Abstract
Telomere length is controlled in part by cis-acting negative regulators that limit telomere extension by telomerase. In budding yeast, the major telomere length regulator scRap1 binds to telomeric DNA and acts to inhibit telomere elongation in cis. Because the human Rap1 ortholog hRap1 does not bind to telomeric DNA directly but is recruited to telomeres by TRF2, we examined its role in telomere length control. The data are consistent with hRap1 being a negative regulator of telomere length, indicating functional conservation. Deletion mapping confirmed that hRap1 is tethered to telomeres through interaction of its C terminus with TRF2. The telomere length phenotypes of hRap1 deletion mutants implicated both the BRCT and Myb domain as protein interaction domains involved in telomere length regulation. By contrast, scRap1 binds to telomeres with its Myb domains and uses its C terminus to recruit the telomere length regulators Rif1 and Rif2. Together, our data show that although the role of Rap1 at telomeres has been largely conserved, the domains of Rap1 have undergone extensive functional changes during eukaryotic evolution. Surprisingly, hRap1 alleles lacking the BRCT domain diminished the heterogeneity of human telomeres, indicating that hRap1 also plays a role in the regulation of telomere length distribution.
INTRODUCTION
The maintenance of human telomeres is regulated at two levels. The primary control of telomere maintenance takes place at the level of telomerase expression. Repression of telomerase in the soma leads to programmed telomere shortening, and this process can limit the replicative life span of primary human cells. However, in the germline and in immortal human cells, telomerase is active and counteracts telomere attrition. Such cells display a second level of control that results in stable maintenance of all telomeres within a limited size range. This telomere length homeostasis is achieved through a negative feedback loop involving cis-acting regulators of telomerase-mediated telomere extension (van Steensel and de Lange, 1997; Kim et al., 1999; Smith and de Lange, 2000; Smogorzewska et al., 2000; Ancelin et al., 2002; Loayza and de Lange, 2003). These negative regulators accumulate along the duplex telomeric repeat array, and their presence at chromosome ends results in increasing inhibition of telomerase as telomeres become elongated.
Studies in yeast and human cells indicate that the principles of telomere length homeostasis are similar in diverged eukaryotes (McEachern and Blackburn, 1995; Krauskopf and Blackburn, 1996; Marcand et al., 1997a; van Steensel and de Lange, 1997). However, the telomeric protein complex responsible for negative length regulation has undergone drastic changes in its composition. A main player in human telomere length homeostasis is the TRF1 complex, composed of the duplex telomeric repeat binding factor TRF1, and its associated proteins tankyrase 1 and 2, TIN2, hPot1, and PINX1 (Chong et al., 1995; Smith et al., 1998; Kim et al., 1999;
Kaminker et al., 2001; Zhou and Lu, 2001; Loayza and de Lange, 2003). In Saccharomyces cerevisiae, an entirely different set of proteins, the telomeric binding protein Rap1 and its interacting factors Rif1 and Rif2, form the major telomere length regulatory complex (Hardy et al., 1992; Marcand et al., 1997b; Wotton and Shore, 1997). Remarkably, there is no TRF-like factor bound to budding yeast telomeres, and the genes for tankyrases and TIN2 seem to be absent from the budding yeast genome.
The human genome encodes a distant ortholog of S. cerevisiae Rap1, hRap1, which differs from its yeast counterpart in that it lacks the ability to directly bind to telomeric DNA (Li et al., 2000; Hanaoka et al., 2001). Instead, hRap1 is recruited to telomeres through interaction with TRF2, a TRF1 paralog (reviewed in de Lange, 2002). The mammalian version of the telomeric complex is likely to represent the ancestral telomere type because Schizosaccharomyces pombe Rap1 also lacks telomeric DNA binding activity and is recruited to telomeres through an interaction with Taz1, which is related to TRF2 both structurally and functionally (Cooper et al., 1997; Chikashige and Hiraoka, 2001; Kanoh and Ishikawa, 2001; de Lange, 2002). Despite these extensive rearrangements within the telomeric complex, Rap1 acts as a negative regulator of telomere length in both fission yeast and two budding yeasts (Conrad et al., 1990; Lustig et al., 1990; Krauskopf and Blackburn, 1996; Marcand et al., 1997a,b; Chikashige and Hiraoka, 2001; Kanoh and Ishikawa, 2001). Here, we further extend the study of these parallels in telomere length regulation and examine the role of hRap1 in telomere length control.
MATERIALS AND METHODS
Retroviral Expression Constructs
A BamHI fragment of hRap1 full-length cDNA was inserted into the BamHI site of pLPC to generate a retroviral construct expressing full-length hRap1 protein. For hRap1 deletion mutant, a FLAG tag (generated by annealing oligos: OM-T-FLAG1 5′-GATCTGCCGCCACCATGGACTACAAAGACGATGACGATAAAG-3′ and OM-T-FLAG2 5′-GATCCTTTATCGTCATCGTCTTTGTAGTCCATGGTGGCGGCA-3′) was first inserted into pLPC at BamHI site. The BamHI site at the N terminus of the FLAG tag was destroyed by ligation to a BglII site. PCR fragments containing different hRap1 deletions were generated, digested with BamHI, and inserted in frame 3′ of the FLAG tag.
Retroviral Infection and Selection
Retroviral gene delivery was performed as described previously (Karlseder et al., 2002). Phoenix amphotropic retroviral packaging cells were transfected with pLPC vectors harboring different alleles of hRap1. At 24, 32, 48, and 56 h posttransfection, HTC75, IMR90, GM847, or HeLa1.2.11 cells were incubated with the virus-containing medium with 4 μg/ml polybrene. Sixteen hours after the last infection, puromycin selection (2 μg/ml) was started and maintained for 5–7 d. Infection efficiencies were typically >90% in HTC75, IMR90, and GM847 cells and ∼20% in HeLa1.2.11 cells. Cells were maintained in DMEM medium with 10% (HTC75 and GM847) or 15% (IMR90) fetal bovine serum or 10% bovine calf serum (HeLa1.2.11), 2 mM l-glutamine, 0.1 mM nonessential amino acids, 100 U/ml penicillin, and 0.1 mg/ml streptomycin. Cells were grown in 10- or 15-cm dishes until 70–80% confluence and split 1:16 (HTC75, HeLa1.2.11, and GM847) or 1:3 (IMR90). IMR90 cells were infected at passage 13 (population doubling [PD]30) and the selection lasted for 2–3 PD. One passage of IMR90 represents a 1:3 split of cells. For telomere elongation experiments in HTC75 and GM847 cells, PD0 was arbitrarily set after infected cells reached 80% confluence after the first split into four 10-cm dishes. All cell populations went through approximately the same number of divisions before reaching “PD0.”
Telomere Length Analysis
Cells from a subconfluent 15-cm dish were trypsinized, washed in cold 1× phosphate-buffered saline (PBS). DNA was isolated from the cell pellet as described previously (de Lange et al., 1990) with inclusion of an RNase-A digestion step. DNA from HTC75 and IMR90 cells was digested with HinfI/RsaI, and genomic blotting for telomeric fragments was carried out according to standard protocols (van Steensel and de Lange, 1997). For telomere length analysis in GM847 cells, DNA was isolated from cells embedded in agarose plugs. DNA embedded in agarose plugs was digested with MboI/AluI and electrophoresed through 1% agarose gels in 0.5× Tris borate-EDTA maintained at 13°C, by using a CHEF DR-II pulsed-field apparatus (Bio-Rad, Hercules, CA). Separation was for 18 h at 5.8 V/cm at a constant pulse time of 5 s. In-gel hybridization was carried out using an end-labeled (CCCTAA)4 oligo as described previously (Karlseder et al., 2002) after in situ denaturation of the DNA. Telomere blots or gels were exposed to PhosphorImager screens for 24 h.
The data was analyzed by ImageQuant and Microsoft Excel. Mean telomeric restriction fragment lengths were calculated assuming a Gaussian distribution. Because of the presence of subtelomeric sequences, the length of the telomeric TTAGGG repeat region is assumed to be 2 kb shorter than the telomeric restriction fragment length. Telomere length distributions and the telomere length heterogeneity index in Table 2 were determined as follows. The signal intensity was determined over the lane as a function of the distance from the slot. A baseline was used that reflected the slight increase in signal background at higher molecular weight (MW). The accumulative sum of signal intensity with baseline correction was calculated as a function of the distance from the slot. The distances from the slot at which the summation of the signal reached 25 and 75% of the total signal were identified and the telomeric restriction fragment sizes corresponding to these positions were calculated based on molecular weight markers. The difference between these MW values is referred to as the 50% signal interval (in kilobases) and functions as a measure for the size distribution of the telomeric signal. The telomere length heterogeneity index is defined as the 50% signal interval (in kilobases) divided by the median telomere length (in kilobases). In control cells, the heterogeneity index is close to 1. For instance, when the mean telomere length is 3 kb, 50% of the telomeric signal is found in a 3-kb size interval.
Table 2.
Effects of hRap1 mutants on telomere length heterogeneity
| hRap1 allele | PDa | Median lengthb | 50% signal intervalc | Heterogeneity indexd |
|---|---|---|---|---|
| hRap1 | 0 | 1.9 | 1.7 | 0.89 |
| hRap1ΔCT | 0 | 3.2 | 3.0 | 1.25 |
| hRap1ΔBr | 0 | 2.8 | 2.6 | 0.93 |
| hRap1ΔBrΔCT | 0 | 2.9 | 3.2 | 1.09 |
| hRap1ΔMΔC | 0 | 2.6 | 2.9 | 1.10 |
| pLPC | 108 | 2.6 | 3.3 | 1.25 |
| pLPC | 120 | 2.6 | 2.9 | 1.13 |
| hRap1 | 120 | 4.8 | 5.2 | 1.10 |
| hRap1ΔCT | 108 | 7.7 | 6.9 | 0.90 |
| hRap1ΔCT | 120 | 7.8 | 6.7 | 0.86 |
| hRap1ΔBr | 120 | 9.0 | 4.8 | 0.53 |
| hRap1ΔBr | 132 | 9.1 | 5.0 | 0.55 |
| hRap1ΔBrΔM | 108 | 9.7 | 4.1 | 0.47 |
| hRap1ΔBrΔM | 180 | 11.8 | 5.6 | 0.47 |
| hRap1ΔMΔC | 96 | 10.0 | 7.5 | 0.75 |
| hRap1ΔMΔC | 108 | 10.9 | 7.3 | 0.67 |
| hRap1ΔM | 132 | 11.6 | 9.0 | 0.78 |
PD 0 represents the first split after selection for retroviral infection. The number of population doublings at this PD could vary slightly for different hRap1 mutants
Median telomere length (in kb) was determined based on telomeric restriction fragment blots by scanning through the lanes. The portion of subtelomeric DNA on telomeric restriction fragments is assumed to be 2 kb, and this length is subtracted from all values
50% signal interval (in kb) represents the size distribution covering 50% of the telomeric signal (MATERIALS AND METHODS)
The heterogeneity index represents the ratio of the 50% signal interval (fourth column) and the median telomere length (third column)
Indirect Immunofluorescence (IF)
For IF, cells were washed with PBS buffer, fixed with 2% formaldehyde/PBS, and permeabilized with 0.5% NP-40/PBS, followed by antibody incubations as described previously (Smogorzewska et al., 2000). For Triton X-100 extraction experiments, cells were washed with PBS buffer, treated with Triton X-100 buffer (0.5% Triton X-100, 20 mM HEPES, pH 7.9, 50 mM NaCl, 3 mM MgCl2, 300 mM sucrose) for 5 min at room temperature (RT), fixed with 3% formaldehyde/2% sucrose/PBS, and permeabilized with the same Triton X-100 buffer, followed by antibody incubations as described in Zhu et al. (2000). Endogenous and retrovirally expressed full-length hRap1 was detected with 765 rabbit anti-hRap1 serum (Li et al., 2000). TRF1 was detected with a polyclonal mouse anti-hTRF1 serum or rabbit 371 anti-hTRF1 serum (van Steensel and de Lange, 1997). hRap1 deletion mutants were detected using anti-FLAG antibody M2 (Sigma-Aldrich, St. Louis, MO).
Cell Extracts and Western Blotting
Whole cell extracts were prepared using a modified radioimmunoprecipitation assay buffer (50 mM Tris-Cl, pH 7.4, 1% Triton X-100, 0.1% SDS, 150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin, 10 μg/ml pepstatin, 1 μg/ml leupeptin, 0.4 M NaCl.). For Western blotting, 60 μg of protein was separated on SDS/10% PAGE and transferred to nitrocellulose by electroblotting. Membranes were stained with Ponceau S to verify equal amount loading of protein samples. Membranes were then blocked with 1× PBS/10% nonfat dry milk/0.5% Tween 20 at RT for 1 h, probed with primary antibodies in 1× PBS/0.1% Tween 20/0.1% nonfat dry milk at 4°C for 16 h and secondary horseradish peroxidaseconjugated donkey anti-rabbit antibody (for antibodies 371, 647, and 765) or sheep anti-mouse antibody (for antibodies M2 and GTU88) at RT for 30 min. Membranes were washed with the same incubation buffer three times before detecting protein bands using an enhanced chemiluminescence kit (Amersham Biosciences, Piscataway, NJ). Expression of hRap1 full-length or mutant proteins was detected using 765 rabbit anti-hRap1 serum or M2 monoclonal antibody against FLAG (see above). Endogenous γ-tubulin was detected with monoclonal antibody GTU88 (Sigma-Aldrich). Endogenous TRF1 and TRF2 proteins were detected by rabbit antibodies 371 and 647, respectively.
RESULTS
Subnuclear Localization of hRap1 Deletion Mutants
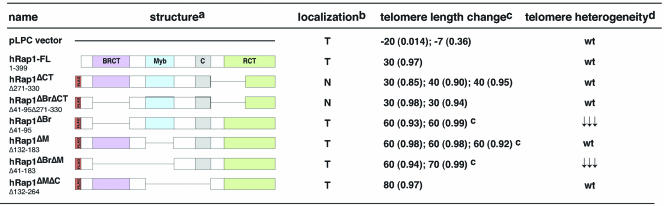
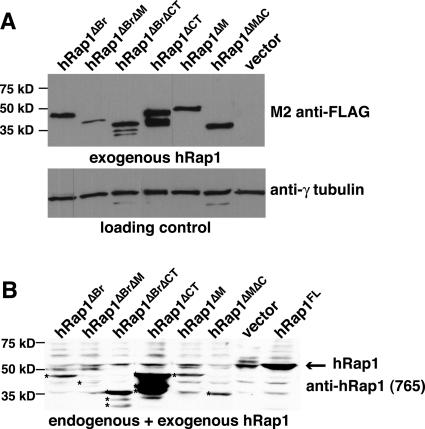
Human Rap1 has four domains, a BRCT domain, a Myb HTH motif, a coiled region, and a C-terminal protein interaction domain (termed RCT) (Table 1) (Li et al., 2000). The RCT region is known to be required for binding to TRF2 and mediates the association of hRap1 with telomeres. The functions of the other domains are not known. Myb domains are more commonly involved in DNA binding, but no such activity has been demonstrated for the hRap1 Myb domain and its structure is not well suited for DNA binding (Li et al., 2000; Hanaoka et al., 2001). Therefore, it was proposed that the hRap1 Myb domain may be involved in protein interaction as has been shown for several other proteins (Li et al., 2000; Hanaoka et al., 2001). We generated 11 hRap1 deletion mutants lacking one or more of its domains and monitored their expression in human cells that also expressed endogenous (wt) hRap1. Each of the alleles was endowed with an N-terminal FLAG tag, allowing detection of the exogenous protein in IF and immunoblotting. Five hRap1 mutants (two lacking the N-terminal 94 or 184 aa, two lacking the C-terminal 25 or 40 aa; and one triple mutant lacking the BRCT domain, Myb domain, and part of the RCT domain) were expressed at extremely low levels (our unpublished data) and not studied further. The other six hRap1 mutants yielded stably expressed proteins as determined by immunoblotting (Figure 1, A and B). Some of the stably expressed hRap1 mutants were overexpressed compared with the endogenous protein, whereas others were expressed at a level comparable with or lower than endogenous hRap1 (Figure 1B and Table 1). Similar expression levels were observed in HeLa1.2.11, IMR90, and GM847 cells (our unpublished data). There was no obvious correlation between the expression levels of the various hRap1 alleles and their telomere length phenotype (see below).
Table 1.
Localization and telomere phenotypes of hRap1 deletion mutants
Figure 1.
Immunoblotting analysis of hRap1 alleles. Extracts were prepared from retrovirally infected HTC75 cells expressing the indicated hRap1 alleles, and immunoblots were probed for the FLAG epitope added to the exogenous hRap1 alleles (M2) and γ-tubulin in A or for hRap1 (antibody 765) in B. The arrow in B indicates the position of endogenous hRap1. Asterisks in B mark bands representing the hRap1 mutant proteins. Some mutant proteins show degradation products.
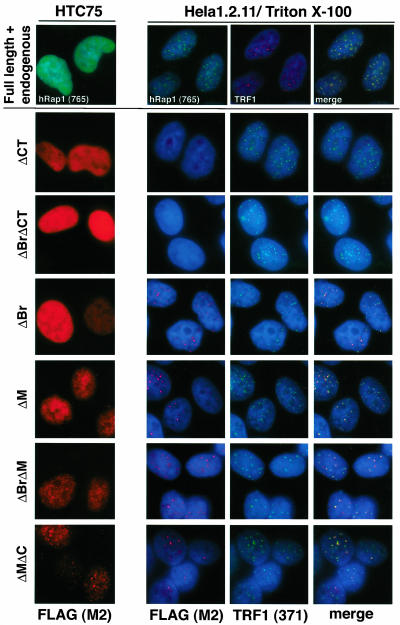
The subnuclear localization of the altered hRap1 proteins was determined by IF in two human tumor cell lines, the HT1080-derived line, HTC75, and HeLa1.2.11, a HeLa subclone with long telomeres (van Steensel and de Lange, 1997). Retroviral expression of full-length hRap1 as well as all mutant alleles resulted in a predominantly nucleoplasmic staining pattern in both HTC75 cells and in HeLa1.2.11 (Figure 2; our unpublished data). Because this pattern is likely due to overexpression and saturation of the telomeric hRap1 binding sites, we examined HeLa1.2.11 cells after extraction of the nucleoplasmic proteins with Triton X-100 (Figure 2). Under these conditions, telomeric association of full-length hRap1 as well as four of the six mutant hRap1 proteins could be demonstrated based on the colocalization with the telomeric protein TRF1, previously shown to be exclusively located at telomeres (Chong et al., 1995). As expected, two hRap1 mutants lacking the C-terminal TRF2 binding domain (hRap1ΔCT and hRap1ΔBrΔCT) were not detectable at telomeres after Triton X-100 extraction, indicating that these alleles were primarily expressed in the nucleoplasm. These results are consistent with previous data indicating that hRap1 depends on its interaction with TRF2 to localize to telomeres (Li et al., 2000) and that other domains, such as the BRCT domain and the Myb domain are not required for the accumulation of hRap1 in the telomeric complex. IF analysis also showed that the hRap1 mutant alleles do not significantly affect the accumulation of TRF1 and TRF2 on telomeres (Figure 2; our unpublished data). In addition, the endogenous protein levels for TRF1 and TRF2 are not affected by expression of exogenous hRap1 alleles (our unpublished data).
Figure 2.
Subnuclear localization of hRap1 alleles. Top, IF for hRap1 (antibody 765) and TRF1 (detected with a mouse polyclonal) in HTC75 and HeLa1.2.11 cells infected with full-length untagged hRap1 retrovirus. The HeLa1.2.11 cells were extracted with Triton X-100 before IF. Bottom, IF for exogenous hRap1 proteins (M2) was carried out in retrovirally infected HTC75 and Triton-extracted HeLa1.2.11 cells. Telomeric loci are identified based on TRF1 IF (antibody 371). DNA was stained by 4,6-diamidino-2-phenylindole. hRap1 mutant alleles are identified to the left of each IF set.
The hRap1 interacting partner TRF2 is required for telomere protection (reviewed in de Lange, 2002). Inhibition of TRF2 by using truncation alleles results in partial loss of the telomeric G-strand overhang, telomere fusions, and cell cycle arrest, accompanied by senescence or apoptosis. None of these phenotypes were observed upon expression of the hRap1 mutants, and there was no obvious effect on the viability and growth characteristics of the cells (our unpublished data). This was true both in short-term cultures and upon long-term growth of tumor cell lines and primary human fibroblasts (our unpublished data).
Telomere Elongation Induced by Expression of hRap1 Alleles in HTC75 Cells
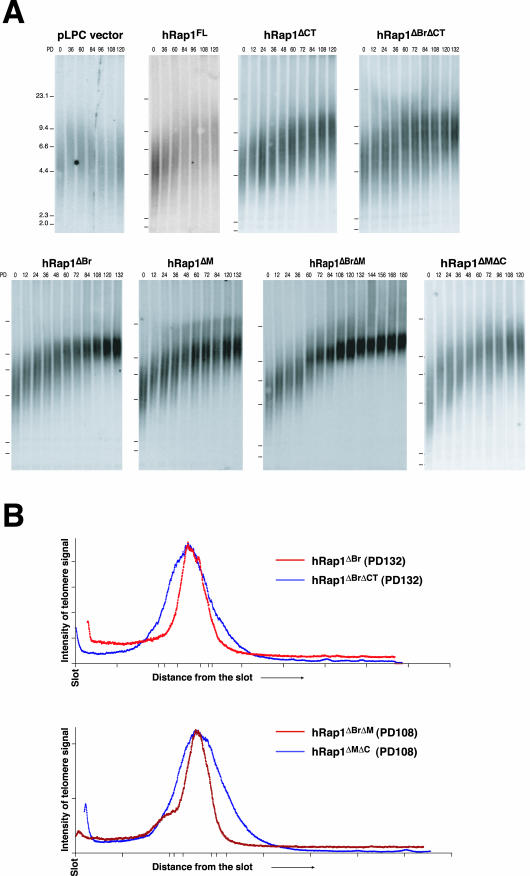
The effect of hRap1 on telomere length homeostasis was determined using the established HTC75 telomere length control test system. HTC75 cells express telomerase and maintain their telomeres at a stable length setting through a homeostasis pathway (van Steensel and de Lange, 1997). Using these cells, it was previously shown that telomere length control is governed by TRF1, tankyrase 1, TIN2, TRF2, and hPot1 (van Steensel and de Lange, 1997; Kim et al., 1999; Smith and de Lange, 2000; Smogorzewska et al., 2000; Loayza and de Lange, 2003). HTC75 cells were used to study the effects of retrovirally expressed hRap1 alleles. Infected cells were selected with puromycin, and a pool of infected cells was examined for telomere length dynamics by genomic blotting. Although HTC75 cells, like other human tumor cell lines, can display a high degree of variation in telomere length and telomere dynamics, the analysis of large pools of infected cells reduces outlier effects, allowing the analysis of the effect of exogenous proteins on telomere length setting. HTC75 cells overexpressing wild-type hRap1 were found to display a telomere elongation phenotype (initial elongation rate of ∼30 base pairs/end/PD) in several independent pools of infected cells (Figure 3A and Table 1). Control HTC75 cells infected with the retroviral vector have stable telomeres, or slight telomere shortening (Figure 3A and Table 1) (van Steensel and de Lange, 1997; Li et al., 2000; Smogorzewska et al., 2000). The elongation phenotype of wild-type hRap1 is consistent with our previous finding that inducible expression of an N-terminally FLAG-tagged hRap1 allele resulted in a slight increase in telomere length in HTC75 cells (Li et al., 2000).
Figure 3.
(facing page). Telomere length changes in HTC75 cells expressing hRap1 mutants. (A) Genomic blotting analysis of telomeric restriction fragments in HinfI/RsaI-digested genomic DNA from retrovirally infected HTC75 cells, probed with a TTAGGG repeat fragment. Population doublings (PDs), expressed hRap1 alleles, and MW markers are indicated with each blot. (B) Charts showing the distribution of signal intensity for telomeric southern blots in lanes representing hRap1ΔBrΔM- and hRap1ΔMΔC-expressing cells at PD108 or hRap1ΔBr- and hRap1ΔBrΔCT-expressing cells at PD132.
We next examined the four mutants of hRap1 that had retained the ability to interact with TRF2 and therefore localized to telomeres (hRap1ΔBr, hRap1ΔM, hRap1ΔBrΔM, and hRap1ΔMΔC). These mutants also induced telomere elongation in multiple independent experiments (Figure 3A and Table 1). For these mutants, the elongation rates seemed higher than for full-length hRap1 (60–80 base pairs/end/PD for the mutants versus 30–40 base pairs/end/PD for wild-type hRap1; Table 1). These differences are not attributable to expression levels because these alleles were expressed at relatively low levels (Figure 1).
Two hRap1 mutants that lacked the TRF2 binding domain and did not localize to telomeres, hRap1ΔCT and hRap1ΔBrΔCT, also induced telomere elongation at a rate of ∼30 base pairs/end/PD (Figure 3A and Table 1). Thus, the telomere elongation phenotype of overexpression of hRap1 was independent of its ability to localize to telomeres.
Telomeric hRap1 Alleles Lacking the BRCT Domain Diminish Telomere Length Heterogeneity
Restriction fragments representing human telomeres are often very heterogeneous in length. This length variability has several potential sources (reviewed in de Lange, 1995). First, individual cells can have different telomere length settings, but this effect is minimized in clonal cell populations such as the HTC75 cell line used in these studies. Second, the length of the telomere repeat tract can vary between individual chromosome ends. Third, each individual telomere is heterogeneous in length even in clonal cell lines (Farr et al., 1991; Hanish et al., 1994). We observed a gradual change in the telomere length heterogeneity in cells expressing hRap1ΔBr and hRap1ΔBrΔM (Figure 3A). After ∼80 PD the telomeric restriction fragments were considerably less heterogeneous, and they continued to become more discrete over the next 40 PD. Scans of the pertinent lanes confirmed that the size distribution of hRap1ΔBr is more narrow than the distribution of hRap1ΔBrΔCT, even though the telomeres have approximately the same mean length (Figure 3B). Similarly scans of pertinent lanes confirmed that the telomeres of hRap1ΔBrΔM cells are less heterogeneous than those of hRap1ΔMΔC infected cells (Figure 3B).
To measure the heterodisperse nature of the telomeric restriction fragment pattern, we determined the size range occupied by 50% of the telomeric hybridization signal and compared this with the median length of the telomeres (the heterogeneity index; see MATERIALS AND METHODS and Table 2). The heterogeneity index for control cells and hRap1 infected cells at PD0 was always close to 1 (mean 1, SD 0.06, n = 5). For instance, control cells with a median telomere length of 2 kb usually showed blots in which 50% of the telomeric signal was spread out over a 2-kb interval. The heterogeneity index did not change during prolonged growth of hRap1 cells (Table 2, compare hRap1 cells at PD0 and PD120), even though the telomeres gradually elongated from 2–4.8 kb. In contrast, cells expressing telomeric hRap1 alleles lacking the BRCT domain showed a significant drop in the heterogeneity index (p = 0.0013). Although these cells started out with a heterogeneity index of close to 1, the index diminished to a mean value of 0.5 (SD 0.02, n = 4) after prolonged culture growth. This reduction is significant (p = 0.01) even compared with the slightly reduced heterogeneity index of late PD cultures of hRap1ΔMΔC that have very long telomeres (Table 2).
Telomere Length Effects of hRap1 Depend on Telomerase Expression
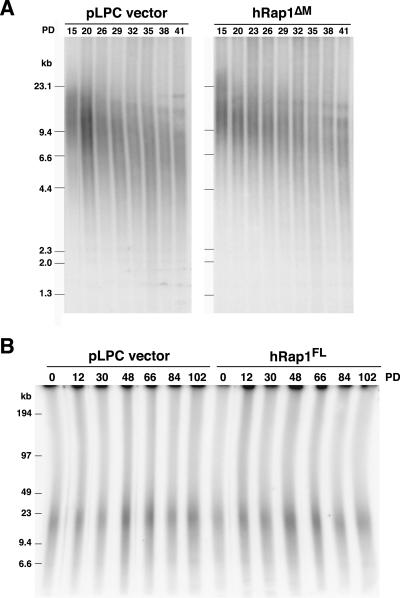
To determine whether the effects of hRap1 were dependent on telomerase, each of the alleles was expressed in primary human IMR90 fibroblasts and cells were passaged until they reached senescence. The telomere shortening rates revealed no discernible differences and the cells entered telomere-directed senescence at approximately the same PD (Figure 4A; our unpublished data). Overexpression of full-length and deletion versions of hRap1 also did not affect telomere length in the ALT-cell line GM847 when analyzed >102 PD (Figure 4B; our unpublished data). These data are consistent with hRap1 affecting the cis-acting telomere length control pathway that affects telomerase-mediated telomere elongation. Furthermore, we did not observe obvious changes in the telomere length distribution of primary cells and ALT cells expressing hRap1ΔBr and hRap1ΔBrΔM.
Figure 4.
Examples of lack of effect of hRap1 alleles on telomere length dynamics in telomerase negative cells. (A) Genomic blotting analysis of telomeric restriction fragments in HinfI/RsaI digested genomic DNA from pLPC vector or hRap1ΔM-infected IMR90 cells, probed with a TTAGGG repeat fragment. Passages and MW markers are indicated. (B) AluI/MboI digested genomic DNA plug from GM847 cells infected with pLPC or hRap1 full-length vector was separated by CHEF gel, and in-gel hybridization analysis of telomeric restriction fragments was carried out with an end-labeled [CCCTAA]4 oligonucleotide after in situ denaturation of the DNA.
DISCUSSION
The data presented here are consistent with Rap1 functioning as a regulator of telomere length in human cells as it does in budding yeast (Conrad et al., 1990; Lustig et al., 1990; Kyrion et al., 1992; Krauskopf and Blackburn, 1996; Marcand et al., 1997a). This conserved role for Rap1 in telomere length regulation is also consistent with fission yeast experiments in which deletion of Rap1 resulted in extensive telomere elongation (Hanaoka et al., 2001; Kanoh and Ishikawa, 2001).
Based on the observation that nucleoplasmic expression of hRap1 induced telomere elongation, we speculate that hRap1, like scRap1, is a cis-acting negative regulator of telomere length. This effect was observed for two hRap1 mutants that lack the TRF2 interacting domain and do not localize to telomeres. The simplest interpretation is that these mutants titrate a limiting hRap1-interacting factor that needs to bind telomeric hRap1 to maintain a stable length setting. The fact that hRap1ΔBrΔCT can have this titration effect suggests that the BRCT domain is not the only domain involved in binding of this putative telomere length regulator. Titration of a limiting factor may also explain why full-length hRap1 induced telomere elongation. Overexpression of wild-type hRap1 in HTC75 cells also results in large amounts of nucleoplasmic protein because the telomeric TRF2 binding sites become saturated. We speculate that this nucleoplasmic pool of hRap1 competes with telomeric hRap1 for the binding of the limiting interacting partner, resulting in diminished telomere length control and longer telomeres. Similarly, an S. cerevisiae Rap1 mutant (ΔBB) that lacks the ability to bind to telomeres induced telomere elongation (Conrad et al., 1990), presumably through titration of Rif1 and Rif2 in the nucleoplasm. However, the data presented here should be interpreted cautiously because they involve overexpression of mutant alleles that may either titrate factors in the nucleoplasm or disrupt telomere length regulatory complexes at the telomere (or both). A direct test of the idea that human Rap1 is a negative regulator of telomere length may be obtained using the recently developed telomere tethering approach (Ancelin et al., 2002). An important challenge is also to identify the putative telomere length control factor that interacts with hRap1. Human Rif1 was recently identified, but there is no indication that this protein binds to hRap1 or even that it is present at telomeres (Silverman and de Lange, unpublished data). Similarly, in fission yeast, Rif1 is not a Rap1 binding factor (Kanoh and Ishikawa, 2001), and it is likely that spRap1 has other interacting partners required for telomere length control. Human or fission yeast orthologs of Rif2 have not been identified so far.
The proposal that hRap1 acts as a negative regulator of telomere length is also consistent with the strong telomere elongation phenotype of four hRap1 mutants that have retained the ability to localize to telomeres. These mutants either lack the BRCT domain or the Myb domain or both. We assume that these hRap1 mutants displace the endogenous hRap1 from telomeres and act as dominant negative alleles. According this view, the BRCT and Myb domains of hRap1 would both be required at telomeres for full telomere length control. To test whether the mutants act as dominant negative alleles, we attempted to generate antibodies specific to the BRCT and Myb domain that would allow us to monitor the displacement of the endogenous hRap1 by the mutants. However, the resulting antibodies did not react with endogenous hRap1 on telomeres. We note that the presumed dominant negative alleles of hRap1 resemble the S. cerevisiae rap1t alleles, which bind to telomeres but fail to control telomere length (Kyrion et al., 1992).
Several hRap1 deletion mutants displayed an unexpected effect on telomere length heterogeneity. Expression of telomere targeted version of hRap1 that lacked the BRCT domain resulted in a more narrow size range of telomeres after prolonged culturing of the cells. We have considered the possibility that this phenotype is due to an effect on telomere elongation by telomerase. In both yeast and mammalian cells, telomerase elongates shorter telomeres in a cell more frequently (or more extensively) than longer telomeres (Zhu et al., 1998; Marcand et al., 1999; Ouellette et al., 2000; Hemann et al., 2001). This preference should ultimately lead to a more narrow size distribution of telomeres. However, this process should be affected equally by all hRap1 telomere elongation mutants and does not explain the specific effects of hRap1ΔBr and hRap1ΔBrΔM. We favor an alternative explanation for the change in telomere length heterogeneity in cells expressing hRap1ΔBr and hRap1ΔBrΔM: an effect on telomere rapid deletion (TRD). TRD has been observed in yeast when telomeres become very long (Kyrion et al., 1992; Li and Lustig, 1996; Bucholc et al., 2001). In this setting, stochastic losses of telomeric DNA take place, shortening the telomeres until they match wild-type telomere length. Li and Lustig (1996) have proposed the occurrence of a t-loop–like structure as an intermediate in TRD. Because t-loops occur at mammalian telomeres (Griffith et al., 1999), recombination within the t-loop may be a major source of constitutive telomere length heterogeneity. If TRD at human telomeres depends on a factor that is brought to the telomere by the BRCT domain of hRap1, expression of these deletion versions will diminish TRD, leading to more discretely sized telomeres.
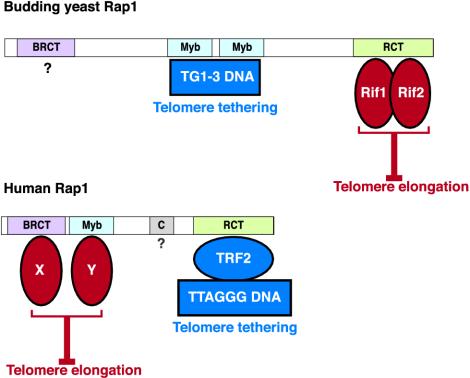
The architecture of hRap1 suggests a function as a protein adaptor that brings different factors into the telomeric complex (Figure 5). Its C-terminal RCT domain functions to interact with TRF2 and tethers hRap1 to telomeres. We propose that the BRCT and Myb domains recruit one or more protein factors, which together are required for the execution of negative telomere length control (Figure 5). Interestingly, in budding yeast Rap1, the Myb domain has gained the role of interacting with DNA (Konig et al., 1996), and the C terminus has become the main site of interaction with limiting telomere length control factors such as Rif1 and Rif2 (Hardy et al., 1992; Wotton and Shore, 1997) (Figure 5). We have previously proposed that this change occurred in conjunction with an alteration of the telomeric DNA sequence and resulted in the loss of the TRF/Taz1-like proteins from yeast telomeres (Li et al., 2000). Proper telomere length control would thus have necessitated a rearrangement of the interaction domains, allowing Rap1 to bind to telomeric DNA and tether Rif1 and Rif2 to the telomere.
Figure 5.
Changes in the function of Rap1 domains. Top, S. cerevisiae Rap1 binds to telomeric DNA with its central Myb domains and tethers the telomere length regulators Rif1 and Rif2 through protein interactions within the RCT domain. The function of the Rap1 BRCT domain is unknown. Bottom, human Rap1 localizes to telomeres through protein interaction of its RCT domain with TRF2. Data reported here suggest that hRap1 is a negative regulator of telomere length and that both the BRCT and Myb domains are required for this function. It is proposed that these domains interact with a factor(s) (either two different proteins, X and Y, or two domains of the same protein) that is required for negative regulation of telomere length. See DISCUSSION for additional details.
Acknowledgments
We thank members of the de Lange laboratory for technical help and discussion. We thank Kristina Hoke for extensive editing of this manuscript. This research is supported by a National Institutes of Health grant to T.dL. (CA 76026). B.L. was in part supported by a fellowship from the Leukemia and Lymphoma Society.
References
- Ancelin, K., Brunori, M., Bauwens, S., Koering, C.E., Brun, C., Ricoul, M., Pommier, J.P., Sabatier, L., and Gilson, E. (2002). Targeting assay to study the cis functions of human telomeric proteins: evidence for inhibition of telomerase by TRF1 and for activation of telomere degradation by TRF2. Mol. Cell Biol. 22, 3474-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucholc, M., Park, Y., and Lustig, A.J. (2001). Intrachromatid excision of telomeric DNA as a mechanism for telomere size control in Saccharomyces cerevisiae. Mol. Cell Biol. 21, 6559-6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikashige, Y., and Hiraoka, Y. (2001). Telomere binding of the Rap1 protein is required for meiosis in fission yeast. Curr. Biol. 11, 1618-1623. [DOI] [PubMed] [Google Scholar]
- Chong, L., van Steensel, B., Broccoli, D., Erdjument-Bromage, H., Hanish, J., Tempst, P., and de Lange, T. (1995). A human telomeric protein. Science 270, 1663-1667. [DOI] [PubMed] [Google Scholar]
- Conrad, M.N., Wright, J.H., Wolf, A.J., and Zakian, V.A. (1990). RAP1 protein interacts with yeast telomeres in vivo: overproduction alters telomere structure and decreases chromosome stability. Cell 63, 739-750. [DOI] [PubMed] [Google Scholar]
- Cooper, J.P., Nimmo, E.R., Allshire, R.C., and Cech, T.R. (1997). Regulation of telomere length and function by a Myb-domain protein in fission yeast [see comments]. Nature 385, 744-747. [DOI] [PubMed] [Google Scholar]
- de Lange, T. (1995). Telomere dynamics and genome instability in human cancer. In: Telomeres, ed. E.H.B.a.C.W. Greider, Cold Spring Harbor, NY: Cold Spring Harbor Press, 265-293.
- de Lange, T. (2002). Protection of mammalian telomeres. Oncogene 21, 532-540. [DOI] [PubMed] [Google Scholar]
- de Lange, T., Shiue, L., Myers, R.M., Cox, D.R., Naylor, S.L., Killery, A.M., and Varmus, H.E. (1990). Structure and variability of human chromosome ends. Mol. Cell Biol. 10, 518-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr, C., Fantes, J., Goodfellow, P., and Cooke, H. (1991). Functional reintroduction of human telomeres into mammalian cells. Proc. Natl. Acad. Sci. USA 88, 7006-7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith, J.D., Comeau, L., Rosenfield, S., Stansel, R.M., Bianchi, A., Moss, H., and de Lange, T. (1999). Mammalian telomeres end in a large duplex loop. Cell 97, 503-514. [DOI] [PubMed] [Google Scholar]
- Hanaoka, S., Nagadoi, A., Yoshimura, S., Aimoto, S., Li, B., de Lange, T., and Nishimura, Y. (2001). NMR structure of the hRap1 Myb motif reveals a canonical three-helix bundle lacking the positive surface charge typical of Myb DNA-binding domains. J. Mol. Biol. 312, 167-175. [DOI] [PubMed] [Google Scholar]
- Hanish, J.P., Yanowitz, J.L., and de Lange, T. (1994). Stringent sequence requirements for the formation of human telomeres. Proc. Natl. Acad. Sci. USA 91, 8861-8865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy, C.F., Sussel, L., and Shore, D. (1992). A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev. 6, 801-814. [DOI] [PubMed] [Google Scholar]
- Hemann, M.T., Strong, M.A., Hao, L.Y., and Greider, C.W. (2001). The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell 107, 67-77. [DOI] [PubMed] [Google Scholar]
- Kaminker, P.G., Kim, S.H., Taylor, R.D., Zebarjadian, Y., Funk, W.D., Morin, G.B., Yaswen, P., and Campisi, J. (2001). TANK2, a new TRF1-associated PARP, causes rapid induction of cell death upon overexpression. J. Biol. Chem. 13, 13. [DOI] [PubMed] [Google Scholar]
- Kanoh, J., and Ishikawa, F. (2001). spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr. Biol. 11, 1624-1630. [DOI] [PubMed] [Google Scholar]
- Karlseder, J., Smogorzewska, A., and de Lange, T. (2002). Senescence induced by altered telomere state, not telomere loss. Science 295, 2446-2449. [DOI] [PubMed] [Google Scholar]
- Kim, S.H., Kaminker, P., and Campisi, J. (1999). TIN2, a new regulator of telomere length in human cells [see comments]. Nat. Genet. 23, 405-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig, P., Giraldo, R., Chapman, L., and Rhodes, D. (1996). The crystal structure of the DNA-binding domain of yeast RAP1 in complex with telomeric DNA. Cell 85, 125-136. [DOI] [PubMed] [Google Scholar]
- Krauskopf, A., and Blackburn, E.H. (1996). Control of telomere growth by interactions of RAP1 with the most distal telomeric repeats. Nature 383, 354-357. [DOI] [PubMed] [Google Scholar]
- Kyrion, G., Boakye, K.A., and Lustig, A.J. (1992). C-terminal truncation of RAP1 results in the deregulation of telomere size, stability, and function in Saccharomyces cerevisiae. Mol. Cell Biol. 12, 5159-5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B., and Lustig, A.J. (1996). A novel mechanism for telomere size control in Saccharomyces cerevisiae. Genes Dev. 10, 1310-1326. [DOI] [PubMed] [Google Scholar]
- Li, B., Oestreich, S., and de Lange, T. (2000). Identification of human Rap1: implications for telomere evolution. Cell 101, 471-483. [DOI] [PubMed] [Google Scholar]
- Loayza, D., and de Lange, T. (2003). hPot1 as a terminal transducer of TRF1 telomere length control. Nature 424, 926-927. [DOI] [PubMed] [Google Scholar]
- Lustig, A.J., Kurtz, S., and Shore, D. (1990). Involvement of the silencer and UAS binding protein RAP1 in regulation of telomere length. Science 250, 549-553. [DOI] [PubMed] [Google Scholar]
- Marcand, S., Brevet, V., and Gilson, E. (1999). Progressive cis-inhibition of telomerase upon telomere elongation. EMBO J. 18, 3509-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcand, S., Gilson, E., and Shore, D. (1997a). A protein-counting mechanism for telomere length regulation in yeast [see comments]. Science 275, 986-990. [DOI] [PubMed] [Google Scholar]
- Marcand, S., Wotton, D., Gilson, E., and Shore, D. (1997b). Rap1p and telomere length regulation in yeast. Ciba Found. Symp. 211, 76-93. [DOI] [PubMed] [Google Scholar]
- McEachern, M.J., and Blackburn, E.H. (1995). Runaway telomere elongation caused by telomerase RNA gene mutations. Nature 376, 403-409. [DOI] [PubMed] [Google Scholar]
- Ouellette, M.M., Liao, M., Herbert, B.S., Johnson, M., Holt, S.E., Liss, H.S., Shay, J.W., and Wright, W.E. (2000). Subsenescent telomere lengths in fibroblasts immortalized by limiting amounts of telomerase. J. Biol. Chem. 275, 10072-10076. [DOI] [PubMed] [Google Scholar]
- Smith, S., and de Lange, T. (2000). Tankyrase promotes telomere elongation in human cells. Curr. Biol. 10, 1299-1302. [DOI] [PubMed] [Google Scholar]
- Smith, S., Giriat, I., Schmitt, A., and de Lange, T. (1998). Tankyrase, a poly-(ADP-ribose) polymerase at human telomeres [see comments]. Science 282, 1484-1487. [DOI] [PubMed] [Google Scholar]
- Smogorzewska, A., van Steensel, B., Bianchi, A., Oelmann, S., Schaefer, M.R., Schnapp, G., and de Lange, T. (2000). Control of human telomere length by TRF1 and TRF2. Mol. Cell Biol. 20, 1659-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel, B., and de Lange, T. (1997). Control of telomere length by the human telomeric protein TRF1 [see comments]. Nature 385, 740-743. [DOI] [PubMed] [Google Scholar]
- Wotton, D., and Shore, D. (1997). A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 11, 748-760. [DOI] [PubMed] [Google Scholar]
- Zhou, X.Z., and Lu, K.P. (2001). The Pin2/TRF1-interacting protein PinX1 is a potent telomerase inhibitor. Cell 107, 347-359. [DOI] [PubMed] [Google Scholar]
- Zhu, L., Hathcock, K.S., Hande, P., Lansdorp, P.M., Seldin, M.F., and Hodes, R.J. (1998). Telomere length regulation in mice is linked to a novel chromosome locus. Proc. Natl. Acad. Sci. USA 95, 8648-8653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, X.D., Kuster, B., Mann, M., Petrini, J.H., and Lange, T. (2000). Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat. Genet. 25, 347-352. [DOI] [PubMed] [Google Scholar]