Abstract
We have purified two new complexes from Saccharomyces cerevisiae, one containing the centromere component Mtw1p together with Nnf1p, Nsl1p, and Dsn1p, which we call the Mtw1p complex, and the other containing Spc105p and Ydr532p, which we call the Spc105p complex. Further purifications using Dsn1p tagged with protein A show, in addition to the other components of the Mtw1p complex, the two components of the Spc105p complex and the four components of the previously described Ndc80p complex, suggesting that all three complexes are closely associated. Fluorescence microscopy and immunoelectron microscopy show that Nnf1p, Nsl1p, Dsn1p, Spc105p, and Ydr532p all localize to the nuclear side of the spindle pole body and along short spindles. Chromatin immunoprecipitation assays show that all five proteins are associated with centromere DNA. Homologues of Nsl1p and Spc105p in Schizosaccharomyces pombe also localize to the centromere. Temperature-sensitive mutations of Nsl1p, Dsn1p, and Spc105p all cause defects in chromosome segregation. Synthetic-lethal interactions are found between temperature-sensitive mutations in proteins from all three complexes, in agreement with their close physical association. These results show an increasingly complex structure for the S. cerevisiae centromere and a probable conservation of structure between parts of the centromeres of S. cerevisiae and S. pombe.
INTRODUCTION
The accurate segregation of chromosomes is a complex and coordinated process mediated by the mitotic spindle. The microtubules of the mitotic spindle are connected to the chromosomes by the kinetochore, a substructure within the centromere of the chromosome (Pollard and Earnshaw, 2002). The yeast Saccharomyces cerevisiae has a relatively simple centromere and kinetochore, and these are likely to be the first characterized in molecular terms, given the large number of components already identified. There is also increasing evidence of conservation at the protein level between yeast and vertebrate centromeres and kinetochores (Kitagawa and Hieter, 2001; Cheeseman et al., 2002a; Biggins and Walczak, 2003; Cleveland et al., 2003). This is despite the large difference in size between the 125-base pair centromere DNA of S. cerevisiae and the centromere DNAs of other species, which are >25 kb in Schizosaccharomyces pombe and several Mb in vertebrate cells (Sullivan et al., 2001). There are also large differences in kinetochore size among these organisms, reflecting the differences in centromere DNA size. Thus, S. cerevisiae probably has one microtubule per kinetochore (Peterson and Ris, 1976; O'Toole et al., 1999), S. pombe three (Ding et al., 1993), and vertebrate cells ∼25 (McDonald et al., 1992). The conservation at the protein level suggests that, despite the difference in size, the study of the simple S. cerevisiae centromere and kinetochore can contribute to a molecular understanding of the more complex mammalian structures.
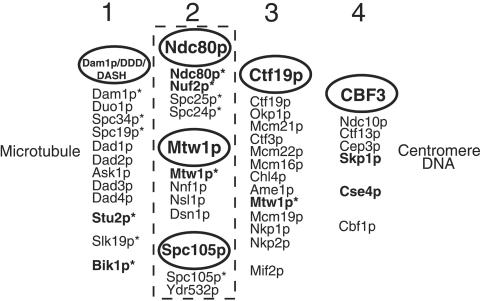
A combination of biochemical, genetic, and one- and two-hybrid approaches have identified many components of the S. cerevisiae centromere/kinetochore (Kitagawa and Hieter, 2001; Cheeseman et al., 2002b; Biggins and Walczak, 2003). The approximate positions of some of these components, in terms of their relationships to centromere DNA or the microtubule, are already becoming clear (see Figure 10 below). Components such as the CBF3 complex (Lechner and Carbon, 1991), Cbf1p (Cai and Davis, 1989; Jiang and Philippsen, 1989), and Cse4p (Meluh et al., 1998), which bind centromere DNA directly, are presumably close to the DNA. Components such as Stu2p (Wang and Huffaker, 1997; He et al., 2001), Slk19p (Zeng et al., 1999), Bik1p (He et al., 2001; Lin et al., 2001), and the Dam1p/DDD/DASH complex (Cheeseman et al., 2001a, 2002a; Janke et al., 2002; Li et al., 2002), which are clearly associated with microtubules and are also associated with centromere DNA, as shown by chromatin immunoprecipitation (ChIP) assays, are probably components of, or closely associated, with the kinetochore. The locations of the other yeast centromere/kinetochore components in relation to the microtubule ends, where the kinetochores presumably reside, have yet to be established. Thus, we will refer to these other yeast centromere/kinetochore components simply as centromere components.
Figure 10.
Possible groupings of S. cerevisiae centromere and kinetochore components. Components in bold type have known human homologues. Groups 1 and 2 contain components (with asterisks) that have been detected in spindle pole preparations by MALDI mass spectrometry (Wigge et al., 1998; Wigge and Kilmartin, 2001). Group 1 components associate laterally with microtubules and have also been detected at the centromere by ChIP assay. Group 2 components localize to the centromere only; the dashed line symbolizes the close association of these complexes. Nearly all group 3 components (Meluh and Koshland, 1995; Cheeseman et al., 2002a; Pot et al., 2003) have been localized to the centromere only and have not been detected in spindle pole preparations (except for Mtw1p; see text). Group 4 components bind centromere DNA directly as judged by band shift assay. References for most of these components can be found in the Introduction.
Our approach to identifying yeast centromere components has been a mass spectrometric analysis of highly enriched spindle poles (Wigge et al., 1998). These contain the yeast equivalent of the centrosome, the spindle pole body (SPB), together with attached spindle microtubules (Rout and Kilmartin, 1990). This spindle pole preparation also contains centromere and kinetochore components because fungi are unusual in having these structures clustered around the SPB through most of the cell cycle, apart from mitosis (Heath, 1980; Funabiki et al., 1993; Jin et al., 2000). However, this preparation contains only a subset of centromere components (Wigge and Kilmartin, 2001), probably because of the DNase I treatment used during the enrichment, which may remove components that are closer to the centromere DNA.
A powerful way of identifying the relationships between some of these components is to tag one component with protein A and then purify the tagged protein together with any associated proteins on an IgG column (Grandi et al., 1993; Knop and Schiebel, 1997). Using this approach, the centromere component Ndc80p (Rout and Kilmartin, 1990; Wigge et al., 1998) was found to be associated with Nuf2p, Spc25p, and Spc24p (Janke et al., 2001; Wigge and Kilmartin, 2001), all three of which had been identified previously in the spindle pole preparation (Wigge et al., 1998). The centromere protein Mtw1p (Goshima and Yanagida, 2000) and also Spc105p, whose localization suggests it might be a centromere component, have also been identified as components of this preparation (Wigge et al., 1998; Wigge and Kilmartin, 2001). Therefore, we decided to use protein A tagging to identify potential interacting partners of Mtw1p and Spc105p.
MATERIALS AND METHODS
Yeast Strains, Plasmids, and Growth Conditions
All S. cerevisiae strains (Table 1) were prepared using standard genetic methods in the K699 background or the isogenic diploid K842 (Nasmyth et al., 1990), and the yeast vectors used (Table 2) were the pRS series (Sikorski and Hieter, 1989). Standard growth conditions were used for both S. cerevisiae (Sherman, 1991) and S. pombe (Moreno et al., 1991). Strains with C-terminal protein A or GFP tags were prepared as before (Wigge et al., 1998), by recombinant PCR using pJK474 and pJK472 as templates, but with Vent polymerase (New England BioLabs, Beverly, MA). A strain containing C-terminal HA-tagged Spc105p (VNY290) marked with Kluyveromyces lactis TRP1 was prepared by recombinant PCR using pVN7.10 1-1 as template. Strains containing SPBs labeled with Spc42p-CFP were prepared by integration of pJK578 at the TRP1 locus as a single copy. pJK578 was prepared from SPC42-GFP (Adams and Kilmartin, 1999) by replacement of the GFP with CFP by exchanging a MscI-MunI fragment from the CFP plasmid pDH3 (Hailey et al., 2002). All tagged strains had the same growth rate as wild type. Yeast genes were cloned by PCR using Vent polymerase or, for larger genes, by gap repair (Orr-Weaver et al., 1983). Deletions were complete removals of the open reading frame by recombinant PCR.
Table 1.
Strain list
| Strain | Description | Source |
|---|---|---|
| S. cerevisiae | ||
| K699 | Mata ho ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3 ssd1 | Nasmyth et al. (1990) |
| K842 | diploid of K699 | Nasmyth et al. (1990) |
| K6445 | as K699 except leu2-3,112::PDS1-18Myc-LEU2 | K. Nasmyth |
| K6475 | as K699 except ura3::3XtetO112-URA3 leu2-3,112::GFP-tetR-LEU2 | Michaelis et al. (1997) |
| K7049 | as K699 except CFIII(CEN3.L.YPH278) TRP1 SUP11 | K. Nasmytha |
| K8572 | as K699 except leu2-3,112::GFP-tetR-LEU2 CEN5(1.4 kb left)::tetO2X112-HIS3 trp1::SPC42-GFP-TRP1 | Tanaka et al. (2000) |
| JK1121 | as K699 except nuf2-61 | This articlea |
| JK1495 | as K699 except YDR532c::GFP-Sphis5+ | This article |
| JK1504 | as K699 except mtw1-11 | This article |
| JK1664 | as K699 except ndc10-1 YDR532c::pA-Sphis5+ | This article |
| JK1667 | as K699 except ndc10-1 DSN1::pA-Sphis5+ | This article |
| JK1669 | as K699 except ndc10-1 SPC105::pA-Sphis5+ | This article |
| JK1670 | as K699 except ndc10-1 NNF1::pA-Sphis5+ | This article |
| JK1673 | as K699 except ndc10-1 NSL1-pA-Sphis5+ | This article |
| JK1676 | as K699 except YDR532c::GFP-Sphis5+ trp1::SPC42-CFP-TRP1 | This article |
| JK1679 | as K699 except SPC105::GFP-Sphis5+ trp1::SPC42-CFP-TRP1 | This article |
| JK1682 | as K699 except NSL1::GFP-Sphis5+ trp1::SPC42-CFP-TRP1 | This article |
| JK1683 | as K699 except DSN1::GFP-Sphis5+ trp1::SPC42-CFP-TRP1 | This article |
| JK1694 | as K699 except NNF1::GFP-Sphis5+ trp1::SPC42-CFP-TRP1 | This article |
| MSY5 | as K699 except MTW1::pA-Sphis5+ | This article |
| MSY48 | as K699 except YDR532c::pA-Sphis5+ | This article |
| MSY52 | as K699 except SPC105::GFP-Sphis5+ | This article |
| MSY72 | as K699 except dam1-31 | This article |
| MSY79 | as K699 except duo1-61 | This article |
| MSY87 | as K699 except ask1-22 | This article |
| MSY90 | as K699 except dad1-13 | This article |
| MSY104 | as K842 except ydr532cΔ::Sphis5+/YDR532c | This article |
| MSY150 | as K699 except spc105-4 + pPAW1(3.10 1-1) | This article |
| MSY151 | as K699 except spc105-4 + pJK19.11-68 | This article |
| MSY152 | as K699 except spc105-4 + pRS426 | This article |
| PWY92 | as K842 except spc105Δ::Sphis5+/SPC105 | Wigge and Kilmartin (2001) |
| PWY199 | as K699 except spc105Δ::Sphis5+ + pPAW1(3.10 1-1) | Wigge and Kilmartin (2001) |
| PWY282 | as K699 except SPC105::pA-Sphis5+ | This article |
| PWY350 | as K699 except NDC80::pA-Sphis5+ | Wigge and Kilmartin (2001) |
| PWY483 | as K699 except spc24-1 | Wigge and Kilmartin (2001) |
| PWY611 | as K699 except ndc10-1 | Wigge and Kilmartin (2001) |
| PWY754 | as K699 except spc25-1 | Wigge and Kilmartin (2001) |
| SHY116 | as K699 except ndc80-1 | Wigge et al. (1998) |
| VNY3 | as K699 except NNF1::GFP-Sphis5+ | This article |
| VNY28 | as K699 except DSN1::GFP-Sphis5+ | This article |
| VNY32 | as K699 except NSL1::pA-Sphis5+ | This article |
| VNY34 | as K699 except NNF1::pA-Sphis5+ | This article |
| VNY38 | as K699 except DSN1::pA-Sphis5+ | This article |
| VNY50 | as K699 except NSL1::GFP-Sphis5+ | This article |
| VNY69 | as K699 except ns11-6 | This article |
| VNY72 | as K699 except ns11-5 | This article |
| VNY87 | as K699 except nnf1-77 | This article |
| VNY96 | as K699 except dsn1-7 | This article |
| VNY99 | as K699 except dsn1-8 | This article |
| VNY116 | as K699 except dsn1-7 leu2-3,112::PDS1-18Myc-LEU2 | This article |
| VNY119 | as K699 except mtw1-11 leu2-3,112::GFP-tetR-LEU2 CEN5(1.4 kb left)::tetO2X112-HIS3 trp1::SPC42-GFP-TRP1 | This article |
| VNY122 | as K699 except nsl1-5 leu2-3,112::GFP-tetR-LEU2 CEN5(1.4 kb left)::tetO2X112-HIS3 trp1::SPC42-GFP-TRP1 | This article |
| VNY130 | as K699 except nsl1-5 leu2-3,112::PDS1-18Myc-LEU2 | This article |
| VNY162 | as K699 except spc105-4 | This article |
| VNY164 | as K699 except spc105-15 | This article |
| VNY198 | as K699 except spc105-15 CFIII(CEN3.L.YPH278) TRP1 SUP11 | This article |
| VNY203 | as K699 except spc105-15 ura3::3XtetO112-URA3 leu2-3,112::GFP-tetR-LEU2 | This article |
| VNY283 | as K699 except dsn1-7 leu2-3,112::PDS1-18Myc-LEU2 mad2Δ::Sphis5+ | This article |
| VNY287 | as K699 except nsl1-5 ura3::3XtetO112-URA3 leu2-3,112::GFP-tetR-LEU2 | This article |
| VNY290 | as K699 except SPC105::3HA-KlTRP1 | This article |
| VNY296 | as K699 except SPC105::3HA-KlTRP1 NDC80::pA-Sphis5+ | This article |
| VNY318 | as K699 except nsl1-5 leu2-3,112::PDS1-18Myc-LEU2 mad2Δ::Sphis5+ | This article |
| S. pombe | ||
| IH365 | h-ura4.d18 leu1.32 | Bridge et al. (1998) |
| JK1527 | h+ura4.d18 leu1.32 bub1+-6HA-ura4+ | This articleb |
| JK1627 | h-ura4.d18 leu1.32 spc105+-GFP-kanRbub1+-6HA-ura4+ | This article |
| JK1636 | h-ura4.d18 leu1.32 nsl1+-GFP-kanRbub1+-6HA-ura4+ | This article |
K7049 and JK1121 were derived by backcrossing of YPH278 (Spencer et al., 1990) and PSY455 (Osborne et al., 1994), respectively, into the K699 background.
JK1527 was derived from a backcross of strain 415 (Bernard et al., 1998).
Table 2.
Plasmid list
| Plasmid | Description | Source |
|---|---|---|
| pRS306 | URA3 (integrating) | Sikorski and Heiter (1989) |
| pRS314 | CEN TRP1 (low copy) | Sikorski and Heiter (1989) |
| pRS316 | CEN URA3 (low copy) | Sikorski and Heiter (1989) |
| pRS426 | 2μ URA3 (high copy) | Christianson et al. (1992) |
| pJK472 | GFP5-Sphis5+ in pBluescript SK- | Wigge et al. (1998) |
| pJK474 | pA-Sphis5+ in pBluescript SK- | Wigge and Kilmartin (2001) |
| pJK544 | GFP5-kanMX4 in pBluescript SK- | This article |
| pJK578 | SPC42-CFP in pRS304 | This article |
| pJK19.9-4 | spc105-4 in pRS306 | This article |
| pJK19.9-15 | spc105-15 in pRS306 | This article |
| pJK19.11-68 | YDR532c in pRS426 | This article |
| pJK4-7a | mtw1-11 in pRS314 | This article |
| pMS27.9 3-1 | ask1-22 in pRS306 | This article |
| pMS27.9 6-1 | duo1-61 in pRS306 | This article |
| pMS2.10 3-1 | dad1-13 in pRS306 | This article |
| pMS18.9 3-1 | dam1-31 in pRS306 | This article |
| pPAW1(3.10 1-1) | SPC105 in pRS316 | Wigge and Kilmartin (2001) |
| pVN7.10 1-1 | 3HA-KITRP1 in pBluescript SK- | This article |
| pVN10.5 137 | nsl1-6 in pRS314 | This article |
| pVN11.4 77-2 | nnf1-77 in pRS314 | This article |
| pVN17.5 157 | nsl1-5 in pRS314 | This article |
| pVN28.5 186-2 | dsn1-8 in pRS306 | This article |
| pVN28.5 187-3 | dsn1-7 in pRS306 | This article |
Sphis5+ is the S. pombe his5+ module from pFA6a-His3MX6 and kanMX4 the kanamycin resistance module from pFA6a-kanMX4 (Wach et al., 1997); KlTrp1 is the Kluyveromyces lactis TRP1 gene (Stark and Milner, 1989).
Temperature-sensitive (Ts– when describing the phenotype, ts when describing the genotype) mutants were prepared by modifications of error-prone PCR and the gapping procedure (Muhlrad et al., 1992; Wigge and Kilmartin, 2001). The procedure evolved during the course of this work. The initial Ts– mutants (mtw1, nsl1, and nnf1) were prepared by replacement of the disrupted locus with the PCR-amplified ts plasmids (Tyers et al., 1993). All the other mutants were prepared by slight modifications of the popin-popout procedure (Rothstein, 1991) described below.
Restriction sites for the gapping were chosen close to the ends of the open reading frames, and oligonucleotide primers for the error-prone PCR (7.5 mM MgCl2, 0.2 mM dATP and dGTP, 1 mM dCTP and dTTP) were 100–150 base pairs before and after these sites. After transformation, the cells were frozen at –70°C after addition of 10% DMSO, and aliquots were plated to give 1000–3000 colonies for each 13-cm-diameter selective plate at 23°C. When transformants were just visible, they were replica plated onto fluoroorotic acid medium (Boeke et al., 1987) at 23°C and then further replica-plated onto warm (37°C) and then ambient (23°C) YEPD plates. Ts– colonies were rechecked by patching on YEPD at 37 and 23°C. The selected strains formed colonies <40 μm in diameter after 2 d at 37°C and had low reversion frequencies; growth at 23°C appeared normal, and colonies were of a uniform size. Yeast DNA was then prepared and electroporated into Escherichia coli. Plasmids were sequenced to identify amino acid changes, and the mutant genes were transferred to pRS306 using the gapping sites to eliminate any changes in the noncoding region. The resulting plasmids, with a single restriction enzyme cut in the coding region, were then transformed into strain K699 for the popin-popout procedure. After restreaking, 16 transformants were patched onto fluoroorotic acid plates and then replica-plated onto warm and ambient YEPD plates. Ts– colonies and the parental Ura+ colonies were checked by PCR to verify the popin and popout events, and they were further checked for rescue by the low-copy plasmid carrying the wild-type gene and by backcrossing to show that a single ts mutation was present.
The changes found for mtw1-11 (JK1504) were E33G, K67E, N74T, V76E, and L144Q; for nnf1-77 (VNY87), R11Q, L57S, M73K, Y124L, and E151G; for nsl1-5 (VNY72), L35P, Q88R, M106T, V131I, I147N, Q158L, and N174I; for nsl1-6 (VNY69), W110R, K136R, and Y182C; for dsn1-7 (VNY96), K73R, K416R, N437I, and V451E; for dsn1-8 (VNY99), Q515P, and T541P; for spc105-4 (VNY162), D55N, V243A, N245D, M376K, Y503H, D553V, N616S, E631G, Y644N, M662T, S680P, L759P, and V856D; for spc105-15 (VNY164), N2Y, S269P, S348P, D363G, I523N, L683S, L736P, D752G, M781V, and K810R (plus the change A-3G in the 5′ noncoding region just before the presumed initiator ATG codon); for dam1-31 (MSY72), G104C, K194Q, K199E and G237S; for dad1-13 (MSY90), T5S, N7K, and L69R; for duo1-61 (MSY79) N32S, C100R, K169E, D178V, and R214G; and for ask1-22 (MSY87), V14D, L60R, L69M, S155A, E176V, and R197S.
The phenotypes of the pairs of nsl1, dsn1, and spc105 alleles were very similar in each case (our unpublished results). In addition the phenotypes of the mtw1-11, nnf1-77, dam1-31, dad1-13, duo1-61, and ask1-22 alleles used here were very similar in each case (our unpublished results) to the previously described alleles (Shan et al., 1997; Goshima and Yanagida, 2000; Cheeseman et al., 2001a; Enquist-Newman et al., 2001; Euskirchen, 2002; Li et al., 2002).
S. pombe strains were constructed by recombinant PCR (Bähler et al., 1998), using pJK544 as template with Vent polymerase and transformation of the PCR products into strain IH365. Transformants were checked as before (Wigge and Kilmartin, 2001), except that the 3′ genomic sequence corresponding to the primer used was not checked because the predicted adjacent 3′ open reading frames are several hundred base pairs away. Transformants were backcrossed into JK1527, which has HA-tagged bub1+ (Bernard et al., 1998).
Isolation of Complexes and Mass Spectrometry
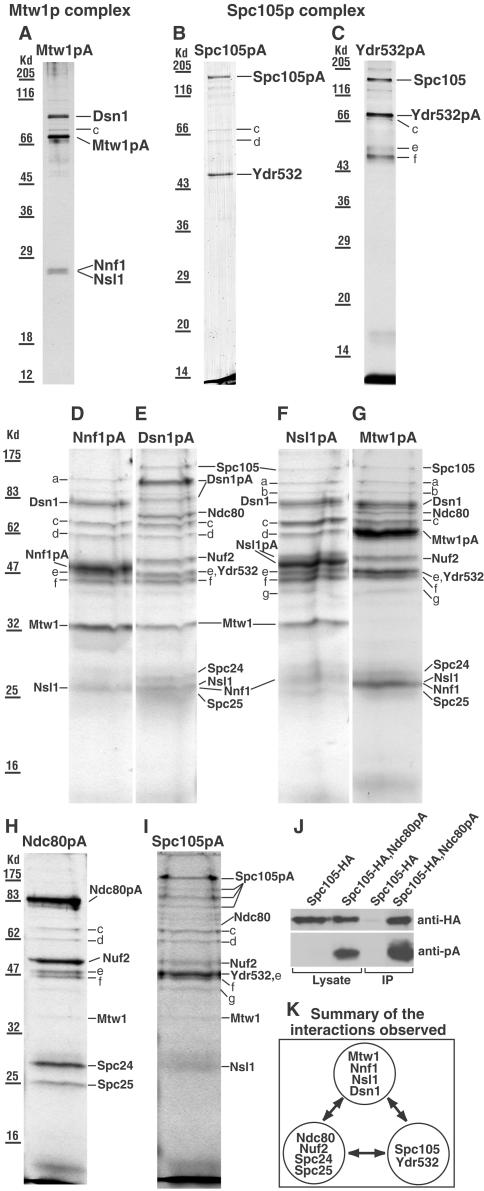
Complexes were isolated as before (Wigge and Kilmartin, 2001). Samples were run in different gels or some distance apart to minimize cross contamination. Proteins in SDS gel bands were identified by MALDI mass spectrometry as before (Wigge and Kilmartin, 2001), except that the entire NCBI database (>1.3 × 106 protein sequences) was searched using Mascot (http://www.matrixscience.com), at between 40 and 100 ppm, allowing for methionine oxidation and up to one missed tryptic cleavage. All identifications reported were the top match found. The MALDI method did not give a clear identification for some proteins, in particular Nnf1p in Figure 1, D–G; Ydr532p in Figure 1, E and G; and Nuf2p in Figure 1I. Liquid chromatography-mass spectrometry/mass spectrometry (LC-MS/MS) was used instead to identify these proteins (Aebersold and Mann, 2003). In LC-MS/MS, the peptides from the digested gel band are separated by HPLC, and the column output is sprayed directly into the QSTAR mass spectrometer (Applied Biosystems [Foster City, CA]/MDS Sciex API QSTAR Pulsar i). The mass spectrometer detects the peptides and selects those for fragmentation and sequencing, based on their intensity and charge state. Thus, in the quite intense Coomassie-stained band (above band e) in Figure 1D, MALDI mass spectrometry only identified protein A and failed to identify the expected Nnf1p. In contrast, LC-MS/MS was able to identify and sequence seven Nnf1p peptides (33% sequence coverage) in this band. In the other gels, LC-MS/MS sequenced 5–6 Nnf1p peptides (25–33% coverage; Figure 1, E–G), 6–7 peptides for Ydr532p (18–23% coverage; Figure 1, E and G), and 11 peptides for Nuf2p (31% coverage; Figure 1I).
Figure 1.
(A–I) SDS gels of the complexes isolated from strains (names in brackets) containing the indicated protein A (pA)-tagged proteins. (A and G) Mtw1p (MSY5), (B and I) Spc105p (PWY282), (C) Ydr532p (MSY48), (D) Nnf1p (VNY34), (E) Dsn1p (VNY38), (F) Nsl1p (VNY32), and (H) Ndc80p (PWY350). Gels A–C were from the first fraction off the columns and gels D–I from the second fraction, which usually contains more material but also more contaminants. All the gels were Coomassie-stained except A and C, which were silver-stained. In gels D–I, the sample wells were loaded as fully as possible, which caused some distortion in the band shape. In E, the lower band labeled Dsn1pA was a proteolytic fragment since it contained peptides from both Dsn1p and protein A. In F, no further proteins were identified in the Nnf1p region of this gel. Bands a-g were identified as Rpn1p (a), Sen3p (b, only identified in gels F and G), Ssa1p, Ssa2p, or both (c), Hsp60p (d), Tef2p (e), Ydj1p (f), and Scj1p (g). Rpn1p and Sen3p are proteosome components, Ssa1p, Ssa2p, Hsp60p, Ydj1p, and Scj1p are heat shock factors, and Tef2p is elongation factor. Further information on these proteins can be found at http://www.yeastgenome.org/. It seems likely that all of these are contaminants since they have been found in isolations with noncentromeric proteins coupled to protein A (our unpublished results), and most are absent from the cleaner gels (A–C). (J) Immunoblot of immunoprecipitates (IP) obtained as described in MATERIALS AND METHODS using strains containing HA-tagged Spc105p only (VNY290), or both HA-tagged Spc105p and protein A-tagged Ndc80p (VNY296). This shows that upon enriching for Ndc80pA, trace amounts of Spc105-HA can be detected. (K) Summary of the interactions observed.
Cytology
Observations of GFP or CFP fluorescence in live S. cerevisiae cells were carried out as described elsewhere (He et al., 2000; Tanaka et al., 2000; Hailey et al., 2002). Immuno-EM was as described previously (Adams and Kilmartin, 1999) using anti-GFP raised in a rabbit against a native GST-GFP fusion, depleted of GST antibodies, and affinity-purified with the fusion protein (our unpublished results). A strain containing GFP-labeled SPBs was included in the immuno-EM to check specificity. Cells for immunofluorescence (Adams and Pringle, 1984; Kilmartin and Adams, 1984) were fixed for 25 min for detecting GFP-labeled chromosomes and tubulin; this period of fixation still allowed detection of GFP with anti-GFP while preserving reasonable microtubule morphology. Fixation for 60 min was used for detection of tubulin, SPBs, and myc-labeled Pds1p. The antibodies used for immunofluorescence were antitubulin YOL1/34 (Kilmartin et al., 1982); anti-Tub4p for SPBs, which was prepared in a rabbit against a GST fusion of residues 447–473 of Tub4p, and absorbed as described for anti-GFP above (our unpublished results); and antimyc 9E10 (Evan et al., 1985), for myc-tagged Pds1p. In classifying the various microtubule morphologies described in the text, 100–150 cells were counted for each time point, unless otherwise stated.
S. pombe GFP fluorescence in live cells was analyzed as before (Wigge and Kilmartin, 2001) except that exposures were 0.2–0.3 s. A different version of GFP, GFP5 (Siemering et al., 1996), was used instead of the S65T GFP used earlier (Wigge and Kilmartin, 2001). The intensity of the GFP5 signal appeared to be more susceptible to formaldehyde fixation; thus, only 3 min of fixation were used for experiments combining immunofluorescence with observations of GFP fluorescence, rather than the 9 min used previously (Wigge and Kilmartin, 2001). Antibodies used for immunofluorescence were anti-Sad1 (Hagan and Yanagida, 1995), antitubulin TAT1 (Woods et al., 1989), and anti-HA 16B12 (Covance, Princeton, NJ) for detecting HA-tagged Bub1.
All images were acquired with a charge-coupled-device camera (model RTEA/CCD-1800-Y, Princeton Instruments, Trenton, NJ).
Genetic Interactions
All of the ts alleles described in this study and four previously described ts alleles of the Ndc80p complex (ndc80-1, nuf2-61, spc24-1, and spc25-1; Osborne et al., 1994; Wigge et al., 1998; Wigge and Kilmartin, 2001) were crossed with each other. The normal replica plating technique had to be changed for crosses involving the spc105 alleles because of microcolony formation (see RESULTS) and reversion. Spore colonies were picked with a toothpick, and most of the adherent cells were wiped off on a YEPD plate. The remaining cells on the toothpick were streaked downwards (to give a variable density of cells) in a patch of ∼15 mm2 and grown for 14 h at 23°C before replica plating. In all the crosses, five tetrads were picked initially to check for any lethality. After that, a total of 20–36 tetrads were picked at 23°C for each cross showing interactions, and spores were checked the next day in the microscope for germination. Tetrads in which one or two of the spores failed to germinate (2%) or where mating type did not segregate 2:2 (0.3%) were not examined further. We also eliminated a small minority of tetrads (4%) in which segregation patterns appeared to show growth of the double ts, in contrast to the bulk of the data for that pair of mutations. For two such tetrads, showing one Ts+ spore and three Ts– spores (one of which should have been a double ts and thus inviable), we backcrossed each Ts– spore with wild type and found that each contained one ts mutation, suggesting that gene conversion had occurred or that a false tetrad (4 loose spores clumped together) had been picked. For the remaining tetrads, colony growth was checked at 23, 30, and 37°C, except that growth at 30°C was not checked for pairs of mutations of the Mtw1p and Ndc80p complexes. In all crosses, with the exceptions outlined above, the spore inviability observed could be entirely explained by the predicted presence of the double ts mutants.
Other Methods
Immunoprecipitations (Figure 1J) were from 25 ml of culture harvested at 5 × 107 cells/ml and broken open with 0.4-mm glass beads in 0.2 ml of lysis buffer (7.5% glycerol, 50 mM Tris-Cl, pH 7.8, 0.1 M NaCl, 5 mM EGTA, 1 mM EDTA, and 1:50 solution Q; Wigge and Kilmartin, 2001). Lysates were precleared by centrifugation at 13,000 rpm for 10 min and then incubated with 5 μl IgG-Sepharose beads (Amersham Pharmacia, Uppsala, Sweden) for 4.5 h at 4°C. Beads were washed three times with lysis buffer and boiled with 50 μl SDS sample buffer; 20 μl was then loaded onto the SDS gel. Immunoblots were stained with 12CA5 anti-HA (Niman et al., 1983; Field et al., 1988) to detect the HA tag or rabbit IgG to detect protein A, followed by appropriate secondary antibodies. Flow cytometry was carried out as described previously (Haase and Lew, 1997). ChIP assays were carried out by multiplex PCR in an ndc10-1 background (Goh and Kilmartin, 1993; Meluh and Koshland, 1997; Ortiz et al., 1999). The regions of chromosome III amplified were 110410–110615, 114322–114558, and 118249–118538. The primers were 23mers except at 114558 where Primer 1 was used (Meluh and Koshland, 1997); the Mg2+ concentration was 4 mM.
RESULTS
Identification of Components of the Mtw1p and Spc105p Complexes and Interactions between Them and the Ndc80p Complex
The centromere protein Mtw1p (Goshima and Yanagida, 2000) and Spc105p have both been identified as components of enriched S. cerevisiae spindle pole preparations (Wigge et al., 1998; Wigge and Kilmartin, 2001). We wished to identify interacting proteins using protein A (pA)-tagging, similar to the way the Ndc80p complex was identified, where four components of the spindle pole preparation were found to be associated in a complex (Janke et al., 2001; Wigge and Kilmartin, 2001). When Mtw1p was coupled to protein A and purified, three other proteins copurified (Figure 1A) and were identified by mass spectrometry as Dsn1p, Nnf1p, and Nsl1p. Mtw1p, Dsn1p, and Nsl1p have recently been identified in genetic screens as either dosage suppressors or synthetically lethal with nnf1-17 (Euskirchen, 2002). These genetic interactions are in agreement with our data showing all four proteins in a complex, which we call the Mtw1p complex. Purification of Spc105p coupled to protein A also copurified Ydr532p, a protein of unknown function (Figure 1B), whereas reciprocally, purification of Ydr532p-protein A also copurified Spc105p (Figure 1C). We call this complex the Spc105p complex. The other bands on the gels, which were identified as heat shock proteins or ribosomal elongation factor, are probably contaminants (see Figure 1 legend). We were unable to identify any proteins in the bands at the bottom of the gel in Figure 1, B and C (or in the gels below); thus, it is possible that additional components of either of these two complexes may exist.
Recently, Mtw1p was found to be associated with another complex, the Ctf19p complex (Cheeseman et al., 2002a), which contains 12 proteins. However, Dsn1p, Nnf1p, and Nsl1p were not found to be associated with the Ctf19p complex. To confirm the composition of the Mtw1p complex that we have found (Figure 1A) and to see whether components of the Ctf19p complex could be isolated in association with Mtw1p, we did reciprocal purifications using Nnf1p, Dsn1p, and Nsl1p tagged with protein A (Figure 1, D–F). Gel bands were analyzed by a combination of MALDI and LC-MS/MS mass spectrometry (see MATERIALS AND METHODS). Here, in order to maximize the amounts of protein so as to detect minor components, we used the fraction eluted later from the column, which contained higher amounts of tagged protein, but also a higher proportion of contaminating proteins (bands a–g; see the Figure 1 legend for identifications). The amounts of these contaminating bands were fairly consistent in the different preparations. The isolation of Mtw1p tagged with protein A under these conditions is also included (Figure 1G).
The result of the Nnf1p-protein A isolation was consistent with the results shown in Figure 1A; i.e., only Dsn1p, Mtw1p, and Nsl1p were found (Figure 1D). A different result was found for the Dsn1p-protein A isolation (Figure 1E). The expected Mtw1p, Nnf1p, and Nsl1p bands were found, but in addition there were extra gel bands, which contained the components of the Spc105p (Figure 1, B and C) and Ndc80p complexes. The composition of all three complexes is summarized in Figure 1K. The Nsl1p-protein A isolation showed the other three Mtw1p complex components and also showed Spc105p (Figure 1F). The Mtw1p-protein A isolations always showed Dsn1p, Nnf1p, and Nsl1p as in Figure 1A, but some preparations (Figure 1G) also showed all of the components of the Spc105p and Ndc80p complexes as well. The reason for this variability was not clear, because Dsn1p-protein A isolations always showed all three complexes. These results suggest that the Mtw1p, Spc105p, and Ndc80p complexes are closely associated.
Why are the Spc105p and Ndc80p complexes mainly absent in the Nnf1p-protein A and Nsl1p-protein A isolations? We think this is probably because of small changes in the binding affinities between the three complexes caused by the protein A tag. These are probably irrelevant inside the cell, because there is no apparent phenotype associated with the tagging, but these differences in affinity may become important during the stringent washing conditions associated with the isolation, so that the Spc105p and Ndc80p complexes can be washed away. It also seems that part of the Dsn1p has a lower affinity for the Mtw1p complex and is partly washed off the column, because it appears to be present in substoichiometric amounts in the Nnf1p-, Nsl1p-, and Mtw1p-protein A isolations (Figure 1, D, F, and G) but not apparently in the Dsn1p-protein A isolation (Figure 1E).
These results show that the Spc105p and Ndc80p complexes can be copurified with the Mtw1p complex. Can the Mtw1p complex be copurified with the Spc105p and Ndc80p complexes? Isolation of both the Ndc80p (Figure 1H) and Spc105p (Figure 1I) complexes to maximize the amounts of protein as above did indeed show the presence of very low amounts of Mtw1p in the Ndc80p complex and Ndc80p, Nuf2p, Mtw1p, and Nsl1p in the Spc105p complex. Again, the presence of these extra proteins in the Spc105p complex was variable, as for the Mtw1p complex above. Although no component of the Spc105p complex was detected by mass spectrometry in the Ndc80p-protein A isolation, Spc105p could be detected in immunoblots of immunoprecipitates of Ndc80p-protein A using less stringent washing (Figure 1J).
Taken as a whole, these results identify two new complexes, the Mtw1p and Spc105p complexes, and suggest that there is a close association between these complexes and the Ndc80p complex (Figure 1K).
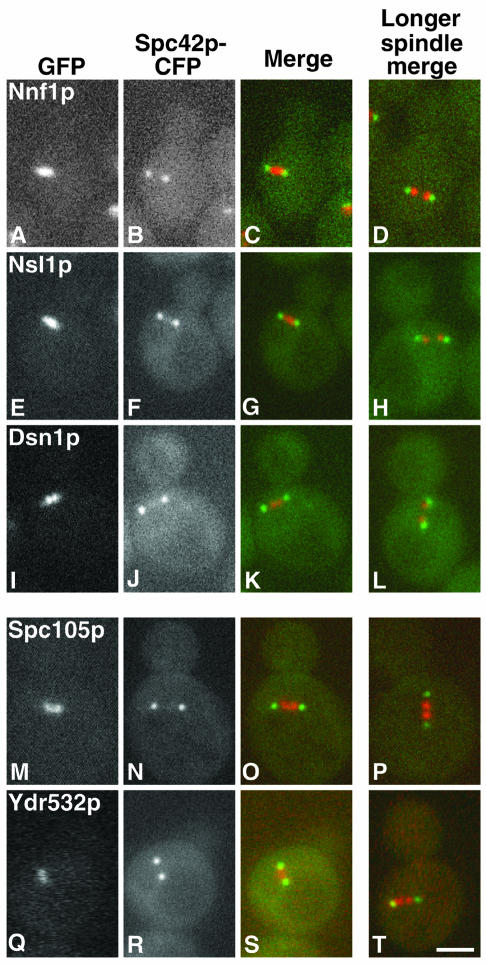
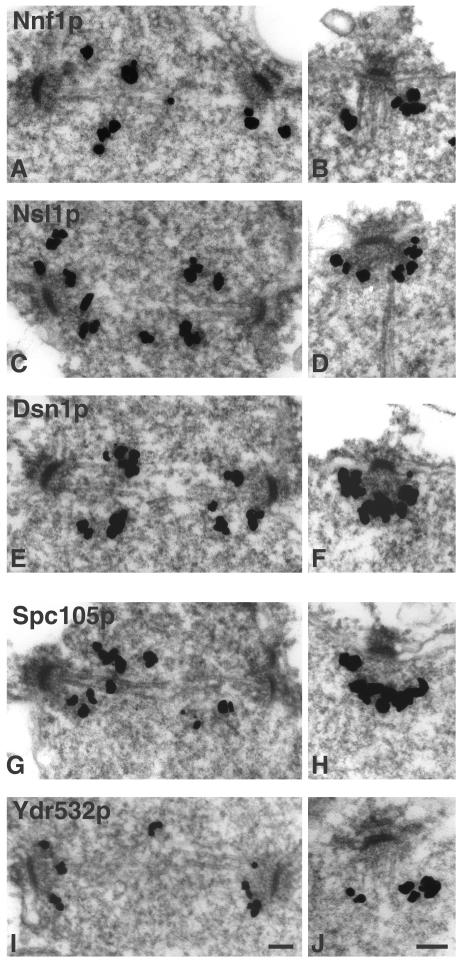
Localization of Nnf1p, Nsl1p, Dsn1p, Spc105p, and Ydr532p
Mtw1p and the components of the Ndc80p complex localize to the centromere (Goshima and Yanagida, 2000; Janke et al., 2001; Wigge and Kilmartin, 2001). This suggests that all of the proteins present in the closely associated Mtw1p and Spc105p complexes should also localize to the centromere. In S. cerevisiae, centromeres are clustered around the nuclear face of the SPB (Goshima and Yanagida, 2000; He et al., 2000; Jin et al., 2000; Tanaka et al., 2000), and indeed HA-tagged Spc105p appeared to localize to the nuclear side of the SPB, although the signal was weak (Wigge et al., 1998). Nnf1p, Nsl1p, and Dsn1p have been localized to the spindle pole region (Euskirchen, 2002), but it was not established whether this was at the SPB or on the spindle close to the nuclear side of the SPB. Thus, we tagged Nnf1p, Nsl1p, Dsn1p, Spc105p, and Ydr532p with GFP to determine their localizations relative to the SPBs both in unfixed cells (Figure 2) and by immuno-EM (Figure 3). All five proteins localized similarly. They were present between the SPBs in short spindles (Figure 2 and left side of Figure 3), and associated with the nuclear face of the SPB in all other cells (right side of Figure 3). This staining pattern is very similar to that found for other centromere components in S. cerevisiae, including Ndc80p (Rout and Kilmartin, 1990), Ndc10p (Goh and Kilmartin, 1993); Ctf19p (Hyland et al., 1999); Mtw1p (Goshima and Yanagida, 2000); Ctf3p, Mcm16p, and Mcm22p (Measday et al., 2002); and Chl4p and Iml3p/Mcm19p (Pot et al., 2003).
Figure 2.
Localization of GFP-tagged proteins in relation to the SPBs (imaged with Spc42p-CFP) in unfixed cells of the indicated strains. The left three columns show the GFP (red), CFP (green) and merged images for a cell of each strain with a short spindle. The right column shows the merged image for a cell with a longer spindle; note the splitting of the GFP image. (A–D) Nnf1p (JK1694), (E–H) Nsl1p (JK1682), (I–L) Dsn1p (JK1683), (M–P) Spc105p (JK1679), and (Q–T) Ydr532p (JK1676). There is a small amount of carryover of the GFP signal into the CFP channel. Bar, 2 μm.
Figure 3.
Immuno-EM of GFP-tagged proteins in the indicated strains. (A and B) Nnf1p (VNY3), (C and D) Nsl1p (VNY50), (E and F) Dsn1p (VNY28), (G and H) Spc105p (MSY52), and (I and J) Ydr532p (JK1495). Staining is seen between SPBs in short spindles (left side) and in association with the nuclear face in all other SPBs (right side). Bar, 0.1 μm.
ChIP Assay
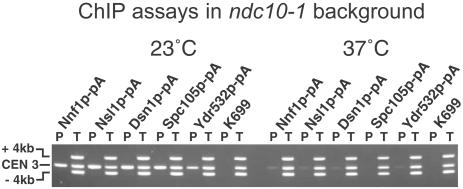
The association of Nnf1p, Nsl1p, Dsn1p, Spc105p, and Ydr532p with the centromere protein Mtw1p and the centromere Ndc80p complex, together with the localization patterns of these proteins, suggest that they are all associated with the centromere. We confirmed this by using the chromatin immunoprecipitation ChIP assay (Meluh and Koshland, 1997). In this assay, PCR is used to detect centromere DNA associated with a tagged protein after formaldehyde cross-linking and sonication to shear the chromosomes and solubilize cross-linked proteins. Additional primers in the same PCR reaction amplify noncentromeric flanking sequences and provide a negative control. Except for the wild-type K699, the strains used carried the ndc10-1 mutation (Goh and Kilmartin, 1993), because the disassembly of the centromere in such a strain at 36°C (Ortiz et al., 1999) is a useful negative control. Nnf1p, Nsl1p, Dsn1p, Spc105p, and Ydr532p all show Ndc10p-dependent association with centromere DNA (Figure 4), indicating that the five proteins are indeed associated with the centromere.
Figure 4.
ChIP assays on protein A-tagged ndc10-1 strains and on the wild-type strain K699 as a negative control. Assays were carried out at 23°C (left), where the centromere is intact in ndc10-1 cells, and at 37°C (right), where the centromere is dissociated. Multiplex PCR was used to detect CEN3 and two regions 4 kb on either side. P is the pellet after enrichment with IgG Sepharose, and T, 1% of the total input, is the starting material. From left to right, the strains used were JK1670, JK1673, JK1667, JK1669, JK1664, and K699.
Localization of S. pombe Homologues of Nsl1p and Spc105p
Because of the relatively large number of 16 chromosomes in S. cerevisiae, the localization pattern of genuine centromere components is not distinguishable from that of microtubule-associated proteins that mainly bind along shorter nuclear microtubules, which are also clustered around the SPB (O'Toole et al., 1999). Moreover, it has not yet been possible to resolve the 16 individual centromeres or kinetochores in S. cerevisiae except in pachytene spreads (Hayashi et al., 1998; Klein et al., 1999; Zeng et al., 1999), where the kinetochores are probably only partly assembled because they lack microtubules (Goetsch and Byers, 1982). In S. pombe, with just three chromosomes, centromere cytology is much clearer, and previously we used this organism to localize homologues of the Ndc80p complex (Wigge and Kilmartin, 2001). Also, comparisons of kinetochores and centromeres of S. cerevisiae and S. pombe are particularly interesting because the two organisms are evolutionarily divergent and there are big differences in centromere DNA content.
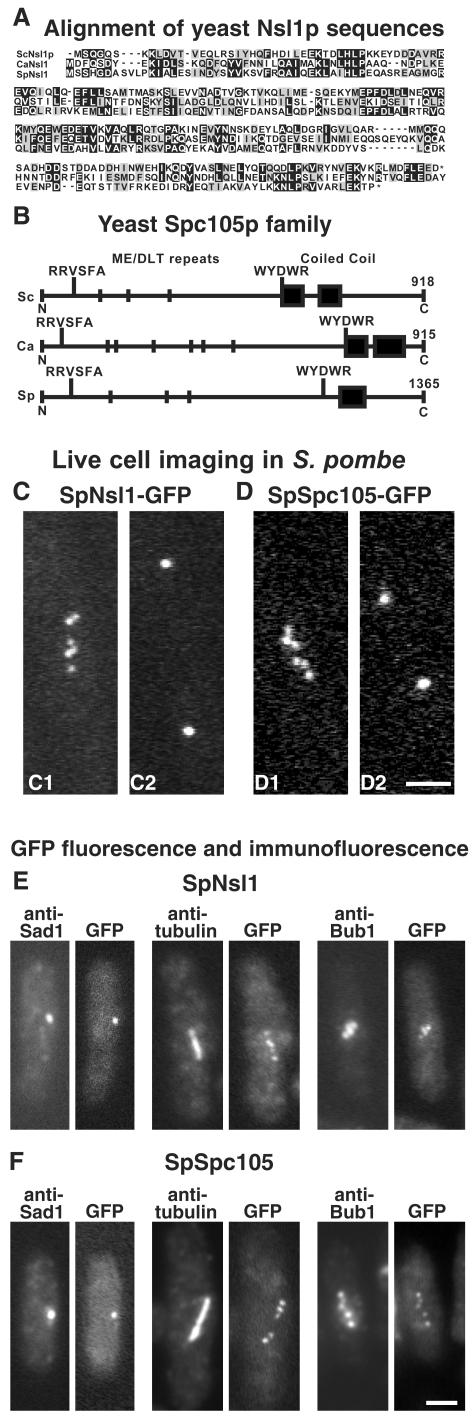
Homologues of both Nsl1p and Spc105p are present in both Candida albicans (http://genolist.pasteur.fr/CandidaDB/, gene names IPF5166 and SPC105) and S. pombe. Blast searches using the S. cerevisiae proteins gave E values of 4 × 10–6 and 3 × 10–14 for the S. pombe homologues of Nsl1p (SPAC688.02c, here named SpNsl1) and Spc105p (SPCC1020.02/SPC7, here named SpSpc105), respectively. An alignment of the three yeast Nsl1p sequences and a comparison of the Spc105p homologues in S. cerevisiae, C. albicans, and S. pombe is shown in Figure 5, A and B. The three Spc105p homologues showed some interesting features. In particular, each has a short region of coiled coil near the C-terminus preceded by the sequence WYD/EWR. In addition, close to the N-terminus, the sequence RRVSFA is followed by repeats of the sequence ME/DLT. At present it is not clear what the functions of these conserved sequences are, but they may be a reliable signature for this protein family. There is also an S. pombe homologue of Nnf1p (SPAC30.08), but we were unable to tag this with GFP.
Figure 5.
(A) Alignment of Nsl1p sequences from S. cerevisiae (Sc), C. albicans (Ca), and S. pombe (Sp); the last nine amino acids of the Candida homologue have been omitted. Black and gray boxes indicate identical and similar residues, respectively. (B) Schematic view of the Spc105p homologues from the same three species. ME/DLT repeats are shown by short vertical lines, and coiled-coil domains found by Paircoil (Berger et al., 1995) are shown by black boxes. (C and D) Localization of GFP-tagged SpNsl1 (C1 and C2) and SpSpc105 (D1 and D2) in live S. pombe cells. Long cells in which the GFP spots had begun to split, indicating entry into mitosis, were followed, and images were recorded when these cells were about to complete anaphase A (C1 and D1). These images show between five and six dots, which then coalesced into two separated dots several minutes later as the cells entered anaphase B (C2 and D2). (E and F) Fluorescence of GFP compared with immunofluorescence with anti-Sad1, antitubulin, and anti-HA-tagged SpBub1 is shown in E for SpNsl1 (strain JK1636) and in F for SpSpc105 (strain JK1627). Bars, 2 μm.
Live S. pombe cells containing GFP-tagged SpNsl1 or SpSpc105 gave similar localization patterns: in mitosis there were up to six separate spots (Figure 5, C1 and D1), which later coalesced into two separated spots, presumably during anaphase B (Figure 5, C2 and D2). This pattern suggests localization to the centromeres. Cells not in mitosis showed a single spot, which in fixed cells (Figure 5, E and F) was coincident with the SPB marker Sad1 (Hagan and Yanagida, 1995). Localization to the centromeres was confirmed by immunofluorescence showing that the staining was positioned along the mitotic spindle and coincided with Bub1 (Figure 5, E and F), a centromere marker (Bernard et al., 1998). Recently, observations on GFP-labeled Bub1 in live cells have questioned its use as a centromere marker because no staining was seen after centromeres had separated (Toyoda et al., 2002). However, our observations on such a strain (our unpublished results) showed multiple dots during mitosis as was observed earlier (Bernard et al., 1998), confirming the usefulness of Bub1 as a centromere marker.
In conclusion, the localization of S. pombe homologues of Nsl1p and Spc105p to the centromeres supports the conclusion that these are indeed centromere proteins in S. cerevisiae.
Phenotype of nsl1-5 Cells
All four genes encoding components of the Mtw1p complex have previously been shown to be essential, and the phenotypes of Ts– mutants in MTW1 and NNF1 have been described (Shan et al., 1997; Goshima and Yanagida, 2000; Euskirchen, 2002). mtw1-1 cells arrest with intermediate-length spindles, with defects in chromosome segregation apparent in about one third of the cells (Goshima and Yanagida, 2000), and nnf1-17 cells arrest with variable-length spindles (Euskirchen, 2002).
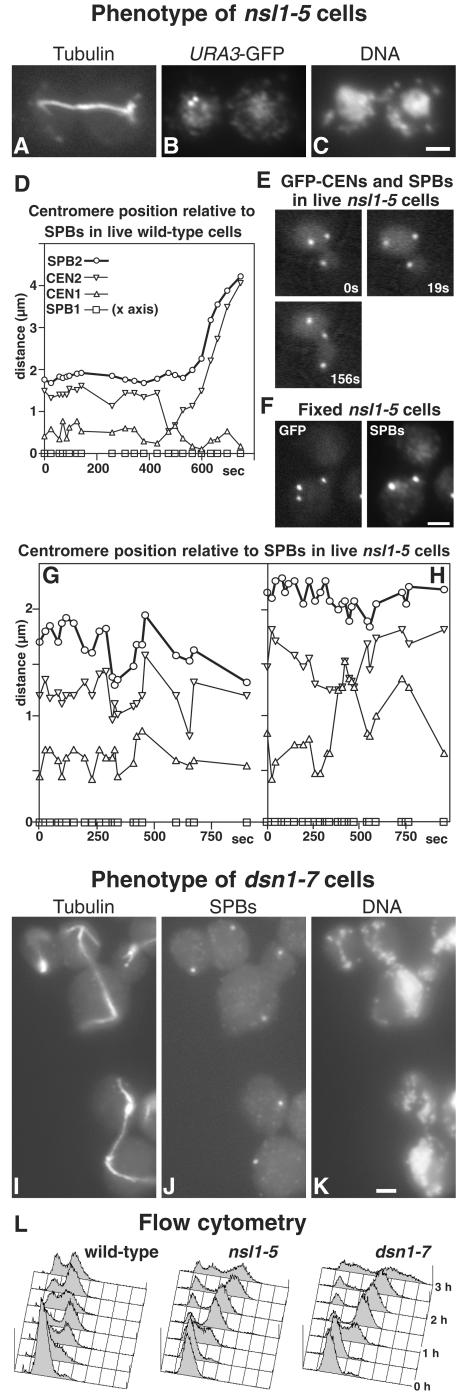
The phenotype of nsl1-5 cells synchronized in G1 and released at 36°C showed some similarities to mtw1-1 and nnf1-17. All cells arrested with either short or intermediate-length spindles and with about a diploid DNA content (Figure 6L); the arrest continued for up to 3 h, during which no defects in chromosome segregation could be seen by DAPI staining (our unpublished results). At later time points (3–4 h), the proportion of cells with spindles decreased, and cells containing apparent single asters with variable DAPI staining increased to 59% at 4 h. These cells may have resulted from spindle breakdown or, to a lesser extent, from the small proportion of cells with long anaphase spindles (5% at 3–3.5 h). These anaphase spindles also showed no clear defect in chromosome segregation by DAPI staining (Figure 6C), but when chromosome V was marked with GFP at the URA3 locus, a clear defect in segregation was seen in 22% of cells with such spindles (Figure 6, A–C). This phenotype is similar to that found in other centromere mutants, such as dam1-11 and duo1-2 mad2Δ (Cheeseman et al., 2001b), mtw1-1 (Goshima and Yanagida, 2000), and ask1-3 mad2Δ (Li et al., 2002).
Figure 6.
Phenotypes of nsl1-5 (A–C and E–H) and dsn1-7 (I–K) mutants, and of a wild-type control (D); flow cytometry for the three strains is also shown (L). Cells were synchronized in G1 with α-factor at 23°C and released at 36°C. (A–C) An nsl1-5 cell in which the URA3 locus is labeled with GFP (strain VNY287) was fixed during anaphase (3 h after release) and processed for immunofluorescence with antitubulin (A), and anti-GFP (B), and stained with DAPI (C). Chromosome V has failed to segregate. (D–H) Imaging of GFP-labeled SPBs and CEN5 in wild-type (D, strain K8572) and nsl1-5 cells (E–H, strain VNY122). (D) A wild-type cell underwent centromere pairing and passed through anaphase during the period of observation. (E) An nsl1-5 cell where the centromeres failed to split. (F) A fixed nsl1-5 cell was processed for immunofluorescence using anti-GFP and anti-Tub4p to identify SPBs. Centromeres (the lowest GFP spot) have not split. (G) One of the majority of nsl1-5 cells, in which the centromeres split but failed to pair during the period of observation. (H) An nsl1-5 cell in which pairing occurred. Symbols in G and H are the same as in D. (I–K) dsn1-7 cells (strain VNY96) were processed for immunofluorescence 2.5 h after release using antitubulin (I), anti-Tub4p for SPBs (J), and DAPI (K). Two anaphase cells that have failed to segregate DNA and one aploid cell (top left) are shown. (L) Flow cytometry of wild-type (K699), nsl1-5 (VNY72), and dsn1-7 (VNY96) cells. Bars, 2 μm.
These defective anaphase cells were a small proportion of the total, so we also looked for a defect at earlier time points, when all the nsl1-5 cells have shorter spindles. Here we observed GFP-labeled centromeres (Goshima and Yanagida, 2000; He et al., 2000; Tanaka et al., 2000) in live cells between 1.5 and 2.5 h after release. Cells were observed in a single focal plane (Pearson et al., 2001), adjusted manually to maintain focus. This has the advantage that data are gathered within 0.2 s but the disadvantage that data can be lost for a period if spindles swivel at an angle to the focal plane. Because of radiation damage, the number of observations was limited to those permitting wild-type cells to pass through anaphase B.
Of seven wild-type cells followed, six passed through anaphase B, and all of these showed separation and transient pairing of centromeres (Figure 6D) before anaphase B (He et al., 2000; Tanaka et al., 2000). In contrast, a clear defect in chromosome segregation was apparent in about one third of nsl1-5 cells (50 cells examined). In these cells, only three GFP spots were observed (Figure 6E), suggesting that the centromeres had failed to split, and this was confirmed by immunofluorescence (Figure 6F). The proportion of such cells was constant through the period after release. In 14 cells whose centromeres did split, transient pairing like that seen in wild-type cells was infrequent (Figure 6G) and was found in only two of the 14 cells (Figure 6H). Although we cannot rule out transient pairing while some of the spots were out of focus or during gaps in the observations, pairing was easy to observe in wild-type cells under similar conditions (Figure 6D). Any inhibition of pairing was not caused by lack of mobility of the centromeres, because they clearly moved relative to the poles during the period of observation (Figure 6G).
We repeated these observations in mtw1-11 (strain VNY119) and obtained similar results. Again, one third of cells failed to split their centromeres, and possible pairing was observed in only one cell out of 13 examined (our unpublished results). This result is in agreement with observations on asynchronous mtw1-1 cells after 3 h of arrest, where just under half the cells contained unsplit centromeres (Goshima and Yanagida, 2000).
In conclusion, these results show that nsl1-5 cells, like mtw1 mutant cells, undergo a prolonged arrest in mitosis and show several defects in chromosome segregation. These include a failure of spindle attachment in some anaphase cells, a failure of centromeres to split in some cells, and a much lowered frequency of centromere pairing.
Phenotype of dsn1-7 Cells
The phenotype of dsn1-7 was initially quite similar to those of mutants in the other Mtw1p complex components: all the cells initially arrested with short or intermediate-length spindles and a mainly diploid DNA content (Figure 6L and our unpublished results). However, after ∼40 min of arrest, clear defects in chromosome segregation were seen by DAPI staining. Spindles in ∼25% of the cells had elongated sufficiently to show that 96% of these had segregated one SPB that was associated with little DNA staining (Figure 6, I-K). There also appeared to be some spindle instability, because nearly half of the intermediate-length spindles had reduced or very little antitubulin staining in the central region. It was difficult to see segregation defects in most of these spindles because the large area of DAPI staining overlapped the spindle (our unpublished results). At 3 h after release from α-factor, half of the cells or buds with single microtubule asters (82% of the total cells) were aploid or contained little DNA (top left cell in Figure 6, I-K). Such aploid cells or buds probably arise in two ways. First, the elongated anaphase spindles that fail to segregate DNA would result in one aploid and one diploid cell. Second, in the case of the unstable intermediate-length spindles, if one of the SPBs was not attached to chromosomes, then it might be segregated by cytoplasmic microtubules as in ndc1-1 (Winey et al., 1993). The failure of chromosomes to attach to one pole in the dsn1-7 mutant is very similar to the phenotype found with Ts– mutants in some other centromere components: ndc10-1 (Goh and Kilmartin, 1993), ndc80-1 (Wigge et al., 1998), and spc24 and spc25 (Janke et al., 2001; Wigge and Kilmartin, 2001).
Activation of the Mad2p-dependent Checkpoint in nsl1-5 and dsn1-7 Mutants
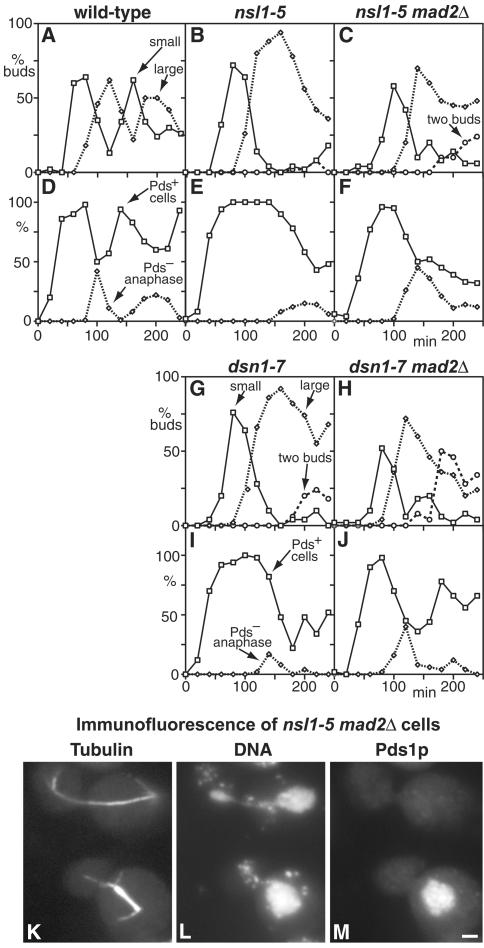
During mitosis, the transition to anaphase is regulated by the Mad2p-dependent checkpoint (Li and Murray, 1991; Wang and Burke, 1995). Defects in mitosis before anaphase can activate this checkpoint and arrest mitosis. Both the nsl1-5 and dsn1-7 mutants showed a delay in mitosis, suggesting Mad2p activation. To test for this, we prepared ts strains containing deletions of MAD2 and expressing myc-tagged Pds1p, which is a convenient marker for metaphase because Pds1p is completely degraded during the metaphase to anaphase transition (Cohen-Fix et al., 1996) by the anaphase-promoting complex. The cells were synchronized, and the budding indices were measured together with the proportions of Pds1p-positive cells and Pds1p-negative anaphase cells containing long spindles. In wild-type cells, the proportion of Pds1p-positive cells corresponds almost exactly to the proportion of short and intermediate-length spindles. This also held true for the two mutants for up to 2–2.5 h of the block. After this there was an increasing proportion of Pds1p-positive cells containing apparent single asters, probably due to spindle breakdown.
As shown in Figure 7, A and D, wild-type cells showed a rapid increase in Pds1p-positive cells as spindles were formed, followed by a decrease between 80 and 100 min, as cells entered anaphase (Yamamoto et al., 1996). In contrast, nsl1-5 cells showed a long delay in Pds1p breakdown, which did not start until around 160 min, followed by the appearance of some Pds1p-negative cells containing anaphase spindles (Figure 7, B and E). This delay in Pds1p breakdown was almost completely abolished when MAD2 was deleted, and there was a concomitant earlier appearance of Pds1p-negative cells containing anaphase spindles (Figure 7, C and F). Interestingly, these spindles, in contrast to those observed in the MAD2+ cells, did show abnormalities in chromosome segregation, with unequal DAPI staining at the two poles in 82% of the spindles examined (Figure 7, K–M). The nsl1-5 mad2Δ cells were also examined by flow cytometry, but no clear conclusions could be made because variations in the amount of DNA per cell caused the signal to smear (our unpublished results). The shorter mitotic delay shown by dsn1-7 cells (Figure 7, G and I) was also Mad2p-dependent, because in the mad2Δ cells the decline in Pds1p staining started at about the same time as in wild-type cells (Figure 7, H and J). These results show that most of the delay in mitosis shown by nsl1-5 and dsn1-7 cells is Mad2p dependent.
Figure 7.
The arrest of nsl1-5 and dsn1-7 mutants during mitosis is Mad2p-dependent. (A–J) Wild-type (A and D; K6445), nsl1-5 (B and E; VNY130), nsl1-5 mad2Δ (C and F; VNY318), dsn1-7 (G and I; VNY116), and dsn1-7 mad2Δ (H and J; VNY283) cells were synchronized in G1 with α-factor and released at 36°C. The percentage of cells with small buds, large buds, or two buds was plotted against time (A–C, G, and H), as were the percentages of Pds1p-positive cells and Pds1p-negative cells containing anaphase spindles as determined by immunofluorescence (D–F, I and J). (K–M) Immunofluorescence of nsl1-5 mad2Δ cells stained with antitubulin (K), DAPI (L), and antimyc for Pds1p (M). Pds1p-negative cells with anaphase spindles show unequal DNA segregation. Bar, 2 μm.
Phenotypes of spc105 and ydr532c Mutants
To study the function of the Spc105p complex, we prepared two ts mutants, spc105-4 and spc105-15. When we observed the phenotypes of these in synchronized cells, as in Figure 6, we could see no difference between the mutants and wild-type cells for up to 4 h at 36°C. To find out what was happening, we examined the phenotype of the deletion (spc105Δ). Our previous study (Wigge et al., 1998) had suggested that SPC105 is probably an essential gene, because sporulation of a heterozygous diploid yielded inviable spc105Δ spores. However, spc105Δ cells can survive, because deletion cells rescued by SPC105 on a URA plasmid (strain PWY199) will grow very slowly on fluoroorotic acid plates, on which the URA plasmid is lost. However, backcrossing these cells showed they had accumulated additional mutations, so they were not studied further (Wigge et al., 1998). In view of the phenotypes of the ts spc105 alleles, we reexamined the growth rates of spc105Δ spores. We observed that all four spores from the heterozygous diploid PWY92 grew at similar rates for up to 14 h at 23°C, when there were between 4 and 8 cells per spore clone (cells were teased apart with a needle for counting). At 17 h, some of the spore clones started to grow more slowly, and by 26 h, all of the spc105Δ clones had arrested with between 9 and 28 cells (the average was 12 ± 6). The finding that spc105Δ spores grow normally for up to three divisions may explain why we saw no phenotype in the Ts– mutants after 4 h at 36°C.
ydr532cΔ spores (from strain MSY104) had a similar phenotype but a larger microcolony size. Mutant spore colonies were indistinguishable from wild-type for up to 26 h of growth at 23°C; growth then slowed, with cells eventually arresting at a microcolony size of between 42 and 114 cells (the average was 71 ± 21). The phenotypes of spc105-4 and spc105-15 were very similar. Growth arrest at 37°C occurred after the formation of microcolonies that were more variable in size, containing 7–174 cells, together with some larger colonies presumably due to reversion. Given the evidence that Spc105p and Ydr532p are centromere proteins, the most likely reason for the eventual growth arrest is cumulative chromosome loss.
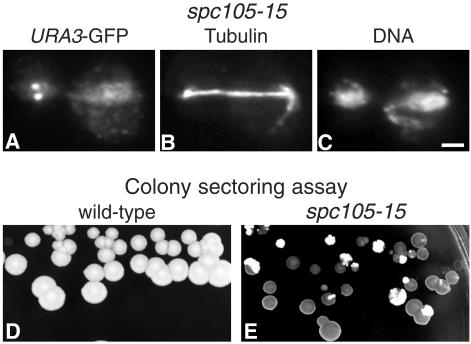
We tested for chromosome loss in spc105 mutants with two methods: first, observing the behavior of GFP-labeled chromosomes (Straight et al., 1996) in synchronized cells at 36°C and second, using a chromosome loss plate assay (Spencer et al., 1990). We used spc105-15 rather than spc105-4 for these experiments because it showed a greater effect. Failures in chromosome segregation (Figure 8A) were observed in 9% of spc105-15 cells (this varied between 5 and 14% in different experiments), compared with 0.7% (1/150) in wild-type cells. We confirmed this phenotype using the chromosome-loss plate assay, in which cells carry a mini-chromosome CFIII containing SUP11, which suppresses ade2-101. If CFIII is lost, the ade2-101 mutation causes accumulation of a red pigment, and colonies are red or sectored red, whereas cells carrying CFIII remain white. At 30°C, spc105-15 cells, in contrast to wild-type cells, showed mainly red or red-sectored colonies (Figure 8, D and E), confirming that Spc105p has a function in chromosome segregation.
Figure 8.
spc105-15 cells are defective in chromosome segregation. (A–C) spc105-15 cells in which the URA3 locus is labeled with GFP (strain VNY203) were synchronized in G1 with α-factor, released at 36°C for 2 h, fixed, and stained with anti-GFP (A), antitubulin (B), and DAPI for DNA (C). (D and E) Colony-sectoring assay for chromosome loss in wild-type (D; strain K7049) and spc105-15 (E; strain VNY198) cells carrying the CFIII mini-chromosome after incubation on low adenine YEPD for 3 d at 30°C. The red colonies (dark in the figure) and sectors show chromosome loss. Bar, 2 μm.
Genetic Interactions within and between the Mtw1p, Spc105p, Ndc80p, and Dam1p/DDD/DASH Complexes
We looked for in vivo evidence for interactions both within and between the Mtw1p and Spc105p complexes that we have characterized here, the Ndc80p complex (Janke et al., 2001; Wigge and Kilmartin, 2001), and the Dam1p/DDD/DASH complex, which has been localized to the spindle and to centromeres by ChIP assay (Cheeseman et al., 2001a; Janke et al., 2002; Li et al., 2002). We restricted ourselves to these four complexes because of the evidence that three of them are closely associated, and because subunits of all four of them have been identified in our enriched spindle pole preparation (Wigge et al., 1998; Wigge and Kilmartin, 2001), suggesting that some of them might be in close proximity.
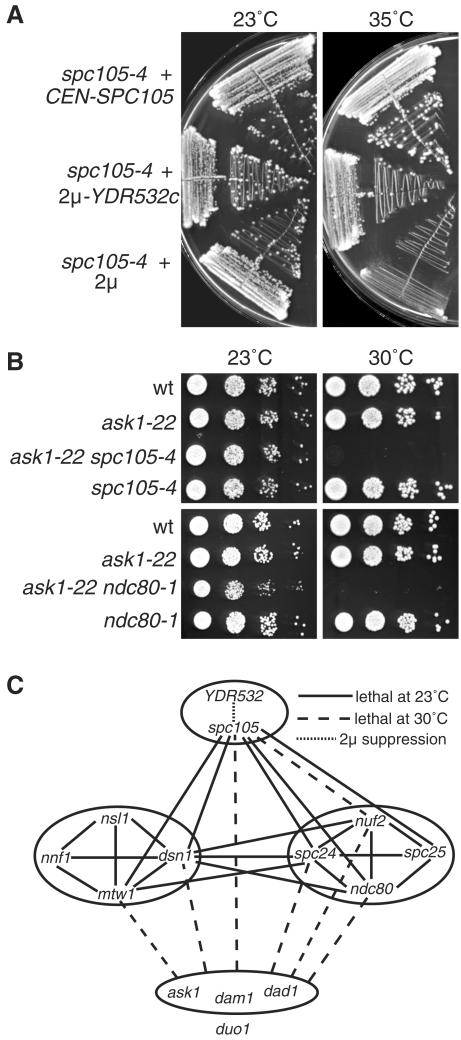
We found a genetic interaction within the Spc105p complex in that overexpression of YDR532c on a 2μ plasmid could suppress both spc105-4 (Figure 9A) and spc105-15 (our unpublished results). We also looked for genetic interactions between ts mutations of components in the four complexes. Examples of some of the interactions found are shown in Figure 9B, and all of the interactions are summarized in Figure 9C. Synthetic-lethal interactions were found between mutations affecting several subunits of the Mtw1p, Ndc80p, and Spc105p complexes (Figure 9C), in agreement with their close association (Figure 1). However, only synthetic growth defects were found between mutations affecting subunits of these three complexes and those affecting three of the subunits of the Dam1p/DDD/DASH complex (Figure 9, B and C). No interactions were found with duo1-61; thus, this allele is excluded from the dam1, ask1, and dad1 group in Figure 9C. These genetic results support the close association between the Mtw1p, Ndc80p, and Spc105p complexes, as shown in Figure 1, and suggest a less close relationship between these three complexes and the Dam1p/DDD/DASH complex. This is in agreement with the clear but weaker interactions recently found between components of the Ndc80p and Dam1p/DDD/DASH complexes (Shang et al., 2003).
Figure 9.
Genetic interactions within and between the Spc105p, Mtw1p, Ndc80p, and Dam1p/DDD/DASH complexes. (A) Suppression of spc105-4 by overexpression of Ydr532p. Strains were MSY150, MSY151, and MSY152. (B) Examples of the synthetic growth defects at 30°C between mutations in the different complexes. (C) A summary of the interactions found. Solid lines show synthetic-lethal interactions at 23°C, dashed lines show synthetic growth defects with lethality at 30°C (see panel B), the dotted line shows dosage suppression (see panel A), and the absence of a line indicates that no genetic interaction was found. All possible combinations between the different mutations were tested; however, the YDR532c dosage suppression was only tested in the spc105 alleles. The synthetic-lethal interactions within the Ndc80p and Mtw1p complexes have been described previously (Wigge and Kilmartin, 2001; Euskirchen, 2002). The ask1, dam1, and dad1 alleles are not distinguished because they gave similar results in all the crosses tested, with the exception of the dam1-31 mtw1-11 cross, in which no interaction was found. duo1 is excluded from the ask1, dad1, and dam1 group because duo1-61 showed no genetic interactions with any of the other alleles. Strains used were JK1121, JK1504, MSY72, MSY79, MSY87, MSY90, PWY483, PWY754, VNY69, VNY87, VNY96, and VNY162.
DISCUSSION
In this article, we describe five components of the S. cerevisiae centromere and show that the homologues of two of these are probably components of the S. pombe centromere. This brings the number of proteins currently identified for the S. cerevisiae centromere to >40 (Figure 10), excluding checkpoint proteins, kinesins, cohesions, and specialized kinases. The picture that emerges is one of increasing complexity for what is probably one of the simplest eukaryote centromeres.
At present, we do not know the precise functions of the centromere components we have identified in this article. However, they have a clear role in chromosome segregation, and Nsl1p may have a role in transient centromere pairing. Our protein A isolations, particularly the Dsn1p-protein A isolation (Figure 1E), suggest that the Mtw1p, Spc105p, and Ndc80p complexes are closely associated. Such an association is supported by the synthetic-lethal interactions found between ts mutations of components of the three complexes (Figure 9). Additional evidence for these associations comes from two-hybrid results, suggesting that the coiled-coil regions of Nnf1p can interact with the coiled-coil regions of Ndc80p, Nuf2p, and Spc105p (Newman et al., 2000).
What is the relationship between these complexes and the other S. cerevisiae centromere components and complexes already identified? We propose a tentative arrangement of S. cerevisiae centromere components into four different groups (Figure 10). The first group contains components that associate laterally with microtubules, as shown by direct binding or immunofluorescence, and in addition associate with the centromere as shown by ChIP assay, indicating that they can be close to the kinetochore. The second group is associated with the centromere but has shown no evidence, so far, of lateral binding to microtubules. These proteins may bind directly or indirectly to the plus ends of microtubules, but this has yet to be shown. The third group has properties very similar to the second group and is only distinguished from it by the fact that none of the proteins in the third group, apart from Mtw1p, has been detected in highly enriched spindle pole preparations (Wigge et al., 1998; Wigge and Kilmartin, 2001). Because these preparations have been digested with DNaseI, this suggests that this third group might be more closely associated with the fourth group, which contains centromere DNA-binding proteins as measured by band-shift assays. Consistent with this hypothesis is evidence for interactions between Ctf19p and both Ndc10p (Ortiz et al., 1999) and Cse1p (Chen et al., 2000).
There is one apparent discrepancy in these groupings, which is the presence of Mtw1p both in group 2, in the Mtw1p complex, and in group 3, in the Ctf19p complex (Cheeseman et al., 2002a). We are confident of our assignment of Mtw1p to group 2 based on the biochemical, genetic, and two-hybrid results summarized above. Moreover, it should be noted that the close association between the Ndc80p, Mtw1p, and Spc105p complexes has been shown using two quite different isolation methods, first by the protein A method used here, and second by the presence of components of the three complexes in the enriched spindle pole preparations (Wigge et al., 1998; Wigge and Kilmartin, 2001). In neither of these preparations have components of the Ctf19p complex other than Mtw1p been detected. One possibility might be that Mtw1p is a bridge between two parts of the centromere (groups 2 and 3 in Figure 10), and depending on the precise conditions of lysis and extraction, either group 2 or group 3 complexes are isolated.
An obvious question with regard to the second group of centromere components, which are presumably associated with the microtubules in the enriched spindle pole preparation, is whether they may be localized to the plus end of the microtubule. The presence of at least one of the group 2 components, Ndc80p, in the spindle pole preparation is microtubule dependent, because we previously showed that removal of the microtubules by DEAE-dextran extraction, which leaves the SPB intact, also removes Ndc80p (Rout and Kilmartin, 1990). Tomographic reconstructions of the S. cerevisiae spindle (O'Toole et al., 1999) show flared ends on most of the discontinuous microtubules, which probably correspond to the plus ends of the kinetochore microtubules. The flared ends could expose the inside of the microtubule, which would in turn expose a unique set of binding sites on tubulin at the plus end of the microtubule. It will be interesting to determine whether some of the group 2 complexes bind to these or other sites on the microtubule, and what other complexes, if any, are necessary for this.
Many of the kinetochore/centromere components listed in Figure 10 are conserved in S. pombe, an evolutionarily distant organism with a structurally different centromere. This suggests a fundamental conservation of at least part of the centromere structure because early in evolution. Does this conservation extend among all eukaryotes? This is still an open question. Vertebrate homologues have been identified for a few of the proteins listed in Figure 10. These now include homologues of three group 2 components, Ndc80p, Nuf2p, and Mtw1p (Chen et al., 1997; Nabetani et al., 2001; Wigge and Kilmartin, 2001; Goshima et al., 2003). RNA interference or antibody depletion experiments have shown a clear role at the centromere for these three homologues (DeLuca et al., 2002; Martin-Lluesma et al., 2002; Goshima et al., 2003; Hori et al., 2003; McCleland et al., 2003), and the Ndc80p and Nuf2p homologues were shown to be associated with each other, suggesting that the Ndc80p complex itself is conserved (Hori et al., 2003; McCleland et al., 2003). These results raise the possibility that all of the group 2 complexes and their interactions, as described in this article, may be conserved in vertebrate centromeres. The main barrier to finding homologues of the other complex components is their high sequence divergence, preventing identification of even S. pombe homologues. However, this barrier may now be overcome by sequence data from other Saccharomyces species (Cliften et al., 2003; Kellis et al., 2003) and fungi, which can establish the conserved residues and allow identification of homologues. An alternative approach is isolation of the human complex, once one human homologue has been identified and tagged with protein A. This has already been done for several other conserved human complexes (Gavin et al., 2002) and has identified the divergent human subunits. In this way, relationships between novel human centromere components can be explored, similar to the way protein-A tagging has been used to establish some of the relationships within the yeast centromere.
Acknowledgments
We are grateful to Farida Begum for assistance with the mass spectrometry, Phil Wigge for the spc105 ts plasmids, Ross Overman for technical help in strain and plasmid construction, the late Douglas Kershaw for cutting the serial thin sections, and Attila Toth for help and advice. Thanks are also due to M.S. Robinson for discussion and to our editor, John Pringle, for editorial help. V.S.N. is supported by the Darwin Trust of Edinburgh.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–06–0419. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-06-0419.
Abbreviations used: SPB, spindle pole body; Ts– or ts, temperature-sensitive; YEPD, yeast extract peptone dextrose; MALDI, matrix-assisted laser desorption/ionization; LC-MS/MS, liquid chromatography mass spectrometry/mass spectrometry; ChIP, chromatin immunoprecipitation; GFP, green fluorescent protein; HA, hemagglutinin; pA, protein A.
References
- Adams, A.E.M., and Pringle, J.R. (1984). Relationship of actin and tubulin distribution to bud growth in wild type and morphogenetic-mutant Saccharomyces cerevisiae. J. Cell Biol. 98, 934–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, I.R., and Kilmartin, J.V. (1999). Localization of core spindle pole body (SPB) components during SPB duplication in Saccharomyces cerevisiae. J. Cell Biol. 145, 809–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aebersold, R., and Mann, M. (2003). Mass spectrometry-based proteomics. Nature 422, 198–207. [DOI] [PubMed] [Google Scholar]
- Bähler, J., Wu, J.-Q., Longtine, M.S., Shah, N.G., McKenzie, A., III, Steever, A.B., Wach, A., Philippsen, P., and Pringle, J.R. (1998). Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943–951. [DOI] [PubMed] [Google Scholar]
- Berger, B., Wilson, D.B., Wolf, E., Tonchev, T., Milla, M., and Kim, P.S. (1995). Predicting coiled coils by use of pairwise residue correlations. Proc. Natl. Acad. Sci. USA 92, 8259–8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard, P., Hardwick, K., and Javerzat, J.P. (1998). Fission yeast Bub1 is a mitotic centromere protein essential for the spindle checkpoint and the preservation of correct ploidy through mitosis. J. Cell Biol. 143, 1775–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins, S., and Walczak, C.E. (2003). Captivating capture: how microtubules attach to kinetochores. Curr. Biol. 13, R449–R460. [DOI] [PubMed] [Google Scholar]
- Boeke, J.D., Trueheart, J., Natsoulis, G., and Fink, G.R. (1987). 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154, 164–175. [DOI] [PubMed] [Google Scholar]
- Bridge, A.J., Morphew, M., Bartlett, R., and Hagan, I.M. (1998). The fission yeast SPB component Cut12 links bipolar spindle formation to mitotic control. Genes Dev. 12, 927–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, M.J., and Davis, R.W. (1989). Purification of a yeast centromere-binding protein that is able to distinguish single base-pair mutations in its recognition site. Mol. Cell. Biol. 9, 2544–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman, I.M., Anderson, S., Jwa, M., Green, E.M., Kang, J., Yates, J.R., 3rd, Chan, C.S., Drubin, D.G., and Barnes, G. (2002a). Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell 111, 163–172. [DOI] [PubMed] [Google Scholar]
- Cheeseman, I.M. et al. (2001a). Implication of a novel multiprotein Dam1p complex in outer kinetochore function. J. Cell Biol. 155, 1137–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman, I.M., Drubin, D.G., and Barnes, G. (2002b). Simple centromere, complex kinetochore: linking spindle microtubules and centromeric DNA in budding yeast. J. Cell Biol. 157, 199–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman, I.M., Enquist-Newman, M., Muller-Reichert, T., Drubin, D.G., and Barnes, G. (2001b). Mitotic spindle integrity and kinetochore function linked by the Duo1p/Dam1p complex. J. Cell Biol. 152, 197–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., Baker, R.E., Keith, K.C., Harris, K., Stoler, S., and Fitzgerald-Hayes, M. (2000). The N terminus of the centromere H3-like protein Cse4p performs an essential function distinct from that of the histone fold domain. Mol. Cell Biol. 20, 7037–7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., Riley, D.J., Chen, P.-L., and Lee, W.-H. (1997). HEC, a novel nuclear protein rich in leucine heptad repeats specifically involved in mitosis. Mol. Cell Biol. 17, 6049–6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson, T.W., Sikorski, R.S., Dante, M., Shero, J.H., and Hieter, P. (1992). Multifunctional yeast high-copy-number shuttle vectors. Gene 110, 119–122. [DOI] [PubMed] [Google Scholar]
- Cleveland, D.W., Mao, Y., and Sullivan, K.F. (2003). Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112, 407–421. [DOI] [PubMed] [Google Scholar]
- Cliften, P., Sudarsanam, P., Desikan, A., Fulton, L., Fulton, B., Majors, J., Waterston, R., Cohen, B.A., and Johnston, M. (2003). Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science 301, 71–76. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix, O., Peters, J.M., Kirschner, M.W., and Koshland, D. (1996). Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 10, 3081–3093. [DOI] [PubMed] [Google Scholar]
- DeLuca, J.G., Moree, B., Hickey, J.M., Kilmartin, J.V., and Salmon, E.D. (2002). hNuf2 inhibition blocks stable kinetochore-microtubule attachment and induces mitotic cell death in HeLa cells. J. Cell Biol. 159, 549–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, R., McDonald, K.L., and McIntosh, J.R. (1993). Three-dimensional reconstruction and analysis of mitotic spindles from the yeast, Schizosaccharomyces pombe. J. Cell Biol. 120, 141–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enquist-Newman, M., Cheeseman, I.M., Van Goor, D., Drubin, D.G., Meluh, P.B., and Barnes, G. (2001). Dad1p, third component of the Duo1p/Dam1p complex involved in kinetochore function and mitotic spindle integrity. Mol. Biol. Cell 12, 2601–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euskirchen, G.M. (2002). Nnf1p, Dsn1p, Mtw1p, and Nsl1p: a new group of proteins important for chromosome segregation in Saccharomyces cerevisiae. Eukaryotic Cell 1, 229–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan, G.I., Lewis, G.K., Ramsey, G., and Bishop, J.M. (1985). Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol. Cell Biol. 5, 3610–3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field, J., Nikawa, J., Broek, D., MacDonald, B., Rodgers, L., Wilson, I.A., Lerner, R.A., and Wigler, M. (1988). Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol. Cell Biol. 8, 2159–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki, H., Hagan, I., Uzawa, S., and Yanagida, M. (1993). Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J. Cell Biol. 121, 961–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin, A.C. et al. (2002). Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141–147. [DOI] [PubMed] [Google Scholar]
- Goetsch, L., and Byers, B. (1982). Meiotic cytology of Saccharomyces cerevisiae in protoplast lysates. Mol. Gen. Genet. 187, 54–60. [DOI] [PubMed] [Google Scholar]
- Goh, P.-Y., and Kilmartin, J.V. (1993). NDC10: a gene involved in chromosome segregation in Saccharomyces cerevisiae. J. Cell Biol. 121, 503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima, G., Kiyomitsu, T., Yoda, K., and Yanagida, M. (2003). Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J. Cell Biol. 160, 25–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima, G., and Yanagida, M. (2000). Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell 100, 619–633. [DOI] [PubMed] [Google Scholar]
- Grandi, P., Doye, V., and Hurt, E.C. (1993). Purification of NSP1 reveals complex formation with `GLFG'nucleoporins and a novel nuclear pore protein NIC96. EMBO J. 12, 3061–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase, S.B., and Lew, D.J. (1997). Flow cytometric analysis of DNA content in budding yeast. Methods Enzymol. 283, 322–332. [DOI] [PubMed] [Google Scholar]
- Hagan, I., and Yanagida, M. (1995). The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J. Cell Biol. 129, 1033–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailey, D.W., Davis, T.N., and Muller, E.G. (2002). Fluorescence resonance energy transfer using color variants of green fluorescent protein. Methods Enzymol. 351, 34–49. [DOI] [PubMed] [Google Scholar]
- Hayashi, A., Ogawa, H., Kohno, K., Gasser, S.M., and Hiraoka, Y. (1998). Meiotic behaviours of chromosomes and microtubules in budding yeast: relocalization of centromeres and telomeres during meiotic prophase. Genes Cells 3, 587–601. [DOI] [PubMed] [Google Scholar]
- He, X., Asthana, S., and Sorger, P.K. (2000). Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell 101, 763–775. [DOI] [PubMed] [Google Scholar]
- He, X., Rines, D.R., Espelin, C.W., and Sorger, P.K. (2001). Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell 106, 195–206. [DOI] [PubMed] [Google Scholar]
- Heath, I.B. (1980). Behavior of kinetochores during mitosis in the fungus Saprolegnia ferax. J. Cell Biol. 84, 531–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori, T., Haraguchi, T., Hiraoka, Y., Kimura, H., and Fukagawa, T. (2003). Dynamic behavior of Nuf2-Hec1 complex that localizes to the centrosome and centromere and is essential for mitotic progression in vertebrate cells. J. Cell Sci. 116, 3347–3362. [DOI] [PubMed] [Google Scholar]
- Hyland, K.W., Kingsbury, J., Koshland, D., and Hieter, P. (1999). Ctf19p: a novel kinetochore protein in Saccharomyces cerevisiae and a potential link between the kinetochore and the mitotic spindle. J. Cell Biol. 145, 15–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke, C., Ortiz, J., Lechner, J., Shevchenko, A., Magiera, M.M., Schramm, C., and Schiebel, E. (2001). The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. EMBO J. 20, 777–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke, C., Ortiz, J., Tanaka, T.U., Lechner, J., and Schiebel, E. (2002). Four new subunits of the Dam1-Duo1 complex reveal novel functions in sister kinetochore biorientation. EMBO J. 21, 181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, W.D., and Philippsen, P. (1989). Purification of a protein binding to the CDEI subregion of Saccharomyces cerevisiae centromere DNA. Mol. Cell. Biol. 9, 5585–5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Q., Fuchs, J., and Loidl, J. (2000). Centromere clustering is a major determinant of yeast interphase nuclear organization. J. Cell Sci. 113, 1903–1912. [DOI] [PubMed] [Google Scholar]
- Kellis, M., Patterson, N., Endrizzi, M., Birren, B., and Lander, E.S. (2003). Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423, 241–254. [DOI] [PubMed] [Google Scholar]
- Kilmartin, J.V., and Adams, A.E.M. (1984). Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J. Cell Biol. 98, 922–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin, J.V., Wright, B., and Milstein, C. (1982). Rat monoclonal antitubulin antibodies derived by using a new nonsecreting rat cell line. J. Cell Biol. 93, 576–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa, K., and Hieter, P. (2001). Evolutionary conservation between budding yeast and human kinetochores. Nat. Rev. Mol. Cell Biol. 2, 678–687. [DOI] [PubMed] [Google Scholar]
- Klein, F., Mahr, P., Galova, M., Buonomo, S.B., Michaelis, C., Nairz, K., and Nasmyth, K. (1999). A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 98, 91–103. [DOI] [PubMed] [Google Scholar]
- Knop, M., and Schiebel, E. (1997). Spc98p and Spc97p of the yeast γ-tubulin complex mediate binding to the spindle pole body via their interaction with Spc110p. EMBO J. 16, 6985–6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner, J., and Carbon, J. (1991). A 240kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell 64, 717–725. [DOI] [PubMed] [Google Scholar]
- Li, R., and Murray, A.W. (1991). Feedback control of mitosis in budding yeast. Cell 66, 519–531. [DOI] [PubMed] [Google Scholar]
- Li, Y., Bachant, J., Alcasabas, A.A., Wang, Y., Qin, J., and Elledge, S.J. (2002). The mitotic spindle is required for loading of the DASH complex onto the kinetochore. Genes Dev. 16, 183–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, H., de Carvalho, P., Kho, D., Tai, C.Y., Pierre, P., Fink, G.R., and Pellman, D. (2001). Polyploids require Bik1 for kinetochore-microtubule attachment. J. Cell Biol. 155, 1173–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Lluesma, S., Stucke, V.M., and Nigg, E.A. (2002). Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science 297, 2267–2270. [DOI] [PubMed] [Google Scholar]
- McCleland, M.L., Gardner, R.D., Kallio, M.J., Daum, J.R., Gorbsky, G.J., Burke, D.J., and Stukenberg, P.T. (2003). The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes Dev. 17, 101–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald, K.L., O'Toole, E.T., Mastronarde, D.N., and McIntosh, J.R. (1992). Kinetochore microtubules in PTK cells. J. Cell Biol. 118, 369–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Measday, V., Hailey, D.W., Pot, I., Givan, S.A., Hyland, K.M., Cagney, G., Fields, S., Davis, T.N., and Hieter, P. (2002). Ctf3p, the Mis6 budding yeast homolog, interacts with Mcm22p and Mcm16p at the yeast outer kinetochore. Genes Dev. 16, 101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh, P.B., and Koshland, D. (1995). Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol. Biol. Cell 6, 793–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh, P.B., and Koshland, D. (1997). Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes Dev. 11, 3401–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh, P.B., Yang, P., Glowczewski, L., Koshland, D., and Smith, M.M. (1998). Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell 94, 607–613. [DOI] [PubMed] [Google Scholar]
- Michaelis, C., Ciosk, R., and Nasmyth, K. (1997). Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91, 35–45. [DOI] [PubMed] [Google Scholar]
- Moreno, S., Klar, A., and Nurse, P. (1991). Molecular biology of the fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795–823. [DOI] [PubMed] [Google Scholar]
- Muhlrad, D., Hunter, R., and Parker, R. (1992). A rapid method for localized mutagenesis of yeast genes. Yeast 8, 79–82. [DOI] [PubMed] [Google Scholar]
- Nabetani, A., Koujin, T., Tsutsumi, C., Haraguchi, T., and Hiraoka, Y. (2001). A conserved protein, Nuf2, is implicated in connecting the centromere to the spindle during chromosome segregation: a link between the kinetochore function and the spindle checkpoint. Chromosoma 110, 322–334. [DOI] [PubMed] [Google Scholar]
- Nasmyth, K., Adolf, G., Lydall, D., and Seddon, A. (1990). The identification of a second cell cycle control on the HO promoter in yeast: cell cycle regulation of SWI5 nuclear entry. Cell 62, 631–647. [DOI] [PubMed] [Google Scholar]
- Newman, J.R., Wolf, E., and Kim, P.S. (2000). A computationally directed screen identifying interacting coiled coils from Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97, 13203–13208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niman, H.L., Houghten, R.A., Walker, L.E., Reisfeld, R.A., Wilson, I.A., Hogle, J.M., and Lerner, R.A. (1983). Generation of protein-reactive antibodies by short peptides is an event of high frequency: implications for the structural basis of immune recognition. Proc. Natl. Acad. Sci. USA 80, 4949–4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver, T.L., Szostak, J.W., and Rothstein, R.J. (1983). Genetic applications of yeast transformation with linear and gapped plasmids. Methods Enzymol. 101, 228–245. [DOI] [PubMed] [Google Scholar]
- Ortiz, J., Stemmann, O., Rank, S., and Lechner, J. (1999). A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev. 13, 1140–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne, M.A., Schlenstedt, G., Jinks, T., and Silver, P.A. (1994). Nuf2, a spindle pole body-associated protein required for nuclear division in yeast. J. Cell Biol. 125, 853–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole, E.T., Winey, M., and McIntosh, J.R. (1999). High-voltage electron tomography of spindle pole bodies and early mitotic spindles in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 10, 2017–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson, C.G., Maddox, P.S., Salmon, E.D., and Bloom, K. (2001). Budding yeast chromosome structure and dynamics during mitosis. J. Cell Biol. 152, 1255–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, J.B., and Ris, H. (1976). Electron-microscopic study of the spindle and chromosome movement in the yeast Saccharomyces cerevisiae. J. Cell Sci. 22, 219–242. [DOI] [PubMed] [Google Scholar]
- Pollard, T.C., and Earnshaw, W.C. (2002). Cell Biology. Philadelphia: Saunders, 805.
- Pot, I., Measday, V., Snydsman, B., Cagney, G., Fields, S., Davis, T.N., Muller, E.G., and Hieter, P. (2003). Chl4p and Iml3p are two new members of the budding yeast outer kinetochore. Mol. Biol. Cell 14, 460–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein, R. (1991). Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 194, 281–301. [DOI] [PubMed] [Google Scholar]
- Rout, M.P., and Kilmartin, J.V. (1990). Components of the yeast spindle and spindle pole body. J. Cell Biol. 111, 1913–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan, X., Xue, Z., Euskirchen, G., and Mélèse, T. (1997). NNF1 is an essential yeast gene required for proper spindle orientation, nucleolar and nuclear envelope structure and mRNA export. J. Cell Sci. 110, 1615–1624. [DOI] [PubMed] [Google Scholar]
- Shang, C., Hazbun, T.R., Cheeseman, I.M., Aranda, J., Fields, S., Drubin, D.G., and Barnes, G. (2003). Kinetochore protein interactions and their regulation by the Aurora kinase Ipl1p. Mol. Biol. Cell 14, 3342–3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F. (1991). Getting started with yeast. Methods Enzymol. 194, 3–21. [DOI] [PubMed] [Google Scholar]
- Siemering, K.R., Golbik, R., and Haseloff, J. (1996). Mutations that suppress the thermosensitivity of green fluorescent protein. Curr. Biol. 6, 1653–1663. [DOI] [PubMed] [Google Scholar]
- Sikorski, R.S., and Hieter, P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer, F., Gerring, S.L., Connelly, C., and Hieter, P. (1990). Mitotic chromosome transmission fidelity mutants in Saccharomyces cerevisiae. Genetics 124, 237–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark, M.J., and Milner, J.S. (1989). Cloning and analysis of the Kluyveromyces lactis TRP1 gene: a chromosomal locus flanked by genes encoding inorganic pyrophosphatase and histone H3. Yeast 5, 35–50. [DOI] [PubMed] [Google Scholar]
- Straight, A.F., Belmont, A.S., Robinett, C.C., and Murray, A.W. (1996). GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr. Biol. 6, 1599–1608. [DOI] [PubMed] [Google Scholar]
- Sullivan, B.A., Blower, M.D., and Karpen, G.H. (2001). Determining centromere identity: cyclical stories and forking paths. Nat. Rev. Genet. 2, 584–596. [DOI] [PubMed] [Google Scholar]
- Tanaka, T., Fuchs, J., Loidl, J., and Nasmyth, K. (2000). Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nat. Cell Biol. 2, 492–499. [DOI] [PubMed] [Google Scholar]
- Toyoda, Y., Furuya, K., Goshima, G., Nagao, K., Takahashi, K., and Yanagida, M. (2002). Requirement of chromatid cohesion proteins Rad21/Scc1 and Mis4/Scc2 for normal spindle-kinetochore interaction in fission yeast. Curr. Biol. 12, 347–358. [DOI] [PubMed] [Google Scholar]
- Tyers, M., Tokiwa, G., and Futcher, B. (1993). Comparison of the Saccharomyces cerevisiae G1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO J. 12, 1955–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach, A., Brachat, A., Alberti-Segui, C., Rebischung, C., and Philippsen, P. (1997). Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast 13, 1065–1075. [DOI] [PubMed] [Google Scholar]
- Wang, P.J., and Huffaker, T.C. (1997). Stu2p: A microtubule-binding protein that is an essential component of the yeast spindle pole body. J. Cell Biol. 139, 1271–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., and Burke, D.J. (1995). Checkpoint genes required to delay cell division in response to nocodazole respond to impaired kinetochore function in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 15, 6838–6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge, P.A., Jensen, O.N., Holmes, S., Souès, S., Mann, M., and Kilmartin, J.V. (1998). Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J. Cell Biol. 141, 967–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge, P.A., and Kilmartin, J.V. (2001). The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J. Cell Biol. 152, 349–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey, M., Hoyt, M.A., Chan, C., Goetsch, L., Botstein, D., and Byers, B. (1993). NDC1: a nuclear periphery component required for yeast spindle pole body duplication. J. Cell Biol. 122, 743–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods, A., Sherwin, T., Sasse, R., MacRae, T.H., Baines, A.J., and Gull, K. (1989). Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J. Cell Sci. 93, 491–500. [DOI] [PubMed] [Google Scholar]
- Yamamoto, A., Guacci, V., and Koshland, D. (1996). Pds1p is required for faithful execution of anaphase in the yeast, Saccharomyces cerevisiae. J. Cell Biol. 133, 85–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, X., Kahana, J.A., Silver, P.A., Morphew, M.K., McIntosh, J.R., Fitch, I.T., Carbon, J., and Saunders, W.S. (1999). Slk19p is a centromere protein that functions to stabilize mitotic spindles. J. Cell Biol. 146, 415–425. [DOI] [PMC free article] [PubMed] [Google Scholar]