Abstract
A number of proteins can be conjugated with both ubiquitin and the small ubiquitin-related modifier (SUMO), with crosstalk between these two post-translational modifications serving to regulate protein function and stability. We previously identified N4BP1 as a substrate for monoubiquitylation by the E3 ubiquitin ligase Nedd4. Here, we describe Nedd4-mediated polyubiquitylation and proteasomal degradation of N4BP1. In addition, we show that N4BP1 can be conjugated with SUMO1 and that this abrogates N4BP1 ubiquitylation. Consistent with this, endogenous N4BP1 is stabilized in primary embryonic fibroblasts from mutants of the desumoylating enzyme SENP1, which show increased steady-state sumoylation levels. We have localized endogenous N4BP1 predominantly to the nucleolus in primary cells. However, a small fraction is found at promyelocytic leukemia (PML) nuclear bodies (NBs). In cells deficient for SENP1 or in wild-type cells treated with the proteasome inhibitor MG132, there is considerable accumulation of N4BP1 at PML NBs. These findings suggest a dynamic interaction between subnuclear compartments, and a role for post-translational modification by ubiquitin and SUMO in the regulation of nucleolar protein turnover.
Keywords: N4BP1, PML nuclear bodies, SUMO, Nucleolus, Ubiquitin
Introduction
The eukaryotic nucleus is organized into discrete domains that are physically and functionally distinct (Dundr and Misteli, 2001; Spector, 2001; Zimber et al., 2004; Handwerger and Gall, 2006; Zaidi et al., 2007). The nucleolus, the most prominent nuclear compartment, has a well-established function in the formation of ribosomes. In addition, the nucleolus is the site of maturation for a number of different small RNAs, including components of the spliceosome and signal recognition particle. It might also play a role in the cell cycle, on the basis of the observed sequestration of several important cellular regulatory proteins (Pederson, 1998; Olson et al., 2000; Raska et al., 2006; Boisvert et al., 2007; Sirri et al., 2008; Pederson and Tsai, 2009). Another much-studied nuclear compartment is the promyelocytic leukemia (PML) nuclear body (NB), also known as the PML oncogenic domain (POD) or nuclear domain 10 (ND10). Although its function is still not completely understood, proteins crucial for apoptosis, transcription and cell-cycle regulation, among other cellular functions, localize to PML NBs, and a number of human diseases are associated with their loss or alteration (Rego et al., 2001; Strudwick and Borden, 2002; Bernardi and Pandolfi, 2007; Bernardi et al., 2008; Borden, 2008; Torok et al., 2009).
Covalent post-translational modification of PML protein by the small ubiquitin-related modifier (SUMO), as well as noncovalent SUMO binding mediated by SUMO interaction motif (SIM) domains, is crucial for the proper formation of PML NBs and the recruitment of other sumoylated proteins to these structures (Ishov et al., 1999; Zhong et al., 2000; Lin et al., 2006; Shen et al., 2006). A number of studies have shown colocalization of PML NBs with proteasomes (reviewed in Wojcik and DeMartino, 2003). The ubiquitin proteasome pathway plays a fundamental role in a number of nuclear processes, including maintenance of chromatin structure and DNA repair, control of cell proliferation and programmed cell death (Hershko and Ciechanover, 1998). It was recently shown that proteasome inhibition leads to the accumulation of both sumoylated and ubiquitylated proteins at PML NBs, providing evidence that PML NBs might function to regulate proteasomal degradation of polyubiquitylated proteins and suggesting an intimate connection with the SUMO pathway in this function (Bailey and O'Hare, 2005). It has also been noted that proteasome inhibition can lead to PML NB component proteins being found in the nucleolus (Mattsson et al., 2001; Matafora et al., 2009). Other recent findings have illustrated a dynamic relationship between the nucleolus and PML NBs (Condemine et al., 2007; Janderova-Rossmeislova et al., 2007; Rokaeus et al., 2007).
We originally identified N4BP1 in a yeast two-hybrid screen for developmentally expressed proteins that interact with the WW and HECT domains of the E3 ubiquitin ligase Nedd4, as part of an effort to discover targets of ubiquitin-mediated protein degradation that might play key roles in normal development (Murillas et al., 2002). We showed that transiently transfected N4BP1 localizes to discrete subnuclear domains and is a substrate for Nedd4-mediated monoubiquitylation. Recent work has shown that N4BP1 also interacts with the related E3 ligase ITCH, but is not a substrate for ITCH-mediated ubiquitylation (Oberst et al., 2007). Rather, N4BP1 binding to ITCH negatively regulates ITCH E3 activity directed toward its substrates, which include the p53-related tumor-suppressor proteins p73 and p63, as well as c-Jun. Whether this is the primary function of N4BP1 is not known, but a recent report predicted that N4BP1 could have ribonuclease activity, on the basis of a newly identified protein fold termed the N4BP1, YacP-like nuclease (NYN) domain, which is shared with previously characterized nucleases (Anantharaman and Aravind, 2006). Indeed, two other NYN-domain-containing proteins, ZC3H12A and KIAA0391, have been shown to have ribonuclease activity (Holzmann et al., 2008; Matsushita et al., 2009).
Here we localize endogenous N4BP1 to the nucleolus in primary cells, suggesting a role in nucleolar RNA processing. We also find that N4BP1 can undergo polyubiquitylation and proteasome-dependent degradation. Several lines of evidence point to N4BP1 turnover not in the nucleolus, but in PML NBs, with this process being regulated by N4BP1 sumoylation. Together our findings provide further support for SUMO and ubiquitin post-translational modification pathways localized specifically at PML NBs regulating nuclear protein levels and extend this paradigm to nucleolar proteins.
Results
Endogenous N4BP1 localizes to the nucleolus in mouse embryonic fibroblasts
We showed previously that N4BP1 transfected into HEK293 cells localizes to the nucleus within discrete circular domains (Murillas et al., 2002). To determine whether N4BP1 colocalizes with any of the well-characterized nuclear bodies, we co-stained with antisera against PML, coilin, SC35 and PSP1, which are signature proteins of PML NBs, Cajal bodies, GEMs and paraspeckles, respectively (Handwerger and Gall, 2006; Spector, 2006). This analysis revealed colocalization only with PML (supplementary material Fig. S1A-C), indicating that overexpressed N4BP1 accumulates at PML NBs. A number of proteins have been found to localize to PML NBs only when expressed above physiological levels (Borden, 2002). To determine the biological significance of the presence of N4BP1 at PML NBs, we asked whether the endogenous protein also localizes there.
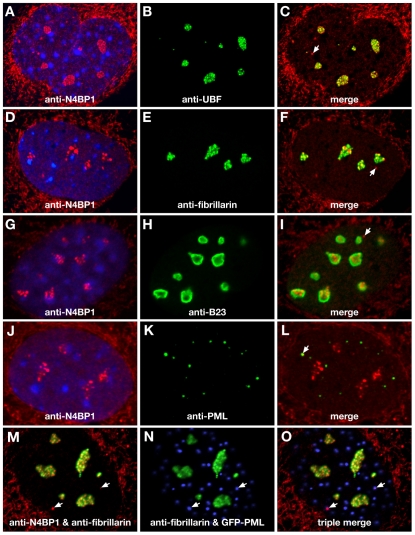
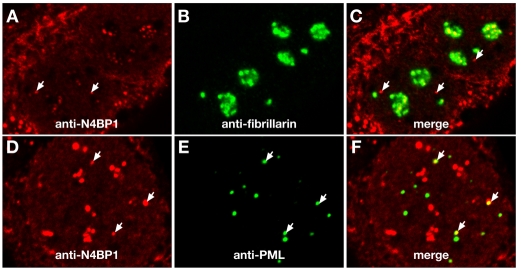
To investigate endogenous N4BP1 localization, we carried out indirect immunofluorescence using a rabbit polyclonal anti-N4BP1 antiserum raised against the mouse protein. We were unable to detect endogenous N4BP1 in HEK293 cells (supplementary material Fig. S2A). However, examination of primary mouse embryonic fibroblasts (MEFs) revealed strong anti-N4BP1 immunostaining in multiple subnuclear domains (Fig. 1A). Some cytoplasmic staining was seen, but this was also found in MEFs derived from N4BP1-knockout mice, indicating non-specific reactivity (supplementary material Fig. S3). The nuclear anti-N4BP1 staining had a distinct clumpy and irregular appearance that was variable in size and number from cell to cell, suggestive of the nucleolus rather than PML NBs. Indeed, we found extensive overlap with upstream binding factor (UBF), which localizes to the fibrillar center of the nucleolus (Fig. 1B,C). Analysis using antibodies against fibrillarin, which localizes to the dense fibrillar component of the nucleolus, also showed overlap with N4BP1 (Fig. 1D-F). B23 (also known as nucleophosmin), found in the granular component of the nucleolus, showed only limited overlap at the edges of the N4BP1-containing domains (Fig. 1G-I). Similar results were found with other mouse primary cells, including tail-tip fibroblasts, trophoblast stem cells and giant cells. These results point to endogenous N4BP1 being a nucleolar protein predominantly resident in the fibrillar regions.
Fig. 1.
N4BP1 localizes predominantly to the nucleolus. MEFs were examined by indirect immunofluorescence using anti-N4BP1 (A,C,D,F,G,I,J,L,M,O), anti-UBF (B,C), anti-fibrillarin (E,F,M-O), anti-B23 (H,I) and anti-PML (K,L), as labeled in each panel. In A,D,G,J, DAPI staining of nuclei is shown. The anti-N4BP1 cytoplasmic staining is non-specific (see supplementary material Fig. S3). N4BP1 is found predominantly in discrete subnuclear domains of varying size that colocalize extensively with UBF (A-C), less so with fibrillarin (D-F) and to only a limited extent with B23 (G-I). In a fraction of cells, N4BP1 domains that do not colocalize with nucleolar markers are seen (arrows). A similar fraction of cells show N4BP1 colocalization with PML (J-L; arrow). (M-O) PML-mutant MEFs reconstituted with GFP-PML fusion. In (M), a merge of anti-N4BP1 (red) and anti-fibrillarin (green) is shown. In (N), a merge of anti-fibrillarin (green) and GFP-PML (blue) is shown. In (O), a triple merge of anti-N4BP1 (red), anti-fibrillarin (green) and GFP-PML (blue) is shown. Non-nucleolar N4BP1 (arrows) colocalizes with GFP-PML (magenta).
We also detected small discrete sites of nuclear anti-N4BP1 staining that did not overlap with nucleolar markers (Fig. 1, white arrows). These had the size and appearance of PML NBs, but were found infrequently, on average less than one per cell (Table 1), whereas there are multiple PML NBs per cell. However, analysis with anti-PML revealed co-localization of N4BP1 with PML NBs at a similar low frequency, with on average less than one PML NB per cell showing co-staining with N4BP1 (Fig. 1J-L, arrow). This suggested that the small fraction of N4BP1 that is non-nucleolar is present at a subset of PML NBs. To confirm this, we examined PML, fibrillarin and N4BP1 in the same cell. We derived MEFs from PML mutant mice and stably expressed a GFP-PML fusion protein. These cells were then co-immunostained for N4BP1 and fibrillarin. This allowed us to determine that non-nucleolar N4BP1 always overlapped with GFP-PML (Fig. 1M-O). Thus, although the majority of N4BP1 is nucleolar, at least in MEFs, there is a small amount of endogenous protein present at PML NBs. This suggests that the localization of transfected N4BP1 at this site might be physiologically relevant.
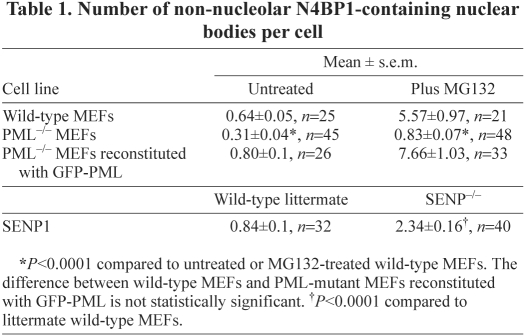
Table 1.
Number of non-nucleolar N4BP1-containing nuclear bodies per cell
Proteasome inhibition leads to increased N4BP1 localization to PML NBs
The inability to detect endogenous N4BP1 in HEK293 cells (supplementary material Fig. S2A) could be due to either low expression levels or limited crossreactivity with the human protein. As an approach to increase endogenous N4BP1 to detectable levels, we treated HEK293 cells with the proteasome inhibitor MG132. This treatment resulted in the appearance of nuclear anti-N4BP1 immunostaining (supplementary material Fig. S2D,G). Interestingly, endogenous N4BP1 did not accumulate in the nucleolus of MG132-treated HEK293 cells (supplementary material Fig. S2E,F), but rather in PML NBs (supplementary material Fig. S2H,I). This suggests that N4BP1 localizes to PML NBs normally for proteasomal turnover. The accumulation of overexpressed exogenous N4BP1 at PML NBs could reflect the same phenomenon.
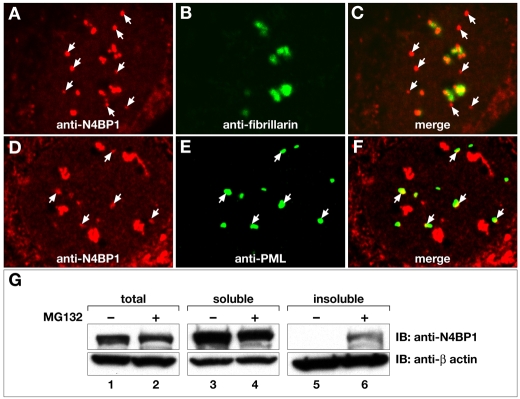
To determine whether the same mechanism is at work in primary cells, in which N4BP1 is predominantly found at the nucleolus, we carried out immunofluorescence on MEFs treated with MG132. We found a significant increase in non-nucleolar N4BP1-containing nuclear bodies (Fig. 2A-C), the frequency of which correlated perfectly with the increase in PML NBs that co-stained with anti-N4BP1 (Fig. 2D-F). There was an approximately tenfold increase from on average 0.6 per wild-type MEF cell to 6 per MG132-treated cell (Table 1). Thus, similar to HEK293 cells, MG132 treatment leads to accumulation of N4BP1 at PML NBs. We repeated the above experiment using PML-mutant MEFs, which lack PML NBs. Although there were non-nucleolar N4BP1-containing nuclear bodies in these cells (not shown), the frequency was significantly lower than in wild-type MEFs and, upon MG132 treatment, there was only a small increase (Table 1). This suggests that N4BP1 can re-localize outside the nucleolus for turnover in the absence of PML, but this is very inefficient compared to the process occurring at PML NBs. In PML-mutant MEFs reconstituted with GFP-PML, we found the usual level of non-nucleolar N4BP1 (Table 1), even though there appeared to be a higher number of PML NBs than in wild-type MEFs. Upon MG132 treatment, there was again an approximately tenfold increase in non-nucleolar N4BP1 co-localizing with PML NBs.
Fig. 2.
Proteasome inhibition leads to accumulation of N4BP1 at PML NBs. (A-F) MEFs treated with MG132 were examined by indirect immunofluorescence using anti-N4BP1 (A,C,D,F), anti-fibrillarin (B,C) and anti-PML (E,F), as labeled in each panel. (A-C) A significant increase in N4BP1 outside the nucleolus was found (arrows point to non-nucleolar N4BP1). (D-F) There is a comparable increase in the colocalization of N4BP1 with PML (arrows point to colocalized N4BP1 and PML). (G) Total protein extracts and detergent-soluble and -insoluble fractions were prepared from either untreated or MG132-treated MEFs, followed by immunoblotting (IB) with anti-N4BP1 to detect N4BP1 levels. MG132 treatment leads to accumulation in the detergent-insoluble fraction. Lower panels show reblotting with anti-β-actin to confirm equivalent sample loading.
The above results are consistent with N4BP1 turnover being mediated by the proteasome and occurring predominantly at PML NBs, which are tightly associated with the nuclear insoluble fraction. To gain additional insight into N4BP1 turnover with respect to cell compartment, we examined N4BP1 levels in the detergent-soluble and -insoluble fractions of untreated MEFs and following proteasomal inhibition. Either whole-cell extracts or separate Triton X-100 soluble and insoluble fractions were prepared. In total protein extracts and in the detergent-soluble fraction, there was no obvious change upon MG132 treatment (Fig. 2G, lanes 1-4). However, there was significant accumulation of N4BP1 in the detergent-insoluble fraction of MG132-treated MEFs (Fig. 2G, lanes 5 and 6). These biochemical experiments show that the majority of endogenous N4BP1 exists as a stable protein and suggest that only a small fraction undergoes proteasomal turnover. Turnover is restricted to the detergent-insoluble fraction, which includes the nuclear-matrix-bound PML NBs.
N4BP1 can be polyubiquitylated
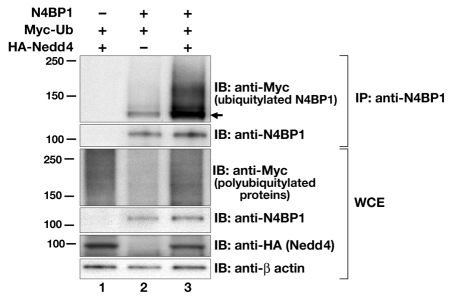
Proteasomal degradation is almost invariably associated with polyubiquitylation of the target protein. However, we previously reported that N4BP1 transfected into HEK293 cells only undergoes monoubiquitylation mediated by the Nedd4 E3 ubiquitin ligase (Murillas et al., 2002), which is not a signal for proteasomal degradation. One possible explanation for our failure to detect polyubiquitylated N4BP1 is that we had examined only the non-ionic detergent-soluble fraction; it is clear from the above results for endogenous N4BP1 that turnover is restricted to the detergent-insoluble fraction. Therefore, we reinvestigated N4BP1 polyubiquitylation by examining total protein extracts from HEK293 cells transfected with N4BP1 and Myc-epitope-tagged ubiquitin, with or without hemagglutinin (HA)-tagged Nedd4. Cells were lysed in buffer containing 3% SDS followed by a tenfold dilution into 0.5% Triton X-100 buffer. This enabled complete extraction of proteins, but still allowed immunoprecipitation using anti-N4BP1 antiserum. Ubiquitylation was then evaluated by immunoblotting with anti-Myc. Under these conditions we detected not only a strong immunoreactive band of 120 kDa, representing monoubiquitylated N4BP1, but also a high molecular mass smear (Fig. 3, lane 2), indicating that N4BP1 can indeed undergo polyubiquitylation. Coexpression with wild-type Nedd4 stimulated a marked increase in N4BP1 polyubiquitylation levels (Fig. 3, lane 3), indicating Nedd4 may mediate both mono- and poly-ubiquitylation of N4BP1.
Fig. 3.
N4BP1 can undergo polyubiquitylation. N4BP1, Myc-tagged ubiquitin (Ub) and HA-tagged Nedd4 were overexpressed in HEK293 cells. Immunoprecipitations (IP) were performed with anti-N4BP1, followed by immunoblotting (IB) either with anti-Myc (upper panel) to detect ubiquitylated forms of N4BP1 (arrow indicates position of monoubiquitylated N4BP1) or with anti-N4BP1. Lower panels show immunoblotting of whole-cell extracts (WCE) with anti-Myc (to show overall polyubiquitylation levels), anti-N4BP1, anti-HA (to detect Nedd4 levels) and anti-β-actin.
N4BP1 undergoes SUMO1 modification and is desumoylated by SENP1
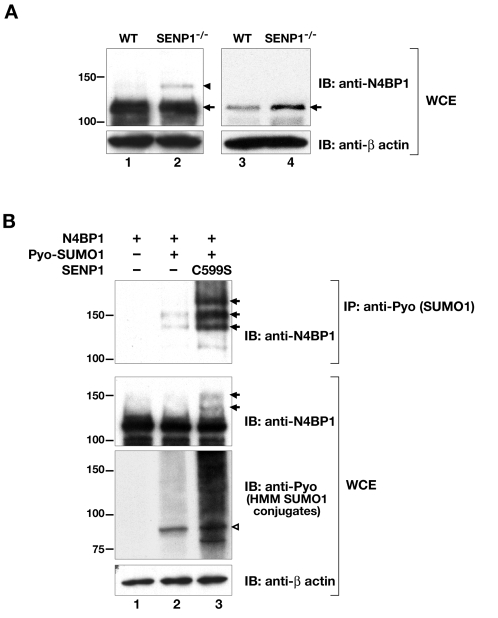
The above results suggest that N4BP1 localizes to PML NBs to undergo ubiquitin-mediated proteasomal degradation. Many of the proteins localizing to PML NBs, including PML, are sumoylated (reviewed in Bernardi and Pandolfi, 2007). To determine whether endogenous N4BP1 is also a SUMO substrate, we first carried out N4BP1 immunoprecipitation from MEFs, followed by anti-SUMO1 or anti-SUMO2 and 3 immunoblotting, but we were unable to detect any sumoylated forms of the protein. It is possible that the anti-N4BP1 antibody does not efficiently immunoprecipitate SUMO-modified N4BP1. As an alternative approach to provide evidence of sumoylation, we took advantage of our previous finding that MEFs derived from embryos deficient for the desumoylating enzyme SENP1 accumulate higher levels of SUMO1-conjugated proteins (Yamaguchi et al., 2005). We carried out direct anti-N4BP1 immunoblotting on total protein extracts from wild-type and SENP1-mutant MEFs, and looked for higher molecular mass species accumulating in the absence of the desumoylating enzyme. Indeed, we were able to detect an approximately 130 kDa form of N4BP1 that was present at a significantly higher level in whole-cell extracts from SENP1-mutant MEFs compared with the wild type (Fig. 4A, compare lanes 1 and 2). This suggests not only that endogenous N4BP1 can be sumoylated, but also that the sumoylated form is normally a substrate for SENP1.
Fig. 4.
N4BP1 is SUMO1 conjugated and desumoylated by SENP1. (A) Immunoblot (IB) analysis of whole-cell extracts (WCE) of wild-type (WT, lanes 1 and 3) and SENP1-mutant (SENP1−/−, lanes 2 and 4) MEFs with anti-N4BP1 to detect endogenous N4BP1. Filters were reblotted with anti-β-actin to ensure equal loading. Arrow indicates position of endogenous N4BP1. Arrowhead indicates position of a higher molecular mass species (obvious in lane 2), presumed to be sumoylated N4BP1, seen in long exposures of immunoblots with at least 100 μg total protein (lanes 1 and 2). Immunoblotting of less total protein (10-20 μg) revealed significantly greater N4BP1 levels in SENP1-mutant cells (lanes 3 and 4). (B) N4BP1 was overexpressed in HEK293 cells with or without Pyo-SUMO1, and with or without SENP1 C599S active-site mutant. Immunoprecipitation (IP) was performed with anti-Pyo to pull down Pyo–SUMO1-conjugated proteins, followed by immunoblotting (IB) with anti-N4BP1 to detect sumoylated N4BP1 isoforms (upper panel). The arrows indicate three N4BP1 isoforms with sizes consistent with conjugation of one, two and three Pyo-SUMO1 moieties, the levels of which were enhanced in the presence of the SENP1 active-site mutant (lane 3). The middle panel shows immunoblotting of whole-cell extracts (WCE) with anti-N4BP1, revealing sumoylated N4BP1 in the presence of SENP1 C599S (lane 3, arrows). The lower panel shows immunoblotting of whole-cell extracts with anti-Pyo to detect high molecular mass (HMM) Pyo-SUMO1 conjugates. The open arrowhead points to the position of SUMO1-conjugated RanGAP1.
To confirm N4BP1 sumoylation, we carried out overexpression studies in HEK293 cells. We transfected expression vectors for N4BP1 and polyoma (Pyo)-epitope-tagged SUMO1, and immunoprecipitated Pyo–SUMO1-conjugated proteins using anti-Pyo antibody followed by immunoblotting with anti-N4BP1. This analysis revealed two higher molecular mass species of approximately 130 kDa and 150 kDa (Fig. 4B, lane 2, upper panel), consistent with N4BP1 having two SUMO conjugation sites. Recently, an active site cysteine to serine mutant form of human SENP1 was shown to block endogenous desumoylation, resulting in the accumulation of high molecular mass SUMO1 conjugates (Bailey and O'Hare, 2004). We generated a similar mutant of the mouse enzyme, SENP1 C599S, and found that it also behaves as a dominant negative. Coexpression of SENP1 C599S with N4BP1 led to enhanced sumoylation levels, providing additional evidence that N4BP1 is normally a substrate for SENP1. The higher molecular mass forms of N4BP1 were more easily detected in the immunoprecipitation and were found even in whole-cell extracts (Fig. 4B, top and middle panels, lane 3). This analysis also revealed a third higher molecular mass species of approximately 170 kDa (Fig. 4B, upper panel, lane 3). Thus, N4BP1 might well have three potential SUMO1 conjugation sites. Inspection of the N4BP1 amino acid sequence revealed three lysine residues (K157, K183 and K607) lying within the ΨKXE sumoylation consensus motif (Rodriguez et al., 2001; Sampson et al., 2001). However, lysine to arginine mutations for each, either singly or in combination, failed to abrogate N4BP1 sumoylation (not shown). Either these three lysines do not serve as SUMO1 acceptor sites or they are used interchangeably for conjugation with other non-consensus sites, as has been reported for Daxx (Lin et al., 2006).
SENP1 regulates endogenous N4BP1 stability
Detection of endogenous sumoylated N4BP1 in SENP1-mutant MEFs required immunoblotting of a minimum of 100 μg of total protein extract. When we analyzed lower amounts (10-20 μg), we could not detect the higher molecular mass form. However, immunoblotting lower amounts of protein did reveal that the steady-state level of N4BP1 protein in SENP1-mutant MEFs was significantly higher than in wild-type cells (Fig. 4A, compare lanes 3 and 4). This was seen consistently over a number of experiments and in independently derived SENP1-mutant MEFs. Further analysis revealed no difference in N4BP1 mRNA levels between wild-type and SENP1-mutant cells (not shown), suggesting that decreased turnover is responsible for the increased protein levels. Sumoylation has been shown to stabilize protein levels by competing with ubiquitylation (reviewed in Ulrich, 2005). Thus, the increase in sumoylation of N4BP1 in SENP1-mutant cells might cause a compensatory decrease in ubiquitylation and proteasomal turnover.
The increased level of N4BP1 in SENP1-mutant MEFs was accompanied by a noticeable increase in the number of N4BP1-containing NBs outside of the nucleolus, on the basis of immunofluorescence with anti-fibrillarin and anti-N4BP1 (Fig. 5A-C). There were, on average, two such bodies in every SENP1-mutant MEF cell examined, compared with approximately one in every other MEF cell derived from wild-type littermates (Table 1). Given our above findings indicating that non-nucleolar N4BP1 localizes to PML NBs in wild-type MEFs, we asked whether there was increased colocalization of N4BP1 with PML in SENP1-mutant MEFs. Indeed, we found that every SENP1-mutant MEF had, on average, two PML NBs that colocalized with N4BP1 (Fig. 5D-F). These numbers are consistent with the increased non-nucleolar N4BP1 staining in SENP1-mutant MEFs representing increased localization to PML NBs. Accumulation at PML NBs might either be due to increased steady-state sumoylation levels driving additional N4BP1 to the PML NBs or indicate that N4BP1 degradation at PML NBs is blocked in the absence of desumoylation, or both.
Fig. 5.
N4BP1 accumulates at PML NBs in SENP1-mutant MEFs. SENP1-mutant MEFs were examined by indirect immunofluorescence using anti-N4BP1 (A,C,D,F), anti-fibrillarin (B,C) and anti-PML (E,F), as labeled in each panel. (A-C) There is an increase in the level of N4BP1 outside of the nucleolus compared to wild-type MEFs (arrows point to non-nucleolar N4BP1). (D-F) There is a comparable increase in the colocalization of N4BP1 with PML (arrows point to co-localized N4BP1 and PML).
SUMO modification negatively regulates N4BP1 polyubiquitylation
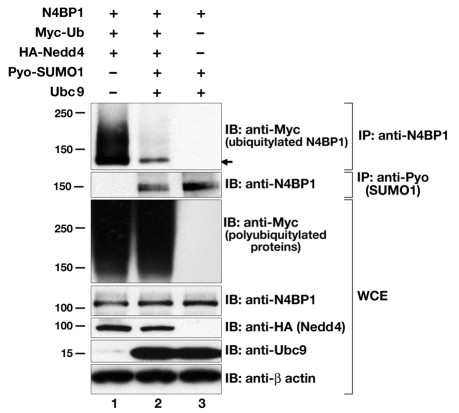
To address our hypothesis that sumoylation of N4BP1 competes with its ubiquitylation, we examined N4BP1 polyubiquitylation in the presence of overexpressed components of the sumoylation pathway. Expression vectors for Pyo-SUMO1 and Ubc9 were cotransfected along with HA-tagged Nedd4 and Myc-tagged ubiquitin. Under these conditions, there was a significant decrease in the level of N4BP1 polyubiquitylation (Fig. 6, compare lanes 1 and 2). This decrease was not due to any general reduction in overall ubiquitylation levels in transfected cells (Fig. 6, WCE, anti-Myc panel). By contrast, immunoprecipitation with anti-Pyo showed that sumoylation levels of N4BP1 were similar in the presence or absence of overexpressed components of the ubiquitylation machinery (Fig. 6, compare lanes 2 and 3). Thus, whereas N4BP1 sumoylation clearly can negatively influence N4BP1 polyubiquitylation, the converse does not appear to be true, at least under these conditions. These results are consistent with sumoylated N4BP1 not being a substrate for polyubiquitylation and suggest that desumoylation of N4BP1 is an obligate step for its ubiquitin-mediated proteasomal turnover.
Fig. 6.
N4BP1 ubiquitylation is inhibited by sumoylation. N4BP1 was overexpressed in HEK293 cells with Myc-tagged ubiquitin (Ub) and HA-tagged Nedd4 (lane 1), or additionally with Pyo-SUMO1 and Ubc9 (lane 2), or with Pyo-SUMO1 and Ubc9 only (lane 3). Immunoprecipitations (IP) were performed with anti-N4BP1, followed by immunoblotting (IB) with anti-Myc (upper panel) to detect ubiquitylated N4BP1 species. There is a significant reduction in ubiquitylated N4BP1 in the presence of SUMO1 and Ubc9. Immunoblotting of whole-cell extracts (WCE) with anti-Myc (third panel down) shows global ubiquitylation is not affected. Immunoprecipitations performed with anti-Pyo, followed by immunoblotting with anti-N4BP1 (second panel down), show no change in sumoylated N4BP1 levels in the presence or absence of ubiquitylation machinery. Lower panels show immunoblotting of whole-cell extracts with anti-N4BP1, anti-HA (to detect Nedd4 levels), anti-Ubc9 and anti-β-actin.
Discussion
We originally identified N4BP1 as a developmentally expressed target of the HECT domain ubiquitin ligase Nedd4 and previously showed, in overexpression studies, that it can undergo Nedd4-mediated monoubiquitylation (Murillas et al., 2002). Here, we have found that N4BP1 can be polyubiquitylated and SUMO conjugated as well. Whereas overexpressed N4BP1 localizes almost exclusively to PML NBs in HEK293 cells, the endogenous protein in MEFs and other primary mouse cells localizes predominantly to the nucleolus. N4BP1 was recently shown to share a domain with known RNases (Anantharaman and Aravind, 2006). Localization in the nucleolus suggests a potential role in nucleolar RNA processing. An important future avenue of research will be to confirm RNase activity and identify substrates.
Although the majority of endogenous N4BP1 is nucleolar, a small fraction localizes to PML NBs. Several lines of evidence point to this fraction specifically undergoing polyubiquitylation and proteasomal degradation, with sumoylation playing a role in the regulation of this process. It was recently shown that proteasome inhibition leads to the accumulation of both sumoylated and ubiquitylated species at discrete foci in the nucleus, in proximity to PML NBs. Thus, one function of PML NBs might be to regulate proteasomal degradation of polyubiquitylated proteins, in concert with the SUMO pathway (Bailey and O'Hare, 2005). A number of other studies have shown that PML NBs partially colocalize with proteasomes (reviewed in Wojcik and DeMartino, 2003), including the 20S proteolytic core complex (Lafarga et al., 2002; Rockel and von Mikecz, 2002), the 19S regulatory complex (Lafarga et al., 2002) and PA28 (the 11S regulatory complex) of the immunoproteasome (Fabunmi et al., 2001; Lallemand-Breitenbach et al., 2001), and might serve as sites of proteolytic degradation for cellular and viral proteins (Anton et al., 1999; Rockel et al., 2005). For N4BP1, proteasome inhibition results in a significant increase in colocalization with PML NBs, suggesting that this is where turnover normally occurs.
It has been known for some time that proteasomes are not found in nucleoli (Wojcik and DeMartino, 2003). Thus, proteasomal turnover of a nucleolar protein requires redistribution to sites outside of nucleoli, as has been shown for fibrillarin (Chen et al., 2002). Although the non-nucleolar sites where fibrillarin undergoes degradation were not examined for colocalization with PML, a similar process to that demonstrated here for N4BP1 might be occurring. Recent findings on the relationship between PML NBs and the nucleolus (Condemine et al., 2007; Janderova-Rossmeislova et al., 2007; Rokaeus et al., 2007) might also be relevant to N4BP1 turnover in MEFs. These reports document a dynamic association between PML and nucleoli, providing a direct conduit for N4BP1 relocalization to PML NBs from the nucleolus. This might also be facilitated or stabilized by sumoylation of N4BP1, similar to other proteins that transiently associate with PML NBs (Weidtkamp-Peters et al., 2008). The recent findings on the intimate relationship of PML and nucleoli have also documented a tight physical linkage with the ubiquitin proteasome pathway, adding to previous work documenting proximity of the proteasomal machinery and PML NBs.
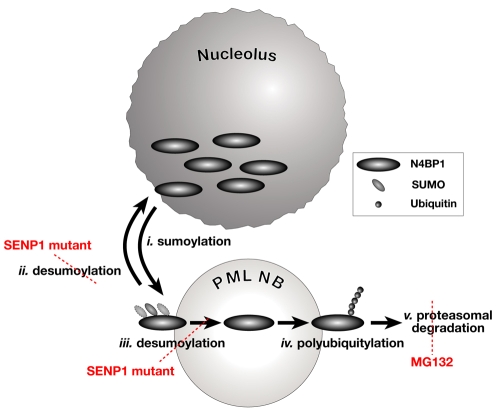
SENP1-mutant cells, which have a significantly higher steady-state level of N4BP1, also show a marked increase in the number of PML NBs containing N4BP1. This result provided the first indication that sumoylation plays a regulatory role in N4BP1 turnover. We subsequently demonstrated using transfected N4BP1 that sumoylation can occur at three sites and that this modification dramatically decreases the potential for polyubiquitylation. The marked inhibitory effect of sumoylation might be due to conjugation to the same lysine residue(s) used for polyubiquitylation, a competition mechanism first proposed for IκBα (Desterro et al., 1998). Ubiquitylation and sumoylation have opposing effects on the stability of other proteins, but apparently through mechanisms that are more complex than simply blocking the acceptor lysine, such as inducing localization or conformational changes that prevent the protein from undergoing the other modification (reviewed in Ulrich, 2005). We favor a model derived from a recent proposal that sumoylated proteins dynamically traffic through PML NBs and must be desumoylated to undergo proteasomal degradation in a process tightly coordinated with the ubiquitylation pathway (Bailey and O'Hare, 2005). This is schematically represented in Fig. 7. Sumoylation might allow stable association of N4BP1 with PML NBs (Fig. 7, i). SENP1-mediated desumoylation might lead to trafficking back to the nucleolus (Fig. 7, ii). Alternatively, desumoylated N4BP1 might remain associated with the PML NB (Fig. 7, iii), as is the case for the PML protein itself (Nefkens et al., 2003). This could explain why only very low levels of endogenous sumoylated N4BP1 can be detected biochemically, whereas the protein is easily seen in PML NBs by immunofluorescence. Desumoylation catalyzed by SENP1 might be a prerequisite, and perhaps a signal, for N4BP1 to undergo polyubiquitylation (Fig. 7, iv), followed by degradation by PML-NB-associated proteasomes (Fig. 7, v). Polyubiquitylation is possibly mediated by Nedd4, which has been shown to function in the nucleus (Hamilton et al., 2001). In the absence of desumoylation, as in the SENP1 mutant, N4BP1 cannot be polyubiquitylated or degraded and thus accumulates at PML NBs. In the absence of proteasomal degradation, as in MG132-treated cells, N4BP1 also accumulates at PML NBs. Thus, the apparent antagonism between sumoylation and ubiquitylation might reflect tightly coordinated sequential action to first direct N4BP1 to the NB, where subsequent desumoylation and ubiquitylation can occur. A similar dynamic for the bulk of nuclear sumoylated proteins has been proposed (Bailey and O'Hare, 2005).
Fig. 7.
Model of N4BP1 post-translational modification and turnover. N4BP1 is predominantly resident in the nucleolus. Sumoylation might promote association with PML NBs (i), where N4BP1 might undergo desumoylation by SENP1. It will then either traffic back to the nucleolus (ii) or be retained (iii) and undergo polyubiquitylation (iv) and degradation at proteasomes physically associated with PML NBs (v). The accumulation of N4BP1 at PML NBs in SENP1-mutant MEFs might be due to reduced relocalization to the nucleolus or because polyubiquitylation is blocked, or both. Treatment with MG132 leads to N4BP1 accumulation as a result of the blocking of degradation at PML-NB-associated proteasomes.
In conclusion, the detailed characterization of N4BP1 presented here indicates that localization to PML NBs and turnover are tightly linked and regulated by the dynamic interplay of post-translational modification by both ubiquitin and SUMO. Our findings provide additional support for PML NBs serving as specialized sites for integrating these two post-translational modifications with proteasomal degradation. N4BP1 is the first nucleolar protein identified whose regulation fits this newly emerging function for PML NBs.
Materials and Methods
Reagents and antibodies
Stock solution of MG132 (carbobenzoxy-L-leucyl-L-leucyl-L-leucinal; Sigma) was prepared in DMSO and applied to cells at 50 μM final concentration for 5 hours. An equivalent concentration of DMSO (0.1%) was used for control cells. Primary antibodies used were: anti-β-actin (clone AC-15; Sigma), anti-Ubc9 (clone 50; BD), anti-HA (clone 3F10; Roche), anti-GMP-1 (anti-SUMO1; clone 21C7; Zymed), anti-Myc (clone 9E10; American Tissue Culture Collection), anti-PML (clone 36.1-104; Millipore), anti-UBF (clone F-9; Santa Cruz Biotechnology) and anti-B23 (Invitrogen). Rabbit polyclonal anti-N4BP1 generated in our laboratory was described previously (Murillas et al., 2002). Rabbit polyclonal anti-SUMO1, mouse monoclonal antibody against the polyoma epitope (anti-Pyo) and mouse monoclonal anti-fibrillarin clone 72B9 were gifts from Mary Dasso (NICHD, Bethesda, MD), Deborah Morrison (NCI, Frederick, MD) and Patrick DiMario (Louisiana State University, LA), respectively.
Expression vectors and in vitro mutagenesis
The mouse N4BP1 cDNA has been described previously (Murillas et al., 2002). The HA-epitope-tagged Nedd4 and N-terminal Myc-epitope-tagged ubiquitin expression constructs were described previously (Magnifico et al., 2003). The expression vector for mouse Ubc9 was provided by Mary Dasso. Double polyoma (Pyo)-epitope-tagged SUMO1 and SENP1 constructs were generated in pcDNA3.1 by PCR (details available upon request). Amino acid changes in SENP1 and N4BP1 were made using the QuikChange site-directed mutagenesis kit (Stratagene). All constructs were confirmed by DNA sequencing.
Cell culture and transfection
HEK293 cells and MEFs were maintained in DMEM medium (Invitrogen) supplemented with 10% heat-inactivated FBS and antibiotics. MEFs were maintained in 3% O2 (Parrinello et al., 2003). All experiments were performed using exponentially growing cells and repeated at least twice. Transfections were performed with Lipofectamine Plus reagent (Invitrogen) according to the manufacturer's protocol. Cells were harvested from 24 to 48 hours after transfection. PML-mutant MEFs were infected with pBabe-GFP-PML virus-containing supernatants generated by transient transfection of Phoenix-Ampho packaging cells. After 24 hours, infected cultures were subjected to selection in the presence of 2 μg/ml puromycin.
Immunoblotting
Whole-cell lysates were prepared by suspending cells in 62.5 mM Tris-HCl (pH 6.8) and then adding an equal amount of 2×SDS sample buffer (10% glycerol, 2% 2-mercaptoethanol, 6% SDS, 62.5 mM Tris-HCl pH 6.8) and boiling for 5 minutes, as described (Ogiso et al., 2000). For preparation of non-ionic detergent-soluble and - insoluble fractions, cells were suspended in ice-cold Triton X-100 lysis buffer [150 mM NaCl, 50 mM Tris-HCl pH 7.5, 1 mM EDTA, 0.5% Triton X-100, 1 mM phenylmethylsulphonylfluoride (PMSF), 10 mM iodoacetamide, protease inhibitor mixture (Roche)] and kept on ice for 45 minutes with occasional mixing, followed by centrifugation. The supernatant represented the detergent-soluble fraction. The pellet, representing the detergent-insoluble fraction, was resuspended in 62.5 mM Tris-HCl (pH 6.8) with an equal volume of 2×SDS sample buffer added, followed by sonication and boiling for 5 minutes. Protein concentrations were determined using a modified Bradford assay (Bio-Rad). For immunoblot analysis, equal amounts of protein (depending on experiment from 10 to 100 μg) were resolved on SDS-polyacrylamide gels, electroblotted onto nitrocellulose membrane (Novex), and probed with specific primary antibodies and appropriate secondary antibodies. Chemiluminescent detection was performed using SuperSignal West Pico chemiluminescent substrate (Pierce).
Immunoprecipitation
For in vivo ubiquitylation and sumoylation experiments, cell lysates were prepared using 2×SDS sample buffer as above. After sonication and boiling for 5 minutes, equal amounts of protein were diluted ten times with the above-described Triton X-100 lysis buffer in the presence of protease inhibitor mixture (Roche), precleared with protein G agarose and immunoprecipitated with the indicated antibodies. Triton X-100 lysis buffer was used for co-immunoprecipitation. Antibodies used for immunoprecipitation were anti-N4BP1 (1:200 dilution) and anti-Pyo (1:10 dilution). After incubation at 4°C overnight, immunoprecipitates were washed five times with 1 ml of the Triton X-100 lysis buffer and subsequently resuspended in 2 × SDS sample buffer. After boiling for 5 minutes, the complexes were evaluated by immunoblotting.
Immunofluorescence
Immunofluorescence analysis of MEFs was done as described previously (Evdokimov et al., 2008). Samples were visualized on a Zeiss LSM510 confocal microscope with all images acquired using identical parameters. N4BP1 antiserum was purified using a Melon gel IgG spin purification kit (Pierce) and used at 1:100 dilution. Other primary antibodies used were anti-UBF (1:50), anti-PML (1:100), anti-fibrillarin (1:75, hybridoma supernatant) and anti-B23 (2.5 μg/ml).
Supplementary Material
Acknowledgments
We thank Margaret Lualdi for expert technical assistance, Mary Dasso and Deborah Morrison for antibodies and reagents, Stephen Lockett for assistance with imaging, and Øyvind Dahle, Alessandra Mazzoni and Allan Weissman for critically reading the manuscript. This research was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services and nor does mention of trade names, commercial products or organizations imply endorsement by the US Government. Deposited in PMC for release after 12 months.
Footnotes
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/123/8/1227/DC1
References
- Anantharaman V., Aravind L. (2006). The NYN domains: novel predicted RNAses with a PIN domain-like fold. RNA Biol. 3, 18-27 [DOI] [PubMed] [Google Scholar]
- Anton L. C., Schubert U., Bacik I., Princiotta M. F., Wearsch P. A., Gibbs J., Day P. M., Realini C., Rechsteiner M. C., Bennink J. R., et al. (1999). Intracellular localization of proteasomal degradation of a viral antigen. J. Cell Biol. 146, 113-124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey D., O'Hare P. (2004). Characterization of the localization and proteolytic activity of the SUMO-specific protease, SENP1. J Biol. Chem. 279, 692-703 [DOI] [PubMed] [Google Scholar]
- Bailey D., O'Hare P. (2005). Comparison of the SUMO1 and ubiquitin conjugation pathways during the inhibition of proteasome activity with evidence of SUMO1 recycling. Biochem. J. 392, 271-281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi R., Pandolfi P. P. (2007). Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat. Rev. Mol. Cell Biol. 8, 1006-1016 [DOI] [PubMed] [Google Scholar]
- Bernardi R., Papa A., Pandolfi P. P. (2008). Regulation of apoptosis by PML and the PML-NBs. Oncogene 27, 6299-6312 [DOI] [PubMed] [Google Scholar]
- Boisvert F. M., van Koningsbruggen S., Navascues J., Lamond A. I. (2007). The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 8, 574-585 [DOI] [PubMed] [Google Scholar]
- Borden K. L. (2002). Pondering the promyelocytic leukemia protein (PML) puzzle: possible functions for PML nuclear bodies. Mol. Cell. Biol. 22, 5259-5269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden K. L. (2008). Pondering the puzzle of PML (promyelocytic leukemia) nuclear bodies: can we fit the pieces together using an RNA regulon? Biochim. Biophys. Acta 1783, 2145-2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Rockel T., Steinweger G., Hemmerich P., Risch J., von Mikecz A. (2002). Subcellular recruitment of fibrillarin to nucleoplasmic proteasomes: implications for processing of a nucleolar autoantigen. Mol. Biol. Cell 13, 3576-3587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condemine W., Takahashi Y., Le Bras M., de The H. (2007). A nucleolar targeting signal in PML-I addresses PML to nucleolar caps in stressed or senescent cells. J. Cell Sci. 120, 3219-3227 [DOI] [PubMed] [Google Scholar]
- Desterro J. M., Rodriguez M. S., Hay R. T. (1998). SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol. Cell 2, 233-239 [DOI] [PubMed] [Google Scholar]
- Dundr M., Misteli T. (2001). Functional architecture in the cell nucleus. Biochem. J. 356, 297-310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evdokimov E., Sharma P., Lockett S. J., Lualdi M., Kuehn M. R. (2008). Loss of SUMO1 in mice affects RanGAP1 localization and formation of PML nuclear bodies, but is not lethal as it can be compensated by SUMO2 or SUMO3. J. Cell Sci. 121, 4106-4113 [DOI] [PubMed] [Google Scholar]
- Fabunmi R. P., Wigley W. C., Thomas P. J., DeMartino G. N. (2001). Interferon gamma regulates accumulation of the proteasome activator PA28 and immunoproteasomes at nuclear PML bodies. J. Cell Sci. 114, 29-36 [DOI] [PubMed] [Google Scholar]
- Hamilton M. H., Tcherepanova I., Huibregtse J. M., McDonnell D. P. (2001). Nuclear import/export of hRPF1/Nedd4 regulates the ubiquitin-dependent degradation of its nuclear substrates. J. Biol. Chem. 276, 26324-26331 [DOI] [PubMed] [Google Scholar]
- Handwerger K. E., Gall J. G. (2006). Subnuclear organelles: new insights into form and function. Trends Cell. Biol. 16, 19-26 [DOI] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. (1998). The ubiquitin system. Annu. Rev. Biochem. 67, 425-479 [DOI] [PubMed] [Google Scholar]
- Holzmann J., Frank P., Loffler E., Bennett K. L., Gerner C., Rossmanith W. (2008). RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell 135, 462-474 [DOI] [PubMed] [Google Scholar]
- Ishov A. M., Sotnikov A. G., Negorev D., Vladimirova O. V., Neff N., Kamitani T., Yeh E. T., Strauss J. F., 3rd, Maul G. G. (1999). PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 147, 221-234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janderova-Rossmeislova L., Novakova Z., Vlasakova J., Philimonenko V., Hozak P., Hodny Z. (2007). PML protein association with specific nucleolar structures differs in normal, tumor and senescent human cells. J. Struct. Biol. 159, 56-70 [DOI] [PubMed] [Google Scholar]
- Lafarga M., Berciano M. T., Pena E., Mayo I., Castano J. G., Bohmann D., Rodrigues J. P., Tavanez J. P., Carmo-Fonseca M. (2002). Clastosome: a subtype of nuclear body enriched in 19S and 20S proteasomes, ubiquitin, and protein substrates of proteasome. Mol. Biol. Cell 13, 2771-2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V., Zhu J., Puvion F., Koken M., Honore N., Doubeikovsky A., Duprez E., Pandolfi P. P., Puvion E., Freemont P., et al. (2001). Role of promyelocytic leukemia (PML) sumolation in nuclear body formation, 11S proteasome recruitment, and As2O3-induced PML or PML/retinoic acid receptor alpha degradation. J. Exp. Med. 193, 1361-1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D. Y., Huang Y. S., Jeng J. C., Kuo H. Y., Chang C. C., Chao T. T., Ho C. C., Chen Y. C., Lin T. P., Fang H. I., et al. (2006). Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol. Cell 24, 341-354 [DOI] [PubMed] [Google Scholar]
- Magnifico A., Ettenberg S., Yang C., Mariano J., Tiwari S., Fang S., Lipkowitz S., Weissman A. M. (2003). WW domain HECT E3s target Cbl RING finger E3s for proteasomal degradation. J. Biol. Chem. 278, 43169-43177 [DOI] [PubMed] [Google Scholar]
- Matafora V., D'Amato A., Mori S., Blasi F., Bachi A. (2009). Proteomic analysis of nucleolar SUMO-1 target proteins upon proteasome inhibition. Mol. Cell Proteomics 8, 2243-2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita K., Takeuchi O., Standley D. M., Kumagai Y., Kawagoe T., Miyake T., Satoh T., Kato H., Tsujimura T., Nakamura H., et al. (2009). Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature 458, 1185-1190 [DOI] [PubMed] [Google Scholar]
- Mattsson K., Pokrovskaja K., Kiss C., Klein G., Szekely L. (2001). Proteins associated with the promyelocytic leukemia gene product (PML)-containing nuclear body move to the nucleolus upon inhibition of proteasome-dependent protein degradation. Proc. Natl. Acad. Sci. USA 98, 1012-1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murillas R., Simms K. S., Hatakeyama S., Weissman A. M., Kuehn M. R. (2002). Identification of developmentally expressed proteins that functionally interact with Nedd4 ubiquitin ligase. J. Biol. Chem. 277, 2897-2907 [DOI] [PubMed] [Google Scholar]
- Nefkens I., Negorev D. G., Ishov A. M., Michaelson J. S., Yeh E. T., Tanguay R. M., Muller W. E., Maul G. G. (2003). Heat shock and Cd2+ exposure regulate PML and Daxx release from ND10 by independent mechanisms that modify the induction of heat-shock proteins 70 and 25 differently. J. Cell Sci. 116, 513-524 [DOI] [PubMed] [Google Scholar]
- Oberst A., Malatesta M., Aqeilan R. I., Rossi M., Salomoni P., Murillas R., Sharma P., Kuehn M. R., Oren M., Croce C. M., et al. (2007). The Nedd4-binding partner 1 (N4BP1) protein is an inhibitor of the E3 ligase Itch. Proc. Natl. Acad. Sci. USA 104, 11280-11285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogiso Y., Tomida A., Lei S., Omura S., Tsuruo T. (2000). Proteasome inhibition circumvents solid tumor resistance to topoisomerase II-directed drugs. Cancer Res. 60, 2429-2434 [PubMed] [Google Scholar]
- Olson M. O., Dundr M., Szebeni A. (2000). The nucleolus: an old factory with unexpected capabilities. Trends Cell. Biol. 10, 189-196 [DOI] [PubMed] [Google Scholar]
- Parrinello S., Samper E., Krtolica A., Goldstein J., Melov S., Campisi J. (2003). Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat. Cell Biol. 5, 741-747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T. (1998). The plurifunctional nucleolus. Nucleic Acids Res. 26, 3871-3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T., Tsai R. Y. (2009). In search of nonribosomal nucleolar protein function and regulation. J. Cell Biol. 184, 771-776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raska I., Shaw P. J., Cmarko D. (2006). Structure and function of the nucleolus in the spotlight. Curr. Opin. Cell Biol. 18, 325-334 [DOI] [PubMed] [Google Scholar]
- Rego E. M., Wang Z. G., Peruzzi D., He L. Z., Cordon-Cardo C., Pandolfi P. P. (2001). Role of promyelocytic leukemia (PML) protein in tumor suppression. J. Exp. Med. 193, 521-529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockel T. D., von Mikecz A. (2002). Proteasome-dependent processing of nuclear proteins is correlated with their subnuclear localization. J. Struct. Biol. 140, 189-199 [DOI] [PubMed] [Google Scholar]
- Rockel T. D., Stuhlmann D., von Mikecz A. (2005). Proteasomes degrade proteins in focal subdomains of the human cell nucleus. J. Cell Sci. 118, 5231-5242 [DOI] [PubMed] [Google Scholar]
- Rodriguez M. S., Dargemont C., Hay R. T. (2001). SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 276, 12654-12659 [DOI] [PubMed] [Google Scholar]
- Rokaeus N., Klein G., Wiman K. G., Szekely L., Mattsson K. (2007). PRIMA-1(MET) induces nucleolar accumulation of mutant p53 and PML nuclear body-associated proteins. Oncogene 26, 982-992 [DOI] [PubMed] [Google Scholar]
- Sampson D. A., Wang M., Matunis M. J. (2001). The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J. Biol. Chem. 276, 21664-21669 [DOI] [PubMed] [Google Scholar]
- Shen T. H., Lin H. K., Scaglioni P. P., Yung T. M., Pandolfi P. P. (2006). The mechanisms of PML-nuclear body formation. Mol. Cell 24, 331-339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirri V., Urcuqui-Inchima S., Roussel P., Hernandez-Verdun D. (2008). Nucleolus: the fascinating nuclear body. Histochem. Cell Biol. 129, 13-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector D. L. (2001). Nuclear domains. J. Cell Sci. 114, 2891-2893 [DOI] [PubMed] [Google Scholar]
- Spector D. L. (2006). SnapShot: cellular bodies. Cell 127, 1071 [DOI] [PubMed] [Google Scholar]
- Strudwick S., Borden K. L. (2002). Finding a role for PML in APL pathogenesis: a critical assessment of potential PML activities. Leukemia 16, 1906-1917 [DOI] [PubMed] [Google Scholar]
- Torok D., Ching R. W., Bazett-Jones D. P. (2009). PML nuclear bodies as sites of epigenetic regulation. Front. Biosci. 14, 1325-1336 [DOI] [PubMed] [Google Scholar]
- Ulrich H. D. (2005). Mutual interactions between the SUMO and ubiquitin systems: a plea of no contest. Trends Cell. Biol. 15, 525-532 [DOI] [PubMed] [Google Scholar]
- Weidtkamp-Peters S., Lenser T., Negorev D., Gerstner N., Hofmann T. G., Schwanitz G., Hoischen C., Maul G., Dittrich P., Hemmerich P. (2008). Dynamics of component exchange at PML nuclear bodies. J. Cell Sci. 121, 2731-2743 [DOI] [PubMed] [Google Scholar]
- Wojcik C., DeMartino G. N. (2003). Intracellular localization of proteasomes. Int. J. Biochem. Cell Biol. 35, 579-589 [DOI] [PubMed] [Google Scholar]
- Yamaguchi T., Sharma P., Athanasiou M., Kumar A., Yamada S., Kuehn M. R. (2005). Mutation of SENP1/SuPr-2 reveals an essential role for desumoylation in mouse development. Mol. Cell. Biol. 25, 5171-5182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi S. K., Young D. W., Javed A., Pratap J., Montecino M., van Wijnen A., Lian J. B., Stein J. L., Stein G. S. (2007). Nuclear microenvironments in biological control and cancer. Nat. Rev. Cancer 7, 454-463 [DOI] [PubMed] [Google Scholar]
- Zhong S., Muller S., Ronchetti S., Freemont P. S., Dejean A., Pandolfi P. P. (2000). Role of SUMO-1-modified PML in nuclear body formation. Blood 95, 2748-2752 [PubMed] [Google Scholar]
- Zimber A., Nguyen Q. D., Gespach C. (2004). Nuclear bodies and compartments: functional roles and cellular signalling in health and disease. Cell Signal 16, 1085-1104 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.