Abstract
Gliding motility and host cell invasion by apicomplexan parasites are empowered by an acto-myosin motor located underneath the parasite plasma membrane. The motor is connected to host cell receptors through trans-membrane invasins belonging to the thrombospondin-related anonymous protein (TRAP) family. A recent study indicates that aldolase bridges the cytoplasmic tail of MIC2, the homologous TRAP protein in Toxoplasma, and actin. Here, we confirm these unexpected findings in Plasmodium sporozoites and identify conserved features of the TRAP family cytoplasmic tail required to bind aldolase: a subterminal tryptophan residue and two noncontiguous stretches of negatively charged amino acids. The aldolase substrate and other compounds that bind to the active site inhibit its interaction with TRAP and with F-actin, suggesting that the function of the motor is metabolically regulated. Ultrastructural studies in salivary gland sporozoites localize aldolase to the periphery of the secretory micronemes containing TRAP. Thus, the interaction between aldolase and the TRAP tail takes place during or preceding the biogenesis of the micronemes. The release of their contents in the anterior pole of the parasite upon contact with the target cells should bring simultaneously aldolase, TRAP and perhaps F-actin to the proper subcellular location where the motor is engaged.
INTRODUCTION
The phylum Apicomplexa is composed of unicellular eukaryotic parasites that include several major pathogens to humans and/or livestock such as Plasmodium, Toxoplasma, Cryptosporidium, Eimeria, and Babesia. They display a common set of organelles known as rhoptries, micronemes, and dense granules that discharge their content in a defined order during the host cell attachment/invasion processes (Preiser et al., 2000; Morrissette and Sibley, 2002). In addition, the apicomplexan invasive forms (termed zoites) are surrounded by a pellicle composed of the plasma membrane and underneath it a continuous layer of flattened vesicles defining the inner membrane complex (IMC).
Even though zoites do not possess cilia or flagella, they exhibit a unique kind of locomotion termed gliding motility, defined by the lack of obvious modification in the moving cell shape, and the need for a supporting substrate (King, 1988). Several lines of evidence indicate that gliding motility and cell invasion are related mechanisms empowered by the same acto-myosin motor located in the cortical space in between the plasma membrane and the IMC (King, 1988; Dobrowolski and Sibley, 1996; Meissner et al., 2002b; Wetzel et al., 2003). According to the current model, trans-membrane proteins displaying adhesive extracellular domains and anchored to the motor trigger either forward locomotion or penetration into the host cells (King, 1988; Menard, 2001; Opitz and Soldati, 2002). One such molecule is the thrombospondin-related anonymous protein (TRAP), whose expression is restricted to Plasmodium sporozoites (Rogers et al., 1992; Robson et al., 1997).
The essential role of TRAP on gliding motility and invasion has been ascertained by genetic approaches (Sultan et al., 1997; Kappe et al., 1999; Matuschewski et al., 2002). TRAP is the founding member of a family of apicomplexan membrane proteins, which includes the TgMIC2 protein of Toxoplasma gondii among others (Menard, 2001; Meissner et al., 2002a). These proteins are stored in the micronemes and rapidly translocated to the cell surface upon parasite activation, likely by Ca2+ signaling (Carruthers and Sibley, 1999; Gantt et al., 2000). The similarities among the ∼45 amino acid-long carboxyl cytoplasmic tail of TRAP family proteins are restricted to a subterminal tryptophan and a high content of acidic residues (Menard, 2001). However, replacement of the cytoplasmic tail of TRAP by that of TgMIC2 did not affect motility or infectivity of P. berghei sporozoites, indicating that notwithstanding their diverse sequence the cytoplasmic tail of these molecules have the same functional properties (Kappe et al., 1999). Several parasite mutants in the TRAP C terminus were either nonmotile or displayed abnormal gliding motility, denoting that this portion of the molecule must interact directly or indirectly with the molecular motor (Kappe et al., 1999). Direct binding of TRAP to cytoskeletal proteins, however, has not been demonstrated so far (Opitz and Soldati, 2002). An indirect kind of TRAP-motor interaction via adapter molecule(s) and/or conformational changes regulated by extracellular ligands, such as that defined for integrins and selectins (Serrador et al., 2002; Liddington and Ginsberg, 2002), seems the most likely possibility.
Indeed, recent evidence indicates that the cytoplasmic tail of both TgMIC2 and TRAP are bound to actin via the tetrameric glycolytic enzyme aldolase (Jewett and Sibley, 2003). It is known that mammalian aldolases bind to actin through defined amino acid motifs (O'Reilly and Clarke, 1993; Wang et al., 1996) and that this binding affects both the enzyme activity and cell contraction/motility (Schindler et al., 2001). Here, we confirm in Plasmodium these unforeseen findings and attempt to define the structural basis and the regulation of the TRAP-aldolase interaction.
MATERIALS AND METHODS
Aldolases
Histidine-tagged P. falciparum aldolase (PfAldo) was expressed in Escherichia coli and purified to crystallographic purposes by means of three chromatography steps, including immobilized metal ion adsorption, ion exchange, and size-exclusion columns (Kim et al., 1998). Partial clones from P. yoelii aldolase obtained from the TIGR P. yoelii Gene Index were assembled by genetic procedures to obtain the full-length sequence in frame with the glutathione S-transferase (GST) protein of the pGEX 4T-1 vector (Amersham Biosciences, Piscataway, NJ). Rabbit aldolase A and spinach aldolase were from Sigma-Aldrich (St. Louis, MO).
Aldolase Activity Assays
Aldolase activity was assayed by either coupling the fructose 1,6-phosphate (F1,6P) cleavage to the triose phosphate isomerase/α-glycerophosphate dehydrogenase reaction and continuous measuring of NADH consumption at 340 nm (Dobeli et al., 1990) or by using the aldolase kit (Sigma-Aldrich). Carbohydrates and aldolase inhibitors were from Sigma-Aldrich. Kinetic values were determined from double-reciprocal plots by using the leastsquares method.
TRAP Proteins
The entire cytoplasmic TRAP C terminus containing 45 residues was amplified from P. berghei (NK65 strain) genomic DNA by using the primers PbTC-t for (5′-ggcGAATTCtataattttatagcaggaagtagcgc-3′) and PbTC-t rev (5′-agcGTCGACtctagattagttccagtcattatcttcagg-3′). The TRAPΔW and TRAPΔacid molecules were constructed using the PbTC-t for primer along with the reverse primers PbTC-tW (5′-agcGTCGACtctagattagttcgcgtcattatcttcaggta-3′) and PbTC-tA (5′-agcGTCGACtctagattagttccaggcattacctgcaggtaatttaaac-3′), respectively. The TRAP25 mutant molecule was constructed using the primers PbTC-tGLUT for (5′-cgGAATTCgatgtaatggcagatgatga-3′) and PbTC-t rev. The amplicons were treated with EcoRI/SalI, purified, and cloned into pGEX 4T-1. Constructs were confirmed by DNA sequence analysis. GST-fusion proteins expressed in E. coli were purified by affinity chromatography on glutathione (GSH)-Sepharose columns (Amersham Biosciences) as described previously (Buscaglia et al., 1998).
Peptides
Peptides derived from P. falciparum TRAP (Rogers et al., 1992) or P. berghei aldolase (Cloonan et al., 2001) were custom-synthesized by Genemed Synthesis Inc. (San Francisco, CA). Their sequences are as follows: TRAP13mer, N-PEQFRLPEENEWN-C; TRAP25mer, N-ETLGEEDKDLDEPEQFRLPEENEWN-C; TRAP34mer, N-CYAGEPAPFDETLGEEDKDLDEPEQFRLPEENEWN-C; and Aldolase C-t, N-GAGGSTAGASLYEKKYVYC-C. In some cases, a cysteine residue (denoted in bold) not present in the original sequence was added for direct conjugation to Imject maleimide-activated keyhole limpet hemocyanin (KLH) and bovine serum albumin (BSA) (both from Pierce Chemical, Rockford, IL) following manufacturer's guidelines.
Antisera
Goat anti-rabbit aldolase antibody was from Chemicon International (Temecula, CA). Anti-GST antibody was from Amersham Biosciences. An antiserum to PfAldo was obtained by immunization of BALB/c mice with 30 μg of enzyme emulsified in complete Freund's adjuvant by the intraperitoneal route, followed by two booster injections of 10 μg in incomplete Freund's adjuvant every 15 d. Two rabbit antisera against P. berghei TRAP were used, one recognizing the cytoplasmic tail and other directed toward the amino acid repeats (Sultan et al., 1997). The specificity of these antisera was confirmed by lack of reactivity toward TRAP null parasites. In addition, an antiserum to the cytoplasmic tail of P. falciparum TRAP was raised in mice immunized as described above with the TRAP34mer peptide coupled to KLH. Anti-KLHAldolase C-t peptide antiserum was raised in rabbits (Covance, Denver, PA). The specificity of the two latter antisera was assessed by reactivity toward the corresponding BSA-coupled peptide by enzyme-linked immunosorbent assay (ELISA).
IgG and Fab Purification
IgG was obtained by protein A-Sepharose chromatography (Amersham Biosciences) following manufacturer's guidelines. To obtain the Fab fragments, IgG was incubated overnight at 37°;C with 0.02 mg/ml papain (Sigma-Aldrich) in phosphate-buffered saline (PBS) containing 20 mM EDTA and 20 mM l-cysteine. Reaction was stopped with 100 μl of 0.3 M iodoacetamide in PBS followed by buffer exchange using NAP-10 desalting columns (Amersham Biosciences). Samples were applied onto protein A-Sepharose columns and the flow-through fractions (containing the Fab) were collected.
Immunoprecipitation
P. berghei sporozoites (5 × 106) were resuspended in 2 ml of 25 mM HEPES, pH 7.3, 1 mM EDTA, 1 mM MgCl2, 50 mM KCl, 0.5% Tween 20, and a protease inhibitor cocktail (Sigma-Aldrich) and subjected to two bursts of sonication (20 sec each) on ice. Every subsequent step was carried out at 4°C. Tubes were kept on ice for 20 min and centrifuged at 14,000 rpm for 20 min. Supernatant was centrifuged again and preadsorbed for 1 h with 200 μl of protein G-Sepharose (Amersham Biosciences) equilibrated in resuspension buffer. Aliquots (500 μl) of the supernatant were incubated for 4 h with 100 μg of the indicated IgG. Protein G-Sepharose (100 μl) was then added, and samples incubated for 1 h. Resins were washed five times in 1 ml each of resuspension buffer and stripped at 100°C in loading buffer (50 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 10% 2-mercaptoethanol, and 0.1% bromphenol blue).
Pull-Down Assays
One hundred micrograms of either PfAldo or as a control rabbit glyceraldehyde-3-phosphate dehydrogenase (Sigma-Aldrich) was mixed with 150 μg of BSA and preadsorbed for 1 h with GSH-Sepharose equilibrated in buffer A (10 mM imidazole acetate, pH 7.3, 50 mM KCl, and 0.2% Tween 20). Supernatants were incubated with 100 μl of GSH-Sepharose loaded with 50 μg of the indicated GST-fusion protein for 3 h at 4°C. Pellets were washed five times in 1 ml each of buffer A and finally stripped at 100°C in loading buffer.
F-Actin Co-sedimentation Assays
Actin was purified from rabbit muscle acetone powder (Sigma-Aldrich) as described previously (Pardee and Spudich, 1982). Nonpolymerized actin (G-actin) was diluted in buffer G (5 mM Tris-HCl, pH 8, 0.2 mM ATP, 0.2 mM dithiothreitol, and 0.2 mM CaCl2) up to 100 μg/ml and centrifuged to remove denatured protein. Actin polymerization was carried out for 2 h at 4°C by the addition of 50 mM KCl, 2 mM MgCl2, and 0.8 mM ATP (Pardee and Spudich, 1982). Twenty microliters of either GST-fusion protein (300 μg/ml) was mixed with 1.5 μl of PfAldo (4 mg/ml) and 180 μl of polymerized actin (F-actin) and incubated at room temperature for 45 min. In some cases, different carbohydrates (100 μM) were added. F-Actin was precipitated at 65,000 × g for 2 h in a Beckman ultracentrifuge. Supernatants were collected and pellets washed twice with 1 ml of buffer G before solubilization in 200 μl of loading buffer diluted 1:5 in 8 M urea. Equivalent aliquots from both pellet and supernatant fractions were analyzed in SDS-PAGE followed by Coomassie Brilliant Blue staining.
Protein Biotinylation
PfAldo and normal rabbit IgG (2 mg/ml) extensively dialyzed against PBS were labeled for 2 h at room temperature with 1 mM EZ-link Sulfo-NHSBiotin (Pierce Chemical). Reaction was stopped with 15 μl of 1 M Tris-HCl, pH 7.2. Samples were applied onto NAP-10 columns to remove the excess of free biotin and stored at 4°C with sodium azide (0.01%). The extent of biotinylation was determined by the 2-hydroxyazobenzene-4′-carboxylic acid dye method (Bayer and Wilchek, 1990).
ELISA
Polystyrene ELISA microplates (NUNC A/S, Roskilde, Denmark) were coated with 1 nmol/well of the TRAP34mer peptide in carbonate buffer, pH 9.5, and blocked in buffer A with 5% BSA for 3 h. Biotin-labeled proteins diluted in the same buffer were incubated for 3 h in the presence of the indicated compounds. After extensive washings in buffer A, NeutrAvidin conjugated to horseradish peroxidase (HRPO) (1:5000; Pierce Chemical) was added for 1 h. Absorbance values at 405 nm were recorded after the addition of 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) (Pierce Chemical) and transformed to protein concentration by using a calibration curve of the corresponding biotin-labeled protein performed in the same plate. Percentage of inhibition displayed by each compound was calculated using the same calibration curves and taking as 0% inhibition the mean value recorded for the biotin-labeled protein without further additions.
Cryoimmunoelectron Microscopy
Salivary glands from P. yoelii-infected Anopheles stephensi mosquitoes were fixed for 2 d at 4°C with 8% paraformaldehyde in 0.25 M HEPES, pH 7.4, infiltrated, frozen, and sectioned as described previously (Folsch et al., 2001). The sections were labeled first with mouse anti-PfAldo antibodies (1:100 in PBS 1% fish skin gelatin) followed by anti-mouse IgG antibodies and 5-nm protein A-gold particles (Department of Cell Biology, Medical School, Utrecht University, Utrecht, The Netherlands). Afterward, sections were incubated with rabbit anti-TRAP repeats antibodies (1:10) followed by 10-nm protein A-gold particles before examination with a Philips 410 electron microscope (Eindhoven, The Netherlands) under 80 kV.
Statistical Analysis
Mean values in every point of the curves were compared with that of the corresponding control by using the Student's t test. p < 0.05 was considered significant.
RESULTS
In Vivo Experiments
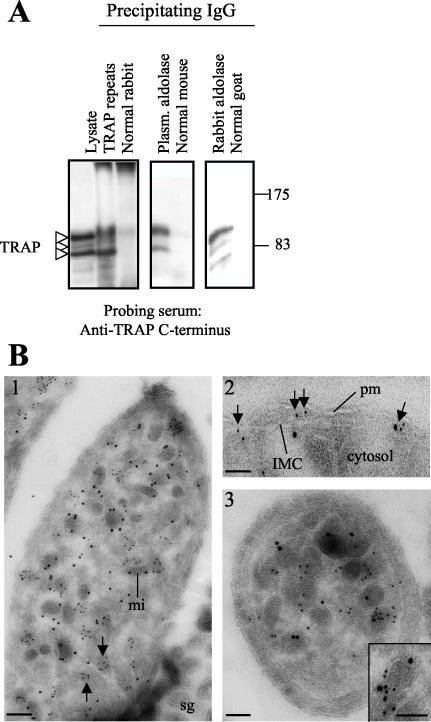
To evaluate the possible in vivo interaction between TRAP and aldolase, extracts from P. berghei sporozoites were immunoprecipitated with purified IgG against either TRAP repeats or different aldolases. Normal IgG purified from the corresponding species was used as control. Immunoprecipitated fractions were analyzed by Western blot. An antiserum raised against the cytoplasmic tail of TRAP reveals a discrete number of bands ranging from ∼100 to ∼75 kDa in both the total parasite lysate and the anti-TRAP immunoprecipitate (Figure 1A). This antiserum also highlights the same bands in both anti-aldolase precipitates but not in the control fractions (Figure 1A), indicating that TRAP and aldolase interact in vivo.
Figure 1.
TRAP and aldolase interact in vivo. (A) Lysates from P. berghei sporozoites were immunoprecipitated using the IgG indicated above each lane and probed with a rabbit anti-TRAP C-terminus antiserum (1:1000). Molecular markers (in kilodaltons) are indicated on the right. (B) Immunoelectron microscopy of P. yoelii sporozoites doubly labeled with mouse anti-PfAldo (5-nm gold particles) and rabbit anti-TRAP repeats (10-nm gold particles). 1, a longitudinal section of the parasite identifies the TRAP-containing micronemes (mi), some of which are peripherally associated with aldolase. Some microneme-like structures are solely labeled by aldolase (arrows). 2, localization of aldolase between the IMC and the plasma membrane (pm) is highlighted by arrows. 3, a transversal section of the parasite pinpoints the intense and specific labeling of micronemes with TRAP and aldolase; few molecules of aldolase are observed in the parasite cytosol. The arrow in the inset of 3 shows the presence of TRAP and aldolase in a single microneme. sg, salivary gland tissue. Bars, 150 nm.
The in vivo aldolase-TRAP interaction was also revealed by cryoimmunoelectron microscopy. For these experiments, the rabbit anti-TRAP repeats antiserum was used along with the mouse anti-PfAldo antiserum. Double labeling of sections obtained from salivary glands from infected mosquitoes colocalizes both proteins mostly to the periphery of certain micronemes (Figure 1B, 1 and 3). Sixty-six of 165 (40%) TRAP-containing organelles analyzed from different sections are labeled with both antisera. In addition, aldolase can be found between the plasma membrane and the IMC (Figure 1B, 2), scattered in the cytosol, and surrounding certain microneme-like structures not labeled by the anti-TRAP antiserum (Figure 1B, 1 and 3). Although these micronemes may not contain TRAP, it is also possible that the absence of TRAP staining is technical in nature. Indeed, using the same antibodies it has been previously shown that most of sporozoite micronemes contain TRAP (Kappe et al., 1999; Bhanot et al., 2003). The same pattern of labeling was obtained when a goat anti-rabbit aldolase antiserum was used (our unpublished data).
In Vitro Binding Assays
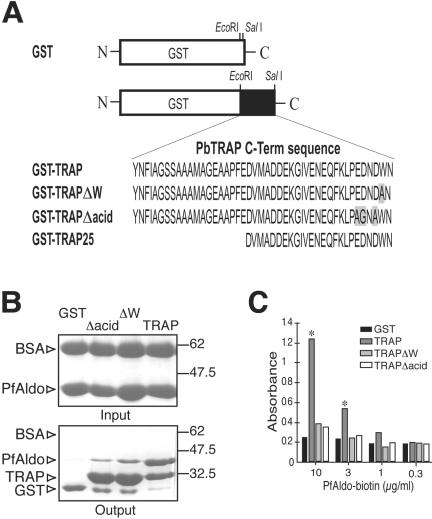
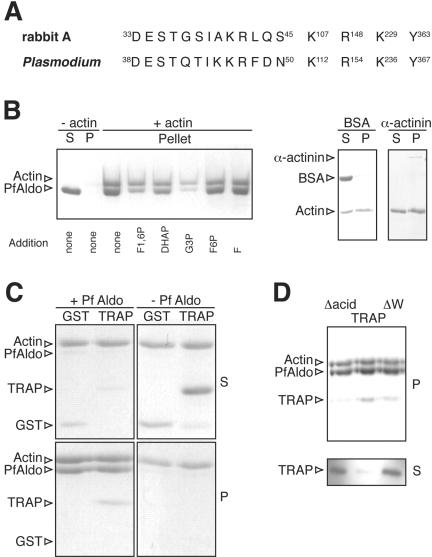
The in vitro interaction between TRAP and PfAldo was initially evaluated by pull-down assays by using as baits three different GST-TRAP proteins (Figure 2A). They include the entire wild-type P. berghei TRAP C-terminus sequence and two mutant molecules with alanine or glycine substitutions in either the penultimate tryptophan or the last tract of acidic residues (termed TRAPΔW and TRAPΔacid, respectively). The parasites bearing these mutant molecules had a profound defect in gliding motility and were not infective (Kappe et al., 1999). These TRAP constructs were immobilized onto GSH-Sepharose and incubated with an equimolar mixture of PfAldo and BSA (Figure 2B, input). The specifically retained proteins were eluted and revealed by Coomassie-stained SDS-PAGE (Figure 2B, output). As shown, PfAldo binds to the wild-type TRAP and to a much lesser extent to the TRAP variant molecules. GST alone displays negligible binding to PfAldo, whereas BSA does not bind to the GST-fusion proteins (Figure 2B, output). Furthermore, the enzyme downstream of aldolase on the glycolytic pathway (glyceraldehyde-3-phosphate dehydrogenase) did not bind to any of the GST-fusion proteins when evaluated by similar pull-down assays (our unpublished data). The effect of the introduced mutations in TRAP was also evaluated by ELISA. Plates coated with either GST-TRAP molecule were incubated with biotin-labeled PfAldo, and the interaction revealed by the addition of HRPO-conjugated avidin. As shown in Figure 2C, only the wild-type TRAP significantly binds to PfAldo by this technique.
Figure 2.
TRAP cytoplasmic tail interacts with PfAldo in vitro. (A) Schematic representation of GST-TRAP variant molecules. Different constructs derived from P. berghei TRAP were cloned in-frame at the C terminus of GST by the unique EcoRI and SalI restriction sites. Amino acid replacements inserted in each construct are shaded. (B) Equal amounts of each GST-fusion protein (indicated above) immobilized onto GSH-Sepharose were incubated with 100 μg of PfAldo and 150 μg of BSA (input). Specifically retained proteins were evaluated by Coomassie Blue-stained SDS-PAGE (output). The identity of the revealed bands is indicated on the left and the molecular markers (in kilodaltons) on the right. (C) ELISA plates coated with 100 ng of the indicated GST-fusion protein were incubated with biotin-labeled PfAldo and revealed by the addition of HRPO-avidin followed by a colorimetric substrate. Samples were tested in triplicate. Mean values of absorbance at 405 nm are indicated (SD did not exceed 5% of mean values; our unpublished data). Asterisks denote significant differences (p < 0.01) with the corresponding GST values.
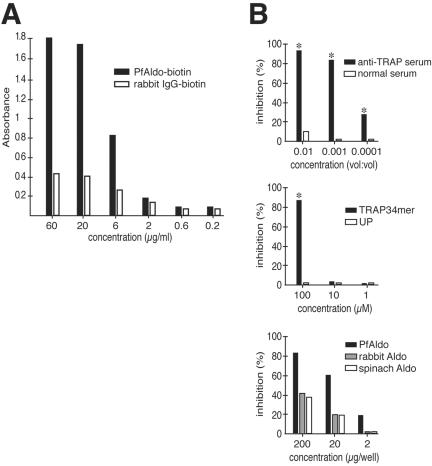
To further characterize this interaction, we coated ELISA plates with a peptide spanning the last 34 residues of P. falciparum TRAP (TRAP34mer) and incubated them with biotin-labeled PfAldo. As shown in Figure 3A, PfAldo binds in a dose-dependent manner to the TRAP34mer-coated plates, whereas purified normal rabbit IgG displaying similar extent of biotinylation exhibits minimal binding capacity in the same conditions. The binding of PfAldo to TRAP-coated plates is strongly and specifically inhibited by a mouse antiserum raised against the TRAP34mer peptide (Figure 3B, top), by the TRAP34mer peptide itself in the fluid phase (Figure 3B, middle) and by nonbiotinylated aldolases (Figure 3B, bottom).
Figure 3.
Specificity of PfAldo-TRAP interaction. (A) ELISA plates coated with the TRAP34mer peptide were probed with either biotin-labeled PfAldo or biotin-labeled rabbit IgG as in legend to Figure 2C. (B) Inhibition of TRAP34mer-biotin-labeled PfAldo (6 μg/ml) interaction displayed by different mouse antisera (top), peptides (middle), or nonbiotinylated aldolases (bottom). Samples were tested in triplicate and the mean values of absorbance at 405 nm are indicated (SD did not exceed 5% of mean values; our unpublished data). Asterisks denote significant differences (p < 0.01) with the mean value recorded for the biotin-labeled PfAldo without further additions. One of three experiments with similar results is shown. UP, peptide unrelated to TRAP.
Mapping of the Aldolase-binding Site in TRAP
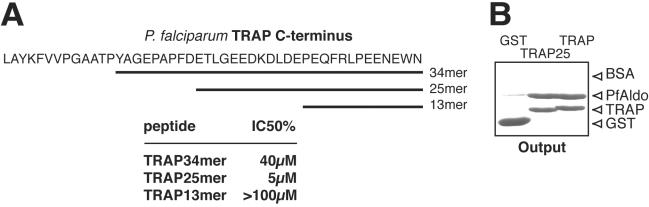
To map the aldolase-binding region in TRAP, we evaluated the extent of inhibition displayed by a series of partially overlapping peptides derived from the TRAP C terminus. As indicated in Figure 4A, the binding of PfAldo to TRAP34mer-coated plates is displaced by the TRAP34mer itself present in the fluid phase (Figure 3B), and by a shorter version of this peptide spanning solely the 25 C-terminal amino acids of TRAP. Surprisingly, the inhibitory capacity of the TRAP25mer peptide is higher than that of the TRAP34mer (50% inhibition achieved at 5 and 40 μM concentration, respectively). A peptide spanning the last 13 residues of TRAP (TRAP13mer), on the other hand, does not render significant inhibition even when assayed at 100 μM concentration (Figure 4A). These results suggest that the last 25 residues of TRAP are sufficient to bind to PfAldo. To confirm these findings in a fluid-phase assay, we performed pull-down experiments by using either the entire TRAP tail or a shorter version spanning just the last 25 residues (GST-TRAP25; Figure 2A). As shown in Figure 4B, the extent of binding to PfAldo is indistinguishable for both proteins.
Figure 4.
Mapping of aldolase-binding site on TRAP. (A) Peptides derived from the P. falciparum TRAP C-terminus sequence were tested for their ability to interfere with the binding between biotin-labeled PfAldo and the TRAP34mer peptide as indicated in legend to Figure 3. The sequence, length, and concentration required to obtain 50% inhibition (IC50%) are shown. (B) Equal amounts of each GST fusion protein were tested for their ability to interact with PfAldo by pull-down assays as described in Figure 2B.
Mapping of the TRAP-binding Site in Aldolase
Compounds That Bind to the Enzyme Active Site Inhibit Aldolase-TRAP Interaction To map the TRAP binding site(s) on Plasmodium aldolase, we initially tried to obtain in E. coli distinct truncated versions of the P. yoelii enzyme. Unfortunately, all of these proteins, except for the full-length molecule, were poorly expressed or targeted to inclusion bodies (our unpublished data).
We then analyzed the effect of catalysis on the aldolase-TRAP interaction. The substrate F1,6P and the products of the reaction catalyzed by aldolase (dihydroxyacetone phosphate and glyceraldehyde-3-phosphate) inhibit at low micromolar concentration the binding of the enzyme to TRAP (Table 1). These values are in the range of the Km for PfAldo (Table 1), suggesting that this inhibition is mediated by occupation of the active site. Related compounds such as fructose 1-phosphate and fructose 6-phosphate display a significantly impaired inhibitory activity (Table 1). The concentrations required to reach 50% of inhibition of the PfAldo-TRAP interaction and 50% inhibition of PfAldo activity on F1,6P are very similar (millimolar range), further supporting the role of the active site occupation in the former effect. Neither fructose nor glucose 6-phosphate inhibits PfAldo-TRAP interaction or PfAldo cleavage of F1,6P (Table 1). We also tested the effect of competitive inhibitors of aldolase in our assay. As shown in Table 1, both suramin and d-myo-inositol 1,4,5-phosphate (but not d-myo-inositol) inhibit both PfAldo-TRAP interaction and F1,6P consumption at roughly similar concentration.
Table 1.
Correlation between inhibition of PfAldo activity and PfAldo-TRAP interaction
| Compound | IC50%a | K1b |
|---|---|---|
| Fructose 1,6-phosphate | 4 μMc | N.A.d |
| Dihydroxyacetone phosphate | 5 μM | N.A. |
| Glyceraldehyde 3-phosphate | 5 μM | N.A. |
| Fructose 1-phosphate | 1 mM | 5 mM |
| Fructose 6-phosphate | 5 mM | 3 mM |
| Glucose 6-phosphate | >50 mM | >50 mM |
| Fructose | >50 mM | >50 mM |
| Suramin | 500 μM | 100 μM |
| D-Myo-inositol | >10 mM | >10 mM |
| D-Myo-inositol 1,4,5-phosphate | 200 μM | 500 μM |
| TRAP34mer | 40 μM | >100 μM |
| TRAP25mer | 5 μM | >100 μM |
Each compound was tested at serial dilutions for the inhibition of biotin-labeled PfAldo-TRAP34mer peptide interaction by ELISA and for the PfAldo inhibition in activity assays using fructose 1,6-phosphate as substrate.
IC50%, concentration required to obtain 50% inhibition of PfAldo-TRAP interaction
Ki, concentration required to obtain 50% inhibition of PfAldo activity. Ki was calculated by plotting 1/Vel against the compound concentration
Km value for PfAldo = 9 μM
N.A., non-applicable
Overall, these results suggest that TRAP is binding either directly to the active site or to another region of the enzyme whose conformation is changed upon occupation of the active site. However, neither the TRAP34mer nor the TRAP25mer inhibit PfAldo enzymatic activity even when assayed at 100 μM concentration (Table 1).
The C Terminus of Aldolase Is Not Involved in the Binding to TRAP Previous site-directed mutagenesis and crystallographic studies (Itin et al., 1993; Kim et al., 1998) indicate that the last ∼20 amino acids of PfAldo play a critical role in the catalytic reaction. It has been speculated that this speciesspecific and flexible segment modulates the PfAldo activity by burrowing into the active site (Kim et al., 1998). In Plasmodium aldolases, this sequence is rich in positively charged residues (Cloonan et al., 2001), suggesting that it might mediate the interaction with the acidic TRAP tail. However, we found that the Aldolase C-t peptide spanning the last 19 residues of P. berghei aldolase (90% identical to the PfAldo sequence) does not inhibit the PfAldo-TRAP binding even at 100 μM concentration (our unpublished data). Furthermore, Fab fragments prepared from an antiserum raised against this peptide are also inactive in preventing PfAldo-TRAP interaction (Figure 5A), although they recognize and partially inhibit PfAldo enzymatic activity (Figure 5B). Overall, our data suggest that the C-terminal region of PfAldo is not critical for its interaction with TRAP.
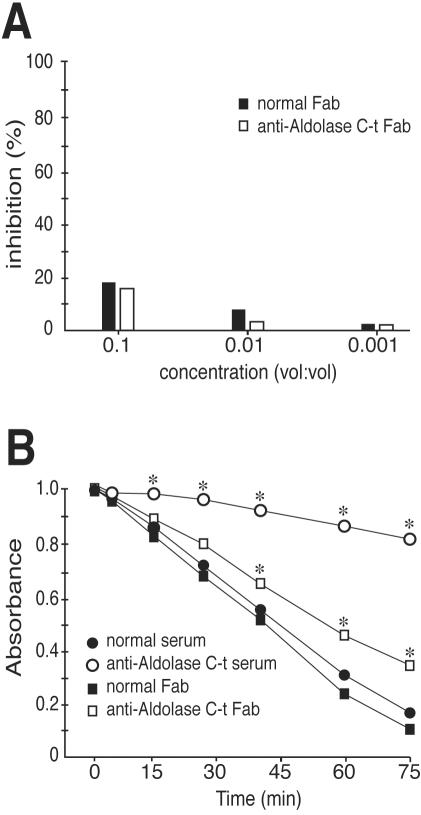
Figure 5.
Effect of aldolase C terminus (c-t) on its interaction with TRAP. (A) The inhibition of biotin-labeled PfAldo-TRAP34mer peptide interaction displayed by different Fab fractions (0.3 mg/ml) was evaluated as indicated in legend to Figure 3. One of two experiments with similar results is shown. (B) PfAldo activity was continuously monitored at 340 nm in the presence of the indicated antisera (1:250 dilution) or Fab fragments (1:50 dilution). Samples were tested in duplicate and the mean values of absorbance are indicated (SD did not exceed 3% of mean values; our unpublished data). Asterisks denote significant differences (p < 0.01) with the mean value recorded for the PfAldo without further additions. One of two experiments with similar results is shown.
The Aldolase-TRAP Complex Interacts with F-Actin
The actin-binding site of mammalian aldolases includes a series of residues (O'Reilly and Clarke, 1993; Wang et al., 1996) highly conserved in the Plasmodium aldolases (Figure 6A) (Cloonan et al., 2001), but the direct interaction of the parasite aldolase with actin has never been demonstrated. Therefore, we first evaluated the in vitro interaction of PfAldo and F-actin by cosedimentation assays. As shown in Figure 6B, PfAldo coprecipitates with actin microfilaments. This interaction is partially inhibited by the aldolase substrate F1,6P and by the aldolase products dihydroxyacetone phosphate and glyceraldehyde-3-phosphate but not by fructose 6-phosphate or fructose (Figure 6B). BSA and α-actinin were used as internal negative and positive F-actin-coprecipitating controls, respectively (Figure 6B) (Hohaus et al., 2002). Additional cosedimentation assays were performed to demonstrate the specific formation of a ternary complex (F-actin-PfAldo-TRAP). As depicted in Figure 6C, GST-TRAP, but not GST, is brought down into the F-actin pellet when in the presence of PfAldo, indicating that the ternary complex is obtained even though TRAP per se is unable to bind F-actin. The intensity of the revealed PfAldo and actin bands in both the supernatant and pellet fractions is very similar in the presence of either GST or GST-TRAP (Figure 6C). Therefore, neither the F-actin-PfAldo interaction, nor the F-actin/G-actin ratio is affected by GST-TRAP.
Figure 6.
An actin-aldolase-TRAP ternary complex is formed in vitro. (A) Sequence alignment of the actin-binding site of rabbit aldolase A, and the corresponding residues in Plasmodium-derived aldolases. The relative position in the primary sequence for each residue is indicated. (B) Left, PfAldo was ultracentrifuged in the presence or absence of F-actin and both supernatant (S) and pellet (P) fractions analyzed by Coomassie-stained SDS-PAGE. The identity of the revealed bands is indicated on the left. The addition of different compounds (100 μM) before centrifugation is indicated below each lane. DHAP, dihydroxyacetone phosphate; G3P, glyceraldehyde-3-phosphate; F6P, fructose 6-phosphate; F, fructose. Right, fractionation of BSA and α-actinin in F-actin cosedimentation assays. Both pellet (P) and supernatant (S) fractions were analyzed as described in A. (C) GST or GST-TRAP molecules were mixed with F-actin in the presence (left) or absence (right) of PfAldo and subjected to ultracentrifugation. Equivalent supernatant (S, top) and pellet (P, bottom) fractions were analyzed as in A. (D) TRAP variants (indicated above each lane) were assayed for their cosedimentation capacity with F-actin in the presence of PfAldo. Equivalent pellet (P, top) and supernatant fractions (S, bottom) were analyzed by Coomassie staining and Western blot by using an anti-GST antibody (1:500), respectively. The identity of the revealed bands is indicated on the left.
Pull-down and ELISA assays revealed the key role of both the penultimate tryptophan and the terminal stretch of acidic residues of TRAP in its association to PfAldo (Figure 2, B and C). Indeed, all of wild-type TRAP is associated via PfAldo to the F-actin pellet, whereas a fraction of both TRAPΔW and TRAPΔacid molecules remain in the supernatant (unbound) fraction as revealed by Western blot by using an anti-GST antibody (Figure 6D).
DISCUSSION
Current evidence strongly suggests that gliding motility and host cell invasion by apicomplexan parasites are empowered by the same acto-myosin motor, a unique machinery restricted to this phylum and highly conserved among its members (Menard, 2001; Opitz and Soldati, 2002). Defining the identity and mode of interaction of the components of this machinery may provide new tools for the prophylaxis or treatment of a wide range of human and livestock diseases. The overall arrangement of the motor and several of its key constituents such as actin (Dobrowolski and Sibley, 1996; Wetzel et al., 2003), myosin A (Pinder et al., 1998; Matuschewski et al., 2001; Meissner et al., 2002b), and a family of surface invasins related to TRAP (Menard, 2001; Matuschewski et al., 2002) have been already characterized. Recent data also indicate participation in the machinery of a protein that binds to the tail of myosin A (Herm-Gotz et al., 2002; Bergman et al., 2003), of a Toxoplasma myosin docking protein (Beckers, personal communication), and of aldolase (Jewett and Sibley, 2003). This latter molecule probably transduces the signals from the parasite surface to the motor by bridging the cytoplasmic tail of invasins to actin.
Here, we use different approaches to study the in vivo and in vitro interaction of Plasmodium aldolase and TRAP. We show that the last 25 residues of the TRAP tail are sufficient to mediate this binding (Figure 4). This region of the TRAP tail harbors most of its charged residues, whereas the remaining sequence close to the trans-membrane domain is biased toward glycines and hydrophobic residues (valines, alanines, prolines, tyrosines; Figure 2A) and promotes the intracellular sorting of the molecule (Bhanot et al., 2003). Within the defined aldolase-binding site on TRAP, our findings reveal the critical role of the penultimate tryptophan in the enzyme recognition (Figures 2, B and C, and 6D). These results are in close agreement with those reported by studying TgMIC2/TRAP and either rabbit or T. gondii aldolase (Jewett and Sibley, 2003), except that we detect a minor binding of aldolase to the TRAPΔW molecule, which is probably of relevance (see below). In addition, we demonstrated that the acidic last stretch of 13 residues of the TRAP tail is required but not sufficient for optimal binding to aldolase (Figures 2, B and C, 4, and 6D). Our binding studies using partially overlapping peptides demonstrate the presence of additional aldolase-contacting residues upstream of the last 13 amino acids of TRAP. Indeed, as shown in Figure 4A, this sequence cannot displace the interaction between aldolase and a longer version of the TRAP tail (TRAP34mer peptide), whereas a 25mer peptide displays full inhibition.
Overall, these data concur with previous in vivo experiments by using TRAP mutants. P. berghei sporozoites harboring a TRAP molecule deleted on its last 14 residues are noninfective but still display residual motility (Kappe et al., 1999). A similar phenomenon was verified for parasites bearing TRAP molecules mutated in either the penultimate tryptophan or the last tract of acidic residues (Kappe et al., 1999). Though aberrant, this residual motility suggests that the tethering of TRAP mutant molecules to the motor is somehow taking place. The comparison of the aldolase-binding sequence in TRAP with the structures of the aldolase-binding motifs of other proteins provides additional supporting evidence for our hypothesis. All of these molecules display a consensus sequence composed of two acidic stretches flanking a single highly hydrophobic residue (Figure 7). Deletion of either acidic tract or posttranslational modification of the hydrophobic residue correlates with a decreased affinity of these molecules for aldolase (Itin et al., 1993; Volker and Knull, 1997; Eisenmesser and Post, 1998). The participation of ionic bonds in the aldolase-TRAP interaction is also supported by the inhibitory effect of salts (IC50% reached at 60 mM) and/or ionic detergents (our unpublished data).
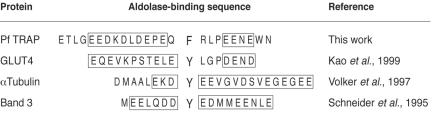
Figure 7.
Sequence comparison of aldolase-binding motifs present in different proteins. Aldolase-binding motifs present in human glucose transporter 4 (GLUT4), bovine α-tubulin, and the anion exchanger of human erythrocyte membrane (band 3) were aligned with the last 25 residues of P. falciparum TRAP cytoplasmic tail (Pf TRAP). Overall acidic regions are indicated by boxes.
Thus, it is very likely that the two noncontiguous acidic stretches in the TRAP tail (Figure 7) interact with positively charged regions scattered throughout the surface of the aldolase. The enzyme active site itself comprises a cluster of basic residues, some of which are involved in the anchoring of the phosphate groups of the substrate/products (Kim et al., 1998). A direct binding of the TRAP tail to the aldolase catalytic region is therefore conceivable. In this case, TRAP and F-actin should be stably accommodated in a small surface to form the ternary complex because both the catalytic pocket and the actin-binding site on PfAldo are closely apposed (Figure 8). This scenario would explain the inhibitory role of aldolase substrate, products, and competitive inhibitors in this interaction (Table 1). Indeed, the association to aldolase of other sequences structurally related to TRAP such as those displayed in Figure 7 is also displaced by aldolase substrate/products (Schneider and Post, 1995; Volker and Knull, 1997; Kao et al., 1999). However, this idea is not supported by the observation that neither the TRAP34mer nor the TRAP25mer peptide inhibited the in vitro PfAldo activity even when tested at 100 μM concentration (Table 1).
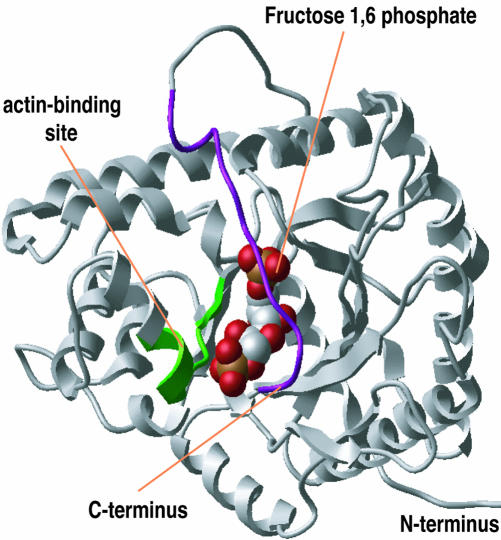
Figure 8.
Proximity of the actin-binding site and the catalytic center in PfAldo. Ribbon diagram of the PfAldo monomer structure based on crystallographic data showing the substrate (fructose 1,6-phosphate) anchored to the active site, the sequence 38DESTQTIKKRFDN50 spanning the core of the actin-binding motif (in green) and the flexible C terminus (in violet) apposed to the catalytic pocket.
Although other explanations can be invoked, it is conceivable that TRAP tail is not interacting with the aldolase active site itself but with another positively charged region whose structure is modified upon the occupancy of the catalytic center. The last ∼20 residues of PfAldo define a highly flexible and positively charged structure that is presumed to modulate the enzymatic activity by its reversible burrowing into the active site (Kim et al., 1998) (Figure 8). We failed, however, to demonstrate a direct interaction between TRAP and the C terminus of PfAldo (Figure 5). If successful, ongoing cocrystallization experiments between PfAldo and the TRAP tail peptide will shed some light on this puzzling issue.
Here, we demonstrated for the first time the specific association of F-actin, aldolase, and TRAP in a ternary complex (Figure 6C). The binding of aldolase to both F-actin and TRAP is, however, inhibited during substrate turnover (Table 1 and Figure 6B). How can the motor bridging properties and the catalytic role of aldolase be reconciled in vivo? Aldolase is a tetramer made up of four identical subunits and F-actin is a multimer that can be assembled in bundles. We favor the idea that actin-aldolase-TRAP form large dissociable dynamic complexes with the rest of the motor beneath the plasma membrane, and that enzymatic turnover will not disrupt all of these links simultaneously. The possibility that the concentration of the aldolase substrate/products in the cortical cytoplasm is lower than that required to inhibit the interaction of the enzyme with TRAP, or even that glycolysis does not take place in the space where the motor is engaged cannot be ruled out. In this case, the ATP required for the assembly and translocation of actin microfilaments should be generated outside the cortical cytoplasm. Further studies dealing with the mechanisms of energy conversion in this parasite stage and/or the in vivo structure/function of the gliding motility machinery may clarify these issues.
It has been previously shown that the aldolase-MIC2 complex is present on the surface of T. gondii tachyzoites (Jewett and Sibley, 2003). Our ultrastructural studies on salivary gland (resting) Plasmodium sporozoites indicate that the formation of this complex occurs at a previous step. As demonstrated for other apicomplexan invasins (Soldati et al., 2001), TRAP is likely to be stored in the micronemes as an integral type I membrane protein, with its adhesive N terminus protruding toward the micronemal lumen and the C-terminus tail facing to the cytoplasm. As shown in Figure 1B, aldolase is localized on the surface of micronemes containing TRAP, most likely bound to its cytoplasmic tail. These observations provide a plausible explanation for the sorting of aldolase (and probably actin) to the narrow, constrained space in between the IMC and the plasma membrane (Ngo et al., 2000). On parasite activation, TRAP-containing micronemes fuse with the parasite membrane (Carruthers and Sibley, 1999; Gantt et al., 2000) bringing the adhesive domains of TRAP to the parasite surface, and piggy back to its tail the aldolase-actin complex. This hypothesis is in agreement with the observation that polymerization of actin in Apicomplexa takes place initially in the anterior parasite pole (Wetzel et al., 2003). During gliding motility and cell invasion, the aldolase-TRAP complex (and perhaps other molecules) would be moved along the locally formed and tightly regulated actin filaments (Jewett and Sibley, 2003), propelled by the myosin A molecules tethered to the IMC (Bergman et al., 2003).
Acknowledgments
We thank Drs. Ute Frevert (New York University School of Medicine, New York, NY) and Ali Sultan (Department of Immunology and Infectious Diseases, Harvard School of Public Health, Boston, MA) for some of the anti-TRAP antibodies and Drs. Karine Kaiser and Stefan Kappe (New York University) for additional Plasmodium peptides and genomic DNA samples. Critical reading of the manuscript by Dr. Stefan Kappe is also appreciated. P. yoelii aldolase clones were kindly provided by Dr. Lawrence Bergman (Drexel University, Philadelphia, PA). We are also indebted to Dr. Matthieu Schapira (New York University), for modeling the aldolases, Dr. Ruth Nussenzweig (New York University) for the sporozoites, Nelly Camargo (New York University) for the purified salivary glands of mosquitoes, Dr. Defne Yarar (Department of Molecular and Cell Biology, University of California at Berkeley) for advice on actin purification, and Brian Krumm (Department of Biochemistry, University of Washington, Seattle, WA) for preparing PfAldo. Our work was supported by a grant from the National Institutes of Health to V.N. C.A.B. is a recipient of a Bernard Levine fellowship.
Abbreviations used: F1,6P, fructose 1,6-phosphate; IMC, inner membrane complex; PfAldo, Plasmodium falciparum aldolase; TRAP, thrombospondin-related anonymous protein.
References
- Bayer, E.A., and Wilchek, M. (1990). Protein biotinylation. Methods Enzymol. 184, 138-160. [DOI] [PubMed] [Google Scholar]
- Bergman, L.W., Kaiser, K., Fujioka, H., Coppens, I., Daly, T.M., Fox, S., Matuschewski, K., Nussenzweig, V., and Kappe, S.H. (2003). Myosin A tail domain interacting protein (MTIP) localizes to the inner membrane complex of Plasmodium sporozoites. J. Cell Sci. 116, 39-49. [DOI] [PubMed] [Google Scholar]
- Bhanot, P., Frevert, U., Nussenzweig, V., and Persson, C. (2003). Defective sorting of the thrombospondin-related anonymous protein (TRAP) inhibits Plasmodium infectivity. Mol. Biochem. Parasitol. 126, 263-273. [DOI] [PubMed] [Google Scholar]
- Buscaglia, C.A., Campetella, O., Leguizamón, M.S., and Frasch, A.C. (1998). The repetitive domain of Trypanosoma cruzi trans-sialidase enhances the immune response against the catalytic domain. J. Infect. Dis. 177, 431-436. [DOI] [PubMed] [Google Scholar]
- Carruthers, V.B., and Sibley, L.D. (1999). Mobilization of intracellular calcium stimulates microneme discharge in Toxoplasma gondii. Mol. Microbiol. 31, 421-428. [DOI] [PubMed] [Google Scholar]
- Cloonan, N., Fischer, K., Cheng, Q., and Saul, A. (2001). Aldolase genes of Plasmodium species. Mol. Biochem. Parasitol. 113, 327-330. [DOI] [PubMed] [Google Scholar]
- Dobeli, H., Trzeciak, A., Gillesen, D., Matile, H., Srivastava, I., Perrin, L., Jakob, P., and Certa, U. (1990). Expression, purification, biochemical characterization and inhibition of recombinant Plasmodium falciparum aldolase. Mol. Biochem. Parasitol. 41, 259-268. [DOI] [PubMed] [Google Scholar]
- Dobrowolski, J.M., and Sibley, L.D. (1996). Toxoplasma invasion of mammalian cells is powered by the actin cytoskeleton of the parasite. Cell 84, 933-939. [DOI] [PubMed] [Google Scholar]
- Eisenmesser, E.Z., and Post, C.B. (1998). Insights into tyrosine phosphorylation control of protein-protein association from the NMR structure of a band 3 peptide inhibitor bound to glyceraldehyde-3-phosphate dehydrogenase. Biochemistry 37, 867-877. [DOI] [PubMed] [Google Scholar]
- Folsch, H., Pypaert, M., Schu, P., and Mellman, I. (2001). Distribution and function of AP-1 clathrin adaptor complexes in polarized epithelial cells. J. Cell Biol. 152, 595-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantt, S., Persson, C., Rose, K., Birkett, A.J., Abagyan, R., and Nussenzweig, V. (2000). Antibodies against thrombospondin-related anonymous protein do not inhibit Plasmodium sporozoite infectivity in vivo. Infect. Immun. 68, 3667-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herm-Gotz, A., Weiss, S., Stratmann, R., Fujita-Becker, S., Ruff, C., Meyhofer, E., Soldati, T., Manstein, D. J., Geeves, M. A., and Soldati, D. (2002). Toxoplasma gondii myosin A and its light chain: a fast, single-headed, plus-enddirected motor. EMBO J. 21, 2149-2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohaus, A., Person, V., Behlke, J., Schaper, J., Morano, I., and Haase, H. (2002). The carboxyl-terminal region of ahnak provides a link between cardiac L-type Ca2+ channels and the actin-based cytoskeleton. FASEB J. 16, 1205-1216. [DOI] [PubMed] [Google Scholar]
- Itin, C., Burki, Y., Certa, U., and Dobeli, H. (1993). Selective inhibition of Plasmodium falciparum aldolase by a tubulin derived peptide and identification of the binding site. Mol. Biochem. Parasitol. 58, 135-143. [DOI] [PubMed] [Google Scholar]
- Jewett, T.J., and Sibley, L.D. (2003). Aldolase forms a bridge between cell surface adhesins and the actin cytoskeleton in apicomplexan parasites. Mol. Cell. 11, 885-894. [DOI] [PubMed] [Google Scholar]
- Kao, A.W., Noda, Y., Johnson, J.H., Pessin, J.E., and Saltiel, A.R. (1999). Aldolase mediates the association of F-actin with the insulin-responsive glucose transporter GLUT4. J. Biol. Chem. 274, 17742-17747. [DOI] [PubMed] [Google Scholar]
- Kappe, S., Bruderer, T., Gantt, S., Fujioka, H., Nussenzweig, V., and Menard, R. (1999). Conservation of a gliding motility and cell invasion machinery in apicomplexan parasites. J. Cell Biol. 147, 937-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H., Certa, U., Dobeli, H., Jakob, P., and Hol, W.G. (1998). Crystal structure of fructose-1, 6-bisphosphate aldolase from the human malaria parasite Plasmodium falciparum. Biochemistry 37, 4388-4396. [DOI] [PubMed] [Google Scholar]
- King, C.A. (1988). Cell motility of sporozoan protozoa. Parasitol. Today 4, 315-319. [DOI] [PubMed] [Google Scholar]
- Liddington, R.C., and Ginsberg, M.H. (2002). Integrin activation takes shape. J. Cell Biol. 158, 833-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matuschewski, K., Mota, M.M., Pinder, J.C., Nussenzweig, V., and Kappe, S.H. (2001). Identification of the class XIV myosins Pb-MyoA and Py-MyoA and expression in Plasmodium sporozoites. Mol. Biochem. Parasitol. 112, 157-161. [DOI] [PubMed] [Google Scholar]
- Matuschewski, K., Nunes, A.C., Nussenzweig, V., and Menard, R. (2002). Plasmodium sporozoite invasion into insect and mammalian cells is directed by the same dual binding system. EMBO J. 21, 1597-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner, M., Reiss, M., Viebig, N., Carruthers, V., Toursel, C., Tomavo, S., Ajioka, J., and Soldati, D. (2002a). A family of transmembrane microneme proteins of Toxoplasma gondii contain EGF-like domains and function as escorters. J. Cell Sci. 115, 563-574. [DOI] [PubMed] [Google Scholar]
- Meissner, M., Schluter, D., and Soldati, D. (2002b). Role of Toxoplasma gondii myosin A in powering parasite gliding and host cell invasion. Science 298, 837-840. [DOI] [PubMed] [Google Scholar]
- Menard, R. (2001). Gliding motility and cell invasion by Apicomplexa: insights from the Plasmodium sporozoite. Cell Microbiol. 3, 63-73. [DOI] [PubMed] [Google Scholar]
- Morrissette, N.S., and Sibley, L.D. (2002). Cytoskeleton of apicomplexan parasites. Microbiol. Mol. Biol. Rev. 66, 21-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo, H.M., Hoppe, H.C., and Joiner, K.A. (2000). Differential sorting and post-secretory targeting of proteins in parasitic invasion. Trends Cell Biol. 10, 67-72. [DOI] [PubMed] [Google Scholar]
- O'Reilly, G., and Clarke, F. (1993). Identification of an actin binding region in aldolase. FEBS Lett. 321, 69-72. [DOI] [PubMed] [Google Scholar]
- Opitz, C., and Soldati, D. (2002). 'The glideosome': a dynamic complex powering gliding motion and host cell invasion by Toxoplasma gondii. Mol. Microbiol. 45, 597-604. [DOI] [PubMed] [Google Scholar]
- Pardee, J.D., and Spudich, J.A. (1982). Purification of muscle actin. Methods Enzymol. 85, 164-181. [DOI] [PubMed] [Google Scholar]
- Pinder, J., Fowler, R., Dluzewski, A., Bannister, L., Lavin, F., Mitchell, G., Wilson, R.J., and Gratzer, W.B. (1998). Actomyosin motor in the merozoite of the malaria parasite, Plasmodium falciparum: implications for red cell invasion. J. Cell Sci. 111, 1831-1839. [DOI] [PubMed] [Google Scholar]
- Preiser, P., Kaviratne, M., Khan, S., Bannister, L., and Jarra, W. (2000). The apical organelles of malaria merozoites: host cell selection, invasion, host immunity and immune evasion. Microbes Infect. 2, 1461-1477. [DOI] [PubMed] [Google Scholar]
- Robson, K.J., Naitza, S., Barker, G., Sinden, R.E., and Crisanti, A. (1997). Cloning and expression of the thrombospondin related adhesive protein gene of Plasmodium berghei. Mol. Biochem. Parasitol. 84, 1-12. [DOI] [PubMed] [Google Scholar]
- Rogers, W., Malik, A., Mellouk, S., Nakamura, K., Rogers, M., Szarfman, A., Gordon, D., Nussler, A., Aikawa, M., and Hoffman, S.L. (1992). Characterization of Plasmodium falciparum sporozoite surface protein 2. Proc. Natl. Acad. Sci. USA 89, 9176-9180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler, R., Weichselsdorfer, E., Wagner, O., and Bereiter-Hahn, J. (2001). Aldolase-localization in cultured cells: cell-type and substrate-specific regulation of cytoskeletal associations. Biochem. Cell Biol. 79, 719-728. [PubMed] [Google Scholar]
- Schneider, M.L., and Post, C.B. (1995). Solution structure of a band 3 peptide inhibitor bound to aldolase: a proposed mechanism for regulating binding by tyrosine phosphorylation. Biochemistry 34, 16574-16584. [DOI] [PubMed] [Google Scholar]
- Serrador, J.M., Urzainqui, A., Alonso-Lebrero, J.L., Cabrero, J.R., Montoya, M.C., Vicente-Manzanares, M., Yanez-Mo, M., and Sanchez-Madrid, F. (2002). A juxta-membrane amino acid sequence of P-selectin glycoprotein ligand-1 is involved in moesin binding and ezrin/radixin/moesin-directed targeting at the trailing edge of migrating lymphocytes. Eur. J. Immunol. 32, 1560-1566. [DOI] [PubMed] [Google Scholar]
- Soldati, D., Dubremetz, J.F., and Lebrun, M. (2001). Microneme proteins: structural and functional requirements to promote adhesion and invasion by the apicomplexan parasite Toxoplasma gondii. Int. J. Parasitol. 31, 1293-1302. [DOI] [PubMed] [Google Scholar]
- Sultan, A.A., Thathy, V., Frevert, U., Robson, K.J., Crisanti, A., Nussenzweig, V., Nussenzweig, R.S., and Menard, R. (1997). TRAP is necessary for gliding motility and infectivity of Plasmodium sporozoites. Cell 90, 511-522. [DOI] [PubMed] [Google Scholar]
- Volker, K.W., and Knull, H. (1997). A glycolytic enzyme binding domain on tubulin. Arch. Biochem. Biophys. 338, 237-243. [DOI] [PubMed] [Google Scholar]
- Wang, J., Morris, A.J., Tolan, D.R., and Pagliaro, L. (1996). The molecular nature of the F-actin binding activity of aldolase revealed with site-directed mutants. J. Biol. Chem. 271, 6861-6865. [PubMed] [Google Scholar]
- Wetzel, D.M., Hakansson, S., Hu, K., Roos, D., and Sibley, L.D. (2003). Actin filament polymerization regulates gliding motility by apicomplexan parasites. Mol. Biol. Cell 14, 396-406. [DOI] [PMC free article] [PubMed] [Google Scholar]