-containing (•) and [N/T/S]xTG-containing (▴) motifs are found in the Pan1p EH domain-binding regions (gray boxes) of each protein, in the vicinity of the NPF motifs (solid boxes). The bars beneath each of the diagrams indicate the region from each protein expressed as GST-fusion proteins. (B and C) Phosphorylation of Yap1801p, Ent1p, and their mutants by Prk1p. The substrates used in each reaction are as indicated. Substrates in C are labeled with the identities at P0 of the motifs arranged in a sequential order. For example, Yap1801AAT (C, lane 4) is a mutant carrying the T-to-A mutations in the first two of the three motifs. Phosphorylation results were shown as autoradiography and the input substrates were visualized by the Coomassie Blue staining.
As expected, a GST-fusion protein containing the VxxQxTG motif of Yap1801p (Yap1801WT) was efficiently phosphorylated by Prk1p (Figure 3B, lane 1). Surprisingly, however, when the T residue within VxxQxTG was mutated to A (T413A), the resulting protein Yap1801T/A could still be phosphorylated by Prk1p (Figure 3B, lane 2). Similar observation was also made with Ent1p. Although GST-fusion proteins containing both, or either one, of the [L/I]xxQxTG motifs of Ent1p could be efficiently phosphorylated by Prk1p (Figure 3B, lanes 3–5), disruption of both motifs by site-directed mutagenesis (T395A and T415A) failed to abolish the Prk1p-dependent phosphorylation (Figure 3B, lane 6). These results indicate that there must be additional Prk1p phosphorylation sites on Yap1801p and Ent1p.
On a closer examination of the sequences of Yap1801p and Ent1p, several motifs approximately resembling the phosphorylation targets of Prk1p were spotted. They were LxxTxT427G and VxxSxT453G on Yap1801p, and LxxNxT346G, LxxTxT366G, and VxxTxT427G on Ent1p (Figure 3A). These motifs differ in that they have residues N, T, and S rather than Q at the P-2 position. Two observations suggested that these motifs may also be phosphorylation sites of Prk1p. First, these motifs are clustered within a short region in both Yap1801p and Ent1p known to be involved in protein–protein interactions, i.e., the Pan1p EH domain-binding region (Wendland and Emr, 1998; Wendland et al., 1999), similarly to the distribution of the LxxQxTG motifs in Pan1p and Sla1p. Second, several such motifs were also found in Pan1p (LxxTxT241G, MxxNxT464G, and LxxTxT944G) and Sla1p (IxxNxT730G and IxxSxT923G). Indeed, as shown in Figure 3C, both VxxQxT413G and LxxTxT427G motifs from Yap1801p could be phosphorylated by Prk1p (Figure 3C, lanes 2 and 3). There was no detectable phosphorylation on the VxxSxT453G motif (Figure 3C, lane 4), although scintillation counting revealed a reading two-fold higher than that of the background (our unpublished data). After all these sites were mutated, the phosphorylation on Yap1801p by Prk1p was completely eliminated (Figure 3C, lane 1). Similarly, any one of these motifs from Ent1p was able to support phosphorylation by Prk1p, albeit to various degrees (Figure 3C, lanes 6–10), whereas the mutant with all the motifs disrupted showed no Prk1p phosphorylation (Figure 3C, lane 5). It is concluded, therefore, that N, T, and S are the additional P-2 residues in the Prk1p recognition motif.
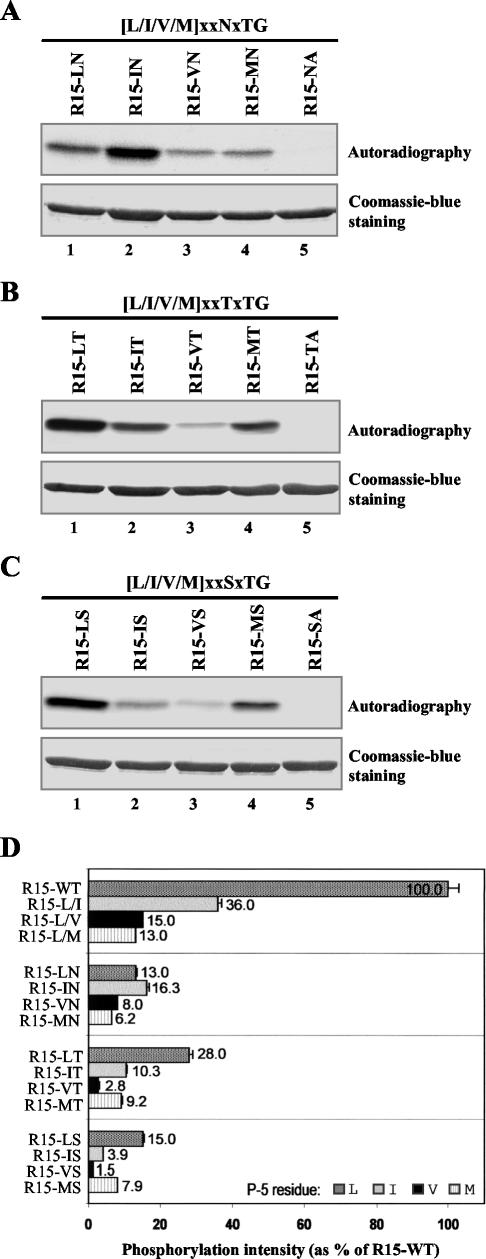
Comparison of Phosphorylation Efficiency of All [L/I/V/M]xx[Q/N/T/S]xTG Motifs
Combining all the sequence requirements at P-2 and P-5, the Prk1p phosphorylation motif is now consolidated to be [L/I/V/M]xx[Q/N/T/S]xTG. Again, using R15 as a template, we generated a series of motifs to compare their efficiency of serving as the target sequence of Prk1p, as we did with the set of mutations on P-5 of LxxQxTG shown in Figure 2B. Figure 4A showed the in vitro kinase assay result of the test on the [L/I/M/V]xxNxTG motifs. It is clear that different combinations gave rise to different phosphorylation efficiencies (Figure 4A, lanes 1–4). Similar tests were also performed on [L/I/M/V]xxTxTG, and [L/I/M/V]xxSxTG, as shown in Figure 4, B and C, respectively. Finally, these results were compared in Figure 4D. Consistent with our result obtained with native substrate, namely, Yap1801AAT (Figure 3C, lane 4), the VxxSxTG motif was the weakest Prk1p phosphorylation site (Figure 4D, R15-VS).
Figure 4.
Comparison of phosphorylation efficiency of the [L/I/V/M]xx[Q/N/T/S]xTG motifs. (A) Phosphorylation of the [L/I/V/M]xxNxTG motifs by Prk1p. (B) Phosphorylation of the [L/I/V/M]xxTxTG motifs by Prk1p. (C) Phosphorylation of the [L/I/V/M]xxSxTG motifs by Prk1p. The substrates used in lanes 1–4 are labeled to show their residue identities at P-5 and P-2. For example, R15-LN (A, lane 1) represents the motif of LxxNxTG. The substrates used in the lanes 5 are the negative controls and labeled with residue identities at P-2 and P0, e.g., R15-NA (A, lane 5) represents the motif of LxxNxAG. (D) Comparison of Prk1p phosphorylation efficiencies on the [L/I/V/M]xx[Q/N/T/S]xTG motifs. The phosphorylation results were measured by scintillation counting and normalized against the quantities of the substrates.
A Homology-based Three-dimensional (3D) Model of Prk1p
Prk1p belongs to a distinct family of kinases and shares significant sequence homology to other kinases in the kinase domain (Cope et al., 1999). The 3D structures of Prk1p or any member of this family have not been resolved. To gain some insights on the structural basis of Prk1p's function, a homology-based three-dimensional model of the Prk1p kinase domain (residues 23–298) was generated as described in MATERIALS AND METHODS. Based on this model, the Prk1p kinase domain comprises two main portions: an N-terminal lobe and a C-terminal lobe (Figure 5A). The N-terminal lobe (residues 23–110) consists of five antiparallel β strands (β1–β5) and one α helix (helix C) and is responsible for binding ATP through a glycine loop located between β1 and β2. The C-terminal lobe (residues 111–298) consists of a four-helix bundle (helices D, E, F, and H) that is flanked by two smaller helices and two short antiparallel β sheets. The catalytic loop with the conserved catalytic ASP158 residue and the activation segment (residues 176–212) are both located in this lobe. Using the sequence of PLTAQKTG (the LxxQxTG motif present in the in vitro substrate R15) as the substrate peptide, a structural model of the kinase–substrate complex was further constructed (Figure 5B). In the docked state, the substrate peptide binds in an extended conformation across the kinase catalytic site, contacting only the C-terminal lobe of the kinase domain (Figure 5B).
Figure 5.
Interactions between Prk1p and its recognition motifs depicted by homology-based modeling. (A) Two views of the Prk1p kinase domain. The left view has ribbons and loops colored by B-factor. Blue indicates regions with high degree of conservation and red low conservation. The major secondary structures such as β strand and α helix are also labeled. The right view highlights the important features of the kinase domain. The light green region represents the N-terminal lobe and the light blue region the C-terminal lobe. The glycine loop responsible for binding the nontransferable phosphates of ATP is located in the N-terminal lobe and colored in dark blue. The C-terminal lobe includes an activation segment in orange, four helices (D, E, F, and H) in dark green and the conserved catalytic residue ASP158 in dark blue. A P-5 binding pocket formed by ILE117, MET120, and ILE238 from the helices D and H is also shown. (B). A general view of the kinase domain of Prk1p with a substrate peptide (PLTAQKTG) bound. The kinase domain is visualized with a MOLCAD surface colored by lipophilic potential. The positions of the activation segment, the ATP-binding pocket, the catalytic residue ASP158, and the P-5 binding pocket are as indicated. (C) Schematic representation of the molecular interactions between Prk1p and its substrate peptide. The substrate peptide colored in black is shown together with the kinase residues essential for the interaction. Residues involved in docking the QxTG motif (ASP158, LYS160, and GLN207) are colored in blue and residues making up the P-5 binding pocket (ILE117, MET120, and ILE238) are colored in yellow. The dashed lines represent the hydrogen bonds between the carboxyl side chain of ASP158 and the hydroxyl group of P0, the amine side chain of LYS160 and the backbone carbonyl group of P-1, and the amide group of GLN207 and the amine or hydroxyl group of P-2, respectively.
Studies on the substrate–kinase complex model revealed four sets of essential interactions between Prk1p and the [L/I/V/M]xx[Q/N/T/S]xTG motif (Figure 5C). Among these interactions, three of them are the hydrogen bonds between the carboxyl side chain of ASP158 and the hydroxyl group of P0 residue T, the amine side chain of LYS160 and the backbone carboxyl group of P-1 residue, and the amine group of GLN207 and the amine group (Q and N) or the hydroxyl group (T and S) of the P-2 residue (Figure 5C, dashed lines). The other interaction is the van der Waals force between a hydrophobic binding pocket consisting of three kinase residues ILE117, MET120, and ILE238 and the P-5 hydrophobic residue (Figure 5C).
The interaction between the hydrophobic pocket of the kinase and the P-5 residue of the substrate explains why only hydrophobic residues should be present at P-5. Among all the hydrophobic residues, i.e., A, I, L, M, F, P, W, and V, only L, I, V, and M are the suitable P-5 residues, because A would be too small and F too bulky to be well docked inside the pocket, whereas both P and W are not flexible enough to be compacted into the pocket due to their rigid structures.
Potential Phosphorylation Targets of Prk1p in the Yeast Genome
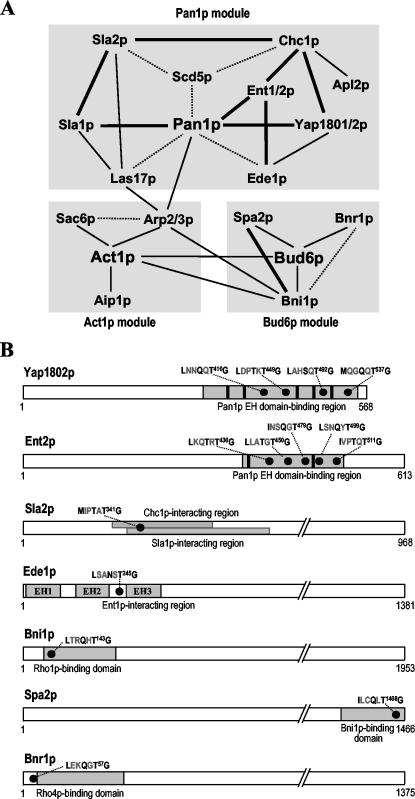
The identification of [L/I/M/V]xx[Q/N/T/S]xTG motifs as the Prk1p recognition sequence has increased the number of the potential Prk1p phosphorylation sites in Pan1p and Sla1p to 19 and 10, respectively. A search of the S. cerevisiae genome database (http://seq.yeastgenome.org/cgi-bin/SGD/PATMATCH/nph-patmatch) with a peptide sequence of [LIMV]xx[QNTS]xTG gave rise to ∼500 proteins that contain one or more copies of the [L/I/M/V]xx[Q/N/T/S]xTG motifs. Some of these proteins stand out as more likely to be potential regulatory targets of Prk1p than others because their functions in actin cytoskeleton organization, cell polarity, or endocytosis have been well documented. Twenty-three of them, including the known Prk1p targets Pan1p and Sla1p, are listed in Table 3. They are tentatively divided into three groups: proteins that are functionally related to Pan1p (Table 3, group I), the ones that are involved in the regulation of actin dynamics (Table 3, group II), and the ones that have important roles in bud site selection and cell polarity (Table 3, group III).
Table 3.
List of confirmed and potential phosphorylation targets of Prk1p in yeast genome
| Proteins | No. of motifs | Important interacting proteins (Physical or *genetic) | |
|---|---|---|---|
| Pan1p | 19 | Sla1p, Ent1/2p, Yap1801/2p, *Scd5p, *Ede1p, *Las17p, Arp2p | |
| Sla1p | 10 | Pan1p, Las17p, Sla2p, Syp1p | |
| Ent1p | 5 | Pan1p, *Ent2p, Chc1p, Ede1p | |
| Ent2p | 5 | Pan1p, *Ent1p | |
| Yap1801p | 3 | Pan1p, Chc1p | |
| I | Yap1802p | 4 | Pan1p, Chc1p, Ede1p |
| Las17p | 1 | *Pan1p, Sla1p, Arp2p, Chc1p, Sla2p, Act1p | |
| Scd5p | 3 | *Pan1p, *Chc1p, *Sla2p | |
| Ede1p | 1 | *Pan1p, Yap1802p, Ent1p, Syp1p, Sla2p | |
| Sla2p | 1 | Sla1p, *Scd5p, Chc1p, *Sac6p, Ede1p, Las17p, *Arp2p | |
| Chc1p | 1 | Yap1801/2p, Ent1p, *Scd5p, Sla2p, Apl2p, Las17p | |
| Apl2p | 1 | Chc1p | |
| Arp2p | 1 | Act1p, Pan1p, Las17p, Arp3p, *Sac6p, *Sla2p, Bni1p | |
| II | Arp3p | 1 | Act1p, Arp2p |
| Aip1p | 1 | Act1p | |
| Sac6p | 2 | Act1p, *Sla2p, *Arp2p | |
| Bni1p | 1 | Act1p, Bud6p, Spa2p, *Bnr1p, Arp2p, *Sla1p | |
| Bnr1p | 1 | *Bni1p, Bud6p | |
| Bud6p | 1 | Act1p, Bni1p, Bnr1p, Spa2p | |
| III | Spa2p | 1 | Bni1p, Bud6p |
| Bud2p | 1 | Cln2p | |
| Bud3p | 1 | Bud4p | |
| Syp1p | 2 | Ede1p, Sla1p |
Scd5p Is a Novel Phosphorylation Target of Prk1p
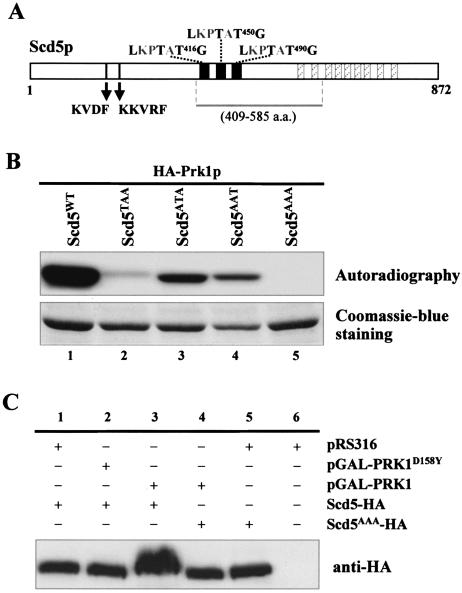
Among the above-mentioned proteins, Scd5p, a protein containing three copies of LxxTxTG, seems particularly promising as a new Prk1p phosphorylation target (Figure 6A). Scd5p was first identified from a genetic screen searching for multicopy suppressors of the clathrin heavy chain-deficient yeast strains (Nelson and Lemmon, 1993; Nelson et al., 1996). Subsequent studies revealed that Scd5p plays an important role in cortical actin organization and endocytosis (Henry etal., 2002). More recently, two potential protein phosphatase-1 (PP1) binding motifs were identified near the N terminus of Scd5p (KVDF and KKVRF; Figure 6A), one of which, the second one, was shown to mediate physical interaction between Scd5p and Glc7p, the yeast PP1 known to have a function in actin organization and endocytosis (Chang et al., 2002). In addition to the PP1-binding motifs, Scd5p also contains a 20-aa sequence (Figure 6A, solid boxes) repeated three times in the central region and a 12-aa sequence (Figure 6A, hatched boxes) repeated nine times at the C-terminal region (Nelson et al., 1996). The newly identified LxxTxTG motifs are embedded in each of the 20-aa repeats (Figure 6A). A resemblance between these central motifs and our previously defined Prk1p phosphorylation site has also been noted by Chang et al. (2002).
Figure 6.
Identification of Scd5p as a new phosphorylation target of Prk1p. (A) Schematic diagram of Scd5p showing the locations of three LxxTxTG motifs. In addition to these motifs, Scd5p also contains two putative PP1-binding motifs (KVDF and KKVRF), three copies of a 20-aa sequence (solid boxes) and nine copies of a 12-aa sequence (hatched boxes). The bar below the diagram represents the region used for GST-fusion protein expression. (B) Phosphorylation of Scd5p and its mutant forms by Prk1p in vitro. The substrates used in each reaction are as indicated. (C) Phosphorylation of Scd5p by Prk1p in vivo. Scd5p from YMC448 (scd5::SCD5-HA) containing vector control pRS316 (lane 1), pGAL-PRK1D158Y (lane 2), or pGAL-PRK1 (lane 3), and Scd5AAAp from YMC449 (scd5::SCD5AAA-HA) containing either pGAL-PRK1 (lane 4) or pRS316 (lane 5) were precipitated by TCA and analyzed by SDS-PAGE and immunoblotting.
To test whether the LxxTxTG motifs in Scd5p could be recognized by Prk1p in vitro, GST-fusion proteins of Scd5p were prepared for kinase assays. As shown in Figure 6B, the motif-containing substrate could indeed be strongly phosphorylated by Prk1p (Figure 6B, lane 1). The phosphorylation was completely abolished when all three motifs were mutated (Figure 6B, lane 5). Any single motif of the three was able to confer phosphorylation (Figure 6B, lanes 2–4), indicating that each of them was recognized by Prk1p.
To further confirm Scd5p as a phosphorylation target of Prk1p, we examined whether Scd5p could be phosphorylated by Prk1p in vivo in a host that overproduced the kinase. Scd5p (tagged with HA) was directly precipitated from cell lysates by TCA to maximally preserve its phosphorylated states, and dissolved in loading buffer for SDS-PAGE analysis. As shown in Figure 6C, in the absence of PRK1 overexpression, wild-type Scd5-HA migrated as a single band on the gel (Figure 6C, lane 1) with a same mobility as that of the nonphosphorylable Scd5AAA-HA mutant (Figure 6C, lane 5). In contrast, the band of Scd5p extracted from cells that had experienced PRK1 overexpression became smeared and retarded (Figure 6C, lane 3). This phenomenon was not observed for Scd5AAA-HA, which showed no changes in gel mobility upon Prk1p overproduction (Figure 6C, lane 4). Similarly, no mobility changes were observed for Scd5-HA precipitated from cells overexpressing the kinase-dead mutant of PRK1, PRK1D158Y (Figure 6C, lane 2). These results indicated that the gel mobility change of Scd5-HA extracted from Prk1p-overproducing cells was due to its phosphorylation by Prk1p on T416, T450, and T490 residues. It is concluded, therefore, that Scd5p could be phosphorylated by Prk1p in vivo.
Suppression of the scd5-1 Mutant by prk1Δ
The above-mentioned result suggested that the function of Scd5p may be subjected to regulation of Prk1p by phosphorylation. Furthermore, because Scd5p is also potentially under the regulation by the phosphatase Glc7p (Chang et al., 2002), it is possible that Glc7p and Prk1p could be counter-acting each other on the LxxTxTG motifs. Scd5p interacts with Glc7p mainly through its second PP1-binding motif (PBM2), and disruption of this motif results in the dissociation of Glc7p from Scd5p accompanied by a phenotype of temperature-sensitive (ts) growth (Chang et al., 2002). Hence, it is likely that the ts phenotype was due to over-phosphorylation of Scd5p by Prk1p and could be expected to be suppressed by deletion of PRK1.
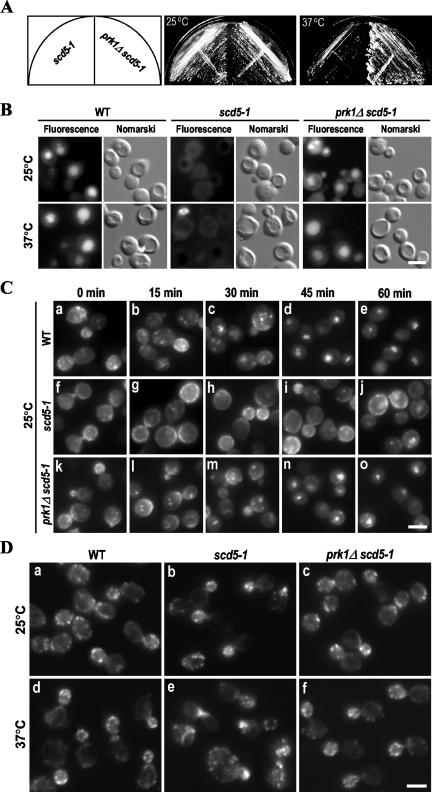
To test this hypothesis, we generated an scd5 mutant (scd5-1, YMC446) that carries the same mutations as reported by Chang et al. (2002) on the second PP1-binding motif of Scd5p. Consistent with the previous report, this PBM2-disrupted scd5 mutant was viable at 25°C and was temperature sensitive for growth at 37°C (Figure 7A). After introduction of the prk1Δ mutation into this mutant, the temperature sensitivity could indeed be suppressed (Figure 7A).
Figure 7.
Suppression of the scd5-1 mutation by prk1Δ. (A) Temperature sensitivity test of the scd5-1 and prk1Δ scd5-1 mutants. The strains YMC446 (scd5-1) and YMC447 (prk1Δ scd5-1) were streaked on YEPD plates and incubated at respective temperatures (25°C and 37°C) for 3 d. (B) LY uptake in wild-type (WT), scd5-1, and prk1Δ scd5-1 cells grown at 25°C or incubated at 37°C for 2 h. After 2 h of incubation with LY, cells were examined under a microscope with fluorescein isothiocyanate and Nomarski optics. (C) Internalization of Ste3-EGFP in WT, scd5-1, and prk1Δ scd5-1 cells grown at 25°C. Cells containing pGAL-STE3-EGFP were induced to express Ste3-EGFP for 1 h and samples were taken for observation at 15-min intervals after shutdown of Ste3p expression. (D) Actin staining of WT, scd5-1, and prk1Δ scd5-1 cells grown at 25°C or incubated at 37°C for 4 h. Cells were fixed and incubated with rhodamine-phalloidin for 30 min before the visualization. Bars, 5 μm.
The prk1Δ mutation not only suppressed the ts phenotype of the scd5-1 mutant but also corrected the deficiencies of the mutant in endocytosis and actin organization characterized previously (Chang et al., 2002; Henry et al., 2002). We performed two types of endocytosis assays. The first one was the fluid-phase endocytosis by using LY as a marker. As shown in Figure 7B, wild-type cells could efficiently internalize the dye at both 25°C and 37°C, and exhibited an unambiguous vacuolar staining (Figure 7B, left), whereas the scd5-1 cells failed to accumulate LY in vacuoles even at 25°C (Figure 7B, middle). In contrast, the prk1Δ scd5-1 cells showed normal LY uptake at both 25°C and 37°C (Figure 7B, right). We next examined the endocytosis of a plasma membrane protein Ste3p. Ste3p is the receptor of a-factor and is known to undergo constitutive endocytosis, leading to degradation in the vacuole in the absence of its ligand (Davis et al., 1993). As shown in Figure 7C, the majority of Ste3-EGFP in the wild-type cells growing at 25°C was found to be in the vacuole 45 min after the shutdown of its expression (Figure 7C, a–e). However, the scd5-1 mutant cells showed severe defects in this process (Figure 7C, f–j). Even at the last time point when no Ste3-EGFP was observed on the membranes of the wild-type cells (Figure 7C, e), the scd5-1 cells maintained significant Ste3-GEFP staining on the plasma membrane (Figure 7C, j). The prk1Δ scd5-1 mutant cells, on the other hand, exhibited a normal pattern of internalization of Ste3-EGFP (Figure 7C, k–o), essentially indistinguishable from that of wild-type cells. Similar results were also obtained with cells incubated at 37°C (our unpublished data). These results indicate that the endocytosis defect of scd5-1 could also be suppressed by prk1Δ.
The organization of the actin cytoskeleton was investigated by rhodamine-phalloidin staining. Wild-type cells exhibited normal actin structures throughout the cell cycle at either 25°C or 37°C (Figure 7D, a and d). At 25°C, the scd5-1 mutant cells exhibited a nearly normal actin organization, as far as the morphology and the distribution pattern of the cortical actin patches were concerned (Figure 7D, b). After temperature shift to 37°Cfor4h, ∼30–40% of the scd5-1 cells obviously showed aberrant actin structures (Figure 7D, e). These include depolarized cortical actin patches and accumulation of abnormal actin aggregates. These aberrant actin structures, however, were no longer visible in the prk1Δ scd5-1 cells either at 25°C or 37°C (Figure 7D, c and f).
DISCUSSION
Previous studies have characterized Prk1p as an important regulator of the actin cytoskeleton organization in yeast (Cope et al., 1999; Zeng and Cai, 1999). One possible mechanism for Prk1p to exert its regulation over the actin cytoskeleton is through modulating the activities of the actin-related Pan1p/Sla1p/End3p complex by phosphorylating Pan1p and Sla1p on the LxxQxTG motifs (Zeng and Cai, 1999; Zeng et al., 2001). Further analysis of this target sequence by systematic mutagenesis in this study has revealed that the Prk1p recognition sequence is less stringent than previously thought. As a consequence of this finding, Scd5p, a protein that contains three LxxTxTG motifs and is known to be involved in actin-related processes in yeast, has been identified to be another regulatory target of Prk1p.
Characterization of the Prk1p Recognition Motifs
Protein kinases generally fall into two major types: the serine/threonine kinases and the tyrosine kinases (Krebs, 1985). The Ser/Thr kinases regulate the functions of their target proteins by recognizing a consensus motif and phosphorylating a serine and/or threonine within it. In some cases, the serine and threonine residues in the recognition motifs are exchangeable. For example, both XS*PX and XT*PX are able to be phosphorylated by the proline-dependent protein kinase (Vulliet et al., 1989). Although Prk1p is a member of the Ser/Thr kinase family, no serine-containing sequences have been identified as its target so far. The finding that Prk1p is unable to phosphorylate the LxxQxSG motif (Figure 1B) suggests that Prk1p could be a threonine-specific kinase. However, the possibility that it may phosphorylate a serine residue within a different sequence context has not been ruled out.
Previously, we identified the target sequence of Prk1p as LxxQxTG with the G at P+1 being essential and the Q at P-2 important for Prk1p recognition (Zeng and Cai, 1999). The requirement for L at P-5 is now investigated in this report. This position is clearly important for Prk1p recognition, because mutation of L to A abolished the Prk1p phosphorylation (Figure 2A). However, the recent demonstration of Prk1p-dependent phosphorylation on the IxxQxTG motif of Ent1p by Watson et al. (2001) already suggested that leucine is not the only P-5 residue to support Prk1p phosphorylation. In addition to isoleucine, our residue scanning experiments had identified V and M as the other P-5 elements (Figure 2B). All of these residues are hydrophobic in nature, implicating a hydrophobic interaction in the recognition of the P-5 residue by the kinase. In the case of P-2, the residues required for Prk1p recognition have been found to contain T, S, and N, in addition to Q. Together, there are 16 distinct motifs that can serve as the Prk1p substrate. Interestingly, 13 of these motifs are found in Pan1p and Pan1p-interacting proteins, including Sla1p, Ent1p, Ent2p, Yap1801p, and Yap1802p, supporting the notion that these motifs are the true Prk1p phosphorylation sites in vivo. It is worth pointing out that, although P-5, P-2, P0, and P+1 are the key positions in the Prk1p recognition motif, other positions within, or adjacent to, the motif may also affect the interaction between the substrates and the kinase to some extent. This may be the reason for a certain degree of variation in the phosphorylation intensity of a same LxxTxTG motif situated in different substrates (Yap1801p and Ent1p; Figure 3C, lanes 3 and 7), or different locations of the same substrate (Scd5p; Figure 6B, lanes 2–4).
The Homology-based 3D Model of Prk1p
The three-dimensional structure of a protein kinase is useful in understanding the mechanism of kinase–substrate recognition. In the absence of a crystal structure of Prk1p, we have attempted to predict the 3D structure of the kinase domain in Prk1p based on known structures of other kinase domains. The notable feature of this model is the presence of a hydrophobic pocket on the kinase that is involved in interacting with the P-5 residue of the substrate. This is in a good agreement with our experimental results that the P-5 residues have to be hydrophobic amino acids. A mutation that changed ILE117, the key residue in the hydrophobic pocket, to Ala resulted in a complete loss of Prk1p kinase activity in vitro (our unpublished data). Nevertheless, due to the low degree of sequence identity of Prk1p (∼30%) with template kinases, significant deviations of this homology-based model from the true three-dimensional structure of the kinase are expected.
Scd5p Is Likely a New Regulatory Target of Prk1p
The knowledge of the recognition sequence of Prk1p allows us to identify potential phosphorylation targets of the kinase. We have chosen Scd5p to perform biochemical and genetic experiments to demonstrate its relationship with Prk1p. Scd5p has recently been shown to genetically interact with Pan1p (Henry et al., 2002). Both Scd5p and Pan1p are involved in the actin cytoskeleton organization and endocytosis (Tang et al., 1997; Henry et al., 2002). Scd5p contains three LxxTxTG motifs in its central region (Figure 6A), which have been noted by Chang et al. (2002) to bear some resemblance to the Prk1p phosphorylation site. It is therefore of great interest to ascertain whether Scd5p is under the regulation of Prk1p through phosphorylation in a way similar to Pan1p. Indeed, our results showed that Scd5p could be recognized and phosphorylated by Prk1p both in vitro and in vivo in a sequence-specific manner (Figure 6, B and C). Additional evidence to support Scd5p as a regulatory target of Prk1p comes from the genetic interaction between scd5-1 and prk1Δ. The suppression of the temperature sensitivity of scd5-1 by prk1Δ (Figure 7A) suggests that the function of Scd5p is under a negative regulation by Prk1p, much as that of Pan1p being regulated by Prk1p (Zeng and Cai, 1999). This notion is strengthened by the observation that the defects of the scd5-1 mutant in endocytosis and actin organization were virtually completely rescued by deletion of the kinase (Figure 7, B–D). Together, these results demonstrate that Scd5p is likely another regulatory target of Prk1p.
The nature of scd5-1 mutation (disruption of a PP1-binding motif) suggests that the suppression of scd5-1 by prk1Δ is probably due to counteracting effects of the kinase Prk1p and the phosphatase (PP1) Glc7p on the Scd5p protein. Presumably, the defects in the scd5-1 cells arise from a loss of access of the protein by Glc7p, because the region required for the interaction with the phosphatase on the mutant protein has been disrupted (Chang et al., 2002). As a consequence, the mutant protein may become persistently phosphorylated by Prk1p, which evidently inhibits its activity. In the absence of the kinase, the mutant Scd5p protein is no longer subject to the phosphorylation inhibition and therefore able to function again. Alternatively, the suppression of scd5-1 by prk1Δ could be mediated via other proteins that are under the negative regulation of Prk1p, such as Pan1p and Sla1p. This possibility, however, is mitigated by our observation that the temperature sensitivity of scd5-1 could not be suppressed by high copy PAN1 or SLA1 (our unpublished data).
Regulation of Protein–Protein Interactions by Prk1p
The negative regulation through phosphorylation by Prk1p as in the cases of Scd5p and Pan1p seems to be a rather general mode of function for Prk1p. There are many yeast proteins that contain the Prk1p phosphorylation motifs. Proteins that have actin-related functions are considered as potential Prk1p regulatory targets and can be classified into three groups, as shown in Table 3. Proteins within each group are not only functionally related but also show either physical or genetic interactions. Therefore, each group can be considered as a modular complex (Figure 8A). In addition, these modules are also linked to one another by the interactions between their members. For example, the Act1p module is connected to the Pan1p module through the interactions between the Arp2/3p complex and its activators, Pan1p and Las17p (Winter et al., 1999; Duncan et al., 2001). Within the Pan1p module, the Sla1p–Pan1p interaction has been shown to be disrupted by Prk1p phosphorylation (Zeng et al., 2001). Interestingly, Prk1p recognition motifs are also present in the Pan1p-interacting regions of the other four proteins, Yap1801/2p and Ent1/2p (Figures 3A and 8B). These proteins have been shown to interact with the EH domains of Pan1p via their NPF motif-containing regions (Wendland and Emr, 1998; Wendland et al., 1999). The intriguing interspacing of Prk1p target motifs with NPF motifs (Figures 3A and 8B) strongly suggests that the interactions of these proteins with Pan1p are likely to be regulated by Prk1p phosphorylation as well. Furthermore, the interactions of Ent1p and Yap1802p with Chc1p may also be affected by Prk1p phosphorylation, because the last Prk1p target motifs of each protein are located closely to their clathrin-binding regions (Wendland and Emr, 1998; Wendland et al., 1999). Similarly, Prk1p phosphorylation may modulate the interactions of Sla2p with Chc1p (Henry et al., 2002), Sla2p with Sla1p (Gourlay et al., 2003), and Ede1p with Ent1p (Aguilar et al., 2003), due to the presence of a Prk1p phosphorylation site in their protein-interacting regions (Figure 8B). The interactions in the Bud6p module that are possibly subjected to the Prk1p regulation are that of Bni1p with Rho1p (Kohno et al., 1996), Spa2p with Bni1p (Fujiwara et al., 1998), and Bnr1p with Rho4p (Imamura et al., 1997). We are currently testing the role of Prk1p phosphorylation in some of these interactions.
Figure 8.
Regulation of protein–protein interactions by Prk1p phosphorylation. (A) Three possible protein modules formed by the potential phosphorylation targets of Prk1p. Solid and dashed lines indicate physical and genetic interactions, respectively. The physical interactions probably regulated by Prk1p phosphorylation are highlighted with thick lines. (B) The location of Prk1p recognition motifs on some of its potential phosphorylation targets. The [L/I/V/M]xx[Q/N/T/S]xTG motifs are shown as solid dots and NPF motifs as solid boxes. The regions involved in protein–protein interactions are indicated as gray boxes.
In summary, the recognition sequence of the yeast actin regulatory kinase Prk1p has been characterized more thoroughly in this report. This information is important for identification of additional regulatory targets of this important kinase. Based on this information, we have identified a new regulatory target of Prk1p, Scd5p, which seems to be negatively regulated by the kinase similarly to the previously studied Pan1p/Sla1p/End3p complex. Further studies of the regulatory network centered on Prk1p will no doubt greatly advance our understanding of the mechanism of regulation of actin dynamics in yeast.
Acknowledgments
We are grateful to Xianwen Yu for providing the plasmid pGAL-STE3-EGFP and to Alan Munn for providing LY. We also thank Agnes L.C. Tan and Serene A.K. Ong for help with the homology-based modeling. We thank Choong Yun Won and Jun Wang for general technical assistance. This work was supported by the Agency for Science, Technology and Research of Singapore. M.C. holds an adjunct faculty appointment from the Department of Biochemistry, Faculty of Medicine, National University of Singapore.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–06–0362. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-06-0362.
References
- Adams, A.E., and Pringle, J.R. (1984). Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. J. Cell Biol. 98, 934–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, A.E., and Pringle, J.R. (1991). Staining of actin with fluorochrome-conjugated phalloidin. Methods Enzymol. 194, 729–731. [DOI] [PubMed] [Google Scholar]
- Aguilar, R.C., Watson, H.A., and Wendland, B. (2003). The yeast Epsin Ent1 is recruited to membranes through multiple independent interactions. J. Biol. Chem. 278, 10737–10743. [DOI] [PubMed] [Google Scholar]
- Chang, F., and Peter, M. (2003). Yeasts make their mark. Nat. Cell Biol. 5, 294–299. [DOI] [PubMed] [Google Scholar]
- Chang, J.S., Henry, K., Wolf, B.L., Geli, M., and Lemmon, S.K. (2002). Protein phosphatase-1 binding to scd5p is important for regulation of actin organization and endocytosis in yeast. J. Biol. Chem. 277, 48002–48008. [DOI] [PubMed] [Google Scholar]
- Conner, S.D., and Schmid, S.L. (2002). Identification of an adaptor-associated kinase, AAK1, as a regulator of clathrin-mediated endocytosis. J. Cell Biol. 156, 921–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope, M.J., Yang, S., Shang, C., and Drubin, D.G. (1999). Novel protein kinases Ark1p and Prk1p associate with and regulate the cortical actin cytoskeleton in budding yeast. J. Cell Biol. 144, 1203–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack, B. (1997). Directed mutagenesis using the polymerase chain reaction. In: Current Protocols in Molecular Biology, vol. 1, ed. F.M. Ausubel, R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smith, and K. Struhl, New York: John Wiley & Sons, 8.5.1–8.5.10. [Google Scholar]
- Davis, N.G., Horecka, J.L., and Sprague, G.F., Jr. (1993). Cis- and trans-acting functions required for endocytosis of the yeast pheromone receptors. J. Cell Biol. 122, 53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulic, V., Egerton, M., Elguindi, I., Raths, S., Singer, B., and Riezman, H. (1991). Yeast endocytosis assays. Methods Enzymol. 194, 697–710. [DOI] [PubMed] [Google Scholar]
- Duncan, M.C., Cope, M.J., Goode, B.L., Wendland, B., and Drubin, D.G. (2001). Yeast Eps15-like endocytic protein, Pan1p, activates the Arp2/3 complex. Nat. Cell Biol. 3, 687–690. [DOI] [PubMed] [Google Scholar]
- Fujiwara, T., Tanaka, K., Mino, A., Kikyo, M., Takahashi, K., Shimizu, K., and Takai, Y. (1998). Rho1p-Bni1p-Spa2p interactions: implication in localization of Bni1p at the bud site and regulation of the actin cytoskeleton in Saccharomyces cerevisiae. Mol. Biol. Cell 9, 1221–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geli, M.I., and Riezman, H. (1998). Endocytic internalization in yeast and animal cells: similar and different. J. Cell Sci. 111, 1031–1037. [DOI] [PubMed] [Google Scholar]
- Gourlay, C.W., Dewar, H., Warren, D.T., Costa, R., Satish, N., and Ayscough, K.R. (2003). An interaction between Sla1p and Sla2p plays a role in regulating actin dynamics and endocytosis in budding yeast. J. Cell Sci. 116, 2551–2564. [DOI] [PubMed] [Google Scholar]
- Guex, N., and Peitsch, M.C. (1997). SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18, 2714–2723. [DOI] [PubMed] [Google Scholar]
- Henry, K.R., D'Hondt, K., Chang, J., Newpher, T., Huang, K., Hudson, R.T., Riezman, H., and Lemmon, S.K. (2002). Scd5p and clathrin function are important for cortical actin organization, endocytosis, and localization of sla2p in yeast. Mol. Biol. Cell 13, 2607–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley, C., Green, E.D., and Dunham, I. (1990). Rapid assessment of S. cerevisiae mating type by PCR. Trends Genet. 6, 236. [DOI] [PubMed] [Google Scholar]
- Imamura, H., Tanaka, K., Hihara, T., Umikawa, M., Kamei, T., Takahashi, K., Sasaki, T., and Takai, Y. (1997). Bni1p and Bnr1p: downstream targets of the Rho family small G-proteins which interact with profilin and regulate actin cytoskeleton in Saccharomyces cerevisiae. EMBO J. 16, 2745–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin, J.V., and Adams, A.E. (1984). Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J. Cell Biol. 98, 922–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno, H., et al. (1996). Bni1p implicated in cytoskeletal control is a putative target of Rho1p small GTP binding protein in Saccharomyces cerevisiae. EMBO J. 15, 6060–6068. [PMC free article] [PubMed] [Google Scholar]
- Krebs, E.G. (1985). The phosphorylation of proteins: a major mechanism for biological regulation. Fourteenth Sir Frederick Gowland Hopkins memorial lecture. Biochem. Soc. Trans. 13, 813–820. [DOI] [PubMed] [Google Scholar]
- Lew, D.J., and Reed, S.I. (1993). Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J. Cell Biol. 120, 1305–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhusudan, Trafny, E.A., Xuong, N.H., Adams, J.A., Ten Eyck, L.F., Taylor, S.S., and Sowadski, J.M. (1994). cAMP-dependent protein kinase: crystallographic insights into substrate recognition and phosphotransfer. Protein Sci. 3, 176–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, K.K., and Lemmon, S.K. (1993). Suppressors of clathrin deficiency: overexpression of ubiquitin rescues lethal strains of clathrin-deficient Saccharomyces cerevisiae. Mol. Cell Biol. 13, 521–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, K.K., Holmer, M., and Lemmon, S.K. (1996). SCD5, a suppressor of clathrin deficiency, encodes a novel protein with a late secretory function in yeast. Mol. Biol. Cell 7, 245–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne, D., and Bretscher, A. (2000). Polarization of cell growth in yeast. J. Cell Sci. 113, 571–585. [DOI] [PubMed] [Google Scholar]
- Ricotta, D., Conner, S.D., Schmid, S.L., von Figura, K., and Honing, S. (2002). Phosphorylation of the AP2 mu subunit by AAK1 mediates high affinity binding to membrane protein sorting signals. J. Cell Biol. 156, 791–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, M.D., Winston, F., and Hieter, P. (1990). Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Smythe, E., and Ayscough, K.R. (2003). The Ark1/Prk1 family of protein kinases. EMBO Rep. 4, 246–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, H.Y., and Cai, M. (1996). The EH-domain-containing protein Pan1 is required for normal organization of the actin cytoskeleton in Saccharomyces cerevisiae. Mol. Cell Biol. 16, 4897–4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, H.Y., Munn, A., and Cai, M. (1997). EH domain proteins Pan1p and End3p are components of a complex that plays a dual role in organization of the cortical actin cytoskeleton and endocytosis in Saccharomyces cerevisiae. Mol. Cell Biol. 17, 4294–4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, H.Y., Xu, J., and Cai, M. (2000). Pan1p, End3p, and S1a1p, three yeast proteins required for normal cortical actin cytoskeleton organization, associate with each other and play essential roles in cell wall morphogenesis. Mol. Cell Biol. 20, 12–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulliet, P.R., Hall, F.L., Mitchell, J.P., and Hardie, D.G. (1989). Identification of a novel proline-directed serine/threonine protein kinase in rat pheochromocytoma. J. Biol. Chem. 264, 16292–16298. [PubMed] [Google Scholar]
- Watson, H.A., Cope, M.J., Groen, A.C., Drubin, D.G., and Wendland, B. (2001). In vivo role for actin-regulating kinases in endocytosis and yeast epsin phosphorylation. Mol. Biol. Cell 12, 3668–3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland, B., McCaffery, J.M., Xiao, Q., and Emr, S.D. (1996). A novel fluorescence-activated cell sorter-based screen for yeast endocytosis mutants identifies a yeast homologue of mammalian eps15. J. Cell Biol. 135, 1485–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland, B., and Emr, S.D. (1998). Pan1p, yeast eps15, functions as a multivalent adaptor that coordinates protein-protein interactions essential for endocytosis. J. Cell Biol. 141, 71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland, B., Steece, K.E., and Emr, S.D. (1999). Yeast epsins contain an essential N-terminal ENTH domain, bind clathrin and are required for endocytosis. EMBO J. 18, 4383–4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter, D., Lechler, T., and Li, R. (1999). Activation of the yeast Arp2/3 complex by Bee1p, a WASP-family protein. Curr. Biol. 9, 501–504. [DOI] [PubMed] [Google Scholar]
- Zeng, G., and Cai, M. (1999). Regulation of the actin cytoskeleton organization in yeast by a novel serine/threonine kinase Prk1p. J. Cell Biol. 144, 71–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, G., Yu, X., and Cai, M. (2001). Regulation of yeast actin cytoskeleton-regulatory complex Pan1p/Sla1p/End3p by serine/threonine kinase Prk1p. Mol. Biol. Cell 12, 3759–3772. [DOI] [PMC free article] [PubMed] [Google Scholar]