Abstract
Transposon-based integration systems have been widely used for genetic manipulation of invertebrate and plant model systems. In the past decade, these powerful tools have begun to be used in vertebrates for transgenesis, insertional mutagenesis and gene therapy applications. Sleeping Beauty (SB) is a member of Tc1/mariner class of transposases and is derived from an inactive form of the gene isolated from Atlantic salmon. SB has been used extensively in human cell lines and in whole animal vertebrate model systems such as the mouse, rat and zebrafish. In this study, we describe the use of SB in the diploid frog Xenopus tropicalis to generate stable transgenic lines. SB transposon transgenes integrate into the X. tropicalis genome by a non-canonical process and are passed through the germline. We compare the activity of SB in this model organism with that of Tol2, a hAT (hobo, Ac1, TAM)-like transposon system.
Keywords: Transposon, Xenopus tropicalis, Xenopus laevis, Sleeping Beauty, SB10, SB11, transgenics, green fluorescent protein, GFP
Introduction
The frog Xenopus laevis (X. laevis) has been used as a model to study early events in vertebrate development for many decades. The large, externally developing embryos allow precise surgical manipulation and modification of gene activity can be readily achieved by microinjection of either messenger RNAs encoding dominant negative constructs or anti-sense morpholino oligonucleotides. Genetic manipulation of this species has not been widely used due to the long generation time (1–2 years) and the tetraploid nature of the X. laevis genome. In recent years another Xenopus species, X. tropicalis, has been introduced as an alternate model for genetic studies in amphibians (Amaya et al., 1998; Hirsch et al., 2002). X. tropicalis embryos share the physical features that allow embryological manipulation in X. laevis, have a shorter generation time (6–9 months) and small diploid genome. Genomic resources for X. tropicalis now available to the Xenopus community include extensive EST databases and a draft sequence with annotation of the entire genome (Klein et al., 2002; Klein et al., 2006; Morin et al., 2006). Our laboratory has focused on using transposons as tools to introduce foreign DNA into the frog genome for standard transgenesis and for insertional mutagenesis approaches (Johnson Hamlet and Mead, 2003; Johnson Hamlet et al., 2006; Yergeau and Mead, 2007). We have recently reported that the Tol2 transposon system functions efficiently in Xenopus tropicalis (Johnson Hamlet et al., 2006). Here, we describe the use of Sleeping Beauty transposon system to stably integrate a GFP reporter under the control of a ubiquitous promoter into the X. tropicalis genome.
Sleeping Beauty (SB) is a member of the Tc1/mariner class of “cut-and-paste” transposases derived from a teleost fish (Ivics et al., 2004). A “common ancestor” cloning strategy was used to predict and then engineer the functional amino acid sequence from an inactive transposase (Ivics et al., 1997). SB has been used to stably integrate DNA into in a wide variety of vertebrate genomes including those of mouse (Dupuy et al., 2001; Dupuy et al., 2002; Collier et al., 2005; Dupuy et al., 2005), zebrafish (Davidson et al., 2003; Balciunas et al., 2004) and X. laevis (Sinzelle et al., 2006; Doherty et al., 2007). Sleeping Beauty integrates the transposon substrate at TA dinucleotides in the host genome and thus results in essentially random integration of the target sequence.
Two recent papers have reported the successful use of Sleeping Beauty (SB) transgenesis in Xenopus laevis (Sinzelle et al., 2006; Doherty et al., 2007). Sinzelle and co-workers first reported the generation of transgenic X. laevis frogs that expressed ubiquitous expression of a green fluorescent protein (GFP) transposon transgene under the control of the β-actin promoter (Sinzelle et al., 2006). Our group reported the use of a tissue-specific promoter (xFlk-1; Xenopus laevis vascular endothelial growth factor receptor, VEGFR2) to drive expression of GFP in the vasculature of Xenopus laevis tadpoles and adults (Doherty et al., 2007). Both groups report similar transgenesis rates in the range of 30 to 40%. The inheritance of the transgenes in the F1 generation did not conform to the expected Mendelian ratios indicating that the germline of the founder animals was mosaic. This is likely to be due to integration of the transposon transgene at early cleavage stages, which results in the developing tadpole being mosaic for the transposon insertion event. We, and Sinzelle and colleagues, demonstrated that integration of the transposon is by a non-canonical process where one or multiple copies of the transgene are integrated in a single locus (Sinzelle et al., 2006; Doherty et al., 2007). This phenomenon appears to be a Xenopus-specific trait and has not been widely reported with Sleeping Beauty transposition in other vertebrate species (zebrafish, mouse, rat (Kitada et al., 2007) and human cell lines (Geurts et al., 2003)). Based on the successful transgenesis of X. laevis, we next investigated the activity of Sleeping Beauty in the closely related Xenopus species, X. tropicalis. Here, we report the generation of germline transgenic Xenopus tropicalis and Xenopus laevis using Sleeping Beauty transposase. Due to the mosaic expression of GFP in the founder (P0) animals, we focused our studies on integration events that are passed through the germline and describe here the insertions generated at a single dose of transposase enzyme and substrate. Our data indicated that the stable integration of the SB transposon in the germline of the frog is not by the anticipated “cut-and-paste” mechanism expected for this enzyme. The non-canonical integration events were observed in both X. laevis and X. tropicalis using two SB enzyme variants (SB10 and SB11) and two SB substrates (pT and pT2).
Results
Sleeping Beauty-mediated germline transgenesis in Xenopus
We used a microinjection strategy that we had successfully employed with SB in X. laevis (Doherty et al., 2007) and with Tol2 in X. tropicalis (Johnson Hamlet et al., 2006) to co-inject a plasmid harboring a SB transposon with synthetic messenger RNA encoding the SB transposase. The SB transposon substrate contained a chicken β-actin promoter and a cytomegalovirus (CMV) enhancer (CAGGS) driving expression of enhanced GFP (Figure 3A). A cocktail of donor plasmid and SB transposase (SB10) mRNA was injected into Xenopus zygotes at the one-cell stage (Figure 1A) and GFP expression was monitored during embryonic development. Injected embryos were scored for GFP fluorescence at approximately two weeks (~stage 50) after injection. Injection of the donor plasmid resulted in mosaic GFP expression due to transcription of the reporter directly off the plasmid (data not shown; Sinzelle et al described a similar phenomena with SB in X. laevis (Sinzelle et al., 2006)). Scoring embryos at early developmental stages is therefore problematic due to the presence of “plasmid-derived” GFP protein. When scored at the swimming tadpole stage we routinely observe robust expression of the GFP reporter in approximately one-quarter of the injected embryos. For example, in one injection set, where one-cell embryos were injected with 75 pg of donor plasmid and 500 pg of SB10 transposase mRNA, 139 tadpoles survived at least two weeks and 26% (n = 36/139) had widespread (non-mosaic) expression of the reporter. The remaining 74% had either mosaic or no expression of the GFP reporter. The rate of apparent transgenesis, that is, robust widespread expression of GFP, in the founder (P0) tadpoles is similar to that we observed using the Tol2 system in X. tropicalis (~25% for SB10 compared with ~30% for Tol2 (Johnson Hamlet et al., 2006)).
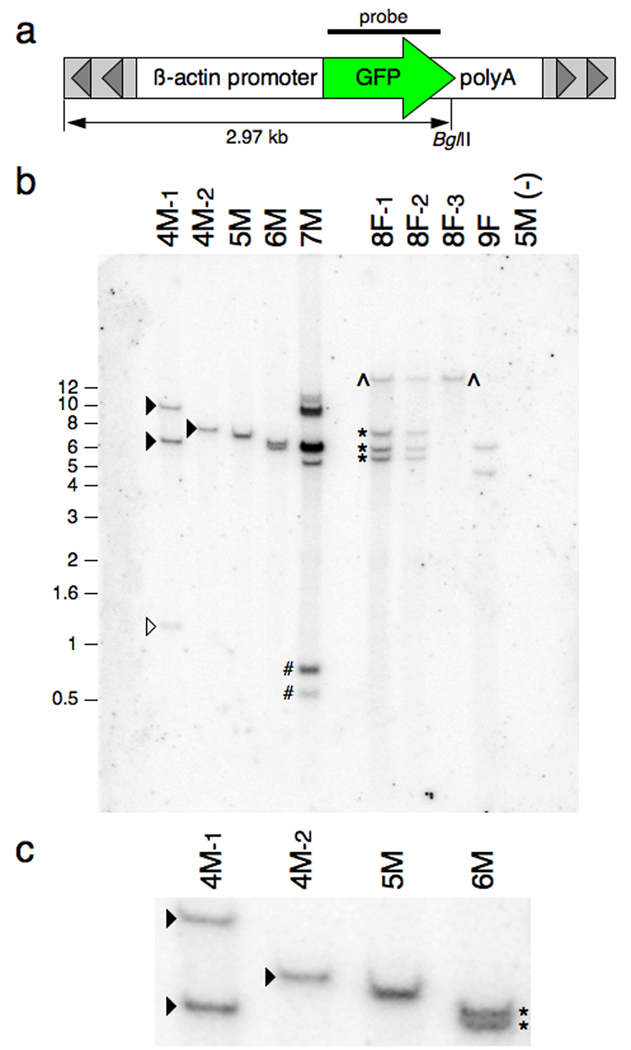
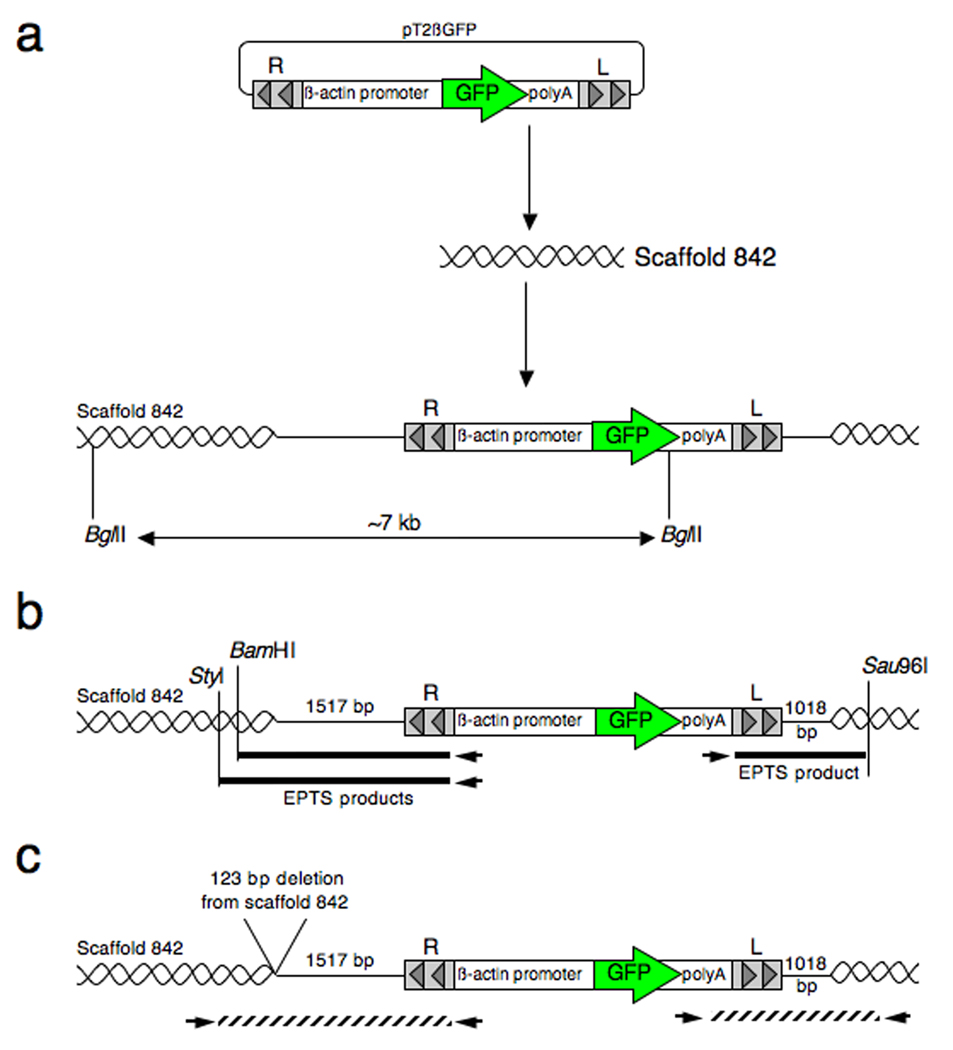
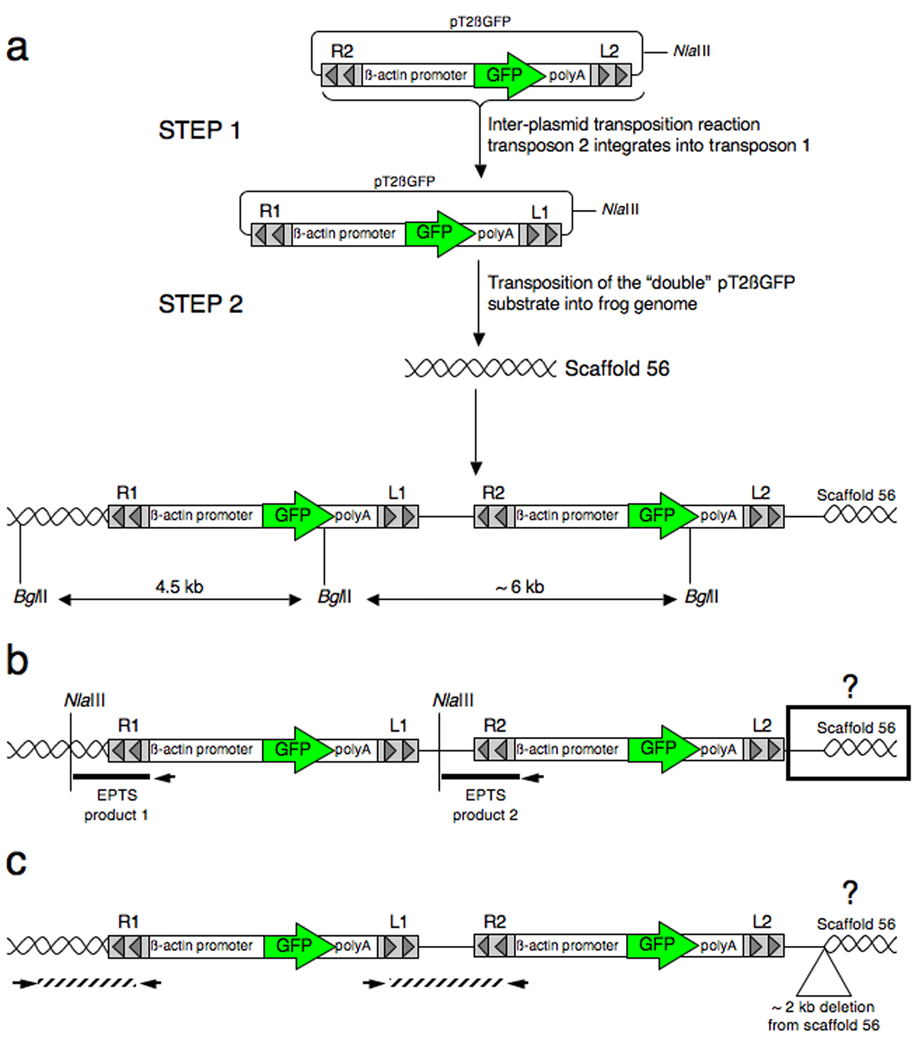
Fig. 3.
Southern blot analysis of genomic DNA harvested from SB-mediated transgenic Xenopus tropicalis and Xenopus laevis. Genomic DNA harvested from progeny of each of the founder animals was digested with BglII and separated by electrophoresis on an agarose gel, transferred to a membrane and probed with a radiolabelled GFP-encoding DNA fragment. a: Schematic representation of the pT2βGFP SB transposon indicating the approximate position of the unique BglII site and region used for the probe (bar). Not to scale. b: Southern blot analysis of pT2βGFP transgenic founders. Outcross of founder 4M resulted in two discrete hybridization patterns indicating independent segregation of the alleles (compare samples 4M-1 and 4M-2). The founder animal contains, at least, four copies of the GFP sequence that are inherited by the progeny (open and closed triangles). Likewise, the progeny of 8F display two different hybridization patterns (compare 8F-1 and 8F-3). Founder 8F also contains at least four copies of the transgene (^ and *s). Progeny from founder 7M have a complex hybridization pattern suggesting the presence of a concatamer of transposon transgenes. Tadpoles 4M-1 and 7M have hybridizing bands (labeled open triangle and #) that migrate faster than the predicted lower limit for the BglII digested transgene (2.97 kb; see Fig. 1a). This indicates that the integration events at these loci are complex and have involved fragmentation of the transposon transgene. Size markers (in kb) are indicated on the left side of the blot. c: Enlarged view of the Southern blot to illustrate that founder 6M has two closely migrating bands (*). d: Southern blot analysis of Xenopus laevis founder lines L2M, L3M and L6M. Genomic DNA samples for three GFP positive F1 animals (#1, 2 and 3) and a GFP negative F1 sibling (#4) from each founder line were digested with BglII, separated on a 0.7% (w/v) agarose gel, and probed with a 700 bp GFP fragment as in figure 3b. L2M founder line has at least 5 hybridizing GFP bands, L3M has at least 3 GFP positive bands and L6M has two GFP positive bands.
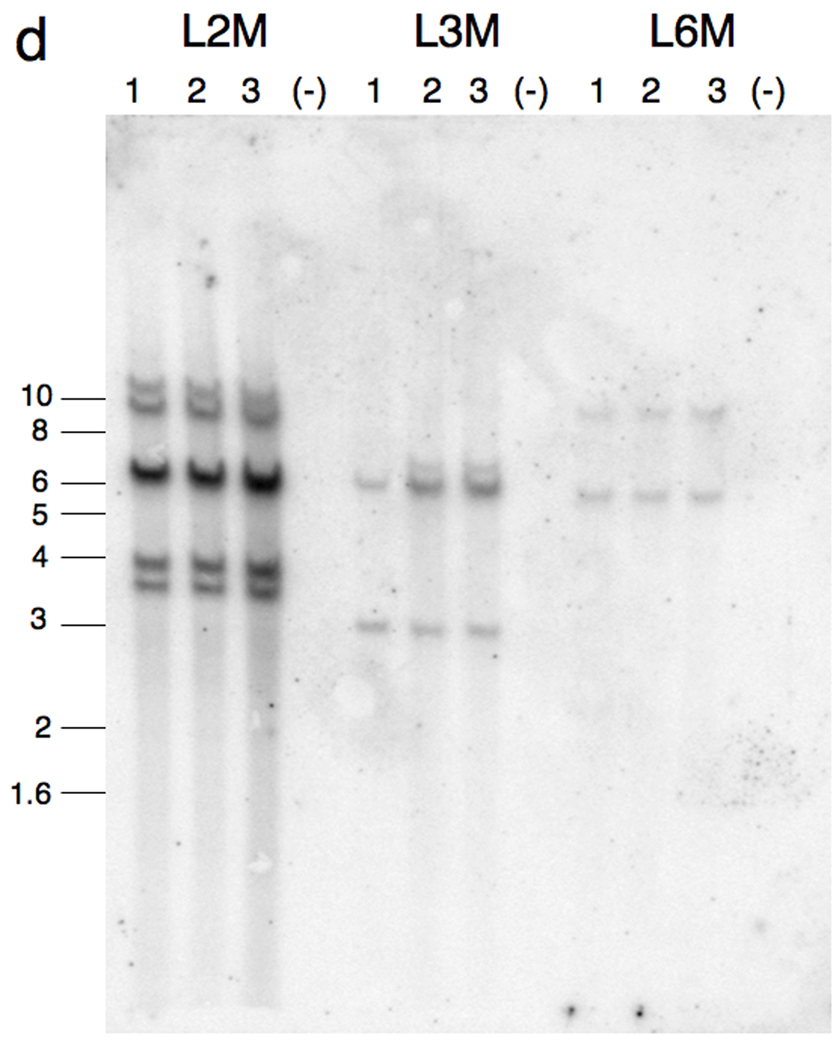
Fig. 1.
Injection-mediated Sleeping Beauty transgenesis in Xenopus. a: Schematic representation of the injection method used to generate the SB transgenic frogs. Xenopus laevis embryos were injected at the one-cell stage with pTβGFP plasmid and mRNA encoding the SB transposase. The cartoon depicts integration of the transposon substrate in one blastomere at the two-cell stage. The resulting embryo is predicted to develop as a “half transgenic” animal. b: “Half transgenic” founder. GFP expression in Xenopus laevis founder L2M is restricted to the left side of the dorsal midline (white dashed line). Outcross of this founder, with one-half of the germline containing the transgene, is predicted to generate GFP-positive F1 progeny at a rate of 25%. The observed rate of GFP transmission in the F1 tadpoles was 22% (n = 376/1733).
Integration of the SB transposon in one blastomere at the two-cell stage will result in a “half” transgenic animal in which one-half of the germline will transmit the dominant GFP allele. Transgenic progeny derived from outcross of this founder would be expected at a frequency of 25%. An example of a one-half transgenic X. laevis founder co-injected with the pTβGFP SB transposon construct together with SB10 transposase mRNA at the one-cell stage is shown in figure 1b. Integration of the transgene is likely to have occurred in one blastomere at the two-cell stage (Figure 1a) as the resulting founder expressed the GFP transgene in only one-half of the body (Figure 1b). Outcross of this founder resulted in approximately 25% of the F1 progeny expressing GFP throughout the body (22%, n = 376/1733). Subsequent outcross of the F1 progeny off this line resulted in the expected Mendelian ratios for a dominant allele (50% GFP-positive, data not shown).
GFP-positive P0 X. tropicalis tadpoles were raised to adulthood and outcrossed with wild type frogs to determine germline transmission of the transposon transgene. To date, we have outcrossed fifteen X. tropicalis P0 adult animals and have recovered six transgenic founders (6/15 = 40%; Table 1). The observed recovery rate of germline transgenic animals is similar to that observed by Sinzelle and coworkers with SB10 in X. laevis (40% = 5/12 (Sinzelle et al., 2006)) and by our group with Tol2 in X. tropicalis (43% = 3/7 (Hamlet et al., 2006)) and SB10 in X. laevis (Table 1 and Doherty et al. (Doherty et al., 2007)). The inheritance of the transposon transgene in the F1 tadpoles was routinely below the expected ratio of 50% for a dominant allele and indicates that the germline of the founder animals is mosaic for the transposon insertion event. We observed transgenic progeny in the F1 tadpoles at frequencies ranging from 1.2% to almost 50% (Table 1). Germline chimerism is most likely due to integration of the transposon at early cleavage stages in the development of the injected P0 tadpoles.
Table 1.
Sleeping Beauty transgenesis in Xenopus tropicalis and Xenopus laevis.
| Founder ID Number/Gender |
F1 Trangenesis rate # GFP-positive/ total # |
F2 Trangenesis rate # GFP-positive/ total # |
|---|---|---|
| Xenopus tropicalis (SB10 + pT2βGFP) | ||
| Transgenic P0 frogs | ||
| 4M | 48% (n = 185/383) | 49% (n = 479/973) |
| 5M | 6% (n = 13/235) | 51% (n = 258/502) |
| 6M | 1.2% (n = 17/1375) | 49% (n = 710/1437) |
| 7M | 22% (n = 69/314) | 51% (n = 514/1014) |
| 8F | 28% (n= 50/176) | 48% (n= 261/547) |
| 9F | 1.6% (n = 25/1534) | 51% (n = 1459/2878) |
| Non-transgenic P0 frogs | ||
| 1M | 0% (n = 0/172) | NA |
| 2M | 0% (n = 0/259) | NA |
| 3M | 0% (n = 0/1088) | NA |
| 10M | 0% (n = 0/173) | NA |
| 11M | 0% (n = 0/2170) | NA |
| 12M | 0% (n = 0/508) | NA |
| 13F | 0% (n = 0/1802) | NA |
| 14M | 0% (n = 0/1873) | NA |
| 15F | 0% (n = 0/479) | NA |
| Xenopus tropicalis (SB11 + pT2βGFP) | ||
| ♀622E | 56.9% (n = 667/1173) | ND |
| ♂623F | 19.5% (n = 81/415) | ND |
| ♂2262 | 9.7% (n = 273/2825) | ND |
| ♂2232 | 14.5% (n = 372/2562) | ND |
| Xenopus laevis (SB10 + pTβGFP) | ||
| L2M | 22% (n = 376/1733) | ND |
| L3M | 2.9% (n = 35/1211) | ND |
| L4M | 0% (n = 0/541) | NA |
| L5M | 0% (n = 0/46) | NA |
| L6M | 53% (n = 233/441) | ND |
| L7F | 27.5% (n = 22/80) | ND |
NA, not applicable; ND, not done.
To demonstrate that the non-Mendelian inheritance of the SB transposon transgenes was a feature of the mosaic founders, we raised transgenic F1 progeny to adulthood and outcrossed them to wild type frogs. For each founder line tested, the observed frequency of GFP-positive F2 offspring conformed to the ratio expected for outcross of a dominant heterozygous allele (1:1 = 50%; Table 1, F2 transgenesis rate). This data indicates that although the germline of the founder P0 animals is mosaic, the germline of the GFP-positive F1 progeny is not. Outcross of successive generations of selected X. tropicalis lines continued to yield the expected Mendelian ratios indicating that the SB transposon transgenes are stable in the frog genome (data not shown). Examples of F1 tadpoles generated by outcross of the X. tropicalis founders with wild type frogs are shown in figure 2.
Fig. 2.
Germline transmission of Sleeping Beauty transposon transgenes in Xenopus tropicalis. Six transgenic founder animals have been identified to date. The β-actin promoter-CMV enhancer (CAGGS) drives robust widespread expression of GFP in F1 progeny from each founder line. The tadpoles were photographed at approximately stage 40, ventral view, 35X magnification. The aperture setting and exposure time were the same for each image. a: non-transgenic 4M F1 tadpole. Note, autofluorescence of the gut. b: GFP-positive 4M F1 sibling of the tadpole depicted in panel a. c: 7M F2 GFP-negative tadpole. d: 7M F2 GFP-positive tadpole sibling of tadpole in panel c. e: 5M F2 GFP-positive tadpole. f: 6M F1 GFP-positive tadpole.
Characterization of Sleeping Beauty transposon transgenic Xenopus
We used Southern blot analysis, fluorescence in situ hybridization (FISH), and PCR-based linker-mediated methodologies to characterize the integration sites for each of the Xenopus tropicalis SB founder lines. Figure 3b shows a representative Southern blot of F1 progeny from the six founder lines identified to date. The presence of hybridizing bands that migrate differently between sibling F1 progeny indicates that the founder has multiple, independently segregating, transposon insertion events. For example, Southern blot analysis of F1 progeny from founder 4M revealed two different patterns (Figure 3b; lanes 4M-1 and 4M-2) and F1 progeny from founder 8F also show independently segregating transposon alleles (Figure 3b; compare lanes 8F-1 and 8F-2 with 8F-3). Southern blot analyses of genomic DNA harvested from multiple individual F1 tadpoles for lines 5M, 6M, 7M and 9F have single, unique hybridization patterns indicating that these lines harbor only one transgene allele (representative blot Figure 3b and data not shown). The presence of multiple bands in each lane suggests that the SB transposon transgenes for these four founder lines are either linked or have formed concatamers at unique loci in each line. The genomic DNA samples used for Southern analysis (Figure 3b) were digested with an enzyme that cuts once within the transposon transgene (BglII; Figure 3a) and the different sized bands observed when the blot was hybridized with a GFP probe result from the random insertion of the transgene into the host genome. The presence of hybridizing bands that are smaller than the transgene fragment (that is, smaller than the expected fragment from the intact transgene, 2.97 kb; Figure 3a) in two of the samples, 4M-1 (Figure 3b; open triangle) and 7M (Figure 3b; #) indicates a complex integration mechanism was used to insert the transposon.
We also performed Southern blot analysis on several X. laevis SB founder lines derived from one-cell embryos injected with the pTβGFP plasmid and SB10 transposase (Figure 3d). Xenopus laevis founder lines L2M, L3M and L6M show multiple integration events indicated by the hybridization of the GFP probe to BglII digested genomic DNA. L3M and L6M show the integration of two discrete bands in the genome whereas L2M shows five GFP-hybridizing bands forming a “ladder-like” pattern. As expected, DNA samples from GFP-negative siblings do not hybridize on the Southern blot (Figure 3d). Our results with X. laevis SB founder lines are consistent with findings from Sinzelle et al and our own work with SB in X. tropicalis (Sinzelle et al., 2006; Doherty et al., 2007). The Southern blot data presented in figures 3b and 3d indicates that integration in Xenopus is via non-canonical transposition. The presence of multiple transposon alleles that do not segregate upon outcross suggest that the transposon has either integrated as a concatamer or as multiple independent integration events occurring in very close proximity to one another.
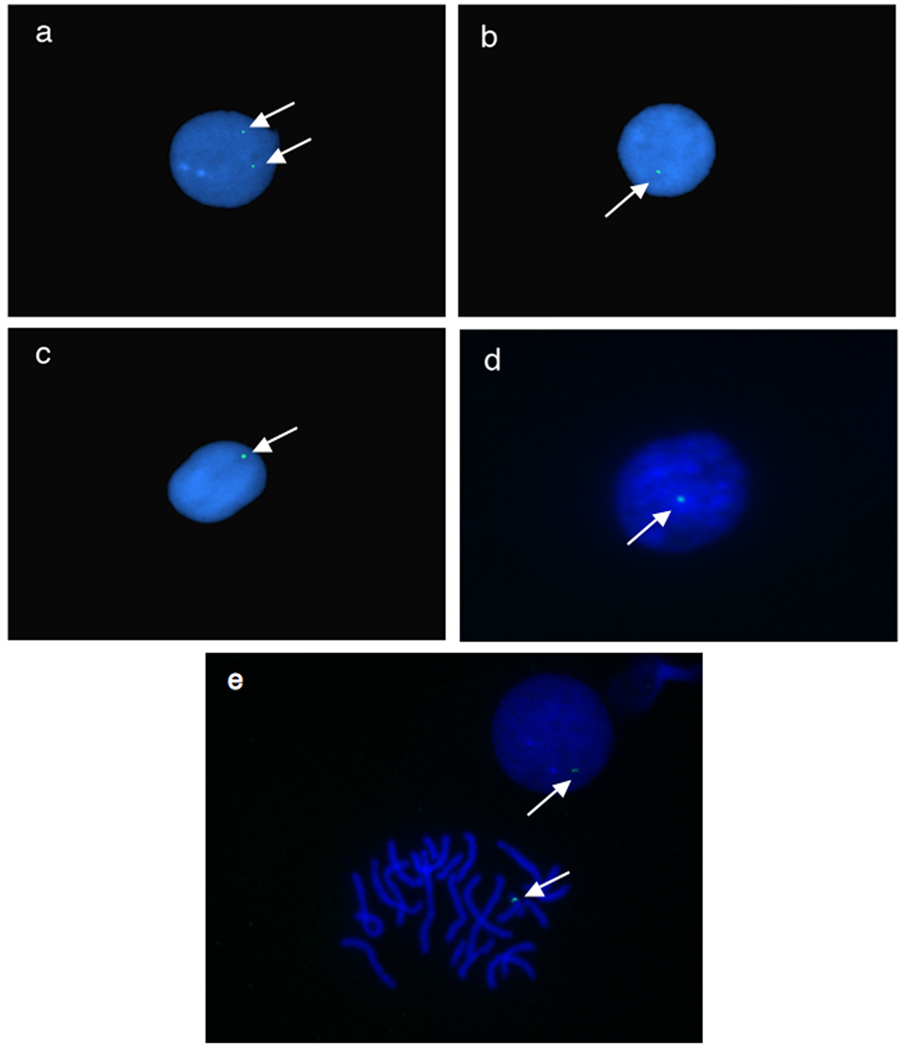
To address the complex integration of the transposon in X. tropicalis founders, fluorescence in situ hybridization (FISH) on interphase nuclei prepared from tadpole blood was used to determine the number of independent integration events in discrete F1 tadpoles for each founder line (Figure 4). The presence of one GFP fluorescent signal would indicate integration of the transgene as a single transposition event whereas multiple GFP foci would suggest multiple independent integrations. Founder line 4M has two independently segregating alleles by Southern blot (Figure 3b) and interphase nuclei (Figure 4a) indicate the presence of two GFP fluorescent loci within blood cells derived from F1 offspring. Other F1 siblings from the 4M founder line contain only one GFP marker in the interphase nuclei, confirming our interpretation of independently segregating alleles in the 4M P0 founder. X. tropicalis founder line 5M shows one transgene integrated into the genome by Southern blot (Figures 3b and 3c) and by FISH analysis (Figure 4b) confirming integration of a single SB transgene in the P0 founder. In contrast, Southern analysis of founder line 6M shows the presence of two GFP-positive bands (Figure 3b and 3c, starred bands) but the FISH results suggest integration of the transgene at a single locus with the genome (Figure 4c). Similarly, founder line 7M shows multiple integrations by Southern blot (Figure 3b, lane 7M) but the transgene appears to be located at a single locus in both metaphase spreads (Figure 4e) and interphase nuclei (Figure 4d and 4e). Both X. tropicalis founder lines 6M and 7M therefore appear to have integrated a duplex (6M) or multiplex (7M) concatamer of SB tranposon transgenes into a single locus. These results confirm the integration of the SB transposon into the genome of X. tropicalis is complex and integration of the transgene appears to have occurred by a non-canonical mechanism.
Fig. 4.
Fluorescence in situ hybidization (FISH) of cells harvested from pT2βGFP transgenic Xenopus tropicalis. Interphase nuclei were prepared from circulating blood cells harvested from individual tadpoles and probed with FITC-labled GFP for detection. White arrows indicate location of the GFP probe in the samples. a: pT2βGFP X. tropicalis founder line 4M. b: pT2βGFP founder line 5M c: pT2βGFP founder line 6M. d: pT2βGFP founder line 7M. e: Interphase nuclei and metaphase spread of founder line 7M.
Identification of the flanking genomic sequence adjacent to the transposon insertion sites using EPTS LM-PCR
Xenopus laevis founder line L2M
Cloning the transposon flanking sequences from X. laevis L2M line provided insight into the possible mechanism for generating the complex Southern blot patterns observed with Sleeping Beauty in Xenopus species. Sequence analysis of LM-PCR products from F1 progeny of L2M indicated that transposons had integrated into plasmid sequence upstream of GFP and within the right indirect/direct repeat (IR/DR) region of another transposon, possibly prior to integration of the transgene into the genome (see Table 2). This is likely to be due to transposition of the pTβGFP transposon from one plasmid to another plasmid. The microinjection strategy used to generate Sleeping Beauty transgenics requires co-injection of plasmid harboring a transposon substrate together with mRNA encoding the transposase enzyme. Approximately 50 pg of the substrate plasmid is injected and, although the zygote’s genomic DNA is the desired target for the transposition reaction, the plasmid DNA itself can also be an effective target for the transposition reaction. Inter-plasmid transposition can result in the generation of high-order transposon substrate plasmids that can then integrate into the genomic DNA. In the example described here, Southern blot analysis of L2M F1 progeny using a GFP probe showed five linked integration events (Figure 3d). The F2, and F3, progeny of L2M adults showed the same Southern blot pattern as the F1 tadpoles indicating that the integrated transposons are linked and do not segregate on outcross (data not shown). LM-PCR analysis identified two independent inter-plasmid transposition events from genomic DNA harvested from a single L2M F1 tadpole (Table 2). In the example shown, one inter-plasmid transposition event targeted the GFP coding sequence and another transposition event targeted the IR/DR of another transposon, indicating that at least three copies of the GFP reporter are present in the substrate that integrated into the genomic DNA of founder L2M. Integration of the high-order complex transposon substrates results in multiple copies of the transgene inserted at a single site in the genome. The inter-plasmid transposition activity also accounts for the presence of plasmid DNA sequences integrated with the SB transposon conglomerate (data not shown; and Sinzelle et al (Sinzelle et al., 2006)).
Table 2.
Identification of flanking scaffold sequence for several founder lines by EPTS LM-PCR
| Founder Line |
Insertion Site (scaffold:base pair) |
Flanking Scaffold Sequence | Flanking Gene (direction) |
|---|---|---|---|
| Xenopus tropicalis | |||
| 4M | 1092:132184 | TCTGCTGACcttgatc‥ggacattTCCCATTCA | Osteoclast Stimulating Factor 1 (3’) |
| 5M | 842:119502 | TTCTGACATcaatctg‥tatcattAGCCCCAAG | Synaptotagmin- 15 (5’) |
| 8F | 57:2456981 | GCAACGCTagtcacg‥caagttATTGATTA | Kielin (3’) |
| 9F | 56:3221468 |
tagggaTCCCCCCACTGAC (right arm only) |
IGF-II mRNA- binding protein 2 (5’) |
| Xenopus laevis | |||
| L2M | pTβGFP plasmid |
AAAACCCTGACCCagcctct | GFP |
| L2M | pTβGFP plasmid |
CCCCCTTTgggtcaaa | Right arm pTβGFP plasmid |
For the X. tropicalis insertion sites, the scaffold sequence is in uppercase and the lowercase italics shows the transposon plasmid sequence. For the X. laevis data, the breakpoints are indicated in the change from uppercase to lowercase italics font.
Integration site analysis of the X. tropicalis SB founder lines also revealed a complex insertion mechanism of the pT2βGFP transposon. DNA sequences flanking the transposon insertion sites were cloned using a modified version of the Extension Primer Tag Selection Linker-Mediated PCR (EPTS LM-PCR) (Yergeau et al., 2007). Transposons integrated into the genome provide an anchor for PCR primers to amplify the flanking genomic sequence and identify the location of the transposon integration site within the genome of the founder lines. Genomic DNA harvested from individual transgenic tadpoles was digested with “frequent-cutter” restriction endonucleases, either NlaIII or AluI, to generate short fragments for primer extension and subsequent PCR amplification. Using this strategy the integration sites of the SB transposon transgene in several founder lines were identified (Table 2). Here, we describe the cloning and characterization of three types of integration events; lines 5M and 4M-2 each contain a single transposon insertion, line 9F has two transposons integrated at the same locus and line 8F-1 at least three complete, and one partial, transposon integrated at a single locus in the frog genome.
X. tropicalis founder line 5M
Initial attempts to clone the flanking DNA sequence using biotinylated primers specific to the right IR/DR of the pT2βGFP transposon in the 5M founder line yielded fragments aligning to the pT2βGFP plasmid upstream of the right IR/DR. We focused on cloning the sequence near the left transposon IR/DR to determine the flanking genomic sequence. EPTS LM-PCR using a biotinylated primer specific to the polyA sequence within the pT2βGFP construct revealed DNA fragments matching the plasmid sequence. We next designed biotinylated primers located within the pT2βGFP plasmid backbone downstream of the left transposon IR/DR and performed EPTS LM-PCR using genomic DNA samples digested with restriction enzymes RsaI, StyI and Sau96I. Using this modified strategy, we were able to clone EPTS LM-PCR fragments from the SB founder line 5M containing flanking sequence identical to Xenopus tropicalis genome sequence (Scaffold 842; base pair position 119502 from the Joint Genome Institute (JGI) Xenopus tropicalis genome sequence assembly version 4.1 (Figure 8a)). The apparent linear integration event does not contain the full-length pT2βGFP plasmid sequence; approximately 320 base pairs of the plasmid backbone were eliminated during integration into the genome. In addition, 125 base pairs of the flanking genomic sequence were deleted from the EPTS LM-PCR product (Figure 8b). The wild type locus, encoded on the non-targeted sister chromosome, was cloned using a simple PCR strategy and the sequence was identical to the published scaffold sequence. This confirmed that the integration event had resulted in a 125 bp deletion of genomic DNA at the insertion site and that the difference in the scaffold sequence and the EPTS LM-PCR product sequence was not simply due to a naturally occurring polymorphism at this locus.
Fig. 8.
Schematic of the integration site of the pT2βGFP transgene into scaffold 842 for the X. tropicalis 5M founder line (not to scale). a: Integration of a single transposon transgene integrated into scaffold 842 as determined by Southern blot analysis showing a single ~7 kb BglII digested GFP-positive band. b: Cloning of the flanking genomic sequence by EPTS LM-PCR. EPTS products identified for both the 5’ and 3’ end of the tranposon are noted. c: Final orientation of the transposon integrated into scaffold 842 for X. tropicalis founder line 5M.
X. tropicalis founder line 4M-2
Cloning of the integration site for founder line pT2βGFP 4M was performed in a similar manner to the pT2βGFP 5M founder line described above. All EPTS LM-PCR fragments from the right IR/DR and left IR/DR from the two segregated F1 lines from the 4M founder corresponded to plasmid sequence adjacent to the right and left transposon IR/DRs, respectively. We focused on cloning the integration site from the 4M-2 line since it had a single integration site (containing only a single GFP band by Southern blot analysis, Figure 3c). EPTS LM-PCR using a biotinylated primer located downstream of the left IR/DR, and digested genomic DNA samples with restriction enzymes RsaI, StyI and Sau96I, revealed integration of the transposon within scaffold 1092 (base pair position 132184) in the X. tropicalis genome sequence v4.1 assembly. EPTS LM-PCR fragments show the breakpoint occurred approximately 160 base pairs upstream of the ampicillin gene within the pT2βGFP backbone. Interestingly, approximately 60 base pairs of the pT2βGFP construct were lost during integration of the SB transgene into the genome of the X. tropicalis 4M-2 founder line. PCR primers designed to the corresponding scaffold sequence in the wild type allele and transposon sequence confirm loss of the sequence from the pT2βGFP construct. Therefore, the locus for the X. tropicalis 4M-2 founder line integrated by a mechanism similar to the 5M founder line and suggests that integration occurred in a non-canonical manner. At this time we have been unable to clone the integration site for the other segregated allele (4M-1) due to complexity of the transposon locus within this line. Nonetheless, we believe that this tranposon site in the pT2βGFP 4M-1 line contains a concatamer of two transposons and a partial third transposon.
X. tropicalis founder line 8F-1
Cloning of the integration site for the X. tropicalis line 8F line was performed using EPTS LM-PCR targeting the right IR/DR. Initial cloning experiments revealed two PCR bands, one corresponding to adjacent plasmid sequence to the right IR/DR and the other to unknown sequence containing a breakpoint upstream, and not at, the expected site in the right IR/DR for SB mediated transposition. BLAST analysis showed integration of the transposon into scaffold 57 (base pair position 2456981). Genomic PCR confirmed this integration (Figure 5a, 5’ end) in F1 animals containing the three GFP-positive bands by Southern analysis (Figure 3b, *’s). The integration event in scaffold 57 was unique to the F1 tadpoles with the three GFP-hybridizing BglII Southern blot banding pattern, and was not found in the segregating allele from the founder (8F-3 (^) in Southern blot Figure 3b and genomic PCR in Figure 5a). In order to clone the left transposon IR/DR and flanking genomic sequence in the 8F-1 embryo, we designed primers specific to the left SB transposon IR/DR and to genomic sequence downstream of the integration site within scaffold 57. We hypothesized that integration of the Sleeping Beauty transgene within the founder line 8F occurred in a canonical process containing the corresponding left and right transposon IR/DR arms and cloning of the flanking DNA sequence adjacent to the opposite arm (left IR/DR in this example) would be rather simple to perform. However, primers designed to the predicted genomic sequence flanking the left IR/DR failed to amplify the expected PCR fragment. PCR primers specific to both the left IR/DR, and to scaffold 57, were able to amplify control DNA fragments (data not shown). Next, we performed standard PCR using primers located throughout the pT2βGFP plasmid construct in combination with primers located within scaffold 57. An amplified PCR fragment of approximately 3.5 kilobases showed sequence identity to both scaffold 57 and the pT2βGFP plasmid construct. The amplified product revealed that the transposon integration site occurred at a breakpoint within the β-actin promoter downstream of the right IR/DR and not at the canonical TA boundary of the left IR/DR (Figure 10b). The breakpoint within the β-actin promoter resulted in the inclusion of a partial tranposon fragment at the end of the multimeric insertion event. To confirm the 3’ end of the transposon integration site in scaffold 57, we designed a biotinylated primer matching to genomic DNA sequence in the flanking region of scaffold 57 to read back into the transposon integration site. EPTS LM-PCR using restriction endonucleases HinDIII, ClaI, VspI, that cut within the transposon but not in the predicted flanking genome sequence, confirmed the 3’ flanking sequence that was identified by standard PCR (data not shown). Standard genomic PCR, with primers designed to sequence beyond the EPTS primer sites in the flanking scaffold, further confirmed the integration site in scaffold 57 (Figure 5a, Figure 3’ end). Southern blot analysis indicated that the 8F line contains at least two independently segregating alleles, one with three hybridizing bands and one with a single hybridizing band. As expected, genomic DNA harvested from 8F tadpoles with a single Southern blot band (Figure 3b, caret) was not positive for PCR amplification of the scaffold 57 integration event, (Figure 5a, sample 8F-3) confirming that there are two independently-segregating transgene alleles in the 8F founder.
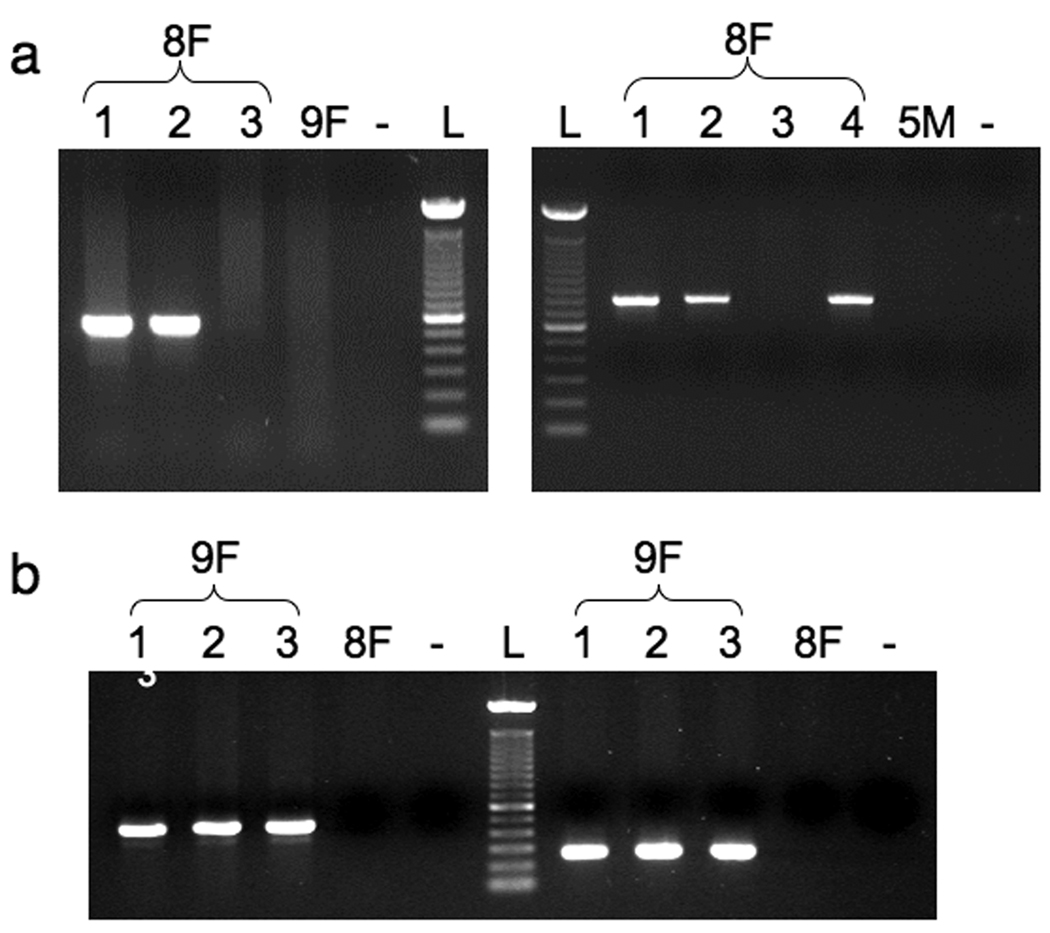
Fig. 5.
Verification of the transposon insertion site in founder lines 8F and 9F. a: Genomic PCR products for both the 5’ (right panel) and 3’ (left panel) ends of the integration sites for founder line 8F. Genomic DNA samples were collected from several GFP-positive F1 8F tadpoles (1, 2, 3, 4) and used in PCR reactions to amplify (3’ end 8Fa/8Fb 517 bp fragment; 5’ end 8Fc/8Fd 797 bp). b. Agarose gel electrophoresis of genomic PCR products for founder line 9F. Genomic DNA prepared from three GFP-positive F1 9F tadpoles (1, 2 and 3) was used to amplify the expected sized fragments using primer pairs 9Fa/9Fb (493 bp product) and 9Fc/9Fd (264 bp product). No products were formed when genomic DNA harvested from either a GFP-positive 8F F1 tadpole (8F) or a GFP-negative 9F F1 tadpole (-) was used as the template. L = 100 bp ladder.
Fig. 10.
Schematic representation of the predicted integration of a multimeric pT2βGFP transposon concatamer in the X. tropicalis founder line 8F-1. a: Stepwise interplasmid integration and insertion of the concatamer into scaffold 57 of the X. tropicalis genome. It is presumed that multiple, interplasmid incorporation of the transgene occurred prior to integration of the entire concatamer into the genome. Southern blot analysis revealed three GFP-hybridizing bands that were mapped according to size and predicted orientation. b: EPTS LM-PCR products from the left and right SB transposon arms. EPTS product 1 confirms integration into scaffold 57. EPTS product 2 corresponds to vector sequence upstream of the right IR/DR in the SB pT2βGFP transgene. c: Predicted orientation of the multimeric transposon concatamer integration site for X. tropicalis founder 8F. The orientation of transposon 3 (R3 and L3) is predicted from Southern blot data. Schematic map is not to scale. Note: plasmid sequences that reside between transposon elements in the integrated DNA have not been fully characterized and are depicted in the schematic map as gaps between the transposon transgenes.
X. tropicalis founder line 9F
We performed EPTS LM-PCR using biotinylated primers targeted to the right IR/DR as previously described for other X. tropicalis pT2βGFP founders. Restriction fragments from NlaIII digested 9F genomic DNA samples showed integration of the right IR/DR within scaffold 56 (base pair position 3221468) of the X. tropicalis genome sequence assembly v4.1. In addition, PCR fragments revealed sequence matching the pT2βGFP plasmid upstream of the right IR/DR suggesting the presence of an additional transposon within the genome of the X. tropicalis founder line 9F. Standard genomic PCR using primers specific to scaffold 56 and the right IR/DR confirmed integration within this scaffold (Figure 5b). Similar to our strategy for X. tropicalis founder line 8F, we first hypothesized the integration into scaffold 56 was canonical and designed PCR primers within the left IR/DR and scaffold 56 to amplify the flanking sequence. However, again we failed to generate the expected amplified products using multiple primer pairs. Biotinylated primers designed within scaffold 56 downstream of the tranposon integration site, and generating primer extension products back towards the integration site revealed only the wild type allele sequence and indicates, as identified with the other SB lines, that some genomic sequence was lost upon insertion of the transposon. We have been unable to identify the 3’ end of the 9F integration site within scaffold 56 using a variety of PCR techniques. Southern blot analysis, however, indicated that at least 2 kb of genomic sequence had been eliminated upon integration of the pT2βGFP concatamer into this locus (data not shown).
We were unable to identify the integration sites for several of the SB10-mediated integration events due to the limitations using a PCR-based strategy, as the primers used for amplification would bind to multiple regions in the complex concatamers. Overall, integration site analysis using the EPTS LM-PCR and standard LM-PCR strategies indicated that in Xenopus tropicalis and Xenopus laevis, SB does not use the precise boundaries of the transposon arm and we were able to identify a variety of “breakpoints” near the predicted boundary of SB (TA*CAG, Table 2). This observation further supports the theory that SB uses a non-canonical integration mechanism in Xenopus.
Comparison of SB10 and SB11 Sleeping Beauty transposase in Xenopus tropicalis
We have demonstrated that the original Sleeping Beauty transposase (SB10) enzyme functions in both Xenopus laevis and Xenopus tropicalis and results in non-canonical integration events in both species. Furthermore, modifications to the transposon substrate (pT) that have been documented to increase transposition efficiency (pT2; (Cui et al., 2002)), do not change the unusual integration patterns observed with SB10 in Xenopus; compare Southern analysis of pTβGFP in X. laevis (Figure 3d) with pT2βGFP in X. tropicalis (Figure 3b). Next, we examined the activity of a modified SB transposase enzyme that contains four amino acid substitutions (SB11; Geurtz et al., 2003) compared to the original reconstructed enzyme (SB10; Ivics et al., 1997). The modified enzyme, SB11, has approximately three-fold higher activity than the original enzyme, SB10. To determine whether the unusual integration mechanism observed with SB10 was unique to the original version of the enzyme, we injected Xenopus tropicalis embryos at the one-cell stage with a combination of pT2βGFP plasmid and mRNA encoding SB11 transposase. GFP-positive tadpoles were raised to adulthood and outcrossed to establish germline transmission of the transgene. To date, we have outcrossed eleven adult “SB11-derived” frogs and have identified four germline transgenic founders (4/11 = 36%). As with SB10, a wide range of germline transmission frequencies were observed with SB11 in X. tropicalis (~12% to ~57% GFP-positive progeny; Table 1), indicating that the founders are mosaic for the integration event(s). Founder ♀622E has a transmission frequency (~57%) that is higher than that expected for Mendelian inheritance of a dominant allele (50%) and indicates the presence of multiple integration events in the germline of this founder. Figure 6 shows GFP fluorescence of representative individual F1 tadpoles from each SB11-derived founder. Similar to that observed with SB10-mediated integration, the CAGGS promoter in pT2βGFP results in widespread expression of the reporter in the transgenic tadpoles. Southern blot analysis (Figure 7) indicates that the integration events observed with SB11 are similar to those identified in SB10 transgenics (compare Southern analyses, Figure 7 and Figure 3b). As determined for the SB10-generated lines, integration site analysis by EPTS LM-PCR indicated the presence of plasmid sequences adjacent to the Sleeping Beauty transposon arms (data not shown). Together, this data suggests that the integration mechanism mediated by the modified Sleeping Beauty enzyme, SB11, is the same as that mediated by the original SB10 transposase.
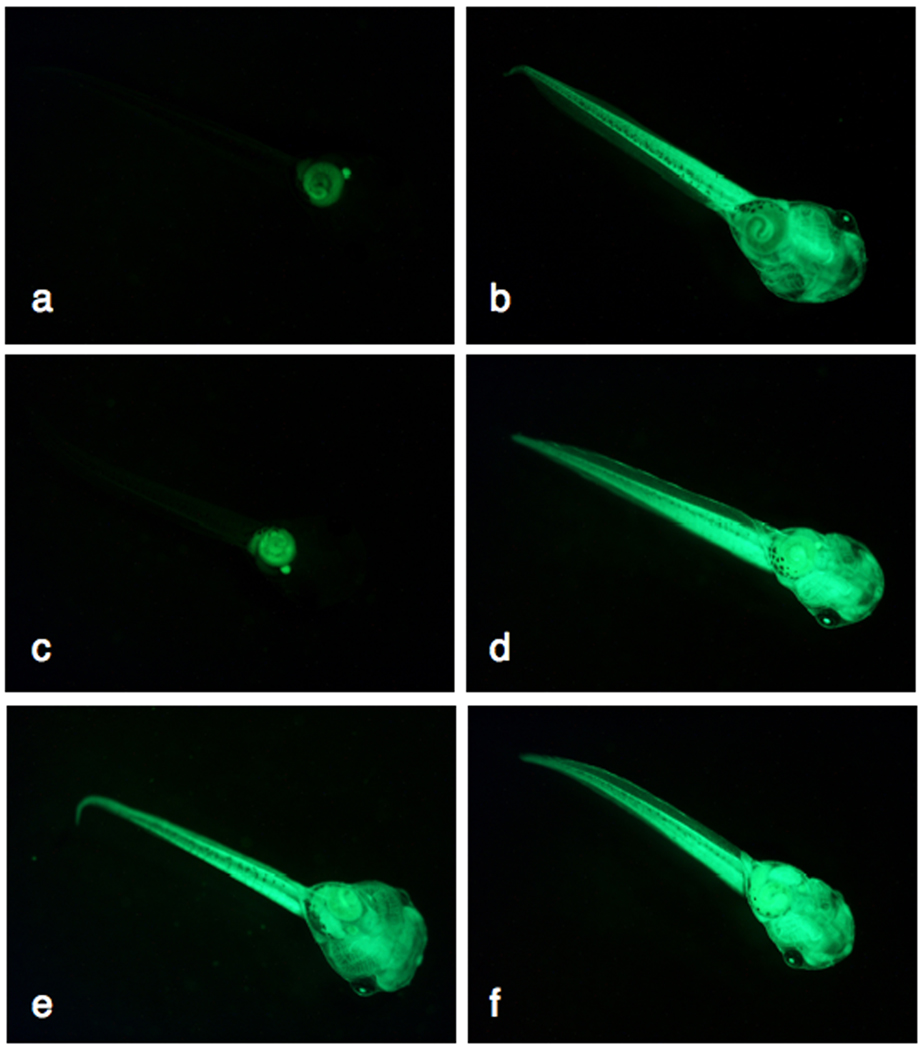
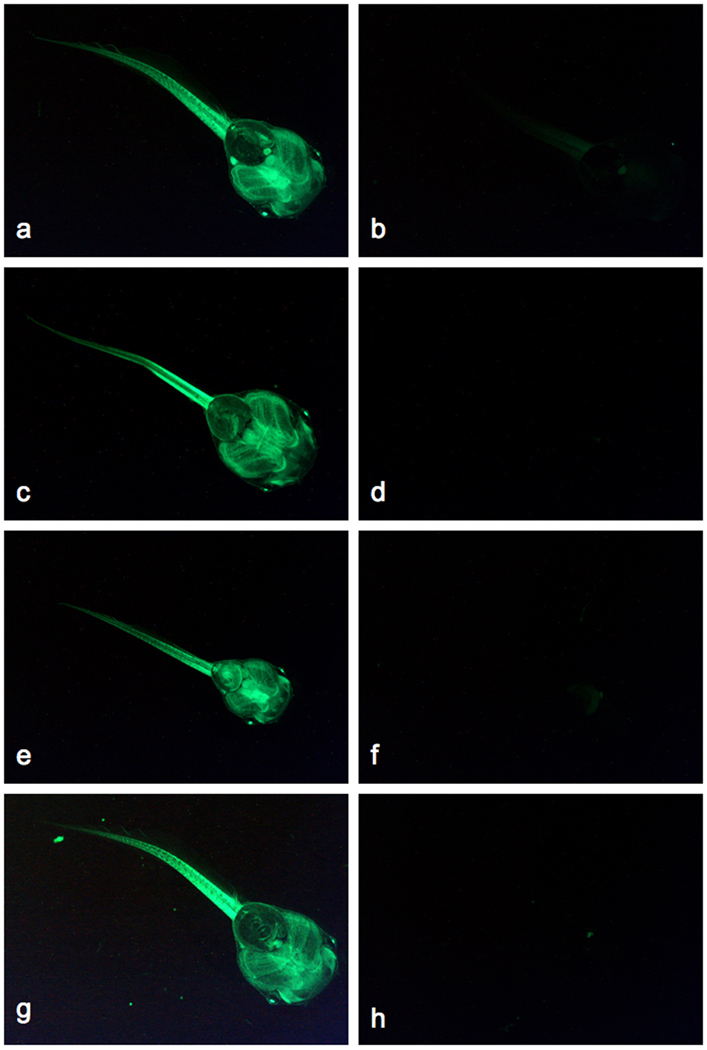
Fig. 6.
Germline transmission of SB11-mediated transposon transgenes in Xenopus tropicalis. The CAGGS promoter in the pT2βGFP transposon drives widespread expression of the GFP reporter in F1 transgenic tadpoles. GFP-positive (a, c, e, g) and GFP-negative (b, d, f, h) F1 individuals are shown for four independent founder lines (♀622E (a, b), ♂623F (c, d), ♂2262 (e, f), ♂2232 (g, h)). Ventral views of each tadpole (approximately stage 45) are shown with the head in the lower right corner. Magnification 25X, same aperture and exposure times were used for each image.
Fig. 7.
Southern blot analysis of F1 tadpoles from SB11-mediated pT2βGFP transgenic Xenopus tropicalis founders. Founder ♀622E has two independently segregating insertion events; compare ♀622E1 (four GFP-positive hybridizing bands) and ♀622E3 (three GFP-positive hybridizing bands). Tadpole ♀622E2 has inherited both integration events. F1 progeny from founders ♂623F, ♂2262 and ♂2232 have two GFP-positive hybridizing bands.
Discussion
We have used the Sleeping Beauty transposon system to generate transgenic Xenopus laevis and Xenopus tropicalis frogs. Sinzelle and coworkers recently described the use of SB in X. laevis to generate germline transmission of transposon transgenes (Sinzelle et al., 2006). Our data also supports the observation that SB transgenesis in Xenopus is via a non-canonical mechanism and results in the integration of high-order transposon transgenes. Integration site analysis suggests that inter-plasmid transposition of the injected transposon substrate is the likely mechanism for the generation of the complex, multimeric transgenes inserted into the frog genome. Other possible mechanisms, with rearrangements before or after integration, cannot, however, be ruled out by our data.
Sleeping Beauty-mediated transgene integration in Xenopus results in the generation of mosaic founder animals. We have observed the same phenomenon when using the Tol2 system in X. tropicalis (Johnson Hamlet et al., 2006) and it may be a feature of the rapidly dividing frog embryo. The injected SB transposase mRNA must be translated at sufficient quantities by the host cell before the transposition reaction can proceed. The rapidly-dividing Xenopus embryo may develop to early cleavage stages before sufficient enzyme is synthesized to catalyze the integration reaction. The mosaic germline of the founders may also account for the observed non-Mendelian transmission frequency for the SB founders. Only 40% of animals that were scored positive for GFP at stage 50 passed the transgene on to the next generation. Transposition during early cleavage stages may result in integration of the transposon transgene in somatic blastomeres that do not contribute to the germline; the resulting mosaic tadpole would express GFP but would fail to pass the transgene on to its progeny. The mosaic integration of the transposon in the injected tadpoles is likely to limit the use of this transgenesis method for studying transgene activity in the founder generation.
Our results provide strong evidence for the non-canonical integration of the transgene into the genome of Xenopus consistent with previous findings from our group as well as others (Sinzelle etal., 2006; Doherty et al., 2007). In addition to previous studies showing the feasibility of SB for transgenesis in the frogs, we identify the transposon integration site by cloning the flanking sequence using a modified EPTS LM-PCR method. Based on our integration site analysis, Southern blots, FISH and amplification of flanking sequences, we present possible schematics for the integration mechanism of the Sleeping Beauty transgene into the genome of three founder lines. The maps depict the non-canonical integration of a single transgene for the 5M line (Figure 8), a concatamer of two transposons in founder line 9F (Figure 9) and a multiplex concatamer in founder line 8F (Figure 10). In figure 8a, a single SB transgene integrated into scaffold 842 of the X. tropicalis genome assembly resulting in a BglII Southern blot fragment of approximately 7 kb (Figure 3b). Cloning the integration sites using biotinylated primers targeted to the pT2βGFP backbone, and digesting the genomic DNA samples with BamHI or StyI for the 5’ end of the transposon locus and Sau96I for the 3’ end, confirmed integration of the transgene into scaffold 842 (Figure 8b). PCR primers, designed to sequences outside those used for the initial characterization of the flanking sequences, were used to validate the integration site and confirmed the loss of 125 bp of genomic sequence from scaffold 842 upon integration of the GFP transgene (Figure 8c).
Fig. 9.
Map of the integration event in X. tropicalis founder line 9F. The SB pT2βGFP transgene is a concatamer of two transposons integrated into one another followed by integration into the X. tropicalis genome on scaffold 56. a: Interplasmid transposition of transposon one into transposon two followed by integration of the complex into scaffold 56. The map was derived from Southern blot data and shows the predicted orientation of the observed GFP-positive bands (4.5 and 6 kb). b: Flanking sequence was determined by EPTS LM-PCR. Two EPTS restriction fragments digested with NlaIII were identified and correspond to genomic DNA (product 1) and vector sequence (product 2). The 3’ end of the integration site has yet to be determined and is noted by a black box and a question mark. c: Predicted orientation of the transposon concatamer integrated on scaffold 56 within the X. tropicalis founder line 9F. Not to scale.
For founder line 9F, two complete SB transposons, as well as some plasmid flanking sequence, have integrated into a single locus. Integration of one transposon into another copy of the plasmid provided a substrate for genomic integration that has two copies of the transgene. Southern blot analysis showed two GFP-positive bands at 4.5 kb and ~6 kb. The 4.5 kb band corresponds to BglII digestion from transposon 1 out to the BglII site present within the genomic scaffold 56 (Figure 9a). EPTS LM-PCR using primers specific to the right transposon IR/DR, and digestion of the genomic DNA with NlaIII, produced two fragments (labeled EPTS product 1 and EPTS product 2; Figure 9b). EPTS product 1 contained the right arm IR/DR and flanking genomic sequence aligned to scaffold 56, while EPTS product 2 contained plasmid vector sequence upstream of the right transposon IR/DR (R2) to the NlaIII restriction site in the pT2βGFP backbone (Figure 9b). Genomic PCR using primers complimentary to the 5’ end of the integration site confirmed EPTS product 1 (Figure 5b). Evidence suggests that two transposons are in a head to tail orientation as depicted in figure 9c.
The proposed map of the complex integration of the SB transposon transgene into scaffold 57 in 8F-1 is shown in figure 10. At least three transposon transgenes integrated into one another (Figure 10a, step 1 and step 2) prior to insertion into the genome. Southern blot analysis of 8F-1 shows three complete copies of the GFP transgene. The size of the individual bands can be accounted for in the proposed map of the integration event (Figure 10). The 3’ end of the insertion event contains a partial transposon substrate that lacks the GFP sequence due to the breakpoint in the β-actin promoter.
In Xenopus, SB uses a complex, non-canonical integration mechanism and often results in the incorporation of multimeric transgenes. We have demonstrated that SB-mediated integration frequently results in small deletions of the genome at the site of insertion. We show that the Sleeping Beauty system can be used in Xenopus with high-efficiency, achieving rates of transgenesis equal to other transposon systems such as Tol2 (Johnson Hamlet et al., 2006). As a tool to create transgenic animals, Sleeping Beauty is reliable and has been shown to recapitulate endogenous gene expression from a tissue specific reporter transposon (Doherty et al., 2007). The complex integration of multiple transposon transgenes as concatamers, however, makes cloning the integration sites using standard PCR-based methodologies time consuming and very difficult and likely precludes the use of SB for insertional mutagenesis studies in the frog. In developing Xenopus tropicalis as a genetic model system, it will be advantageous to have multiple transgenic strategies for integrating foreign DNA into the germline. For example, SB can be used in combination with other transposon systems to develop transgenic frogs for transposon remobilization strategies. Dual transposon systems can also potentially be used to integrate, and remobilize, loxP containing transposon vectors for chromosome engineering strategies.
The unusual SB transposition mechanism in this genus may be due to host factors specific to Xenopus, as this phenomenon has not been described in other model species. We have not performed extensive dose-response curves for injected enzyme and substrate combinations and it is possible that further testing of transposon and transposase dosing may reveal conditions in which SB may integrate into the Xenopus genome in a canonical manner. Our analysis of SB10 and the hyperactive SB11 enzyme variant, however, produced similar non-canonical integration events indicating that, in Xenopus tropicalis, SB transgenesis is not occurring by the expected “cut-and-paste” mechanism.
The non-canonical integration mechanism used by SB in Xenopus is not a general feature of “cut-and-paste” transposases in this species. We have used Tol2, a member of the hAT (hobo, Ac and Tam) family of DNA transposases, to stably integrate transgenes into the frog genome and have demonstrated that this system uses a standard integration mechanism in Xenopus tropicalis; the transposition events are characterized by precise integration at the terminal repeat boundaries and are flanked by 8-base pair tandem site duplications (TSDs) (Johnson Hamlet et al., 2006). In contrast, the predicted transposon boundaries for SB integration in Xenopus are not at the expected ends of the IR/DR’s and not flanked by the 2-base pair (TA) TSDs characteristic of SB integration events in other species. It will be interesting, in future studies, to determine whether other transposable elements, such as PiggyBac (a TTAA-specific Class II short inverted repeat family member derived from the cabbage looper, Trichoplusia ni (Finnegan, 1990)) and Frog Prince (a Tc1/mariner element derived from Rana pipiens (Miskey et al., 2003)), share the non-canonical integration mechanism used by SB, or whether this unusual mechanism is unique to SB and not a general property of transposon-mediated transgenesis in Xenopus.
Experimental Procedures
Plasmids and mRNA synthesis
The Sleeping Beauty reagents (pT and pSBRNAX (SB10) and SB11) were kindly provided by Dr. Perry Hackett (Ivics et al., 1997; Geurtz et al., 2003). PCR mutagenesis was used to modify the original pT substrate plasmid to generate the pT2 version described by Cui et al. (Cui et al., 2002). PCR mutagenesis was performed with the Quick Change Kit (Qiagen). The left and right arms were cloned into the SacI and KpnI sites of pBS SK+ to generate a transposon donor plasmid with multiple cloning sites to facilitate subcloning (Cui et al., 2002). The chicken β-actin promoter and CMV enhancer (CAGGS) was cloned upstream of enhanced GFP and a polyadenylation signal from the rabbit β1-globin gene was added at the 3’-end to enhance stability of the transgene mRNA. This reporter construct was cloned into both the original pT and the modified pT2 transposon substrate plasmids. Plasmids were isolated using Qiagen Midi-prep kits and further purified by phenol/chloroform extraction and ethanol precipitation. Messenger RNA encoding the SB transposase (either SB10 or SB11) was transcribed from linearized plasmids using mMessage mMachine kits (Ambion, TX) according to the manufacturers instructions.
GFP Expression Analysis
A Leica FLIII fluorescent dissecting microscope was used to analyze GFP expression. Digital images were captured using a Nikon D5-5M color digital camera.
Microinjection of Xenopus tropicalis embryos
Xenopus tropicalis females were induced to ovulate by injection of 80 U of chorionic gonadotropin (hCG; Novarel, Ferring Pharmaceuticals) one day before use. On the day of use, pre-primed females were injected with 400 U of hCG. Eggs were fertilized in vitro using testis homogenate prepared in Leibovitz L15 medium/10% (v/v) fetal goat serum. Fertilized eggs were de-jellied with 2% (w/v) cysteine made in 0.05X MMR. Embryos were injected with a cocktail of 500 pg mRNA and 75 pg circular plasmid DNA. Embryos were raised in 0.05X MMR supplemented with penicillin/streptomycin (Gibco) at 23°C (1X MMR = 100 mM NaCl, 2 mM KCl, 1 mM MgSO4, 2 mM CaCl2, 5 mM HEPES, pH 7.6, 0.1 mM EDTA). Embryos were staged according to Xenopus laevis normal tables of development (Nieuwkoop and Faber, 1994).
Southern Blot Hybridization
Genomic DNA was harvested from individual tadpoles by overnight proteinase K digestion at 55°C followed by phenol:chloroform:isoamyl alcohol extraction and alcohol precipitation. DNA was resuspended in water overnight at 4°C. 3–5 µg of genomic DNA were digested with BglII, separated on a 0.7% (w/v) agarose gel and transferred to Hybond N+ hybridization transfer membranes (Amersham Life Sciences). The hybridization membranes were probed with a 32P-radiolabeled fragment of the GFP open reading frame (~700 bp) and exposed on a Molecular Dynamics phosphoimager screen for detection.
Genomic PCR and Transposon Integration Site Analysis
For integration site analysis of each founder line, the PCR-based EPTS LM-PCR was used as described with modifications (Yergeau et al., 2007). A biotinylated primer located downstream of the right IR/DR of the pT2βGFP construct (5’-BIO GGG CGG GGG TCG TTG GGC GGT CAG C-3’) or within scaffold 57 (5’-BIO TGC CCC TCT TAG AGT TCA CCA TTT G) was used for primer extension on NlaIII or AluI digested genomic DNA samples (~1 ug per sample). Internal primers specific to the right IR/DR pT2B1 (5’ CCG GTA CCT CAC AAA GCT), pT2B2 (5’ TAA AGG CAA TGC TAC CAA A), pT2B3 (5’ GTG GGA AGC TTG TGG AAG G) and pT2B4 (5’ CAC TGG GAA TGT GAT GAA) were used for primary PCR and if needed, nested PCR reactions in combination with the OCI and OCII linker primer set (Schmidt et al., 2001a; Schmidt et al., 2001b). For EPTS from the left arm the biotinylated primer BIOpTpolyA (5’-BIO ATG AAG CCC CTT GAG CAT CTG ACT T) was used to generate the primer extension product and primary and nested left arm-specific primers were polyAp3 (5’ TGT GTC TCT CAC TCG GAA GG) and polyAp4 (5’ GGG AGG GCA AAT CAT TTA). Linker-mediated PCR was used to isolate the 5’ junction site of X. laevis L2M line as described by Dupuy et al. (Dupuy et al., 2005) using the NlaIII enzyme with minor modifications. We used the same NlaIII-derived double-stranded linker for ligation to digested genomic DNA. We also used the same linker primer and linker nested primers for the primary and secondary PCR reactions, respectively. For genomic PCR, ~250 ng of genomic DNA was used for each PCR reaction. To confirm the integration sites in founder lines, PCR primers were designed to flank the PCR fragment identified by EPTS LM-PCR. For the 8F and 9F founder lines, primers were designed with compatibility to both the scaffold and the transposon IR/DR (3’ end of integration site 8F-1, 5’-TTA TCC TAG GCA AAG TCA AGC and transposon specific primer 5’-CTT GGG TCA AAC ATT TCG, anneal at 52°C; 5’ end of junction 8F-2, 5’-AAA GGC AAT GCT ACC AAA TAC with transposon specific primer 5’-GTT CAT ATC GTC GCA CAA A, anneal at 50°C)(9F primers set 1 5’-TAA ATG TAT TTG GCT AAG GTG and 5’-GGC AAC TAA TAT CCC TGA AAC, anneal at 51°C; set 2 5’-TAT GTA AAC TTC CGA CTT CAA and 5’-CGC CTA CA GAGA TTT AAC C, anneal at 51°C). PCR was performed using Hot Start Taq polymerase (Qiagen) under conditions 95°C 15’, 94°C 30”, anneal temp 30”, 72°C 1’ for 35 cycles with extension for 5’ at 72°C. All PCR fragments were subcloned into either pCR4TOPO (Invitrogen) or pGEM T Easy (Promega) for sequencing. Sequences were aligned with the JGI X. tropicalis (version 4.1) genomic scaffold database and NCBI databases.
Acknowledgments
We thank Kevin Bergeron and Shelby Benson of the SJCRH Animal Resource Center for expert animal husbandry, Dr. Perry Hackett for kindly providing the Sleeping Beauty reagents (pT, SB10 and SB11) and the following SJCRH Shared Resources; the Hartwell Center of Bioinformatics and Biotechnology for DNA sequencing and bioinformatics support and the Cytogenetics Lab for FISH analysis. Support for this study was provided by the NIH (RO1HD42994 to PEM) and by the American Lebanese and Syrian Associated Charities (ALSAC) of St. Jude Children’s Research Hospital. APS and TDA were supported, in part, by NCI 5R25CA023944.
References
- Amaya E, Offield MF, Grainger RM. Frog genetics: Xenopus tropicalis jumps into the future. Trends Genet. 1998;14:253–255. doi: 10.1016/s0168-9525(98)01506-6. [DOI] [PubMed] [Google Scholar]
- Balciunas D, Davidson AE, Sivasubbu S, Hermanson SB, Welle Z, Ekker SC. Enhancer trapping in zebrafish using the Sleeping Beauty transposon. BMC Genomics. 2004;5:62. doi: 10.1186/1471-2164-5-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier LS, Carlson CM, Ravimohan S, Dupuy AJ, Largaespada DA. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature. 2005;436:272–276. doi: 10.1038/nature03681. [DOI] [PubMed] [Google Scholar]
- Cui Z, Geurts AM, Liu G, Kaufman CD, Hackett PB. Structure-function analysis of the inverted terminal repeats of the Sleeping Beauty transposon. J Mol Biol. 2002;318:1221–1235. doi: 10.1016/s0022-2836(02)00237-1. [DOI] [PubMed] [Google Scholar]
- Davidson AE, Balciunas D, Mohn D, Shaffer J, Hermanson S, Sivasubbu S, Cliff MP, Hackett PB, Ekker SC. Efficient gene delivery and gene expression in zebrafish using the Sleeping Beauty transposon. Dev Biol. 2003;263:191–202. doi: 10.1016/j.ydbio.2003.07.013. [DOI] [PubMed] [Google Scholar]
- Doherty JR, Johnson Hamlet MR, Kuliyev E, Mead PE. A flk-1 promoter/enhancer reporter transgenic Xenopus laevis generated using the Sleeping Beauty transposon system: an in vivo model for vascular studies. Dev Dyn. 2007;236:2808–2817. doi: 10.1002/dvdy.21321. [DOI] [PubMed] [Google Scholar]
- Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature. 2005;436:221–226. doi: 10.1038/nature03691. [DOI] [PubMed] [Google Scholar]
- Dupuy AJ, Clark K, Carlson CM, Fritz S, Davidson AE, Markley KM, Finley K, Fletcher CF, Ekker SC, Hackett PB, Horn S, Largaespada DA. Mammalian germ-line transgenesis by transposition. Proc Natl Acad Sci U S A. 2002;99:4495–4499. doi: 10.1073/pnas.062630599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuy AJ, Fritz S, Largaespada DA. Transposition and gene disruption in the male germline of the mouse. Genesis. 2001;30:82–88. doi: 10.1002/gene.1037. [DOI] [PubMed] [Google Scholar]
- Finnegan DJ. Transposable elements and DNA transposition in eukaryotes. Curr Opin Cell Biol. 1990;2:471–477. doi: 10.1016/0955-0674(90)90130-7. [DOI] [PubMed] [Google Scholar]
- Geurts AM, Yang Y, Clark KJ, Liu G, Cui Z, Dupuy AJ, Bell JB, Largaespada DA, Hackett PB. Gene transfer into genomes of human cells by the Sleeping Beauty transposon system. Mol Ther. 2003;8:108–117. doi: 10.1016/s1525-0016(03)00099-6. [DOI] [PubMed] [Google Scholar]
- Hamlet MR, Yergeau DA, Kuliyev E, Takeda M, Taira M, Kawakami K, Mead PE. Tol2 transposon-mediated transgenesis in Xenopus tropicalis. Genesis. 2006;44:438–445. doi: 10.1002/dvg.20234. [DOI] [PubMed] [Google Scholar]
- Hirsch N, Zimmerman LB, Grainger RM. Xenopus, the next generation: X. tropicalis genetics and genomics. Dev Dyn. 2002;225:422–433. doi: 10.1002/dvdy.10178. [DOI] [PubMed] [Google Scholar]
- Ivics Z, Hackett PB, Plasterk RH, Izsvak Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- Ivics Z, Kaufman CD, Zayed H, Miskey C, Walisko O, Izsvak Z. The Sleeping Beauty transposable element: evolution, regulation and genetic applications. Curr Issues Mol Biol. 2004;6:43–55. [PubMed] [Google Scholar]
- Johnson Hamlet MR, Mead PE. Sleeping Beauty and Xenopus: Transposons as genetic tools. Current Genomics. 2003;4:687–697. [Google Scholar]
- Johnson Hamlet MR, Yergeau DA, Kuliyev E, Takeda M, Taira M, Kawakami K, Mead PE. Tol2 transposon-mediated transgenesis in Xenopus tropicalis. Genesis. 2006;44:438–445. doi: 10.1002/dvg.20234. [DOI] [PubMed] [Google Scholar]
- Kitada K, Ishishita S, Tosaka K, Takahashi R, Ueda M, Keng VW, Horie K, Takeda J. Transposon-tagged mutagenesis in the rat. Nat Methods. 2007;4:131–133. doi: 10.1038/nmeth1002. [DOI] [PubMed] [Google Scholar]
- Klein SL, Gerhard DS, Wagner L, Richardson P, Schriml LM, Sater AK, Warren WC, McPherson JD. Resources for genetic and genomic studies of Xenopus. Methods Mol Biol. 2006;322:1–16. doi: 10.1007/978-1-59745-000-3_1. [DOI] [PubMed] [Google Scholar]
- Klein SL, Strausberg RL, Wagner L, Pontius J, Clifton SW, Richardson P. Genetic and genomic tools for Xenopus research: The NIH Xenopus initiative. Dev Dyn. 2002;225:384–391. doi: 10.1002/dvdy.10174. [DOI] [PubMed] [Google Scholar]
- Miskey C, Izsvák Z, Plasterk RH, Ivics Z. The Frog Prince: a reconstructed transposon from Rana pipiens with high transpositional activity in vertebrate cells. Nucleic Acids Res. 2003;31(23):6873–6881. doi: 10.1093/nar/gkg910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, Chang E, Petrescu A, Liao N, Griffith M, Chow W, Kirkpatrick R, Butterfield YS, Young AC, Stott J, Barber S, Babakaiff R, Dickson MC, Matsuo C, Wong D, Yang GS, Smailus DE, Wetherby KD, Kwong PN, Grimwood J, Brinkley CP, 3rd, Brown-John M, Reddix-Dugue ND, Mayo M, Schmutz J, Beland J, Park M, Gibson S, Olson T, Bouffard GG, Tsai M, Featherstone R, Chand S, Siddiqui AS, Jang W, Lee E, Klein SL, Blakesley RW, Zeeberg BR, Narasimhan S, Weinstein JN, Pennacchio CP, Myers RM, Green ED, Wagner L, Gerhard DS, Marra MA, Jones SJ, Holt RA. Sequencing and analysis of 10,967 full-length cDNA clones from Xenopus laevis and Xenopus tropicalis reveals post-tetraploidization transcriptome remodeling. Genome Res. 2006;16:796–803. doi: 10.1101/gr.4871006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber F. A systematical and chronological survey of the development from the fertilized egg till the end of metamorphosis. New York & London: Garland Publishing, Inc; 1994. Normal Table of Xenopus laevis (Daudin) p. 193. [Google Scholar]
- Schmidt M, Glimm H, Lemke N, Muessig A, Speckmann C, Haas S, Zickler P, Hoffmann G, Von Kalle C. A model for the detection of clonality in marked hematopoietic stem cells. Ann N Y Acad Sci. 2001a;938:146–155. doi: 10.1111/j.1749-6632.2001.tb03584.x. discussion 155–146. [DOI] [PubMed] [Google Scholar]
- Schmidt M, Hoffmann G, Wissler M, Lemke N, Mussig A, Glimm H, Williams DA, Ragg S, Hesemann CU, von Kalle C. Detection and direct genomic sequencing of multiple rare unknown flanking DNA in highly complex samples. Hum Gene Ther. 2001b;12:743–749. doi: 10.1089/104303401750148649. [DOI] [PubMed] [Google Scholar]
- Sinzelle L, Vallin J, Coen L, Chesneau A, Du Pasquier D, Pollet N, Demeneix B, Mazabraud A. Generation of transgenic Xenopus laevis using the Sleeping Beauty transposon system. Transgenic Res. 2006;15:751–760. doi: 10.1007/s11248-006-9014-6. [DOI] [PubMed] [Google Scholar]
- Yergeau DA, Kuliyev E, Mead PE. Injection-mediated transposon transgenesis in Xenopus tropicalis and the identification of integration sites by modified extension primer tag selection (EPTS) linker-mediated PCR. Nat Protocols. 2007;2:2975–2986. doi: 10.1038/nprot.2007.428. [DOI] [PubMed] [Google Scholar]
- Yergeau DA, Mead PE. Manipulating the Xenopus genome with transposable elements. Genome Biol. 2007;8 Suppl 1:S11. doi: 10.1186/gb-2007-8-s1-s11. [DOI] [PMC free article] [PubMed] [Google Scholar]