Abstract
It is unclear whether the mammalian Golgi apparatus can form de novo from the ER or whether it requires a preassembled Golgi matrix. As a test, we assayed Golgi reassembly after forced redistribution of Golgi matrix proteins into the ER. Two conditions were used. In one, ER redistribution was achieved using a combination of brefeldin A (BFA) to cause Golgi collapse and H89 to block ER export. Unlike brefeldin A alone, which leaves matrix proteins in relatively large remnant structures outside the ER, the addition of H89 to BFA-treated cells caused ER accumulation of all Golgi markers tested. In the other, clofibrate treatment induced ER redistribution of matrix and nonmatrix proteins. Significantly, Golgi reassembly after either treatment was robust, implying that the Golgi has the capacity to form de novo from the ER. Furthermore, matrix proteins reemerged from the ER with faster ER exit rates. This, together with the sensitivity of BFA remnants to ER export blockade, suggests that presence of matrix proteins in BFA remnants is due to cycling via the ER and preferential ER export rather than their stable assembly in a matrix outside the ER. In summary, the Golgi apparatus appears capable of efficient self-assembly.
INTRODUCTION
The Golgi apparatus is the central processing and sorting station of the secretory pathway. The structural organization of the mammalian Golgi is complex: hundreds of stacks, each containing several cis-to-trans cisternae, are laterally linked to form a ribbon-like membrane system next to the microtubule organizing center. Despite its complex structure, the Golgi undergoes substantial and reversible structural transformations under a variety of conditions, most notably at mitosis when the organelle extensively vesiculates.
Most other examples of Golgi disassembly come from experimentally induced perturbations. The extent and nature of disassembly varies. Presumably, these experimentally induced perturbations reflect a constant flux through the organelle mediated in large part by vesicle formation and vesicle docking reactions that are regulated to allow Golgi growth and maintenance. Thus, perturbations that favor output over input cause dramatic Golgi disassembly. Despite their questionable physiological relevance, these perturbations and their consequences tell us much about the capacity of the organelle for dynamic behavior. For instance, the fact that the Golgi apparatus undergoes disassembly followed by efficient reassembly raises the fundamental question of whether it requires or uses a preexisting template for reassembly (Seemann et al., 2000, 2002).
Evidence favoring a role for a template derives from the discovery of a detergent insoluble extract of purified Golgi membranes, which yielded a pattern in electron microscope images reminiscent of stacked cisternae (Slusarewicz et al., 1994). The material was called the Golgi “matrix,” and a component identified in the matrix, the Golgi protein GM130, was termed a Golgi matrix protein (Nakamura et al., 1995). GM130 is a peripheral membrane protein that interacts with lipid anchored GRASP65 (Barr et al., 1997). Significantly, GM130 and GRASP65, together with the Golgi proteins giantin, GRASP55 and golgin-45, are collectively being called matrix proteins because they exhibit a behavior distinct from other Golgi proteins during treatment of cells with brefeldin A (BFA). BFA treatment, which blocks activation of the GTPase Arf1 (Peyroche et al., 1999), induces redistribution of most Golgi-localized proteins to the ER, but matrix proteins end up in membranes called BFA remnants that are distinct from the ER (Seemann et al., 2000). Because BFA-induced Golgi disassembly is reversed upon drug washout, this finding is consistent with the view that these proteins remain in intact assemblies that then mediate reassembly upon drug washout. In fact, each of these putative matrix proteins is required for Golgi stacking as measured using an in vitro assay (Barr et al., 1997; Shorter et al., 1999; Shorter and Warren, 1999).
Paradoxically, evidence in support of the contrasting view—that the Golgi has and uses a capacity to self-assemble—is also based in large part on work with BFA. Before the full realization that a subset of Golgi proteins accumulate outside the ER during BFA treatment, the reversibility of BFA-induced Golgi collapse suggested that the Golgi apparatus has the capacity to undergo de novo biogenesis from the ER. Although the BFA remnants undermine this conclusion, several lines of evidence suggest that many, if not all Golgi proteins, continuously cycle through the ER. Golgi proteins redistribute via the ER to peripheral mini-stacks in response to microtubule depolymerization (Cole et al., 1996; Storrie et al., 1998). Golgi proteins redistribute to the ER in response to ER-export blockade (Miles et al., 2001; Ward et al., 2001). And, a Golgi-localized chimeric form of tsO45 VSVG redistributes to the ER in response to temperature shifts that alter its folding (Cole et al., 1998). Further, BFA remnants costain with ERGIC-53, which is a marker of the intermediate compartment, and BFA remnants localize adjacent to ER export sites (Nakamura et al., 1995; Ward et al., 2001). Thus, GM130 and partners may reach their position in BFA remnants by cycling through the ER rather than by staying intact in a matrix (Cole et al., 1998; Ward et al., 2001). Finally, in the yeast Pichia pastoris, a GFP-labeled Golgi marker appears, seemingly out of nowhere, adjacent to ER export sites (Bevis et al., 2002). This suggests that it cycles via the ER, is concentrated during ER export, and then participates in de novo formation of the Golgi.
A major shortcoming of the evidence in support of de novo biogenesis is that the Golgi apparatus has not been demonstrated to reform from the ER. In fact, the one published test of this capacity concluded that the Golgi apparatus requires matrix components maintained in an assembled state outside the ER (Pelletier et al., 2000). Herein, we used a reversible ER export block to cause redistribution of both matrix and nonmatrix markers into the ER before a test for reassembly. Our results indicate that the Golgi has the capacity to reform from the ER in the absence of a preassembled Golgi matrix.
MATERIALS AND METHODS
Cell Culture and Staining
Growth medium for NRK cells (DMEM, Life Technologies, Grand Island, NY) and HeLa cells (Minimal essential medium, Sigma-Aldrich, St. Louis, MO) contained 10% fetal bovine serum and 100 IU/ml penicillin-streptomycin. Where indicated, the growth medium was supplemented with 10 μg/ml BFA (Sigma), 100 μM H89 (Toronto Research, Toronto, ON), or 1 mM clofibrate (Sigma). For immunofluorescence, cells were grown on 12-mm glass coverslips, treated where indicated, and either fixed in methanol (-20°C for 10 min) or 3% paraformaldehyde (20 min at room temperature). Paraformaldehyde-fixed cells were stained as previously described (Jesch and Linstedt, 1998). After three washes with PBS, methanol-fixed cells were incubated for 30 min in blocking buffer (PBS containing 2.5% calf serum and 0.1% Tween-20), followed by consecutive 30-min incubations in primary and secondary antibodies diluted in blocking buffer. Each antibody incubation was followed by five washes with PBS. The antibodies and their dilutions were: mouse anti-giantin at 1:100 (Linstedt and Hauri, 1993); rabbit anti-giantin at 1:500 (Puthenveedu and Linstedt, 2001), rabbit anti-GM130 at 1:500 (Puthenveedu and Linstedt, 2001), rabbit anti-p115 at 1:500 (Puthenveedu and Linstedt, 2001); rabbit anti-GRASP65 at 1:2000 in NRK and 1:500 in HeLa (Sutterlin et al., 2002); rabbit anti-GRASP55 at 1:2000 (a gift from Dr. Vivek Malhotra); mouse anti-mannosidase II at 1:10,000 (Sigma); rabbit anti-Sec31 at 1:500 (Tang et al., 2000; a gift from Dr. B.L. Tang); rabbit anti-β-COP at 1:200 (Affinity BioReagents Inc., Golden, CO); fluorescein-labeled goat anti-mouse at 1:200 (Zymed, San Francisco, CA); rhodamine-labeled goat anti-rabbit at 1:200 (Zymed); and Cy5-labeled goat anti-mouse at 1:200 (Zymed).
Image Analysis
Microscopy was performed using either a conventional fluorescence microscope (Linstedt et al., 1997) or a Deltavision system (Applied Precision Inc., Issaquah, WA) essentially as previously described (Jesch et al., 2001). In the latter case either a 100×, 1.35 NA oil immersion objective yielding a pixel dimension of 0.0671 μm or a 60×, 1.40 NA lens with pixel size 1.114 μm was used. Images were deconvolved using SoftWorx (Applied Precision Inc.). For Sec31 quantification a fixed threshold (123) was applied to images acquired from control and drug-treated cells. The threshold level was set to recover fluorescence corresponding to bona fide ER export sites and remove fluorescence corresponding to the cytoplasmic pool of Sec31. The total fluorescence in objects was then determined for each cell. Electron microscopy was carried out as described (Linstedt et al., 1997) except that cells were fixed in PBS containing 2% glutaraldehyde for 60 min at room temperature and then after three PBS washes were directly transferred to 1% OsO4.
Density Gradient Analysis
Cells, 80% confluent in 15-cm dishes, were either incubated with growth medium, with 10 μg/ml BFA for 30 min followed by a 10-min treatment with 100 μM H89 or with 1 mM clofibrate for 10 min. After two washes with cold PBS, the cells were removed by scraping and collected at 1000 × g for 3 min. The cell pellet was washed in 10 ml 0.25 M sucrose, 1 mM EDTA, and 10 mM TEA, pH 7.4, resuspended and homogenized in 50 mM NaCl, 1 mM EDTA, and 10 mM TEA, pH 7.4, using a 25-gauge needle. After adjustment to 250 mM sucrose a postnuclear supernatant was collected at 1000 × g for 1 min. This was adjusted to 1.6 M sucrose (1.1 ml) and covered with three sucrose layers (1.1 ml each: 1.4 M, 1.2 M, 0.8 M), followed by centrifugation at 105,000 × g for 105 min in a SW50.1 rotor (Beckman), and 0.4 ml fractions were collected from the top (Nagata-Kuno et al., 1990). All steps were carried out at 0-4°C in the presence of protease inhibitors. Immunoblotting was as described (Linstedt and Hauri, 1993) and used rabbit antibodies against GM130 (1:5000) and calreticulin (Stressgen Biotechnologies, Victoria, BC; 1:5000). Detection was by enhanced chemiluminescence (Pierce, Rockford, IL) followed by densitometric scanning.
RESULTS
To test the capacity of the Golgi apparatus for de novo biogenesis from the ER we sought reversible treatments that cause complete collapse of the Golgi—including matrix components—into the ER. Two were identified, and similar results were obtained for each. The first involved a combined BFA and H89 treatment. As stated above, BFA induces redistribution of nonmatrix components to the ER and of matrix markers to remnant structures that costain for ERGIC-53 (Nakamura et al., 1995; Ward et al., 2001). The protein kinase inhibitor H89 blocks COPII recruitment and ER export, thereby causing collapse of ERGIC membranes into the ER (Aridor and Balch, 2000; Lee and Linstedt, 2000). Therefore, we expected the combined treatment to cause ER localization of both matrix and nonmatrix Golgi markers.
Before any treatment, the matrix marker GM130 and the enzyme mannosidase II were coincident in the Golgi complex (Figure 1, A and B). BFA treatment alone induced redistribution of GM130 to peripheral punctate structures, whereas mannosidase II became ER localized (Figure 1, C and D). Signifi-cantly, the GM130-positive peripheral punctate structures appeared to collapse into the ER in cells treated with BFA and H89 (our unpublished results) or BFA followed by H89 (Figure 1, E and F). This implies that GM130 cycles via the ER during the BFA treatment and becomes trapped in the ER upon the ER export blockade induced by H89. Note that the H89 treatment was effective in blocking ER export because GM130 did not move out of the ER upon BFA removal even after extended incubations in the continued presence of H89 (our unpublished results). ER localization was also observed under these conditions for matrix markers giantin and GRASP65, and the Golgi protein GPP130.
Figure 1.
ER localization of Golgi matrix markers after ER export blockade. NRK cells were left untreated (A and B), treated with BFA for 30 min (C and D), or treated sequentially with BFA for 30 min followed by H89 for 10 min (E and F). The paraformaldehyde-fixed cells were stained with GM130 (A, C, and E) and mannosidase II (B, D, and F) antibodies. Note that upon H89 treatment, GM130 staining collapsed from a punctate BFA remnant pattern to a dispersed ER pattern. Bar, 10 μm.
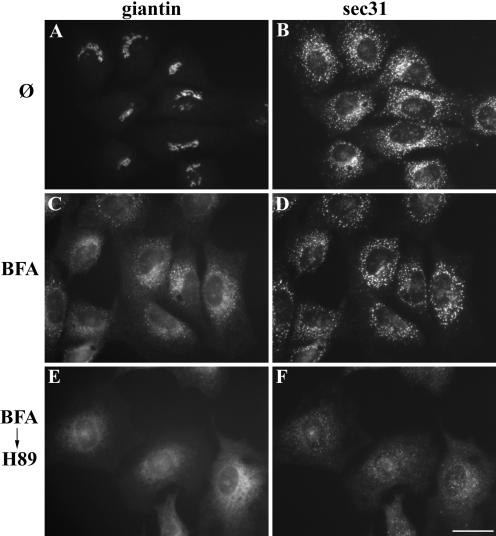
Furthermore, inhibition of COPII recruitment was confirmed by localizing the COPII component Sec31 (Tang et al., 2000) in cells subjected to the combined BFA and H89 treatment. In untreated cells, giantin was Golgi localized and Sec31 was localized to distributed ER exit sites (Figure 2, A and B). After BFA treatment, giantin was present in peripheral punctate structures that were in close apposition to the Sec31-positive ER export sites (Figure 2, C and D). Note that, as previously observed (Linstedt and Hauri, 1993) giantin was also present in the ER in BFA-treated cells. A quantitative analysis suggests that matrix markers giantin, GM130, GRASP65, and GRASP55 are partially ER localized after BFA treatment with giantin, showing the greatest fraction in the ER (Puri et al., 2004). In cells treated with BFA followed by H89, giantin collapsed into the ER and Sec31 was significantly lost from ER export sites (Figure 2, E and F). Quantification after thresholding to remove the contribution of the cytoplasmic Sec31 pool, revealed that the level of Sec31 staining at ER export sites was reduced by more than 95% under these conditions.
Figure 2.
Inhibition of COPII recruitment upon H89 treatment. NRK cells were left untreated (A and B), treated with BFA for 30 min (C and D), or treated sequentially with BFA for 30 min followed by H89 for 10 min (E and F). The methanol-fixed cells were stained with anti-giantin (A, C, and E) and anti-Sec31 (B, D, and F) antibodies. BFA treatment yielded giantin in Sec31 adjacent remnant structures that collapsed into the ER upon H89 addition. Bar, 10 μm.
To confirm ER localization of GM130 before washout, we used density gradient fractionation. In accordance with previous work (Jesch and Linstedt, 1998), Golgi markers including GM130 were recovered in a membrane peak at the top of the gradients that was well resolved from the ER membrane peak recovered at the bottom of the gradients and marked by calreticulin (Figure 3A). Significantly, analysis of cells that had been treated with BFA followed by H89 yielded a GM130 fractionation pattern that was consistent with complete GM130 redistribution to the ER. That is, GM130 was recovered in the ER fractions in relative amounts that closely coincided with those of calreticulin (Figure 3B).
Figure 3.
Cofractionation of GM130 with ER membranes. NRK cells were either untreated (A) or treated sequentially with BFA for 30 min followed by H89 for 10 min (B). The cells were then subjected to sucrose density gradient fractionation. Recovery of GM130 (□) and the ER marker calreticulin (♦) in each fraction was determined by immunoblotting. In untreated cells, Golgi membranes are recovered near the top (peak at fraction 3), ER membranes near the bottom (fraction 9), and soluble proteins remain at the bottom (fractions 10-11). After treatment, GM130 was recovered together with the calreticulin in the ER position.
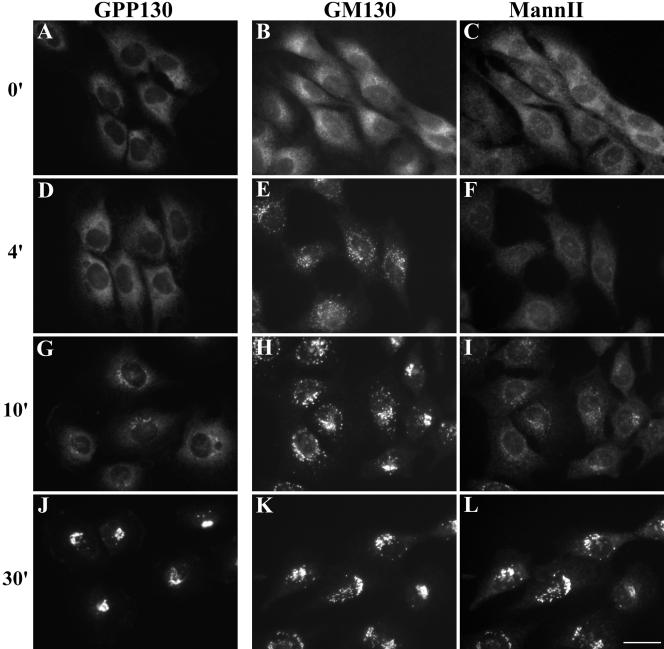
Having established the ER localization of matrix and nonmatrix Golgi markers, we tested for Golgi reassembly upon H89 washout. After a 30-min washout incubation, all Golgi markers that we tested (including GM130, giantin, GRASP65, GPP130, and mannosidase II) had emerged from the ER and reestablished normal Golgi patterns. Intermediates in Golgi biogenesis in cells stained for GPP130 or cells costained for GM130 and mannosidase II were revealed by analysis of time points after washout (Figure 4, A-L). As expected, each marker was first present in punctate structures likely to correspond to peripheral and centrally localized ERGIC membranes. The staining then increasingly exhibited loop patterns, which are typical of isolated Golgi stacks. Finally, the staining became restricted to juxta-nuclear regions and exhibited typical Golgi ribbon patterns.
Figure 4.
Reemergence of the Golgi from the ER. NRK cells were sequentially treated with BFA for 30 min followed by H89 for 10 min and then analyzed after the indicated times of H89 washout. The methanol-fixed cells were stained with anti-GPP130 (A, D, G, and J) or costained with anti-GM130 (B, E, H, and K) and anti-mannosidase II (C, F, I, and L) antibodies. Bar, 10 μm.
Strikingly, the kinetics of emergence from the ER differed for each marker. Of those tested, GM130 was fastest exhibiting a t1/2 of 6 min and GPP130 was slowest exhibiting a t1/2 of 17 min (Figure 5). Distinct ER export rates for Golgi proteins provides an explanation for their presence or absence in BFA remnants. That is, in BFA-treated cells, all Golgi proteins may cycle to and from BFA remnants to some extent. Those with rapid ER exit would then exhibit significant staining in remnants, whereas those with slow exit would not. Consistent with this, giantin—which yielded an intermediate ER export rate— was associated with BFA remnants to an extent that was weak relative to GM130, yet strong relative to GPP130 (Figure 2 and our unpublished results).
Figure 5.
Kinetics of reemergence of Golgi markers from the ER. Object fluorescence, which corresponded to GM130 (□) or GPP130 (⋄) fluorescence in ERGIC and Golgi structures, was determined at various time points of washout. Objects were defined by using constant thresholds for each marker on background subtracted images. This effectively removed fluorescence that was dispersed in the ER. Data points are averages of all cells present in three fields expressed as percent of total fluorescence at 30 min.
To confirm that the reassembled Golgi ribbon visualized by fluorescence microscopy corresponded to a stacked structure, we carried out electron microscopy. As expected, untreated cells yielded readily identifiable Golgi stacks characterized by uniform and close spacing of adjacent cisternal membranes enclosing slightly distended lumens (Figure 6A). Cells treated with BFA followed by H89 lacked any distinguishable Golgi membranes (Figure 6B). Swollen rough ER membranes were observed under these conditions consistent with previous work examining BFA-treated cells (Alvarez and Sztul, 1999). Significantly, after drug washout, Golgi stacks that were indistinguishable from those in control cells were observed (Figure 6C). In a random sampling of at least 15 cells for each condition, all untreated and washout cells contained stacked Golgi complexes, whereas no such structures were observed in drug treated cells. These results indicate that intact Golgi stacks had reassembled.
Figure 6.
Cisternal stacking of reassembled Golgi membranes. NRK cells were left untreated (A) or treated with BFA for 30 min followed by H89 for 10 min (B) or treated with BFA for 30 min followed by H89 for 10 min followed by a 30 min washout (C). The cells were processed and examined by transmission electron microscopy. Juxtanuclear regions viewed at 120,000× are shown and Golgi stacks (arrowheads) and nuclei (n) are indicated. Golgi stacks were not observed in treated cells until after drug washout. ER membranes, identified on the basis of associated ribosomes, were frequently dilated in treated cells (*). Bar, 0.2 μm.
The membrane localization of β-COP, a component of the ERGIC and Golgi associated COPI coat complex, was also examined during biogenesis. Giantin-positive punctate structures evident at 5 min were positive for β-COP (Figure 7, A and B), and a β-COP localization pattern typical of untreated cells was established by 30 min (Figure 7, C and D). Therefore, COPI function is likely active early during biogenesis. This is not surprising in that a major COPI function is to support recycling from the ERGIC (Klumperman et al., 1998). However, COPI-mediated recycling reactions are also likely to establish and maintain Golgi subcompartments. Therefore, it was of interest if, and when, evidence for subcompartmentalization of Golgi markers could be detected.
Figure 7.
Recruitment of COPI to Golgi membranes undergoing biogenesis. NRK cells were sequentially treated with BFA for 30 min followed by H89 for 10 min and then analyzed after the indicated times of H89 washout. The methanol-fixed cells were costained with anti-β-COP (A and C) and anti-giantin (B and D) antibodies. Bar, 10 μm.
Despite the observed differences in ER export kinetics, the various Golgi proteins ultimately appeared to colocalize in newly formed pre-Golgi intermediates. However, analysis of costained deconvolved optical sections indicated that even the earliest punctate structures positive for both GM130 and mannosidase II (Figure 8A) or GPP130 and giantin (Figure 8B) yielded adjacent rather than coincident staining. Given that coincident staining is detected under these conditions when costained patterns correspond to the same membrane (Puri et al., 2002), this indicates that the collection of membranes that comprise these structures did not completely coalesce. Further, these same marker pairs also yielded adjacent rather than coincident staining in untreated Golgi ribbons (Puri et al., 2002, and our unpublished results) suggesting that marker compartmentalization in the Golgi stack is established early during Golgi biogenesis from the ER.
Figure 8.
Evidence of marker segregation during Golgi biogenesis. NRK cells were costained for mannosidase II (green) and GM130 (red) after 5 min of H89 washout (A) or giantin (green) and GPP130 (red) after 10 min of H89 washout (B). Adjacent rather than coincident staining suggests marker compartmentalization early during Golgi biogenesis from the ER. The images shown are 2D projections from the deconvolved 3D data set using summation of pixel values in each plane. Bar, 10 μm.
As mentioned above, a second treatment of cells was identified that also caused ER redistribution of all Golgi markers that we tested, including the matrix markers GM130, giantin, GRASP55, and GRASP65. This was with the drug clofibrate, which had previously been demonstrated to cause, by an unknown mechanism, ER redistribution of mannosidase II (de Figueiredo et al., 1999; Nakamura et al., 2001). As expected, mannosidase II collapsed from a typical Golgi ribbon pattern (Figure 9A) to an ER pattern (Figure 9B) upon clofibrate treatment, and the Golgi pattern was restored upon drug washout (Figure 9C). In the same cells, GM130 also collapsed from the Golgi (Figure 9D) to a dispersed ER pattern (Figure 9E) and back to a Golgi ribbon pattern upon washout (Figure 9F). Density gradient analysis confirmed ER redistribution of GM130 before drug washout (Figure 9G). Thus, both treatments, BFA/H89 and clofibrate, caused a rapid loss of the Golgi apparatus accompanied by redistribution of Golgi-localized proteins, including matrix markers, into the ER. Despite the apparent complete collapse of the Golgi into the ER, a normal Golgi ribbon reappeared upon drug washout.
Figure 9.
Reversible ER redistribution of Golgi markers during clofibrate treatment. NRK cells were left untreated (A and D) or treated with clofibrate for 10 min (B and E) or treated with clofibrate for 10 min followed by a 30 min washout (C and F). The cells were then costained with antimannosidase II (A-C) and anti-GM130 (D-F) antibodies. The images shown are 2D projections from the deconvolved 3D data set using summation of pixel values in each plane. Bar, 5 μm. Clofibrate-treated cells were also subjected to sucrose density gradient fractionation (G). Recovery of GM130 (♦) and calreticulin (▪) in each fraction was determined by immunoblotting. The sedimentation positions for Golgi and ER membranes in untreated cells are indicated (see also Figure 3).
DISCUSSION
Disassembly of the Golgi apparatus occurs at mitosis (Shorter and Warren, 2002), in response to osmotic shock (Lee and Linstedt, 1999), and under numerous experimental conditions. Despite its complex stereotypical organization, Golgi reassembly can be rapid consistent with the involvement of a template. Further, Golgi components are accurately partitioned during cell division, so a maternally derived preassembled template might always be present. On the other hand, a capacity for self-assembly is common in cell biology. Most Golgi components are not only synthesized at the ER, but also constitutively cycle via the ER. Even peripheral membrane proteins, such as GM130, that are initially targeted by directly binding the Golgi (Yoshimura et al., 2001) or those that associate with the membrane dynamically (Ward et al., 2001) appear to cycle via the ER once associated with their membrane receptors. Thus, it is reasonable that the Golgi apparatus has the capacity to form de novo from the ER, even if this capacity is not always used.
By causing Golgi collapse under conditions of reversible ER export blockade, we were able to provide the first demonstration that the Golgi can reform from the ER under conditions where both matrix and nonmatrix components start from the ER. Previous work used a dominant negative version of Sar1 to block ER export (Storrie et al., 1998; Miles et al., 2001; Ward et al., 2001). The ensuing Golgi collapse may reflect normal cycling of all Golgi proteins through the ER or it might reflect an abnormal process triggered by ER export blockade. Even if it is assumed that all Golgi proteins cycle via the ER, the percent in the ER at any given time would have to be relatively minor. It remained possible that a major fraction of matrix components exist in an assembled state outside the ER and mediate Golgi assembly. Thus, it was critical to test whether the Golgi apparatus can assemble from the ER after an essentially quantitative collapse of matrix components into the ER. Cells from which the Golgi has been removed by cytoplast preparation fail to reform identifiable Golgi structures (Pelletier et al., 2000). However, it is unclear whether the Golgi-minus cytoplasts were capable of restoring normal levels of Golgi components after their formation. Based on our results, it seems likely that if such cytoplasts could survive long enough to synthesize near-normal amounts of Golgi components, they would regenerate the Golgi.
Interestingly, upon restoration of ER export, the kinetics of emergence from the ER differed for each Golgi protein tested. As mentioned above, the rapid sorting of matrix proteins during ER export that we observed provides an explanation for appearance of these proteins in BFA remnants. This is based on the idea that BFA-induced redistribution involves cycling of Golgi proteins through the ER followed by subsequent accumulation of some golgins and the GRASPs in post-ER structures such as the ERGIC (Ward et al., 2001). Indeed, H89-induced ER export blockade during or after BFA treatment caused accumulation of golgins and GRASPs in the ER. Although the physiological relevance of BFA remnants is unclear, it is presence in BFA remnants that largely defines proteins as matrix markers. If presence in remnants is due to preferential ER exit, why do GM130, the GRASPs, and several other golgins exhibit this behavior? It may be that different explanations apply to different members of this group. What follows are three nonmutually exclusive possibilities.
It could be that some of these components form a dynamic matrix—one that is dispersed and then reassembles to mediate Golgi organization. This is particularly relevant in the case of golgin-45 in that the Golgi is fragmented in its absence (Short et al., 2001). Of course, until there is direct evidence indicating participation of golgin-45 in a structural scaffold, it must be acknowledged that other roles for golgin-45 could readily explain the Golgi fragmentation phenotype. In the case of GM130 and giantin, the matrix idea is considerably less likely as these proteins appear dispensable for Golgi biogenesis. Maintenance and reformation of the Golgi ribbon takes place in cells in which microinjected antibodies cause giantin degradation or prevent GM130 binding to p115 (Puthenveedu and Linstedt, 2001). The Golgi appears normal in a CHO cell line with an unknown defect causing a knockdown of GM130 expression (Vasile et al., 2003). Also, Golgi assembly takes place in cells after RNAi-mediated knockdown of GM130 or giantin expression (Puri et al., unpublished results). Therefore, despite the interesting attribute that it is the only Golgi protein specifically identified in the insoluble material termed Golgi matrix, GM130 does not perform a required template function during assembly of the Golgi ribbon.
Another possible explanation for rapid ER exit is if “matrix” markers are actually trafficking components (Sonnichsen et al., 1998; Shorter et al., 1999). Rapid ER exit of matrix proteins is likely due to the presence of sorting signals that bind directly or indirectly, and with relatively high affinity, to the COPII complex. Assuming a role in trafficking, the presence of such signals could be rationalized because, unlike proteins with functions unrelated to trafficking, every vesicle must have appropriate trafficking machinery. Nevertheless, roles in trafficking could be either required or facilitating. If only facilitating, the in vitro Golgi stacking reaction, in which several matrix proteins are required (Barr et al., 1997; Shorter et al., 1999; Shorter and Warren, 1999), may have created dependencies for these proteins—through dilution etc.—which are not evident in vivo.
A third possibility is that ER exit kinetics of Golgi proteins reflects cis-to-trans subcompartmental localization. Although matrix markers were not examined, a cis-to-trans order was previously observed for Golgi reassembly after BFA washout (Alcalde et al., 1992). With one notable exception, the order we observed (GM130 > giantin > mannosidase II > GPP130) was consistent with a cis-to-trans ordering. The exception, cis-localized GPP130, might be explainable as it has an unusual propensity for trafficking to compartments well distal of its steady state localization (Puri et al., 2002). If ER export of Golgi proteins approximates a cis-to-trans order, then this may contribute to Golgi subcompartmentalization. During Golgi biogenesis from the ER, the cis subcompartment may form first, followed by later compartments. Indeed, early in Golgi biogenesis, we observed segregation rather than coalescence of GM130, which is localized to the cis Golgi, and mannosidase II, which is localized to the medial Golgi. A COPII bias for early Golgi proteins might also aid subcompartmentalization at steady state because it would tend to generate membranes for input to the Golgi with higher concentrations of early Golgi proteins. Together with local recycling between established cisternae, this would support maintenance of Golgi subcompartments during cisternal progression.
A final discussion point concerns mitotic Golgi disassembly. Given that the Golgi exhibits a capacity for biogenesis from the ER, why does it breakdown at M-phase into vesicles that remain independent of the ER (Lucocq et al., 1989; Jesch et al., 2001; Jokitalo et al., 2001) as opposed to collapse into the ER (Zaal et al., 1999; Terasaki, 2000)? Our working hypothesis is that cells evolved so that different physiological conditions impact reactions mediating Golgi structure to yield distinct changes in Golgi organization. These may confer advantages appropriate to the condition. In the case of mitotic Golgi breakdown a reaction might have been selected that yields the most efficient reassembly. One such reaction is inhibition of docking and fusion of Golgi derived retrieval vesicles leading to nearly complete conversion of the Golgi into vesicles. In contrast to ER redistribution, Golgi vesiculation would conserve the sorting, i.e., concentration, of Golgi resident proteins, that took place to build the interphase organelle. The net effect would be accurate partitioning of the organelle coupled with reassembly that does not involve rebuilding the organelle from scratch.
Acknowledgments
We thank Dr. Tina Lee and Manojkumar Puthenveedu for their essential assistance, Joe Suhan for expert assistance with the electron microscopy, lab members for helpful comments, Dr. Vivek Malhotra for anti-GRASP antibodies, Dr. Amy Csink for the use of the Delta Vision system, and Dr. Bor Luen Tang for anti-Sec31. This work was supported by a National Institutes of Health Grant GM-56779-02 to A.D.L.
References
- Alcalde, J., Bonay, P., Roa, A., Vilaro, S., and Sandoval, I.V. (1992). Assembly and disassembly of the Golgi complex: two processes arranged in a cis-trans direction. J. Cell Biol. 116, 69-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez, C., and Sztul, E.S. (1999). Brefeldin A (BFA) disrupts the organization of the microtubule and the actin cytoskeletons. Eur. J. Cell Biol. 78, 1-14. [DOI] [PubMed] [Google Scholar]
- Aridor, M., and Balch, W.E. (2000). Kinase signaling initiates coat complex II (COPII) recruitment and export from the mammalian endoplasmic reticulum. J. Biol. Chem. 275, 35673-35676. [DOI] [PubMed] [Google Scholar]
- Barr, F.A., Puype, M., Vandekerckhove, J., and Warren, G. (1997). GRASP65, a protein involved in the stacking of Golgi cisternae. Cell 91, 253-262. [DOI] [PubMed] [Google Scholar]
- Bevis, B.J., Hammond, A.T., Reinke, C.A., and Glick, B.S. (2002). De novo formation of transitional ER sites and Golgi structures in Pichia pastoris. Nat. Cell Biol. 4, 750-756. [DOI] [PubMed] [Google Scholar]
- Cole, N.B., Sciaky, N., Marotta, A., Song, J., and Lippincott-Schwartz, J. (1996). Golgi dispersal during microtubule disruption: regeneration of Golgi stacks at peripheral endoplasmic reticulum exit sites. Mol. Biol. Cell 7, 631-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, N.B., Ellenberg, J., Song, J., DiEuliis, D., and Lippincott-Schwartz, J. (1998). Retrograde transport of Golgi-localized proteins to the ER. J. Cell Biol. 140, 1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Figueiredo, P., and Brown, W.J. (1999). Clofibrate inhibits membrane trafficking to the Golgi complex and induces its retrograde movement to the endoplasmic reticulum. Cell Biol. Toxicol. 15, 311-323. [DOI] [PubMed] [Google Scholar]
- Jesch, S.A., and Linstedt, A.D. (1998). The Golgi and endoplasmic reticulum remain independent during mitosis in HeLa cells. Mol. Biol. Cell 9, 623-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jesch, S.A., Mehta, A.J., Velliste, M., Murphy, R.F., and Linstedt, A.D. (2001). Mitotic Golgi is in a dynamic equilibrium between clustered and free vesicles independent of the ER. Traffic 2, 873-884. [DOI] [PubMed] [Google Scholar]
- Jokitalo, E., Cabrera-Poch, N., Warren, G., and Shima, D.T. (2001). Golgi clusters and vesicles mediate mitotic inheritance independently of the endoplasmic reticulum. J. Cell Biol. 154, 317-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumperman, J., Schweizer, A., Clausen, H., Tang, B.L., Hong, W., Oorschot, V., and Hauri, H.P. (1998). The recycling pathway of protein ERGIC-53 and dynamics of the ER-Golgi intermediate compartment. J. Cell Sci. 111, 3411-3425. [DOI] [PubMed] [Google Scholar]
- Lee, T.H., and Linstedt, A.D. (1999). Osmotically induced cell volume changes alter anterograde and retrograde transport, Golgi structure, and COPI dissociation. Mol. Biol. Cell 10, 1445-1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, T.H., and Linstedt, A.D. (2000). Potential role for protein kinases in regulation of bidirectional endoplasmic reticulum-to-Golgi transport revealed by protein kinase inhibitor H89. Mol. Biol. Cell 11, 2577-2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linstedt, A.D., and Hauri, H.P. (1993). Giantin, a novel conserved Golgi membrane protein containing a cytoplasmic domain of at least 350 kDa. Mol. Biol. Cell 4, 679-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linstedt, A.D., Mehta, A., Suhan, J., Reggio, H., and Hauri, H.P. (1997). Sequence and overexpression of GPP130/GIMPc: evidence for saturable pH-sensitive targeting of a type II early Golgi membrane protein. Mol. Biol. Cell 8, 1073-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucocq, J.M., Berger, E.G., and Warren, G. (1989). Mitotic Golgi fragments in HeLa cells and their role in the reassembly pathway. J. Cell Biol. 109, 463-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles, S., McManus, H., Forsten, K.E., and Storrie, B. (2001). Evidence that the entire Golgi apparatus cycles in interphase HeLa cells: sensitivity of Golgi matrix proteins to an ER exit block. J. Cell Biol. 155, 543-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata-Kuno, K., Hino, Y., Nanri, H., Shibata, Y., and Minakami, S. (1990). Fragmentation of re-formation of mitotic Golgi apparatus detected by a centrifugal method. Exp. Cell Res. 191, 273-277. [DOI] [PubMed] [Google Scholar]
- Nakamura, N., Rabouille, C., Watson, R., Nilsson, T., Hui, N., Slusarewicz, P., Kreis, T.E., and Warren, G. (1995). Characterization of a cis-Golgi matrix protein, GM130. J. Cell Biol. 131, 1715-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, M., Kuroiwa, N., Kono, Y., and Takatsuki, A. (2001). Characterization of clofibrate-induced retrograde Golgi membrane movement to the endoplasmic reticulum: clofibrate distinguishes the Golgi from the trans Golgi network. Biosci. Biotechnol. Biochem. 65, 1812-1823. [DOI] [PubMed] [Google Scholar]
- Pelletier, L., Jokitalo, E., and Warren, G. (2000). The effect of Golgi depletion on exocytic transport. Nat. Cell Biol. 2, 840-846. [DOI] [PubMed] [Google Scholar]
- Peyroche, A., Antonny, B., Robineau, S., Acker, J., Cherfils, J., and Jackson, C.L. (1999). Brefeldin A acts to stabilize an abortive ARF-GDP-Sec7 domain protein complex: involvement of specific residues of the Sec7 domain. Mol. Cell 3, 275-285. [DOI] [PubMed] [Google Scholar]
- Puri, S., Bachert, C., Fimmel, C.J., and Linstedt, A.D. (2002). Cycling of early Golgi proteins via the cell surface and endosomes upon lumenal pH disruption. Traffic 3, 641-653. [DOI] [PubMed] [Google Scholar]
- Puri, S., Telfer, H., Velliste, M., Murphy, R., and Linstedt, A.D. (2004). Dispersal of Golgi matrix proteins during mitotic Golgi disassembly. J. Cell Sci., (in press). [DOI] [PubMed]
- Puthenveedu, M.A., and Linstedt, A.D. (2001). Evidence that Golgi structure depends on a p115 activity that is independent of the vesicle tether components giantin and GM130. J. Cell Biol. 155, 227-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann, J., Jokitalo, E., Pypaert, M., and Warren, G. (2000). Matrix proteins can generate the higher order architecture of the Golgi apparatus. Nature 407, 1022-1026. [DOI] [PubMed] [Google Scholar]
- Seemann, J., Pypaert, M., Taguchi, T., Malsam, J., and Warren, G. (2002). Partitioning of the matrix fraction of the Golgi apparatus during mitosis in animal cells. Science 295, 848-851. [DOI] [PubMed] [Google Scholar]
- Short, B., Preisinger, C., Korner, R., Kopajtich, R., Byron, O., and Barr, F.A. (2001). A GRASP55-rab2 effector complex linking Golgi structure to membrane traffic. J. Cell Biol. 155, 877-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter, J., Watson, R., Giannakou, M.E., Clarke, M., Warren, G., and Barr, F.A. (1999). GRASP55, a second mammalian GRASP protein involved in the stacking of Golgi cisternae in a cell-free system. EMBO J. 18, 4949-4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter, J., and Warren, G. (1999). A role for the vesicle tethering protein, p115, in the post-mitotic stacking of reassembling Golgi cisternae in a cell-free system. J. Cell Biol. 146, 57-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter, J., and Warren, G. (2002). Golgi architecture and inheritance. Annu. Rev. Cell Dev. Biol. 18, 379-420. [DOI] [PubMed] [Google Scholar]
- Slusarewicz, P., Nilsson, T., Hui, N., Watson, R., and Warren, G. (1994). Isolation of a matrix that binds medial Golgi enzymes. J. Cell Biol. 124, 405-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnichsen, B., Lowe, M., Levine, T., Jamsa, E., Dirac-Svejstrup, B., and Warren, G. (1998). A role for Giantin in docking COPI vesicles to Golgi membranes. J. Cell Biol. 140, 1013-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storrie, B., White, J., Rottger, S., Stelzer, E.H., Suganuma, T., and Nilsson, T. (1998). Recycling of Golgi-resident glycosyltransferases through the ER reveals a novel pathway and provides an explanation for nocodazole-induced Golgi scattering. J. Cell Biol. 143, 1505-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutterlin, C., Hsu, P., Mallabiabarrena, A., and Malhotra, V. (2002). Fragmentation and dispersal of the pericentriolar Golgi complex is required for entry into mitosis in mammalian cells. Cell 109, 359-369. [DOI] [PubMed] [Google Scholar]
- Tang, B.L., Zhang, T., Low, D.Y., Wong, E.T., Horstmann, H., and Hong, W. (2000). Mammalian homologues of yeast sec31p. An ubiquitously expressed form is localized to endoplasmic reticulum (ER) exit sites and is essential for ER-Golgi transport. J. Biol. Chem. 275, 13597-13604. [DOI] [PubMed] [Google Scholar]
- Terasaki, M. (2000). Dynamics of the endoplasmic reticulum and Golgi apparatus during early sea urchin development. Mol. Biol. Cell 11, 897-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasile, E., Perez, T., Nakamura, N., and Krieger, M. (2003). Structural integrity of the Golgi is temperature sensitive in conditional-lethal mutants with no detectable GM130. Traffic 4, 254-272. [DOI] [PubMed] [Google Scholar]
- Ward, T.H., Polishchuk, R.S., Caplan, S., Hirschberg, K., and Lippincott-Schwartz, J. (2001). Maintenance of Golgi structure and function depends on the integrity of ER export. J. Cell Biol. 155, 557-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura, S.I., Nakamura, N., Barr, F.A., Misumi, Y., Ikehara, Y., Ohno, H. Sakaguchi, M., and Mihara, K. (2001). Direct targeting of cis-Golgi matrix proteins to the Golgi apparatus. J. Cell Sci. 114, 4105-4115. [DOI] [PubMed] [Google Scholar]
- Zaal, K.J. et al. (1999). Golgi membranes are absorbed into and reemerge from the ER during mitosis. Cell 99, 589-601. [DOI] [PubMed] [Google Scholar]