Abstract
Background
The mouse skeletal muscle is composed of four distinct fiber types that differ in contractile function, number of mitochondria and metabolism. Every muscle type has a specific composition and distribution of the four fiber types. To find novel genes involved in specifying muscle types, we used microarray analysis to compare the gastrocnemius with the quadriceps from mice fed a low fat diet (LFD) or high fat diet (HFD) for 8 weeks. Additional qPCR analysis were performed in the gastrocnemius, quadriceps and soleus muscle from mice fed an LFD or HFD for 20 weeks.
Results
In mice fed the 8-week LFD 162 genes were differentially expressed in the gastrocnemius vs. the quadriceps. Genes with the strongest differences in expression were markers for oxidative fiber types (e.g. Tnni1) and genes which are known to be involved in embryogenesis (Dkk3, Hoxd8,Hoxd9 and Tbx1). Also Dkk2, Hoxa5, Hoxa10, Hoxc9, Hoxc10, Hoxc6 and Tbx15 were detectably, but not differentially expressed in adult muscle tissue. Expression of differentially expressed genes was not influenced by an 8-week or 20-week HFD. Comparing gastrocnemius, quadriceps and soleus, expression of Hoxd8 and Hoxd9 was not related with expression of markers for the four different fiber types. We found that the expression of both Hoxd8 and Hoxd9 was much higher in the gastrocnemius than in the quadriceps or soleus, whereas the expression of Dkk3 was high in quadriceps, but low in both gastrocnemius and soleus. Finally, expression of Tbx1 was high in quadriceps, intermediate in soleus and low in gastrocnemius.
Conclusions
We found that genes from the Dkk family, Hox family and Tbx family are detectably expressed in adult mouse muscle. Interestingly, expression of Dkk3, Hoxd8, Hoxd9 and Tbx1 was highly different between gastrocnemius, quadriceps and soleus. In fact, every muscle type showed a unique combination of expression of these four genes which was not influenced by diet. Altogether, we conclude that genes important for embryogenesis identify mouse muscle types in a diet-independent and fiber type-unrelated manner.
Background
The mouse skeletal muscle is composed of four distinct fiber types, i.e., the type I, IIa, IIx and IIb fibers that differ in respect to contractile function, the number of mitochondria and metabolism. The slow type I fibers can sustain prolonged low power work, contain more mitochondria and exhibit higher rates of fat oxidative metabolism. The fast type IIx and IIb fibers are adapted to brief and intense contractions, contain fewer mitochondria and generate energy mainly through glycolysis. The type IIa fibers exhibit an intermediate contractile function and are oxidoglycolytic [1-4]. Every muscle type has a specific composition and distribution of the four different fiber types. The soleus contains the highest number of type I fibers, whereas both the quadriceps and gastrocnemius are muscle groups that contain predominantly type II fibers [5,6]. However, varying percentages of type I fibers ranging from 0-45% for the quadriceps and 1-8% for the gastrocnemius are reported [7].
Numerous genes have been identified playing a role in the generation of more oxidative muscle types [6,8-13]. The most extensively studied gene is calcineurin (CnA). In the C2C12 cell line, CnA up-regulates the mRNA levels of genes that are markers for the slow fiber types. Additionally, studies with CnA transgenic mice have reported an increase of oxidative fibers in the skeletal muscle [8,9]. PGC1α is another well-established factor that induces remodeling of skeletal muscle fiber type composition. Lin et al. [10] showed that when PGC1α is expressed at physiological levels in muscle groups normally rich in type II fibers a fiber type conversion occurs from type II fibers to type I fibers [10]. In line with this, Mortensen et al. [11] showed that PGC1α overexpression in primary rat skeletal muscle cells induces a switch towards a more oxidative fiber type. Finally, muscle-specific overexpression of PPARδ resulted in the increase of the number of oxidative fibers and in higher expression levels of several markers of oxidative metabolism [6,12].
The first aim of the present study was to find novel genes that may play a role in specifying muscle types. Therefore we compared gene expression profiles of the gastrocnemius with the quadriceps at the level of the whole-transcriptome. Functional implications were assessed by the analyses of predefined gene sets based on Gene Ontology, biochemical, metabolic and signaling pathways. Recently, we studied the effects of a short-term high fat diet (HFD) on skeletal muscle gene and protein expression. Since both gene and protein levels of markers of the more oxidative fiber types were increased in the quadriceps of HFD mice we hypothesized that an HFD can induce a conversion towards a more oxidative fiber type via a transcriptional mechanism [14]. The second aim of the present study therefore was to explore the diet-sensitivity of genes that are involved in the determination of muscle types. We compared expression patterns of genes that are differentially expressed between muscle types in mice fed a low fat diet (LFD) with mice fed an HFD.
Results
Microarray analysis shows that the majority of genes involved in the determination of muscle types are not influenced by diet
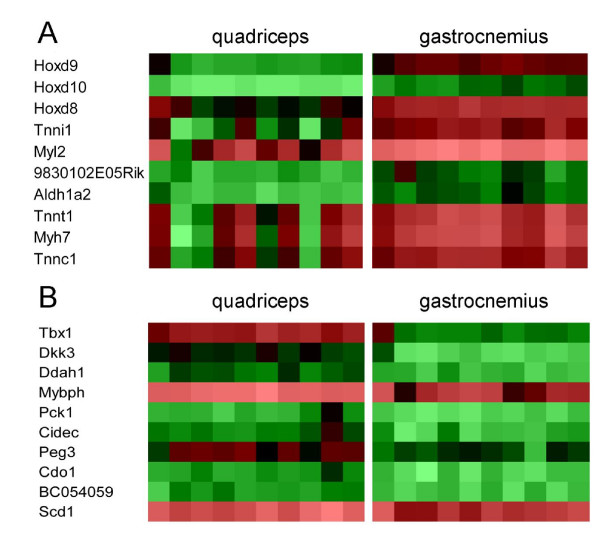
To obtain more insight in genes involved in specifying muscle types we compared the gastrocnemius transcriptome with the quadriceps transcriptome of 8-week LFD mice. When using the criteria of a fold change > 1.3 and an FDR < 0.05, we found 91 genes with an increased expression and 71 genes with a decreased expression in the gastrocnemius as compared to the quadriceps. The ten genes with the strongest increased and decreased expression levels are shown in Table 1 and Fig. 1. The ten genes with the strongest increased expression included markers of oxidative fiber types (Tnnc1, Tnni1, Tnnt1 and Myh7), but also genes from the Hox family (Hoxd8, Hoxd9 and Hoxd10). In fact, Hoxd8, Hoxd9 and Hoxd10 were the genes with the strongest increased expression in the gastrocnemius. The ten genes with the strongest decreased expression contained a variety of genes such as Scd1 and Cidec, which are involved in lipogenesis, and Pck1, which plays a role in gluconeogenesis, but also the Dkk3 and Tbx1 genes, which are important during embryogenesis. A complete list of all differentially expressed genes is available in supplement 1. qPCR analyses were performed for 13 genes validating the microarray results (supplement 2).
Table 1.
Ten genes that showed the strongest increased or decreased expression in gastrocnemius compared to quadriceps
| probe set | gene name | gene description | Accession (EntrezID) | Fold change | FDR |
|---|---|---|---|---|---|
| Strongest up-regulated genes | |||||
| 15438_at | Hoxd9 | homeo box D9 | 15438 | 7.70 | 0.000 |
| 15430_at | Hoxd10 | homeo box D10 | 15430 | 7.36 | 0.000 |
| 15437_at | Hoxd8 | homeo box D8 | 15437 | 4.78 | 0.000 |
| 21952_at | Tnni1 | troponin I, skeletal, slow 1 | 21952 | 4.46 | 0.005 |
| 17906_at | Myl2 | myosin, light polypeptide 2, regulatory, cardiac, slow | 17906 | 3.89 | 0.004 |
| 277898_at | 9830102E05Rik | RIKEN cDNA 9830102E05 gene | 277898 | 3.69 | 0.002 |
| 19378_at | Aldh1a2 | aldehyde dehydrogenase family 1, subfamily A2 | 19378 | 3.61 | 0.000 |
| 21955_at | Tnnt1 | troponin T1, skeletal, slow | 21955 | 3.56 | 0.024 |
| 140781_at | Myh7 | myosin, heavy polypeptide 7, cardiac muscle, beta | 140781 | 3.26 | 0.039 |
| 21924_at | Tnnc1 | troponin C, cardiac/slow skeletal | 21924 | 3.05 | 0.020 |
| Strongest down-regulated genes | |||||
| 21380_at | Tbx1 | T-box 1 | 21380 | -7.27 | 0.000 |
| 50781_at | Dkk3 | dickkopf homolog 3 (Xenopus laevis) | 50781 | -5.40 | 0.000 |
| 69219_at | Ddah1 | dimethylarginine dimethylaminohydrolase 1 | 69219 | -2.91 | 0.000 |
| 53311_at | Mybph | myosin binding protein H | 53311 | -2.66 | 0.002 |
| 18534_at | Pck1 | phosphoenolpyruvate carboxykinase 1, cytosolic | 18534 | -2.62 | 0.015 |
| 14311_at | Cidec | cell death-inducing DFFA-like effector c | 14311 | -2.57 | 0.009 |
| 18616_at | Peg3 | paternally expressed 3 | 18616 | -2.49 | 0.005 |
| 12583_at | Cdo1 | cysteine dioxygenase 1, cytosolic | 12583 | -2.45 | 0.004 |
| 246747_at | BC054059 | cDNA sequence BC054059 | 246747 | -1.96 | 0.010 |
| 20249_at | Scd1 | stearoyl-Coenzyme A desaturase 1 | 20249 | -1.79 | 0.018 |
By microarray analysis we found 161 that were differentially expressed in the gastrocnemius as compared with the quadriceps from mice fed an 8-week LFD. This table shows the ten strongest up-regulated and ten strongest down-regulated genes in the gastrocnemius as compared with the quadriceps (n = 10).
Figure 1.
Heatmap of the ten genes with the strongest difference in expression between gastrocnemius and quadriceps. Heatmap of the log-transformed microarray signal intensity values of the ten genes that showed the strongest increased (A) and decreased (B) expression level in the gastrocnemius as compared to the quadriceps of 10 LFD mice. The heatmap was generated by using GeneMaths XT software. Signal intensities are shown by a color range; bright red, black, and bright green represent high, average, and low levels of gene expression, respectively.
To relate changes in gene expression to biological changes we applied ErmineJ and GSEA. By using Ermine J we identified 68 GO classes that were overrepresented in the gastrocnemius (Table 2). These overrepresented GO classes were mainly descriptors for contraction, morphogenesis, remodeling and development. Other overrepresented GO classes included descriptors for calcium homeostasis, immune function and metabolic processes. A parallel GSEA was used to identify up- or down-regulated processes. GSEA revealed that ten gene sets were down-regulated including three gene sets related to the cell cycle, two gene sets describing amino acid metabolism and two gene sets corresponding to immune function. Only the gene sets "striated muscle contraction" and "ribosome" were up-regulated in the gastrocnemius as compared with the quadriceps. Among the core-enriched genes in the gene set "striated muscle contraction" were genes that are markers for oxidative fiber types, whereas all core-enriched genes in the gene set "ribosome" were ribosomal proteins which are involved in translation (Table 3).
Table 2.
Overrepresented Gene Ontology classes in the gastrocnemius as compared to the quadriceps
| GO ID | GO class | N | Raw Score | FDR |
|---|---|---|---|---|
| GO:0001974 | blood vessel remodeling | 15 | 1.99 | 2.39E-10 |
| GO:0006776 | vitamin A metabolic process | 15 | 1.92 | 9.95E-11 |
| GO:0051705 | behavioral interaction between organisms | 18 | 1.63 | 3.89E-04 |
| GO:0006775 | fat-soluble vitamin metabolic process | 20 | 1.57 | 4.17E-04 |
| GO:0048705 | skeletal morphogenesis | 17 | 1.41 | 8.14E-04 |
| GO:0009855 | determination of bilateral symmetry | 31 | 1.30 | 5.97E-11 |
| GO:0003007 | heart morphogenesis | 29 | 1.28 | 8.53E-11 |
| GO:0014033 | neural crest cell differentiation | 31 | 1.28 | 3.98E-10 |
| GO:0006936 | muscle contraction | 69 | 1.19 | 1.49E-10 |
| GO:0006937 | regulation of muscle contraction | 25 | 1.19 | 7.35E-04 |
| GO:0043010 | camera-type eye development | 56 | 1.14 | 7.02E-11 |
| GO:0007368 | determination of left/right symmetry | 30 | 1.14 | 8.02E-04 |
| GO:0048762 | mesenchymal cell differentiation | 38 | 1.13 | 9.19E-11 |
| GO:0008016 | regulation of heart contraction | 34 | 1.13 | 3.23E-04 |
| GO:0060047 | heart contraction | 41 | 1.11 | 1.09E-10 |
| GO:0007498 | mesoderm development | 44 | 1.06 | 1.33E-10 |
| GO:0001654 | eye development | 82 | 1.03 | 7.46E-11 |
| GO:0048592 | eye morphogenesis | 37 | 1.02 | 6.97E-04 |
| GO:0006766 | vitamin metabolic process | 56 | 1.01 | 6.28E-11 |
| GO:0009952 | anterior/posterior pattern formation | 86 | 0.96 | 4.59E-11 |
| GO:0016055 | Wnt receptor signaling pathway | 107 | 0.94 | 4.98E-11 |
| GO:0042445 | hormone metabolic process | 71 | 0.92 | 1.71E-04 |
| GO:0048771 | tissue remodeling | 104 | 0.92 | 1.71E-10 |
| GO:0006875 | cellular metal ion homeostasis | 57 | 0.92 | 2.99E-04 |
| GO:0048637 | skeletal muscle development | 61 | 0.92 | 7.96E-11 |
| GO:0006959 | humoral immune response | 52 | 0.92 | 7.46E-04 |
| GO:0007519 | striated muscle development | 103 | 0.91 | 1.19E-10 |
| GO:0055065 | metal ion homeostasis | 58 | 0.91 | 1.99E-04 |
| GO:0030324 | lung development | 59 | 0.89 | 4.07E-04 |
| GO:0007178 | transmembrane receptor protein serine/threonine kinase signaling pathway | 67 | 0.89 | 1.93E-04 |
| GO:0006575 | amino acid derivative metabolic process | 70 | 0.88 | 5.69E-11 |
| GO:0030323 | respiratory tube development | 60 | 0.88 | 4.78E-04 |
| GO:0007229 | integrin-mediated signaling pathway | 74 | 0.88 | 2.91E-04 |
| GO:0048469 | cell maturation | 66 | 0.87 | 4.68E-04 |
| GO:0006817 | phosphate transport | 61 | 0.86 | 2.84E-04 |
| GO:0055074 | calcium ion homeostasis | 53 | 0.86 | 7.58E-04 |
| GO:0045786 | negative regulation of progression through cell cycle | 101 | 0.85 | 5.19E-11 |
| GO:0048732 | gland development | 63 | 0.85 | 6.85E-04 |
| GO:0044271 | nitrogen compound biosynthetic process | 61 | 0.84 | 3.98E-04 |
| GO:0030003 | cellular cation homeostasis | 87 | 0.84 | 1.99E-10 |
| GO:0021700 | developmental maturation | 79 | 0.84 | 2.21E-04 |
| GO:0016311 | dephosphorylation | 105 | 0.83 | 5.97E-10 |
| GO:0055080 | cation homeostasis | 88 | 0.83 | 2.13E-04 |
| GO:0055082 | cellular chemical homeostasis | 90 | 0.82 | 5.43E-11 |
| GO:0006470 | protein amino acid dephosphorylation | 93 | 0.82 | 6.63E-11 |
| GO:0050778 | positive regulation of immune response | 75 | 0.81 | 3.32E-04 |
| GO:0051240 | positive regulation of multicellular organismal process | 94 | 0.81 | 1.76E-04 |
| GO:0008015 | circulation | 84 | 0.81 | 5.43E-04 |
| GO:0050776 | regulation of immune response | 88 | 0.80 | 1.87E-04 |
| GO:0002684 | positive regulation of immune system process | 77 | 0.80 | 4.59E-04 |
| GO:0050801 | ion homeostasis | 103 | 0.79 | 2.99E-10 |
| GO:0002682 | regulation of immune system process | 90 | 0.79 | 2.06E-04 |
| GO:0001503 | ossification | 80 | 0.79 | 4.51E-04 |
| GO:0045165 | cell fate commitment | 94 | 0.78 | 4.42E-04 |
| GO:0015674 | di-, tri-valent inorganic cation transport | 112 | 0.78 | 1.19E-09 |
| GO:0031214 | biomineral formation | 81 | 0.78 | 7.21E-04 |
| GO:0046849 | bone remodeling | 89 | 0.78 | 3.81E-04 |
| GO:0008361 | regulation of cell size | 91 | 0.77 | 4.78E-11 |
| GO:0030005 | cellular di-, tri-valent inorganic cation homeostasis | 81 | 0.77 | 7.08E-04 |
| GO:0043062 | extracellular structure organization and biogenesis | 85 | 0.77 | 6.28E-04 |
| GO:0009968 | negative regulation of signal transduction | 81 | 0.77 | 6.74E-04 |
| GO:0040008 | regulation of growth | 117 | 0.77 | 1.81E-04 |
| GO:0006816 | calcium ion transport | 85 | 0.75 | 8.78E-04 |
| GO:0002009 | morphogenesis of an epithelium | 103 | 0.73 | 4.87E-04 |
| GO:0002252 | immune effector process | 106 | 0.73 | 6.40E-04 |
| GO:0043549 | regulation of kinase activity | 120 | 0.72 | 3.14E-04 |
| GO:0045859 | regulation of protein kinase activity | 115 | 0.72 | 3.73E-04 |
| GO:0051338 | regulation of transferase activity | 121 | 0.72 | 3.06E-04 |
ErmineJ was used to identify significantly overrepresented GO classes in the gastrocnemius as compared with the quadriceps from LFD mice (n = 10). For the concept biological process we selected GO classes with a FDR < 0.001. For this analysis only classes containing 8 through 125 genes were taken into account. N, number of genes in GO class
Table 3.
Changed gene sets in the gastrocnemius versus quadriceps.
| N | ES | NES | FDR | |
|---|---|---|---|---|
| up-regulated cellular processes | ||||
| Striated muscle contraction1 | 41 | 0.68 | 2.37 | 0.000 |
| Ribosome2 | 20 | 0.71 | 2.05 | 0.005 |
| down-regulated cellular processes | ||||
| Tissues, muscle, fat, bone and connective 21 | 47 | -0.57 | -2.14 | 0.003 |
| Cell cycle1 | 85 | -0.52 | -2.14 | 0.003 |
| CTCF first multivalent nuclear factor3 | 17 | -0.71 | -2.07 | 0.006 |
| Valine, leucine and isoleucine degradation2 | 40 | -0.59 | -2.16 | 0.006 |
| Cell cycle G1 to S control reactome2 | 69 | -0.49 | -2.01 | 0.013 |
| Antigen processing and presentation2 | 51 | -0.52 | -2.01 | 0.014 |
| Mets affect on macrophage differentiation3 | 16 | -0.69 | -1.97 | 0.014 |
| Amino acid metabolism1 | 46 | -0.52 | -1.95 | 0.016 |
| Porphyrin and Chlorophyll metabolism2 | 19 | -0.64 | -1.88 | 0.033 |
| DNA replication reactome1 | 40 | -0.51 | -1.86 | 0.038 |
GSEA was applied to identify up-regulated and down-regulated processes in the gastrocnemius as compared with the quadriceps in LFD mice. Presented are regulated processes with a false discovery rate (FDR) < 0.05. An FDR was calculated to adjust for multiple hypothesis testing. Sources of the gene sets: 1GenMAPP, 2KEGG, 3Biocarta. N, number of genes; ES, enrichment score for the gene set, that reflects the degree to which a gene set is overrepresented at the top or bottom of the ranked list; FDR, NES, normalized enrichment score, that is, the normalized ES to account for the size of the set.
The diet-sensitivity of differentially expressed genes, overrepresented GO classes and regulated gene sets was studied by comparing the gastrocnemius with the quadriceps under HFD conditions (supplement 3). In the HFD mice we found 215 differentially expressed genes in the gastrocnemius as compared to the quadriceps. A total of 128 genes was overlapping with the differentially expressed genes in the LFD mice. Interestingly, these overlapping genes were all regulated in the same direction with a comparable fold change. A total of 87 genes was only differentially expressed in HFD mice, whereas 34 genes were differentially expressed in LFD mice suggesting that these genes are diet-dependent. Under HFD conditions, ErmineJ revealed that 193 GO classes were overrepresented in the gastrocnemius as compared to the quadriceps. A total of 67 GO classes showed an overlap with the overrepresented GO classes under LFD conditions. The other overrepresented GO classes corresponded also to contraction, morphogenesis, remodeling, development, calcium homeostasis, immune function and metabolic processes. GSEA showed that 11 gene sets were down-regulated including five gene sets describing cell cycle and four gene sets related to amino acid metabolism. Only one gene set "striated muscle contraction" was up-regulated in the gastrocnemius vs. the quadriceps. A total of six gene sets (five down-regulated and one up-regulated) were overlapping between the LFD and HFD condition.
To summarize, we showed that a considerable number of genes is differentially expressed in the gastrocnemius as compared with the quadriceps. The genes with the strongest regulations were markers for oxidative fiber types (Tnnc1, Tnni1, Tnnt1 and Myh7) and genes which are known to be involved in embryogenesis (Dkk3, Hoxd8, Hoxd9, Hoxd10 and Tbx1). The differentially expressed genes corresponded to a variety of process including morphogenesis and contraction, but also amino acid metabolism, cell cycle and immune function. Finally, we found more differentially expressed genes and more overrepresented GO classes under HFD conditions than under LFD conditions. However, this did not result in the identification of other regulated processes indicating that diet has only a very small effect on the difference between gastrocnemius and quadriceps.
Numerous genes involved in embryogenesis are detectably expressed in adult mouse muscle tissue
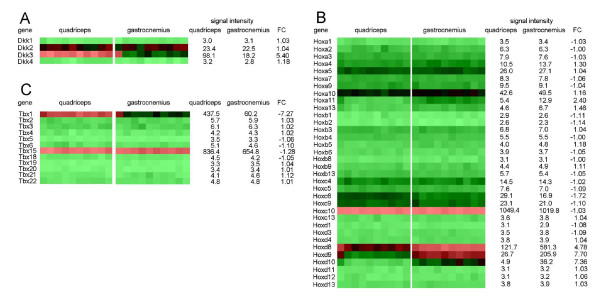
One of the most interesting findings of the comparison between gastrocnemius and quadriceps was the fact that genes of the Dkk family (Dkk3), Hox family (Hoxd8, Hoxd9 and Hoxd10) and Tbx family (Tbx1) were detectably expressed in adult mouse muscle tissue. To find out if also other genes from these three gene families are detectably expressed in adult mouse muscle tissue we analyzed the microarray signal intensities of all members, which were present on the microarray, in the gastrocnemius and quadriceps. An average microarray signal intensity > 20 was used as cut off. Fig. 2A shows that four members of the Dkk gene family were present on the microarray. In addition to Dkk3, we found that also Dkk2 was detectably expressed in both gastrocnemius and quadriceps (22.5 vs. 23.4 in gastrocnemius vs. quadriceps). From the Hox family 34 members were present on the microarray. Eight genes of the Hox family, including Hoxd8, Hoxd9 and Hoxd10, were expressed in adult muscle tissue. Hoxa5, Hoxa10, Hoxc9, Hoxc10, Hoxd8 and Hoxd9 were detectably expressed in both the gastrocnemius and quadriceps. Furthermore, Hoxc6 was detectably expressed in the quadriceps and not in the gastrocnemius (29.1 versus 16.9), whereas Hoxd10 was detectably expressed in the gastrocnemius and not in the quadriceps (36.2 versus 4.9). The gene with the highest expression level was identified as the Hoxc10 gene (Fig. 2B). Fig. 2C shows that from the Tbx family 12 members were present on the microarray. Additionally to Tbx1, we found that Tbx15 was highly expressed in the gastrocnemius as well as in the quadriceps (654.8 vs. 836.4, respectively). Altogether, these results show that some genes that are known to play an important role in embryogenesis are also detectably expressed in adult mouse muscle tissue.
Figure 2.
Heatmap of Dkk genes, Hox genes and Tbx genes in gastrocnemius and quadriceps. Heatmap of the log-transformed microarray signal intensity values of (A) Dkk genes, (B) Hox genes and (C) Tbx genes in the quadriceps and the gastrocnemius of 10 LFD mice. A heatmap was generated by using GeneMaths XT software. Signal intensities are shown by a color range; bright red, black, and bright green represent high, average, and low levels of gene expression, respectively. Values are means (n = 10)
The Tbx1 gene is a muscle-type specific marker
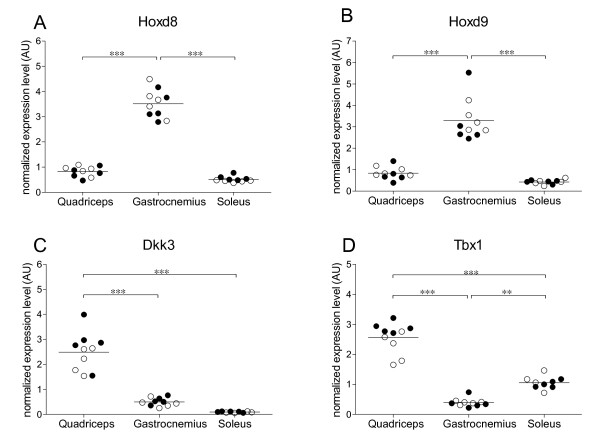
Dkk3, Hoxd8, Hoxd9 and Tbx1 were relatively high and differentially expressed in the gastrocnemius as compared to the quadriceps of mice fed an 8-week LFD. To extrapolate our result to a third muscle type we analyzed the expression level of these genes in quadriceps, gastrocnemius and soleus muscle of 20-week LFD mice and 20-week HFD mice. None of the genes was significantly influenced by diet which is in line with the earlier described observations (Fig. 3A-D). Hoxd8 and Hoxd9 were highly expressed in the gastrocnemius, but low gene expression levels were observed in the quadriceps and soleus (Fig. 3A-B). Dkk3 was significantly higher expressed in the quadriceps than in the gastrocnemius and soleus. Although not significantly, Dkk3 gene expression was lower in the soleus than in the gastrocnemius (Fig. 3C). The highest expression levels of the Tbx1 gene was found in the quadriceps. Intermediate expression levels were measured in the soleus and the lowest expression level was found in the gastrocnemius. The Tbx1 expression levels between the three muscle types were all significantly different (Fig. 3D).
Figure 3.
Expression of Hoxd8, Hoxd9, Dkk3 and Tbx1 in quadriceps, gastrocnemius and soleus. Gene expression levels of (A) Hoxd8, (B) Hoxd9, (C) Dkk3 and (D) Tbx1 in quadriceps, gastrocnemius and soleus of 20-week LFD mice and 20-week HFD mice. Black dots represent the 20-week LFD mice (n = 5); open dots represent the 20-week HFD mice (n = 5), lines represent grand mean values. *** Indicate significant differences with p < 0.001 obtained with two-way ANOVA
Gene expression patterns of Hoxd8 and Hoxd9 do not correlate with expression patterns of markers for the different fiber types
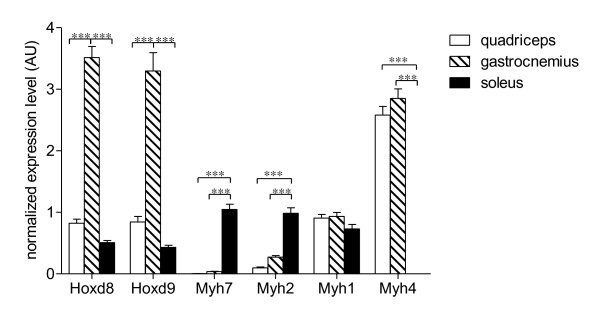
We found that markers for the oxidative fiber types as well as members of the Hoxd gene cluster were higher expressed in the gastrocnemius than in the quadriceps. To find out if these Hoxd genes play a role in specifying more oxidative muscle types we compared genes expression levels of Hoxd8, Hoxd9 and markers for different fiber types (type I: Myh7; type IIa: Myh2; type IIx: Myh1 and type IIb: Myh4) in quadriceps, gastrocnemius and soleus muscle of 20-week LFD mice and 20-week HFD mice. Expression levels of markers for the different fiber types were not affected by diet. Whereas Hoxd8 and Hoxd9 showed increased expression in the gastrocnemius as compared to quadriceps and soleus, expression levels of Myh7 was significantly higher in the soleus than in the quadriceps and gastrocnemius, respectively. Also the Myh2 expression level was higher in the soleus than in the quadriceps and gastrocnemius. For both Myh7 and Myh2 no differences between the quadriceps and the gastrocnemius were observed. Myh1 gene expression levels were comparable between the three muscle types. Finally, the expression level of the Myh4 gene was significantly lower in the soleus than in the quadriceps and gastrocnemius, respectively. Myh4 gene expression levels were similar in the quadriceps and gastrocnemius (Fig. 4). Altogether, the expression patterns of Hoxd8 and Hoxd9 did not correspond with the expression patterns of one of the markers for the different fiber types.
Figure 4.
Expression of Hoxd8, Hoxd9, Myh7, Myh2, Myh1 and Myh4 in quadriceps, gastrocnemius and soleus. Gene expression levels of Hoxd8, Hoxd9, Myh7, Myh2, Myh1 and Myh4 in quadriceps, gastrocnemius and soleus of 20-week LFD and HFD mice. The genes Myh7, Myh2, Myh1 and Myh4 are markers for type I, IIa, IIx and IIb fiber types, respectively. White bars, dashed bars and black bars represent quadriceps, gastrocnemius and soleus, respectively. Bars represent grand mean values of 20-week LFD mice and 20-week HFD mice. *** Indicate significant differences with p < 0.001 obtained with two-way ANOVA
Discussion
In the present study we searched for novel genes that are involved in specifying muscle types. Additionally, we studied the diet-sensitivity of differentially expressed genes. By comparing the gastrocnemius with the quadriceps we identified 162 differentially expressed genes corresponding to a variety of biological processes. Especially processes involved in cell cycle, contraction, development, differentiation, morphogenesis and remodeling were differentially regulated in the gastrocnemius vs. the quadriceps. Both the gastrocnemius and the quadriceps are muscle groups that consist of predominantly type II fibers. In the present study we found that Myh7, Tnnc1, Tnni1 and Tnnt1, which are all markers for the more oxidative fiber types, are strongly up-regulated in the gastrocnemius as compared to the quadriceps. Western blotting showed that protein levels of myosin heavy chain, slow fiber type protein (marker for the oxidative type I fibers) were similar between the gastrocnemius and the quadriceps, whereas the protein level of Myh2 (marker for the oxidoglycolytic type IIA fibers) was higher in the gastrocnemius than in the quadriceps (data not shown). Therefore, we propose that in this study the gastrocnemius had a more oxidative character than the quadriceps.
Recently, we showed that a 4-week HFD intervention results in increased gene expression and protein levels of markers for the oxidative fiber types in the quadriceps of mice. These findings suggested that the skeletal muscle can adapt to an increased lipid load by inducing a switch to a more oxidative phenotype [14]. Although the adult skeletal muscle is known to have the capacity to adapt to functional demands including exercise, hormones, innervation and mechanical loading [15,16], a link between HFD and a switch of fiber type was not yet described. However, in the present study we showed that an 8-week HFD has none or only minor effects on expression levels of genes that play a role in specifying different muscle types including the markers for oxidative fiber types. Therefore we speculate that the up-regulation of markers for the oxidative fiber types is an early response of the skeletal muscle to the HFD which is not maintained in the long run.
Among the strongest regulated genes we found five genes (Dkk3, Hoxd8, Hoxd9, Hoxd10 and Tbx1) which are especially known for their critical role during embryogenesis. The skeletal muscle is a dynamic tissue containing different types of stem cells such as satellite cells, side-population (SP) cells and muscle-derived stem cells (MDSC) [17,18]. To exclude the possibility that expression of these five embryogenesis-related genes originated from other cell types than myocytes we analyzed the expression of genes that specify satellite cells, SP cells and MDSC (satellite cells, pax7, myf5, cd34 and c-met; SP cells, sca1, sdc4 and pax3; MDSC, cd34 and Blc2 [17,18]). We found that only three genes (cd34, sca1 and sdc4) were detectably expressed (microarray signal intensity > 20) in the skeletal muscle of these mice. However, none of these detectably expressed markers differed more than 1.3-fold between the gastrocnemius and the quadriceps. Altogether, we cannot exclude the presence of cells other than myocytes, however, it is most unlikely that these cells have contributed to the different expression levels of Dkk3, Hoxd8, Hoxd9, Hoxd10 and Tbx1.
Two genes, Dkk3 and Tbx1, were expressed at a lower level in the gastrocnemius than in the quadriceps. Extrapolating our data to the soleus we found that Dkk3 expression in the soleus was lower than in the quadriceps or gastrocnemius. Interestingly, Tbx1 expression levels in the soleus were lower than in the quadriceps, but higher than in the gastrocnemius. During embryogenesis Dkk3 and Tbx1 are expressed in a variety of organs including the heart and limb buds [19,20]. Whereas the function of Dkk3 is still poorly understood [21], Tbx1 is well-studied especially in relation to heart development [20]. Recently it was shown that Tbx1 is also involved in development of the limb buds by regulating the number of myocytes [22]. Since further research revealed that Tbx1 is not necessary for skeletal muscle specification, differentiation, patterning or activation of the embryonic myogenic program, this is the only described function of Tbx1 in relation to embryonic muscle development [23]. Both Dkk3 and Tbx1 are reported to be detectably expressed in the adult skeletal muscle [19,24], but until now nothing is known about their possible function. We speculate that the relatively high expression levels of Dkk3 and Tbx1 in the quadriceps is involved in maintaining identity of this muscle type.
Three genes of the Hoxd gene cluster (Hoxd8, Hoxd9 and Hoxd10) were up-regulated in the gastrocnemius as compared with the quadriceps. The Hox gene family consists of at least 39 Hox genes organized in four gene clusters (A, B, C and D) which are subdivided into parallel groups numbered 1 to 13. The major function of Hox genes is the regulation of the formation of anterior-posterior patterning during embryonic development [25]. Different groups have studied the expression levels of the Hox gene family in adult tissues of human origin. They found that each tissue displays a unique combination of detectable Hox gene expression levels which is altered when normal cells changes to malignant cancer cells [26-28]. Thus, in addition to their critical role in embryonic development, Hox genes play an important role in adult cells by controlling critical processes like cellular identity and differentiation [29]. Houghton et al. [30]. have studied expression patterns of the Hox gene family in skeletal muscle tissue of adult mice. In line with our observations they reported that the Hoxa10, Hoxc6, Hoxc9 and Hoxc10 genes were detectably expressed. Furthermore, they could not detect Hoxd11 which also is consistent with our findings. Other members of the Hoxd gene cluster were not studied [30]. Thus to our knowledge, we are the first to show that Hoxd8, Hoxd9 and Hoxd10 are detectably expressed in adult murine muscle tissue, with highly increased expression levels in the gastrocnemius as compared with the quadriceps.
Together with the higher Hoxd8 and Hoxd9 gene expression levels in the gastrocnemius we found increased gene expression levels of markers for the more oxidative fiber types. Therefore we hypothesized that Hoxd8 and Hoxd9 might be involved in the determination of more oxidative muscle types. However, when we extrapolated our data to the soleus muscle we could not find a corresponding expression pattern between Hoxd8, Hoxd9 and Myh7 (marker for type I fibers) and Tnni1 (marker for type I fibers; data not shown). In fact, Myh7 and Tnni1 were highly expressed in the soleus, whereas Hoxd8 and Hoxd9 expression in the soleus was as low as in the quadriceps. Also the expression patterns of Myh2, Myh1 and Myh4, which are markers for type IIA, IIX and IIB fibers, respectively, did not show any resemblance with the expression pattern of Hoxd8 and Hoxd9. Although we did not find any resemblances between expression patterns of Hoxd8, Hoxd9 and any of the fiber type-specific markers we could distinguish the gastrocnemius, quadriceps and soleus from each other. Altogether, we suggest that the high expression levels of the Hoxd8, Hoxd9 and possibly Hoxd10 genes are a specific characteristic of the molecular profile of the gastrocnemius and are not related to an oxidative phenotype.
Conclusion
We found that especially genes that are markers for oxidative fiber types and genes known to be important for embryogenesis were differentially expressed in the gastrocnemius vs. the quadriceps. The expression levels of these genes were not influenced by diet. Furthermore, expression of Hoxd8 and Hoxd9 was low and expression of Dkk3 and Tbx1 was high in the quadriceps. In the gastrocnemius we observed high expression of Hoxd8 and Hoxd9 and low expression of Dkk3 and Tbx1. Finally, in the soleus we found low expression of Hoxd8, Hoxd9 and Dkk3, but intermediate expression of Tbx1. Comparing gastrocnemius, quadriceps and soleus, the gene expression pattern of Hoxd8 and Hoxd9 did not correspond to the gene expression pattern of any of the fiber type-specific markers. Therefore, we conclude that the highly expressed embryonic genes Dkk3, Hoxd8, Hoxd9 and Tbx1 are involved in identifying muscle types in a diet-independent and fiber type-unrelated manner.
Methods
Mice and diet
Male C57BL/6J mice were obtained from Harlan (Horst, The Netherlands). At 9 weeks of age mice were switched to a run-in diet consisting of an LFD (10 kcal% fat) for 3 weeks. Following the run-in period mice were randomly assigned to the LFD or the HFD (45 kcal% fat). Both diets contained fat in the form of palm oil (based on D12450B and D12451; Research Diet Services, Wijk bij Duurstede, the Netherlands) as described previously [31].
Study 1
After 8 weeks of diet intervention, mice (n = 10 per diet) were fasted for 6 hours, anesthetized with a mixture of isofluorane (1.5%), nitrous oxide (70%) and oxygen (30%) and killed by cervical dislocation. Gastrocnemius and quadriceps muscles were dissected, snap-frozen in liquid nitrogen and stored at -80°C until further analysis.
Study 2
After 20 weeks of diet intervention, mice (n = 5 per diet) were fasted for 6 hour, anaesthetized and killed as described. Quadriceps, gastrocnemius and soleus muscles were dissected, snap-frozen in liquid nitrogen and stored at -80°C until further analysis.
Both animal studies were approved by the Local Committee for Care and Use of Laboratory Animals at Wageningen University.
RNA isolation
Mouse total RNA was isolated from gastrocnemius, quadriceps and soleus muscles by mechanically homogenization in Trizol reagent (Invitrogen, Breda, the Netherlands). RNA was purified with the RNeasy Mini Kit (Qiagen, Venlo, the Netherlands) and contaminating genomic DNA was removed with the RNase-free DNase set (Qiagen). RNA quantity and purity was measured with the ND-1000 spectrophotometer (Isogen Life Science B.V., IJsselstein, the Netherlands). An A260/A280 ratio between 2.04 and 2.16 was found for all RNA samples. RNA integrity was checked on an Agilent 2100 BioAnalyzer (Agilent Technologies, Amsterdam, The Netherlands) using nanochips according to the manufacturer's instructions. All RNA samples showed an RIN value between 7.5 and 9.0.
Affymetrix microarray analysis
RNA isolated from the gastrocnemius and quadriceps of mice fed an 8-week LFD or HFD was used for performing Affymetrix GeneChip® Mouse Genome 430 2.0 arrays as described previously [14]. Array images were processed using packages from the Bioconductor project [32] and probe sets were redefined according to Dai et al. [33]. In this method probes are annotated using up-to-date databases and assigned to unique gene identifiers, in this case Entrez ID's, instead of the 'classic' GeneChip probe sets. This results in a less ambiguous and more accurate annotation. Arrays were normalized using quantile normalization, and expression estimates were calculated using GC-RMA, implementing the empirical Bayes estimate for non-specific binding [34]. Differentially expressed probe sets were identified using linear models, applying moderated t-statistics that implement empirical Bayes regularization of standard errors [35]. Comparisons were made between the gastrocnemius and quadriceps muscles using the fold change threshold method. In this method, most false-positives are caused by genes with low signal intensity and/or absent call (~50% of all genes). Thus, filtering out this constant noise before performing data analysis will improve the power to discriminate true changes from noise in the fold-change threshold method [36]. We manually set the signal threshold at 20, across all arrays, filtering out 60% of the genes. Probe sets that satisfied the criteria of a fold change > 1.3 and an FDR < 0.05, correcting for multiple-testing [37], were considered as differentially expressed.
To relate changes in gene expression to functional changes two complementary methods were used. The first method is based on overrepresentation of Gene Ontology (GO) terms, which uses a gene score resampling (GSR) method [38]. Full resampling was run with 200,000 iterations. Only classes with a false discovery rate (FDR) < 0.001 and with minimal 8 and maximal 125 genes were taken into account. The second method, gene set enrichment analysis (GSEA), is focused on predefined gene sets, that is, groups of genes that share biological function, chromosomal location or regulation [39]. The 'functional catalogue' constructed by Subramanian et al. was modified to contain only 505 well-defined murine, biochemical, metabolic and signal pathways compiled from the following publicly available, curated databases: BioCarta (BioCarta. BioCarta, 2005, p. http://www.biocarta.com.), GenMAPP [40], Kyoto Encyclopedia of Genes and Genomes (KEGG) [41], Sigma-Aldrich pathways (Sigma-Aldrich. Sigma-Aldrich Metabolic and Cell Signaling pathways, 2005.) and Signal Transduction Knowledge Environment (STKE; Signal Transduction Knowledge Environment. In: http://stke.sciencemag.org/. 2005.). The analysis was run using 1000 permutations per gene set. Gene sets with a FDR < 0.05 were considered as significantly regulated. The advantage is that both methods are unbiased and a score is computed based on all genes in a GO term or gene set.
The diet-sensitivity of differentially expressed genes, overrepresented GO classes and regulated gene sets was studied by comparing the gastrocnemius with the quadriceps under HFD conditions. Also in this comparison we used the criteria of a fold change > 1.3 and an FDR < 0.05. A heatmap of log-transformed microarray signal intensities was generated by using GeneMaths XT software (Sint-Martens-Latem, Belgium). Array data have been submitted to the Gene Expression Omnibus, accession number GSE18127.
Quantitative real-time PCR for RNA
To validate microarray-detected changes as well as extrapolating microarray-detected changes to the gastrocnemius, quadriceps and soleus of 20-week LFD mice and HFD mice quantitative real-time PCR (qPCR) was performed using individual cDNA samples. RNA (1 μg in 20 μl) was reverse transcribed using the iScript cDNA synthesis kit (Biorad, Veenendaal, the Netherlands) containing RNase H+ iScript reverse transcriptase, a premixed RNase inhibitor to prevent indiscriminate degradation of RNA template, and a unique blend of oligo(dT) and random primers. cDNA was synthesized using a 3-step program (5 minutes (min) at 25°C, 30 min at 42°C and 5 min at 85°C). Primer sequences were retrieved from the online PrimerBank database [42], or otherwise designed using Beacon Designer5 (Biorad). Primers were tested for specificity by BLAST analysis. The qPCR reactions were performed in a volume of 25 μl containing 12.5 ng cDNA, 1× IQ SYBR Green Supermix (100 mM KCl, 40 mM Tris-HCI, 6 mM MgCl2, 0.4 mM of each dNTP, 50 units/ml iTaq DNA polymerase, SYBR Green I and 20 nM fluoresein; Biorad) and 400 nM of gene-specific forward and reverse primers (Biolegio, Nijmegen, the Netherlands). cDNA was amplified using a 2-step program (40 cycles of 10 seconds (sec) at 95°C and 45 sec at 60°C) with a MyiQ system (Biorad). Specificity of amplification was verified by melt curve analysis and evaluation of efficiency of PCR amplification. PCR reactions were performed in duplicate and gene expression levels were determined using iQ5 software (Biorad) using a Δ-Cq relative quantification model with PCR efficiency correction and multiple reference gene normalization [43,44]. As the Canx, Hprt1 and Arbp genes were identified as most stably expressed reference genes, we used the geometric mean of these three reference genes as normalization factor. Supplement 4 shows all relevant primer information.
Statistical analyses
All data are expressed as means ± SE. Microarray data were analyzed as described above. All other statistical analyses were performed using Prism software (GraphPad Software, San Diego, CA, USA). Two-way ANOVA was used to test gene expression levels of Dkk3, Hoxd8, Hoxd9, Myh1, Myh2, Myh4, Myh7 and Tbx1 between muscle types and diets. When significant differences were found, a Bonferroni post hoc test was used to determine the exact location of the differences. A p value ≤ 0.05 was considered as statistically significant.
Authors' contributions
JdW, ES and EM participated in the design and supervision of the study. JdW and MH performed the RNA isolations and qPCRs. PDG and MB assessed the quality control of the microarrays and provided support for microarray analysis. JDW performed microarray analysis and drafted the manuscript. EM and ES provided valuable feedback on the initial draft. All authors read and approved the final manuscript
Supplementary Material
List of differentially expressed genes. Additional file 1 gives the differentially expressed genes in the gastrocnemius as compared to the quadriceps from 10 LFD mice. To find these differentially expressed genes we used the criteria of an average signal intensity > 20 over all arrays, a fold change > 1.3 and an FDR < 0.05.
Overview of qPCR analyses for 13 genes validating the microarray results. Additional file 2 gives an overview of the microarray signal intensity, relative expression (qPCR), normalized expression (qPCR) and corresponding fold changes of 13 genes.
Differentially expressed genes, overrepresented GO classes (ErmineJ) and regulated gene sets (GSEA) between gastrocnemius and quadriceps in either LFD or HFD conditions. Additional file 3 gives an overview of the differentially expressed genes, overrepresented GO classes (ErmineJ) and regulated gene sets (GSEA) in the gastrocnemius as compared to the quadriceps in HFD and LFD conditions.
Primer information. Additional file 4 gives the sequences, start, end and amplicon length of the used primer pairs.
Contributor Information
Janneke de Wilde, Email: j.dewilde@hb.unimaas.nl.
Martijn FM Hulshof, Email: Martijn.Hulshof@HB.unimaas.nl.
Mark V Boekschoten, Email: Mark.Boekschoten@wur.nl.
Philip de Groot, Email: Philip.deGroot@wur.nl.
Egbert Smit, Email: E.smit@HB.unimaas.nl.
Edwin CM Mariman, Email: E.Mariman@HB.unimaas.nl.
Acknowledgements
We thank Freek Bouwman, Mechteld Grootte Bromhaar, Shohreh Keshtkar Ghiasabadi and Bert Weijers for excellent technical assistance. This study was funded by the Top Institute Food and Nutrition, with financial support by the Dutch government.
References
- Liang H, Ward WF. PGC-1alpha: a key regulator of energy metabolism. Advances in physiology education. 2006;30(4):145–151. doi: 10.1152/advan.00052.2006. [DOI] [PubMed] [Google Scholar]
- Scott W, Stevens J, Binder-Macleod SA. Human skeletal muscle fiber type classifications. Physical therapy. 2001;81(11):1810–1816. [PubMed] [Google Scholar]
- Berchtold MW, Brinkmeier H, Muntener M. Calcium ion in skeletal muscle: its crucial role for muscle function, plasticity, and disease. Physiological reviews. 2000;80(3):1215–1265. doi: 10.1152/physrev.2000.80.3.1215. [DOI] [PubMed] [Google Scholar]
- Booth FW, Thomason DB. Molecular and cellular adaptation of muscle in response to exercise: perspectives of various models. Physiological reviews. 1991;71(2):541–585. doi: 10.1152/physrev.1991.71.2.541. [DOI] [PubMed] [Google Scholar]
- Chanseaume E, Malpuech-Brugere C, Patrac V, Bielicki G, Rousset P, Couturier K, Salles J, Renou JP, Boirie Y, Morio B. Diets high in sugar, fat, and energy induce muscle type-specific adaptations in mitochondrial functions in rats. The Journal of nutrition. 2006;136(8):2194–2200. doi: 10.1093/jn/136.8.2194. [DOI] [PubMed] [Google Scholar]
- Wang YX, Zhang CL, Yu RT, Cho HK, Nelson MC, Bayuga-Ocampo CR, Ham J, Kang H, Evans RM. Regulation of muscle fiber type and running endurance by PPARdelta. PLoS biology. 2004;2(10):e294. doi: 10.1371/journal.pbio.0020294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kho AT, Kang PB, Kohane IS, Kunkel LM. Transcriptome-scale similarities between mouse and human skeletal muscles with normal and myopathic phenotypes. BMC musculoskeletal disorders. 2006;7:23. doi: 10.1186/1471-2474-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naya FJ, Mercer B, Shelton J, Richardson JA, Williams RS, Olson EN. Stimulation of slow skeletal muscle fiber gene expression by calcineurin in vivo. The Journal of biological chemistry. 2000;275(7):4545–4548. doi: 10.1074/jbc.275.7.4545. [DOI] [PubMed] [Google Scholar]
- Talmadge RJ, Otis JS, Rittler MR, Garcia ND, Spencer SR, Lees SJ, Naya FJ. Calcineurin activation influences muscle phenotype in a muscle-specific fashion. BMC cell biology. 2004;5:28. doi: 10.1186/1471-2121-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN. et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418(6899):797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- Mortensen OH, Frandsen L, Schjerling P, Nishimura E, Grunnet N. PGC-1alpha and PGC-1beta have both similar and distinct effects on myofiber switching toward an oxidative phenotype. Am J Physiol Endocrinol Metab. 2006;291(4):E807–816. doi: 10.1152/ajpendo.00591.2005. [DOI] [PubMed] [Google Scholar]
- Luquet S, Lopez-Soriano J, Holst D, Fredenrich A, Melki J, Rassoulzadegan M, Grimaldi PA. Peroxisome proliferator-activated receptor delta controls muscle development and oxidative capability. Faseb J. 2003;17(15):2299–2301. doi: 10.1096/fj.03-0269fje. [DOI] [PubMed] [Google Scholar]
- Hakimi P, Yang J, Casadesus G, Massillon D, Tolentino-Silva F, Nye CK, Cabrera ME, Hagen DR, Utter CB, Baghdy Y. et al. Overexpression of the Cytosolic Form of Phosphoenolpyruvate Carboxykinase (GTP) in Skeletal Muscle Repatterns Energy Metabolism in the Mouse. Journal of Biological Chemistry. 2007;282(45):32844–32855. doi: 10.1074/jbc.M706127200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wilde J, Mohren R, Berg S van den, Boekschoten M, Dijk KW, de Groot P, Muller M, Mariman E, Smit E. Short-term high fat-feeding results in morphological and metabolic adaptations in the skeletal muscle of C57BL/6J mice. Physiol Genomics. 2008;32(3):360–369. doi: 10.1152/physiolgenomics.00219.2007. [DOI] [PubMed] [Google Scholar]
- Pette D, Staron RS. Myosin isoforms, muscle fiber types, and transitions. Microscopy research and technique. 2000;50(6):500–509. doi: 10.1002/1097-0029(20000915)50:6<500::AID-JEMT7>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Pette D, Staron RS. Transitions of muscle fiber phenotypic profiles. Histochemistry and cell biology. 2001;115(5):359–372. doi: 10.1007/s004180100268. [DOI] [PubMed] [Google Scholar]
- Otto A, Collins-Hooper H, Patel K. The origin, molecular regulation and therapeutic potential of myogenic stem cell populations. Journal of anatomy. 2009;215(5):477–497. doi: 10.1111/j.1469-7580.2009.01138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figeac N, Daczewska M, Marcelle C, Jagla K. Muscle stem cells and model systems for their investigation. Development dynamics. 2007;236(12):3332–3342. doi: 10.1002/dvdy.21345. [DOI] [PubMed] [Google Scholar]
- Monaghan AP, Kioschis P, Wu W, Zuniga A, Bock D, Poustka A, Delius H, Niehrs C. Dickkopf genes are co-ordinately expressed in mesodermal lineages. Mech Dev. 1999;87(1-2):45–56. doi: 10.1016/S0925-4773(99)00138-0. [DOI] [PubMed] [Google Scholar]
- Stennard FA, Harvey RP. T-box transcription factors and their roles in regulatory hierarchies in the developing heart. Development (Cambridge, England) 2005;132(22):4897–4910. doi: 10.1242/dev.02099. [DOI] [PubMed] [Google Scholar]
- Niehrs C. Function and biological roles of the Dickkopf family of Wnt modulators. Oncogene. 2006;25(57):7469–7481. doi: 10.1038/sj.onc.1210054. [DOI] [PubMed] [Google Scholar]
- Dastjerdi A, Robson L, Walker R, Hadley J, Zhang Z, Rodriguez-Niedenfuhr M, Ataliotis P, Baldini A, Scambler P, Francis-West P. Tbx1 regulation of myogenic differentiation in the limb and cranial mesoderm. Dev Dyn. 2007;236(2):353–363. doi: 10.1002/dvdy.21010. [DOI] [PubMed] [Google Scholar]
- Grifone R, Jarry T, Dandonneau M, Grenier J, Duprez D, Kelly RG. Properties of branchiomeric and somite-derived muscle development in Tbx1 mutant embryos. Dev Dyn. 2008;237(10):3071–3078. doi: 10.1002/dvdy.21718. [DOI] [PubMed] [Google Scholar]
- Chieffo C, Garvey N, Gong W, Roe B, Zhang G, Silver L, Emanuel BS, Budarf ML. Isolation and characterization of a gene from the DiGeorge chromosomal region homologous to the mouse Tbx1 gene. Genomics. 1997;43(3):267–277. doi: 10.1006/geno.1997.4829. [DOI] [PubMed] [Google Scholar]
- Svingen T, Tonissen KF. Hox transcription factors and their elusive mammalian gene targets. Heredity. 2006;97(2):88–96. doi: 10.1038/sj.hdy.6800847. [DOI] [PubMed] [Google Scholar]
- Calvo R, West J, Franklin W, Erickson P, Bemis L, Li E, Helfrich B, Bunn P, Roche J, Brambilla E. et al. Altered HOX and WNT7A expression in human lung cancer. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(23):12776–12781. doi: 10.1073/pnas.97.23.12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantile M, Pettinato G, Procino A, Feliciello I, Cindolo L, Cillo C. In vivo expression of the whole HOX gene network in human breast cancer. Eur J Cancer. 2003;39(2):257–264. doi: 10.1016/S0959-8049(02)00599-3. [DOI] [PubMed] [Google Scholar]
- Cillo C. HOX genes in human cancers. Invasion & metastasis. 1994;14(1-6):38–49. [PubMed] [Google Scholar]
- Cillo C, Cantile M, Faiella A, Boncinelli E. Homeobox genes in normal and malignant cells. Journal of cellular physiology. 2001;188(2):161–169. doi: 10.1002/jcp.1115. [DOI] [PubMed] [Google Scholar]
- Houghton L, Rosenthal N. Regulation of a muscle-specific transgene by persistent expression of Hox genes in postnatal murine limb muscle. Dev Dyn. 1999;216(4-5):385–397. doi: 10.1002/(SICI)1097-0177(199912)216:4/5<385::AID-DVDY7>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- de Wit NJ, Bosch-Vermeulen H, de Groot PJ, Hooiveld GJ, Bromhaar MM, Jansen J, Muller M, Meer R van der. The role of the small intestine in the development of dietary fat-induced obesity and insulin resistance in C57BL/6J mice. BMC medical genomics. 2008;1:14. doi: 10.1186/1755-8794-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J. et al. Bioconductor: open software development for computational biology and bioinformatics. Genome biology. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H. et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33(20):e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Irizarry R, Gentleman R, Martinez-Murillo F, Spencer F. A model-based background adjustment for oligonucleotide expression arrays. Journal of the American Statistical Association. 2004;99(468):909–917. doi: 10.1198/016214504000000683. [DOI] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- Xinmin L, Kim J, Zhou J, Gu W, Quigg R. Use of signal thresholds to determine significant changes in microarray data analyses. Genetics and Molecular Biology. 2005;28:191–200. doi: 10.1590/S1415-47572005000200002. [DOI] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HK, Braynen W, Keshav K, Pavlidis P. ErmineJ: tool for functional analysis of gene expression data sets. BMC Bioinformatics. 2005;6:269. doi: 10.1186/1471-2105-6-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlquist KD, Salomonis N, Vranizan K, Lawlor SC, Conklin BR. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat Genet. 2002;31(1):19–20. doi: 10.1038/ng0502-19. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Seed B. A PCR primer bank for quantitative gene expression analysis. 2003. p. e154. [DOI] [PMC free article] [PubMed]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome biology. 2002;3(7):RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome biology. 2007;8(2):R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of differentially expressed genes. Additional file 1 gives the differentially expressed genes in the gastrocnemius as compared to the quadriceps from 10 LFD mice. To find these differentially expressed genes we used the criteria of an average signal intensity > 20 over all arrays, a fold change > 1.3 and an FDR < 0.05.
Overview of qPCR analyses for 13 genes validating the microarray results. Additional file 2 gives an overview of the microarray signal intensity, relative expression (qPCR), normalized expression (qPCR) and corresponding fold changes of 13 genes.
Differentially expressed genes, overrepresented GO classes (ErmineJ) and regulated gene sets (GSEA) between gastrocnemius and quadriceps in either LFD or HFD conditions. Additional file 3 gives an overview of the differentially expressed genes, overrepresented GO classes (ErmineJ) and regulated gene sets (GSEA) in the gastrocnemius as compared to the quadriceps in HFD and LFD conditions.
Primer information. Additional file 4 gives the sequences, start, end and amplicon length of the used primer pairs.