Abstract
Our group identified angiotensin II type 1 (AT1) receptor-associated protein (ATRAP) in a yeast two-hybrid screen for proteins that bind to the carboxyl-terminal cytoplasmic domain of the AT1. In this work, we characterize ATRAP as a transmembrane protein localized in intracellular trafficking vesicles and plasma membrane that functions as a modulator of angiotensin II-induced signal transduction. ATRAP contains three hydrophobic domains at the amino-terminal end of the protein, encompassing the amino acid residues 14–36, 55–77, and 88–108 and a hydrophilic cytoplasmic carboxyl-terminal tail from residues 109–161. Endogenous and transfected ATRAP cDNA shows a particulate distribution; electron microscopy reveals the presence of ATRAP in prominent perinuclear vesicular membranes; and colocalization analysis by immunofluorescence shows that ATRAP colocalizes in an intracellular vesicular compartment corresponding to endoplasmic reticulum, Golgi, and endocytic vesicles. Real-time tracking of ATRAP vesicles shows constitutive translocation toward the plasma membrane. Using epitope-tagged forms of ATRAP at either the amino or carboxyl end of the molecule, we determined the orientation of the amino end as being outside the cell. Mutant forms of ATRAP lacking the carboxyl end are unable to bind to the AT1 receptor, leading to the formation of prominent perinuclear vesicle clusters. Functional analysis of the effects of ATRAP on angiotensin II-induced AT1 receptor signaling reveals a moderate decrease in the generation of inositol lipids, a marked decrease in the angiotensin II-stimulated transcriptional activity of the c-fos promoter luciferase reporter gene, and a decrease in cell proliferation.
INTRODUCTION
The active peptide angiotensin II (Ang II) is the most prominent regulator of blood pressure, exerting its effects through the control of vasoconstriction and regulation of water and salt balance (Audoly et al., 2000). In addition, Ang II has important effects on diverse cellular systems, including neuromodulation, cellular growth, and proliferation (Sayeski et al., 1998; Allen et al., 2000). Ang II exerts its functions through activation of type 1 (AT1) and type 2 (AT2) angiotensin receptors. AT1 receptor (which in mice has two subtypes, AT1a and AT1b) is a member of the superfamily of G protein-coupled receptors (GPCR) and activates G proteins through regions of the third intracellular loop and the intracellular carboxyl-terminal tail of the receptor (Blume et al., 1999; Inagami, 1999; Guo et al., 2001).
A number of proteins have been shown to be involved in the molecular mechanism of AT1 receptor actions. The best characterized chain of events is the rapid activation of heterotrimeric G proteins (Gαq/11, Gα12/13, and Gβγ) that subsequently activate phospholipase Cβ and phospholipase Cγ to induce the generation of inositol-1,4,5-triphosphate (IP3) and diacylglycerol (DAG), increase in cytosolic Ca2+, activation of cytosolic kinases, and transcriptional activation (Blume et al., 1999; Inagami, 1999). Additional mediators have been identified, including cytosolic and transmembrane tyrosine kinases, phosphatases, serine-threonine kinases, and others (Sayeski et al., 1998; Blume et al., 1999; Hall et al., 1999).
A global mechanism for GPCR signaling independent of G proteins has emerged during the past few years and involves the recruitment of a number of proteins to the receptor (Brzostowski and Kimmel, 2001; von Zastrow, 2001; Perry and Lefkowitz, 2002). After binding of ligand to the receptor and activation of heterotrimeric G proteins, the responsiveness of the receptor is determined by additional steps of phosphorylation of intracellular loops and the cytoplasmic tail of AT1 receptor mediated by G protein receptor kinases (GRKs) and followed by binding of receptor to β-arrestin, internalization of the receptor, and the subsequent dephosphorylation and recycling of receptors to the cell surface. AT1 receptor is internalized via clathrin-coated and caveolae vesicles (Hunyady et al., 2000). AT1 receptor remains associated with β-arrestin after internalizing, and both receptor and β-arrestin are subsequently trafficked to endosomes (Gaborik et al., 2001; Perry and Lefkowitz, 2002; Tohgo et al., 2002).
Studies with mutants of AT1 receptors have found that the C-terminal cytoplasmic end is involved in the control of receptor internalization independently of G protein coupling (Conchon et al., 1997; Hein et al., 1997). Serial deletions of the C-terminal tail lead to a reduction in internalization, an impairment of desensitization, and a decrease in the coupling to Gq proteins (Hunyady et al., 1994; Hunyady, 1999); in addition, phospholipase D activation is diminished (Thomas et al., 1995) and Janus tyrosine kinase 2 activation is attenuated (Liang et al., 1999). The precise mechanisms of control of these biochemical activities are not yet completely understood, but what is known suggests direct protein–protein interactions of the receptor with different proteins as effectors. This suggests that the C-terminal tail of AT1 receptor may act as a scaffold for the assembly of molecules that are important in linking receptor-mediated signal transduction to the specific biological response to Ang II (Guo and Inagami, 2001).
Using a diverse set of approaches, such as the yeast two-hybrid system, phage display, and fusion protein overlays, several investigators have identified associations of G protein–coupled receptors with a variety of intracellular partners other than heterotrimeric G proteins (Sayeski et al., 1998; Hall et al., 1999; Brzostowski and Kimmel, 2001; Marinissen and Gutkind, 2001; von Zastrow, 2001). A novel protein recently identified in our laboratory, angiotensin II type I receptor-associated protein (ATRAP), has been shown to be a partner of the AT1 receptor, interacting specifically with this receptor both in vitro and in vivo (Daviet et al., 1999). Functionally, ATRAP is able to induce a decrease in the generation of IP3 in an agonist-dependent manner and to enhance AT1 internalization (Daviet et al., 1999; Cui et al., 2000; Wang et al., 2002). In this work, we analyzed the function of ATRAP in cell signaling through its structural characterization, identified the cellular compartments in which ATRAP acts, and analyzed the nature of ATRAP interaction with AT1 receptor both structurally and functionally.
MATERIALS AND METHODS
Reagents and Plasmids
DMEM restriction enzymes for molecular biology, and polymerase chain reaction (PCR) primers were purchased from Invitrogen (Carlsbad, CA). Fetal calf serum and l-glutamine were obtained from Invitrogen. Leupeptin and aprotinin were from Sigma-Aldrich (St. Louis, MO). DiOC6(3) and Alexa secondary fluorescent antibodies were from Molecular Probes (Eugene, OR). The full-length ATRAP was PCR amplified and subcloned in a modified pcDNA3.1-HA vector (Invitrogen). In a similar manner, 3′ carboxyl-terminal deletions at the indicated amino acids (C-ter Δ142, Δ132, Δ122 Δ112, Δ102, Δ92, Δ82, and Δ82) and 5′ amino-terminal deletions of ATRAP (N-ter Δ40, Δ80, and Δ110) were generated by PCR with reverse primers containing stop codons at the desired locations, and the deleted cDNAs were subcloned into pcDNA3.1-HA, pcDNA3.1-EE, pGEX-4T1 (Amersham Biosciences, Piscataway, NJ), pEGFP-C2 (BD Biosciences Clontech, Palo Alto, CA), or pRFP-N (BD Biosciences Clontech). The primer sequences are available upon request. All the constructs were verified by DNA sequencing by the Sanger dideoxy termination method adapted to model 373S automated DNA sequencer (Applied Biosystems, Foster City, CA).
Subcellular localization vectors for visualizing Golgi apparatus, peroxisomes, and endosomes were obtained from BD Biosciences Clontech. The Golgi marker vector encodes a fusion protein consisting of enhanced cyan fluorescent protein (ECFP) and a sequence encoding the N-terminal 81 amino acids of human β-1,4-galactosyltransferase containing the membrane-anchoring signal peptide that targets the fusion protein to the trans-medial region of the Golgi apparatus. Enhanced green fluorescent protein (EGFP)-peroxisome vector encodes a fusion protein consisting of the EGFP and the peroximal targeting signal 1 (PTS1). The endosome marker encodes a fusion protein consisting of the human RhoB GTPase containing an N-terminal c-Myc epitope tag and the EGFP.
Cell Culture and Transient Transfection Procedures
Cells were kept at logarithmic growth phase in DMEM supplemented with 10% heat-inactivated fetal calf serum and 2 ml of l-glutamine in a humidified atmosphere containing 5% CO2. The N-terminal hemagglutinin (HA) epitope-tagged ATRAP in pcDNA3 constructs were transiently transfected in human embryonic kidney (HEK)-293 cells according to the FuGENE protocol (Roche Diagnostics, Indianapolis, IN). In cotransfections with AT1 receptor, the ratio of AT1 receptor to ATRAP cDNA was 1:3. Cells were harvested 48 h after transfection, and membrane fractions prepared from the transfected cells were solubilized in 50 mM Tris-HCl (pH 7.5), 140 mM NaCl, 1 mM CaCl2, 1 mM phenylmethylsulfonyl fluoride, and 1 μg of aprotinin/ml in the presence of 1% Triton X-100. The mixture was gently agitated for 30 min at 4°C and thereafter centrifuged at 13,000 × g for 20 min. Either supernatants or pellets were resuspended in loading buffer; the samples were then subjected to SDS-PAGE, transferred to nitrocellulose membrane (Hybond-ECL; Amersham Biosciences), and probed with anti-HA monoclonal antibody 12CA5 (Covance, Princeton, NJ). Epitope-tagged ATRAP was detected with peroxidase-conjugated sheep anti-mouse secondary antibody (Amersham Biosciences) and enzyme-linked chemiluminescence (Amersham Biosciences).
Immunofluorescence Microscopy
HEK-293 cells were seeded in glass coverslips and cotransfected with N-terminal HA-tagged ATRAP and FLAG epitope-tagged AT1 with the method described above. The cells were then fixed and permeabilized with 4% paraformaldehyde (PFA) for 5 min. HA-ATRAP was detected with either the monoclonal mouse antibody 12CA5 (Covance) and an Alexa-conjugated goat anti-mouse IgG2b monoclonal antibody (Molecular Probes) or an HA-specific rabbit antiserum (Covance) and Alexa-goat anti-rabbit antibody (Molecular Probes). For the real-time tracking of ATRAP-red fluorescent protein (RFP), we located a selected cell and after marking its localization, we took several snapshots at the indicated times. All images were acquired with a scientific-grade, cooled, charge-coupled camera by using a microscopy system from Carl Zeiss (Thornwood, NY). Imaging processing was carried out with the image processing software ImagePro (Media Cybernetics, Silver Spring, MD).
Electron Microscopy
Stably transfected ATRAP-RFP HEK-293 cells were washed and then fixed in 2.5% glutaraldehyde (1 h at 4°C) and postfixed in 1% osmium tetroxide. Cells were then embedded in Epon by standard procedures. Ultrathin sections were stained with or without anti-RFP polyclonal antibodies (BD Biosciences Clontech) and then with 10-nm colloidal gold-conjugated anti-rabbit IgG (BB International, Cardiff, United Kingdom). The sections were examined with a Philips CM-10 transmission electron microscope.
Bioluminescence Resonance Energy Transfer (BRET) Assays
ATRAP full-length and AT1 receptors were cloned in frame into the expression vectors pRluc and pGFP2 (Biosignal; PerkinElmer-Cetus, Montreal, ON, Canada). Their identity was verified by DNA sequencing, and expression in HEK-293 cells was checked by Western blot and luciferase assays. For the experiments of interaction of ATRAP with AT1, HEK-293 cells were transiently transfected with FuGENE reagent at a ratio of 3:1 to green fluorescent protein (GFP):luciferase constructs. Forty-eight hours posttransfection, HEK-293 cells were detached with phosphate-buffered saline (PBS)/EDTA and washed twice in PBS. Approximately 50,000 cells/well were distributed in a 96-well microplate (white Optiplate; PerkinElmer Life Sciences, Boston, MA). The DeepBlue coelenterazine substrate (PerkinElmer Life Sciences) was added at a final concentration of 5 μM, and readings were collected by using a Victor2 microplate reader (PerkinElmer-Cetus, Wellesley, MA) that permits detection of signals using filters at 410- and 515-nm wavelengths.
Yeast and Mammalian Two-Hybrid Assays
For the yeast two-hybrid assay, the yeast reporter strain AH109 (BD Biosciences Clontech) was cotransformed with the deleted versions of the ATRAP two-hybrid expression plasmid pGADT7 and the C-terminal tail of AT1 cloned in the two-hybrid expression plasmid pGBKT7. The cotransformants were selected in SD medium lacking leucine, tryptophan, histidine, and adenine (QDO). The yeast α-galactosidase activity, expressed from the MEL1 gene in response to GAL4 activation, was determined in plates containing X-α-Gal (2 mg/ml) as a chromogenic substrate. For the mammalian two-hybrid analysis, various deletion versions of ATRAP were cloned in the target vector pCMV-NF-κB-AD and AT1 C-terminal end in the bait vector pCMVGAL4-BD (Stratagene, La Jolla, CA). HEK-293 cells were cotransfected with the target and bait constructs together with the reporter plasmid pFR-luciferase at a ratio of 3:3:1. After 48 h, the transfected cells were harvested and lysed, and the cell lysate was used to run the luciferase assay (Promega, Wisconsin, WI) as described above.
Glutathione S-Transferase (GST) and Maltose Binding Protein (MBP) Pulldown Assays
The full-length and deletion versions of ATRAP cloned in the pGEX vector (Amersham Biosciences) and the cytoplasmic AT1a tail cloned into a pMal-c2 prokaryotic expression vector (New England Biolabs, Beverly, MA) were expressed in Escherichia coli BL21 and purified according to the manufacturer's instructions. Equimolar amounts of GST and MBP fusion proteins were incubated at 4°C for 2 h. For affinity chromatography, the protein mixture was passed through a 2-ml volume of the glutathione-Sepharose beads (Amersham Biosciences). Columns were washed six times with washing buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, and 1 mg of aprotinin/ml) and eluted with SDS-PAGE sample buffer. Samples were then subjected to SDS-PAGE and visualized with Coomassie Blue staining.
Inositol Phosphate Determination
HEK-293 cells were transiently cotransfected with N-terminal HA epitope-tagged ATRAP and FLAG epitope-tagged AT1 receptor or with the FuGENE reagent. The ratio of receptor and ATRAP DNA was 1:3. Transfected cells plated in 12-well plates (2 × 105 cells/well) were labeled overnight with myo-[3H]inositol (5 μCi/ml; PerkinElmer Life Sciences) in serum-free DMEM. After stimulation with increasing concentrations of Ang II in the presence of 10 mM LiCl, inositol phosphate was extracted and separated on Dowex AG1-X8 columns (Bio-Rad, Hercules, CA). Total inositol phosphate was eluted with 2 M ammonium formate, 0.1 M formic acid.
GTP-binding Assay
Determinations of GTP loading into membrane preparations of HEK-293 cells overexpressing AT1 receptors were performed using a time-resolved fluorescence protocol (PerkinElmer Life Sciences) according to the manufacturer's instructions. Briefly, 10 μg of membrane preparation was incubated in 50 mM HEPES (pH 7.4), 10 μM GDP, 10 mM MgCl2, 20 mM NaCl, and 500 μg/ml saponin in the presence and absence of different concentrations of Ang II for 30 min. GTP-Europium (GTP-Eu) (30 nM) was added for an additional 30 min before filtration, washing, and measurement at 615 nm on a microplate reader Victor2 (Perkin Elmer Life Sciences).
Transcriptional fos-Luciferase Assay
Chinese hamster ovary (CHO) K1 cells overexpressing FLAG-AT1 receptor (Daviet et al., 1999) were used; 3.5 × 105 CHO AT1 cells were seeded in six-well plates and transiently cotransfected with pcDNA3/HA-ATRAP, fos-luciferase reporter gene, and β-galactosidase reporter gene with the FuGENE reagent. The ratio of HA-ATRAP, fos-luciferase, and β-gal DNA was 3:1:1. Forty-eight hours after transfection, cells were incubated in serum-free medium for 16 h. Quiescent cells were then treated with 100 nM Ang II for 3.5 h, washed with PBS, and lysed for 10 min with 250 μl of lysis buffer (luciferase assay system; Promega)) at 4°C. Cell extract (10 μl) was mixed with 100 μl of luciferase reagent, and the light produced was measured for 10 s with a LUMAT LB 9507 luminometer (Berthold Technologies, Bad Wildbad, Germany). Results were normalized to the β-gal activity with a β-galactosidase enzyme assay system (Promega).
Statistics
For the BRET, inositol phosphate, and luciferase assays, results are expressed as mean ± SD. Statistical significance was assessed by t test.
RESULTS
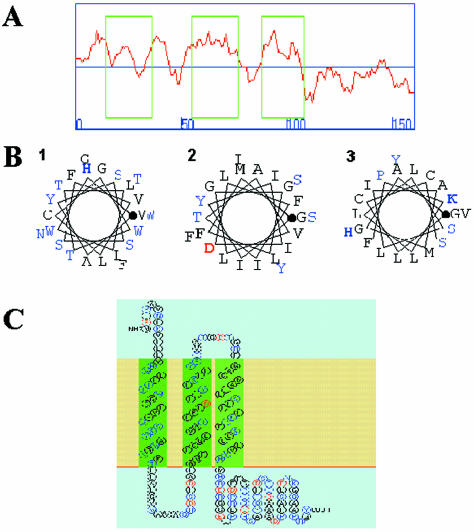
Full-length ATRAP cDNA was cloned from a yeast two-hybrid genetic screen with the C-terminal domain of the AT1 receptor used as bait. Previously, we reported the association of ATRAP and AT1 receptor both in yeast genetic assays and in mammalian cells overexpressing these two proteins (Daviet et al., 1999). The analysis of the ATRAP sequence did not reveal any obvious catalytic or functional signature or significant homology to any previously characterized protein. As an initial approach to understanding the functional role of ATRAP, we performed an analysis of the structural and topological characteristics of this protein. Using the SOSUI algorithm (Hirokawa et al., 1998) and the Chou and Fastman and Kyte and Doolittle indexes (Kyte and Doolittle, 1982), we found a consensus prediction of three prominent regions of hydrophobicity that encode for putative transmembrane domains at the N-terminal end of the protein and a hydrophilic cytoplasmic C-terminal tail. To identify transmembrane helices, the SOSUI system uses two kinds of physical-chemical indices, an index by Kyte and Doolittle (Kyte and Doolittle, 1982) and a new amphiphilicity index (Hirokawa et al., 1998).
Figure 1A shows the output for the hydrophobicity analysis of the SOSUI algorithm that predicts three hydrophobic regions encompassing the amino acid residues 14–36, 55–77, and 88–108 and a hydrophilic region from residues 109–161. The first transmembrane domain consists of a mixture of apolar and polar amino acid residues; the second and third transmembrane domains are composed mainly of hydrophobic residues with some polar amino acid residues. Figure 1B shows the prediction of transmembrane helical conformation as a helical wheel diagram of the segments. The accuracy of the SOSUI system in terms of discrimination of membrane proteins, existence of transmembrane helices, and transmembrane helical regions is ∼99, 96, and 85%, respectively (Hirokawa et al., 1998). Figure 1C shows a representation of the putative ATRAP topology after the consensus prediction of the algorithms.
Figure 1.
Predicted structural organization of ATRAP. (A) Hydrophobicity plot with the SOSUI program showing three boxed hydrophobic regions in the N-terminal end of ATRAP comprising the amino acid residues 14–36, 55–77, and 88–108 and a hydrophilic region from residues 109–161. (B) α-Helix conformation of the hydrophobic segments with the hydrophobic residues is shown in black; the polar residues, in blue; positively charged residues, in bold blue; and negatively charged residues, in bold red; the initial residue for the helix is marked with a bold dot. Numbers indicate the putative transmembrane domains. (C) Proposed structure of ATRAP showing the two first transmembrane domains in green as “primary” hydrophobic domains and the third transmembrane domain in light green as a “secondary” hydrophobic domain.
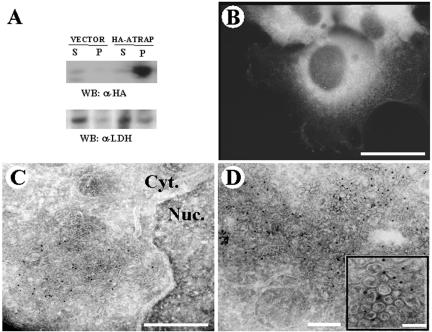
Because the bioinformatic analysis suggested that ATRAP is a membrane protein with specific transmembrane domains, we searched for biochemical and biological evidence of such predictions. HEK-293 cells transiently transfected with HA-epitope–tagged ATRAP were lysed in a Triton X-100 buffer, and the detergent-soluble and -insoluble fractions were subjected to Western blot analysis with monoclonal antibodies to the HA-epitope tag. The Triton X-100–insoluble fraction (pellet) contained >95% of the expressed protein, whereas a Western blot of the cytosolic marker lactate dehydrogenase showed the opposite (Figure 2A). These fractionation results suggest that ATRAP is strongly attached to a detergent-insoluble particulate fraction of the cell and is probably not free at the cytoplasm.
Figure 2.
Detergent insolubility, immunofluorescence, and electron microscopy of HEK-293 cells expressing ATRAP. (A) HEK-293 cells were transiently transfected with HA-ATRAP or empty vector. Western blots were performed using monoclonal antibodies to HA-epitope tag. (B) Endogenous ATRAP in HEK-293 cells was stained with polyclonal antibodies raised to the C-terminal region of ATRAP and observed by using Alexa-labeled secondary antibodies. Bar, 10 μm. (C and D) HEK-293 cells stably transfected with ATRAP-RFP were used for the electron microscopy studies and visualized with RFP polyclonal antibodies and 10-nm colloidal gold-conjugated secondary antibodies. Numerous gold beads labeled the vesicular structures, whereas no significant labeling was obtained on control cells or when anti-RFP serum was omitted. Bars (electron micrograph), 1 μm (C), 250 nm (D), and 100 nm (D, inset).
Determining the cellular localization of ATRAP may help define the kind of functions in which it is involved. Evidence derived from cellular studies suggests that ATRAP is intimately associated with a particulate structure inside the cell. Immunochemical staining of cells transfected with ATRAP shows a specific particulate localization of this protein both at the cell periphery and in a prominent perinuclear region of the cell (Figure 4A). This specific localization of ATRAP was observed in cells transfected with ATRAP tagged with HA, poly-Glu (EE), or GFP or RFP epitopes, located either at the NH2 or COOH ends of the molecule (our unpublished data), suggesting that the particulate localization is an intrinsic property of the molecule. The detergent insolubility and the perinuclear clusters observed in transfected cells suggest the localization of ATRAP in a specific membrane compartment. To demonstrate the intracellular distribution of endogenous ATRAP, we generated affinity-purified polyclonal rabbit antibodies directed at the C-terminal region of the molecule. Immunofluorescence staining of endogenous ATRAP in HEK-293 cells showed a similar particulate distribution (Figure 2B).
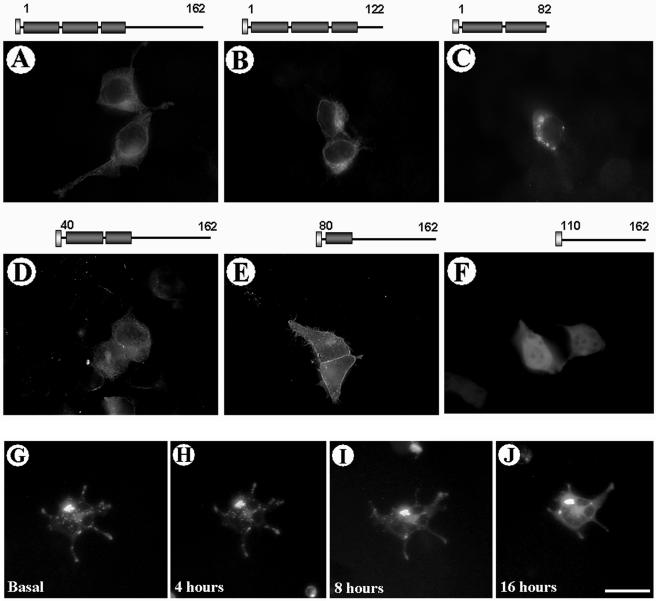
Figure 4.
Expression of N-terminal and C-terminal deletion mutants of ATRAP. HEK-293 cells were transiently transfected with HAATRAP full length or the indicated mutants. After 48 h of expression, the cells were fixed in 4% PFA, stained with antibodies to HA, and detected by using Alexa-labeled secondary antibodies. The schematic diagram of the constructs is shown on the top of each image. The deletion of the C-terminal domain of ATRAP 1–82 aa, leads to the formation of prominent perinuclear vesicle clusters (C), whereas the deletion of the two first transmembrane domains leads to a sharp peripheral membrane localization of the protein (E). The deletion of the N-terminal domain containing the transmembrane domains leads to a diffuse cytoplasmic distribution of the protein (F). (G–J) Real-time tracking of ATRAP vesicles trafficking from the perinuclear compartment to the periphery of the cell. HEK-293 cells were transiently transfected with ATRAP-RFP; target cells were identified 24 h after plating in glass coverslips and followed in an inverted epifluorescence microscope for the indicated times. Bar, 30 μm.
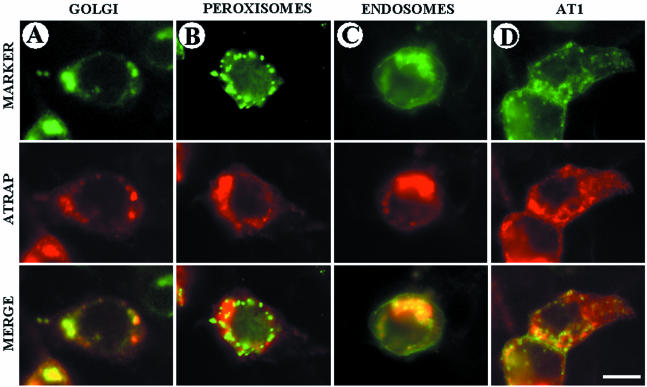
To identify the particulate structure containing immunoreactivity to ATRAP, we performed immunogold electron microscopy analysis of HEK-293 cells stably transfected with ATRAP-RFP. Using specific antibodies directed at the RFP-epitope, we were able to document that ATRAP is localized in perinuclear vesicles. Most of the signal is observed in prominent perinuclear vesicles; with the immunoreactivity distributed in cytoplasmic vesicular compartments and in peripheral regions of the cell (Figure 2, C and D). The insolubility in Triton X-100 suggested membrane lipid rafts as candidates for ATRAP localization; however, staining of ATRAP-expressing cells with specific antibodies to caveolin-coated pits did not show colocalization of these proteins (our unpublished data). Further analysis to determine the exact identity of these vesicles showed that ATRAP colocalizes with the trans-medial Golgi marker ECFP-β-1,4-galactosyltransferase and the vesicle marker of the endocytic pathway GFP-rhoB (Figure 3, A and C). Additionally, we observed colocalization of endogenous ATRAP with markers of the endoplasmic reticulum (ER) such as the dye DiOC6(3) and partial colocalization with the protein calreticulin (our unpublished data), giving further support for the localization of ATRAP in a trafficking vesicular structure. Markers of peroxisome or lysosomal compartments were unable to colocalize with ATRAP (Figure 3B; our unpublished data), further suggesting a specific membrane compartmentalization. The specific colocalization of ATRAP with the indicated vesicular markers was observed in cells transfected with ATRAP labeled with different epitope tags, such as HA, poly-Glu (EE), GFP, or RFP located either at the N- or C-terminal ends of the molecule. Furthermore, immunolocalization of endogenous ATRAP with the use of polyclonal antibodies directed at ATRAP gave the same result (our unpublished data). On the other hand, ATRAP colocalizes to a great extent with AT1 (Figure 3D); from immunoprecipitation experiments, we observed that ∼10% of total ATRAP is complexed with AT1 (Daviet et al., 1999). These results are in agreement with the cellular colocalization analysis that shows a significant but not complete colocalization of these proteins. Together, these analyses suggest that ATRAP localizes in a specific intracellular trafficking structure involving ER, Golgi, and endocytic vesicles.
Figure 3.
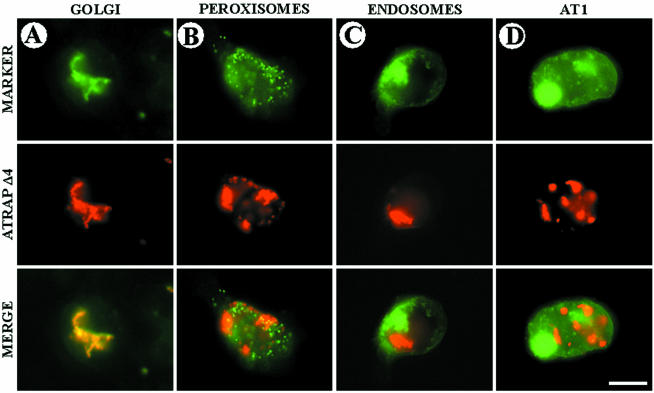
Subcellular distribution of ATRAP in vesicular structures. The indicated fluorescent markers were cotransfected with full-length ATRAP-RFP in HEK-293 cells plated on glass coverslips. After 48 h of expression, the cells were fixed in PFA, washed, and mounted for microscope visualization. (A, C, and D) Colocalization of ATRAP with the Golgi marker vector encoding CFP-β-1,4-galactosyltransferase, the endosome marker encoding a fusion protein consisting of the human RhoB GTPase and the EGFP. FLAG-AT1 construct was visualized with polyclonal anti-FLAG antibodies and Alexa-goat anti-rabbit secondary antibodies. The EGFP-peroxisome marker vector (B) encodes a fusion protein consisting of the green fluorescent protein (EGFP) and the peroximal targeting signal 1 (PTS1). Bar, 20 μm.
To test whether the putative ATRAP domains confer specificity in the localization of the protein, we generated a set of deletions of HA epitope-tagged ATRAP, including both the N-terminal and the C-terminal domains of the protein. Figure 4 shows the expression of these constructs in HEK-293 cells after immunostaining with anti-HA antibodies and the schematic diagram of the constructs on the top of each image. Overexpression of ATRAP deletion mutants confirm the predicted topology obtained in the previous bioinformatics analysis, with the deletion of the C-terminal domain of ATRAP (Figure 4C, ATRAP 1–82 aa), leading to the formation of dramatic perinuclear vesicle clusters, whereas the deletion of the N-terminal domain containing the transmembrane domains leads to a diffuse cytoplasmic distribution of the protein (Figure 4F, ATRAP ΔTM3, 110–162 aa). The deletion of the two first transmembrane domains leads to a sharp peripheral membrane localization of the protein (Figure 4E). Together, these results indicate that ATRAP has specific domains responsible for anchoring the protein to lipid membranes and suggests the existence of a dynamic translocation of ATRAP from intracellular vesicle compartments to plasma membrane regions. In support of this proposal, using fluorescent forms of ATRAP, we were able to follow in real-time the trafficking of ATRAP vesicles from the perinuclear compartment to the periphery of the cell. Figure 4G shows several snapshots of a selected cell expressing ATRAP-RFP, 24 h after plating, in the presence of regular medium culture and followed for the indicated times. At the time indicated as zero (Figure 4G, basal), most of the fluorescence is concentrated in prominent perinuclear vesicles and after 4, 8, or 16 h of tracking the cell, we were able to observe the translocation of the vesicles to the cell periphery (Figure 4G), ending in some cases in fusion of the vesicles to the peripheral plasma membrane. The treatment with Ang II seems not to induce any significant change in the distribution of ATRAP (our unpublished data).
The appearance of dramatic perinuclear vesicular structures in cells transfected with the ATRAP mutant containing only the two first transmembrane domains prompted us to investigate in more detail the characteristics of this construct. Figure 5 shows the colocalization of ATRAP Δ4 (aa 1–82) with a panel of vesicular markers. ATRAP Δ4 is unable to colocalize with AT1 receptor or with the endosomal and peroxisome markers (Figure 5, B and C); this construct remains concentrated in tight perinuclear clusters, whereas the AT1 molecule is located at the plasma membrane and in a few endocytic vesicles (Figure 5D). In contrast, most of the ATRAP clusters colocalize with the Golgi marker (Figure 5A) and ER (our unpublished data), suggesting that the C-terminal end of ATRAP is essential for correct trafficking along the vesicular system.
Figure 5.
Subcellular distribution of mutant ATRAP Δ4 (1–82 aa) in vesicular structures. The indicated fluorescent markers were cotransfected with mutant ATRAP-RFP in HEK-293 cells plated on glass coverslips. After 48 h of expression, the cells were fixed in PFA, washed, and mounted for microscope visualization. (A, C, and D) Colocalization of ATRAP with the Golgi marker vector encoding CFP-β-1,4-galactosyltransferase, the endosome marker encoding a fusion protein consisting of the human RhoB GTPase, and the EGFP. FLAG-AT1 construct was visualized with polyclonal anti-FLAG antibodies and Alexa goat anti-rabbit secondary antibodies. The EGFP-peroxisome marker vector (B) encodes a fusion protein consisting of EGFP and the peroximal targeting signal 1 (PTS1). Bar, 20 μm.
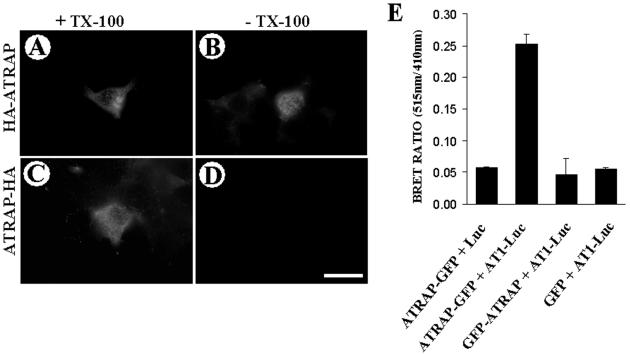
Another aspect of the transmembrane analysis of ATRAP concerns the orientation of the molecule in relation to the plasma membrane. Using epitope-tagged forms of ATRAP either at the N-terminal or C-terminal end, we were able to show that the orientation of the molecule is with the N-terminal end outside of the cell and the C-terminal end in the cytoplasmic face. The HA-epitope tag located at the N-terminal end of ATRAP is accessible to the antibodies both in Triton X-100–permeabilized (Figure 6A) and –nonpermeabilized (Figure 6B) cells; in contrast, cells expressing ATRAP with the epitope tag at the C-terminal end are accessible to the antibody only when the cells are permeabilized with detergent (Figure 6, C and D). Additionally, in intact cells, after transfection of chimeric forms of ATRAP fused to GFP either at the N- or C-terminal end and the AT1 receptor fused to luciferase in the C-terminal cytoplasmic end, we were able to detect BRET signal only when ATRAP was tagged with GFP at the C-terminal end (Figure 6E). These observations give further support to the proposal that the C-terminal end of ATRAP is oriented toward the cytoplasmic face of the membranes.
Figure 6.
Determination of the orientation of the N-terminal end of ATRAP. (A–D) HEK-293 cells were transiently transfected with HA-ATRAP full-length tagged at either the N-terminal or C-terminal end and processed for immunofluorescence with or without Triton X-100 membrane solubilization after fixation. The HA-epitope tag located at the N-terminal end of ATRAP is accessible to the antibodies both in Triton X-100–permeabilized (A) and –nonpermeabilized cells (B); in contrast, cells expressing ATRAP with the epitope tag at the C-terminal end are accessible to the antibody only when the cells are permeabilized with detergent (C and D). Bar, 30 μm. (E) BRET assay showing that only ATRAP epitope tagged at its C-terminal end is able to give BRET a signal with AT1 receptor tagged with luciferase at the C-terminal tail. HEK-293 cells were transiently transfected with GFP and luciferase constructs at a ratio of 3:1. Forty-eight hours posttransfection, HEK-293 cells were detached with PBS/EDTA and washed twice in PBS. Approximately 50,000 cells/well were distributed in a 96-well microplate; the DeepBlue coelenterazine substrate was added at a final concentration of 5 μM, and readings were collected in a Victor2 microplate reader with filters at 410- and 515-nm wavelengths.
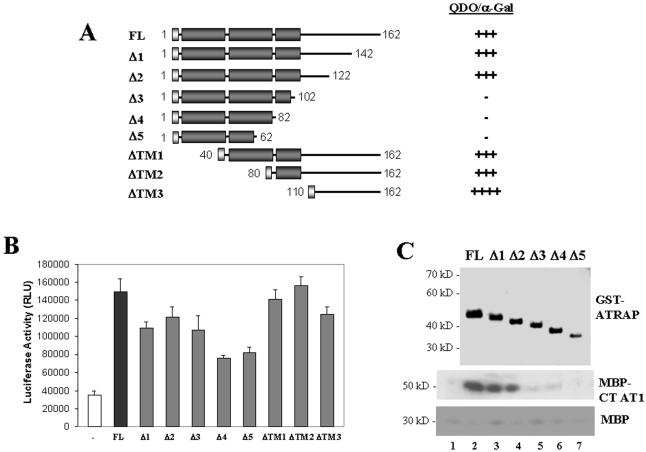
This structural and topological analysis of ATRAP gave us some hints about the biological function of ATRAP; however, to understand the specific role of this protein in AT1 signaling, we characterized the biochemical determinants of the AT1–ATRAP interactions and the consequences of this interaction in AT1 signaling. Our initial report of ATRAP interaction with AT1 showed that the last 20 amino acid residues of the receptor are the target of interaction with ATRAP (Daviet et al., 1999). The other way around, having defined some structural domains in ATRAP, we sought to identify the domains of interaction present in ATRAP responsible for the binding to AT1. Using a yeast two-hybrid assay, we determined that the region of contact between ATRAP and AT1 receptor lies in the C-terminal domain of ATRAP. By using a set of deletions both from the C-terminal and N-terminal ends of ATRAP, we found that the amino acid residues 110–120 seem to be critical for interaction with the receptor (Figure 7A). To demonstrate that this interaction happens in the absence of any additional factor that could be present in the cellular assays, we performed an in vitro pulldown assay with recombinant proteins. The top panel of Figure 7C shows the expression of GST-ATRAP constructs, and the bottom panels show the recovery after pulldown with glutathione beads of the C-terminal tail of AT1 fused to MBP or of MBP alone as a control. The C-terminal deletions of ATRAP lose the ability to interact with AT1, suggesting that the domain of interaction with the receptor is in the cytoplasmic tail of ATRAP. Additionally, we obtained similar results by using a modified two-hybrid assay for mammalian cells that allows full posttranscriptional processing of the transgenes (Figure 7B). Overall, these three independent approaches of domain interaction agree in ruling out an involvement of the ATRAP first two transmembrane domains in the interaction with AT1.
Figure 7.
Domains of interaction of ATRAP with AT1. (A) Schematic representation of the yeast two-hybrid constructs used in the interaction assay with the C-terminal tail of AT1 receptor. The yeast reporter strain AH109 was cotransformed with the deleted versions of the ATRAP two-hybrid expression plasmid pGADT7 and the C-terminal tail of AT1 cloned in the two-hybrid expression plasmid pGBKT7. The cotransformants were selected in SD medium lacking leucine, tryptophan, histidine, and adenine (QDO). The yeast α-galactosidase activity was determined in plates containing X-α-Gal (2 mg/ml) as a chromogenic substrate. The strength of interaction was evaluated qualitatively as the intensity of the development of blue color in yeast colonies growing in selective medium. (B) Results of the mammalian two-hybrid analysis of ATRAP–AT1 interaction. HEK-293 cells were transiently transfected with the indicated deletion versions of ATRAP cloned in the target vector pCMV-NF-κB-AD together with the C-terminal end of AT1 cloned in the vector pCMV-GAL4-BD. Cells were lysed after 48 h of expression, and the luciferase activities were determined. (C) Coomassie Blue staining of the GST-fusion proteins purified from bacteria (top) and the results of the pulldown of the MBP fusion of AT1 (middle) or of MBP alone as a control (bottom). The lane number 1 shows the pulldown with GST alone.
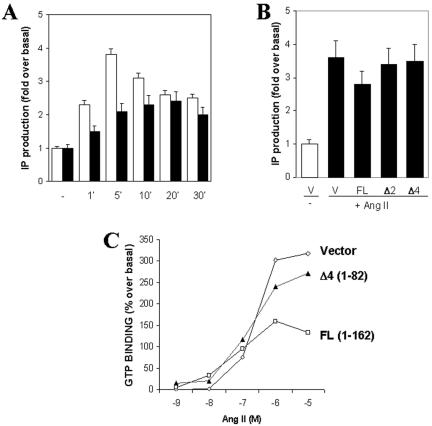
To evaluate the functional consequences of ATRAP overexpression, we performed an analysis of the effects of ATRAP on Ang II-induced AT1 receptor signaling (Figure 8). Our preliminary characterization of the ATRAP effect on inositol lipid generation showed a moderate decrease in the generation of these lipids after treatment with increasing concentrations of Ang II (Daviet et al., 1999). CHO K1 cells stably expressing AT1 receptor were transiently transfected with full-length ATRAP or the panel of C-terminal and N-terminal deletions; after cells were labeled overnight with myo-[3H]inositol in serum-free medium, they were incubated with 10 mM LiCl, stimulated with Ang II for different times and concentrations, and the production of inositol phosphate determined. Figure 8A shows the effect of ATRAP on the generation of IP3 after stimulation with Ang II at different time intervals. The expression of C-terminal deletion mutants had no effect in the Ang II-induced IP3 generation (Figure 8B). To analyze the biochemical mechanism of this effect, we evaluated the ability of ATRAP to affect the coupling to G proteins in GTP-binding assays by using a time-resolved fluorescence protocol. Membrane preparations of HEK-293 cells overexpressing AT1 with or without ATRAP were tested in the presence of increasing concentrations of Ang II. The overexpression of full-length ATRAP led to a decrease in the GTP binding after Ang II stimulation (Figure 8C) in comparison with the transfection of vector alone or the ATRAP mutant (Δ4, aa 1–82), which is unable to bind to the receptor.
Figure 8.
Functional effects of the overexpression of ATRAP in Ang II-induced signaling. (A) Inositol phosphate production in CHO-K1 cells stably expressing AT1 receptors and transiently transfected with full-length ATRAP. Transfected cells were labeled overnight with myo-[3H]inositol in serum-free DMEM. After stimulation with Ang II 1 × 106 M, cell lysates were extracted and separated on Dowex AG1-X8 columns. Bars in white are cells transfected with empty vector, and bars in black are cells transfected with ATRAP and treated with Ang II, 1 × 106 M for the indicated times. (B) Inositol phosphate production in CHO-K1 cells transfected with deletion mutants of ATRAP after 5 min of treatment with Ang II 1 × 106 M. (C) Determination of GTP-loading into membrane preparations of HEK-293 cells overexpressing AT1 receptors and the indicated ATRAP constructs. The determinations were performed using a time-resolved fluorescence protocol. Membrane preparations (10 μg) were incubated in the presence or absence of Ang II for 30 min; GTP-Eu (30 nM) was added for an additional 30 min before filtration, washing, and measurement at 615 nm on a microplate reader.
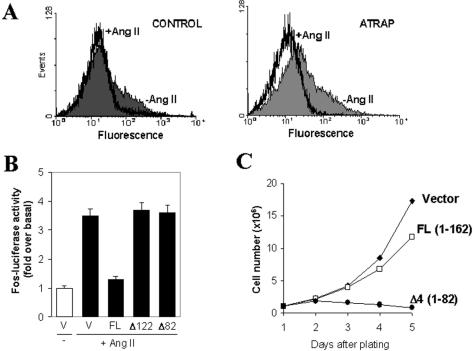
In contrast to the proximal effectors, such as G proteins, phospholipase C (PLC), or kinases that interact directly with the AT1 receptor, additional downstream signaling events that can be affected by agonist binding to AT1 are internalization and Ang II-induced transcriptional activation. The effect of ATRAP on internalization of AT1 receptor was evaluated in HEK-293 cells stably overexpressing both epitope-tagged AT1 receptor and full-length ATRAP. After 5 min of stimulation with 1 × 10-6 Ang II, the cells were fixed in PFA, washed, and stained with monoclonal antibodies to the FLAG epitope tag. After cells were incubated for 30 min, washed, and incubated with the secondary antibody, they were subjected to flow cytometry to detect the number of cells expressing the receptor on their surface. The fluorescence of cells overexpressing ATRAP shifted to the left (Figure 9A), indicating an enhancement of receptor internalization. We also were able to show that transfection of ATRAP in CHO-K1 cell stably expressing AT1 is able to induce a decrease in both the basal and Ang II-stimulated transcriptional activity of the c-fos promoter luciferase reporter gene (Figure 9B). This effect seems to be dependent on the C-terminal domain of ATRAP, as the transfection of C-terminal deletion mutants of ATRAP (Δ2, 1–122 aa; Δ4, 1–82 aa) has no effect on this reporter. The down-regulation of the transcriptional activity suggested a general effect of ATRAP on cell proliferation. Cells transfected with full-length ATRAP-RFP and sorted to obtain a homogenous population were plated in equal numbers; using these cells, we observed a moderate decrease in the rate of proliferation (Figure 9C). In contrast, the cells expressing the C-terminal deletion of ATRAP (Δ4, 1–82 aa) showed a dramatic decrease in the rate of proliferation, became rounded, and lost attachment to the plate surface after a few passages. The behavior of cells expressing a shorter C-terminal deletion (Δ2, 1–122 aa) is similar to that of the full-length expressing cells. Together, these results identify ATRAP as a transmembrane protein with potential involvement in vesicular trafficking and cell signaling induced by Ang II.
Figure 9.
(A) Effect of ATRAP on internalization of AT1 receptor. HEK-293 cells stably expressing FLAG-epitope–tagged AT1 receptor and cotransfected with control vector (left) or full-length ATRAP (right) were stimulated with 1 × 10-6 M Ang II for 5 min and labeled with monoclonal antibodies specific to the FLAG epitope tag. Fluorescence analysis of 20,000 cells was performed in a fluorescence-activated cell sorting scan flow cytometer. (B) Fos-luciferase activity in CHO-K1 cells stably expressing AT1 receptors and transiently transfected with full-length ATRAP or the indicated deletion mutants and stimulated with Ang II 1 × 106 M for 2 h. (B) Cell proliferation in HEK-293 cells stably transfected with ATRAP-RFP. After transfection and sorting, the cells were maintained in cell culture and subsequently plated in equal numbers (1 × 106 cells/well) to respective plates; at each respective time cells were detached with PBS and counted in a hemocytometer.
DISCUSSION
The identification of novel interacting partners of GPCRs in general and of AT1 receptor in particular has shown the dynamic and complex nature of the molecular signaling mechanisms of these receptors. The identification of these novel protein–protein interactions has led to the realization that the mechanisms of receptor signaling are more diverse than previously thought (Brzostowski and Kimmel, 2001; Marinissen and Gutkind, 2001; Pierce et al., 2001; Hall and Lefkowitz, 2002). From the complex network of GPCR-interacting proteins, it seems that some of these interactions are likely to be general, e.g., arrestins and GRKs can bind to many seven-transmembrane receptors; other interactive mechanisms, such as the activation of small GTP-binding proteins or the formation of SH2-based signaling complexes, may be relevant to only a number of GPCRs (Hall et al., 1999; Pierce et al., 2001). Still other mechanisms are likely to be highly receptor-specific, such as for the binding of sodium-hydrogen exchanger regulatory factor (NHERF) to β-adrenergic receptors and of Homer to metabotropic glutamate receptors that depend on the presence of specific motifs that are likely to be found in few other seven-transmembrane receptors (Hall et al., 1999; Tang et al., 1999). The identification of ATRAP as an interacting partner of the AT1 receptor provides us with the opportunity to explore the role of this protein in AT1 receptor signaling.
The initial primary structure characterization of ATRAP did not show any obvious catalytic domain, functional signature, or significant homology to any previously described protein. It seems that ATRAP is evolutionarily recent in the phylogenetic tree, because we have not found any ATRAP homolog in the fully sequenced genomes of yeast, Caenorhabditis elegans, or Drosophila melanogaster; however, ATRAP seems to be widely expressed in mammalian tissues. Using affinity-purified rabbit antiserum raised to the C-terminal end of ATRAP, we have found immunoreactivity to ATRAP in a variety of cells and tissues and prominently in heart, kidney, and brain (our unpublished observations). The lack of any significant homology or known signature domain in ATRAP prompted us to examine in more detail the primary and secondary structure of ATRAP. Using a number of computer algorithms, we found a consensus prediction of three hydrophobic domains in the N-terminal end of the molecule that encodes for putative transmembrane domains. The insolubility in Triton X-100, the particulate localization of both endogenous and overexpressed protein in a transient transfection system, and the electron microscopy analysis agree with the bioinformatic predictions. The last two transmembrane domains are rich in apolar residues, whereas the first transmembrane domain contains some polar amino acids that generate a less hydrophobic domain. Such less hydrophobic helices involving polar residues are often functionally significant in the interior of membrane proteins, for example, in ion pairs of channel proteins (Hirokawa et al., 1998). Membrane proteins destined for organelles of the secretory pathway or the plasma membrane are initially inserted in the ER membrane (Goder and Spietz, 2001). These proteins contain stretches of amino acids that target them to the ER; in fact, the first transmembrane domain of ATRAP contains a sequence compatible with a signal peptide sequence. The short hydrophilic stretches flanking the signal peptide sequence can affect the processing of ATRAP by a signal peptidase; in Western blot analysis we have not found evidence for processed forms of ATRAP, suggesting that the first transmembrane domain can act as a signal anchor domain.
Subcellular localization is critical for the action and regulation of the AT1 receptor (Hunyady, 1999). Thus, an important way to explore ATRAP function is to determine whether it localizes to cellular areas compatible with receptor activity. The finding of ATRAP both at the cell periphery and in intracellular vesicles is compatible with the cellular distribution of the AT1 receptor. AT1 receptor is known to follow a dynamic membrane traffic, becoming part of early endocytic vesicles rapidly after agonist stimulation and recycling back to the plasma membrane after desensitization (Hunyady, 1999; Inagami, 1999); a fraction of the internalized receptor also becomes a target for degradation in lysosomal vesicles. In immunostaining analysis of the intracellular vesicles that harbor ATRAP, we have found significant colocalization of ATRAP with known markers of the early endocytic pathway, Golgi, and ER. These immunostaining results are in agreement with the results of electron microscopy analysis showing prominent perinuclear vesicles. On the other hand, we were able to show colocalization of ATRAP with the ER marker DiOc(6) and partial colocalization with the ER protein calreticulin. In addition, in yeast two-hybrid screens in which ATRAP was used as a bait, we found a number of interacting candidates that also locate at the ER (our unpublished observations). In neonatal rat cardiomyocytes, we were able to localize endogenous ATRAP in endoplasmic/sarcoplasmic structures, giving support to the intracellular membrane localization of this protein and implicating ATRAP in a sarcoplasmic reticulum-related function.
The C-terminal tail of the AT1 receptor is clearly involved in the control of intracellular recycling of the receptor. ATRAP as a novel AT1 receptor C-terminal–interacting protein may be involved in some of the events responsible for the trafficking of the receptor. A question that arises from the vesicular localization of ATRAP is whether this localization is regulated by Ang II. In preliminary experiments, we have not found any significant modification of ATRAP trafficking before and after stimulation with Ang II. The real-time tracking of fluorescent forms of ATRAP-containing vesicles showing the fusion to the peripheral plasma membrane suggests that these vesicles travel through the exocytic arm of the vesicular trafficking process. Because ATRAP is ubiquitously expressed and is particularly highly expressed in kidney, it is interesting to speculate whether ATRAP can have a particular localization and function in kidney polarized cells.
Another fact that underlines the notion of a specific role of ATRAP in vesicle biology is the dramatic formation of gigantic vesicular clusters in cells expressing a mutant of ATRAP that lacks the whole cytoplasmic domain plus the third transmembrane domain. On the other hand, the complementary mutant that lacks the two first transmembrane domains showed sharp plasma membrane localization. These results suggest that there are specific domains responsible for the anchorage and mobilization of the protein; we propose that the third transmembrane domain together with the carboxyl tail contains information that makes possible the translocation of the vesicle to the peripheral plasma membrane, whereas the first and second transmembrane domains alone are unable to travel to the periphery and become trapped in a intracellular vesicular compartment or contain information that allows fusion of vesicles. The specific mechanisms of such control are currently being characterized in our laboratory.
A key question in our study relates to the influence of ATRAP on the actions of the AT1 receptor. It has recently been shown that ER resident proteins, such as the tyrosine phosphatase PTP1B and the phosphatidylinositol kinase hVps34, play a role in the regulation of signal transduction of tyrosine kinase receptors in a step after internalization and before the routing of the receptors to lysosomes (Gill, 2002; Haj et al., 2002). Whether ATRAP participates in a specific step of the internalization or in a step in the synthetic pathway of AT1 receptor warrants further investigation.
Having defined some structural domains in ATRAP, we sought to identify the domains of interaction present in ATRAP responsible for the binding to AT1. Using three independent approaches (yeast two-hybrid, mammalian two-hybrid, and recombinant pulldown assays), we found that the region of amino acids 110–122 located at the C-terminal domain seems to be critical for interaction with the receptor. The construct Δ3 (aa 1–102) shows a positive signal only in the mammalian two-hybrid analysis, although at lower levels than that of full-length ATRAP. The difference in the assay systems (e.g., different cell systems, expression vectors, conditions of assays) could explain some of the differences observed; on the other hand, it is remarkable that the three independent approaches indicate that the N-terminal ATRAP construct (aa 1–82) is unable to bind to AT1 receptor.
From our previous report (Daviet et al., 1999) and the additional analysis performed in the present study, we were able to observe that ∼10% of total ATRAP is complexed with AT1 receptor in immunoprecipitation experiments. This result is in agreement with the cellular colocalization analysis that shows a significant but not complete colocalization of these proteins, suggesting that ATRAP and AT1 can have other effectors. Additionally, yeast two-hybrid screens for both AT1 and ATRAP reveal the existence of additional partners for these molecules that may act cooperatively or independently both in time and in a specific cellular location. These questions are interesting, and our laboratory is actively characterizing these interactions.
The hallmarks of activation of the AT1 receptor at the biochemical level include the activation of the phosphoinositide pathway, increase in intracellular calcium, inhibition of adenylate cyclase (depending on the cell type and tissue), and activation of transcription of c-fos, c-myc, AP1, tissue inhibitors of metalloproteinase (TIMP), and α2-macroglobulin promoters (Blume et al., 1999). In addition, it has been shown a variety of other possible ways of signal transduction for Ang II, with tyrosine and serine/threonine phosphorylation playing important roles in these processes. We analyzed the role of ATRAP in the signaling by the AT1 receptor by examining the best characterized pathways of activation. Ang II induces phosphatydylinositide hydrolysis via G protein-mediated stimulation of PLC, leading to the generation of DAG and IP3, which transiently activate protein kinase (protein kinase C) and the release of calcium from intracellular stores. Our studies suggest a modest but consistent effect of ATRAP on the inhibition of PLC response to Ang II and a corresponding ability to decrease GTP binding to membrane preparations. The magnitude of the inhibitory effect of ATRAP overexpression may be influenced by the expression of endogenous ATRAP in cells and additional partners that may be present in limiting amounts relative to overexpressed ATRAP. The lack of effect of the C-terminal end deletion mutants suggests that the interaction of ATRAP with the AT1 receptor is critical for IP3 generation, because these mutants are unable to fully interact with the receptor. Interestingly, the construct 1–122 of ATRAP is unable to suppress Ang II-dependent IP3 production but is still sufficient to interact with AT1 receptor. The exact mechanism of this effect is unknown, but probably conformational changes, strength of interactions in comparison with the full-length construct, binding, or stabilization of additional effectors could explain these results. The increase in internalized AT1 in the presence of ATRAP in vascular smooth muscle cells has been shown by others (Cui et al., 2000) and could be due to an enhancement of internalization or a decrease in recycling back to the plasma membrane. These alternatives and the molecular mechanisms of this effect deserve additional investigation.
In contrast to the proximal effectors that interact directly with AT1 receptor, a downstream signaling event that is a hallmark of the receptor activation is the Ang II-induced transcriptional activation (Blume et al., 1999; Inagami et al., 1999). In fact, one of the effects we have observed in our studies is the inhibition of both basal and stimulated c-fos gene expression. Consistent with the role of c-fos gene expression in cell proliferation, we observed a decrease in the rate of cellular proliferation in stable cell lines overexpressing ATRAP. In particular, the overexpression of the ATRAP mutant lacking the cytosolic tail and the third transmembrane domain seems to have profound effects on the rate of cell proliferation. We observed signs of toxicity in cells expressing this mutant, with rounding, detachment, and loss of the cell from the cell-culture plate. These effects seem to be independent of AT1 interaction and show that ATRAP could have additional effector targets.
A number of additional questions regarding to ATRAPAT1 interaction remain to be analyzed. How does ATRAP interact with the AT1 receptor in the presence of other binding partners of the receptor C-terminal tail such as β-arrestin, GRK, and protein kinase C, for example? The AT1 receptor interacts with ATRAP via the last 20 amino acids; however, it is not known whether ATRAP can influence the interaction of other molecules with the AT1 receptor. This analysis will allow us to understand how proteins from various signaling pathways bind to the C terminus of the AT1 receptor and thereby influence, independently or cooperatively, the biochemical actions of Ang II, e.g., activation of kinase cascades, calcium mobilization, transcriptional activation, and other read-outs yet to be determined.
Our identification of ATRAP as a novel interacting partner of the AT1 receptor adds to the list a potentially interesting molecule able to regulate the actions of seven transmembrane receptors. These results identify a novel interacting partner of the AT1 receptor with the potential ability to modulate some of the biological actions of Ang II. The results of our studies should improve our understanding of the molecular mechanism of action of G protein-coupled receptors in general and, more specifically, how the AT1 receptor controls the cellular machinery responsible for the biological actions of Ang II and may lead to the development of novel pharmacological interventions in the control of the actions of Ang II.
Acknowledgments
We thank Dr. Richard E. Pratt for useful discussions and Maria Ericsson for help with the electron microscopy. This work was supported by National Institutes of Health grant HL058516 (to V.J.D.)
Abbreviations used: Ang II, angiotensin II; AT1, angiotensin II type 1 receptor; ATRAP, AT1 receptor-associated protein; BRET, bioluminescence resonance energy transfer; CFP, cyan fluorescent protein; CHO, Chinese hamster ovary; DAG, diacylglycerol; ER, endoplasmic reticulum; GFP, green fluorescent protein; GST, glutathione S-transferase; GPCR, G protein-coupled receptor; GRK, G protein receptor kinase; HA, hemagglutinin; HEK, human embryonic kidney; MBP, IP3 inositol-1,4,5-triphosphate; MBP, maltose-binding protein; PFA, paraformaldehyde; PLC, phospholipase C; RFP, red fluorescent protein.
References
- Allen, A.M., Zhuo, J., and Mendelsohn, F.A. (2000). Localization and function of angiotensin AT1 receptors. Am. J. Hypertens. 13, 31S-38S. [DOI] [PubMed] [Google Scholar]
- Audoly, L.P., Oliverio, M.I., and Coffman, T.M. (2000). Insights into the functions of type 1 (AT1) angiotensin II receptors provided by gene targeting. Trends Endocrinol. Metab. 11, 263-269. [DOI] [PubMed] [Google Scholar]
- Blume, A., Herdegen, T., and Unger, T. (1999). Angiotensin peptides and inducible transcription factors. J. Mol. Med. 77, 339-357. [DOI] [PubMed] [Google Scholar]
- Brzostowski, J.A., and Kimmel, A.R. (2001). Signaling at zero G: G-protein-independent functions for 7-TM receptors. Trends Biochem. Sci. 26, 291-297. [DOI] [PubMed] [Google Scholar]
- Conchon, S., Barrault, M.B., Miserey, S., Corvol, P., and Clauser, E. (1997). The C-terminal third intracellular loop of the rat AT1 angiotensin II receptor plays a key role in G-protein coupling specificity and transduction of the mitogenic signal. J. Biol. Chem. 272, 25566-25572. [DOI] [PubMed] [Google Scholar]
- Cui, T., Nakagami, H., Iwai, M., Takeda, Y., Shiuchi, T., Tamura, K., Daviet, L., and Horiuchi, M. (2000). ATRAP, novel AT1 receptor associated protein, enhances internalization of AT1 receptor and inhibits vascular smooth muscle cell growth. Biochem. Biophys. Res. Commun. 279, 938-941. [DOI] [PubMed] [Google Scholar]
- Daviet, L., Lehtonen, J.Y., Tamura, K., Griese, D.P., Horiuchi, M., and Dzau, V.J. (1999). Cloning and characterization of ATRAP, a novel protein that interacts with the angiotensin II type 1 receptor. J. Biol. Chem. 274, 17058-17062. [DOI] [PubMed] [Google Scholar]
- Gaborik, Z., Szaszak, M., Szidonya, L., Balla, B., Paku, S., Catt, K.J., Clark, A.J., and Hunyady, L. (2001). Beta-arrestin- and dynamin-dependent endocytosis of the AT1 angiotensin receptor. Mol. Pharmacol. 59, 239-247. [DOI] [PubMed] [Google Scholar]
- Goder, V., and Spietz, M. (2001). Topogenesis of membrane proteins: determinants and dynamics. FEBS Lett. 504, 87-93. [DOI] [PubMed] [Google Scholar]
- Guo, D.F., Sun, Y.L., Hamet, P., and Inagami, T. (2001). The angiotensin II type 1 receptor and receptor-associated proteins. Cell Res. 11, 165-180. [DOI] [PubMed] [Google Scholar]
- Gill, G.N. (2002). A pit stop at the ER. Science 295, 1654-1655. [DOI] [PubMed] [Google Scholar]
- Haj, F.G., Verveer, P.J., Squire, A., Neel, B.G., and Bastiaens, P.I. (2002). Imaging sites of receptor dephosphorylation by PTP1B on the surface of the endoplasmic reticulum. Science 295, 1708-1712. [DOI] [PubMed] [Google Scholar]
- Hall, R.A., and Lefkowitz, R.J. (2002). Regulation of G protein-coupled receptor signaling by scaffold proteins. Circ. Res. 91, 672-680. [DOI] [PubMed] [Google Scholar]
- Hall, R.A., Premont, R.T., and Lefkowitz, R.J. (1999). Heptahelical receptor signaling: beyond the G protein paradigm. J. Cell Biol. 145, 927-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein, L., Meinel, L., Pratt, R.E., Dzau, V.J., and Kobilka, B.K. (1997). Intracellular trafficking of angiotensin II and its AT1 and AT2 receptors: evidence for selective sorting of receptor and ligand. Mol. Endocrinol. 11, 1266-1277. [DOI] [PubMed] [Google Scholar]
- Hirokawa, T., Boon-Chieng, S., and Mitaku, S. (1998). SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14, 378-379. [DOI] [PubMed] [Google Scholar]
- Hunyady, L. (1999). Molecular mechanisms of angiotensin II receptor internalization. J. Am. Soc. Nephrol. 10, S47-S56. [PubMed] [Google Scholar]
- Hunyady, L., Bor, M., Balla, T., and Catt, K.J. (1994). Identification of a cytoplasmic Ser-Thr-Leu motif that determines agonist-induced internalization of the AT1 angiotensin receptor. J. Biol. Chem. 269, 31378-31382. [PubMed] [Google Scholar]
- Hunyady, L., Catt, K.J., Clark, A.J., and Gaborik, Z. (2000). Mechanisms and functions of AT(1) angiotensin receptor internalization. Regul. Pept. 91, 29-44. [DOI] [PubMed] [Google Scholar]
- Inagami, T. (1999). Molecular biology and signaling of angiotensin receptors: an overview. J. Am. Soc. Nephrol. 10, S2-S7. [PubMed] [Google Scholar]
- Inagami, T., Kambayashi, Y., Ichiki, T., Tsuzuki, S., Eguchi, S., and Yamakawa, T. (1999). Angiotensin receptors: molecular biology and signaling. Clin. Exp. Pharmacol. Physiol. 26, 544-549. [DOI] [PubMed] [Google Scholar]
- Kyte, J., and Doolittle, R.F. (1982). A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105-132. [DOI] [PubMed] [Google Scholar]
- Liang, H., Venema, V.J., Wang, X., Ju, H., Venema, R.C., and Marrero, M.B. (1999). Regulation of angiotensin II-induced phosphorylation of STAT3 in vascular smooth muscle cells. J. Biol. Chem. 274, 19846-19851. [DOI] [PubMed] [Google Scholar]
- Marinissen, M.J., and Gutkind, J.S. (2001). G-protein-coupled receptors and signaling networks: emerging paradigms. Trends Pharmacol. Sci. 22, 368-376. [DOI] [PubMed] [Google Scholar]
- Perry, S.J. and Lefkowitz, R.J. (2002). Arresting developments in heptahelical receptor signaling and regulation. Trends. Cell. Biol. 12, 130-138. [DOI] [PubMed] [Google Scholar]
- Pierce, K.L., Luttrell, L.M., and Lefkowitz, R.J. (2001). New mechanisms in heptahelical receptor signaling to mitogen activated protein kinase cascades. Oncogene 20, 1532-1539. [DOI] [PubMed] [Google Scholar]
- Sayeski, P.P., Showkat, A., Semeniuk, D.J., Thanh, N.D., and Bernstein, K.E. (1998). Angiotensin II signal transduction pathways. Reg. Peptides 78, 19-29. [DOI] [PubMed] [Google Scholar]
- Tang, Y., Hu, L.A., Miller, W.E., Ringstad, N., Hall, R.A., Pitcher, J.A., DeCamilli, P., and Lefkowitz, R.J. (1999). Identification of the endophilins (SH3p4/p8/p13) as novel binding partners for the β1-adrenergic receptor. Proc. Natl. Acad. Sci. USA 96, 12559-12564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, W.G., Thekkumkara, T.J., Motel, T.J., and Baker, K.M. (1995). Stable expression of a truncated AT1 receptor in CHO. K1 cells. J. Biol. Chem. 270, 207-213. [DOI] [PubMed] [Google Scholar]
- Tohgo, A., Pierce, K.L., Choy, E.W., Lefkowitz, R.J., and Luttrell, L.M. (2002). Beta-Arrestin scaffolding of the ERK cascade enhances cytosolic ERK activity but inhibits ERK-mediated transcription following angiotensin AT1a receptor stimulation. J. Biol. Chem. 277, 9429-9436. [DOI] [PubMed] [Google Scholar]
- von Zastrow, M. (2001). Role of endocytosis in signalling and regulation of G-protein-coupled receptors. Biochem. Soc. Trans. 29, 500-4. [DOI] [PubMed] [Google Scholar]
- Wang, W., et al. (2002). Identification and characterization of AGTRAP, a human homolog of murine angiotensin II receptor-associated protein (AGTRAP). Int. J. Biochem. Cell. Biol. 34, 93-102. [DOI] [PubMed] [Google Scholar]